Abstract
For most multigenic disorders, clinical manifestation (penetrance) and presentation (expressivity) are likely to be an outcome of genetic interaction between multiple susceptibility genes. Here, using gene knockouts in mice, we evaluated genetic interaction between loss of Ret and loss of Sema3d, two Hirschsprung disease susceptibility genes. We intercrossed Ret and Sema3d double null heterozygotes to generate mice with the nine possible genotypes and assessed survival by counting various genotypes, myenteric plexus presence by acetylcholinesterase staining and embryonic day 12.5 (E12.5) intestine transcriptome by RNA-sequencing. Survival rates of Ret wild-type, null heterozygote and null homozygote mice at E12.5, birth and weaning were not influenced by the genotypes at Sema3d locus and vice versa. Loss of myenteric plexus was observed only in all Ret null homozygotes, irrespective of the genotypes at Sema3d locus, and Sema3d null heterozygote and homozygote mice had normal intestinal innervation. As compared with wild-type mice intestinal gene expression, loss of Ret in null homozygotes led to differential expression of ∼300 genes, whereas loss of Sema3d in null homozygotes had no major consequence and there was no evidence supporting major interaction between the two genes influencing intestine transcriptome. Overall, given the null alleles and phenotypic assays used, we did not find evidence for genetic interaction between Ret and Sema3d affecting survival, presence of myenteric plexus or intestine transcriptome.
Introduction
Hirschsprung disease (HSCR: MIM 142623) is the most common developmental defect of the enteric nervous system (ENS), and is characterized by the lack of ganglion cells in the Auerbach’s (myenteric) and Meissner’s (submucosal) plexuses in the distal rectum and variable lengths along the gastrointestinal (GI) tract (1). HSCR has an incidence of ∼15 in 100 000 newborns among European-ancestry individuals and is the most frequent cause of functional bowel obstruction among newborns (2,3). Abnormal migration, proliferation, differentiation, survival or colonization by the enteric neural crest cells (ENCCs) during GI tract development is considered to be the primary defect (4). Based on the length of the contiguous aganglionic GI tract, the disorder is classified into short-segment HSCR (S-HSCR: aganglionosis limited to the rectosigmoid colon), long-segment HSCR (L-HSCR: aganglionosis extending to the splenic flexure), total colonic aganglionosis (TCA) or total intestinal aganglionosis (aganglionosis extending into the small intestine) (1).
HSCR displays characteristics of a multifactorial genetic disorder, including high heritability (>80%), high recurrence risk among siblings (4%), non-Mendelian inheritance patterns, significant sex bias in incidence and variation in clinical expression (length of aganglionosis, associated syndromes), incomplete penetrance and variable expressivity (1,2). Human genetic studies have identified rare, high-penetrance coding variants in 15 genes that primarily lead to isolated and/or syndromic forms of HSCR (5–12) (isolated and syndromic HSCR: RET [MIM 164761], EDNRB [MIM 131244] and EDN3 [MIM 131242]; isolated HSCR only: GDNF [MIM 600837], NRTN [MIM 602018], SEMA3C [MIM 602645], SEMA3D [MIM 609907] and NRG1 [MIM 142445]; syndromic HSCR only: SOX10 [MIM 602229], ECE1 [MIM 600423], ZEB2 [MIM 605802], TCF4 [MIM 602272], PHOX2B [MIM 603851], KIF1BP [MIM 609367] and L1CAM [MIM 308840]). Collectively, these rare coding variants occur in less than 20% of HSCR cases and are most frequently (>80%) observed in RET that encodes a receptor tyrosine kinase (5,6). Expression of all known HSCR genes in enteric neuroblasts or mesenchymal cells during ENS development is indicative of their critical roles in ENCC fate determination and consequent proliferation, migration, survival or colonization and differentiation into enteric neurons (4). However, in the vast majority of the HSCR cases (∼80%), which are isolated and non-syndromic, multiple common, low-penetrance non-coding variants at RET (13–15), NRG1 (16–18) and the SEMA3 (12,19) gene cluster have been identified through association studies. While the causal molecular basis of the genetic associations at the NRG1 and SEMA3 loci remain unknown, functional in vitro and in vivo experiments have shown that some of the HSCR associated risk alleles at the RET locus, such as rs2435357T (14,20,21) and rs2506030G (21), lead to partial loss-of-function (hypomorphs) at hindgut-specific RET enhancers that reduce RET expression.
Although several HSCR-associated coding and non-coding variants at multiple genes/loci have been identified, none of them, however, are individually necessary and sufficient to cause clinical disease, suggesting that disease manifestation is the result of the combined effects of mutant alleles at multiple loci, environmental factors and/or stochastic factors, features common to many human diseases. Indeed, we have previously shown that genetic interaction, both in humans and in mice, between mutant alleles of RET and EDNRB can be necessary and sufficient for the manifestation of HSCR (22,23). In an Old Order Mennonite kindred, we had identified statistically significant joint transmission of RET and EDNRB alleles in HSCR cases (22). And, in an oligogenic mutant mouse model, we had observed complete penetrance of the Ednrbs/s genotype (homozygotes for the hypomorphic piebald allele of Ednrb (Ednrbs)) for developing megacolon and aganglionosis in conjunction with Ret kinase-null heterozygotes (Rettm1C°s/+), which is otherwise ∼10% in the Ret wild-type background (23). Other such HSCR genetic interactions have also been detected. First, we and others have shown genetic interaction between the RET and NRG1 polymorphisms in HSCR risk among Asian ancestry subjects (9,18). Second, we have demonstrated significant association between HSCR and common variants at the SEMA3 locus, following which, we demonstrated, using morpholino-based gene expression knockdowns in zebrafish, that genetic interaction between loss-of-function of class 3 semaphorins, sema3c or sema3d, and ret led to aganglionosis in the developing zebrafish intestine (12). Specifically, injection of morpholinos for either sema3c or sema3d alone in zebrafish embryos led to hypoganglionosis but co-injection with a ret morpholino led to complete loss of migrating ENCCs (12). These findings indicate the presence of genetic, if not biochemical, interactions between apparently independent signaling pathways. In this study, we have examined the nature of this interaction during mouse intestine development.
We generated mice by intercrossing Ret and Sema3d double null heterozygotes (Rettm2.1Hen°/+ or RetCFP/+; Sema3d−/+), to assess genetic interaction between Ret and Sema3d null alleles by studying the, (a) survival at embryonic day 12.5, at birth (P0) and at three weeks of age (P21), (b) presence of the myenteric plexus, as evaluated by acetylcholinesterase (AchE) staining, at P0 and at 2 weeks of age (P14) and (c) E12.5 intestine transcriptome profile. We demonstrate that (i) survival rates of Ret+/+, RetCFP/+ and RetCFP/CFP at E12.5, at P0 and at P21 are not affected by the genotype at the Sema3d locus and vice versa, (ii) aganglionosis was observed only in RetCFP/CFP mice, irrespective of the genotype at the Sema3d locus, (iii) Sema3d−/− mice have normal intestinal innervation in both Ret+/+ and RetCFP/+ genetic backgrounds, (iv) the set of differentially expressed genes in RetCFP/CFP; Sema3d+/+ E12.5 intestine largely overlaps the same from RetCFP/CFP; Sema3d−/− intestine, in comparison with the wild-type intestine and (v) loss of Sema3d alone (Ret+/+; Sema3d−/−) has no major impact on the E12.5 intestine transcriptome. Our results fail to detect and support genetic interaction between Ret and Sema3d in murine ENS development and maintenance, at least with the null alleles and the phenotypic assays used, despite replicated associations at the corresponding human loci.
