Abstract
A challenge for evolutionary research is to uncover how new morphological traits evolve the coordinated spatial and temporal expression patterns of genes that govern their formation during development. Detailed studies are often limited to characterizing how one or a few genes contributed to a trait’s emergence, and thus our knowledge of how entire GRNs evolve their coordinated expression of each gene remains unresolved. The melanic color patterns decorating the male abdominal tergites of Drosophila (D.) melanogaster evolved in part by novel expression patterns for genes acting at the terminus of a pigment metabolic pathway, driven by cis-regulatory elements (CREs) with distinct mechanisms of Hox regulation. Here, we examined the expression and evolutionary histories of two important enzymes in this pathway, encoded by the pale and Ddc genes. We found that while both genes exhibit dynamic patterns of expression, a robust pattern of Ddc expression specifically evolved in the lineage of fruit flies with pronounced melanic abdomens. Derived Ddc expression requires the activity of a CRE previously shown to activate expression in response to epidermal wounding. We show that a binding site for the Grainy head transcription factor that promotes the ancestral wound healing function of this CRE is also required for abdominal activity. Together with previous findings in this system, our work shows how the GRN for a novel trait emerged by assembling unique yet similarly functioning CREs from heterogeneous starting points.
Keywords: cis-regulatory element, transcription factor, gene regulatory network, morphological evolution, Hox, Drosophila
Introduction
A major challenge for evolutionary developmental biology research is to explain how novel morphological traits come into existence through genetic changes and their underlying molecular mechanisms. The reigning paradigm for trait development involves the action of a gene regulatory network (GRN) in which a hierarchy of regulatory genes, encoding signaling pathway and transcription factor proteins, drive the expression of a set of realizator genes whose encoded proteins drive the physical manifestation of the trait (Davidson, 2006; Levine and Davidson, 2005). The expression patterns of a GRN’s genes are specified by the binding of transcription factors to cis-regulatory elements (CREs) that function as enhancers or silencers of transcription (Arnone and Davidson, 1997; Gray and Levine, 1996; Johnson et al., 2015; Stanojevic et al., 1991). As GRNs are generally complex in realizator and regulatory gene number (Bonn and Furlong, 2008a), the origin of a novel trait requires an explanation regarding how the full set of realizator genes arrived at their appropriate patterns of expression (García-Bellido, 1975).
A multitude of possible scenarios could explain the origination of complex GRNs, and these differ in terms of their number and distribution of necessary genetic modifications. For instance, if a top-level regulatory gene of an ancestral GRN evolves a novel domain of expression, possibly by changing a single CRE, this could co-opt an entire GRN through the pleiotropic actions of this single gene (Monteiro and Gupta, 2016; Rebeiz and Tsiantis, 2017). In contrast, changes may be distributed throughout a GRN, involving the gain, modification, or loss of dedicated CREs for each realizator gene in the network. In order to distinguish whether the origins of novelty tend to emerge through such short or long evolutionary paths, it is necessary to characterize how GRNs and their constituent CREs evolve for a myriad of novel morphological traits in diverse lineages.
The coloration patterns adorning the cuticle of insects present ideal traits for investigation as a great diversity phenotypes exist within a tractable phylogenetic framework in which ancestral and derived character states can be reasonably inferred (Jeong et al., 2006; Wittkopp et al., 2003). One experimentally tractable coloration trait is the male-specific melanism of the dorsal tergites that cover the 5th and 6th abdominal segments of Drosophila (D.) melanogaster (Rebeiz and Williams, 2017). This dimorphic trait is suspected to have evolved from a monomorphic ancestral state in the Sophophora subgenus (Fig. 1A) (Jeong et al., 2006). In D. melanogaster, cuticle pigmentation requires the activities of several realizator genes whose protein products function in a tyrosine metabolic pathway that can produce yellow sclerotin or a black melanic cuticle (Fig. 1B) (Wright, 1987). In males, the realizator genes yellow and tan are highly expressed in the A5 and A6 segment epidermis underlying the developing tergites, expression of which is necessary for the formation of the black tergite color (Camino et al., 2015). In females, the realizator gene ebony is highly expressed in the A5 and A6 segment epidermis (Rebeiz et al., 2009) which leads to broadly yellow tergites. The CREs responsible for the sex- and segment-specific expression patterns of yellow, tan, and ebony have been identified (Jeong et al., 2008; Rebeiz et al., 2009; Wittkopp et al., 2002), as well as some of the direct interacting transcription factors (Camino et al., 2015; Jeong et al., 2006; Roeske et al., 2018). Notably, yellow expression is directly activated by the Hox factor Abd-B binding to sites within the body element CRE, whereas the limitation of tan expression to 5th and 6th segments involves the direct binding of Abd-A and Hth to sites within the t_MSE to facilitate repression in the more anterior abdomen segments. Although tan, yellow, and ebony play essential roles in making black and yellow pigments, the pigment metabolism pathway involves several other key genes whose expression and regulation need to be understood within the context of this trait’s development and evolution.
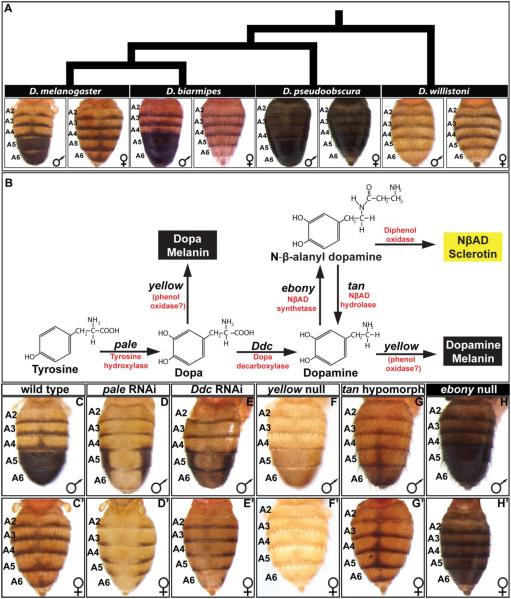
Fig. 1. Pigmentation phenotypes and enzymatic pathway in the Sophophora subgenus.
(A) Extant species that represent the inferred diversification of abdominal pigmentation from an ancestral monomorphic state to a derived dimorphic state in the Sophophora subgenus. The tergites covering the A5 and A6 abdominal segments of D. melanogaster and D. biarmipes are sexually dimorphic for a melanic pigment color, with the male tergites being fully pigmented and pigmentation limited to posterior stripes in females. The more distantly related species such as D. pseudoobscura and D. willistoni are monomorphic, though differ greatly in the extent of their melanization. (B) The production of yellow and black cuticle depends on the activity of a metabolic pathway converting Tyrosine into NβAD sclerotin or Dopa/Dopamine melanin. (C and C’) The wild type abdomen pigmentation pathway includes a male-specific pattern of fully melanic A5 and A6 segment tergites. (D and D’) RNA-interference for pale results in a near complete absence of black cuticle and a reduction in yellow cuticle color. (E and E’) RNA-interference for Ddc results in a stark reduction in black cuticle color and a reduction in yellow color. (F and F’) The null allele phenotype for yellow is a loss of black cuticle color. (G and G’) The hypomorphic allele phenotype for tan is a reduction in black cuticle color though the stripe and midline spot region pigmentation remains. (H and H’) The null allele phenotype for ebony is a broadening of black cuticle to all abdomen segments. RNA-interference was achieved by driving a UAS-regulated dsRNA transgene for pale and Ddc by the GAL4 transcription factor that was expressed in the dorsal-medial pattern of the pnr gene.
The first steps to forming catecholamine pigments are the conversion of Tyrosine to Dopa and then to Dopamine through the activity of Tyrosine Hydroxylase (encoded by the gene pale) (Neckameyer and White, 1993), and Dopa Decarboxylase (encoded by Ddc), respectively (Fig. 1B) (Wright, 1987). pale and Ddc differ from yellow, tan, and ebony in that their activities are seemingly needed to make both black and yellow pigments (Fig. 1B). Both proteins have been shown to accumulate transcripts before eclosion, but appear to be under translational control that governs the timing of tanning (Davis et al., 2007). However, the spatial distributions of pale and Ddc expression remain largely uncharacterized during pupal and adult stages when tergite pigmentation is developing, and the evolutionary origin of their CREs remain unresolved.
