Abstract
The skin is a complex and dynamic ecosystem that is inhabited by bacteria, archaea, fungi and viruses. These microbes—collectively referred to as the skin microbiota—are fundamental to skin physiology and immunity. Interactions between skin microbes and the host can fall anywhere along the continuum between mutualism and pathogenicity. In this Review, we highlight how host–microbe interactions depend heavily on context, including the state of immune activation, host genetic predisposition, barrier status, microbe localization, and microbe–microbe interactions. We focus on how context shapes the complex dialogue between skin microbes and the host, and the consequences of this dialogue for health and disease.
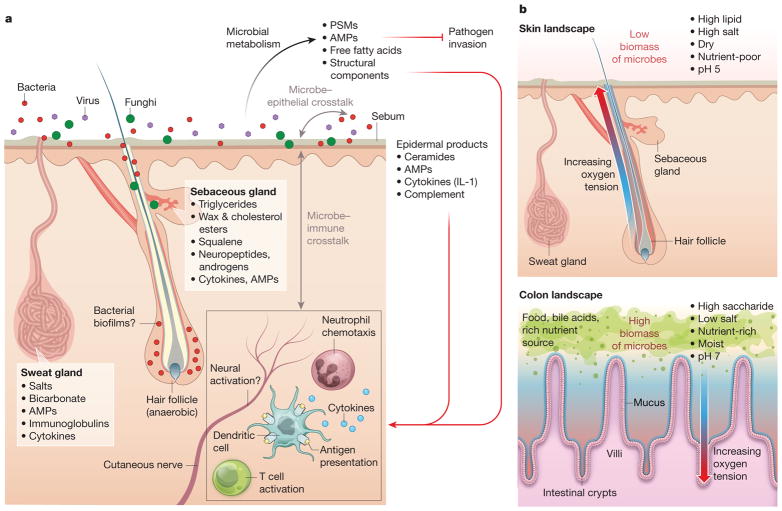
The skin’s outermost aspect consists of a lipid- and protein-laden cornified layer dotted with hair follicles and glands that secrete lipids, antimicrobial peptides, enzymes, salts, and many other compounds (Fig. 1a). Whereas the skin surface is an acidic, high-salt, dessicated, aerobic environment, the invaginations that form folliculo-sebaceous units are comparatively anaerobic and even more lipid-rich7,8 (Fig. 1b). The skin surface and follicles are physically and chemically distinct from another microbe-rich barrier site: the small and large intestines. The intestine is moist, polysaccharide-rich, neutral in pH, and full of diverse carbon and nitrogen sources9–11. Additionally, in contrast to the hair follicle, deeper aspects of intestinal crypts that are closer to the epithelium become more aerobic while the lumen is more anaerobic12,13. The skin, on the other hand, is replete in diverse and unusual lipids not found elsewhere in the body14,15 (Fig. 2). Some of these lipids, such as sapienic acid, can have antimicrobial activities16, while others, such as triglycerides, can be metabolized by microbes17 into free fatty acids and di- and monoglycerides that can be bioactive against other microbes or stimulatory to host cells18.
Figure 1. Crosstalk between skin microbiota and the host.
a, Diverse microbes (viruses, fungi and bacteria) cover the skin surface and associated structures (hair follicles, sebaceous glands and sweat glands), possibly forming biofilms at some sites. These microbes metabolize host proteins and lipids and produce bioactive molecules, such as free fatty acids, AMPs, phenol-soluble modulins (PSMs), cell wall components, and antibiotics158,159. These products act on other microbes to inhibit pathogen invasion, on the host epithelium to stimulate keratinocyte-derived immune mediators such as complement and IL-1, and on immune cells in the epidermis and dermis. In turn, host products and immune cell activity influence microbial composition on the skin. b, The skin differs from the gut in its physical and chemical properties. The skin is a dry, acidic, lipid-rich, high-salt environment without exogenous nutrient sources, and therefore has low microbial biomass. By contrast, the gut is moist and has abundant nutrients and a thick layer of mucin9,159, enabling it to support much greater microbial biomass. While hair follicles become more anaerobic deeper into the follicle, crypts become more aerobic closer to the epithelium7,11. In addition, material within crypts regularly exchanges with material in the gut lumen, owing to peristalsis, whereas hair follicles have narrow openings filled with sebum and keratinocytic debris, making them more isolated.
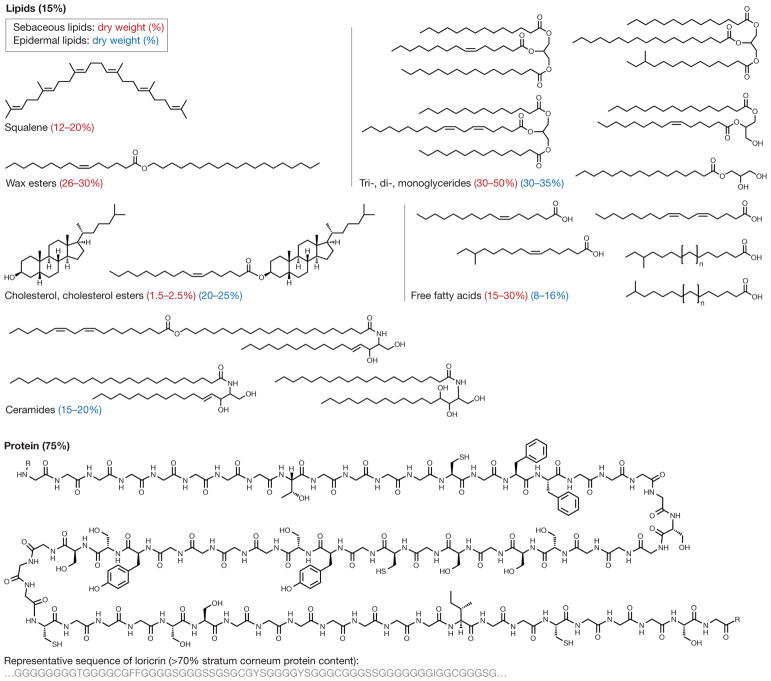
Figure 2. Chemistry of the skin.
The skin surface consists of a highly organized basketweave structure of keratinocytic proteins and lipids, which are produced by keratinocytes (epidermal lipids) and sebaceous glands (sebaceous lipids)160. Ceramides are unique to epidermal origin, whereas squalene and wax esters are unique to sebaceous origin, with variable composition under endocrine control14. Other dominant skin lipids are cholesterol, triglycerides and free fatty acids (which are often microbial products). Some lipids, such as sphingosine and free fatty acids, demonstrate antimicrobial activity against bacteria, fungi, and viruses66 and may have immunomodulatory effects144. Of keratinocytic proteins, more than 70% of the dry protein weight consists of loricrin, a glycine-rich protein that is thought to have important barrier properties160.
Across skin regions, the density and variety of glands and hair follicles vary considerably, creating a complex physical and chemical landscape of geographically distinct niches for bacterial growth. For example, Cutibacterium (formerly Propionibacterium)19 and Staphylococcus species dominate sebaceous areas (such as the face and torso), while Corynebacterium, Staphylococcus, and beta-Proteobacteria are found in moist areas (such as the armpits and the elbow and knee creases)20.
In broad terms, the chemistry of a skin niche drives its microbiome composition, but unknown microbial and host factors contribute to important species- and strain-level differences in composition. For some species, such as Cutibacterium acnes19, the same strain tends to colonize multiple body sites of the same individual; others, such as Staphylococcus epidermidis, differ among body sites of an individual (but tend to be similar in, for example, the axillae of different individuals)21. Most metagenomic cataloguing of the human microbiome has focused on species composition. However, recent work demonstrates that, even within the same species, different strains can differ markedly in their effects on the host22. Strain-level differences have been largely unexplored and remain a frontier for studies of the skin microbiota.