Results
Absence of genetic interaction for survival between Ret and Sema3d null alleles
Mice heterozygous for the Ret cyan fluorescent protein (CFP) knock-in allele, Rettm2.1Hen° (RetCFP/+), a reporter-tagged null allele (24), were crossed with mice heterozygous for the Sema3d lacZ-tagged null allele (Sema3d−/+) to generate Ret-Sema3d double null heterozygote mice (RetCFP/+; Sema3d−/+). To evaluate genetic interaction between the RetCFP null allele and the Sema3d− null allele at the level of survival, we intercrossed RetCFP/+; Sema3d−/+ mice and first counted the nine possible genotypes in 389 mice at P21 (Table 1). There was no significant difference in the sex distribution in these weaned mice with 183 males and 206 females (χ2=1.36, df = 1, P = 0.24). For the Ret locus, we observed 143 Ret+/+, 246 RetCFP/+ and 0 RetCFP/CFP mice, showing that complete loss of Ret (RetCFP/CFP) leads to fully-penetrant lethality by this age, as reported previously (24). Single copy loss of Ret (RetCFP/+) had no significant impact on survival (24) and a near-expected ratio of Ret+/+ to RetCFP/+ mice, adjusted for RetCFP/CFP lethality, was observed (χ2=2.05, df = 1, P = 0.15). The proportions of the three Sema3d locus genotypes were not significantly different between the Ret+/+ and RetCFP/+ mice (χ2=0.28, df = 2, P = 0.87). For the Sema3d locus, we observed 133 Sema3d+/+, 230 Sema3d−/+ and 26 Sema3d−/− mice, a significant departure from those expected under Mendelian segregation (χ2=71.82, df = 2, P = 2.5 × 10−16) and pointing to partial lethality of Sema3d−/− mice by this age (26.7% survival rate). The survival rate of Sema3d−/− mice was, however, not influenced by Ret locus genotype (27.8% in Ret+/+ versus 26.2% in RetCFP/+), thus indicating the absence of genetic interaction between Sema3d and Ret null alleles for survival in P21 mice (Table 1).
Table 1.
Observed and expected genotype counts in P21, P0 and E12.5 mice from RetCFP/+; Sema3d−/+ intercrosses
| P21 |
P0 |
E12.5 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| observed |
Exp.a | observed |
Exp. | observed |
Exp. | |||||||
| Genotype | M | F | All | M | F | All | M | F | All | |||
| Ret+/+; Sema3d+/+ | 20 | 27 | 47 | 32 | 5 | 6 | 11 | 14 | 8 | 12 | 20 | 13 |
| Ret+/+; Sema3d−/+ | 45 | 42 | 87 | 65 | 13 | 13 | 26 | 27 | 13 | 22 | 35 | 26 |
| Ret+/+; Sema3d−/− | 5 | 4 | 9 | 32 | 4 | 3 | 7 | 14 | 5 | 7 | 12 | 13 |
| RetCFP/+; Sema3d+/+ | 43 | 43 | 86 | 65 | 17 | 24 | 41 | 27 | 12 | 9 | 21 | 26 |
| RetCFP/+; Sema3d−/+ | 61 | 82 | 143 | 130 | 35 | 28 | 63 | 54 | 30 | 21 | 51 | 52 |
| RetCFP/+; Sema3d−/− | 9 | 8 | 17 | 65 | 8 | 6 | 14 | 27 | 11 | 15 | 26 | 26 |
| RetCFP/CFP; Sema3d+/+ | 0 | 0 | 0 | 5 | 7 | 12 | 14 | 6 | 6 | 12 | 13 | |
| RetCFP/CFP; Sema3d−/+ | 0 | 0 | 0 | 19 | 16 | 35 | 27 | 13 | 6 | 19 | 26 | |
| RetCFP/CFP; Sema3d−/− | 0 | 0 | 0 | 4 | 3 | 7 | 14 | 8 | 5 | 13 | 13 | |
| Total | 183 | 206 | 389 | 389 | 110 | 106 | 216 | 216 | 106 | 103 | 209 | 209 |
Exp. expected; M, male; F, female.
aExpectations calculated based on a modified ratio (1/12:2/12:1/12:2/12:4/12:2/12).
Having observed partial lethality in Sema3d−/− adult mice, we wanted to assess whether partial lethality was present at birth and whether there was genetic interaction between the RetCFP null allele and the Sema3d− null allele at the level of survival. So, we intercrossed RetCFP/+; Sema3d−/+ mice and counted the nine possible genotypes at P0. A total of 216 mice were studied at for the Ret and Sema3d autosomal loci and the Kdm5 and Sry sex-linked loci (Table 1). There was no significant difference in the sex distribution in these newborn mice with 110 males and 106 females (χ2=0.07, df = 1, P = 0.79). For the Ret locus, we observed 44 Ret+/+, 118 RetCFP/+ and 54 RetCFP/CFP mice, close to the distribution expected under Mendelian transmission (χ2=2.78, df = 2, P = 0.25). The proportions of the three Sema3d locus genotypes were not significantly different between the Ret+/+, RetCFP/+ and RetCFP/CFP mice (χ2=3.63, df = 4, P = 0.46). RetCFP/CFP have been reported to be born in expected ratios, as observed here, but die within 24–48 h of birth. For the Sema3d locus, we observed 64 Sema3d+/+, 124 Sema3d−/+ and 28 Sema3d−/− mice, a significant departure from that expected under Mendelian segregation (χ2=16.74, df = 2, P = 2.3 × 10−4), and again pointing to partial lethality of Sema3d−/− in mice by this age (51.9% survival rate), although the survival rate was approximately double of that in adult mice (see above). The survival rate of Sema3d−/− mice was not influenced by Ret locus genotype and was identical across the three Ret locus genotypes (51.9% in Ret+/+, RetCFP/+ and RetCFP/CFP), again indicating the absence of genetic interaction between Sema3d and Ret null alleles for survival in P0 mice (Table 1).
The above observations prompted us to look earlier in embryonic development, at E12.5, for partial lethality and genetic interactions. So, we performed timed-matings by intercrossing RetCFP/+; Sema3d−/+ mice and counted the nine possible genotypes at E12.5. A total of 209 embryos were studied at for the Ret and Sema3d autosomal loci and for the Kdm5 and Sry sex-linked loci (Table 1). There was no significant difference in the sex distribution in these embryos with 106 males and 103 females (χ2=0.04, df = 1, P = 0.84). For the Ret locus, we observed 67 Ret+/+, 98 RetCFP/+ and 44 RetCFP/CFP embryos, not significantly different from the distribution expected under Mendelian transmission (χ2=5.87, df = 2, P = 0.053). The small decrease in RetCFP/CFP embryos than expected appears to be a chance event (21.1% versus the expected 25%), especially since the observed proportion of RetCFP/CFP mice at a later time (P0) was 25%, as expected. The proportions of the three Sema3d locus genotypes were not significantly different between the Ret+/+, RetCFP/+ and RetCFP/CFP embryos (χ2=3.56, df = 4, P = 0.47). For the Sema3d locus, we observed 53 Sema3d+/+, 105 Sema3d−/+ and 51 Sema3d−/− embryos, close to expectations under Mendelian transmission (χ2=0.04, df = 2, P = 0.98), indicating no lethality for the Sema3d−/− embryos at E12.5 (97.6% survival rate). These observations indicate absence of genetic interaction between Sema3d and Ret null alleles for survival at this earlier stage as well (Table 1).
Collectively, these results show that loss of Sema3d (Sema3d−/−) leads to embryonic lethality, starting after E12.5 and by birth only half of the expected Sema3d−/− mice are observed, further dropping to a quarter by the weaning age. Also, the distributions of wild-type, null heterozygote and null homozygote genotypes at one locus are not influenced by the genotypes at the other locus in E12.5, P0 and P21 mice, showing the absence of genetic interaction between Ret and Sema3d null alleles.