Here, we show that both pale and Ddc exhibit spatial and temporal patterns of expression during pupal and adult life stages in D. melanogaster. While pale expression has been broadly conserved among Sophophora species exhibiting both derived and ancestral pigmentation patterns, we show that high levels of epidermal Ddc expression evolved in the lineage of species with male-limited patterns of melanic pigmentation. This gain of abdominal Ddc expression requires the activity of a pleiotropic CRE that additionally regulates Ddc expression in the larval epidermis and in response to epidermis wounding (Mace et al., 2005; Scholnick et al., 1993). We find that the abdominal function of this CRE requires an ancestral binding site for Grainy head (Grh) necessary for its wound-response function. However, robust pupal abdomen CRE activity required changes elsewhere to this CRE. Together with previous results, our findings illustrate how a novel trait evolved through a lengthy evolutionary path, which involved independent gains of new expression patterns for an entire pathway of realizator genes through varied mechanisms.
Materials and Methods
Fly Stocks and Genetic Crosses
Fly stocks used in this study were maintained at 22°C on a previously published sugar food medium (Salomone et al., 2013). Species stocks used in this study were D. melanogaster (14021-0231.04), and D. willistoni (14030-0811.24) from the National Drosophila Species Stock Center, and D. melanogaster yellow and white (yw) gene mutant, D. biarmipes, and D. pseudoobscura stocks that were obtained from Dr. Sean B. Carroll. Mutant allele stocks used to visualize abdomen pigmentation phenotypes were the null mutant y1 (Bloomington Drosophila Stock Center or BDSC #169) allele for yellow, the hypomorphic t5 (BDSC #133) allele for tan, and the null e1 (BDSC #1658) allele for ebony. UAS-regulated RNA-interference (RNAi) stocks were generated by the Transgenic RNAi Project at Harvard Medical School, and acquired from the BDSC. These stocks express double stranded RNAs that specifically target mCherry (BDSC #35785), grh (BDSC #28820), abd-A (BDSC #28739), hth (BDSC #27655), Ddc (BDSC #51462), and pale (BDSC #65875). Experiments requiring ectopic expression of Abd-B in the A4 abdominal segment were achieved through crosses with a stock possessing the Abd-BMcp mutant allele (BDSC #24618). In order to use the GAL4/UAS system (Brand and Perrimon, 1993) in the dorsal abdomen midline, a stock with the genotype pnr-GAL4/TM3, Ser1 (BDSC #3039) was used that possesses a 3rd chromosome from which GAL4 is expressed in the dorsal midline pattern of the pannier gene (Calleja et al., 2000). In order to make qualitative, and in some cases quantitative, comparisons of reporter transgene activities, transgenes were integrated into the attP2 and/or 51D attP sites via phiC31 integrase-mediated transgenesis (Best Gene Inc.) (Bischof et al., 2007; Groth and Calos, 2004).
Gain- and loss-of-function experiments
To assess adult pigmentation phenotypes following reduction in either pale, Ddc, and grh expression, males from the respective UAS-RNAi stocks were crossed to pnr-GAL4/TM3, Ser1 virgin female flies. Adult progeny inheriting the pnr-GAL4 chromosome and the UAS-RNAi transgene bearing chromosome express the double stranded (ds)RNA hairpin in the pnr gene’s dorsal-medial pattern, including the pupal dorsal abdominal epidermis. The following crosses were made in order to investigate the effects of reduced transcription factor gene expression on the ability of the t_MSE and Ddc-MEE1 CREs to drive the expression of the Enhanced Green Fluorescent Protein (EGFP) reporter transgene in P14-P15(i) stage pupae. Virgin females from the RNAi stocks for UAS-mCherry, UAS-abd-A, UAS-hth, and UAS-grh were collected and separately crossed to males bearing a CRE reporter transgene in the 51D attP site of the 2nd chromosome (Bischof et al., 2007) and a pnr-GAL4 bearing 3rd chromosome. As the mCherry gene does not exist in the D. melanogaster genome, expression of a dsRNA targeting this gene served as a negative control.
In order to compare the responsiveness of the Ddc-MEE1 and t_MSE CREs to Abd-B in pupae, adult males possessing the Abd-BMcp allele were crossed to virgin females possessing an EGFP reporter transgene under the regulatory control of either the t_MSE or the Ddc-MEE1 integrated into the 51D attP site (Bischof et al., 2007). As a negative control, similar crosses were made in which males from a yw stock that exhibit wild type expression of Abd-B (+) were utilized in place of the Abd-BMcp allele stock. EGFP reporter expression was assessed in P14-P15(i) stage pupae that inherited the reporter transgene and either the wild type or Abd-B mutant genotype.
in situ hybridization and immunohistochemistry
A previously describe protocol was used in the in situ hybridization studies (Jeong et al., 2008). In brief, digoxigenin labeled riboprobes for Ddc and pale were prepared through in vitro transcription of PCR templates amplified from each species’ genomic DNA (Table S1 for antisense probe primers and Table S2 for sense probe primers). Abdomens were dissected at the pupal developmental stages P10, P11, P12, P13, P14-15(i) and the newly eclosed adult fly stage P15(i). These different stages were identified by inspection for the presence and absence of various morphological markers (Ashburner et al., 2005) (Fig. S1). Probe hybridizations were made visible through the use of an anti-digoxigenin antibody (Roche Diagnostics), whose presence was detected by an alkaline phosphatase reaction using BCIP/NBT (Promega).
Dorsal abdomens were dissected for immunohistochemistry at the P12 and P14-P15(i) stages for D. melanogaster and D. willistoni. These stages are when we found Ddc transcription to be robust (P12) and declining P14-P15(i) in the D. melanogaster abdominal epidermis (Fig. 2). Qualitative comparisons of Grh expression patterns were made through immunohistochemical analysis using an antibody specific for Grh (Kim and Mcginnis, 2010). Abdomens were dissected to isolate the dorsal epidermis. Samples were fixed for 35 min in PBST (phosphate-buffered saline with 0.3% Triton X-100) with 4% paraformaldehyde, and then blocked in blocking buffer (PBST with 1% bovine serum albumin) for 1 hour at room temperature. The abdomens were then incubated overnight with guinea pig anti-Grh primary antibody at a dilution of 1:200 in PBST. After four washes in PBST and then an hour incubation in blocking buffer, specimens were incubated with goat anti-guinea pig Alexa Fluor 647 (Invitrogen) secondary antibody at a dilution of 1:500. After four washes in PBST, samples were incubated for 10 minutes in a 1:1 solution of glycerol mount (80% glycerol, 0.1M Tris pH 8.0) and PBST. Samples were transferred to glycerol mount, and finally situated between a cover slip and slide for imaging with a confocal microscope.
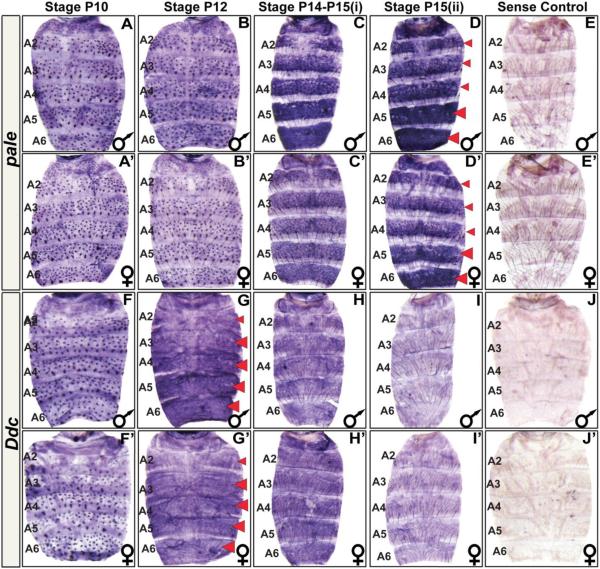
Fig. 2. The spatial and temporal expression pattern of D. melanogaster pale and Ddc.
(A–J) Samples for D. melanogaster males and (A’–J’) females. (A and A’) pale is expressed in the mechansosensory bristle cells as early as the P10 stage of pupal development. (B and B’) Mechanosensory cell expression continues through the P12 stage. (C and C’) By the P14-P15(i) stage, pale expression has dramatically increased in the abdominal epidermis. Most notably in the A5 and A6 segments and in the epidermal cells underlying where tergites will form. (D and D’) The epidermis pattern of pale expression can still be detected in newly eclosed flies. (E and E’) A sense probe control shows the background pattern of signal observed in an eclosed fly. (F and F’) Ddc is expressed in the mechansosensory bristle cells as early as the P10 stage of pupal development. (G and G’) By the P12 stage, Ddc expression has dramatically increased in the abdominal epidermis, most notably cells in the A3–A6 segment regions underlying where tergites will form. (H and H’) By the P14-P15(i) stage, Ddc expression persists but at an apparently reduced level. (I and I’) Epidermis expression has become difficult to detect in newly eclosed flies. (J and J’) A Ddc sense probe control shows background signal observed in eclosed fly. Red arrowheads indicate stages and segments with conspicuous expression detected in the epidermis. These stages were prioritized for investigations of expression in related fruit fly species.