The process of skin microbiota assembly begins during birth and proceeds primarily according to body site over several weeks23. The microbiota shifts notably during puberty, with increased predominance of Corynebacterium and Cutibacterium (formerly Propionibacterium) and decreased abundance of Firmicutes (including Staphylococcus and Streptococcus species)24. In adulthood, despite the skin’s continuous exposure to the environment, the microbial composition remains surprisingly stable over time25. This suggests that stabilizing, mutually beneficial interactions exist among commensal microbes and between microbes and the host.
The composition of the skin microbiome can shift markedly during inflammation26. It is not yet understood how pathogens and skin inflammation contribute to a vicious cycle, how homeostasis is re-established, or how pathogens interact with the existing commensal population. The critical role of context to the outcome of a microbe–host interaction animates this review. For example, pathogens such as Staphylococcus aureus often colonize the skin asymptomatically, whereas mutualists such as S. epidermidis can at times promote disease27,28. In this Review, we highlight recent work demonstrating that host–microbe interactions fall along a continuum in which pure pathogenicity and mutualism are at extreme ends, and are rarely useful descriptors. We discuss the importance of context—genetic predisposition, the level of host immune activation, the physical and chemical landscape of the niche, and mitigating or activating microbe–microbe interactions—to the outcome of a host–microbe interaction, and consider how colonization extends from the skin’s surface into the follicles and even into subcutaneous tissues.
Host–mutualist interactions
Most microbes living on the skin behave as commensal or mutualistic under steady-state conditions. In contrast to the gut of germ-free mice, which shows grossly altered lymphoid organ development, the skin of germ-free mice does not show marked morphological defects29,30. Nonetheless, skin-resident microbes play important roles in the maturation and homeostasis of cutaneous immunity. The skin microbiota modulate the expression of various innate factors, including interleukin 1a (IL-1α)29; components of complement31; and antimicrobial peptides (AMPs), which are produced by keratinocytes and sebocytes (Fig. 1a). Skin-derived AMPs constitute a diverse array of protein families, but cathelicidins and β-defensins predominate. Although some AMPs are constitutively expressed, others can be stimulated by specific members of the microbiota such as Cutibacterium5,32 or produced by microbes themselves (including Cutibacterium thiopeptides33 and S. epidermidis AMPs34,35). It is not yet known how the combination of microbiota-induced and microbiota-produced AMPs shape microbial communities, but this multidirectional signalling is likely to play an important role in the ecology of skin microbial communities.
One major genus of skin-resident bacteria is Corynebacterium, members of which are present at all body sites and dominate in moist sites. Interestingly, corynebacteria share many microbiological features with the closely related mycobacteria, but these two genera interact very differently with the host. It remains a challenge to understand how the immune system distinguishes between bacteria with such similar surface and cellular structures (Fig. 3), and to determine which factors unique to Corynebacterium might be responsible for its commensalism. These questions will help to define the molecular-level differences between mutualism and pathogenicity, and explain how commensal bacteria ‘educate’ the cutaneous immune system.
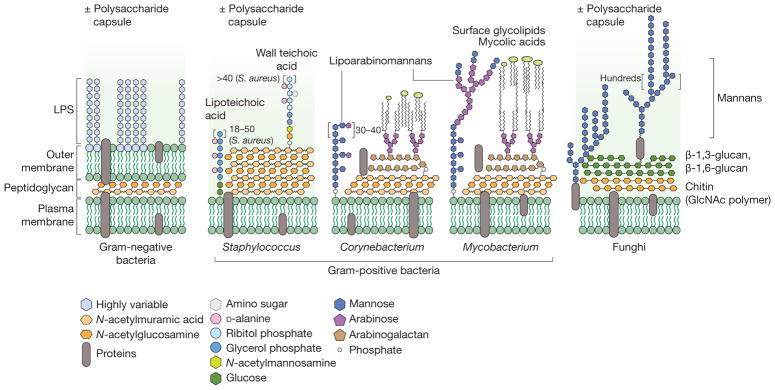
Figure 3. Chemistry of microbial surfaces.
Bacteria and fungi have diverse cell envelopes loaded with immunogenic molecules. Gram-negative bacteria (left) have two lipid bilayers separated by a peptidoglycan cell wall. The outer leaflet of the outer membrane is studded with an immunogenic lipoglycan called lipopolysaccharide (LPS), which has a lipid anchor and highly variable polysaccharide region called the O-antigen. In Escherichia coli, for example, 184 different O-antigen structures are known161. Gram-positive bacteria (middle) lack an outer lipid bilayer but have a thicker peptidoglycan cell wall. Staphylococcus species have wall-bound and membrane-anchored teichoic acids, which are analogous to Gram-negative LPS, with a host-facing strain-specific branched polysaccharide. Corynebacterium and Mycobacterium species also have complex lipoglycans, called lipoarabinomannans. They also have a unique lipid outer layer made up of cell-wall-bound mycolic acids and other noncovalently bound glycolipids. Like Gram-positive bacteria, fungi (right) have only one lipid bilayer membrane. This is covered by a cell wall usually consisting of chitin and a β-D-glucan mesh. The outer layer of the fungal cell wall often contains heavily mannosylated proteins and sometimes a capsule of various polysaccharides. GlcNAc, N-acetylglucosamine.
Corynebacterium minutissimum (erythrasma) and Corynebacterium tenuis (trichomycosis axillaris) have been associated with superficial skin pathology, but most Corynebacterium species present in surveys of the skin microbiome do not cause any known disease. Corynebacteria and mycobacteria share the unusual feature among Gram-positive bacteria of having an outer membrane, analogous to that of Gram-negative bacteria (Fig. 3). This outer membrane consists of an outer lipid bilayer of long α-branched fatty acids called mycolic acids, which envelop (and are covalently linked to) the meshwork of peptidoglycan underneath. The Corynebacterium cell wall features additional lipoglycans termed lipomannans and lipoarabinomannans, which are anchored to the plasma membrane and have long oligosaccharide chains that emanate from the cell surface. Lipomannans and lipoarabinomannans are ligands for host glycan receptors such as Toll-like receptors (TLRs) and C-type lectin receptors, driving either pro- or anti-inflammatory responses depending on their structure and the immunological context in which they are sensed36–42. In mycobacteria, lipomannans and lipoarabinomannans play important roles in immune recognition and evasion43. It remains to be determined whether similar structures in corynebacteria engage cutaneous immune cells, and whether immune recognition of corynebacteria may protect against future mycobacterial infections.
In addition to microbe–host interactions, many reports have suggested that microbe–microbe interactions also impact human health. For example, a common skin resident, Corynebacterium accolens, was recently shown to inhibit the growth of Streptococcus pneumoniae, a common respiratory tract pathogen44. The active principle of this interaction is an essential corynebacterial lipase that hydrolyses triolein to release oleic acid, which inhibits pneumococcal growth44. Another common skin resident, Corynebacterium striatum, shifts the global transcriptional program of co-cultured S. aureus in a way that suppresses virulence-related genes and stimulates genes assocated with commensalism28. These data suggest that the role of skin-resident microbes goes beyond competitive exclusion; these microbes are likely to engage in a web of microbe–microbe interactions that help to tune the behaviour of their co-residents in subtle and context-specific ways.
Another dominant group of skin colonists are the coagulase-negative Staphylococcus species, the most prominent of which is S. epidermidis. Although S. epidermidis can be an opportunisitic pathogen in the context of primary or iatrogenic immunosuppression, it functions predominantly as a mutualist. Skin-resident Staphylococcus species engage in microbe–microbe interactions that are beneficial to the host. For example, S. epidermidis and Staphylococcus hominis have been shown to secrete antimicrobial peptides that kill S. aureus, and transplantation of these species onto the skin of patients with atopic dermatitis led to decreased colonization by S. aureus35.