Loss of Sema3d does not lead to hypoganglionosis or aganglionosis in the ENS
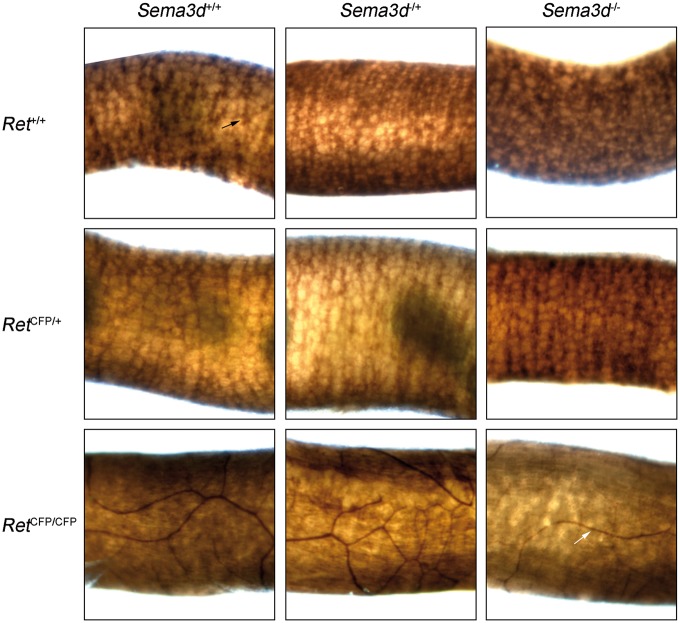
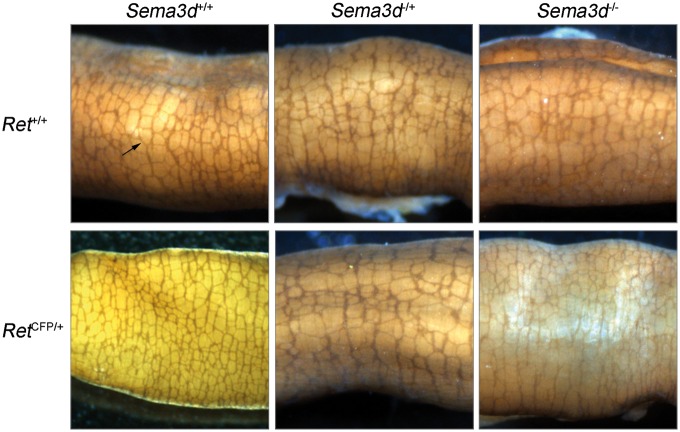
HSCR is usually diagnosed by the absence of ENCC-derived enteric ganglia from the Auerbach’s (myenteric) and Meissner’s (submucosal) plexuses in the colon wall. Therefore, to assess genetic interactions at this level, we performed acetylcholinesterase (AchE) staining of whole-mount small and large intestines dissected out from P0 and P14 mice generated by intercrossing RetCFP/+; Sema3d−/+ mice. In P0 mice, we performed AchE staining from all nine possible genotypes (a total of 112 mice), and in P14 mice, we performed AchE staining from all the six viable genotypes (43 mice; we did not observe any RetCFP/CFP mice) (Figs. 1 and 2 and Table 2; Supplementary Material, Fig. S1). In P0 mice, the normal reticulate plexus of enteric ganglia was present in the small and large intestines of Ret+/+; Sema3d+/+ (n = 7), Ret+/+; Sema3d−/+ (n = 15), Ret+/+; Sema3d−/− (n = 4), RetCFP/+; Sema3d+/+ (n = 23), RetCFP/+; Sema3d−/+ (n = 35) and RetCFP/+; Sema3d−/− (n = 4) mice, thereby indicating that loss of one copy of Sema3d or loss of two copies of Sema3d, both in Ret+/+ and RetCFP/+ background, did not lead to abnormal enteric ganglia (Fig. 1 andTable 2). One copy loss of Ret also did not affect intestinal innervation, as reported previously (24). However, in all RetCFP/CFP mice assessed (n = 24), the normal reticulate plexus of enteric ganglia was missing in the small and large intestines and was replaced by large AchE-positive hypertrophic nerve fibers, especially in the distal large intestine (Fig. 1 and Table 2; Supplementary Material, Fig. S1). Moreover, this Ret loss-of-function effect was independent of their Sema3d genotype (RetCFP/CFP; Sema3d+/+ (n = 9), RetCFP/CFP; Sema3d−/+ (n = 14) and RetCFP/CFP; Sema3d−/− (n = 1)). In P14 mice, the normal reticulate plexus of enteric ganglia was present throughout the intestine in Ret+/+; Sema3d+/+ (n = 7), Ret+/+; Sema3d−/+ (n = 11), Ret+/+; Sema3d−/− (n = 1), RetCFP/+; Sema3d+/+ (n = 7), RetCFP/+; Sema3d−/+ (n = 15) and RetCFP/+; Sema3d−/− (n = 2) mice, identical to that observed in P0 mice (Fig. 2 and Table 2). Taken together, these results show that loss of Sema3d (Sema3d−/− or Sema3d−/+) in a Ret+/+ or RetCFP/+ background does not affect the presence of enteric ganglia as assessed by AchE staining, thereby indicating absence of genetic interaction between Ret and Sema3d null alleles in intestinal innervation at this level of resolution.
Figure 1.
Acetylcholinesterase staining of the distal large intestine in P0 mice. Representative images of acetylcholinesterase-stained whole mounts of distal large intestine from the following genotypes are shown: Ret+/+; Sema3d+/+ (top, left), Ret+/+; Sema3d−/+ (top, center), Ret+/+; Sema3d−/− (top, right), RetCFP/+; Sema3d+/+ (middle, left), RetCFP/+; Sema3d−/+ (middle, center), RetCFP/+; Sema3d−/− (middle, right), RetCFP/CFP; Sema3d+/+ (bottom, left), RetCFP/CFP; Sema3d−/+ (bottom, center) and RetCFP/CFP; Sema3d−/− (bottom, right). Normal myenteric plexus (black arrow) was observed in Ret+/+; Sema3d+/+, Ret+/+; Sema3d−/+, Ret+/+; Sema3d−/−, RetCFP/+; Sema3d+/+, RetCFP/+; Sema3d−/+, and RetCFP/+; Sema3d−/− mice. RetCFP/CFP; Sema3d+/+, RetCFP/CFP; Sema3d−/+ and RetCFP/CFP; Sema3d−/− mice display an absence of enteric ganglia and presence of acetylcholinesterase-positive hypertrophic nerve fibers (white arrow).
Figure 2.
Acetylcholinesterase staining of the distal large intestine in P14 mice. Representative images of acetylcholinesterase-stained whole mounts of distal large intestine from the following genotypes are shown: Ret+/+; Sema3d+/+ (top, left), Ret+/+; Sema3d−/+ (top, center), Ret+/+; Sema3d−/− (top, right), RetCFP/+; Sema3d+/+ (bottom, left), RetCFP/+; Sema3d−/+ (bottom, center), and RetCFP/+; Sema3d−/− (bottom, right). Normal myenteric plexus (black arrow) was observed in Ret+/+; Sema3d+/+, Ret+/+; Sema3d−/+, Ret+/+; Sema3d−/−, RetCFP/+; Sema3d+/+, RetCFP/+; Sema3d−/+, and RetCFP/+; Sema3d−/− mice.
Table 2.