EGFP reporter transgenes and their quantitation
EGFP reporter transgenes were used to study the gene regulatory activity (Rebeiz and Williams, 2011) of sequences controlling Ddc expression and to make comparisons with the t_MSE that controls male-specific tan expression (Camino et al., 2015). All reporter transgene were made by cloning CREs into the AscI and SbfI restriction enzyme sites for the S3aG vector (Rogers and Williams, 2011). In this position each CRE is placed 5’ of a minimal hsp70 promoter and the coding sequence for the EGFP-NLS reporter protein (Barolo et al., 2004). The primer pairs used to amplify D. melanogaster Ddc sequences are presented in Table S3. The primer pairs used to amplify sequences orthologous to the D. melanogaster Ddc-MEE1 CRE are presented in Table S4. The details on the construction of the t_MSE EGFP reporter transgenes were discussed previously (Camino et al., 2015). Ddc-MEE1 transcription factor binding site mutant sequences (Fig. S2), and scanning mutant sequences (Fig. S3) were synthesized by GenScript USA Inc. Each synthesized sequence was flanked by an AscI and SbfI restriction enzyme site for subsequent cloning of the mutant CREs into the S3aG vector. Reporter transgenes containing truncated version of the Ddc upstream region, and those possessing Ddc-MEE1 scanning or binding site mutant sequences were each integrated in the attP2 site on the 3rd chromosome (Groth et al., 2000). The t_MSE, Ddc-MEE1, Ddc-MEE364, Ddc-MEE130, and orthologous Ddc-MEE1 CREs were integrated into the 51D attP site on the 2nd chromosome (Bischof et al., 2007).
Quantitative comparisons of the levels of EGFP reporter gene expression driven by sequences orthologous to the D. melanogaster Ddc-MEE1 were achieved following a previously described approach (Camino et al., 2015; Rebeiz and Williams, 2011; Roeske et al., 2018; Rogers et al., 2013; Rogers and Williams, 2011). In brief, for each transgene EGFP expression was imaged from four or five biological replicate specimens using a confocal microscope with settings for which few pixels were saturated when EGFP expression was driven by the D. melanogaster Ddc-MEE1. For each confocal image, a separate pixel value statistic was measured for the dorsal epidermis of either the A5 or A4 abdominal segment using the Image J program (Abràmoff et al., 2004). For each reporter transgene, the regulatory activity was calculated as the mean pixel value and standard error of the mean (SEM). CRE activities in the A5 abdominal segment for binding site mutant, scanning mutant, and orthologous Ddc-MEE1 sequences were normalized to the activity for the wild type Ddc-MEE1 in the same attP2 site, which was considered 100%. CRE activities in the A5 abdomen segment for the Ddc-MEE364 and Ddc-MEE130 were normalized to the activity for the wild type Ddc-MEE1 in the same 51D site, which was considered 100%. CRE activities in the A4 abdomen segment reported in the Abd-BMcp mutant background were normalized to the A4 activity of the species’ CRE in an Abd-B wild type background, in which CRE activity was considered 100%.
Imaging of fly abdomens and experimental replication
Images of fruit fly abdomen pigmentation patterns were taken using an Olympus SZX16 Zoom Stereoscope and Olympus DP72 digital camera. Specimens were prepared from four day old flies. Projection images for EGFP-NLS reporter transgene expression were generated with an Olympus Fluoview FV 1000 confocal microscope and software. In each figure panel reporting a pigmentation pattern, gene expression pattern, or CRE activity, a representative image was selected from biological replicate specimens (n≥4). In all figures where comparisons were made between images, each image was processed through the same sequence of modifications using Photoshop CS3 (Adobe).
DNA sequence representations and alignments
Sequence visualizations for the Ddc loci of D. melanogaster was made using the GenePalette tool (Rebeiz and Posakony, 2004; Smith et al., 2017). DNA sequence alignments of wild type, mutant, and orthologous Ddc-MEE1 sequences were made using the Chaos and Dialign software (Brudno et al., 2004).
Results
Melanic tergite pigmentation depends on pale and Ddc activity
The genetic and enzymatic pathway for the synthesis of the D. melanogaster cuticle pigments has been studied for several decades (Rebeiz and Williams, 2017; Wittkopp et al., 2003; Wright, 1987), though in recent years, much focus has been devoted to the expression and regulatory mechanisms of tan, ebony, and yellow genes which act at later steps in the pathway (Fig. 1B). Though the precise molecular mechanism of Yellow protein function remains unknown (Ferguson et al., 2011; Hinaux et al., 2018), the loss of yellow gene function results in a loss of black melanin pigments (Fig. 1F and F’). ebony encodes a protein with NβAD synthetase activity which shunts Dopamine to form a yellow colored NβAD sclerotin. The loss of ebony function results in a broadly melanic phenotype (Fig. 1H and H’). tan encodes an enzyme with NβAD hydrolase activity, which converts NβAD back to Dopamine (True et al., 2005). Reduced tan activity results in a broad loss of melanin pigments (Fig. 1G and G’). pale encodes an enzyme with Tyrosine hydroxylase activity, which is the initial step in the pigmentation pathway converting Tyrosine to Dopa. RNA-interference (RNAi) of pale expression in the dorsal midline of D. melanogaster results in a dramatic loss of melanic pigments and to a lesser extent yellow cuticle color (Fig. 1D and D’). Ddc encodes an enzyme with Dopa decarboxylase activity and is responsible for converting Dopa to Dopamine. RNAi for Ddc in the dorsal midline results in a stark reduction in melanic pigments in the abdominal tergites (Fig. 1E and E’).
Collectively, these results show that pale and Ddc are essential for the formation of the sexually dimorphic pattern of D. melanogaster abdomen pigmentation. The dramatic loss of black colored cuticle following Ddc RNAi indicates that most black color is that of Dopamine melanin rather than Dopa melanin (Fig. 1B). ebony, tan, and yellow have been established to be expressed in complex spatial, temporal, and sex-specific manners. Thus, we sought to reveal whether the expression patterns for pale and Ddc are similarly complex.
The spatial and temporal pattern of abdominal pale expression
During the approximately 100 hour time course of D. melanogaster pupal development at 25°C, pupae transition through roughly 15 distinct stages that can be identified by morphological markers (Fig. S1) (Ashburner et al., 2005). By the P10 stage, pale is expressed robustly in the abdomen mechanosensory bristle cells, whereas little to no expression is seen in the surrounding epidermis (Fig. 2A and A’). This pattern of expression is maintained through the P12 stage when some bristles have developed clear projections (Fig. 2B and B’). By the P14-P15(i) stage, pale expression has dramatically increased in the abdominal epidermis region underlying where tergites will form, and is especially elevated in the A5 and A6 segments. Expression of pale is comparatively absent from the epidermal cells that will be covered by flexible un-pigmented inter-segmental membrane that will ultimately fold underneath each tergite (Fig. 2C and C’). This epidermal pattern of expression is sustained in newly eclosed, P15(ii) stage, flies (Fig. 2D and D’). Collectively, these results show that pale undergoes a complex temporal and spatial pattern of expression like yellow, tan, and ebony. However, pale differs from the other pigmentation genes in that its expression is not conspicuously dimorphic.
The spatial and temporal pattern of abdominal Ddc expression
By the P10 stage, D. melanogasterDdc is expressed robustly in the abdomen mechanosensory bristle cells, whereas the surrounding epidermis lacks detectible expression (Fig. 2F and F’). By the P12 stage, Ddc expression has dramatically increased in the abdominal epidermis region underlying where tergites will form, notably in the A3–A6 segments, while expression is comparatively absent from the epidermal cells that will be covered by flexible un-pigmented inter-segmental membrane (Fig. 2G and G’). By the P14-P15(i) stage, Ddc expression remains in the abdominal epidermis though the level of expression has declined (Fig. 2H and H’). In newly eclosed flies, Ddc expression has become difficult to detect (Fig. 2I and I’). Collectively, these results show that Ddc undergoes a complex temporal and spatial pattern of expression like yellow, tan, and ebony. However, like pale, Ddc expression is not conspicuously dimorphic. As the pattern of D. melanogaster tergite pigmentation represents a novel trait that evolved from a monomorphic ancestral state, we were curious whether the Ddc and pale expression patterns were modified during this trait’s origins.