Recent studies of S. epidermidis provided the first evidence that skin-resident bacteria are not just passive residents; they actively engage host immunity through an intact skin barrier, and activate specific immune cell populations in a species- and strain-dependent manner29,45. For instance, some strains of S. epidermidis induce activation of S. epidermidis-specific IL-17+CD8+ T cells that protect against cutaneous infections by inducing keratinocytes to produce AMPs, a phenomenon called heterologous protection45. In addition to their protective role, these commensal-specific T cells also promote wound repair46. Of interest, S. epidermidis can elicit T cell responses restricted to non-classical major histocompatibility complex (MHC) class I molecules46. Thus, non-classical MHC class I molecules, an evolutionarily ancient arm of the immune system, may play an important role in promoting homeostatic immunity to the microbiota. These data show that skin-resident bacteria can have myriad effects on the host; in addition to promoting immune barrier responses, commensal–immune interactions can also affect epithelial biology. The effects of commensal–immune interactions on many other cutaneous processes, including adnexal development, tumorigenesis, ageing, and sensory nerve function, remain to be determined. Additionally, whether immune responses against the skin microbiota also influence microbiota composition or function remains unexplored. In most settings, the skin flora controls skin immunity in an autonomous manner and independently of the gut flora29. This compartmentalization and specialization of responses may have evolved as a mechanism to constrain the adjuvant properties of commensals and unwanted consequences associated with systemic increases in inflammatory responses.
Notably, adaptive responses to members of the skin community develop in the absence of inflammation45, in contrast to the response to invading pathogens. This process, termed ‘homeostatic immunity’, may be induced (at least in part) by the endogenous network of skin-resident antigen-presenting cells45. Under steady-state conditions, the skin is populated by highly diverse T cells47. Thus, because of the extraordinary number of potential antigens expressed by the microbiota, a substantial fraction of these skin-resident T cells are expected to be microbiota-specific. As a result, primary exposure to a pathogen in the skin or exposure during an injury is likely to occur in the context of a much broader recall response against diverse microbial antigens. The consequences of this phenomenon for tissue responses remain unclear.
Although B cell dynamics in the skin and the role of antibodies in controlling skin microbes are not well understood, it is known that IgA is secreted on the skin surface by eccrine and sebaceous glands48,49. In the gut, IgA has substantial effects on microbiota composition50–52 through a process that involves coating commensal bacteria53; in turn, commensal microbes are essential to development of this antibody response54,55 and may protect against autoimmunity56. Skin commensals are also likely to affect the B cell repertoire, but the extent of this interaction and its effects on the microbiota are not yet known.
In light of the finding that S. epidermidis potently and specifically activates a unique branch of adaptive immunity, one major challenge is to dissect mechanistically how specific host cells and receptors recognize molecular features of S. epidermidis. Staphylococci produce a variety of immune modulatory molecules, such as teichoic acids, capsular polysaccharides57,58, and dipeptide aldehydes59,60. Just as Corynebacterium and Mycobacterium share features but can be distinguished by the host, S. epidermidis shares many of these molecular features with the contextual pathogen S. aureus. Further studies of how, at the molecular level, S. aureus differs from S. epidermidis will help us understand how these two important human skin residents are distinguished by the immune system, and will highlight key host targets for more effective and specific therapeutic approaches.
Microbial sensing by the immune system is also likely to be controlled by the developmental stage of the host. Although little is understood about the factors that regulate the acquisition of skin microbes at birth, regulatory T cells, which are highly enriched in the skin tissue, have been proposed to control early dialogue with the microbiota. Indeed, colonization of mouse skin with S. epidermidis early in life (but not later) induces tolerance to the same microbe in adulthood61 and promotes accumulation of S. epidermidis-specific regulatory T cells in neonatal skin62.
As well as modulating immune cells, S. epidermidis and other commensals promote epithelial integrity, especially during tissue repair. For example, an S. epidermidis cell wall component mitigates inflammation by binding to TLR2, limiting tissue damage and promoting wound healing63. Other commensal microbiota are also likely to contribute to wound healing, which is a dynamic process associated with global shifts in the skin microbiome; wound bed microbiomes that fail to shift are associated with chronic ulcers64,65. Within chronic wounds, fungi and bacteria form mixed biofilms and certain fungal taxa, such as the phylum Ascomycota, are predictive of wounds that take more than eight weeks to heal. The fungal mycobiome and its interactions with commensal bacteria may therefore be important contributors to chronic wounds, via mechanisms that have not been explored64,65.
The skin microbiota are likely to affect many immune-related and immune-independent properties of epithelial health that are not yet appreciated. In the skin, epithelial homeostasis is a constant, active, and energy-intensive process that involves the secretion of complex lipids for signalling and barrier purposes2,66, maintenance of tight junctions67,68 and production of a lipid–protein coat to prevent trans-epidermal water loss69,70, repair of UV-mediated and oxidative damage to epithelial cells to prevent malignant transformation71,72, and constant remediation of accidental trauma (for example, scrapes, cuts, and nicks). Any disruption of even a small component of these complex processes can result in extreme phenotypes, such as ichthyoses, blistering disorders, progerias, and diffuse fibrosis69,73,74. How the skin community contributes to these processes remains to be addressed.
Host–pathogen interactions
Microbe–host interactions that drive (or result from) infectious processes have historically received the most investigative attention. A canonical host–pathogen interaction in the skin involves a one-to-one mapping of microbe to disease and an easily identified phenotype of inflammation. Most of what is known about the skin immune system has been discovered by studying interactions of this sort, highlighting the utility of this simplistic paradigm and foreshadowing its limitations. As we will discuss, most microbe–host interactions on the skin are more nuanced; a threshold example is the observation that traditional pathogens often reside on the skin surface in an asymptomatic manner.
In terms of cost and prevalence, one of the most important pathogens of the skin is S. aureus. Although more than 30% of healthy individuals are colonized asymptomatically by S. aureus75,76, it can cause a wide spectrum of infections: some are limited to a single hair follicle (furuncle), others involve subcutaneous tissues (cellulitis), and the most serious feature potentially fatal penetration into any organ in the body, including bone (osteomyelitis), bloodstream (bacterial sepsis), and heart valves (bacterial endocarditis). S. aureus has also been implicated in the pathogenesis of chronic diseases such as atopic dermatitis22,77–79, and more recently in systemic lupus erythematosus with renal and skin involvement80.
S. aureus is a versatile pathogen with a broad array of virulence factors81,82, including neutrophil-killing toxins83, chemotaxis inhibitors84, anti-phagocytic and anti-killing surface molecules57,85–88, superantigens, and immune evasion proteins89,90. In patients with atopic dermatitis, S. aureus isolates grow as biofilms on the skin and produce proteases that degrade host AMPs, such as cathelicidin LL-3791. The host has evolved mechanisms to ward off invasion by S. aureus at every level of the skin and subcutaneous tissue. In addition to a diverse arsenal of AMPs covering the epidermis, there is also evidence that adipose tissue under the dermis contributes to the innate immune response. Following breach of the skin barrier and subsequent S. aureus infection, local pre-adipocytes proliferate rapidly, expanding the subdermal fat layer and increasing production of the AMP cathelicidin92.
Although some virulence and immune evasion elements are conserved across all species of S. aureus, there are important strain-level differences. For example, the arginine catabolic mobile element contributes to the ability of USA300, a methicillin-resistant S. aureus strain, to thrive in the acidic environment of human skin and resist host polyamines, helping to explain this strain’s prevalence in skin and soft tissue infections93,94. Recent work has also shown that certain strains of S. aureus are not only associated with more severe atopic dermatitis, but are also sufficient to induce skin inflammation in mice independent of host genetic predisposition22. This work revealed that the most common method of describing microbiome composition, with genus- or species-level data, fails to resolve important functional differences among strains, which can result from modest gene gain or loss events or even differences in gene expression among strains.