Acetylcholinesterase (AchE) staining of intestine in P0 and P14 mice from RetCFP/+; Sema3d−/+ intercrosses
| Genotype | #at P0 | #at P14 | AchE staininga |
|---|---|---|---|
| Ret+/+; Sema3d+/+ | 7 | 7 | Normal innervation |
| Ret+/+; Sema3d−/+ | 15 | 11 | Normal innervation |
| Ret+/+; Sema3d−/− | 4 | 1 | Normal innervation |
| RetCFP/+; Sema3d+/+ | 23 | 7 | Normal innervation |
| RetCFP/+; Sema3d−/+ | 35 | 15 | Normal innervation |
| RetCFP/+; Sema3d−/− | 4 | 2 | Normal innervation |
| RetCFP/CFP; Sema3d+/+ | 9 | 0 | Aganglionosis of small & large intestine |
| RetCFP/CFP; Sema3d−/+ | 14 | 0 | Aganglionosis of small & large intestine |
| RetCFP/CFP; Sema3d−/− | 1 | 0 | Aganglionosis of small & large intestine |
| Total | 112 | 43 |
aPhenotypes observed in all mice.
Loss of Sema3d has no major impact on embryonic intestine transcriptome
Despite the absence of measureable genetic interaction between Ret and Sema3d null alleles at a gross phenotypic level (survival and presence of enteric ganglia), we evaluated whether such an interaction existed at a molecular level by comparing the E12.5 intestine transcriptomes and assessing differential gene expression. We looked at E12.5 as it represents a mid-point in development of the enteric neurons in the mouse gut, between embryonic days 10.5 and 14.5, where the intestine is undergoing major morphological and transcriptional changes. We performed stranded mRNA-seq in intestinal tissues dissected from three E12.5 males and three E12.5 females with the following genotypes: Ret+/+; Sema3d+/+, RetCFP/CFP; Sema3d+/+, Ret+/+; Sema3d−/− and RetCFP/CFP; Sema3d−/−. Paired-end 100 bp sequencing was performed on an Illumina HiSeq 2500 with a desired sequencing depth of ∼30 million paired-end reads per sample. The sequencing paired-end reads were processed to remove Illumina adapter sequences and low quality bases using Trimmomatic (25). Gene and isoform expression level estimation was performed using RSEM (26), which included mapping of trimmed paired-end reads to the mouse genome and GENCODE annotated transcripts using STAR aligner (27). Supplementary Material Table S1 lists the number of read pairs in each sample at each step of RNA-seq analysis.
Differential gene expression analyses were performed using DESeq (28) by comparing RSEM-generated gene-level read count data from six mutants (three males and three females) and six wild-type (three males and three females) mice after adjusting for gender effects based on generalized linear models (Supplementary Material, Table S2–S4 and Fig. S2). As compared with E12.5 wild-type intestine, 275 protein-coding genes were differentially expressed (DE) (false discovery rate (FDR) <0.01 and |log2 fold-change|>1) in E12.5 RetCFP/CFP; Sema3d+/+ intestine, showing that loss of Ret leads to a major impact on the E12.5 intestine transcriptome (Supplementary Material, Table S5; see Table 3 for top 50 DE genes). In contrast, only Sema3d was differentially expressed in E12.5 Ret+/+; Sema3d−/− intestine, showing that loss of Sema3d has no major impact on E12.5 intestine transcriptome (Supplementary Material, Table S5). In E12.5 double null homozygote RetCFP/CFP; Sema3d−/− intestine, 244 protein coding genes were DE, with these genes largely overlapping those identified in RetCFP/CFP; Sema3d+/+ intestine (226 (93%) DE genes shared between RetCFP/CFP; Sema3d+/+ and RetCFP/CFP; Sema3d−/− genotypes), indicating that the transcriptional effect from Ret loss is nearly independent of the same from Sema3d loss (Supplementary Material, Table S5). Pathway enrichment analysis based on the union of RetCFP/CFP; Sema3d+/+ and RetCFP/CFP; Sema3d−/− DE genes shows that these 293 DE genes are overrepresented in various pathways related to nervous system function (Supplementary Material, Table S6).
Table 3.
Top 50 differentially expressed genes between Ret+/+; Sema3d+/+ and RetCFP/CFP; Sema3d+/+
| Gene_name | log2 fold change |
|---|---|
| Ret | −8.34 |
| Dbh | −6.28 |
| Chrna3 | −6.18 |
| Stmn3 | −5.83 |
| Elavl3 | −5.42 |
| Phox2b | −5.35 |
| Ascl1 | −5.34 |
| Prph | −4.50 |
| Gal | −4.34 |
| Phox2a | −4.18 |
| Dlx2 | −4.16 |
| Grik3 | −4.06 |
| Nos1 | −4.06 |
| Cartpt | −4.05 |
| Stmn4 | −3.91 |
| Celf3 | −3.91 |
| L1cam | −3.82 |
| Tnr | −3.65 |
| Myt1 | −3.54 |
| Celsr3 | −3.46 |
| Fam163a | −3.43 |
| Dll3 | −3.39 |
| Kcnq2 | −3.37 |
| Slc10a4 | −3.36 |
| Elavl4 | −3.21 |
| Scg3 | −2.62 |
| Fam184b | −2.55 |
| Ptpro | −2.44 |
| Nfasc | −2.39 |
| Stmn2 | −2.30 |
| Nalcn | −2.25 |
| Serpini1 | −2.21 |
| Sox8 | −2.16 |
| Rtn1 | −2.13 |
| Mapt | −1.97 |
| Rundc3a | −1.93 |
| Chrnb2 | −1.93 |
| Tmem132e | −1.82 |
| Slc18a2 | −1.75 |
| Eya1 | −1.72 |
| Adcyap1r1 | −1.57 |
| Nrcam | −1.53 |
| Kif5c | −1.52 |
| Plxna4 | −1.45 |
| Ngfr | −1.23 |
| Apc2 | −1.22 |
| Dclk1 | −1.18 |
| Ednrb | −1.17 |
| Nefl | −1.12 |
| Dpysl3 | −1.07 |
In addition to these pair-wise comparisons of main effects from Ret and Sema3d loss, we also assessed possible interactions between Ret and Sema3d loss on intestine transcriptome. We regressed gene expression across all samples using generalized linear models implemented within DESeq (28). Under one model (main effects only), the regression fit was based on Ret and Sema3d locus genotypes and gender (three variables), while under the other model (main effects plus interaction), the regression fit was based on Ret locus genotype, Sema3d locus genotype, Ret-Sema3d interaction and gender (four variables). On a gene-by-gene basis, the two models were compared, and only one gene (Rpl27-ps3) at FDR 1% or 20 genes (Rpl27-ps3, Lyst, Pdia4, Klhl11, Col12a1, Farp2, Zfp398, Vps13c, Birc6, Itgav, Zfp369, Wdfy2, Tuba1c, Plxna4, Itpripl2, Prex2, Samd5, Rpl29, Herc2 and Zfp729a; sorted by descending rank) at FDR5%, had a significantly better fit under the four variable model (main effects plus interaction), providing evidence for possible genetic interaction. It is important to note that none of these 20 genes, except Plxna4 in RetCFP/CFP; Sema3d+/+, are differentially expressed in RetCFP/CFP; Sema3d+/+, Ret+/+; Sema3d−/− or RetCFP/CFP; Sema3d−/− E12.5 intestines (Supplementary Material, Table S7). Collectively, these results highlight the absence of widespread genetic interactions between Ret and Sema3d null alleles on the intestine transcriptome.
