Abstract
Chromogranins are pro-hormone secretory proteins released from neuroendocrine cells, with effects on control of blood pressure. We conducted a genome-wide association study for plasma catestatin, the catecholamine release inhibitory peptide derived from chromogranin A (CHGA), and other CHGA- or chromogranin B (CHGB)-related peptides, in 545 US and 1252 Australian subjects. This identified loci on chromosomes 4q35 and 5q34 affecting catestatin concentration (P = 3.40 × 10−30 for rs4253311 and 1.85 × 10−19 for rs2731672, respectively). Genes in these regions include the proteolytic enzymes kallikrein (KLKB1) and Factor XII (F12). In chromaffin cells, CHGA and KLKB1 proteins co-localized in catecholamine storage granules. In vitro, kallikrein cleaved recombinant human CHGA to catestatin, verified by mass spectrometry. The peptide identified from this digestion (CHGA360–373) selectively inhibited nicotinic cholinergic stimulated catecholamine release from chromaffin cells. A proteolytic cascade involving kallikrein and Factor XII cleaves chromogranins to active compounds both in vivo and in vitro.
Introduction
The chromogranin/secretogranin pro-hormone family (1,2) constitutes a source of active peptides with a spectrum of biological activities, including the catecholamine release inhibitory fragment catestatin derived from chromogranin A (CHGA) and a peptide with similar activity derived from chromogranin B (CHGB) (3). CHGA plays a necessary role in the formation of catecholamine storage vesicles, and its absence results in unregulated release of vesicle contents, including catecholamines, both in vivo (4) and in cultured cells (5). Targeted ablation of the Chga locus in the mouse results in higher blood pressure (4) and increased fat deposition (6), which can be reversed by administration of catestatin. Catestatin, and other CHGA-derived peptides, may provide cardioprotection following ischaemia through activation of nitrous oxide pathways (7–10), and non-cardiovascular functions or associations of CHGA and related peptides have also been reported (11,12).
Because the catestatin fragment of CHGA exerts both antihypertensive (4) and vasodilatory (13) actions in vivo, we have studied genetic influences on human plasma catestatin concentration, beginning with genome wide linkage in two cohorts of twins and sibling pairs in the USA and Australia. We previously found genetic linkage evidence for a locus on chromosome 4q (14), but the wide confidence interval for this method precluded identification of the causative gene. We have now turned to genome-wide association analysis (GWAS) through SNP genotyping. Our results show an effect of variation in previously unsuspected regions of chromosomes 4q35 and 5q34 on the catestatin trait. These regions contain genes for the proteases KLKB1 (kallikrein B, plasma (Fletcher factor) 1; EC 3.4.21.34) and F12 (coagulation factor XII (Hageman factor), EC 3.4.21.38), suggesting an enzymatic cascade, F12 → KLKB1, for proteolytic activation of peptides derived from CHGA and CHGB. Following from this, we have characterized a product of CHGA digestion by kallikrein as catestatin and shown its functional activity.
Results
GWAS: effects of the KLKB1 locus on chromosome 4q35 on CHGA- and CHGB-related peptide concentrations
Significant allelic associations were found in the UCSD and QIMR cohorts, and confirmed in a meta-analysis of the two datasets. These are summarized in Table 1 and Fig. 1.
Table 1.
Summary of allelic effects at KLKB1 and F12 on circulating chromogranin A (CHGA) and B (CHGB) fragments, showing results for the lead SNPs at the chromosome 4 and 5 loci for the UCSD and QIMR cohorts and results from meta-analysis.
| SNP | Chr | BP (Build 37) | Gene | Phenotype | Subjects | A1/A2 | Freq A1 | Beta | SE beta | P-value | p Meta | p Het |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4253311 | 4 | 187,174,683 | KLKB1 | CHGA 361–372 | UCSD | G/A | 0.501 | 0.439 | 0.065 | 5.65 × 10−13 | 3.40 × 10−30 | 0.347 |
| CHGA 361–372 | QIMR | G/A | 0.511 | 0.447 | 0.047 | 9.07 × 10−22 | ||||||
| CHGA 116–439 | UCSD | G/A | 0.501 | −0.205 | 0.066 | 0.0014 | 0.0011 | 0.346 | ||||
| CHGA 116–439 | QIMR | G/A | 0.511 | −0.116 | 0.060 | 0.054 | ||||||
| CHGA 116–130 | UCSD | G/A | 0.501 | −0.030 | 0.088 | 0.820 | – | – | ||||
| CHGB 312–331 | UCSD | G/A | 0.501 | 0.029 | 0.066 | 0.555 | – | – | ||||
| CHGB 439–451 | UCSD | G/A | 0.501 | 0.331 | 0.071 | 1.45 × 10−6 | 1.39 × 10−7 | 0.044 | ||||
| CHGB 439–451 | QIMR | G/A | 0.511 | 0.156 | 0.046 | 6.5 × 10−4 | ||||||
| CHGB 568–577 | UCSD | G/A | 0.501 | 0.642 | 0.089 | 1.53 × 10−12 | – | – | ||||
| rs2731672 | 5 | 176,842,474 | F12 | CHGA 361–372 | UCSD | C/T | 0.759 | 0.327 | 0.075 | 7.35 × 10−6 | 1.85 × 10−19 | 0.038 |
| CHGA 361–372 | QIMR | C/T | 0.747 | 0.462 | 0.056 | 8.58 × 10−17 | ||||||
| CHGA 116–439 | UCSD | C/T | 0.759 | −0.180 | 0.074 | 0.016 | 0.0022 | 0.843 | ||||
| CHGA 116–439 | QIMR | C/T | 0.747 | −0.158 | 0.073 | 0.031 | ||||||
| CHGA 116–130 | UCSD | C/T | 0.759 | 0.190 | 0.106 | 0.095 | – | – | ||||
| CHGB 312–331 | UCSD | C/T | 0.759 | 0.058 | 0.081 | 0.437 | – | – | ||||
| CHGB 439–451 | UCSD | C/T | 0.759 | 0.252 | 0.087 | 0.0034 | 2.93 × 10−4 | 0.278 | ||||
| CHGB 439–451 | QIMR | C/T | 0.747 | 0.138 | 0.054 | 0.011 | ||||||
| CHGB 568–577 | UCSD | C/T | 0.759 | 0.464 | 0.107 | 2.70 × 10−5 | – | – |
Figure 1.
