Abstract
Fragile X related protein 1 (FXR1P) is a member of the fragile X family of RNA-binding proteins, which includes FMRP and FXR2P. Both FMRP and FXR2P regulate neurogenesis, a process affected in a number of neurological and neuropsychiatric disorders, including fragile X syndrome. Although FXR1P has been implicated in various developmental processes and neuropsychiatric diseases, its role in neurodevelopment is not well understood. The goal of the present study was to elucidate the function of FXR1P in adult neurogenesis. We used an inducible mouse model that allows us to investigate how FXR1P deficiency in adult neural stem cells (aNSCs) affects proliferation and neuronal differentiation. Deletion of FXR1 in aNSCs resulted in fewer adult-born cells in the dentate gyrus (DG) overall, reducing populations across different stages of neurogenesis, including radial glia-like cells, intermediate progenitors, neuroblasts, immature neurons and neurons. We hypothesized that this reduction in new cell numbers resulted from impaired proliferation, which we confirmed both in vivo and in vitro. We discovered that FXR1P-deficient aNSCs have altered expression of a select number of cell-cycle genes, and we identified the mRNA of cyclin-dependent kinase inhibitor 1A (Cdkn1a, p21) as a direct target of FXR1P. Restoration of p21 mRNA to wild-type levels rescued the proliferation deficit in cells lacking FXR1P, demonstrating that p21 is a mediator of FXR1P in aNSCs. These results indicate that FXR1P plays an important role in regulating aNSC self-renewal and maintenance in the adult brain, which may have implications for a number of neurodevelopmental and psychiatric disorders.
Introduction
Fragile X related protein 1 (FXR1P) is linked to a growing number of neurological disorders, but its function in the brain remains unclear. The FXR1 gene was first identified due to its high homology to the fragile X mental retardation protein (FMRP) and may play a functional role in regulating many of FMRP’s targets (1–9). This is consistent with the identification of FXR1P as a modulator of autistic phenotypes, and the location of the FXR1 gene is within a major susceptibility locus for autism (10,11). Looking at protein interactome data, FXR1P is highly interconnected with other autism-associated proteins (12). In addition to its connection with autism, single nucleotide polymorphisms (SNPs) in FXR1P are also associated with bipolar disorder and schizophrenia (13–16). This mounting evidence linking FXR1P with human neurological disease merits further study of its role in the brain. Despite all this, FXR1P’s function and mechanism in the brain remain largely unstudied.
FXR1P, FXR2P and FMRP are part of a family of RNA-binding proteins (FXRs) highly expressed in neurons. All three proteins contain two highly conserved RNA-binding domains, a polyribosome binding domain and a nuclear localization sequence (17). Although studies using whole brain homogenate or immortalized cell lines have proposed that the FXRs have overlapping mRNA targets, suggesting functional redundancy, we and others have identified unique roles and distinct underlying mechanisms for these proteins in specific cell types (4,18–27). FXR1P is unique among the FXRs, in that it is the only protein that is neonatal lethal when deleted in mice (28). Its importance is further underscored by the fact that it is highly conserved in vertebrates and has been implicated in several developmental processes, including oocyte maturation, eye and neural crest development and myoblast differentiation (25,29–34). How functionally similar the FXRs are in the brain is not known.
In the adult human hippocampus, over a third of neurons are replaced through neurogenesis (35). These new neurons are crucial for complex learning, including episodic memory and contextual learning. Dysregulation of this process is linked to a wide spectrum of diseases, among them psychiatric diseases such as schizophrenia and developmental diseases such as autism (36). Previous studies from our group and others have found that FMRP and FXR2P play important and distinct regulatory roles in adult neurogenesis (20,21,37–39). Deletion of FXR1P in excitatory hippocampal neurons is known to alter spatial learning in adult mice (40). The role of FXR1P in adult neurogenesis, however, has yet to be assessed.
Here, we show that specific deletion of FXR1P in adult neural stem cells (aNSCs) leads to decreased generation of new cells, including new neurons. These decreases are a result of reduced cell proliferation both in aNSCs and neural progenitors. We found that lack of FXR1P altered gene expression for proteins important in cell cycle regulation. Specifically, FXR1P binds the mRNA of the cell cycle inhibitor p21 (Cdkn1A, P21Cip1 or P21Waf1). FXR1P deficiency leads to elevated p21 mRNA expression, and restoration of p21 to wild-type levels rescued the proliferation deficit of Fxr1-deficient NSCs. Therefore, FXR1P is an important regulator of p21 in aNSCs, which is important for aNSC proliferation during adult neurogenesis.
Results
FXR1P is expressed in adult-born cells throughout adult neurogenesis
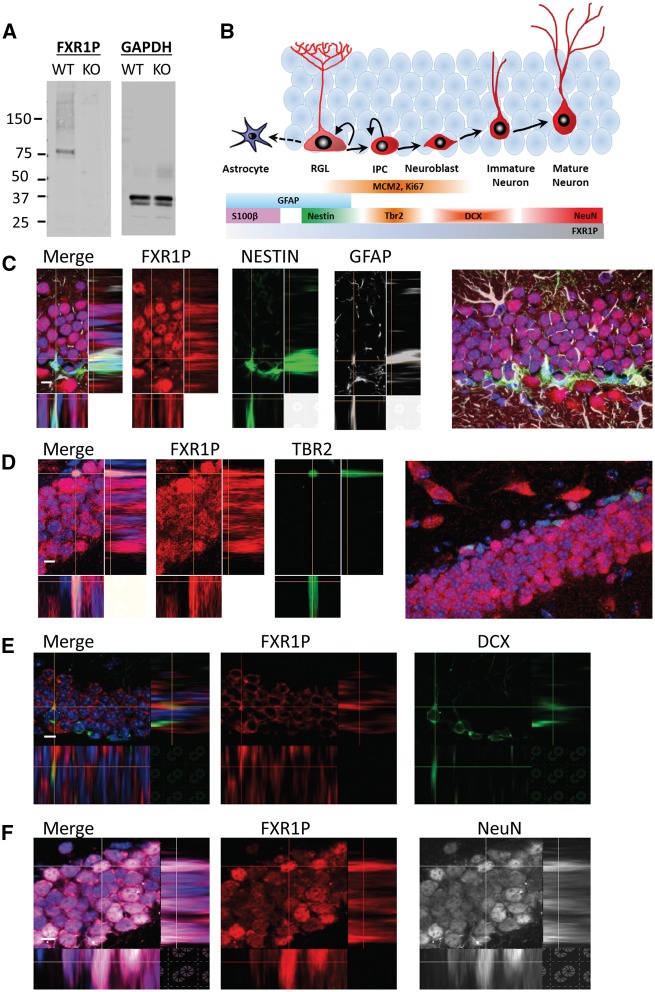
FXR proteins exhibit strong homology, making it critical to use an antibody that can discriminate among them. Since commercially available antibodies for FXR1P proved to be ineffectual, we created a mouse monoclonal antibody (mAb) based on the peptide sequence used to generate the highly successful polyclonal antibodies 830 and mL13 (32,41). We validated the specificity of our antibody using cortical tissue from neonatal brains of FXR1P knockout (Fxr1-/-, KO) mice and compared our mAb to the 830 antibody (Fig. 1A) (28). We found that, similar to 830 antibody, our mAb recognized a specific band corresponding to the molecular weight of FXR1P in the wildtype (WT) but not Fxr1-/- tissues (Fig. 1A). We did not see any bands corresponding to the molecular weight of FMRP or FXR2P in Fxr1-/- tissues; therefore, our mAb appears to be specific to FXR1P.
Figure 1.
FXR1P is expressed in adult-born cells throughout adult neurogenesis. (A) Immunoblot analysis using the FXR1P mAb and a control GAPDH antibody in wildtype (WT) and Fxr1 knockout (Fxr1-/-, KO) neonatal brain tissue. Protein standards in kilo daltons (kDa) are indicated to the left. (B) Schematic illustration of FXR1P expression throughout adult hippocampal neurogenesis with markers representing each cell type. RGL, radial glia-like cell; IPC, intermediate progenitor cell. (C) FXR1P (red) expression in Nestin (green) and GFAP (white) double-positive RGLs. Staining performed on coronal sections from a Nestin-GFP transgenic mouse. Expanded view of image shown at the far right. DAPI (blue); scale bar = 10 µM. (D) FXR1P (red) expression in TBR2-positive (green) IPC. Expanded view of image shown at the far right. DAPI (blue); scale bar = 10 µM. (E) FXR1P (red) expression in DCX-positive (green) neuroblasts and immature neurons. DAPI (blue); scale bar = 10 µM. (F) FXR1P (red) expression in NeuN-positive (white) mature neurons. DAPI (blue); scale bar = 10 µM.