Discussion
Previous studies from our laboratory prompted this examination of genetic interaction between Ret and Sema3d in mice, based on the following four observations: (1) genome-wide association studies (GWAS) had mapped a novel HSCR susceptibility locus, in addition to the RET locus, to variants within the SEMA3 gene cluster at chromosome 7q21, (2) among the four genes in the cluster, Sema3c and Sema3d were expressed in a subset of developing ENS cells in mice overlapping with Ret, (3) combined knockdown of sema3c or sema3d led to aganglionosis in the ENS of zebrafish embryos and showed gene interactions with ret and (4) rare loss-of-function variants in SEMA3C and SEMA3D are more frequent in HSCR subjects as compared with controls (12).
In this study, null homozygotes at Ret lead to complete lethality within 24 h of birth arising from the lack of enteric neurons in addition to renal agenesis or severe dysgenesis, as previously reported (24,29). Although Sema3d null alleles have also been characterized with respect to defects in the patterning of pulmonary veins during embryogenesis, their effect on survival has not been reported (30). Here we show that Sema3d null homozygotes show no detectable lethality (survival 96.7%) during embryogenesis (E12.5) with decreasing levels of survival post-birth (51.9%) and post-weaning (26.7%). We have not investigated the cause of this lethality but, based on the anomalous pulmonary vein connections reported (30), speculate that this arises from cardiac defects. The genetic interaction between ret and sema3c/d loss-of-function we previously observed in the zebrafish (12) could be manifested in many ways in mice. We first tested for an effect of compound genotypes at Ret and Sema3d on survival but failed to detect any effect. However, this assessment does not rule out the possibility of a genetic or biochemical interaction between these genes in ENS development and function without differential consequences on survivability.
Our second approach was to use AchE histochemical staining in the mouse intestine to examine the reticulate plexus of enteric ganglia, because such staining is extensively used in HSCR diagnosis (31) and for characterizing mouse models of HSCR (23,24). In Ret null homozygote mice, the reticulate plexus of enteric ganglia, observed in wild-type mice, are missing and are replaced by large AchE-positive hypertrophic nerve fibers, as also reported previously (24). In contrast, similar studies on intestine tissue from Sema3d null homozygote mice, showed the normal pattern indistinguishable from wild-type mice. Although these results indicate that loss of Sema3d has no apparent effect on mouse ENS formation, it is important to note that not all myenteric neurons are acetylcholine positive. Indeed, in the human colon, ∼50% of myenteric neurons are positive for choline acetyltransferase only, ∼40% are positive for nitric oxide synthase only, and, ∼5% each are positive and negative for both enzymes (32). Similarly, in guinea pig distal colon, a model in which most extensive knowledge of different enteric neuron types has been obtained, ∼57% of myenteric neurons are positive for choline acetyltransferase (33). Thus, AchE staining is unlikely to detect all major enteric neuronal subclasses (34), suggesting that the Sema3d effect, either alone or in conjunction with Ret loss-of-function, needs to be studied by more detailed examination of myenteric neuron subtypes. It is also important to note that we have not evaluated the submucosal plexus in these assays and thus cannot rule out possible loss-of-function defects in the submucosal plexus.
As a third approach, we tested for differences in the intestine transcriptome during mouse development (E12.5) in compound Ret and Sema3d null allele genotypes. Our data clearly show that Ret loss-of-function induces significant changes in the intestine transcriptome (∼300 genes) but not that by Sema3d loss-of-function (1 gene). Moreover, the genes’ with altered expression in RetCFP/CFP; Sema3d+/+ and RetCFP/CFP; Sema3d−/− intestine tissues were substantially overlapping and statistical analysis provided little support for Ret-Sema3d interaction. Once again, these negative results need to be interpreted in the light of knowledge that only ∼5% of all cells in the mouse intestine tissue comprise the ENS and that tissue-level RNA-seq may not be sensitive to uncover the molecular consequences of loss-of-function at cellular level. Such tests will require transcriptome analysis of isolated ENS cells from mice of various genotypes. Also, we have evaluated the effect on intestine transcriptome at only one time-point during embryonic development (E12.5) and it is possible that the loss of Sema3d mediated molecular effects set in later during development and would require such comparisons to be performed at multiple time points during development. In this regard, it is important to note that conditional loss of key molecules such as Ret (24) and Gfra1 (35) at E14.5 in post-migratory enteric neurons has been shown to induce death of enteric neurons in embryonic colon.
Other caveats should be noted to account for the mouse-zebrafish differences we observe. First, the zebrafish data came from morpholino-based somatic knockdown experiments (12) while the mouse data are from germline null alleles. There is now increasing evidence to support true differences between knockdown and targeted mutation phenotypes across many genes in the mouse, zebrafish and other model systems (36). One of the likely reasons is that gene knockout, but not knockdown, can lead to activation of compensatory genes and pathways, and subsequent phenotype rescue or diminution of effect (36). Second, it is not surprising to see differences in phenotypes for at least some gene knockouts versus knockdowns between mouse and zebrafish given the ∼400 million years of evolutionary divergence between the two species (37). We speculate, particularly given the multiple closely related genes encoding class 3 semaphorins at the Sema3 locus that the critical gene(s) affecting ENS development and function may have changed during evolution. Despite our negative data in mice, we believe that the class 3 semaphorins remain attractive candidates for ENS development and HSCR for the following reasons.
The HSCR GWAS signal in the SEMA3 locus is not a false positive signal having been replicated by us (15) and others (Bjarke Feenstra, personal communication). Within the association signal at 7q21.11, there are only two protein-coding genes, SEMA3A and SEMA3D, both of which are expressed in the ENS. Semaphorins are a family of secreted, transmembrane or glycophosphatidylinositol-anchored proteins implicated in diverse biological processes (38). Since their initial discovery as axon guidance molecules (39), semaphorins have been known to be involved in migration, proliferation and survival of neurons, patterning of vasculature, immune cell regulation and tumor progression (38). Specifically, SEMA3A has been shown to be involved in development of microvasculature (40), bone development (41), tumor growth and metastasis (42), inflammatory and immune response (42) and hindgut innervation (43), and SEMA3D has been shown to be involved in cardiovascular development (30) and tumor growth and metastasis (42). There are two more class 3 semaphorins in this gene cluster, SEMA3C and SEMA3E, but these map outside the association signal, ∼4Mb and ∼1.2Mb upstream of the GWAS hit, respectively, and are not likely candidates. The literature suggests that SEMA3A is a promising HSCR candidate. Sema3a has been shown, using Sema3a null mice, to regulate the entry of sacral enteric neural precursors into the distal hindgut in mice by acting as a repulsive signal secreted from the intestine mesenchyme (43). Increased expression of SEMA3A in aganglionic smooth muscle layer of the colon has also been reported in some HSCR patients (44). Finally, by targeted sequencing in HSCR subjects, rare coding deleterious variants have been identified in SEMA3C and SEMA3D by us (12) and in SEMA3A and SEMA3D by others (11). Although in our previous work, we did not detect Sema3a expression in the mouse ENS during development by RNA in situ hybridization and did not observe loss of enteric neurons upon sema3aa/sema3ab knockdown in zebrafish embryos (12), we do detect Sema3a expression in E12.5 intestine by RNA-seq here, which is a higher sensitivity assay.
Based on the physiological roles inferred for class 3 semaphorins from various knockout mice, as outlined above, one could argue that assessing axonal projections of enteric neurons to their targets (circular muscle, longitudinal muscle, mucosa, blood vessels) in knockout/knockdown models would be more appropriate than assessing the myenteric plexus by AchE staining (34), especially when class 3 semaphorins have been shown to pattern axonal projections in spinal cord by selectively repelling one class of axons but not the others (45). A more direct approach to identifying the human susceptibility gene within the SEMA3 cluster would be to identify the functional basis of the risk association through identification of the cis regulatory element and the target gene it regulates.