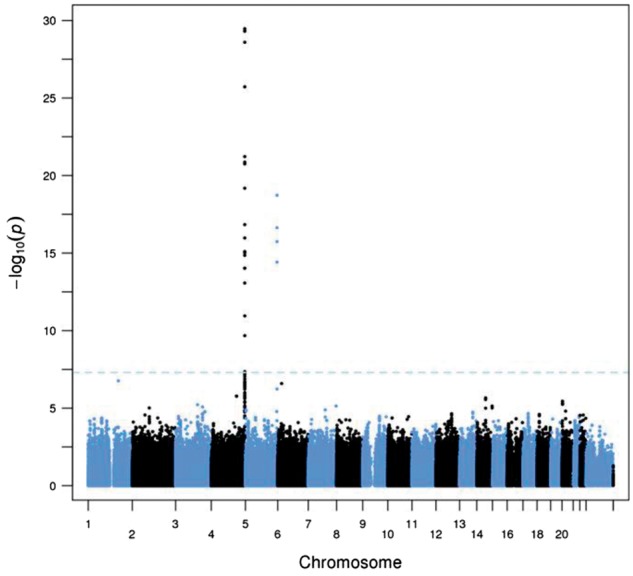
Discovery of novel loci on chromosome 4q35 (KLKB1, Fletcher factor) and chromosome 5 (F12, Hageman factor) that influence plasma concentrations of CHGA fragments. Results from meta-analysis of UCSD and QIMR GWAS results for CHGA 361–372 (catestatin).
The catestatin trait (epitope: CHGA361–372) was initially evaluated in the UCSD subjects, and a significant association was found on chromosome 4q. Local analysis of this region positioned the peak association within a linkage disequilibrium (LD) block of ∼100 kbp, centred on KLKB1 (Table 1, Fig. 2, Supplementary Material, Fig. S1). The most significant SNP was rs4253311 (P = 5.65 × 10−13), with a group of five other SNPs showing P < 10−12 (see Supplementary Material, Table S2). One of the SNPs with substantial effects on catestatin concentration (P = 1.10 × 10−12) is the non-synonymous coding variant rs3733402, Asn124Ser. As discussed below, there is reason to believe that this variant affects the enzymatic activity of kallikrein.
Figure 2.
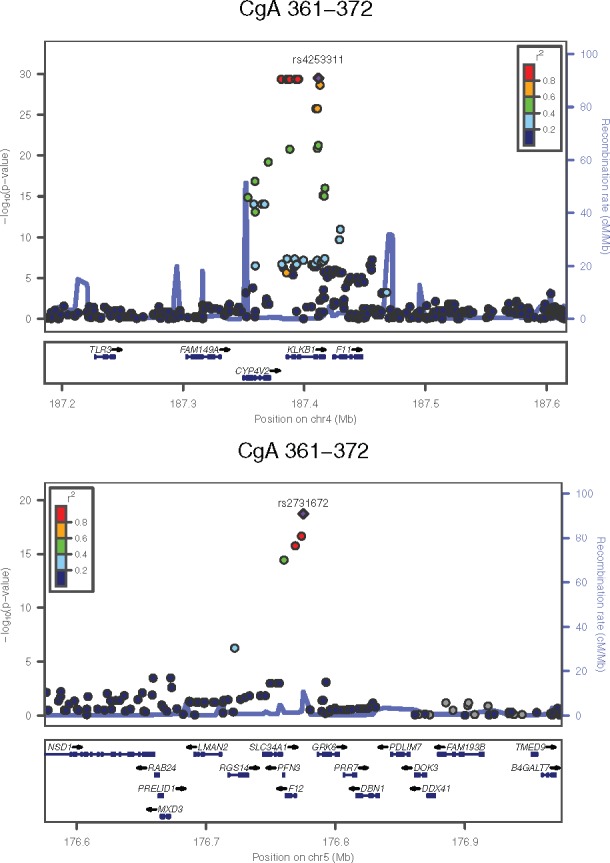
Regional plots for the chromosome 4 and chromosome 5 loci; combined data from UCSD and QIMR for CHGA 361–372 (catestatin).
We checked for documented effects of SNPs in this region on KLKB1 gene expression, and found multiple SNPs with highly significant effects on KLKB1 expression (Supplementary Material, Fig. S2). However, the SNPs with strong effects on gene expression showed only weak effects on plasma catestatin concentration, and vice versa, contrary to the hypothesis that the allelic associations with catestatin concentration are mediated through effects on KLKB1 expression.
Two other regions, on chromosomes 1 and 6, showed significant SNP associations with catestatin in the UCSD data. The lead SNPs were rs12127550 (P = 2.36 × 10−8) and rs7771424 (P = 4.51 × 10−8), respectively, but the minor allele frequencies of these SNPs are less than 1% and therefore they did not survive quality control procedures in the QIMR data. There were no significant associations in regions containing genes for other enzymes known to cleave CHGA, such as prohormone convertases PCSK1 (chromosome 5q15) and PCSK2 (chromosome 20p11), FUR (furin, chromosome 15q25), cathepsin L (CTSL on chromosome 9q21), or plasmin (PLG, chromosome 6q26).
We also measured a CHGB proteolytic fragment, using an assay directed against the epitope human CHGB568–577, and found that SNPs at the chromosome 4 locus had a strong effect (P = 1.53 × 10−12 at rs4253311) on plasma concentration of this fragment (Table 1, Supplementary Material, Fig. S3). Once again, the peak centred on KLKB1. This locus did not have genome-wide-significant effects on the other chromogranin-related peptides measured (see Table 1) but a near-significant effect was detected in the meta-analysis for CHGB439–451 (P = 7.21 × 10−8 for rs2048).
In the QIMR twin-family cohort, the lead SNP rs4253311 was also significantly associated with the plasma concentrations of catestatin (CHGA361–372P = 9.07 × 10−22). For CHGA116–439 the association P-value was 0.054, and for CHGB439–451P = 6.5 × 10−4 (Table 1).
Discovery of a second locus on chromosome 5q34 centred on the protease F12
The larger number of subjects (n = 1267) in the QIMR sample revealed a second locus on chromosome 5q34 (Table 1, Fig. 1, Supplementary Material, Fig. S4), centred on an LD block containing the gene encoding the protease F12 (Hageman Factor; see Fig. 2). The peak association (P = 8.58 × 10−17) for catestatin was in the F12 promoter region, at rs2731672. Re-examination of the UCSD twins’ data confirmed this finding (P = 7.35 × 10−6 for rs2731672, pmeta = 6.27 × 10−19).
Proportion of variance explained
The proportion of phenotypic variance in plasma catestatin concentration explained by the lead SNP at the chromosome 4 KLKB1 locus (rs4253311) was 12.8% in the UCSD subjects and 9.9% in the QIMR subjects. At the chromosome 5 F12 locus the proportions were 5.1% and 8.0%, respectively. When the effects of the peak SNPs at KLKB1 or F12 were controlled for in a separate analysis, no independent effects of other SNPs at these loci were found.