To investigate the possible role of FXR1P in adult neurogenesis, we examined the expression pattern of FXR1P protein in the adult neurogenic regions. We distinguished the phases of neurogenesis using a variety of established cell lineage and cell stage-specific markers (Fig. 1B). In the DG, FXR1P was expressed in NESTIN- and GFAP-double-positive radial glia-like cells (RGLs) (Fig. 1C), as well as TBR2+ intermediate neural progenitors (IPCs) (Fig. 1D) and DCX+ neuroblasts and immature neurons (Fig. 1E). As previously published, FXR1P was also highly expressed in both granule neurons of the DG and interneurons within the hilar region (Fig. 1F). Similar to FMRP and FXR2, FXR1P was not detected strongly in S100β+ astrocytes (Supplementary Material, Fig. S1A). A similar expression pattern was also seen in the subventricular zone (SVZ), the other adult neurogenic zone in the brain (Supplementary Material, Fig. S1B–D). Therefore, FXR1P is expressed in new cells generated in the adult DG throughout the progression of adult neurogenesis and could play important roles in neurogenic regulation.
Selective deletion of FXR1P in adult neural stem cells leads to fewer adult-born cells
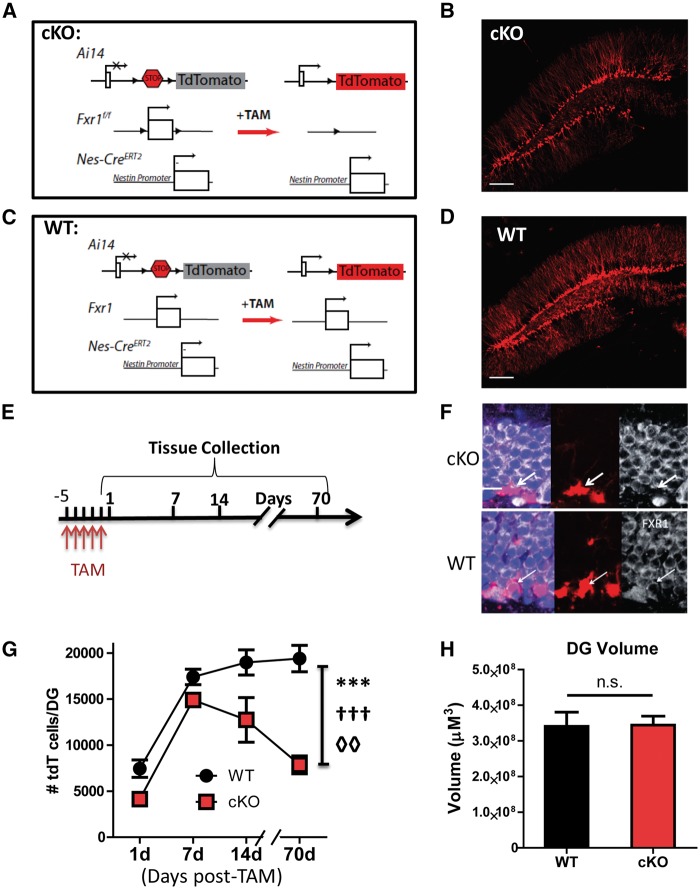
To specifically examine the intrinsic effects of FXR1P on adult neurogenesis, we generated an inducible conditional knockout mouse line (cKO: Fxr1f/f;Nes-CreERT2;Ai14) by crossing Fxr1 floxed mice (Fxr1f/f) with a tamoxifen-inducible Nestin promoter-driven Cre line (Nes-CreERT2) and the Rosa26-STOP-tdTomato reporter line (Ai14) (Fig. 2A and B) (28,42,43). The control line (WT: Nes-Cre ERT2;Ai14) lacks the floxed Fxr1 allele but has both Nes-CreERT2 and Ai14 (Fig. 2C and D). Following the published protocol, 8-week-old male mice were injected with tamoxifen for 5 days to achieve recombination in 95% of NESTIN-expressing cells (37,42,44). We collected brain tissues at 1, 7, 14 and 70 days post-injection, which allowed us to assess both the early stages and final terminal differentiation in adult neurogenesis (Fig. 2E). Upon injection of tamoxifen into adult mice, the first exon of the Fxr1 gene was deleted in NESTIN-expressing cells, and the stop codon in front of the tdTomato reporter gene was removed, causing these cells and their progeny to be tdTomato+ (tdT+, red) (Fig. 2A, B and F). Control animals (WT) underwent injections but lacked the floxed Fxr1 allele, allowing FXR1P expression to continue (Fig. 2C, D and F). Since FXR1P was efficiently deleted in tdT+ cells in the DG of FXR1P-cKO mice, we used this model to study the impact of FXR1P loss in aNSCs (Fig. 2F).
Figure 2.
Selective deletion of FXR1P in adult neural stem cells leads to fewer adult-born cells. (A) Schematic representation of Fxr1f/f;Nestin-CreERT2;Ai14 (cKO) mouse model. Tamoxifen injection (TAM) leads to Cre-mediated removal of the first exon of Fxr1, stopping its expression in Nestin-positive aNSCs. At the same time, a stop codon is removed before tdTomato (tdT), marking these cells red. (B) Example cKO image of a 70-day post-injection brain. Scale bar = 150 µM. (C) Schematic representation of control Nestin-CreERT2;Ai14::tdTomato (WT) mouse model. In wildtype brains, the Fxr1 gene lacks loxP sites, allowing continuous FXR1P expression in the tdTomato-positive (tdT+) cells. (D) Example WT image of a 70-day post-injection brain. Scale bar = 150 µM. (E) Eight-week-old male mice were injected with tamoxifen (180 mg/kg) for five days. WT and cKO mice brains were collected at 1, 7, 14 and 70 days post-injection to correspond to stages of neurogenesis. (F) The deletion efficiency of FXR1P was assessed in tdT cells by immunofluorescence. FXR1P (white), tdT (red), DAPI (blue). Scale bar = 10 µM. (G) Quantitative analyses of tdT+ cells produced in the adult DG over time (n = 3–6). Error bars indicate mean ± SEM. Symbols indicate statistical differences determined by Two-way ANOVA. ***: Significantly different between genotypes, F1,27 = 46.38, P < 0.0001; †††: Significantly different among time points, F3,27 = 38.54, P < 0.0001; ◊◊: Significant interaction between genotype and time points, F3,27 = 4.75, P = 0.0087. (H) The volume of the DG at the 70-day time point was assessed by stereological measurement (n = 3). Error bars indicate mean ± SEM.
We first quantified the number of tdT+ cells in the DG. The cKO mice contained significantly lower numbers of tdT+ cells than WT animals (Fig. 2G; genotype, F1,27 = 46.38, P < 0.0001). In particular, cKO mice and WT mice exhibited significant differences in the dynamics of cell number change over time (genotype and time interaction, F3,27 = 4.75, P = 0.0087). The number of new cells in cKO mice was modestly reduced at 1 and 7 days post-injection compared to WT mice; however, this difference became more pronounced at 14 days post-injection. While the number of tdT+ cells continued to increase in WT mice, the cell numbers in cKO mice declined. Since it takes 30–60 days for adult NSCs to become new neurons, we analyzed mice at 70 days post-injection and found that the number of new cells further declined compared to cKO mice at 14 days post-injection, whereas new cell numbers continued to increase in WT mice (Fig. 2G). Despite this difference, examining the animals at the 70-day time point, we found no significant decrease in DG volume between WT and cKO mice (Fig. 2H). Therefore, the loss of FXR1P in NESTIN-expressing stem cells leads to fewer new cells produced in the adult DG over time.