Conserved pale expression is a fixture of evolved pigmentation traits and GRNs
The working model for pigmentation evolution in the Sophophora subgenus places a monomorphic pattern of tergite pigmentation as ancestral, and dimorphic pigmentation being a derived state (Jeong et al., 2006; Rebeiz and Williams, 2017). Moreover, many species within the obscura species group, including D. pseudoobscura (Fig. 1A), are darkly pigmented across the entire abdomen. It has been shown that yellow and tan expression correlate with these patterns of pigmentation, and ebony expression has an inverse correlation (Camino et al., 2015; Jeong et al., 2008; Johnson et al., 2015; Ordway et al., 2014; Rebeiz et al., 2009). We sought to reveal whether the earliest acting gene in the Drosophila pigmentation pathway, pale, has similarly evolved diverse patterns of expression to shape the visible pigmentation patterns seen among Sophophora (Fig. 1A).
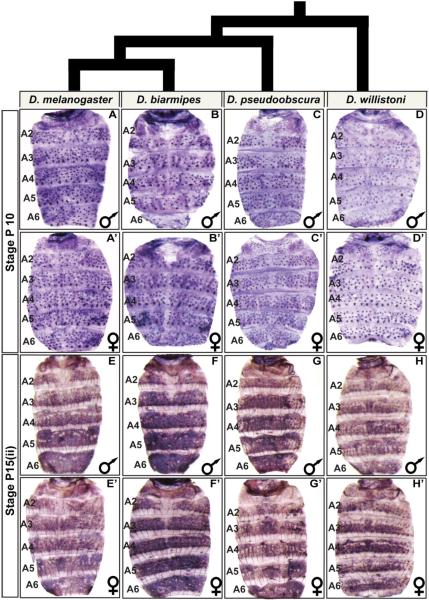
We focused our attention on the P10, and P15(ii) developmental stages for which we found D. melanogasterpale expression to transition from a mechanosensory-specific expression pattern to a pattern including the abdominal epidermis. We observed that expression at the P10 and P15(ii) stages were conserved between species with dimorphic and monomorphic patterns of pigmentation (Fig. 3). This outcome suggests that the expression pattern for pale has been a conserved feature of the evolving pigmentation GRNs for abdominal tergite pigmentation.
Fig. 3. pale abdominal epidermis expression is broadly conserved in Sophophora.
(A–H) pale expression in the dorsal abdomens of male and (A’–H’) female pupae. At the P10 stage, pale expression occurs robustly in the mechanosensory bristle cells of (A and A’) D. melanogaster, (B and B’) D. biarmipes, (C and C’) D. pseudoobscura, and (D and D’) D. willistoni. At the P15(ii) stage, pale expression is robust in the abdominal epidermis of (E and E’) D. melanogaster, (F and F’) D. biarmipes, (G and G’) D. pseudoobscura, and (H and H’) D. willistoni.
Ddc is broadly expressed in the abdomen epidermis of highly pigmented species
The conserved abdominal epidermis pattern of pale expression represents a departure from the evolved patterns observed with yellow, tan, and ebony. We sought to determine whether the second gene in the pigmentation pathway, Ddc, is characterized by a conserved pattern of abdominal epidermis expression. We focused our attention on the P10 and P12 stages of pupal development where we respectively observed a conspicuous pattern of expression in the mechanosensory bristle cells and abdominal epidermis for D. melanogaster (Fig. 2).
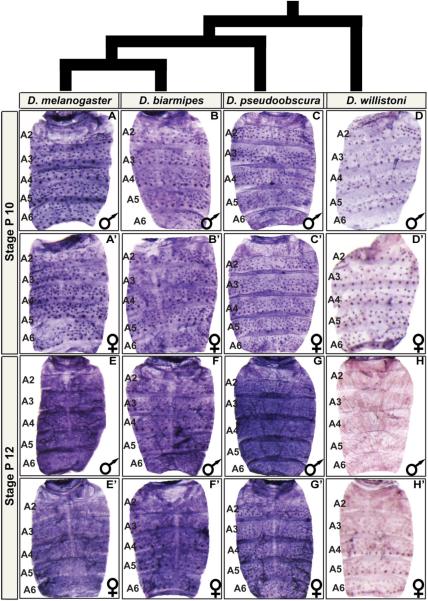
At the P10 stage, robust mechanosensory expression was found in all species evaluated (Fig. 4A–D and A’–D’), consistent with their melanic appearance. At the P12 stage, Ddc expression occurs in a monomorphic pan-abdomen epidermal pattern for D. biarmipes and D. pseudoobscura (Fig. 4E–4G and E’–G’). D. biarmipes has a male-specific pattern of tergite pigmentation like D. melanogaster, and D. pseudoobscura has a monomorphic melanic pattern that spans the tergites of the A1–A6 segments (Fig. 1A). Interestingly though, at the P12 stage we detected little to no Ddc expression in the abdominal epidermis of D. willistoni (Fig. 4H and H’). Expanding our search to a range of developmental stages, we failed to detect strong epidermal expression in D. willistoni, despite the clear expression detected in bristle cells (Fig. 5). Unlike the other species evaluated, D. willistoni lacks notable pigmentation on its abdominal tergites (Fig. 1A), and this pigmentation pattern is considered to descend from a lineage bearing the ancestral pattern from which dimorphism evolved (Jeong et al., 2006). These results suggest that the origin of dimorphic pigmentation involved the gain of a robust pattern of Ddc expression in the abdominal epidermis. Thus, we next sought to understand how Ddc expression is regulated in the abdominal epidermis and elucidate how this expression pattern evolved.
Fig. 4. Ddc is broadly expressed in the abdominal epidermis of species with elaborate patterns of tergite pigmentation.
Ddc expression in the dorsal abdomens of (A–H) male and (A’–H’) female pupae. (A–D and A’–D’) Expression at the P10 and (E–H and E’–H’) P12 developmental stages. At P10, Ddc expression occurs robustly in the mechanosensory bristle cells of (A and A’) D. melanogaster, (B and B’) D. biarmipes, (C and C’), D. pseudoobscura, and (D and D’) D. willistoni. At P12, expression has expanded to include the abdominal epidermis of (E and E’) D. melanogaster, (F and F’) D. biarmipes, and (G and G’), and D. pseudoobscura. (H and H’) For D. willistoni, a similar robust pattern of Ddc expression was not observed at the P12 stage.
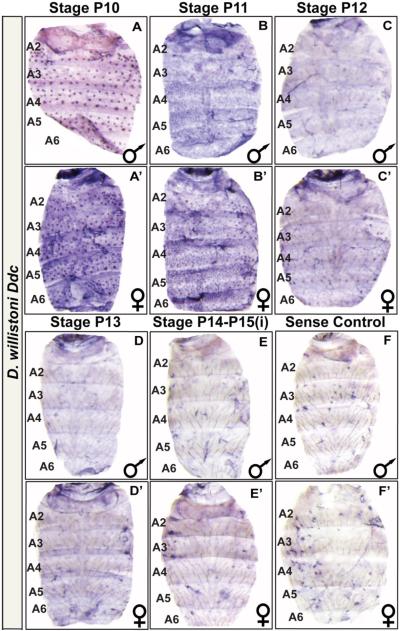
Fig. 5. D. willistoni exhibits little to no Ddc expression in the abdominal epidermis of various pupal stages through eclosion.
In situ hybridization for Ddc expression was performed on the dorsal abdominal epidermis of D. willistoni (A–F) male and (A’–F’) female specimens. (A and A’) Ddc is expressed in the mechansosensory bristle cells as early as the P10 stage of pupal development. (B and B’) By the P11 stage, a low level of Ddc expression can be observed in the epidermis regions underlying where tergites will form. (C and C’) By the P12 stage, little Ddc expression is observed in the epidermis regions underlying where tergites will form. (D and D’) At the P13 and (E and E’) P14-P15(i) stages, little to no expression was observed in the epidermis. (F and F’) A Ddc sense probe control shows background signal observed in eclosed flies.