Functional variation at KLKB1 and F12
Functional genetic variation is already understood at both KLKB1 and F12. At KLKB1 Asn124Ser (rs3733402), serine at position 124 results in diminished substrate binding. This variant was in near-complete linkage disequilibrium with eight other SNPs which showed the strongest, and almost identical, allelic association results. This non-synonymous variant is within the substrate-binding (or ‘apple’) domain, in which the Ser allele is known to impair substrate binding by this enzyme (15). Inter-species sequence alignment in primates indicates that the local region is highly conserved; indeed, all primates except humans are monomorphic for the Asn allele. We note the directionally coordinate effects of Asn124Ser on both catestatin/CHGA361–372 and CHGB568–577. Effects of Asn124Ser on concentration of a second CHGB fragment, assayed by the CHGB439–451 epitope, were also substantial (P = 1.45 × 10−6). As predicted, the relatively inactive Ser allele of the enzyme (15) was associated with lower formation of both CHGA and CHGB fragments . We also observed reciprocal effects of KLKB1 SNPs on concentrations of the CHGA116–439 precursor and catestatin (Table 1), further suggesting a differential effect of the enzyme’s alleles on CHGA cleavage to its catestatin product.
At the other significant locus, at F12 in 5’-UTR C46T (rs1801020, pmeta = 1.82 × 10−16), the T-allele creates an alternative translational start codon, thereby diminishing formation of the Factor XII protein (16).
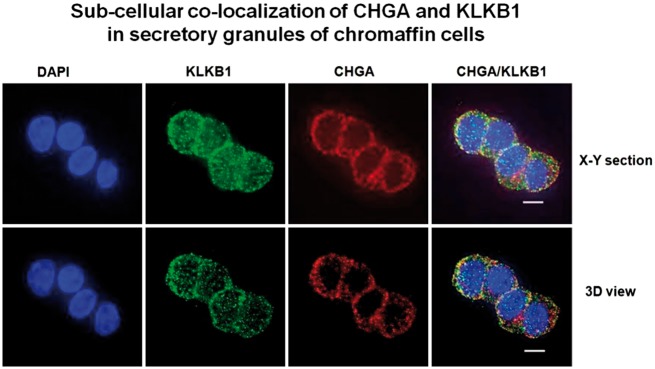
Subcellular co-localization of CHGA and KLKB1
Co-immunostaining revealed both CHGA and KLKB1 proteins within chromaffin cells, with immunoreactivity clustered just beneath the plasma membrane (Fig. 3), a location typical for docked secretory granules (catecholamine storage vesicles, or chromaffin granules). Optical overlap of the two probes (CHGA in red, KLKB1 in green) revealed substantial co-localization (coefficient = 0.67), as evidenced by the resulting yellow absorbance. Substantial overlap was noted on both shallow x/y sections, and deeper (3D, x/y/z) sections.
Figure 3.
Sub-cellular co-localization of CHGA and KLKB1 in catecholamine storage vesicles of chromaffin cells. Experiments were conducted in PC12 cells, with immuno-staining of KLKB1 (green conjugate) and CHGA (red conjugate). A series of x/y optical sections along z-axis were acquired with increments of 0.2 µm. Data were processed to generate pseudo-three-dimensional (3D) or representative x/y sections. Co-localization of KLKB1 (green) and CHGA (red) was shown by yellow fluorescence, with a Pearson coefficient of overlap = 0.67.
Generation of active catestatin peptide by KLKB1 digestion of human CHGA
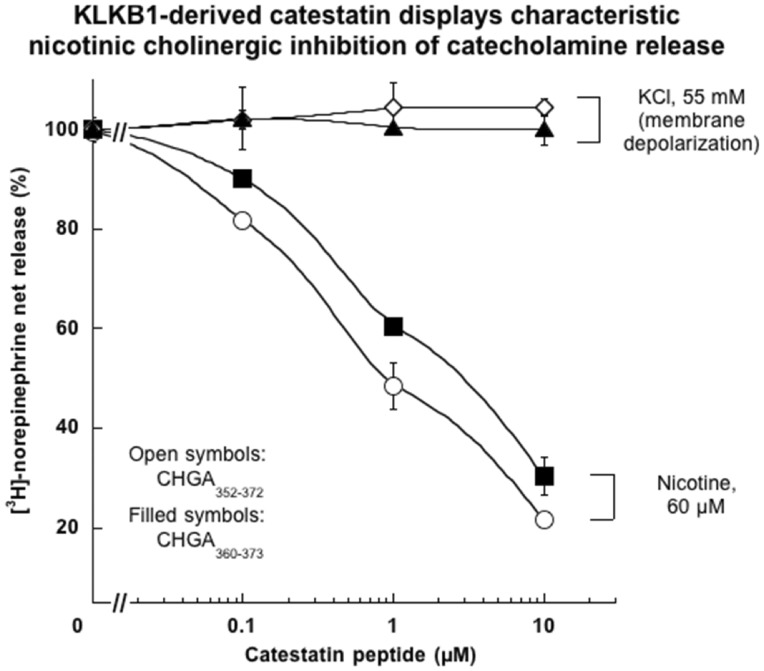
To understand how KLKB1 variation might influence catestatin concentration, recombinant human CHGA was digested in vitro by active human kallikrein (purified from human plasma), after which tandem mass spectrometry (MS/MS) revealed post-Arg cleavage yielding a 1548.5 m/z fragment containing the catestatin-region sequence R↓A360RAYGFRGPGPQLR373↓R (CHGA360–373; Supplementary Material, Fig. S5). We therefore synthesized this peptide (RAYGFRGPGPQLR), and tested it on catecholamine secretion, in comparison with the customary longer catestatin sequence CHGA352–372 (Fig. 4). Each of the peptides effectively inhibited catecholamine secretion when triggered by the physiological (nicotinic cholinergic) pathway, with similar potency, but did not affect secretion in response to membrane depolarization. Thus, each of the two peptides exhibited the typical pharmacological actions of catestatin.
Figure 4.
Catestatin derived by KLKB1 digestion of recombinant human CHGA: Synthesis potency and selective effects on catecholamine secretion triggered by the nicotinic cholinergic pathway in chromaffin cells. PC12 cells were labelled with [3H]-norepinephrine and then incubated with either 60 µM nicotine, or 55 mM KCl (for membrane depolarization), or vehicle. Secretory stimulation occurred either alone or in combination with ascending doses (0.1, 1 and 10 µM) of each catestatin peptide, either the KLKB1-derived (and then synthesized) version ARAYGFRGPGPQLR (hCgA360–373), or the usual longer version (hCgA352–372). Control (100%) net norepinephrine release represents the release in the presence of nicotine or KCl (without inhibitor).