Another prominent genus of skin pathogens is Mycobacterium, a Gram-positive rod within the phylum Actinobacteria. Mycobacteria are a diverse genus of organisms that includes the causative agents of tuberculosis (Mycobacterium tuberculosis) and leprosy (Mycobacterium leprae), and other species that cause infections at surgical sites or sites of accidental trauma (for example, Mycobacterium kansasii, Mycobacterium chelonae, and Mycobacterium marinum). M. tuberculosis, which generally causes pulmonary or systemic infections, is well known; however, skin and soft tissue infections caused by other Mycobacterium species are increasing in prevalence in developed countries95 and continue to be serious problems in developing countries96. We highlight mycobacteria because they generate an especially broad spectrum of pathologies, from acute systemic illness to skin manifestations to inert granulomas that persist throughout the lifetime of a host. In addition, mycobacteria are closely related to skin-resident corynebacteria but have very different effects on the host. The similarities and differences between Mycobacterium and Corynebacterium will probably yield insights into a broad swath of fundamentally important host–microbiota interactions.
M. tuberculosis is one of the most successful pathogens on the planet: it colonizes one-quarter of the world’s population (1.7 billion people) in the form of a latent infection97, with the World Health Organization estimating that 10.4 million new infections occurred in 201598. Among humans who are latent carriers of M. tuberculosis, only 10% will suffer reactivation into active tuberculosis during their lifetime99. A related pathogen, M. leprae, also causes a wide range of diseases, including diverse skin manifestations that can involve the nerves, liver, and bones100,101. The time period between M. leprae inoculation and clinical manifestation of infection is typically 2–12 years and can be up to a few decades; during this time, the bacterium handily evades host immunity. It is particularly notable that the majority of humans who harbour M. tuberculosis and M. leprae neither display obvious pathology nor die from their infection. This suggests that we have much to discover about the mechanisms of long-term immune evasion, and that the traditional definition of ‘colonist’ may need to expand to include an organism such as M. tuberculosis, which on the one hand is a pathogen that kills more than a million people each year102, and on the other hand lives asymptomatically in billions of people and results in the death of only a small percentage of its hosts.
One recently discovered mechanism of mycobacterial host evasion involves the nervous system. Mycobacterium ulcerans causes the Buruli ulcer, a progressive, necrotic ulcer that is the third most common mycobacterial disease worldwide103. The Buruli ulcer is painless, which contributes to delays in treatment and therefore increases the requirement for more drastic interventions at later infectious stages, when the only available treatment is surgical resection. M. ulcerans produces a polyketide toxin, mycolactone, that is essential for virulence. Recent work has shown that mycolactone induces analgesia via the angiotensin II receptor, COX-1, and prostaglandin E2, ultimately resulting in activation of TRAAK potassium channels and cell hyperpolarization104. Not only does this work provide possible avenues to therapeutic biomimetics for pain relief and to the development of therapies against M. ulcerans, but it also demonstrates a novel mode of pathogen–host interaction in which the peripheral nervous system is targeted directly.
Involvement of the peripheral nervous system may be more general and integral to skin immunity than has been previously recognized. Candida albicans, a fungal pathogen, also triggers sensory neurons directly; these neurons then stimulate host immunity and activate protective IL-17-producing dermal T cells105. S. aureus also activates cutaneous neurons directly via N-formylated peptides and the pore-forming toxin α-haemolysin, inducing pain and neuropeptide-mediated induction of vasodilation and inflammation106. More recently, a direct mechanistic link between neurons and immune cells has been discovered. For instance, in the gut, mucosal neurons were found to produce a neuropeptide, neuromedin U (NMU), that binds an NMU receptor on group 2 innate lymphoid cells (ILC2s) and triggers a protective immune response107. Direct microbiota–nervous system interactions appear to be a broader phenomenon than was previously appreciated, with examples emerging in other body sites; in a recent case, a microbiota-derived metabolite (isovaleric acid) was shown to trigger a receptor enriched in enterchromaffin cells, resulting in the basolateral release of serotonin, which stimulated sub-epithelial enteric nervous system afferents108.
As well as evading immunity so that an infection can establish, mycobacteria can also persist for decades inside granulomas, which are organized aggregates of macrophages. Bacteria living within a granuloma, in equilibrium with the host, can be considered a form of tissue colonization. In the gut, for example, certain subsets of commensal bacteria intracellularly colonize CD11c+ dendritic cells in healthy mice and promote innate lymphoid cell responses that prevent systemic dissemination of these bacteria, as well as IL-10 production that protects against intestinal inflammation and damage109,110. These data demonstrate that commensal colonists can reside not only on the surface of the host, but also within host cells and tissues, and that these bacteria can elicit a balance of immune-reactive and immunosuppressive responses in the host. For mycobacteria, granulomas may seed the infection of new macrophages, leading to dissemination111. However, in 80–90% of healthy patients with latent tuberculosis, reactivation does not occur during the lifetime of the host112. In these patients, M. tuberculosis persists within macrophages, using a variety of strategies including efflux pumps that promote drug tolerance113–115, increased cell wall biosynthesis116, and global transcriptional changes to survive in anaerobic conditions117–119. Within a single host, granulomas can resolve variably: some become sterile, others harbour latent bacteria, and some allow bacteria to escape120 in a manner that does not depend on host or bacterial genetic features. Therefore, even for an individual pathogen within a single host, it is not yet understood how the specifics of context affect the outcome of a microbe–host encounter.
As well as bacterial pathogens, there are numerous viral and fungal pathogens of the skin. Some viruses, such as human papillomaviruses and herpesviruses, can cause acute pathology but generally persist in a latent manner for a lifetime. Other viruses, such as the orthopoxvirus vaccinia virus, are cleared after epicutaneous infection. Although our current understanding of these viruses encompasses only their pathogenic behaviour, analyses of the double-stranded DNA virome have shown that papillomaviruses and poxviruses can also colonize hosts asymptomatically21,121. Even in the ‘simple’ case of vaccinia virus, intravital multiphoton microscopy has shown a complex spatial orchestration of immune events; although viral infection occurs in keratinocytes, responding CD8+ T cells do not target the infected keratinocytes but rather innate immune cells122. Counterintuitively, high local production of the anti-inflammatory cytokine IL-10 helps to limit vaccinia replication and dissemination123.
An immune cell population that has received attention for its ability to promote memory responses to viruses (and, potentially, other members of the microbiota) is resident memory T (TRM) cells—a population of lymphocytes that occupies tissues without recirculating124. While most studies have focused on virally induced CD8+ TRM cells, CD4+ TRM cells (or at least cells with a resident phenotype) have also been shown to accumulate in the skin in response to a large array of infections. Notable differences in anatomical localization exist for memory CD4+ and CD8+ T cells, with CD4+ T cells maintained primarily within the dermis and CD8+ T cells within the epidermal compartment125. Given that herpesviruses, papillomaviruses and polyomaviruses that infect epithelial cells generate a high burden of chronic skin disease and, in some cases, aggressive skin cancers, there is an urgent need to better understand how these infections are contained by or escape cutaneous immunity.
Contextual pathogenicity
Traditionally, the term ‘pathobiont’ has been applied to organisms that have the potential to cause disease but often colonize a host without inducing pathology. Two prominent gut pathobionts are segmented filamentous bacterium (SFB), which stimulates T helper 17 (TH17) cells in the mouse gut to confer protection against pathogens but can also induce severe colitis; and Helicobacter pylori, which colonizes half of the human population, but in a small percentage can cause peptic ulcer disease and potentiate gastric adenocarcinoma126–129. However, in more extreme contexts of host predisposition, many other microbial species have the potential to cause diease. Patients with primary immunodeficiency (PID), for example, develop chronic, severe skin infections, many of them induced by normal constituents of the microbiota or environmental microbes130.
Transitioning from commensalism (for example, on the skin surface or in a follicle) to pathogenicity (for example, in the bloodstream) is a complex and potentially costly process for the microbe. The skin is an imposing physical barrier, and even if it is breached, the microbe must fend off numerous layers of innate responses, including AMPs, proteases, and reactive oxygen species. In addition, would-be pathogens need to induce the expression of genes that enable adhesion, invasion, and immune evasion (that is, virulence factors). As a result, most commensal microbes coexist peacefully with their host, and will exhibit pathogenic potential only in specific settings. Conversely, microbes that are traditionally considered pathogens do not indiscriminately display aggressive behaviour. As discussed above, the term pathogen does not encompass the range of phenotypes displayed by S. aureus or M. tuberculosis. Viewed in this light, the term pathobiont becomes unhelpfully broad. Therefore, we suggest that the concept of a continuum of contextual pathogenicity and mutualism may be more useful (Fig. 4). In this section, we discuss examples of microbes that might traditionally be referred to as pathobionts: those that represent the middle of the spectrum between aggressive and mutualistic behaviour.