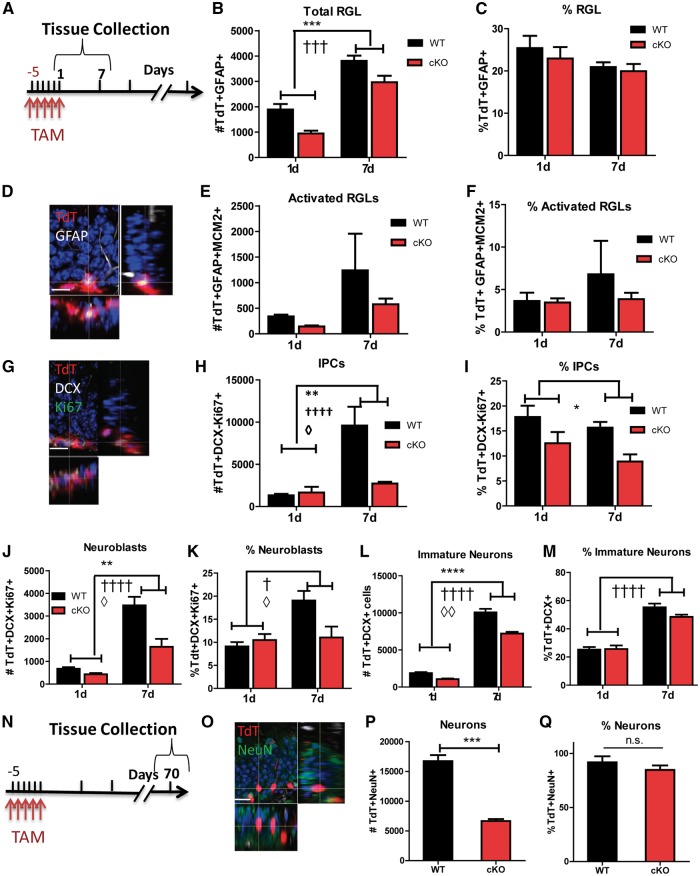
The reduction in new cell numbers in Fxr1-cKO animals is due to decreased proliferative cell types during adult neurogenesis
The reduction in cell numbers in cKO mice could either be due to a decrease in proliferation or an increase in cell death. To distinguish these two potential mechanisms, we assessed cell death at seven days post-injection and found no significant difference in the percentage of tdT+ cells that were activated caspase3-positive between cKO and WT mice, suggesting that cell death may not be behind the reduced cell numbers in cKO mice (data not shown). Next, we used cell cycle and lineage-specific markers, as well as morphological features to quantify individual cell populations within the total tdT+ cells (Fig. 1B). We first examined the total RGL (tdT + GFAP+) and activated RGL (tdT + GFAP + MCM2+) populations at the early time points (Fig. 3A–F). We found that the total number of RGLs (genotype F1,16 = 19.17, P = 0.0005) but not activated RGLs (F1,15 = 1.239, P = 0.2832) was significantly decreased in cKO compared to WT animals at the 1- and 7-day time points (Fig. 3B and E). The proportion of total and activated RGLs among tdTomato cells, however, was not significantly different between WT and cKO animals (total RGLs: genotype, F1,16 = 0.5871, P = 0.4547; activated RGLs: genotype F1,16 = 0.5601, P = 0.4651) (Fig. 3C and F). In addition, at day 1 and day 7, the number of proliferating neuroblasts (tdT + DCX + ki67+; genotype: F1,17 = 11.57, P = 0.0034) and IPCs (tdT + DCX-ki67+; genotype: F1,16 = 7.713, P = 0.0135) were significantly lower in cKO mice compared to WT mice (Fig. 3G, H and J). There were also significant differences in the proportion of neuroblasts over time (genotype and time: F1,17 = 5.627, P = 0.0298) and proportion of IPCs (genotype: F1,16 = 10.73, P = 0 .0048) (Fig. 3I and K). The overall number of immature neurons was lower in cKO mice compared to WT mice (genotype: F1,17 = 46.82, P < 0.0001), although the proportion of immature neurons (DCX + tdT+/tdT + total) remained unchanged between genotypes (genotype: F1,17 = 2.039, P = 0.1714) (Fig. 3L and M). Finally, we examined the terminal differentiation of these cells at 70 days post-injection and found that the numbers of tdT + NeuN+ neurons created were significantly decreased (n = 3, P < 0.001), despite the fact that the proportion of neurons among the tdT+ population was no different between WT and cKO animals (n = 3, P > 0.1) (Fig. 3N–Q). Thus, although the total numbers of each cell type were decreased in cKO animals throughout the timeline, this reduction was not due to reduced differentiation or increased cell death but rather a decrease in the proportions of proliferating neuroblasts and IPCs. This indicates that the strongest impact of FXR1P deficiency is on proliferation rather than cell death or differentiation.
Figure 3.
The reduction in new cell numbers in Fxr1-cKO animals is due to decreased proliferative cell types during adult neurogenesis. (A) Timeline of tissue collection for B-M. Eight-week-old male mice were injected with tamoxifen (TAM) (180 mg/kg) for five days. WT and cKO brains were collected at 1 or 7 days after the last injection. (B) Quantitative comparison of the numbers of tdT + GFAP+ RGLs among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (C) Comparison of the percentage of tdT + GFAP+ RGLs among total tdT+ cells in the DG of cKO (red) and WT control (black) mice. (D) Representative staining of a tdT + GFAP+ RGL. tdT (red); MCM2 (green); GFAP (white); DAPI (blue). Scale bar = 20 µM. (E) Quantitative comparison of the numbers of tdT + MCM2 + GFAP+ activated RGLs among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (F) Comparison of the percentage of tdT + MCM2 + GFAP+ activated RGLs among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (G) Representative staining of tdT + Ki67 + DCX+ neuroblasts. tdT (red); Ki67 (green); DCX (white); DAPI (blue). Scale bars = 20 µM. (H) Quantitative comparison of the numbers of tdTomato + Ki67 + DCX- IPCs in the DG of cKO (red) and WT control mice (black). (I) Comparison of the percentage of tdT + Ki67 + DCX- IPCs among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (J) Quantitative comparison of the numbers of tdTomato + DCX+ Ki67+ neuroblasts in the DG of cKO (red) and WT control mice (black). (K) Comparison of the percentage of tdT + DCX+ Ki67+ neuroblasts among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (L) Quantitative comparison of the numbers of tdT + DCX+ immature neurons in the DG of cKO (red) and WT control mice (black). (M) Comparison of the percentage of tdT + DCX+ immature neurons among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (N) Timeline of tissue collection for O-Q. Eight-week-old male mice were injected with tamoxifen (180 mg/kg) for 5 days. WT and cKO mice brains were collected at day 70 to assess for mature neurons. (O) Representative staining of a tdT + NeuN+ mature neurons. tdT (red); NeuN (green); DAPI (blue). Scale bars = 20 µM. (P) Quantitative comparison of the numbers of tdT + NeuN+ mature neurons among total tdT+ cells in the DG of cKO (red) and WT control mice (black). (Q) Comparison of the percentage of tdT + NeuN+ neurons among total tdT+ cells in the DG of cKO (red) and WT control mice (black). For all graphs the bar represents ± SEM. ANOVA symbols: Significantly different between genotypes: *P < 0.05; **P < 0.01; ***P < 0.001; Significantly different between time points: †P < 0.05; ††P < 0.01; †††P < 0.001; Significant interaction between genotypes and time points, ◊P < 0.05; ◊◊P < 0.01; ◊◊◊P < 0.001.
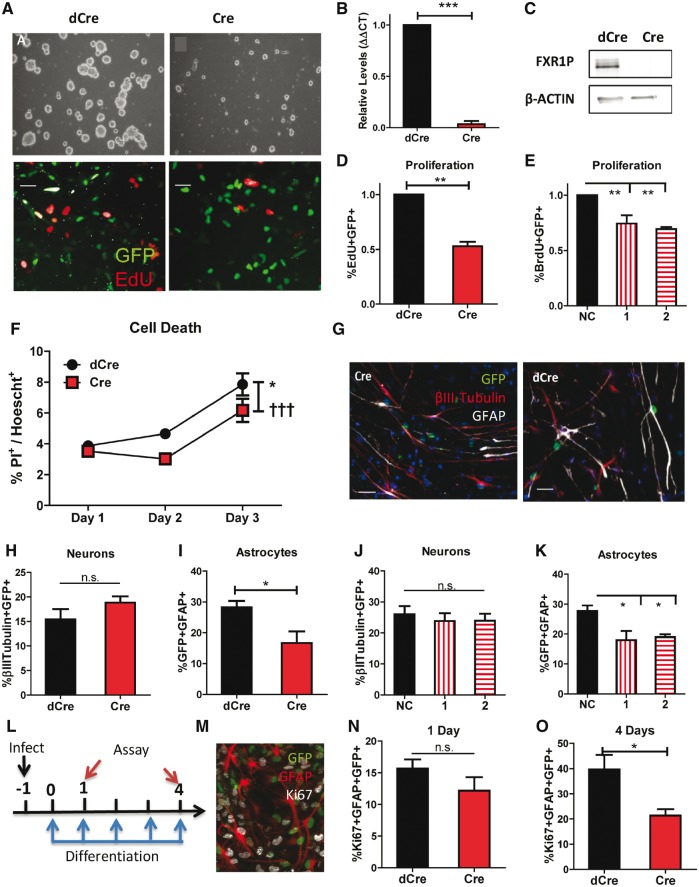
FXR1P deletion in primary aNSCs leads to decreases in proliferation and cell death
To confirm our in vivo results, we assessed the impact of FXR1P loss in primary adult neural stem/progenitor cells (aNSCs) isolated from 8-week-old Fxr1f/f male mice (45). These cells grow as neurospheres and are similar to the fast-proliferating IPCs in the DG. We infected the aNSCs with lentivirus expressing either a nuclear localized Cre-GFP fusion protein (Cre) to delete Fxr1 or a non-functional delta Cre-GFP fusion protein control (dCre) (Fig. 4A) (46). The Cre virus-infected cells exhibited significantly reduced Fxr1 mRNA and FXR1P protein levels, demonstrating that this is an effective Fxr1 deletion model (Fig. 4B and C).
Figure 4.