Differential regulation of CHGA and KLKB1 in rodent models of hereditary hypertension
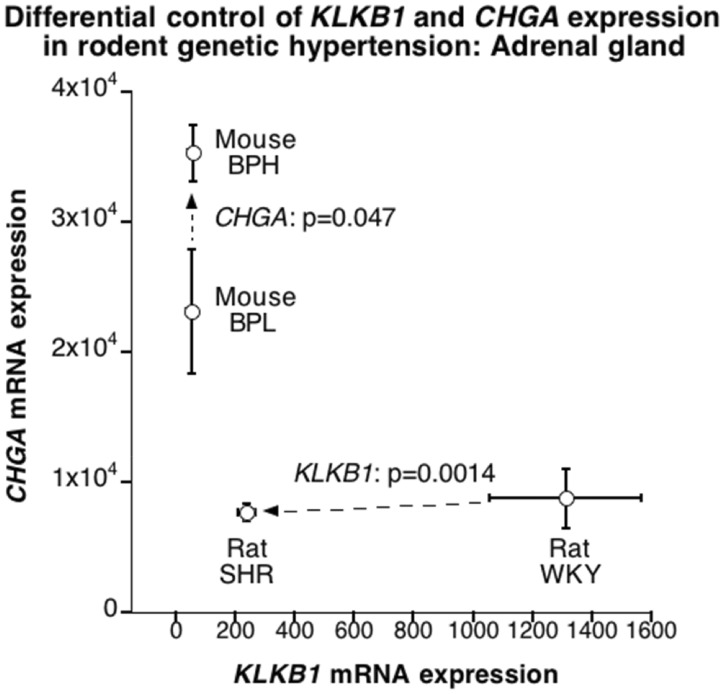
We examined the potential for differential adrenal mRNA expression of Chga and Klkb1 in two rodent models of human essential hypertension: the mouse BPH (versus BPL) model and the rat SHR (versus WKY) model (Fig. 5). In the mouse BPH, Chga (though not Klkb1) was over-expressed, while in the rat SHR, Klkb1 (though not Chga) was under-expressed. F12 mRNA was also expressed in mouse BPH/BPL adrenal glands, without difference by strain. F12 was not included on the rat SHR/WKY transcriptome chip.
Figure 5.
Differential control of expression of Chga versus Klkb1 mRNA in the adrenal gland for two rodent models of human essential (genetic) hypertension: rat SHR (Spontaneously Hypertensive rat, versus its WKY [Wistar-Kyoto] control); and mouse BPH (Blood Pressure High, versus its BPL [Blood Pressure Low] control).
Discussion
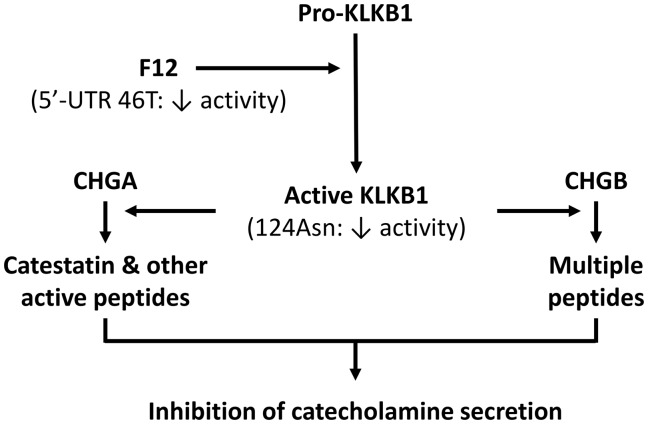
When triggered by the physiological nicotinic cholinergic pathway, the catestatin peptide derived from CHGA (CHGA352–372) is a potent and specific antagonist of catecholamine release (17). When administered intravenously, catestatin reverses the profound hypertension of Chga deficiency in a knockout mouse model (4), and dilates human veins in vivo (13). Genetic variation within the catestatin motif also perturbs human autonomic function and blood pressure in vivo (18). Catestatin, and other CHGA-derived peptides, may provide cardioprotection following ischaemia (7–10). Given these diverse effects on the vasculature, we sought genetic determinants of the formation and secretion of catestatin, employing a genome wide association design. From this, we discovered a novel influence on the processing of chromogranins: genetic variation at the protease KLKB1 (plasma kallikrein) locus, which encodes an enzyme mainly known for its roles in coagulation and allergy. Identification of a second significant association at the protease F12 (Factor XII) locus implicates a cascade of enzymatic events (F12→KLKB1) leading to catestatin formation (Fig. 6). These variants account for a high proportion (around 15% to 20%) of the phenotypic variation in circulating catestatin concentration, and assuming heritability of 40–60% for plasma catestatin concentration (14) the KLKB1 and F12 variants account for a third to half of genetic variation.
Figure 6.
Proposed schema for the effects of functional genetic variation at sequential serine proteases F12 (Hageman Factor; 5’-UTR C46T, rs2731672) and KLKB1 (Fletcher factor; Asn124Ser, rs3733402), on formation of active peptides by proteolytic cleavage of CHGA or CHGB.
After discovery of the effects of variation in the KLKB1 and F12 genes on plasma catestatin concentration, we went on to confirm that catestatin is produced by the action of kallikrein on chromogranin A. Our functional studies have confirmed the structure and pharmacological activity of the catestatin peptide produced by this mechanism.
The GWAS results should be seen in the context of proteolytic processing of pre-hormones to active forms, as the combined action of kallikrein and Factor XII is known to produce other bioactive peptides. Factor XII activates prokallikrein to kallikrein, which in turn activates Factor XII. This sequence of events was first established for the intrinsic or contact activation pathway of coagulation (19), and in relation to formation of bradykinin (20). Proteolytic action of kallikrein on chromogranin A in vitro was suggested over 20 years ago (21,22), although the products were not characterized. Processing of other pro-hormones to active products by kallikrein and Factor XII was found in a recent GWAS which focused on the generation of endothelin-1 and adrenomedullin from pre-proendothelin-1 and pre-pro-adrenomedullin (23). Other GWAS have identified associations between rs4253252 in KLKB1 and serum bradykinin (24) and renin (25). Plasma kallikrein also plays a role, but in this case an inactivating one, in the processing of neuropeptide Y (NPY) (26).
Human catestatin has been defined as CHGA352–372 with the amino acid sequence SSMKLSFRARAYGFRGPGPQL (amino acids numbered 370–390 by Uniprot, http://www.uniprot.org/blast/?about=5[370-390]&key = Peptide&id = PRO_0000432682; date last accessed December 12, 2016) and measurement of the catestatin phenotype for our study was by an immunoassay directed against CHGA361–372 (RAYGFRGPGPQL). The product identified by mass spectrometry after digestion of full-length CHGA by kallikrein was CHGA360–373, A360RAYGFRGPGPQLR373 (Supplementary Material, Fig. S5). When this fragment was synthesized and tested in vivo it showed the same pharmacological effects as CHGA352–372. We therefore suggest that there are different but overlapping peptides with catestatin activity (CHGA352–372 and CHGA360–373), produced by the action of different proteolytic enzymes.
The genome-wide-significant effects of KLKB1 and F12 variation on formation of peptides from CHGA and CHGB are supplemented by evidence that the alleles of rs3733402 (KLKB1 Ser124Asn) associated with higher catestatin concentration are also associated with lower concentration of CHGA, at least in the UCSD data.