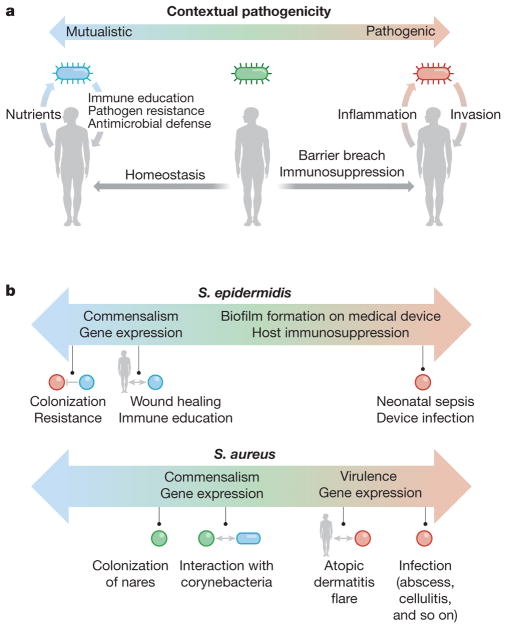
Figure 4. Contextual pathogenicity.
a, Microbes exhibit contextual pathogenicity along a spectrum. Host factors such as barrier breaches and immunosuppression bias microbes towards pathogenic behaviour, whereas homeostatic conditions bias them towards mutualistic behaviour. On the microbial side, virulence gene expression and microbe–microbe interactions can also push microbial behaviour to be mutualistic or pathogenic. In a mutualistic host–microbe relationship, the host provides nutrients, while the microbe promotes epithelial and immune homeostasis as well as pathogen resistance through microbial products and occupation of metabolic niches. In a pathogenic relationship, the microbe invades past the epithelium, causing inflammation, and sometimes also benefiting from a host inflammatory response. b, Both S. epidermidis and S. aureus are examples of contextual pathogenicity; S. epidermidis is biased towards mutualistic behaviour whereas S. aureus displays more pathogenic character.
One example is C. acnes, which has been implicated in acne131–133 and hidradenitis suppurativa134. However, the contribution of C. acnes to the pathogenesis of acne is unclear131. On the one hand, C. acnes produces coproporphyrin III, which has been shown to induce the formation of S. aureus biofilms135—generally seen as a negative consequence for the host. On the other hand, C. acnes has also been shown to ferment glycerol into short-chain fatty acids, which suppress the growth of virulent methicillin-resistant S. aureus USA300136. These data suggest that rich networks of microbe–microbe interactions may govern host inflammation and disease in a strain- and context-dependent manner.
A similar range of harmful and beneficial effects have been demonstrated for other microbes. Herpesviruses are frequent pathogens of the skin, with notable examples including chickenpox (varicella zoster virus) and recurrent labial ulcers (HSV1 and HSV2); however, after the acute infection has resolved, herpesviruses persist within the host as dormant, latent viruses for the host’s lifetime. In mice, this herpesvirus latency has been shown to stimulate the immune system in a way that is protective against bacterial pathogens for months137,138. As noted above, H. pylori colonizes the stomach and can cause ulcers and promote gastric cancer. However, it has long evolved to live within human hosts, and is also thought to protect against tuberculosis139 and allergic diseases such as childhood asthma140.
Although S. epidermidis is generally beneficial to the host, it is also a leading cause of death in premature infants and nosocomial infections27,141. Conversely, Mycobacterium species such as M. leprae and M. tuberculosis are known for their ability to cause serious systemic illness but can persist subclinically inside a granuloma for the lifetime of a host. To generalize, all microbes that reside on or inside a host fall along a spectrum, with some displaying almost no aggressive behaviour and others displaying primarily virulent, invasive phenotypes. The majority of skin residents probably lie somewhere in the middle of the spectrum, and an important challenge in future work will be to better understand which host and environmental factors govern a microbe’s switch between passive and aggressive behaviours.
Discussion
The examples discussed herein illustrate that a scheme in which skin-resident microbes are classified as ‘full-time’ pathogens, mutualists or pathobionts may need to be updated to include the effects of context (Fig. 4). Future work will need to investigate the context-dependent behaviour of resident microbes—how microbe–microbe interactions, host–microbe interactions, and strain-specific differences may govern a microbe’s tendency towards cooperation or aggression.
As we learn more about how commensal bacterial strains activate specific immune cell populations, we may be able to harness this specificity by engineering microbes to deliver cytokines, small molecules, or vaccines to specific, activated immune cell populations across an intact skin barrier. A clearer understanding of the dense network of microbe–microbe interactions will also allow us to provide more targeted therapies for dysbiosis, which has been implicated in atopic dermatitis but is also being explored as a pathogenic contributor to many other skin diseases, including psoriasis, hidradenitis suppurativa, and lupus erythematosus. In the gut, using microbes to correct dysbiosis has been successful in the case of faecal transplants for Clostridium difficile infections142, and more recently in the use of a Lactobacillus plantarum synbiotic to prevent neonatal sepsis143. Similar live microbial therapies for the skin have not yet been developed. However, harnessing the immunomodulatory45 or antimicrobial properties of skin commensal bacteria144 has great potential. Furthermore, because of the unique chemical milieu of the skin (Fig. 2), local alterations in defined nutrients may have a marked impact on the composition or function of skin microbiota and—when rationally designed—could promote the expansion of microbes endowed with regulatory or protective properties.
Another important category of microbe–host interactions that has not been well explored consists of distant effects between microbes at one site, for example in the gut, and host responses at another site, such as the skin. Recently, immune checkpoint blockade has achieved unprecedented success in the treatment of multiple skin cancers that were previously associated with high rates of mortality, including metastatic melanoma, squamous cell carcinoma, and Merkel cell carcinoma145–148. Although these are cutaneous cancers, commensal gut bacteria have been implicated in the efficacy of anti-tumour immunotherapies at distant sites149,150. Conversely, how skin-resident microbes influence immune responses systemically or at distant sites is an important area for further research. Processes that were previously thought to involve skin-limited inflammation, such as plaque psoriasis, have now been linked to an increase in systemic inflammatory co-morbidities, such as atheroscle-rotic cardiovascular disease151–154. Indeed, many types of immune cell are known to traffic in and out of skin during both homeostasis and inflammation155–157. These observations suggest that the effects of the skin microbiota on the immune system may have wide-ranging systemic sequelae that are ripe for exploration in the near future.
These findings add another layer of complexity to microbe–host interactions, suggesting that research should not only focus on interactions within the local microenvironment but also encompass trafficking of microbiota-educated immune cell populations, or microbial products and metabolites, to other body sites upon stimulation by microbes at diverse barrier sites.
Acknowledgments
We apologize for not having cited all papers relevant to this expanding field of research (in particular, older literature) because of space constraints and editorial limits. This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Y.B.), DP1 DK113598 (M.A.F.), R01 DK110174 (M.A.F.), an HHMI-Simons Faculty Scholars Award (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.), a Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award (M.A.F.) and the Dermatology Foundation (Y.E.C.).