FXR1P deletion in primary aNSCs leads to decreases in proliferation and cell death. (A) Primary aNSCs from a Fxr1f/f mouse, infected with either a control Lenti-dCre (left panels) or Lenti-Cre to delete Fxr1 (right panels). Top panels: Neurospheres four days post-infection after trypsinizing cells at day 2. Bottom panels: Representative proliferation assays, virus (green), EdU (red). (B) Relative expression of Fxr1 mRNA in control dCre- (black) and Cre-infected cells (red). (C) Immunoblot of FXR1P in control dCre- (black) and Cre-infected cells (red). (D) Comparison of the percentage of EdU + GFP+ cells out of total GFP+ cells, relative to dCre control (black) in Fxr1f/f aNSCs. (E) Comparison of the percentage of BrdU + GFP+ cells out of total GFP+ cells, for Fxr1-shRNAs (1 and 2, vertical and horizontal red stripe, respectively) relative to non-coding (NC) shRNA control (black). (F) Comparison of the percentage of propidium iodide-positive (PI+) cells out of total Hoechst-positive cells for dCre- (black circles) and Cre-infected (red squares) Fxr1f/f cells. *Significantly different between dCre and Cre, F1,8 = 9.376, P < 0.05; †††: Significantly different among time points, F2,8 = 34.93, P = 0.0001 (G) Representative images for four-day differentiation into neurons (red, βIII-Tubulin) and astrocytes (white, GFAP) for dCre- and Cre-infected Fxr1f/f aNSCs (green). DAPI = blue. (H) To evaluate neuronal differentiation, the percentage of βIII-tubulin + GFP+ cells among total GFP+ cells was assessed for dCre- (black) and Cre-infected (red) Fxr1f/f cells. (I) To evaluate astrocyte differentiation, the percentage GFAP + GFP+ cells among total GFP+ cells was assessed for dCre- (black) and Cre-infected (red) Fxr1f/f aNSCs. (J) The percentage of βIII-tubulin + GFP+ among total GFP+ cells was assessed for non-coding (NC) shRNA control (black) and Fxr1-shRNAs (1 and 2, vertical and horizontal red stripe, respectively)-infected Fxr1f/f aNSCs. (K) The percentage GFAP + GFP+ among total GFP+ cells was assessed for non-coding (NC) shRNA control (black) and Fxr1-shRNAs (1 and 2, vertical and horizontal red stripe, respectively) infected aNSCs. (L) Timeline for astrocyte proliferation assay. Fxr1f/f aNSCs were infected on day -1, then treated with retinoic acid (RA) and forskolin (FSK) for 1 to 4 days before fixation and staining. (M) Representative image for 4-day astrocyte proliferation assay. Virus (green), GFAP (red), Ki67 (white). (N) Astrocyte proliferation assay after 1 day of differentiation; comparison of the percentage of Ki67 + GFAP + GFP+/GFAP + GFP+ cells for dCre- (black) and Cre-infected (red) Fxr1f/f aNSCs. (O) Assessment of astrocyte proliferation after 4 days of differentiation; comparison of the percentage of Ki67 + GFAP + GFP+ cells over GFAP + GFP+ cells for dCre- (black) and Cre-infected (red) Fxr1f/f aNSCs. For all graphs, the bar represents mean ± SEM. See text for statistical analysis.
We discovered that FXR1P-deficient aNSCs exhibited a drastic reduction in cell numbers in culture. In fact, we had difficulty maintaining these cells for more than 4 or 5 days, whereas Fxr1f/f cells infected with dCre virus could be propagated for many generations in culture (Fig. 1A, upper panels). This observation is consistent with the reduced number of tdT+ new cells that we found in the DG of adult cKO mice. Since FXR1P deficiency led to significant reductions in proliferative cell types in vivo, we first assessed aNSC proliferation using pulse labeling by a thymidine analog, EdU. Cre virus-infected aNSCs exhibited significantly reduced EdU labeling, indicative of reduced proliferation (n = 4, P = 0.0013) (Fig. 4A, lower panels, 4D). To ensure that this result was not due to a nonspecific effect of the Cre protein, we knocked down FXR1P in aNSCs using lentivirus expressing small hairpin RNAs against Fxr1 (shFxr1-2). We found that shFxr1-infected aNSCs exhibited significantly reduced EdU labeling compared to control shRNA (shNC)-infected aNSCs (genotype, F = 13.47, P = 0.006) (Fig. 4E). We again investigated whether FXR1P deficiency in primary aNSCs affected cell death. To do this, we used propidium iodide (PI), which can enter dead cells but is excluded from live cells. After 2 days post-Cre viral infection of Fxr1f/f aNSCs, we added PI to the cells and quantified the percentage of PI-positive cells among total cells during a 3-day culture period (Fig. 4F). The percentages of PI-positive cells were similar on day 1, but to our surprise, cell death was significantly lower overall in Cre viral-infected compared to dCre viral-infected cells (genotype, F = 9.38, P = 0.0376). Therefore, consistent with our in vivo findings, the drastic reduction in the numbers of FXR1P-deficient aNSCs was a result of reduced cell proliferation rather than increased cell death.
We next assessed whether FXR1P deficiency affected the differentiation of aNSCs using cell lineage markers βIII-tubulin for neurons and GFAP for astrocytes (Fig. 4G). We found that FXR1P-deficient (Cre) aNSCs showed no significant difference in neuronal differentiation compared to control aNSCs (dCre) (Fig. 4H). However, the percentage of astrocytes produced from FXR1P-deficient cells was decreased by a third compared with those formed from WT cells (Fig. 4I). These findings were further confirmed by acute knockdown of FXR1P using shFxr1 (astrocytes, F = 7.645, P = 0.0224) (Fig. 4J and K). Since we saw no change in neuronal differentiation and astrocytes are known to be proliferative even after differentiation, we postulated that this reduction in astrocyte production was not necessarily due to altered fate specification but rather altered proliferation of astrocytes. To test this theory, we assessed proliferation of astrocytes using proliferative marker Ki67 after 1 and 4 days of aNSC differentiation (Fig. 4L and M). The proportion of proliferative astrocytes (Ki67 + GFAP+) trended downward for Cre-infected cells after 1 day of differentiation and was significantly lower compared to dCre-infected cells at day 4 (n = 3, P < 0.05) (Fig. 4N and O). This is consistent with our in vivo observation that the deletion of FXR1P causes a strong proliferative deficit rather than changes in cell fate, leading to decreases in astrocyte numbers.
FXR1P directly binds and regulates p21 mRNA levels
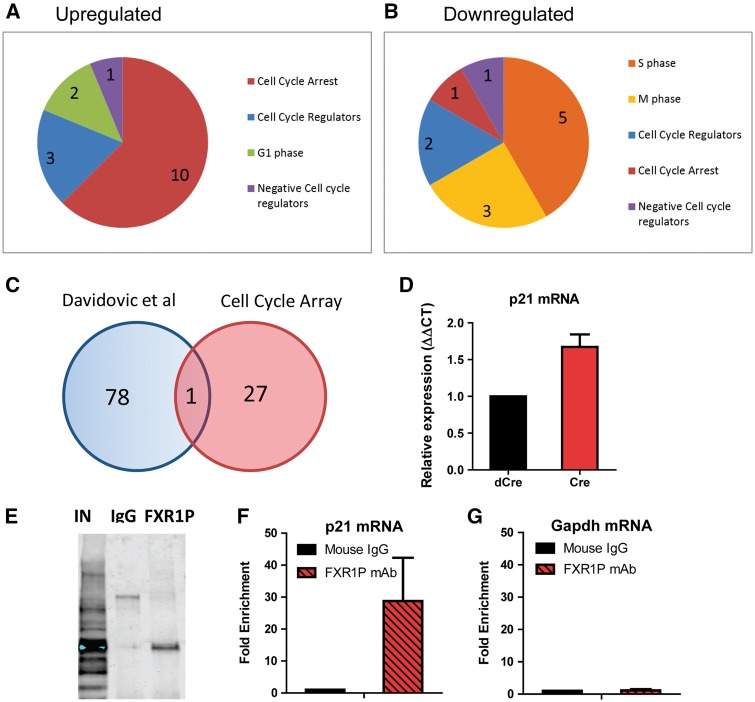
Since deletion of FXR1P in primary aNSCs recapitulated our findings in vivo, we decided to use these cells to further examine the mechanism underlying FXR1P’s regulation of aNSCs. Because FXR1P-deficient aNSCs exhibited significant changes in cell proliferation, we compared Cre and dCre virus-infected aNSCs using Cell Cycle qPCR pathway arrays (Qiagen; Supplementary Material, Table S1). We found that the most commonly upregulated mRNAs in Cre-infected aNSCs were for cell cycle arrest factors, whereas the most common downregulated genes were factors for cell cycle progression (S and M phase), which is consistent with the reduced proliferation phenotype of Cre-infected cells (Fig. 5A and B). We next compared our list of differentially expressed mRNAs to a published list of mRNAs differentially expressed in an FXR1P-deficient muscle stem cell line (Fig. 5C) (33). From this comparison, we found a single factor in common, p21 (P21Cip1 or P21Waf1), a well-known cell cycle inhibitor (47). We further confirmed that Cre virus-infected aNSCs indeed exhibited increased p21 mRNA levels (Fig. 5D). Since p21 acts to arrest the cell cycle and inhibit cell death, the upregulation of p21 mRNA is consistent with the decreases in proliferation and cell death. Therefore, FXR1P may regulate aNSC proliferation through direct regulation of p21 mRNA.