Thus, KLKB1 seems to be an authentic proteolytic enzyme for both CHGA and CHGB as substrates. Indeed, the codon 124 Ser allele of KLKB1 occurs in the substrate binding ‘Apple-2’ domain of the enzyme, resulting in diminished proteolytic activity towards its best-characterized substrate, HMWK (15). Likewise, within the F12 5’-UTR C46T variant, the T-allele is associated with diminished formation of the functional F12 protein (16). Here we found that, in each case, the loss-of-function allele was associated with a lower concentration, and presumably decreased formation, of catestatin.
From our results and previous publications, it is clear that genetic variation in KLKB1 and F12 leads to variation in concentrations of catestatin and probably other chromogranin-related peptides, and also of adrenomedullin, bradykinin, endothelin, neuropeptide Y and renin. This has possible implications for control of blood vessels and blood pressure. These two loci do not show up in published GWAS for blood pressure or hypertension (27); rs4253311 was non-significant and rs2731672 was not included in summary results for systolic and diastolic blood pressure from the International Consortium for Blood Pressure (downloaded from ftp://ftp.ncbi.nlm.nih.gov/dbgap/studies/phs000585/analyses/phs000585.pha003588.txt.gz and ftp://ftp.ncbi.nlm.nih.gov/dbgap/studies/phs000585/analyses/phs000585.pha003589.txt.gz; date last accessed December 12, 2016). We speculate that this pathway may have particular relevance for either systemic or local blood pressure control when clotting processes are activated, for example after trauma.
Because of the reported protective effects of catestatin against injury from ischaemia or reperfusion in experimental animals (7–10), we also checked for associations between the lead SNPs at the KLKB1 and F12 loci and ischaemic heart disease in humans. Results from the CARDIoGRAM and C4D consortia do show some evidence for these loci affecting coronary artery disease and myocardial infarction. For the CARDIoGRAM data alone (http://www.cardiogramplusc4d.org/media/cardiogramplusc4d-consortium/data-downloads/cardiogram_gwas_results.zip; date last accessed December 12, 2016) P = 0.0053 for rs4253311 (the chromosome 4 KLKB1 locus) and P = 0.626 for rs2731672 (the chromosome 5 F12 locus). For the combined CARDIoGRAM and C4D data, 1000-Genomes imputation (28), corresponding P-values were 0.017 and 0.205 for coronary artery disease and 0.019 and 0.099 for myocardial infarction. Therefore, there is nominal significance (P < 0.05) for the rs4253311-coronary artery disease association. The G allele for rs4253311 is associated with lower risk, and it is associated in our data with higher plasma catestatin levels, consistent with catesatin having some protective action in relation to coronary artery disease. However the KLKB1 gene product has activating effects on multiple pro-hormones and on the coagulation pathway, and any effect of this locus on coronary artery disease risk may not be mediated through conversion of chromogranin A to catestatin.
Although the kallikrein-Factor XII pathway for conversion of pre-hormone to active peptides has been demonstrated for several systems, it is not necessarily the only one for any of them. Because of differences in sequence specificity, different proteolytic enzymes will lead to slightly different peptides. For CHGA/catestatin, the peptide produced by kallikrein is shorter than the previously recognized fragment but has the same biological activity. Formation of peptides with catestatin activity by different enzymes may have physiological implications, although at present this is speculative. Previous studies showed substantial effects of polymorphic variation in KLKB1 and F12 on the activation of other peptide hormones. Our results show that these are the major genetic determinants for several granin-derived peptides, including a novel catestatin. The roles of different pre-hormone-processing enzymes in generating active peptides, and the potential for functional or pharmacokinetic differences between peptides produced by different enzymes, may repay further examination.
In conclusion, our GWAS of CHGA (catestatin)- and CHGB-related traits discovered a protease cascade, involving F12 (Hageman factor) → KLKB1 (Fletcher factor), as a major determinant of CHGA/CHGB cleavage. This was confirmed by replication and meta-analysis, microanatomic co-localization of CHGA and KLKB1 in catecholamine secretory vesicles, and generation of active catestatin by KLKB1 cleavage of CHGA with sequence verification. The results document a novel pathway of pro-hormone cleavage to catestatin, similar to that known for other pro-hormones and with potential implications for endocrine and homeostatic pathways.
Materials and methods
UCSD twin and sibling cohort for GWAS
Twin and sibling participants were recruited from southern California by access to a population birth record-based twin registry (29), and by newspaper advertisement, as described previously (30). The protocol was approved by the University of California-San Diego (UCSD) Institutional Review Board, and each subject gave written informed consent. Subjects included in the data analysis were those who had genotyping and measurement of at least one of the chromogranin peptides, a total of 545 individuals (144 males, 401 females) from 260 nuclear families, including 60 DZ and 161 MZ twin pairs. Zygosity of twins was confirmed by use of microsatellite and SNP markers (30). Ethnicity was initially established by self-identification, including information on the geographic origin of both parents and all four grandparents, and only individuals of Caucasian (European-American) or Hispanic (Mexican-American) ancestry/ethnicity are included here. The age of the subjects ranged from 14 to 78 years, with a median of 39. Phenotyping (biochemical and physiological) was conducted as previously described (30).
Australian twin cohort
Study participants were from twin pairs from studies conducted at QIMR Berghofer Medical Research Institute (QIMR), Brisbane, Queensland, Australia, and were ascertained and studied as previously described (14). These studies were approved by the QIMR Human Research Ethics Committee, and participants gave informed consent. The samples analysed come from the SSAGA Blood (31) and Anxiety (32) studies. Chromogranin or chromogranin-peptide measurements were made on up to 4106 subjects, with genotyping available on 1294 of these (see Supplementary Material, Table S1 for details).
Biochemical assays
For the UCSD participants, plasma concentrations of circulating CHGA (measured by immunoassay against epitope CHGA116–439), and two of its peptides (epitopes CHGA116–130 and CHGA361–372, catestatin); and three CHGB peptides (epitopes CHGB312–331, CHGB439–451, CHGB568–577) were quantified by radioimmunoassay with region-specific peptides and rabbit polyclonal antibodies, as previously described (33). For the Australian twins, only CHGA (epitope CHGA116–439), catestatin (epitope CHGA361–372,), and one CHGB peptide (epitope CHGB439–451) were measured.
For UCSD data, phenotypes were transformed to approximate a normal distribution through the removal of outliers, exclusion of data points more than three standard deviations from the mean, and/or base-10 log transformation. In the QIMR data, there were significant batch effects, both between and within studies, which were identified and adjusted for before log-transformation of phenotypes with skewed distributions. Summary statistics for chromogranin and peptide results for each cohort are given in Supplementary Material, Table S1.