Footnotes
Author Contributions Y.E.C., M.A.F. and Y.B. conceptualized the article structure, content, and figures, and wrote and edited the manuscript and figures.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.De Luca C, Valacchi G. Surface lipids as multifunctional mediators of skin responses to environmental stimuli. Mediators Inflamm. 2010;2010:321494. doi: 10.1155/2010/321494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Smeden J, Bouwstra JA. Stratum corneum lipids: their role for the skin barrier function in healthy subjects and atopic dermatitis patients. Curr Probl Dermatol. 2016;49:8–26. doi: 10.1159/000441540. [DOI] [PubMed] [Google Scholar]
- 3.Niyonsaba F, Kiatsurayanon C, Chieosilapatham P, Ogawa H. Friends or foes? Host defense (antimicrobial) peptides and proteins in human skin diseases. Exp Dermatol. 2017;26:989–998. doi: 10.1111/exd.13314. [DOI] [PubMed] [Google Scholar]
- 4.Bek-Thomsen M, Lomholt HB, Scavenius C, Enghild JJ, Brüggemann H. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS One. 2014;9:e107908. doi: 10.1371/journal.pone.0107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee DY, et al. Sebocytes express functional cathelicidin antimicrobial peptides and can act to kill propionibacterium acnes. J Invest Dermatol. 2008;128:1863–1866. doi: 10.1038/sj.jid.5701235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boncheva M. The physical chemistry of the stratum corneum lipids. Int J Cosmet Sci. 2014;36:505–515. doi: 10.1111/ics.12162. [DOI] [PubMed] [Google Scholar]
- 7.Matard B, et al. First evidence of bacterial biofilms in the anaerobe part of scalp hair follicles: a pilot comparative study in folliculitis decalvans. J Eur Acad Dermatol Venereol. 2013;27:853–860. doi: 10.1111/j.1468-3083.2012.04591.x. [DOI] [PubMed] [Google Scholar]
- 8.Puhvel SM, Reisner RM, Amirian DA. Quantification of bacteria in isolated pilosebaceous follicles in normal skin. J Invest Dermatol. 1975;65:525–531. doi: 10.1111/1523-1747.ep12610239. [DOI] [PubMed] [Google Scholar]
- 9.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 11.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lind Due V, Bonde J, Kann T, Perner A. Extremely low oxygen tension in the rectal lumen of human subjects. Acta Anaesthesiol Scand. 2003;47:372. doi: 10.1034/j.1399-6576.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- 13.Crompton DWT, Shrimpton DH, Silver IA. Measurements of the oxygen tension in the lumen of the small intestine of the domestic duck. J Exp Biol. 1965;43:473–478. doi: 10.1242/jeb.43.3.473. [DOI] [PubMed] [Google Scholar]
- 14.Strauss JS, Pochi PE, Downing DT. The sebaceous glands: twenty-five years of progress. J Invest Dermatol. 1976;67:90–97. doi: 10.1111/1523-1747.ep12512506. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaides N. Skin lipids: their biochemical uniqueness. Science. 1974;186:19–26. doi: 10.1126/science.186.4158.19. [DOI] [PubMed] [Google Scholar]
- 16.Drake DR, Brogden KA, Dawson DV, Wertz PW. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res. 2008;49:4–11. doi: 10.1194/jlr.R700016-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Puhvel SM, Reisner RM, Sakamoto M. Analysis of lipid composition of isolated human sebaceous gland homogenates after incubation with cutaneous bacteria Thin-layer chromatography. J Invest Dermatol. 1975;64:406–411. doi: 10.1111/1523-1747.ep12512337. This study showed that common skin bacteria, such as Cutibacterium (Propionibacterium) species and Staphylococcus epidermidis, can modify skin lipids through hydrolysis of triglycerides and esterification of cholesterol, and that these enzymatic activities can be modified by other skin features, such as pH. [DOI] [PubMed] [Google Scholar]
- 18.Sanford JA, et al. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol. 2016;1:eaah4609. doi: 10.1126/sciimmunol.aah4609. [DOI] [PubMed] [Google Scholar]
- 19.Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66:4422–4432. doi: 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 20.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 324:1190–1192. doi: 10.1126/science.1171700. Using 16S ribosomal RNA sequencing, this study provided a metagenomic analysis of the human skin microbiome and described previously unappreciated bacterial diversity at different skin sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh J, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrd AL, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9:eaal4651. doi: 10.1126/scitranslmed.aal4651. This study highlights the utility of shotgun metagenomic sequencing over 16S ribosomal RNA sequencing to assess how strain differences within the same Staphylococcus epidermidis species can contribute to disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu DM, et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh J, Byrd AL, Park M, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. This study was one of the first to use metagenomic sequencing to characterize dysbiosis in inflammatory skin diseases, showing that atopic dermatitis flares are associated not only with blooms of Staphylococcus aureus but also with significant decreases in overall skin microbial diversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otto M. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol. 2016;7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. This study demonstrated that skin-resident commensal bacteria are critical for establishing skin immune homeostasis and that this process occurs through an intact, uninflamed skin barrier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 31.Chehoud C, et al. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc Natl Acad Sci USA. 2013;110:15061–15066. doi: 10.1073/pnas.1307855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy I, et al. Propionibacterium acnes and lipopolysaccharide induce the expression of antimicrobial peptides and proinflammatory cytokines/chemokines in human sebocytes. Microbes Infect. 2006;8:2195–2205. doi: 10.1016/j.micinf.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Christensen GJM, et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics. 2016;17:152. doi: 10.1186/s12864-016-2489-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cogen AL, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatsuji T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Källenius G, Correia-Neves M, Buteme H, Hamasur B, Svenson SB. Lipoarabinomannan, and its related glycolipids, induce divergent and opposing immune responses to Mycobacterium tuberculosis depending on structural diversity and experimental variations. Tuberculosis (Edinb ) 2016;96:120–130. doi: 10.1016/j.tube.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Afonso-Barroso A, et al. Lipoarabinomannan mannose caps do not affect mycobacterial virulence or the induction of protective immunity in experimental animal models of infection and have minimal impact on in vitro inflammatory responses. Cell Microbiol. 2013;15:660–674. doi: 10.1111/cmi.12065. [DOI] [PubMed] [Google Scholar]
- 38.Briken V, Porcelli SA, Besra GS, Kremer L. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol Microbiol. 2004;53:391–403. doi: 10.1111/j.1365-2958.2004.04183.x. [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee D, Khoo KH. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology. 1998;8:113–120. doi: 10.1093/glycob/8.2.113. [DOI] [PubMed] [Google Scholar]
- 40.Dao DN, et al. Mycobacterium tuberculosis lipomannan induces apoptosis and interleukin-12 production in macrophages. Infect Immun. 2004;72:2067–2074. doi: 10.1128/IAI.72.4.2067-2074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doz E, et al. Acylation determines the toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor-, and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J Biol Chem. 2007;282:26014–26025. doi: 10.1074/jbc.M702690200. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda T, et al. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. MBio. 2013;4:e00472–e12. doi: 10.1128/mBio.00472-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishikawa E, Mori D, Yamasaki S. Recognition of mycobacterial lipids by immune receptors. Trends Immunol. 2017;38:66–76. doi: 10.1016/j.it.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio. 2016;7:e01725–e15. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naik S, et al. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linehan JL, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. doi: 10.1016/j.cell.2017.12.033. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark RA, et al. The vast majority of CLA+ T cells are resident in normal skin. J Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 48.Metze D, et al. Immunohistochemical demonstration of immunoglobulin A in human sebaceous and sweat glands. J Invest Dermatol. 1989;92:13–17. doi: 10.1111/1523-1747.ep13070402. [DOI] [PubMed] [Google Scholar]