Figure 5.
FXR1P directly binds and regulates p21 mRNA levels. (A) Pie graph categorizing mRNAs upregulated on the Qiagen Cell Cycle qPCR arrays in Cre-infected Fxr1f/f aNSCs relative to dCre-infected cells. (B) Pie graph categorizing mRNAs downregulated on the Qiagen Cell Cycle qPCR arrays in Cre-infected Fxr1f/f aNSCs relative to dCre-infected cells. (C) Venn diagram of published FXR1P targets (Davidovic et al. 2013 (33)) compared to mRNAs represented on Qiagen cell cycle arrays. (D) Relative expression of p21 mRNA in control dCre- (black) and Cre-infected Fxr1f/f aNSCs (red). (E) Immunoblot using 830 FXR1P polyclonal antibody, representing RNA immunoprecipitation (RIP) input (IN), mouse IgG control (IgG), and FXR1P mAb (FXR1P) pulldowns. (F) Comparison of RIP enrichment of p21 mRNA relative to input for IgG control (black) and FXR1P mAb (red). (G) Comparison of RIP enrichment of non-target Gapdh mRNA relative to input for IgG control (black) and FXR1P mAb (red). For all graphs, the bar represents mean ± SEM.
In muscle stem cells, FXR1P is found to bind p21 mRNA (33). Knowing this, we then determined whether FXR1P binds p21 mRNA in WT aNSCs using RNA immunoprecipitation (RNA-IP) comparing our FXR1P mAb and a control mouse IgG. Our antibody specifically pulled down FXR1P, versus little to none in the IgG control (Fig. 5E). When p21 mRNA levels were quantified using qPCR, they were found to be enriched over 30-fold in RIP samples compared to the IgG control samples (Fig. 5F); on the other hand, the negative control Gapdh mRNA was not significantly enriched in RIP samples compared to IgG samples (Fig. 5G). Therefore, FXR1P directly binds p21 mRNA in aNSCs, making p21 a likely modulator of FXR1P regulation of aNSCs.
Downregulation of p21 mRNA rescues the proliferation deficits in FXR1P-deficient cells
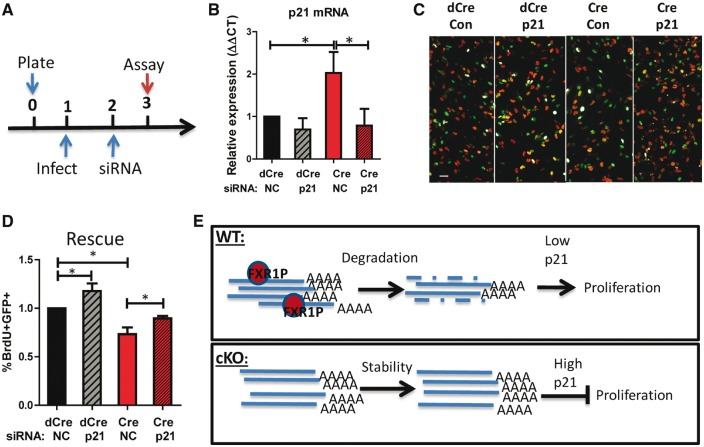
FXR1P deletion altered the levels of multiple cell cycle regulators. To determine whether p21 is a key mediator for FXR1P, we investigated whether reducing p21 levels would have a functional impact on FXR1P-deficient aNSCs. We first infected Fxr1f/f aNSCs with either Cre or dCre virus, and then we transfected small inhibitor RNA (siRNA) against p21 (siP21) or a non-silencing control siRNA (siNC) into these cells. At 2 days post-transfection, we assayed cell proliferation (Fig. 6A) and confirmed that siP21 siRNA could effectively knock down p21 expression using qPCR (Fig. 6B). We found that the acute knockdown of p21 restored proliferation of Cre-infected aNSCs (Cre, siP21) to near WT (dCre, siNC) levels (Fig. 6C and D). Therefore, p21 is a mediator of FXR1P’s regulation of aNSC proliferation.
Figure 6.
Downregulation of p21 mRNA rescues the proliferation deficits in FXR1P-deficient cells. (A) Timeline for the rescue experiment. Fxr1f/f aNSCs were infected with Cre or dCre viruses on day 1, transfected with siRNA on day 2 and assayed for proliferation on day 3. (B) Relative expression levels of p21 mRNA in Fxr1f/f aNSCs infected with either Cre or dCre lentiviruses followed by transfection with either non-silencing control siRNA (siNC) or p21 siRNA (siP21) transfected. Statistical analysis was performed using one-way ANOVA with a Bonferroni post-hoc test using dCre and siNC condition as the control, *P < 0.05. (C) Representative images of proliferating aNSCs infected with either Cre or dCre lentiviruses (green), transfected with either siNC or siP21, and pulse-labeled with BrdU (red). Scale bar = 20µM. (D) Comparison of the percentage of BrdU+GFP+ cells among total GFP+ cells, relative to dCre infected and with siNC transfected control condition (black bar) in Fxr1f/f aNSCs. Statistical analysis was performed using one-way ANOVA with a Bonferroni post-hoc test. *P < 0.05. (E) Model of FXR1P regulation of p21. WT: FXR1P normally binds p21 mRNA, leading to its instability and degradation. This leads to low levels of p21 and allows proliferation. cKO: without FXR1P, p21 mRNA is more stable, leading to higher p21 protein levels, which inhibit proliferation.
Discussion
The neonatal lethality of FXR1P-null mice clearly demonstrates that FXR1P plays an important role in development. Although most research has focused on FXR1P’s role in heart and muscle (48), FXR1P is also important in eye and neural crest development (34); however, despite being highly expressed in mouse and human neurons, particularly in the hippocampus, little is known about its role in the brain (6,49,50). In this study, we have shown a clear and important role for FXR1P in sustaining neural stem and progenitor cell proliferation during adult neurogenesis. Although FXR1P regulates multiple mRNA targets in aNSCs, here we examined only one. Others should be further explored to fully understand FXR1P's impact on adult neurogenesis.
In our model in wild-type cells, FXR1P binds to p21 mRNA (Fig. 6E). Previously, FXR1P was found to regulate p21 mRNA stability in myoblasts and cancer cells by binding an ARE in the 3’UTR, leading us to speculate that this is also the case in aNSCs (33,51). Decreased levels of p21 mRNA keep p21 protein levels low and allow proliferation to occur. When FXR1P is absent, the stability of p21 mRNA increases, leading to increased p21 protein levels (Fig. 6E). This results in the inhibition of proliferation. This cell cycle regulator is known to inhibit cell cycle progression and short-term cell death, in keeping with our phenotypic results. p21 is also known to be an important regulator of adult neurogenesis (52–54). Knockout animals exhibit increased numbers of proliferative cells, until the stem cell pool becomes depleted (52). This factor was also shown to be a key effector in antidepressant drug effects on adult neurogenesis (54,55).
FXR1P’s function in promoting proliferation is supported by research in the cancer field. FXR1P was identified as a key driver in the 3q26-27 amplicon for lung squamous cell carcinoma, and its overexpression is essential for non-small cell lung cancer growth (56,57). FXR1P is also elevated in the plasma of colorectal cancer patients and acts as an oncogene to promote the proliferation, invasion and migration of cancer cells (58). Its overexpression is also associated with poor survival outcomes (56,58). Functionally, this is likely due to FXR1P’s participation in an RNP complex that regulates translation in metastasis (59). FXR1P was also found to destabilize p21 mRNA in cancer cells, and deletion of Fxr1 in these cells rescues cell cycle control (51). This evidence supports a regulatory role for FXR1P on the cell cycle that is important in cancer as well as stem cells.
The process of adult neurogenesis relies on a largely quiescent pool of radial glia-like stem cells (RGLs) that expand into fast proliferating cell types (neuroblasts and IPCs), which mature into neurons. Changes in the proliferation and cell death of this pool can dramatically impact the maintenance of stem cell numbers throughout life. These processes can be affected by a variety of cellular factors including FMRP, FXR2 and, as demonstrated, FXR1 (21,37) . Because of the high homology of the FXRs, there has been debate about the unique and overlapping roles of these proteins. Their functional differences in the brain were first indicated in Drosophila melanogaster, which has a single dFMR1; of the three human FXRs, only the FMRP could rescue the mutant’s neuronal deficits (23). In comparing our findings to past studies, these proteins also have unique functions in neurogenesis. We found that the protein expression patterns of FXR1P, FXR2P and FMRP are highly similar within cell types in the DG, yet each has distinct functions in adult neurogenesis. Both FMRP and FXR2P inhibit proliferation of aNSCs, whereas we demonstrate here that FXR1P enhances proliferation (21,37). In terms of differentiation, FMRP promotes neurogenesis, whereas FXR2P promotes astrogenesis (21,37). Uniquely, we find that FXR1P has no demonstrated preference in aNSC differentiation. In considering all three proteins of this family, differential regulation of distinct and similar targets throughout neurogenesis could determine cell fate. It is possible that the balance and upstream regulation of the fragile X proteins control major aspects of adult neurogenesis, making an intriguing avenue for future study.