A multifunctional CRE drives Ddc expression in the abdominal epidermis
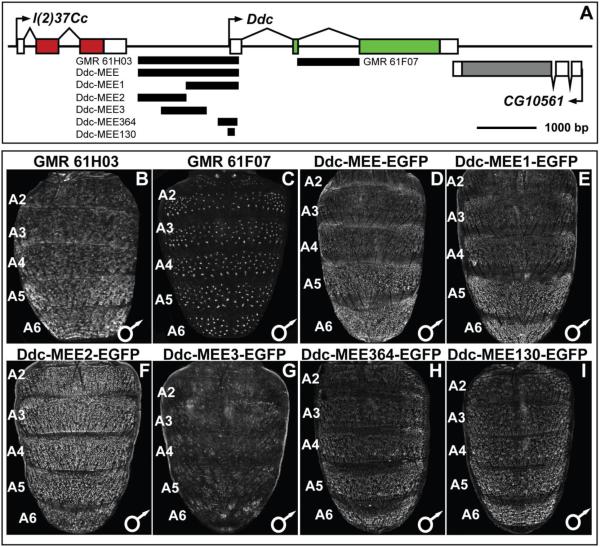
The Ddc locus resides on D. melanogaster chromosome 2, between the genes l(2)37Cc and CG10561 (Fig. 6A). Ddc has three transcribed exons, although a splice variant exists that includes an additional non-coding exon situated in the first intron. As a first pass at identifying CREs active in the abdomen during development and pigmentation pattering, we crossed virgin females carrying a UAS-GFP-nls transgene to males of the GMR61H03 and GMR601F07 GAL4 lines (Pfeiffer et al., 2008). These GAL4 lines respectively possess a 1,692 base pair (bp) piece that spans most of the sequence between the Ddc 1st exon and the last exon of the upstream l(2)37Cc gene, and a 1,115 bp sequence spanning the entire Ddc 2nd intron (Fig. 6A). In pupae inheriting the UAS-GFP-nls and GMR61H03-GAL4 transgenes, we observed GFP expression in the abdominal epidermis of the A1–A6 segments which was noticeably elevated in more posterior segments (Fig. 6B). On the other hand, GMR61F07-GAL4 drove GFP expression in the developing mechanosensory bristle cells (Fig. 6C). Collectively, these patterns of GFP expression recapitulated our observed patterns Ddc mRNA expression detected by in situ hybridization experiments (Fig. 2), and demonstrates that the two phases of pupal abdomen expression are under the control of multiple modular CREs.
Fig. 6. An abdominal epidermis cis-regulatory element is located 5’ of the endogenous Ddc promoter.
(A) To scale representation of the Ddc locus showing regions that were evaluated for CRE activity. (B–I) CRE activity observed by the expression of the EGFP reporter gene in the dorsal abdomens of P14-P15(i) stage transgenic male D. melanogaster pupae. (B and C) UAS-EGFP reporter expression driven by GAL4 expressed under the control of Ddc locus subregions. (B) The entire upstream region of Ddc, named the Ddc-MEE, drove reporter expression in the A2–A6 abdominal regions underlying where tergites develop, a pattern that mimics the endogenous Ddc expression. (C). The 2nd intron region of Ddc drove reporter expression in the mechanosensory bristle cells, a pattern that mimics the endogenous Ddc expression. (D–I) Expression of direct CRE-GFP fusion transgenes. (D) The entire Ddc upstream region drove reporter transgene expression in an abdominal epidermis patter similar to endogenous Ddc expression and that driven by the same region using the GAL4/UAS system. (E) The promoter-proximal Ddc-MEE1 subdivision of the Ddc-MEE retained the patterned abdominal epidermis activity. (F) The Ddc-MEE2 reporter drove expression more broadly in the abdominal epidermis than the Ddc-MEE CRE and endogenous Ddc expression. (G) The Ddc-MEE3 reporter lacks noteworthy abdominal activity. (H and I) Extreme truncated versions of the Ddc-MEE1 region to 364 and 130 base pairs had regulatory activities similar in pattern to that of the Ddc-MEE1 sequence.
To examine the molecular basis for the evolved patterns of Ddc expression in the abdominal epidermis, we focused on the GMR61H03 fragment. Generating direct fusions of sub-fragments to Enhanced Green Fluorescent Protein (EGFP) reporter gene, we found this Ddc sequence, named Ddc-MEE, to drive a similar abdominal epidermis pattern of reporter expression (Compare Fig. 6D to B). To better resolve which sequences within this Ddc-MEE region drives the pupal abdominal epidermis reporter expression, we created three reporter transgenes that subdivided this region (Fig. 6A).
We found that the reporter gene containing the promoter-proximal region (Ddc-MEE1) drives EGFP expression in a pattern indistinguishable from that of the Ddc-MEE (compare Fig. 6E to B and D). EGFP expression was more robust in the male abdomen than in the female abdomen when the Ddc-MEE1 reporter transgene resides in the attP2 insertion site, though this disparity was less dramatic when the same reporter resides in the 51D attP site (Fig. S4). This modest dimorphism may have at least two explanations. The first is that the increased activity is due to the reporter transgene vector exhibiting a dosage-compensation boost in males due to the vectors inclusion of the mini-white version of the X-chromosome white gene (Ashburner et al., 2005; Qian and Pirrotta, 1995). The other possibility is that CRE reporter transgenes make expression differences more apparent than in situ hybridization. We found the central (Ddc-MEE3) region to lack any noteworthy pupal abdomen regulatory activity (Fig. 6G). In contrast, the distal (Ddc-MEE2) region drove strong epidermis activity (Fig. 6F), which poorly matched expression of the larger fragment or endogenous expression. We interpret this fragment to either be a secondary CRE (Hong et al., 2008; Lagha et al., 2012), a CRE regulating another gene, or artefactual activity. Since the Ddc-MEE1 region generally drives a pattern of expression similar to the evolved pattern of Ddc expression (Fig. 4), we next sought to determine how this CRE is regulated and evolved.
The Ddc-MEE1 pigmentation patterning activity is regulated by Grainy head
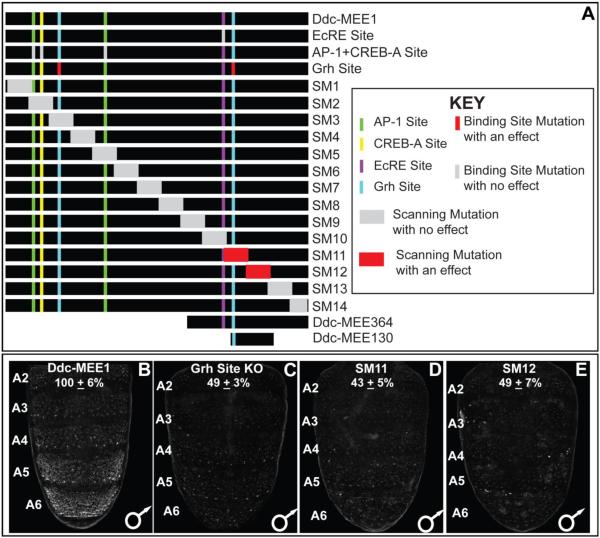
The Ddc-MEE1 sequence 5’ of the Ddc first exon possesses binding sites (Fig. 7A) that contribute to multiple previously described regulatory activities. Within this sequence, a single ecdysone response element (EcRE) is required for the spike in Ddc expression that occurs at pupariation (Chen et al., 2002). For the wound response activity, binding sites for the AP-1, CREB-A, and Grh transcription factors were necessary (Mace et al., 2005). Hence, we referred to this sequence as the Ddc-Multifunctional Epidermis Element 1 or Ddc-MEE1. We sought to determine whether any of these binding sites were required for the tergite pigmentation patterning activity of Ddc, by introducing the previously characterized mutations that disrupt these sites into the Ddc-MEE1. We observed no noticeable alteration in Ddc-MEE1 activity when the EcRE site was mutated, nor when the two AP-1 and the CREB-A sites were mutated (Figs. S2 and S5). However, activity of the Ddc-MEE1 was reduced to 49±3% of the activity of the wild type element when the two Grh sites were mutated (compare Fig. 7C to B).
Fig. 7. A Grh binding site is a required input in the regulatory logic driving Ddc abdominal epidermis CRE activity.
(A) To scale annotation of the Ddc MEE1 region, and the mutant forms of the Ddc-MEE1 that were evaluated for alterations in regulatory activity. (B–E) Regulatory activities of CREs as seen by EGFP reporter gene expression in transgenic D. melanogaster abdomens. (B) Patterned CRE activity driven by the wild type Ddc-MEE1. (C) Compared to the wild type Ddc-MEE1, reporter activity was reduced to 49±3% when the two Grh binding sites were mutated. (D and E) Similarly, the scanning mutant 11 and 12 alterations respectively lowered Ddc-MEE1 reporter activity to 43±5% and 49±7% of the wild type element. Regulatory activity measurements are represented as the % of the wild type D. melanogaster Ddc-MEE1 mean A5 segment intensity plus the Standard Error of the Mean (SEM). Each activity measurement and SEM were derived using images for five biological replicates.