- 49.Okada T, Konishi H, Ito M, Nagura H, Asai J. Identification of secretory immunoglobulin A in human sweat and sweat glands. J Invest Dermatol. 1988;90:648–651. doi: 10.1111/1523-1747.ep12560807. This study used immunohistochemistry to show that secretory IgA was associated with human sweat glands, and was probably being actively transported in a way similar to the intestine. This study raises the question of how IgA on the skin influences microbiota composition and whether commensal microbes stimulate IgA secretion similarly to gut commensal flora. [DOI] [PubMed] [Google Scholar]
- 50.Fagarasan S, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 51.Macpherson AJ, Hunziker L, McCoy K, Lamarre A. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect. 2001;3:1021–1035. doi: 10.1016/s1286-4579(01)01460-5. [DOI] [PubMed] [Google Scholar]
- 52.Kawamoto S, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 53.van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–3913. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 56.Vossenkämper A, et al. A role for gut-associated lymphoid tissue in shaping the human B cell repertoire. J Exp Med. 2013;210:1665–1674. doi: 10.1084/jem.20122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng BL, et al. Evaluation of serotypes 5 and 8 capsular polysaccharides in protection against Staphylococcus aureus in murine models of infection. Hum Vaccin Immunother. 2017;13:1609–1614. doi: 10.1080/21645515.2017.1304334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann M, Fischbach MA. A family of pyrazinone natural products from a conserved nonribosomal peptide synthetase in Staphylococcus aureus. Chem Biol. 2010;17:925–930. doi: 10.1016/j.chembiol.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Wyatt MA, et al. Staphylococcus aureus nonribosomal peptide secondary metabolites regulate virulence. Science. 2010;329:294–296. doi: 10.1126/science.1188888. [DOI] [PubMed] [Google Scholar]
- 61.Scharschmidt TC, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scharschmidt TC, et al. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell Host Microbe. 2017;21:467–477.e5. doi: 10.1016/j.chom.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loesche M, et al. Temporal stability in chronic wound microbiota is associated with poor healing. J Invest Dermatol. 2017;137:237–244. doi: 10.1016/j.jid.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalan L, et al. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. MBio. 2016;7:e01058–e16. doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feingold KR. The outer frontier: the importance of lipid metabolism in the skin. J Lipid Res. 2009;50(Suppl):S417–S422. doi: 10.1194/jlr.R800039-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brandner JM. Importance of tight junctions in relation to skin barrier function. Curr Probl Dermatol. 2016;49:27–37. doi: 10.1159/000441541. [DOI] [PubMed] [Google Scholar]
- 68.Natsuga K. Epidermal barriers. Cold Spring Harb Perspect Med. 2014;4:a018218. doi: 10.1101/cshperspect.a018218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McLean WHI. Filaggrin failure—from ichthyosis vulgaris to atopic eczema and beyond. Br J Dermatol. 2016;175(Suppl 2):4–7. doi: 10.1111/bjd.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madison KC. Barrier function of the skin: “la raison d’être” of the epidermis. J Invest Dermatol. 2003;121:231–241. doi: 10.1046/j.1523-1747.2003.12359.x. [DOI] [PubMed] [Google Scholar]
- 71.Cleaver JE. Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum. J Dermatol Sci. 2000;23:1–11. doi: 10.1016/s0923-1811(99)00088-2. [DOI] [PubMed] [Google Scholar]
- 72.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Has C, Bruckner-Tuderman L. The genetics of skin fragility. Annu Rev Genomics Hum Genet. 2014;15:245–268. doi: 10.1146/annurev-genom-090413-025540. [DOI] [PubMed] [Google Scholar]
- 74.Capell BC, Tlougan BE, Orlow SJ. From the rarest to the most common: insights from progeroid syndromes into skin cancer and aging. J Invest Dermatol. 2009;129:2340–2350. doi: 10.1038/jid.2009.103. [DOI] [PubMed] [Google Scholar]
- 75.Totté JE, et al. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. 2016;175:687–695. doi: 10.1111/bjd.14566. [DOI] [PubMed] [Google Scholar]
- 76.Totté JEE, et al. A systematic review and meta-analysis on Staphylococcus aureus carriage in psoriasis, acne and rosacea. Eur J Clin Microbiol Infect Dis. 2016;35:1069–1077. doi: 10.1007/s10096-016-2647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–e814. doi: 10.1542/peds.2008-2217. [DOI] [PubMed] [Google Scholar]
- 78.Kobayashi T, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 42:756–766. doi: 10.1016/j.immuni.2015.03.014. This study demonstrated potential mechanistic links between dysbiotic skin flora and inflammation in atopic dermatitis by using a mouse model of eczema with ADAM17 deficiency that recapitulates spontaneous development of dysbiotic flora and skin inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol. 1974;90:525–530. doi: 10.1111/j.1365-2133.1974.tb06447.x. This was one of the first studies to demonstrate abundant Staphylococcus aureus colonization of patients with atopic dermatitis, even in areas of normal-appearing skin, and established the concept that colonizing microbes can have pathogenic effects without overt infection. [DOI] [PubMed] [Google Scholar]
- 80.Conti F, et al. Association between Staphylococcus aureus nasal carriage and disease phenotype in patients affected by systemic lupus erythematosus. Arthritis Res Ther. 2016;18:177. doi: 10.1186/s13075-016-1079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakagawa S, et al. Staphylococcus aureus virulent PSMα peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe. 2017;22:667–677.e5. doi: 10.1016/j.chom.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu H, et al. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe. 2017;22:653–666.e5. doi: 10.1016/j.chom.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 84.de Haas CJC, et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luong TT, Lee CY. Overproduction of type 8 capsular polysaccharide augments Staphylococcus aureus virulence. Infect Immun. 2002;70:3389–3395. doi: 10.1128/IAI.70.7.3389-3395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uhlén M, et al. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 87.Palmqvist N, Patti JM, Tarkowski A, Josefsson E. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 2004;6:188–195. doi: 10.1016/j.micinf.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 88.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 89.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 90.Rooijakkers SHM, et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 91.Sonesson A, et al. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci Rep. 2017;7:8689. doi: 10.1038/s41598-017-08046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang LJ, et al. Innate immunity Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science. 2015;347:67–71. doi: 10.1126/science.1260972. This study showed that adipogenesis and adipocyte production of AMPs help to protect against Staphylococcus aureus infection via intradermal injection, demonstrating that in addition to keratinocytes and sebocytes, subcutaneous tissues can participate in the immue response to microbes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joshi GS, Spontak JS, Klapper DG, Richardson AR. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol. 2011;82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thurlow LR, et al. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe. 2013;13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wentworth AB, Drage LA, Wengenack NL, Wilson JW, Lohse CM. Increased incidence of cutaneous nontuberculous mycobacterial infection, 1980 to 2009: a population-based study. Mayo Clin Proc. 2013;88:38–45. doi: 10.1016/j.mayocp.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Merritt RW, et al. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl Trop Dis. 2010;4:e911. doi: 10.1371/journal.pntd.0000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Houben RMGJ, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 2016;13:e1002152. doi: 10.1371/journal.pmed.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.World Health Organization. Global Tuberculosis Report 2016. 2016 http://apps.who.int/iris/bitstream/10665/250441/1/9789241565394-eng.pdf?ua=1.