FXR1P is considered an autism-associated protein due to its high homology with FMRP (5). The silencing of the FMRP gene (FMR1) in fragile X syndrome (FXS) is the largest single-gene contributor to autism (60). FXR1P may be a key regulator in FXS as it is also capable of hetero-dimerizing with FMRP and is believed to have many overlapping mRNA targets (1–4,18). Understanding the role of FXR1P may allow us to better understand FXS. In support of this, SNPs in FXR1 are found to enhance the severity of phenotypic autistic behaviors (10). Hippocampal deletion of FXR1P also alters spatial learning and memory in adult mice (40). In addition to its interaction with FMRP, FXR1P can also bind other autism-associated proteins (12). Since FXR1P can interact with a large number of autism-associated factors, insights into its function in the brain may help explain this genetically complex disease.
Beyond its association with autism, FXR1P has ties to bipolar disorder and schizophrenia (13,14,61). Schizophrenia, unlike autism, has more direct ties to adult neurogenesis. The schizophrenia susceptibility gene, Disrupted-In-Schizophrenia 1 (Disc1), regulates both the speed and migration of new neurons in adult neurogenesis (62,63). DISC1 is a scaffolding protein that interacts with multiple pathways, including glycogen synthase kinase 3b (GSK3β) (64). FXR1P is a substrate for GSK3β, and their interaction affects emotional stability for human subjects (65). Interestingly, inhibition of GSK3β also rescues adult neurogenesis and hippocampal learning in a FXS mouse model (66). This regulation is likely important to FXR1P’s role in neurogenesis and schizophrenia and, in addition to neuronal morphology, should be explored further. In conclusion, FXR1P plays an important regulatory role in adult neurogenesis. Its function in this process may elucidate its importance in the brain and provide important therapeutic insights for a variety of neurological disorders.
Materials and Methods
Mice
All animal procedures were performed according to protocols approved by the University of Wisconsin-Madison Animal Care and Use Committee. All mice were maintained on a C57B/L6 genetic background. Nestin-GFP mice were used solely for immunohistochemistry (67). The Fxr1f/f mice and Fxr1-/-, KO tissue were obtained from Baylor College of Medicine and used for mAb validation (28). We generated the inducible conditional knockout and wildtype control mice by breeding Nes-CreERT2 with the Ai14 reporter line (42,43). Double heterozygous offspring were then crossed with the Fxr1f/f line and then further crossed to create Fxr1f/f::Nes-CreERT2::Ai14 (cKO) and Nes-CreERT2::Ai14 (WT) homozygous lines used for in vivo fate mapping. Tamoxifen (Sigma-Aldrich) injection and preparation were performed based on a previously published procedure (42).
Tissue preparation, immunohistochemistry and confocal imaging
Brain tissue processing and histological analysis of mouse brains were performed as described in our previous publications (21,37,44). FXR1P mAb was created in collaboration with Neoclone against amino acids 543-562, which had generated previously successful polyclonal antibodies (32,41). The primary antibodies used were: chicken-anti-GFP (1:500, Invitrogen, A10262), rabbit anti-MCM2 (1:500, Cell Signaling, 4007), rabbit anti-GFAP (1:1000, Dako, Z0334), mouse anti-Ki67 (1:500, Thermo, 9106S), mouse anti-NeuN (1:500, Millipore, MAB377), rabbit anti-S100β (1:1000, Dako, Z0334), rabbit DCX (1:500, Cell Signaling, 4604) and chicken anti-TBR2 (1:1000, Millipore, AB15894). Fluorescent secondary antibodies used were: goat anti-mouse 568 (1:500, Invitrogen, A11004), goat anti-rat 568 (1:500, Invitrogen, A11077), goat anti-rabbit 568 (1:500, Invitrogen, A11011), goat anti-rabbit 647 (1:500, Invitrogen, A21245), goat anti-mouse 647 (1:500, Invitrogen, A21235), goat anti-chicken 488 (1:500, Invitrogen, A11039) and goat anti-mouse 488 (1:500, Invitrogen, A11029).
I n vivo cell fate mapping
For in vivo fate mapping of tdT+ cells, at 1, 7, 14 or 70 d after the last tamoxifen injection, we killed mice by intraperitoneal injection of sodium pentobarbital, and then transcardial perfusion of 4% paraformaldehyde. We performed histological analysis of mouse brains as previously described (68,69). For quantification of total tdT+ DG cells, we used 1 in 6 serial sections from the beginning to the end of the hippocampus and stained with DAPI and determined the tdT+ cell count and volume (three-dimensional size) of the dentate gyrus using unbiased stereology (Stereo Investigator, MBF Biosciences). For individual cell populations, 2–5 sections were stained (see above for antibody details), and we used a Nikon A1 confocal microscope. Then 100+ tdT+ cells were randomly selected, and their colocalization with cell lineage markers was determined and quantified using ImageJ software and the Cell Counter plugin [National Institutes of Health (NIH)]. This number was used as a percentage of a specific population. These numbers were then multiplied by total cell counts from Stereo Investigator to calculate the cell type-specific population.
Isolation and analysis of aNSCs
Primary aNSCs were isolated and cultured from 8-week-old Fxr1f/f or WT mice as previously described (45). Three lines of independently isolated cells served as biological replicates. Proliferation and differentiation of NPCs were analyzed for proliferation, differentiation and cell death as described (21,37,44). Proliferation was assessed using BrdU or EdU pulse labeling and cell death was determined using propidium iodide (Sigma). Both proliferation and cell death were quantified using a Operetta high content imaging system and the Harmony software (PerkinElmer). We used only early passage cells (P4-10), and 2–3 replicate wells of cells were analyzed and averaged as one data point (n = 1). Fxr1f/f cells were infected with a lentivirus expressing a Cre-GFP fusion protein or a nonfunctional dCre-Cre GFP control. Constructs were previously published and a generous gift from Lu Chen (Stanford) (46). Lentiviral vectors expressing shFxr1 were created from purchased SureSilencing shFxr1 vectors (Qiagen, KM35574G) as described (21). Lentiviruses were prepared as previously described (70). aNSCs were assayed at 2 days post-infection. For siRNA rescue experiments, aNSCs were first infected with lentivirues, and at 1 day post-infection, transfected with double-stranded scrambled siRNA control (IDT, 51-01-19-08) or previously publishes p21 siRNA (AGACC AGCCUGACAGAUUU) using SiLentFect (BioRad) as described in the literature (71,72). At least three independent biological replicates were used (n = 3) for statistical analyses.
RNA immunoprecipitation
RNA-IP was performed on WT aNSCs as described, using 1 mg of monoclonal antibody against immunoglobulin G (IgG) (5415S, Cell Signaling) or FXR1 mAb (Neoclone) (21,44). An aliquot of precleared input was saved for RNA extraction (200 ml) and protein analysis (100 ml). The precleared lysates were incubated with antibody for 2 h and immunoprecipitated with recombinant G Dynabeads (Invitrogen) at 4 °C for 1 h. After 3 washes with lysis buffer, 10% of immunoprecipitate was saved for protein analysis. The remainder was re-suspended in 1 mL TRIzol (Invitrogen) for RNA isolation.
Real-time quantitative PCR and qPCR pathway arrays
Real-time PCR was performed using standard methods as described (60). The first-strand complementary DNA (cDNA) was generated by reverse transcription with oligo(dT) primer (Roche). To quantify the mRNA expression using real-time PCR, aliquots of first-strand cDNA were amplified with gene-specific primers and SYBR Green PCR Master Mix (Bio-Rad) using a StepOne Real-Time PCR System (Applied Biosystems). The PCR reactions contained 1 mg cDNA (except the cDNA for immunoprecipitation, for which 5% of the cDNA was used for each gene examined), Universal Master Mix (Applied Biosystems) and 10 mM of forward and reverse primers in a final reaction volume of 20 ml. The mRNA expression of different samples was calculated by the data analysis software built in with the 7300 Real-Time PCR System. For RNA immunoprecipitation/real-time PCR, cDNA from immunoprecipitation and input were used and immunoprecipitated samples were normalized to input samples. Fxr1-f: GATGATCGAGAGACTCGACATC; Fxr1-R: TTA G T A CTGTGGTGAGATTCGC; cdkn1a(p21)-f: GGAA CATCTCAGG GC CG AAA; cdkn1a(p21)-R: GAAGATGGGGAAGAGGCCTC; gapdh-f: GCTCCTCCCTGTTCCAGAGACGG; gapdh-f: ACA ATCT CCAC TTT G CCACTGC.
Immunoblot
Protein samples were separated on SDS–polyacrylamide gel electrophoresis gels (Bio-Rad), transferred to polyvinylidene difluoride membranes (Millipore) and incubated with primary antibodies. Primary antibodies include anti-actin (1:5000; Sigma), anti-GAPDH (1:5000; Thermo Scientific, MA5-15738), anti-FXR1P rabbit polyclonal 830 antibody (1:5000; gift) and FXR1P monoclonal antibody (1:500; created in collaboration with Neoclone, Inc). After incubation with fluorescence-labeled secondary antibodies (LI-COR), the membranes were imaged using a LI-COR Odyssey.