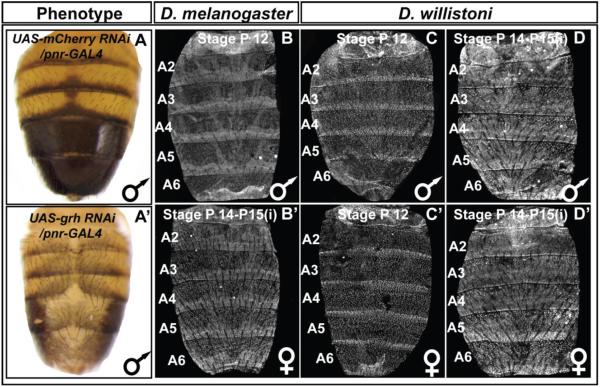
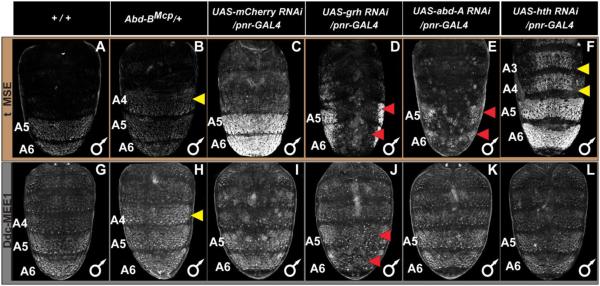
To further validate a role for grh in tergite pigmentation patterning, we used the GAL4/UAS system (Brand and Perrimon, 1993) to drive a dsRNA specific for grh in the dorsal midline region. Compared to the normal tergite pigmentation phenotype observed when RNAi targeted the negative control mCherry gene, pigmentation was greatly reduced upon grh knockdown (compare Fig. 8A and A’). Relatedly, when we tested the Ddc-MEE1 reporter under these conditions, its activity was dramatically reduced (compare Fig. 9J to 9I). Collectively, these data reveal that melanic tergite pigmentation is positively-regulated by Grh, a result that can be explained by the direct regulation of Ddc in the pupal abdomen through one or both of the pleiotropic binding sites in the Ddc-MEE1 CRE.
Fig. 8. Male-specific pigmentation requires the activity of Grh whose expression has remained conserved.
(A) A wild type pattern of pigmentation is exhibited in D. melanogaster males following RNA-interference, RNAi, for the negative control mCherry gene in the dorsal midline pattern of the pnr gene. (A’) Melanic pigmentation was altogether lost in male abdomens following dorsal midline RNAi for the grh gene. Grh protein is expressed in the abdominal epidermis of (B) P12 and (B’) P14-15(i) stage D. melanogaster. Similar patterns of Grh expression occur in (C and C’) P12 and (D and D’) P14-15(i) stage D. willistoni.
Fig. 9. Genetic interactions between pigmentation network transcription factors and CREs regulating tan and Ddc expression.
(A–F) EGFP reporter expression driven by the t_MSE was imaged at the P14-15(i) stage. (G–L) EGFP reporter expression driven by the Ddc-MEE1 was imaged at the P14-15(i) stage. The relevant genetic background genotypes are listed at the top of each column. All specimens are hemizygous for the EGFP reporter transgene. Compared to the (A and G) wild type genetic background, (B) t_MSE, and (H) Ddc-MEE1 regulatory activity expands into the A4 segment in which Abd-B is ectopically expressed. Compared to a (C) control genetic background, suppression of (D) grh expression resulted in a stark loss of t_MSE activity. In contrast, the suppression of (E) abd-A and (F) hth expression respectively led to reduced A5 and A6 segment t_MSE activity and an ectopic t_MSE activity in the A4 and A3 segments. Compared to Ddc-MEE1 activity in a (I) control genetic background, suppression of (J) grh expression resulted in a conspicuous loss of Ddc-MEE1 activity in the A5 and A6 segments, whereas suppression of (K) abd-A and (L) hth respectively had little to no effect on A5 and A6 segment Ddc-MEE1 activity, nor any ectopic activity in the A4 and A3 segments. Yellow arrowheads indicate segments where the genetic background alterations resulted in conspicuous ectopic EGFP reporter expression. Red arrowheads indicate segments where the genetic background alterations resulted in a reduced EGFP reporter expression.
Previously, the Ddc wound response element was resolved down to the immediate 472 bp upstream of the gene’s transcriptional start site (Mace et al., 2005). We wanted to determine whether the wound and tergite pigmentation patterning activities shared additional function-driving sequences, or whether sharing was limited to the Grh binding sites. We created a set of 14 scanning mutant Ddc-MEE1 sequences, in which a different block of ~100 bp was mutated at every other bp by a non-complementary transversion mutation (Figs. 7A, S3, and S6) (Camino et al., 2015; Rogers et al., 2013). For twelve of the fourteen mutant CREs, we saw no conspicuous alteration in the expression of the EGFP reporter gene (Fig. S6). However, reporter expression observed in the abdominal epidermis for the Ddc-MEE1 CREs with either the SM11 and SM12 mutations were reduced respectively to 43±5% and 49±7% of the activity measured for the wild type element (compare Fig. 7D and E to B). The sequences disrupted by SM11 and SM12 mutations include sequences necessary for the wound response activity, and the more evolutionarily conserved of the two Grh biding sites (Mace et al., 2005) in the larger wound element CRE (Fig. S7). As the SM11 and SM12 sequences were necessary for the pupal abdomen epidermis regulatory activity, we tested whether these sequences were sufficient in the context of two truncated versions, named the Ddc-MEE364 and Ddc-MEE130 sequences (Figs. 6E, H and I, and S8). The Ddc-MEE364 contains the non-mutant sequence for the SM11 and SM12 regions. This truncated sequence retained the abdominal epidermis pattern of activity, albeit with modestly-reduced levels of EGFP reporter expression compared to the larger Ddc-MEE1 sequence (82±3%, Fig. S8). The Ddc-MEE130 sequence contains the non-mutant sequence for the SM11 region and the more conserved Grh binding site, but lacks the adjacent EcRE. Similar to the Ddc-MEE364, the Ddc-MEE130 also had a moderated activity compared to that of the Ddc-MEE1 sequence, though it’s pattern resembled the Ddc-MEE1 and Ddc-MEE364 reporters (86±8%, Fig. S8). These experiments localize the bulk of the Ddc-MEE1’s patterning and activity to the MEE130 region, though additional sequences elsewhere within the Ddc-MEE1 are required for full activity. We next sought to investigate which transcription factors in addition to Grh shape the pupal abdomen activity of the Ddc-MEE1 CRE.
The abdominal epidermis activity of the Ddc-MEE1 is Hox-regulated
Previous studies revealed that the D. melanogaster A5 and A6 tergite pigmentation and expression of yellow and tan genes are regulated by Hox transcription factors and their cofactors (Jeong et al., 2006; Rogers et al., 2014). We were curious whether Ddc-MEE1 regulatory activity is Hox-regulated as well. Abd-B, the Hox gene with the most posterior-limited expression, is present in the A5 segment and more posterior segments (Kopp and Duncan, 2002; Wang and Yoder, 2012). In the Abd-BMcp mutant background, Abd-B expression additionally includes the A4 segment. Like the t_MSE, which controls tan expression, ectopic A4 segment reporter expression occurs when the Ddc-MEE1 is assayed in the Abd-BMcp background, indicating that this element is downstream of Abd-B (Compare Fig. 9B to A for tan, and H to G for Ddc).
Ddc expression and Ddc-MEE1 activity extends anterior of the A5 segment, and thus other transcription factors must govern expression and CRE activity in these segments. Plausible candidates include the Hox protein Abd-A which is expressed up through the A2 segment (Kopp and Duncan, 2002; Rogers et al., 2014) and the Hox cofactor Hth which directly represses t_MSE activity in the anterior abdomen segments (Camino et al., 2015). Given the similarities of Ddc expression with tan, we compared the expression of the Ddc-MEE1 with that of the t_MSE under RNAi conditions for factors that disrupt t_MSE function. The t_MSE activity is reduced upon abd-A RNAi (Fig. 9E), and shows ectopic expression in the A3 and A4 segments when hth expression is antagonized by RNAi (Fig. 9F). In contrast, we observed only minor reductions in midline reporter expression of the Ddc-MEE1 when abd-A was reduced (Fig. 9K). Similarly, RNAi for hth did not increase the A3 and A4 segment reporter expression of the Ddc-MEE1 (Fig. 9L). These results suggest that the pupal abdominal epidermis regulatory activity of the D. melanogaster Ddc-MEE1 depends upon a unique set of factors, compared to the t_MSE. The discovery that Ddc expression is Hox-regulated inspired us to examine whether the gain in pupal abdomen Ddc expression for D. melanogaster and its melanic relatives (Fig. 4) involved cis-regulatory evolution at the Ddc-MEE1.