Genotyping, imputation and association analysis
For participants in the San Diego study, genomic DNA was extracted from leukocytes in EDTA-anticoagulated blood after Proteinase-K digestion of proteins, by adsorption/elution from Qiagen columns, as previously described (30). Four hundred and eighty-one UCSD subjects were genotyped at 592,312 SNPs using the Illumina 610-Quad genotyping array (Illumina Inc., San Diego, CA 92122). For each of 161 MZ twin pairs, only one individual underwent genotyping, and the genotype information was used for both members of an MZ twinship. During analysis, family structure was accounted for in MERLIN (see below). Subjects with >5% missing genotypes were excluded (N = 3). SNPs missing in >5% of subjects (n = 2505 SNPs), with minor allele frequency <0.01 (n = 28,851 SNPs), not in Hardy-Weinberg Equilibrium (n = 15), SNPs with >3 Mendelian errors (n = 15), SNPs showing a genotyping ‘plate’ effect (n = 1530), or perfectly correlated with gender (n = 5) were excluded, leaving 559,400 SNPs. In addition, subjects' gender was confirmed based on X-chromosomal markers, and family structure and zygosity of DZ pairs was confirmed based on genotypic information. Recorded status was corrected where necessary. A total of 455 subjects’ genotyping passed QC, with the addition of 161 MZ co-twins, resulting in a final dataset comprising 616 subjects genotyped for 559,400 markers, though only 545 of the genotyped subjects had one or more chromogranin peptide measurements.
To control for additional genetic background heterogeneity in this predominantly Caucasian cohort we performed a multidimensional scaling analysis using PLINK (34) including all autosomal SNPs. We then included the first MDS dimension, which corresponded to the Native American admixture of Hispanic subjects (35,36), as covariate in the association analysis.
Genotype imputation for additional SNPs was performed using MACH v. 1.0.16 (http://www.sph.umich.edu/csg/abecasis/MaCH/; date last accessed December 12, 2016). Phased haplotypes of 60 unrelated HapMap II CEU founders were used as the reference data (for autosomes: (CEU_r22_nr.b36_fwd.phased; http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2007-08_rel22/phased/, for X chromosome and the pseudo-autosomal regions PAR1/PAR2: CEU_r21_nr_fwd_phased; http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2006-07_phaseII/phased/; these two URLs can no longer be accessed since June 2016 for security reasons). Imputations were based on 537,371 genotyped SNPs in common with the reference data (G-C and A-T SNPs were excluded). To allow imputation with MACH for the haploid non-PAR regions of the male X chromosome, the phased haplotypes were duplicated. A two-step imputation approach was used: step 1 estimated per SNP error and per interval crossover rates at 50 iterations from a random sample of 200 genotyped individuals, while step 2 used these model parameters to assign allele dosages and genotypes based on a maximum likelihood approach. A total of 2,072,428 SNPs were imputed, with 2,028,122 high-confidence SNPs remaining after removal of SNPs with low imputation quality (r2 <0.3). Adding SNPs typed but not used for imputation purposes resulted in a total of 2,587,527 genotyped or imputed SNPs available for association analyses.
For participants in the Australian Study, genotype data were derived from several genotyping projects with Illumina 317K, 370K or 610K chips. After quality control of sample and SNP data, imputation of HapMap2 SNP genotypes was performed using SNPs common to these platforms; these procedures were as previously described (37).
Association analysis and meta-analysis
To test SNP on phenotype effects within a family structure, MERLIN v1.1.2 (http://www.sph.umich.edu/csg/abecasis/merlin/; date last accessed December 12, 2016) was used. Age, gender, and the first MDS component (see above) were included as covariates. GWAS results for 3 traits that were available in both San Diego and QIMR cohorts were meta-analysed using Metal package (38). A standard criterion of P < 5 × 10−8 was used to indicate significant SNP (allelic) effects on traits.
Subcellular co-localization of CHGA and KLKB1 in chromaffin cells
PC12 cells (originally derived from a rat adrenal medulla pheochromocytoma) were grown on cover slips, washed with phosphate-buffered saline (PBS) and fixed with 2.5% paraformaldehyde in PBS for 20 min at room temperature. Cells were then permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. Cells were blocked using 5% bovine serum albumin (BSA) in PBS for 30 min followed by primary antibody incubation with rabbit anti-KLKB1 (1:100, catalogue number bs-5872R, Bioss) and goat anti CHGA (1:100, Santa Cruz Biotechnology, Dallas TX 75220) in 2% BSA for 2 h at room temperature. Coverslips were washed 3 times 5 min each and then incubated with secondary antibody Alexa Fluor 488 donkey anti rabbit (1:250, Invitrogen, Grand Island NY 14072) and Alexa Fluor 594 donkey anti goat (1:350, Invitrogen) along with Hoechst 33342 (1 µg/mL) in 1% BSA for 1 h at room temperature. Coverslips were washed and mounted on glass slide using Slowfade-antifade (Molecular Probes, Thermo Fisher Scientific, Waltham MA 02451). Images were acquired on a Delta Vision deconvolution microscope and SoftWorx software (Applied Precision, Issaquah, WA 98027), using 60x objective as described previously (39,40).
KLKB1 proteolytic cleavage of CHGA
Activated human KLKB1, purified from human plasma to ≥95% homogeneity (by SDS-PAGE), was purchased from Calbiochem (#420307; Merck, Darmstadt, Germany), at a specific activity of ≥15 units/mg protein, and stored at -20ºC. Recombinant human CHGA (minus the 18-amino acid signal peptide sequence; with a carboxy-terminal 6-His tag) was expressed in E. coli, isolated, and purified to homogeneity on SDS-polyacrylamide gel electrophoresis (by Ni-NTA affinity chromatography followed by concentration over a Microcon YM-30 centrifugal filter with molecular weight cutoff >30 kDa (Amicon/Millipore; Merck, Darmstadt, Germany)) as previously described (41). Human CHGA (10 µM substrate) was digested with human KLKB1 (0.2 µM enzyme) for 15 min at 37 °C in 10 mM Tris pH8, 150 mM NaCl; the reaction was terminated with aprotinin (2.5 µM; Calbiochem). The peptide fragments were purified by adsorption/elution on a 10 µL ZipTip C-18 resin (Millipore), and were analyzed on a 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems, Foster City CA 94404) with CID (collision induced dissociation) yielding amino acid sequence, interpreted by Mascot-2.1 software (Matrix Science Inc, Boston MA 02110), as described before (42).