Statistical analysis
All experiments were randomized and blinded to scientists who performed quantification. Statistical analyses were performed using ANOVA and Student’s t-test, unless otherwise specified, with the GraphPad software. Two-tailed and unpaired t-tests were used to compare two conditions. One-way ANOVA was used for comparison among multiple experimental conditions. Bonferroni's post-hoc test was then used to determine significance between paired conditions. Two-way ANOVAs were used to compare multiple time points and genotypes during in vivo cell fate mapping.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to thank David Wassarman, Erik Dent, Randall Tibbetts and Avtar Roopa (UW-Madison) for critical discussion and manuscript feedback. Special thanks to Laetitia Davidovic (Center National De La Recherché Scientifique) and Edward Khandjian (Universite Laval) for 830 and mL13 FXR1P antibodies; Ruiting Zong and David Nelson (Baylor) for Fxr1f/f mice and Fxr1-/- neonatal tissue; Amelia Eisch (UT-Southwestern) for Nes-CreERT2 mice; and Lu Chen (Stanford) for lentivectors expressing Cre and dCre. We would like to thank Cheryl Strauss for editing, Aimee Teo Broman for statistical consultation, Yina Xing for technical assistance, Karla Knobel at the Waisman Cell and Molecular Neuroscience Core, and Jason Pinnow, Dawna Bollig and Megan Eastwood at the Waisman Rodent Models Core.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (MH080434, MH07897, R21NS095632) to X.Z.; National Institutes of Health (P30HD03352; U54 HD090256) to Waisman Center IDDRC; Molecular and Cellular Pharmacology Training Grant National Institutes of Health (T32 GM008688) to N.E.P.; a National Institutes of Health NSRA (F31MH103945) to N.E.P.; the Wayne and Jean Roper Memorial Wisconsin Distinguished Graduate Student Fellowship to N.E.P. and the Hilldale Undergraduate Fellowship to K.N.
References
- 1. Tamanini F., Unen L.V.A.N., Bakker C., Sacchi N., Galjaard H., Oostra B.A., Hoogeveen A.T. (1999) Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. Biochem. J., 523, 517–523. [PMC free article] [PubMed] [Google Scholar]
- 2. Siomi M.C., Zhang Y. (1996) Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol., 16, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y., O’Connor J.P., Siomi M.C., Srinivasan S., Dutra A., Nussbaum R.L., Dreyfuss G. (1995) The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J., 14, 5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ascano M., Mukherjee N., Bandaru P. (2012) FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature, 492, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siomi M.C., Siomi H., Sauer W.H., Srinivasan S., Nussbaum R.L., Dreyfuss G. (1995) FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J., 14, 2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakker C., de Diego Otero Y., Bontekoe C., Raghoe P., Luteijn T., Hoogeveen A.T., Oostra B.A., Willemsen R. (2000) Immunocytochemical and biochemical characterization of FMRP, FXR1P, and FXR2P in the mouse. Exp. Cell Res., 258, 162–170. [DOI] [PubMed] [Google Scholar]
- 7. Christie S.B., Akins M.R., Schwob J.E., Fallon J.R. (2009) The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J. Neurosci., 29, 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akins M.R., Leblanc H.F., Stackpole E.E., Chyung E., Fallon J.R. (2012) Systematic mapping of Fragile X granules in the developing mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. J. Comp. Neurol., 520, 3687–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levenga J., Buijsen R.A.M., Rifé M., Moine H., Nelson D., Oostra B.A., Willemsen R., de Vrij F.M.S. (2009) Ultrastructural analysis of the functional domains in FMRP using primary hippocampal mouse neurons. Neurobiol. Dis., 35, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stepniak B., Kästner A., Poggi G., Mitjans M., Begemann M., Hartmann A., Van Der Auwera S., Sananbenesi F., Krueger-burg D., Matuszko G.. et al. (2015) Accumulated common variants in the broader fragile X gene family modulate autistic phenotypes. EMBO Mol. Med., 7, 1565–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Auranen M., Vanhala R., Varilo T., Ayers K., Kempas E., Ylisaukko-Oja T., Sinsheimer J.S., Peltonen L., Järvelä I. (2002) A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am. J. Hum. Genet., 71, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sakai Y., Shaw C.A., Dawson B.C., Dugas D.V., Al-Mohtaseb Z., Hill D.E., Zoghbi H. (2011) Protein interactome reveals converging molecular pathways among autism disorders. Sci. Transl. Med., 3, 86ra49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu X., Kelsoe J.R., Greenwood T.A. (2016) A genome-wide association study of bipolar disorder with comorbid eating disorder replicates the SOX2-OT region. J. Affect. Disord., 189, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H.. et al. (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hauberg M.E., Roussos P., Grove J., Børglum A.D., Mattheisen M. (2016) Analyzing the role of microRNAs in schizophrenia in the context of common genetic risk variants. JAMA Psychiat., 73, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O’Donovan M.C., Sullivan P.F., Sklar P. (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kirkpatrick L.L., McIlwain K.A., Nelson D. (2001) Comparative genomic sequence analysis of the FXR gene family: FMR1, FXR1, and FXR2. Genomics, 78, 169–177. [DOI] [PubMed] [Google Scholar]
- 18. Darnell J.C., Fraser C.E., Mostovetsky O., Darnell R. (2009) Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum. Mol. Genet., 18, 3164–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bechara E.G., Davidovic L., Melko M., Bensaid M., Tremblay S., Grosgeorge J., Khandjian E.W., Lalli E., Bardoni B. (2007) Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Res., 35, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo W., Polich E.D., Su J., Gao Y., Christopher D.M., Allan A.M., Wang M., Wang F., Wang G., Zhao X. (2015) Fragile X proteins FMRP and FXR2P control synaptic GluA1 expression and neuronal maturation via distinct mechanisms. Cell Rep., 11, 1651–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo W., Zhang L., Christopher D.M., Teng Z.Q., Fausett S.R., Liu C., George O.L., Klingensmith J., Jin P., Zhao X. (2011) RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron, 70, 924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cavallaro S., Paratore S., Fradale F., de Vrij F.M.S., Willemsen R., Oostra B.A. (2008) Genes and pathways differentially expressed in the brains of Fxr2 knockout mice. Neurobiol. Dis., 32, 510–520. [DOI] [PubMed] [Google Scholar]
- 23. Coffee R.L., Tessier C.R., Woodruff E.A., Broadie K.S. (2010) Fragile X mental retardation protein has a unique, evolutionarily conserved neuronal function not shared with FXR1P or FXR2P. Dis. Model. Mech., 3, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu X.L., Zong R., Li Z., Biswas M.H.U., Fang Z., Nelson D., Gao F.B. (2011) FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J. Neurosci., 31, 13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubé M., Huot M.E., Khandjian E.W. (2000) Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet., 1, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheever A., Blackwell E., Ceman S. (2010) Fragile X protein family member FXR1P is regulated by microRNAs. RNA, 16, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahishi L., Usdin K. (2006) NF-Y, AP2, Nrf1 and Sp1 regulate the fragile X-related gene 2 (FXR2). Biochem. J., 400, 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mientjes E., Willemsen R., Kirkpatrick L.L., Nieuwenhuizen I., Hoogeveen-Westerveld M., Verweij M., Reis S., Bardoni B., Hoogeveen A.T., Oostra B.A.. et al. (2004) Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum. Mol. Genet., 13, 1291–12302. [DOI] [PubMed] [Google Scholar]
- 29. Yang C.X., Wright E.C., Ross J.W. (2012) Expression of RNA-binding proteins DND1 and FXR1 in the porcine ovary, and during oocyte maturation and early embryo development. Mol. Reprod. Dev., 79, 541–552. [DOI] [PubMed] [Google Scholar]
- 30. Truesdell S.S., Mortensen R.D., Seo M., Schroeder J.C., Lee J.H., Letonqueze O., Vasudevan S. (2012) MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep., 2, 842.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mortensen R.D., Serra M., Steitz J. a., Vasudevan S. (2011) Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc. Natl. Acad. Sci., 108, 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khandjian E.W., Bardoni B., Corbin F., Sittler A., Giroux S., Heitz D., Tremblay S., Pinset C., Montarras D., Rousseau F.. et al. (1998) Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet., 7, 2121–2128. [DOI] [PubMed] [Google Scholar]
- 33. Davidovic L., Durand N., Khalfallah O., Tabet R., Barbry P., Mari B., Sacconi S., Moine H., Bardoni B. (2013) A novel role for the RNA–binding protein FXR1P in myoblasts cell-cycle progression by modulating p21/Cdkn1a/Cip1/Waf1 mRNA stability. PLoS Genet., 9, e1003367.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gessert S., Bugner V., Tecza A., Pinker M., Kühl M. (2010) FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev. Biol., 341, 222–235. [DOI] [PubMed] [Google Scholar]