The activity of the Ddc-MEE1 was shaped by cis-evolution
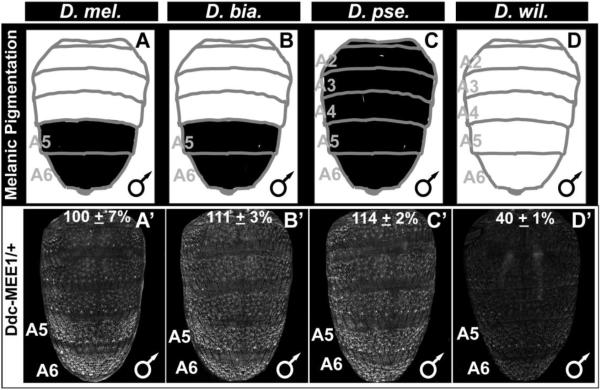
To this point, our results suggested that the transition from an ancestral monomorphic pigmentation to derived melanic patterns involved the gain of a robust pattern of epidermal Ddc expression during the P12 developmental stage (Fig. 4). One possible explanation for the derived expression pattern is through the gain or modification of a CRE activity driving Ddc expression. To test this possibility, we investigated the regulatory activities of sequences orthologous to the D. melanogaster Ddc-MEE1 from species with dimorphic (D. biarmipes), monomorphic melanic (D. pseudoobscura), and monomorphic non-melanic (D. willistoni) patterns of tergite pigmentation (Fig. 10A–D). We saw that each of the orthologous sequences were capable of driving reporter expression in the abdominal epidermis of D. melanogaster (Fig. 10A’–D’), indicating that the origin of the P12 stage epidermal Ddc expression was not due to the evolution of an altogether new CRE activity. Importantly though, EGFP reporter expression driven by the D. willistoni sequence was consistently more modest in biological replicate specimens (Fig. 10D’). Quantification of EGFP intensity in the A5 segment revealed that the D. willistoni CRE drove 40±1% of the D. melanogaster Ddc-MEE1 activity (100±7%; compare Fig. 10D’ to A’). In contrast, the D. biarmipes and D. pseudoobscura activities respectively were 111±3% and 114±2% of the D. melanogaster CRE’s activity (Fig. 10B’ and C’). In a genetic background with ectopic Abd-B expression in the A4 abdomen segment, the activity of the D. melanogaster Ddc-MEE1 increased to 180±14% of the activity in the A4 segment of a Abd-B wild type background (Fig. S9). In comparison, the activity of D. willistoni sequence orthologous to the D. melanogaster Ddc-MEE1 increased to 148±10% A4 segment activity of the D. willistoni CRE in a wild type background (Fig. S9). These results are consistent with a scenario in which the enhanced expression of Ddc in the pupal abdomen epidermis of D. melanogaster involved cis-evolution. This regulatory evolution seems to have included an increased responsiveness of the Ddc-MEE1 to Hox-regulation compared to the antecedent CRE.
Fig. 10. The ancestry of the robust abdominal epidermis regulatory activity of the CRE controlling Ddc expression.
(A–D) A schematic representation of the male abdomen pigmentation patterns for several Sophophora species. (A’–D’) EGFP-reporter transgene expression driven by sequences orthologous to the D. melanogaster Ddc-MEE1 in transgenic D. melanogaster male pupae at the P14-15(i) stage. (A’–C’) Sequences from species with conspicuous melanic pigmentation phenotypes drive reporter expression throughout the abdomen, though the activity appears most pronounced in the A5 and A6 segments. (D’) The sequence from the most distantly-related species with monomorphic tergite pigmentation, D. willistoni, has comparatively less abdominal regulatory activity. Regulatory activity measurements are represented as the % of the wild type D. melanogaster Ddc-MEE1 mean A5 segment intensity plus the Standard Error of the Mean (SEM). Each activity measurement and SEM were derived using images for five biological replicates.
While our results implicate a role for cis-evolution in shaping the activity of the Ddc-MEE1 CRE, it is also possible that changes to the expressions or activities of trans-regulators were additionally required. One plausible candidate would be spatially and/or temporally expanded expression of Grh, the direct regulator of the Ddc-MEE1. However, we observed a conserved pattern of pan-abdominal epidermis Grh expression in the abdomens of D. melanogaster and the monomorphic D. willistoni (Fig. 8). Thus, future studies are needed to resolve whether trans-evolution played a role in the derived expression pattern of Ddc.
Discussion
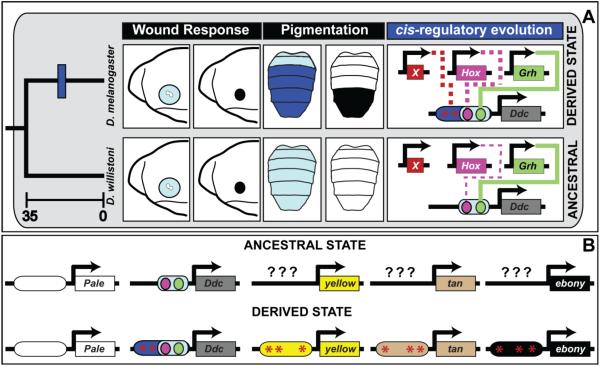
Our findings illustrate how an important realizator gene was recruited to a novel trait through the alteration of a presumably ancient CRE that functions in an entirely unrelated task. By characterizing the expression of two genes whose protein products catalyze early steps of melanin synthesis, we found that strong pale expression is conserved broadly across the Sophophora subgenus, while high levels of Ddc expression were only observed in species that had derived dark melanic pigments. We uncovered how abdominal Ddc expression in D. melanogaster is driven by a pleiotropic CRE that plays a presumably ancient role in activating Ddc expression following epidermal wounding (Fig. 11A). Notably, a Grh binding site required for wound healing is also required for its abdominal function. Comparative analysis of this CRE revealed that its augmented pigment associated activity evolved more recently in the lineage of species with melanic abdomens. These results contribute to a body of literature showing that the origin of dimorphic pigmentation required cis-evolution at several realizator genes in the network (Figure 11B), providing an example in which an entire network was assembled through widespread cis-regulatory modifications.
Fig. 11. The evolution of a novel pigmentation trait required cis-regulatory evolution at several realizator loci.
(A) In the embryo, Ddc expression (light blue) is upregulated around sites of epidermal wounding as part of a response that forms a melanized plug (black circle). Schematic depicts the anterior embryo. This gene function is conserved, and encoded in the activity of a CRE directly regulated by the Grh transcription factor. In D. willistoni, which serves as a proxy for the ancestrally monomorphic trait, Ddc expression is driven at a low level by the pleiotropic activity of the wound response CRE. This ancestral CRE possesses a direct binding site for Grh and is modestly-responsive to the Hox transcription factor Abd-B. In species with derived melanic abdomen pigmentation phenotypes, Ddc expression is robust throughout the abdomen (dark blue). This expression is in part due to cis-evolution at Ddc, which evolved a stronger, Hox-responsive CRE activity that is apparent for D. melanogaster, and this CRE seemingly evolved to be under the control of some trans-regulator(s) whose identity remains unknown and is represented here as factor “X”. Time scale is depicted in millions of years. Solid lines in cis-regulatory evolution indicate cases where the transcription factor directly binds to the Ddc CRE, and dashed lines indicate cases where the mechanism of transcription factor regulation remains unknown. (B) Model for the origin of the derived dimorphic pigmentation trait of D. melanogaster from an ancestral monomorphic state. Here, abdominal pale expression was ancestral, whereas Ddc, yellow, tan, and ebony each evolved novel expression through changes in CREs controlling their expression. The ancestral states upon which novel CREs evolved for yellow, tan, and ebony remain unknown (question marks). For Ddc, cis-evolution augmented an ancestral wound response CRE to have a robust pleiotropic activity.
The novel deployment of a realizator gene pathway by widespread CRE evolution
The development of most well-studied animal morphological traits relies upon dozens of regulatory genes that specify the spatial and temporal expression of pathways of realizator genes (Bonn and Furlong, 2008b; Levine and Davidson, 2005). One of the most difficult challenges in evolutionary biology is to develop molecular explanations of how such vast networks of regulators and realizators emerge to control novel traits. A critical feature that must be resolved is whether these novel traits are built up gradually through evolution at each realizator, or if networks evolve primarily through the co-option of large batteries of realizators from one developmental setting to another by expressing their upstream regulators in new locations (Monteiro and Gupta, 2016; Rebeiz et al., 2015). To address this distinction, the pigmentation of the Drosophila abdomen is one of the most extensively studied traits for which the evolutionary trajectories of key components of the network have been traced. This study expands a growing perspective of how its network was assembled.