Functional secretory properties of catestatin generated by KLKB1 digestion of CHGA
Human catestatin peptides were synthesized by the solid-phase F-moc method, then purified by reverse phase HPLC, with documentation by electrospray mass spectrometry as well as repeat HPLC. Catecholamine secretory studies were accomplished in rat PC12 chromaffin cells, with cellular catecholamine stores pre-labelled by uptake of [3H]-L-norepinephrine (PerkinElmer Life Sciences, Waltham, MA 02451), and secretion triggered by stimulation of either the nicotinic cholinergic pathway (nicotine, 60 µM) or membrane depolarization (55 mM KCl), as previously described (17,42). Peptides were tested over a range of concentrations (typically from 0.1 to 10 µM) for their ability to antagonize the secretory stimuli on PC12 cells. The cells were treated with nicotine (60 µM) or membrane depolarizer KCl (55 mM) in secretion buffer either alone or in combination with three ascending doses (0.1, 1 and 10 µM) of synthetic peptide 1 (full-length catestatin, SSMKLSFRARAYGFRGPGPQL, hCgA352–372) or peptide 2 (ARAYGFRGPGPQLR hCgA360–373). The release medium and the cell lysates were assayed for [3H]-norepinephrine by liquid scintillation counting. Results were expressed as percent secretion: [amount released/(amount released + amount in lysate)] × 100. Net secretion was calculated as agonist-stimulated release minus basal release.
Gene expression
Measurements of expression of Chga, Klkb1, and F12 mRNAs in the adrenal glands of rodents with hereditary models of human hypertension (mouse BPH/BPL model, rat SHR/WKY model) were undertaken as previously described (43,44).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
Funding
Support for the US study came from the National Institutes of Health Grants P01HL058120-10 (to DTO’C) and R01MH093500 (to CMN). Australian sample collection and genotyping were supported by the US National Institutes of Health (AA007535) and the Australian National Health and Medical Research Council (971232). BB was supported by the Australian National Health and Medical Research Council (APP1084417 and APP1079583). GWM was supported by the Australian National Health and Medical Research Council Fellowship scheme.
References
- 1. Taupenot L., Harper K.L., O'Connor D.T. (2003) The chromogranin-secretogranin family. N. Engl. J. Med., 348, 1134–1149. [DOI] [PubMed] [Google Scholar]
- 2. Loh Y.P., Cheng Y., Mahata S.K., Corti A., Tota B. (2012) Chromogranin A and derived peptides in health and disease. J. Mol. Neurosci., 48, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang K., Biswas N., Gayen J.R., Miramontes-Gonzalez J.P., Hightower C.M., Mustapic M., Mahata M., Huang C.T., Hook V.Y., Mahata S.K., et al. (2014) Chromogranin B: intra- and extra-cellular mechanisms to regulate catecholamine storage and release, in catecholaminergic cells and organisms. J. Neurochem., 129, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahapatra N.R., O'Connor D.T., Vaingankar S.M., Hikim A.P., Mahata M., Ray S., Staite E., Wu H., Gu Y., Dalton N., et al. (2005) Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog. J. Clin. Invest., 115, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y., Rao F., Rodriguez-Flores J.L., Mahata M., Fung M.M., Stridsberg M., Vaingankar S.M., Wen G., Salem R.M., Das M., et al. (2008) Naturally occurring human genetic variation in the 3'-untranslated region of the secretory protein chromogranin A is associated with autonomic blood pressure regulation and hypertension in a sex-dependent fashion. J. Am. Coll. Cardiol., 52, 1468–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bandyopadhyay G.K., Vu C.U., Gentile S., Lee H., Biswas N., Chi N.W., O'Connor D.T., Mahata S.K. (2012) Catestatin (chromogranin A(352-372)) and novel effects on mobilization of fat from adipose tissue through regulation of adrenergic and leptin signaling. J. Biol. Chem., 287, 23141–23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tota B., Angelone T., Cerra M.C. (2014) The surging role of Chromogranin A in cardiovascular homeostasis. Front. Chem., 2, 64.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bassino E., Fornero S., Gallo M.P., Gallina C., Femmino S., Levi R., Tota B., Alloatti G. (2015) Catestatin exerts direct protective effects on rat cardiomyocytes undergoing ischemia/reperfusion by stimulating PI3K-Akt-GSK3beta pathway and preserving mitochondrial membrane potential. PLoS One, 10, e0119790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao F., Zheng Y., Cai J., Fan J., Wang J., Yang J., Cui Q., Xu G., Tang C., Geng B. (2015) Catestatin attenuates endoplasmic reticulum induced cell apoptosis by activation type 2 muscarinic acetylcholine receptor in cardiac ischemia/reperfusion. Sci.Rep., 5, 16590.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D., Liu T., Shi S., Li R., Shan Y., Huang Y., Hu D., Huang C. (2016) Chronic administration of catestatin improves autonomic function and exerts cardioprotective effects in myocardial infarction rats. J. Cardiovasc. Pharmacol. Ther., 21, 526–535. [DOI] [PubMed] [Google Scholar]
- 11. Li R.F., Lu Y.L., Lu Y.B., Zhang H.R., Huang L., Yin Y., Zhang L., Liu S., Lu Z., Sun Y. (2015) Antiproliferative effect and characterization of a novel antifungal peptide derived from human Chromogranin A. Exp. Ther. Med., 10, 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T., Mujagic Z., Vila A.V., Falony G., Vieira-Silva S., et al. (2016) Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science, 352, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fung M.M., Salem R.M., Mehtani P., Thomas B., Lu C.F., Perez B., Rao F., Stridsberg M., Ziegler M.G., Mahata S.K., et al. (2010) Direct vasoactive effects of the chromogranin A (CHGA) peptide catestatin in humans in vivo. Clin. Exp. Hypertens., 32, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Connor D.T., Zhu G., Rao F., Taupenot L., Fung M.M., Das M., Mahata S.K., Mahata M., Wang L., Zhang K., et al. (2008) Heritability and genome-wide linkage in US and australian twins identify novel genomic regions controlling chromogranin a: implications for secretion and blood pressure. Circulation, 118, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katsuda I., Maruyama F., Ezaki K., Sawamura T., Ichihara Y. (2007) A new type of plasma prekallikrein deficiency associated with homozygosity for Gly104Arg and Asn124Ser in apple domain 2 of the heavy-chain region. Eur. J. Haematol., 79, 59–68. [DOI] [PubMed] [Google Scholar]
- 16. Kanaji T., Okamura T., Osaki K., Kuroiwa M., Shimoda K., Hamasaki N., Niho Y. (1998) A common genetic polymorphism (46 C to T substitution) in the 5'-untranslated region of the coagulation factor XII gene is associated with low translation efficiency and decrease in plasma factor XII level. Blood, 91, 2010–2014. [PubMed] [Google Scholar]