- 35. Spalding K.L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H.B., Boström E., Westerlund I., Vial C., Buchholz B.A.. et al. (2013) Dynamics of hippocampal neurogenesis in adult humans. Cell, 153, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin X. (2016) The role of neurogenesis during development and in the adult brain. Eur. J. Neurosci., 44, 2291–2299. [DOI] [PubMed] [Google Scholar]
- 37. Guo W., Allan A.M., Zong R., Zhang L., Johnson E.B., Schaller E.G., Murthy A.C., Goggin S.L., Eisch A.J., Oostra B.A.. et al. (2011) Ablation of FMRP in adult neural stem cells disrupts hippocampus-dependent learning. Nat. Med., 17, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eadie B.D., Zhang W.N., Boehme F., Gil-Mohapel J., Kainer L., Simpson J.M., Christie B.R. (2009) Fmr1 knockout mice show reduced anxiety and alterations in neurogenesis that are specific to the ventral dentate gyrus. Neurobiol. Dis., 36, 361–373. [DOI] [PubMed] [Google Scholar]
- 39. Saffary R., Xie Z. (2011) FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci., 31, 1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cook D., Nuro E., Jones E.V., Altimimi H.F., Farmer W.T., Gandin V., Hanna E., Zong R., Barbon A., Nelson D.L.. et al. (2014) FXR1P limits long-term memory, long-lasting synaptic potentiation, and de novo GluA2 translation. Cell Rep., 9, 1402–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazroui R., Huot M.E., Tremblay S., Boilard N., Labelle Y., Khandjian E.W. (2003) Fragile X mental retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet., 12, 3087–3096. [DOI] [PubMed] [Google Scholar]
- 42. Lagace D.C., Whitman M.C., Noonan M.A., Ables J.L., DeCarolis N.A., Arguello A.A., Donovan M.H., Fischer S.J., Farnbauch L.A., Beech R.D.. et al. (2007) Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci., 27, 12623–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R.. et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci., 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y., Stockton M.E., Bhuiyan I., Eisinger B.E., Gao Y., Miller J.L., Bhattacharyya A., Zhao X. (2016) MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Sci. Transl. Med., 8, 336ra61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo W., Patzlaff N.E., Jobe E.M., Zhao X. (2012) Isolation of multipotent neural stem or progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nat. Protoc., 7, 2005–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaeser P.S., Deng L., Wang Y., Dulubova I., Liu X., Rizo J., Südhof T.C. (2011) RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell, 144, 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kreis N.N., Louwen F., Yuan J. (2014) Less understood issues: p21(Cip1) in mitosis and its therapeutic potential. Oncogene, 0, 1–10. [DOI] [PubMed] [Google Scholar]
- 48. Zarnescu D.C., Gregorio C.C. (2013) Fragile hearts: new insights into translational control in cardiac muscle. Trends Cardiovasc. Med., 23, 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamanini F., Willemsen R., van Unen L., Bontekoe C., Galjaard H., Oostra B.A., Hoogeveen A.T. (1997) Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum. Mol. Genet., 6, 1315–1322. [DOI] [PubMed] [Google Scholar]
- 50. de Diego Otero Y., Bakker C., Raghoe P., Severijnen L.A.W.F.M., Hoogeveen A.T., Oostra B.A., Willemsen R. (2000) Immunocytochemical characterization of FMRP, FXR1P and FXR2P during embryonic development in the mouse. Gene Funct. Dis., 1, 28–37. [DOI] [PubMed] [Google Scholar]
- 51. Majumder M., House R., Palanisamy N., Qie S., Day T.A., Neskey D., Diehl J.A., Palanisamy V. (2016) RNA-binding protein FXR1 regulates p21 and TERC RNA to bypass p53-mediated cellular senescence in OSCC. PLoS Genet., 12, e1006306.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pechnick R.N., Zonis S., Wawrowsky K., Pourmorady J., Chesnokova V. (2008) p21 Cip1 restricts neuronal proliferation in the subgranular zone of the dentate gyrus of the hippocampus. PNAS, 105, 1358–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ángeles M.T.M., Porlan E., Banito A., Gómez-Ibarlucea E., Lopez-Contreras A.J., Fernández-Capetillo O., Vidal A., Gil J., Torres J., Fariñas I. (2013) Cyclin-dependent kinase inhibitor p21 controls adult neural stem cell expansion by regulating Sox2 gene expression. Cell Stem Cell, 12, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patrício P., Mateus-Pinheiro A., Sousa N., Pinto L. (2013) Re-cycling paradigms: cell cycle regulation in adult hippocampal neurogenesis and implications for depression. Mol. Neurobiol., 48, 84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pechnick R.N., Zonis S., Wawrowsky K., Cosgayon R., Farrokhi C., Lacayo L., Chesnokova V. (2011) Antidepressants stimulate hippocampal neurogenesis by inhibiting p21 expression in the subgranular zone of the hipppocampus. PLoS One, 6, e27290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qian J., Hassanein M., Hoeksema M.D., Harris B.K., Zou Y., Chen H., Lu P., Eisenberg R., Wang J., Espinosa A.. et al. (2015) The RNA binding protein FXR1 is a new driver in the 3q26-29 amplicon and predicts poor prognosis in human cancers. Proc. Natl. Acad. Sci., 112, 3469–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Comtesse N., Keller A., Diesinger I., Bauer C., Kayser K., Huwer H., Lenhof H.P., Meese E. (2007) Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26-27 in squamous cell carcinoma of the lung. Int. J. Cancer, 120, 2538–2544. [DOI] [PubMed] [Google Scholar]
- 58. Jin X., Zhai B., Fang T., Guo X., Xu L. (2016) FXR1 is elevated in colorectal cancer and acts as an oncogene. Tumor Biol., 37, 2683–2690. [DOI] [PubMed] [Google Scholar]
- 59. Gumireddy K., Li A., Yan J., Setoyama T., Johannes G.J., Ørom U.A., Tchou J., Liu Q., Zhang L., Speicher D.W.. et al. (2013) Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBOJ., 32, 2672–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Coffee B., Keith K., Albizua I., Malone T., Mowrey J., Sherman S.L., Warren S.T. (2009) Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am. J. Hum. Genet., 85, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O’Dushlaine C., Chambert K.D., Bergen S.E., Kähler A.. et al. (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duan X., Chang J.H., Ge S., Faulkner R.L., Kim J.Y., Kitabatake Y., Liu X., Yang C.H., Jordan J.D., Ma D.. et al. (2007) Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell, 130, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim J.Y., Liu C.Y., Zhang F., Duan X., Wen Z., Song J., Feighery E., Lu B., Rujescu D., St Clair D.. et al. (2012) Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell, 148, 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu Q., Li Y., Xiao B. (2013) DISC1-related signaling pathways in adult neurogenesis of the hippocampus. Gene, 518, 223–230. [DOI] [PubMed] [Google Scholar]
- 65. Del’Guidice T., Latapy C., Rampino A., Khlghatyan J., Lemasson M., Gelao B., Quarto T., Rizzo G., Barbeau A., Lamarre C.. et al. (2015) FXR1P is a GSK3β substrate regulating mood and emotion processing. Proc. Natl. Acad. Sci., 112, E4610–E4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guo W., Murthy A.C., Zhang L., Johnson E.B., Schaller E.G., Allan A.M., Zhao X. (2012) Inhibition of GSK3β improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum. Mol. Genet., 21, 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamaguchi M., Saito H., Suzuki M., Mori K. (2000) Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport, 11, 1991–1996. [DOI] [PubMed] [Google Scholar]
- 68. Luo Y., Shakir K., Guo W., Smrt R.D., Johnson E.B., Li X., Pfeiffer R.L., Szulwach K.E., Duan R., Barkho B.Z.. et al. (2010) Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet., 6, e1000898.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smrt R.D., Pfeiffer R.L., Zhao X. (2011) Age-dependent expression of MeCP2 in a heterozygous mosaic mouse model. Hum. Mol. Genet., 20, 1834–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barkho B.Z., Munoz A.E., Li X., Li L., Cunningham L.A., Zhao X. (2008) Endogenous matrix metalloproteinase (MMP)-3 and MMP-9 promote the differentiation and migration of adult neural progenitor cells in response to chemokines. Stem Cells, 26, 3139–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang F., Tidei J.J., Polich E.D., Gao Y., Zhao H., Perrone-Bizzozero N.I., Guo W., Zhao X. (2015) Positive feedback between RNA-binding protein HuD and transcription factor SATB1 promotes neurogenesis. Proc. Natl. Acad. Sci. U. S. A., 10.1073/Pnas.1513780112, E4995–E5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fujiwara K., Daido S., Yamamoto A., Kobayashi R., Yokoyama T., Aoki H., Iwado E., Shinojima N., Kondo Y., Kondo S. (2008) Pivotal role of the cyclin-dependent kinase inhibitor p21 WAF1/CIP1 in apoptosis and autophagy. J. Biol. Chem., 283, 388–397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.