Materials And Methods
Generation of Ret and Sema3d null mice
Mice heterozygous for the Ret CFP knock-in allele, Rettm2.1Hen° (RetCFP/+), a reporter-tagged null allele, were obtained by David Ginty (Johns Hopkins University, Baltimore, MD, USA) from Hideki Enomoto (RIKEN Center for Developmental Biology, Kobe, Japan); their generation has been previously described (24). Mice heterozygous for the Sema3d lacZ-tagged null allele (Sema3d−/+) were generated by David Ginty's laboratory; their generation will be described elsewhere. The Sema3d loss-of-function allele (Sema3d−) leads to a western null (data not shown, David Ginty, personal communication). Both RetCFP/+ and Sema3d−/+ mice were maintained on a mixed genetic background (C57BL/6, 129/Sv and C3H). Double heterozygote mice (RetCFP/+; Sema3d−/+) were generated by David Ginty’s laboratory by crossing RetCFP/+ mice to Sema3d−/+ mice. We received the double heterozygote mice (RetCFP/+; Sema3d−/+) from David Ginty and then intercrossed them to generate mice with the expected nine genotypes (Ret+/+; Sema3d+/+, Ret+/+; Sema3d−/+, Ret+/+; Sema3d−/−, RetCFP/+; Sema3d+/+, RetCFP/+; Sema3d−/+, RetCFP/+; Sema3d−/−, RetCFP/CFP; Sema3d+/+, RetCFP/CFP; Sema3d−/+, RetCFP/CFP; Sema3d−/−). Timed matings were evaluated by checking females for vaginal plugs and matings were assumed to occur at the midpoint of the dark cycle.
Genotyping
For newborns (P0), 2 weeks (P14) and 3 weeks (P21) old mice, genomic DNA was isolated from tail-tips, and for embryonic day 12.5 (E12.5) mice, genomic DNA was isolated from one hind limb, following standard methods. For the Ret locus, mice were genotyped using two PCRs with the following primers: Ret+ allele forward 5′-CAGCGCAGGTCTCTCATCAGTACCGCA-3′ and Ret+ allele reverse 5′-CAGCTAGCCGCAGCGACCCGGTTC-3′ (modified from (46)) mapping to the 5′ UTR and the first intron of Ret, respectively (449 bp PCR product); RetCFP allele forward 5′-ATGGTGAGCAAGGGCGAGGAGCTGTT-3′ and RetCFP allele reverse 5′-CTGGGTGCTCAGGTAGTGGTTGTC-3′ mapping to CFP cDNA (615 bp PCR product). For the Sema3d locus, mice were genotyped using a three primer PCR: a common forward 5′-CTCCTGATCCCTCCTCACAG-3′, a wild-type-specific (Sema3d+) reverse 5’-GAGCACACAGCATCTGCATT-3’ (367 bp PCR product) and a mutant-specific (Sema3d−) reverse 5′-TCCCAAG GGCAG T A G AGTTC-3′ (523 bp PCR product). The sex of the embryos was determined using two PCRs with the following primers: Kdm5 forward 5′-CTGAAGCTTTTGGCTTTGAG-3′ and Kdm5 reverse 5′-CCGCTGCCAAATTCTTTGG-3′ mapping to exons 9 and 10 of the Kdm5c/d genes, respectively (331/302 bp PCR product from X/Y chromosome) (47); Sry forward 5′-TTGTCTAGAGAGC ATGGA GGGCCATGTCAA-3′ and Sry reverse 5′-CCACTCCTC TGTGAC AC T T T AGCCCTCCGA-3′ mapping to Sry on the Y chromosome (273 bp PCR product in males only). All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Johns Hopkins University (Protocol MO15M328) and were in accordance with Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines. All animals were fed a standard rodent chow ad libitum.
Acetylcholinesterase histochemistry
The acetylcholinesterase (AchE) staining protocol was adapted from published methods (48,49). Briefly, P0 and P14 mice were euthanized using inhaled isoflurane in a closed chamber. The entire intestine was dissected out, followed by removal of the mesentery in cold phosphate buffered saline (PBS, ThermoFisher Scientific) under a stereo microscope. Intestine tissue was irrigated with cold PBS to flush out fecal contents using a 30 gauge needle syringe. Tissues were fixed in excess of fresh 4% paraformaldehyde (PFA, Electron Microscopy Sciences) in PBS for 1 h at 4 °C and then stored overnight at 4 °C in saturated sodium sulfate (24.5 g sodium sulfate in 100 mL water, Sigma). Tissues were then incubated in substrate buffer, pH 5.5 (Ethopropazine HCl 0.2 mM, Acetylthiocholine iodide 4 mM, glycine 10 mM, cupric sulfate pentahydrate 2 mM, sodium acetate 65 mM in water, Sigma) at room temperature with gentle rocking for 2 h (P14) or 4 h (P0). AchE staining was developed by incubating tissues in 1.25% sodium sulfide, pH 6.0, (1.25 g sodium sulfide non-ahydrate in water, Sigma), at room temperature with gentle rocking for 2 min (P14) or 6 min (P0). Tissues were then rinsed in excess water at room temperature with gentle rocking for 6 × 5 min. Stained tissues were photographed (trans-illumination for P0 and epi-illumination for P14) as whole mounts in water using a stereo microscope.
RNA isolation, library preparation and sequencing
RNA-seq was performed from E12.5 intestine tissue from three males and three females from the following genotypes: Ret+/+; Sema3d+/+, RetCFP/CFP; Sema3d+/+, Ret+/+; Sema3d−/− and RetCFP/CFP; Sema3d−/−. Total RNA was isolated from each E12.5 intestine using RNeasy Mini Kit following manufacturers’ recommendations (Qiagen) that included the on-column DNase digestion using RNase-Free DNase set (Qiagen). The quantity and quality of total RNA was assessed by NanoDrop spectrophotometer (Thermo Scientific). KAPA Stranded mRNA-Seq kit (KAPA Biosystems) was used to generate indexed Illumina platform sequencing libraries. Briefly, polyA RNA was captured from 1 µg total RNA using magnetic oligo-dT beads. After elution from the magnetic beads, polyA RNA was fragmented to generate inserts ranging in size from 100 to 200 bp, followed by random priming and reverse transcription to generate double-stranded cDNA. Next, after performing a 1.8× SPRI cleanup using AMPure XP beads (Agencourt), dAMP was added to 3′ ends of the cDNA fragments followed by ligation with indexed 3′ dTMP Illumina TruSeq adapters. Ligated fragments were subsequently size selected using PEG/NaCl SPRI solution and underwent PCR amplification (12 cycles) to generate the sequencing libraries. After performing a 1× SPRI cleanup using AMPure XP beads (Agencourt), a sample from each library was used to assess library fragment size distribution by electrophoresis using BioAnalyzer High Sensitivity DNA Assay (Agilent Technologies) and to assess library concentration by qPCR using KAPA library quantification kit (KAPA Biosystems). Equimolar amounts of libraries were pooled and sequenced on an Illumina HiSeq 2500 instrument using standard protocols for paired-end 100 bp sequencing with a desired sequencing depth of ∼30 million paired end reads per library.