- 17. Mahata S.K., O'Connor D.T., Mahata M., Yoo S.H., Taupenot L., Wu H., Gill B.M., Parmer R.J. (1997) Novel autocrine feedback control of catecholamine release. A discrete chromogranin a fragment is a noncompetitive nicotinic cholinergic antagonist. J. Clin. Invest., 100, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rao F., Wen G., Gayen J.R., Das M., Vaingankar S.M., Rana B.K., Mahata M., Kennedy B.P., Salem R.M., Stridsberg M., et al. (2007) Catecholamine release-inhibitory peptide catestatin (chromogranin A(352-372)): naturally occurring amino acid variant Gly364Ser causes profound changes in human autonomic activity and alters risk for hypertension. Circulation, 115, 2271–2281. [DOI] [PubMed] [Google Scholar]
- 19. Renne T. (2012) The procoagulant and proinflammatory plasma contact system. Semin. Immunopathol., 34, 31–41. [DOI] [PubMed] [Google Scholar]
- 20. Joseph K., Kaplan A.P. (2005) Formation of bradykinin: a major contributor to the innate inflammatory response. Adv. Immunol., 86, 159–208. [DOI] [PubMed] [Google Scholar]
- 21. Leduc R., Hendy G.N., Seidah N.G., Chretien M., Lazure C. (1990) Fragmentation of bovine chromogranin A by plasma kallikrein. Life Sci., 46, 1427–1433. [DOI] [PubMed] [Google Scholar]
- 22. Parmer R.J., Miles L.A., Xi X.P., Gill B.M., Wu H.J., O'Connor D.T. (1993) Processing of chromaffin granule proteins: a profusion of proteases?. Neurochem. Int., 22, 361–367. [DOI] [PubMed] [Google Scholar]
- 23. Verweij N., Mahmud H., Mateo Leach I., de Boer R.A., Brouwers F.P., Yu H., Asselbergs F.W., Struck J., Bakker S.J., Gansevoort R.T., et al. (2013) Genome-wide association study on plasma levels of midregional-proadrenomedullin and C-terminal-pro-endothelin-1. Hypertension, 61, 602–608. [DOI] [PubMed] [Google Scholar]
- 24. Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B., Altmaier E., Deloukas P., Erdmann J., Grundberg E., et al. (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature, 477, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lieb W., Chen M.H., Teumer A., de Boer R.A., Lin H., Fox E.R., Musani S.K., Wilson J.G., Wang T.J., Volzke H., et al. (2015) Genome-wide meta-analyses of plasma renin activity and concentration reveal association with the kininogen 1 and prekallikrein genes. Circ. Cardiovasc. Genet., 8, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abid K., Rochat B., Lassahn P.G., Stocklin R., Michalet S., Brakch N., Aubert J.F., Vatansever B., Tella P., De Meester I., et al. (2009) Kinetic study of neuropeptide Y (NPY) proteolysis in blood and identification of NPY3-35: a new peptide generated by plasma kallikrein. J. Biol. Chem., 284, 24715–24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simino J., Shi G., Bis J.C., Chasman D.I., Ehret G.B., Gu X., Guo X., Hwang S.J., Sijbrands E., Smith A.V., et al. (2014) Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am. J. Hum. Genet., 95, 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. (2015) A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet., 47, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cockburn M., Hamilton A., Zadnick J., Cozen W., Mack T.M. (2002) The occurrence of chronic disease and other conditions in a large population-based cohort of native Californian twins. Twin Res., 5, 460–467. [DOI] [PubMed] [Google Scholar]
- 30. Zhang L., Rao F., Wessel J., Kennedy B.P., Rana B.K., Taupenot L., Lillie E.O., Cockburn M., Schork N.J., Ziegler M.G., et al. (2004) Functional allelic heterogeneity and pleiotropy of a repeat polymorphism in tyrosine hydroxylase: prediction of catecholamines and response to stress in twins. Physiol. Genomics, 19, 277–291. [DOI] [PubMed] [Google Scholar]
- 31. Whitfield J.B., Zhu G., Nestler J.E., Heath A.C., Martin N.G. (2002) Genetic covariation between serum gamma-glutamyltransferase activity and cardiovascular risk factors. Clin. Chem., 48, 1426–1431. [PubMed] [Google Scholar]
- 32. Kirk K.M., Birley A.J., Statham D.J., Haddon B., Lake R.I., Andrews J.G., Martin N.G. (2000) Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: prodromus to a linkage study. Twin Res., 3, 299–309. [DOI] [PubMed] [Google Scholar]
- 33. Greenwood T.A., Rao F., Stridsberg M., Mahapatra N.R., Mahata M., Lillie E.O., Mahata S.K., Taupenot L., Schork N.J., O'Connor D.T. (2006) Pleiotropic effects of novel trans-acting loci influencing human sympathochromaffin secretion. Physiol. Genomics, 25, 470–479. [DOI] [PubMed] [Google Scholar]
- 34. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nievergelt C.M., Libiger O., Schork N.J. (2007) Generalized analysis of molecular variance. PLoS Genet., 3, e51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Libiger O., Nievergelt C.M., Schork N.J. (2009) Comparison of genetic distance measures using human SNP genotype data. Hum. Biol., 81, 389–406. [DOI] [PubMed] [Google Scholar]
- 37. Medland S.E., Nyholt D.R., Painter J.N., McEvoy B.P., McRae A.F., Zhu G., Gordon S.D., Ferreira M.A., Wright M.J., Henders A.K., et al. (2009) Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet., 85, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Courel M., Vasquez M.S., Hook V.Y., Mahata S.K., Taupenot L. (2008) Sorting of the neuroendocrine secretory protein Secretogranin II into the regulated secretory pathway: role of N- and C-terminal alpha-helical domains. J. Biol. Chem., 283, 11807–11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Biswas N., Rodriguez-Flores J.L., Courel M., Gayen J.R., Vaingankar S.M., Mahata M., Torpey J.W., Taupenot L., O'Connor D.T., Mahata S.K. (2009) Cathepsin L colocalizes with chromogranin a in chromaffin vesicles to generate active peptides. Endocrinology, 150, 3547–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mosley C.A., Taupenot L., Biswas N., Taulane J.P., Olson N.H., Vaingankar S.M., Wen G., Schork N.J., Ziegler M.G., Mahata S.K., et al. (2007) Biogenesis of the secretory granule: chromogranin A coiled-coil structure results in unusual physical properties and suggests a mechanism for granule core condensation. Biochemistry, 46, 10999–11012. [DOI] [PubMed] [Google Scholar]
- 42. Biswas N., Vaingankar S.M., Mahata M., Das M., Gayen J.R., Taupenot L., Torpey J.W., O'Connor D.T., Mahata S.K. (2008) Proteolytic cleavage of human chromogranin a containing naturally occurring catestatin variants: differential processing at catestatin region by plasmin. Endocrinology, 149, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fries R.S., Mahboubi P., Mahapatra N.R., Mahata S.K., Schork N.J., Schmid-Schoenbein G.W., O'Connor D.T. (2004) Neuroendocrine transcriptome in genetic hypertension: multiple changes in diverse adrenal physiological systems. Hypertension, 43, 1301–1311. [DOI] [PubMed] [Google Scholar]
- 44. Friese R.S., Mahboubi P., Mahapatra N.R., Mahata S.K., Schork N.J., Schmid-Schonbein G.W., O'Connor D.T. (2005) Common genetic mechanisms of blood pressure elevation in two independent rodent models of human essential hypertension. Am. J. Hypertens., 18, 633–652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.