- 99.Haley CA. Treatment of latent tuberculosis infection. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.tnmi7-0039-2016. TNMI7-0039–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sehgal VN. Leprosy. Dermatol Clin. 1994;12:629–644. [PubMed] [Google Scholar]
- 101.Talhari C, Talhari S, Penna GO. Clinical aspects of leprosy. Clin Dermatol. 2015;33:26–37. doi: 10.1016/j.clindermatol.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 102.GBD 2016 Causes of Death Collaborators. Global regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wansbrough-Jones M, Phillips R. Buruli ulcer: emerging from obscurity. Lancet. 2006;367:1849–1858. doi: 10.1016/S0140-6736(06)68807-7. [DOI] [PubMed] [Google Scholar]
- 104.Marion E, et al. Mycobacterial toxin induces analgesia in Buruli ulcer by targeting the angiotensin pathways. Cell. 2014;157:1565–1576. doi: 10.1016/j.cell.2014.04.040. This study shows that mycolactone, a virulence factor produced by the cutaneous pathogen Mycobacterium ulcerans, causes analgesia by directly binding to the angiotensin II receptor on nerve cells and triggering downstream potassium channel activation and resultant cell hyperpolarization. [DOI] [PubMed] [Google Scholar]
- 105.Kashem SW, et al. Nociceptive sensory fibers drive interleukin-23 production from CD301b+ dermal dendritic cells and drive protective cutaneous immunity. Immunity. 2015;43:515–526. doi: 10.1016/j.immuni.2015.08.016. This study shows that neurons can participate directly in the immune response to microbes. Cutaneous sensory neurons are directly activated by Candida albicans and subsequently stimulate dermal dendritic cells to produce IL-23, thus driving protective immunity by IL-17A-producing dermal T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chiu IM, et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 2013;501:52–57. doi: 10.1038/nature12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cardoso V, et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature. 2017;549:277–281. doi: 10.1038/nature23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bellono NW, et al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell. 2017;170:185–198. doi: 10.1016/j.cell.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fung TC, et al. Lymphoid tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity. 2016;44:634–646. doi: 10.1016/j.immuni.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sonnenberg GF, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Horsburgh CRJ., Jr Priorities for the treatment of latent tuberculosis infection in the United States. N Engl J Med. 2004;350:2060–2067. doi: 10.1056/NEJMsa031667. [DOI] [PubMed] [Google Scholar]
- 113.Adams KN, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schnappinger D, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rittershaus ESC, Baek SH, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bartek IL, et al. Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. MBio. 2014;5:e01106–e01114. doi: 10.1128/mBio.01106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Eoh H, et al. Metabolic anticipation in Mycobacterium tuberculosis. Nat Microbiol. 2017;2:201784. doi: 10.1038/nmicrobiol.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Galagan JE, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin PL, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hannigan GD, et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio. 2015;6:e01578–e15. doi: 10.1128/mBio.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hickman HD, et al. Anatomically restricted synergistic antiviral activities of innate and adaptive immune cells in the skin. Cell Host Microbe. 2013;13:155–168. doi: 10.1016/j.chom.2013.01.004. This study used intravital multiphoton microscopy to demonstrate that effector CD8+ T cells respond to cutaneous vaccinia virus infection by killing infected monocytes in the periphery but not infected keratinocytes in the center, thus highlighting the spatial complexity and specificity of immune cell dynamics in the skin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cush SS, et al. Locally produced IL-10 limits cutaneous vaccinia virus spread. PLoS Pathog. 2016;12:e1005493. doi: 10.1371/journal.ppat.1005493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carbone FR. Tissue-resident memory T cells and fixed immune surveillance in nonlymphoid organs. J Immunol. 2015;195:17–22. doi: 10.4049/jimmunol.1500515. [DOI] [PubMed] [Google Scholar]
- 125.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 126.Lanas A, Chan FKL. Peptic ulcer disease. Lancet. 2017;390:613–624. doi: 10.1016/S0140-6736(16)32404-7. [DOI] [PubMed] [Google Scholar]
- 127.Kalisperati P, et al. Inflammation, DNA damage, Helicobacter pylori and gastric tumorigenesis. Front Genet. 2017;8:20. doi: 10.3389/fgene.2017.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 129.Kienesberger S, et al. Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Reports. 2016;14:1395–1407. doi: 10.1016/j.celrep.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lehman H. Skin manifestations of primary immune deficiency. Clin Rev Allergy Immunol. 2014;46:112–119. doi: 10.1007/s12016-013-8377-8. [DOI] [PubMed] [Google Scholar]
- 131.Barnard E, Shi B, Kang D, Craft N, Li H. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep. 2016;6:39491. doi: 10.1038/srep39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Agak GW, et al. Propionibacterium acnes Induces an IL-17 response in acne vulgaris that is regulated by vitamin A and vitamin D. J Invest Dermatol. 2014;134:366–373. doi: 10.1038/jid.2013.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fitz-Gibbon S, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ring HC, et al. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 2017;153:897–905. doi: 10.1001/jamadermatol.2017.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wollenberg MS, et al. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. MBio. 2014;5:e01286–e14. doi: 10.1128/mBio.01286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shu M, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8:e55380. doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 138.Yager EJ, et al. γ-Herpesvirus-induced protection against bacterial infection is transient. Viral Immunol. 2009;22:67–72. doi: 10.1089/vim.2008.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Perry S, et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One. 2010;5:e8804. doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Arnold IC, et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121:3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheung GYC, Otto M. Understanding the significance of Staphylococcus epidermidis bacteremia in babies and children. Curr Opin Infect Dis. 2010;23:208–216. doi: 10.1097/QCO.0b013e328337fecb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rohlke F, Stollman N. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Therap Adv Gastroenterol. 2012;5:403–420. doi: 10.1177/1756283X12453637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Panigrahi P, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi: 10.1038/nature23480. This study was the first, to our knowledge, to use an oral synbiotic (Lactobacillus plantarum and fructooligosaccharide) to promote effective gut colonization of the inoculated bacterium and reduce neonatal sepsis. [DOI] [PubMed] [Google Scholar]
- 144.Nakatsuji T, et al. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating β-defensin-2 expression. J Invest Dermatol. 2010;130:985–994. doi: 10.1038/jid.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Falchook GS, et al. Responses of metastatic basal cell and cutaneous squamous cell carcinomas to anti-PD1 monoclonal antibody REGN2810. J Immunother Cancer. 2016;4:70. doi: 10.1186/s40425-016-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morris VK, et al. Nivolumab for previously treated unresectable metastatic anal cancer (NCI9673): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18:446–453. doi: 10.1016/S1470-2045(17)30104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mahoney KM, Freeman GJ, McDermott DF. The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in melanoma. Clin Ther. 2015;37:764–782. doi: 10.1016/j.clinthera.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cassler NM, Brownell I. PD-1 checkpoint blockade is an emerging treatment for Merkel cell carcinoma. Br J Dermatol. 2017;176:18. doi: 10.1111/bjd.14828. [DOI] [PubMed] [Google Scholar]
- 149.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. This study and the next one demonstrated that the gut microbial composition can alter immunotherapies at distant sites, suggesting that microbe–immune interactions at barrier sites can have far-reaching effects at other barrier sites or systemically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Golden JB, et al. Chronic, not acute, skin-specific inflammation promotes thrombosis in psoriasis murine models. J Transl Med. 2015;13:382. doi: 10.1186/s12967-015-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Santilli S, et al. Visualization of atherosclerosis as detected by coronary artery calcium and carotid intima-media thickness reveals significant atherosclerosis in a cross-sectional study of psoriasis patients in a tertiary care center. J Transl Med. 2016;14:217. doi: 10.1186/s12967-016-0947-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Y, et al. Chronic skin-specific inflammation promotes vascular inflammation and thrombosis. J Invest Dermatol. 2012;132:2067–2075. doi: 10.1038/jid.2012.112. This study used a mouse model of psoriasis (keratinocyte-specific Tie2 transgene expression) to mechanistically link skin inflammation to the development of resultant aortic root inflammation and also to show that subsequent treatment of the skin disease can eliminate not only skin inflammation but also systemic vascular inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Evensen K, et al. Increased subclinical atherosclerosis in patients with chronic plaque psoriasis. Atherosclerosis. 2014;237:499–503. doi: 10.1016/j.atherosclerosis.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 155.Tomura M, et al. Activated regulatory T cells are the major T cell type emigrating from the skin during a cutaneous immune response in mice. J Clin Invest. 2010;120:883–893. doi: 10.1172/JCI40926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tomura M, et al. Tracking and quantification of dendritic cell migration and antigen trafficking between the skin and lymph nodes. Sci Rep. 2014;4:6030. doi: 10.1038/srep06030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Geherin SA, et al. The skin, a novel niche for recirculating B cells. J Immunol. 2012;188:6027–6035. doi: 10.4049/jimmunol.1102639. This study was one of the first to demonstrate that B cells actively traffic in and out of the skin, even in uninflamed skin, suggesting that B cells play an active and previously underappreciated role in skin homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 159.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nithya S, Radhika T, Jeddy N. Loricrin—an overview. J Oral Maxillofac Pathol. 2015;19:64–68. doi: 10.4103/0973-029X.157204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Iguchi A, et al. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 2015;22:101–107. doi: 10.1093/dnares/dsu043. [DOI] [PMC free article] [PubMed] [Google Scholar]