RNA-seq analyses
Illumina RNA-seq paired-end read fastq files were quality checked using FASTQC (version: 0.11.5) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; date last accessed January 10, 2016) and then processed using Trimmomatic (version: 0.36) (25), for removing adapters and other Illumina-specific sequences from the reads, and, for performing a sliding-window based trimming of low quality bases from each read (ILLUMINACLIP:TruSeq3-PE-2.fa:2:30: 10:1: TRUE LEADING:3 TRAILING:3 SLIDINGWIND OW: 4:15 MINLE N:36). For estimating gene and isoform expression levels, we first extracted reference transcript sequences from the mouse genome (GRCm38, primary assembly) based on the GENCODE (http://www.gencodegenes.org/mouse_releases/current.html; date last accessed January 10, 2016) primary assembly gene annotation (release M10) and built STAR aligner (27) indices using the RSEM software package (version: 1.2.31) (26). Trimmed paired-end reads from each sample were then aligned to the reference transcript sequences by calling the STAR aligner within RSEM and using alignment parameters from the ENCODE STAR-RSEM long RNA-seq pipeline (–outSAMunmapped Within –outFilterType BySJout –out SA Mattributes NH HI AS NM MD –outFilterMultimapNmax 20 –outFilterMismatchNmax 999 –outFilterMismatchNoverLmax 0.04 –alignIntronMin 20 –alignIntronMax 1 000 000 –align Mates GapMax 1 000 000 –alignSJoverhangMin 8 –alignSJD Bover hangMin 1 –sjdbScore 1 –quantMode TranscriptomeSAM). Gene and isoform expression levels were then estimated in each sample from these transcriptome alignments using RSEM keeping in mind the strandedness of the prepared RNA-seq libraries (–forward-prob 0.0). Gene-level read count data generated by RSEM was compared between various wild-type and mutant mice to assess differential gene expression using the DESeq software (version: 1.24.0) (28). Only those genes where the sum of read counts across the 12 samples in each comparison (three male and three female wild-type and the three male and three female mutant replicates) was >1, were retained for differential gene expression analysis. Although, release M10 of the GENCODE primary assembly gene annotation has 48 526 genes, we limited differential gene expression comparison to only protein coding genes (22 098) and calculated false-discovery rates (FDR) based on the number of protein coding genes within each comparison. FDR were calculated from P-values using the Benjamini–Hochberg procedure (50). All data have been deposited in NCBI’s GEO and are accessible at GEO Series accession number GSE89627.
Pathway enrichment analysis
Pathway over-representation or enrichment analysis was performed for the set of differentially expressed genes using the online ConsensusPathDB tool (51) (http://cpdb.molgen.mpg.de/; date last accessed January 10, 2016). Among all the available pathway databases, the enrichment analysis was limited to KEGG (52) database and was restricted to report only those pathways with a minimum overlap of two genes among the differentially expressed genes and a P-value <0.01.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to Prof. Hideki Enomoto (RIKEN Center for Developmental Biology, Kobe, Japan) for providing RetCFP/+ mice, to Prof. David Ginty and Dr. Qingguang Jiang (Johns Hopkins University, now at Harvard Medical School) for providing RetCFP/+; Sema3d−/+ mice, to Prof. Raj Kapur (University of Washington) for evaluating AchE staining images, to Prof. Alex Kolodkin (Johns Hopkins University) for critical comments and to the members of the Chakravarti laboratory for discussions.
Conflict of Interest statement. None declared.
Funding
U.S. National Institutes of Health (Grant RO1 HD28088 to A.C.).
References
- 1. Chakravarti A., Lyonnet S. (2001) Hirschsprung disease In Scriver C.R., Beaudet A.L., Valle D., Sly W.S., Childs B., Kinzler K., Vogelstein B. (eds.), The Metabolic and Molecular Bases of Inherited Disease (8th Edition).McGraw-Hill, New York, pp. 6231–6255. [Google Scholar]
- 2. Badner J.A., Sieber W.K., Garver K.L., Chakravarti A. (1990) A genetic study of hirschsprung disease. Am. J. Hum. Genet, 46, 568–580. [PMC free article] [PubMed] [Google Scholar]
- 3. Parisi M.A., Kapur R.P. (2000) Genetics of hirschsprung disease. Curr. Opin. Pediatr., 12, 610–617. [DOI] [PubMed] [Google Scholar]
- 4. Heanue T.A., Pachnis V. (2007) Enteric nervous system development and hirschsprung's disease: advances in genetic and stem cell studies. Nat. Rev. Neurosci., 8, 466–479. [DOI] [PubMed] [Google Scholar]
- 5. Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., et al. (2008) Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet., 45, 1–14. [DOI] [PubMed] [Google Scholar]
- 6. Alves M.M., Sribudiani Y., Brouwer R.W., Amiel J., Antinolo G., Borrego S., Ceccherini I., Chakravarti A., Fernandez R.M., Garcia-Barcelo M.M., et al. (2013) Contribution of rare and common variants determine complex diseases-hirschsprung disease as a model. Dev. Biol., 382, 320–329. [DOI] [PubMed] [Google Scholar]
- 7. McKeown S.J., Stamp L., Hao M.M., Young H.M. (2013) Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip. Rev. Dev. Biol., 2, 113–129. [DOI] [PubMed] [Google Scholar]
- 8. Luzon-Toro B., Torroglosa A., Nunez-Torres R., Enguix-Riego M.V., Fernandez R.M., de Agustin J.C., Antinolo G., Borrego S. (2012) Comprehensive analysis of NRG1 common and rare variants in hirschsprung patients. PLoS One, 7, e36524.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gui H., Tang W.K., So M.T., Proitsi P., Sham P.C., Tam P.K., Ngan E.S., Cherny S.S., Garcia-Barcelo M.M. (2013) RET and NRG1 interplay in hirschsprung disease. Hum. Genet., 132, 591–600. [DOI] [PubMed] [Google Scholar]
- 10. Jiang Q., Turner T., Sosa M.X., Rakha A., Arnold S., Chakravarti A. (2012) Rapid and efficient human mutation detection using a bench-top next-generation DNA sequencer. Hum. Mutat., 33, 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luzon-Toro B., Fernandez R.M., Torroglosa A., de Agustin J.C., Mendez-Vidal C., Segura D.I., Antinolo G., Borrego S. (2013) Mutational spectrum of semaphorin 3A and semaphorin 3D genes in spanish hirschsprung patients. PLoS One, 8, e54800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang Q., Arnold S., Heanue T., Kilambi K.P., Doan B., Kapoor A., Ling A.Y., Sosa M.X., Guy M., Jiang Q., et al. (2015) Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to hirschsprung disease liability. Am. J. Hum. Genet., 96, 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emison E.S., McCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., Portnoy M.E., Cutler D.J., Green E.D., Chakravarti A. (2005) A common sex-dependent mutation in a RET enhancer underlies hirschsprung disease risk. Nature, 434, 857–863. [DOI] [PubMed] [Google Scholar]
- 14. Emison E.S., Garcia-Barcelo M., Grice E.A., Lantieri F., Amiel J., Burzynski G., Fernandez R.M., Hao L., Kashuk C., West K., et al. (2010) Differential contributions of rare and common, coding and noncoding ret mutations to multifactorial hirschsprung disease liability. Am. J. Hum. Genet., 87, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kapoor A., Jiang Q., Chatterjee S., Chakraborty P., Sosa M.X., Berrios C., Chakravarti A. (2015) Population variation in total genetic risk of hirschsprung disease from common RET, SEMA3 and NRG1 susceptibility polymorphisms. Hum. Mol. Genet., 24, 2997–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia-Barcelo M.M., Tang C.S., Ngan E.S., Lui V.C., Chen Y., So M.T., Leon T.Y., Miao X.P., Shum C.K., Liu F.Q., et al. (2009) Genome-wide association study identifies NRG1 as a susceptibility locus for hirschsprung's disease. Proc. Natl. Acad. Sci. U. S. A., 106, 2694–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phusantisampan T., Sangkhathat S., Phongdara A., Chiengkriwate P., Patrapinyokul S., Mahasirimongkol S. (2012) Association of genetic polymorphisms in the RET-protooncogene and NRG1 with hirschsprung disease in Thai patients. J. Hum. Genet., 57, 286–293. [DOI] [PubMed] [Google Scholar]
- 18. Gunadi, Kapoor A., Ling A.Y., Rochadi, Makhmudi A., Herini E.S., Sosa M.X., Chatterjee S., Chakravarti A. (2014) Effects of RET and NRG1 polymorphisms in indonesian patients with hirschsprung disease. J. Pediatr. Surg., 49, 1614–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L.L., Zhang Y., Fan Y., Li H., Zhou F.H., Miao J.N., Gu H., Huang T.C., Yuan Z.W. (2012) SEMA3A rs7804122 polymorphism is associated with hirschsprung disease in the northeastern region of china. Birth Defects Res. Clin. Mol. Teratol., 94, 91–95. [DOI] [PubMed] [Google Scholar]
- 20. Grice E.A., Rochelle E.S., Green E.D., Chakravarti A., McCallion A.S. (2005) Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum. Mol. Genet., 14, 3837–3845. [DOI] [PubMed] [Google Scholar]
- 21. Chatterjee S., Kapoor A., Akiyama J.A., Auer D.R., Lee D., Gabriel S., Berrios C., Pennacchio L.A., Chakravarti A. (2016) Enhancer variants synergistically drive dysfunction of a gene regulatory network in hirschsprung disease. Cell, 167, 355–368.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carrasquillo M.M., McCallion A.S., Puffenberger E.G., Kashuk C.S., Nouri N., Chakravarti A. (2002) Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in hirschsprung disease. Nat. Genet., 32, 237–244. [DOI] [PubMed] [Google Scholar]
- 23. McCallion A.S., Stames E., Conlon R.A., Chakravarti A. (2003) Phenotype variation in two-locus mouse models of hirschsprung disease: tissue-specific interaction between ret and ednrb. Proc. Natl. Acad. Sci. U. S. A, 100, 1826–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uesaka T., Nagashimada M., Yonemura S., Enomoto H. (2008) Diminished ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J. Clin. Invest., 118, 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bolger A.M., Lohse M., Usadel B. (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li B., Dewey C.N. (2011) RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics, 12, 323-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106-2010-11-10-r106. Epub 2010 Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schuchardt A., D'Agati V., Larsson-Blomberg L., Costantini F., Pachnis V. (1994) Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor ret. Nature, 367, 380–383. [DOI] [PubMed] [Google Scholar]
- 30. Degenhardt K., Singh M.K., Aghajanian H., Massera D., Wang Q., Li J., Li L., Choi C., Yzaguirre A.D., Francey L.J., et al. (2013) Semaphorin 3d signaling defects are associated with anomalous pulmonary venous connections. Nat. Med., 19, 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moore S.W., Johnson G. (2005) Acetylcholinesterase in hirschsprung's disease. Pediatr. Surg. Int., 21, 255–263. [DOI] [PubMed] [Google Scholar]
- 32. Murphy E.M., Defontgalland D., Costa M., Brookes S.J., Wattchow D.A. (2007) Quantification of subclasses of human colonic myenteric neurons by immunoreactivity to hu, choline acetyltransferase and nitric oxide synthase. Neurogastroenterol. Motil., 19, 126–134. [DOI] [PubMed] [Google Scholar]
- 33. Lomax A.E., Furness J.B. (2000) Neurochemical classification of enteric neurons in the guinea-pig distal colon. Cell Tissue Res., 302, 59–72. [DOI] [PubMed] [Google Scholar]
- 34. Hao M.M., Young H.M. (2009) Development of enteric neuron diversity. J. Cell. Mol. Med., 13, 1193–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uesaka T., Jain S., Yonemura S., Uchiyama Y., Milbrandt J., Enomoto H. (2007) Conditional ablation of GFRalpha1 in postmigratory enteric neurons triggers unconventional neuronal death in the colon and causes a hirschsprung's disease phenotype. Development, 134, 2171–2181. [DOI] [PubMed] [Google Scholar]
- 36. Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M., Stainier D.Y. (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature, 524, 230–233. [DOI] [PubMed] [Google Scholar]
- 37. Kumar S., Hedges S.B. (1998) A molecular timescale for vertebrate evolution. Nature, 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 38. Kruger R.P., Aurandt J., Guan K.L. (2005) Semaphorins command cells to move. Nat. Rev. Mol. Cell Biol., 6, 789–800. [DOI] [PubMed] [Google Scholar]
- 39. Kolodkin A.L., Matthes D.J., Goodman C.S. (1993) The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell, 75, 1389–1399. [DOI] [PubMed] [Google Scholar]
- 40. Cerani A., Tetreault N., Menard C., Lapalme E., Patel C., Sitaras N., Beaudoin F., Leboeuf D., De Guire V., Binet F., et al. (2013) Neuron-derived semaphorin 3A is an early inducer of vascular permeability in diabetic retinopathy via neuropilin-1. Cell. Metab., 18, 505–518. [DOI] [PubMed] [Google Scholar]
- 41. Hayashi M., Nakashima T., Taniguchi M., Kodama T., Kumanogoh A., Takayanagi H. (2012) Osteoprotection by semaphorin 3A. Nature, 485, 69–74. [DOI] [PubMed] [Google Scholar]
- 42. Worzfeld T., Offermanns S. (2014) Semaphorins and plexins as therapeutic targets. Nat. Rev. Drug Discov, 13, 603–621. [DOI] [PubMed] [Google Scholar]
- 43. Anderson R.B., Bergner A.J., Taniguchi M., Fujisawa H., Forrai A., Robb L., Young H.M. (2007) Effects of different regions of the developing gut on the migration of enteric neural crest-derived cells: a role for Sema3A, but not Sema3F. Dev. Biol., 305, 287–299. [DOI] [PubMed] [Google Scholar]
- 44. Wang L.L., Fan Y., Zhou F.H., Li H., Zhang Y., Miao J.N., Gu H., Huang T.C., Yuan Z.W. (2011) Semaphorin 3A expression in the colon of hirschsprung disease. Birth Defects Res. Clin. Mol. Teratol., 91, 842–847. [DOI] [PubMed] [Google Scholar]
- 45. Messersmith E.K., Leonardo E.D., Shatz C.J., Tessier-Lavigne M., Goodman C.S., Kolodkin A.L. (1995) Semaphorin III can function as a selective chemorepellent to pattern sensory projections in the spinal cord. Neuron, 14, 949–959. [DOI] [PubMed] [Google Scholar]
- 46. Jain S., Naughton C.K., Yang M., Strickland A., Vij K., Encinas M., Golden J., Gupta A., Heuckeroth R., Johnson E.M. Jr, et al. (2004) Mice expressing a dominant-negative ret mutation phenocopy human hirschsprung disease and delineate a direct role of ret in spermatogenesis. Development, 131, 5503–5513. [DOI] [PubMed] [Google Scholar]
- 47. Clapcote S.J., Roder J.C. (2005) Simplex PCR assay for sex determination in mice. BioTechniques, 38, 702, 704, 706.. [DOI] [PubMed] [Google Scholar]
- 48. Enomoto H., Araki T., Jackman A., Heuckeroth R.O., Snider W.D., Johnson E.M. Jr., Milbrandt J. (1998) GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron, 21, 317–324. [DOI] [PubMed] [Google Scholar]
- 49. Howe D.G., Clarke C.M., Yan H., Willis B.S., Schneider D.A., McKnight G.S., Kapur R.P. (2006) Inhibition of protein kinase A in murine enteric neurons causes lethal intestinal pseudo-obstruction. J. Neurobiol., 66, 256–272. [DOI] [PubMed] [Google Scholar]
- 50. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological), 57, 289–300. [Google Scholar]
- 51. Herwig R., Hardt C., Lienhard M., Kamburov A. (2016) Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc., 11, 1889–1907. [DOI] [PubMed] [Google Scholar]
- 52. Kanehisa M., Goto S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.