The current favored model for pigment evolution in the Sophophora subgenus places monomorphic, non-melanic pigmentation as the ancestral character state, as exemplified by D. willistoni, and male-specific melanic pigmentation as the derived state, as seen for D. melanogaster and D. biarmipes (Fig. 1A) (Rebeiz and Williams, 2017). In the obscura species group, a broadly monomorphic melanic character state evolved. The expression of yellow, tan, and ebony evolved to fit these different pigmentation patterns. For D. willistoni, the dorsal abdominal epidermis experiences little to no yellow and tan expression, whereas expression occurs robustly throughout the D. pseudoobscura abdomen and in the male-specific pigmented regions of D. melanogaster (Camino et al., 2015). Thus, the origin of melanic phenotypes required gains in yellow and tan expression. In this study, we show that while pale expression throughout the abdominal epidermis has maintained high levels of ancestral expression, Ddc evolved a robust pattern of expression in the abdomen epidermis in the lineage of melanic species. Thus, a critical question is whether this network evolved its associated expression domains through co-option or independent evolutionary changes.
If the origin of male-specific tergite pigmentation in the D. melanogaster lineage originated via a co-option mechanism, we would anticipate two hallmark signs of this event. First, co-option would likely involve a master regulator whose activity is sufficient to deploy all of the underlying regulatory and realizator genes. Second, we would expect these CREs to all share an ancestral activity in a second tissue or developmental setting from which the abdomen melanization function was co-opted (Glassford et al., 2015). The case we presented here of the Ddc-MEE1 illustrates a clear ancestral function at Ddc that is unique among genes of the pigmentation pathway. To our knowledge, none of the other CREs in the pathway are associated with a wound response function. Further, we have yet to identify a master regulator of the network that would make sense as a source of the co-option event. Of all the transcription factors thus far identified in the abdominal pigmentation network, the most likely master regulator is Abd-B. CREs of four realizator enzymes of the network respond to Abd-B in trans (Camino et al., 2015; Jeong et al., 2006), and in the case of yellow, Abd-B is a direct regulator (Jeong et al., 2006). However, the major problem with considering Abd-B as a master regulator that mediated the redeployment of this network, is that its pattern of expression is likely ancestral, and an Abd-B dependent ancestral network that drives all of these genes elsewhere in the posterior of the embryo, larvae, or pupae seems unlikely. Indeed, upon discovering the direct binding of the Bric-á-brac (Bab) transcription factors to the CRE controlling abdominal epidermis yellow expression, we found no evidence of co-option from an ancestral CRE function (Roeske et al., 2018). Thus, considering the lack of a shared ancestral network or master regulatory gene, we propose that the network of pigmentation realizator CREs was assembled in a piecemeal fashion. The path by which Ddc independently evolved its abdominal activity is a curious one.
Gene expression evolution through the augmentation of a pleiotropic CRE activity
To understand how GRNs of regulators and realizators emerged to generate novel traits, we must precisely examine how their CRE activities originated. Several mechanisms have been posited, including the gain of a CRE by transposition, the de novo evolution of a CRE activity by gains in transcription binding sites, and the co-option of an existing CRE with an ancestral function (Rebeiz et al., 2015). Problematically though, differences in traits between closely-related species often represent losses, and experimental models for trait gains often require comparisons between more distantly-related taxa whose cis-regulatory regions may be difficult to compare due to extensive sequence divergence.
The gain of a fruit fly male-specific wing pigment spot and abdomen tergite pigmentation have been two productive models for investigating the origins of CREs associated with novel traits. In D. biarmipes, a spot of pigmentation on the wing requires the expression of yellow that is driven by a CRE that possesses binding sites for the activator Dll and the repressor En (Arnoult et al., 2013; Gompel et al., 2005). The spot CRE resides adjacent to a wing element CRE that drives a low level of yellow expression throughout the developing wing, and comparative analyses of this region suggested that the spot CRE evolved from this weakly active element. Similarly, the origin of the D. melanogaster abdominal tergite pigmentation required gains of male-specific patterns of yellow and tan expression (Camino et al., 2015; Jeong et al., 2008, 2006). In D. melanogaster, yellow expression is driven by an Abd-B and Bab regulated CRE known as the body element (Jeong et al., 2006; Roeske et al., 2018), and tan is driven by a Hox and Hox cofactor regulated CRE known as the t_MSE (Camino et al., 2015). The origin of these CRE activities remain difficult to resolve, as the surrogate for the ancestral trait, D. willistoni, lacks these CRE activities and any sequence with noteworthy conservation. Thus, the genetic events and molecular details responsible for the origin of the yellow body element and the tan t_MSE remain nebulous.
In contrast, we found clear evidence for the evolutionary path taken by Ddc, as it gained a robust pattern of abdominal epidermis expression in melanic species. Unlike the CREs of yellow and tan, the orthologous sequence of the Ddc-MEE1 is highly conserved (Fig. S7), and the ancestral function can be reasonably inferred as this region is known to drive Ddc expression following epidermal wounding between fruit fly species more distantly-related than those studied here (Mace et al., 2005). Moreover, some of the molecular details regarding the evolution of the Ddc-MEE1 can be inferred. Both the wound response (Mace et al., 2005) and abdominal epidermis activities require a conserved binding site for the Grh transcription factor (Fig. S7). This indicates that the ancestral wound response CRE was modified, resulting in the augmentation of its pleiotropic function in tergite pigmentation (Fig. 11A). While the precise details on the mechanism of augmentation remains to be elucidated, part of the explanation requires the gain of an increased responsiveness to the Hox gene, Abd-B. Importantly, our work provides an example whereby part of a trait’s origin required the gain of a realizator gene’s expression through the modification of an ancient CRE for an augmented pleiotropic use. Our ability to discern an evolutionary past to this element highlights how different CREs, even in the same network, will contrast greatly in their rates of divergence. Finally, our findings reinforce how CRE activities can emerge from unexpected developmental contexts that provide a rich molecular explanation for the origin of traits and the CREs that enable them.
Supplementary Material
Milestones during Drosophila pupal development can be identified by the presence of conspicuous morphological markers. The earliest stage analyzed in this study was P10, which can be identified by the appearance of red-colored eyes, and anteriorly-positioned malpighian tubes on the abdomen. Pupae at the P11 stage can be distinguished by the addition of light gray wing pigmentation, and thin head and thorax bristles. Pupae at the slightly more advanced P12 stage were identified by grey wing pigmentation, and gray bristles on the dorsal head, abdomen and thorax. Pupae at the P13 stage can be identified by the appearance of black wing and bristle pigmentation. At the even more advanced P14-15(i) stage, pupae were identified by black wing pigmentation, and black bristles on the abdomen and thorax, disappearance of the malpighian tubes, and meconium visible at the posterior-tip of the abdomen. Stage P15(ii) is not represented here, but these samples were identified as flies that recently eclosed from their puparium.
(A) Key detailing the names, locations, and effects of Ddc-MEE1 scanning mutations (SM) in transgenic male D. melanogaster pupae. Scanning mutations are indicated as gray blocks when abdomen activity appeared unaltered, and as red blocks when activity was conspicuously reduced. (B–M) The EGFP expression pattern in the male abdomen at the P14-P15(i) developmental stage driven by the Ddc-MEE1 scan mutant sequences that had no apparent effect on abdomen CRE activity. Representative results for SM11 and SM12 appear in Figure 7.
Puple background indicates the AscI (GGCGCGCC) and SbfI (CCTGCAGG) restriction enzymes sites that were added for cloning CRE versions into an EGFP reporter transgene vector. Sequences characteristic of binding sites for the AP-1+CREB, Grh, and EcRE transcription factors that were previously studied for the Ddc wound response enhancer activity are indicated by the yellow, teal, and blue background colors. The orthologous sequence from D. virilis, MEE1(vir), was included as an outgroup non-Sophophora species. The D. virilis sequence possesses wound response CRE activity (Mace et al. 2005).
(A) Reporter expression driven by the Ddc-MEE1 CRE in the 51D attP insertion site. (B and C) Reporter expression driven respectively by the truncated Ddc-MEE364 and Ddc-MEE130 CREs in the 51D attP site. Compared to the larger CRE, the truncated forms drove a similar pattern of expression, yet modestly-reduced activities. Regulatory activity measurements are represented as the % of the wild type D. melanogaster Ddc-MEE1 mean A5 segment intensity plus the Standard Error of the Mean (SEM). Each activity measurement and SEM were derived using images for five biological replicates.
(A) EGFP reporter expression for a representative male pupae hemizygous for the D. melanogaster Ddc-MEE1-EGFP reporter transgene in a wild type Abd-B expressing genetic background. (A’) EGFP reporter expression for a representative male pupae hemizygous for the D. melanogaster Ddc-MEE1-EGFP reporter transgene in an Abd-BMcp genetic background with ectopic Abd-B expression in the A4 segment. (B) EGFP reporter expression for a representative male pupae hemizygous for the D. willistoni Ddc-MEE1-EGFP reporter transgene in a wild type Abd-B expressing genetic background. (B’) EGFP reporter expression for a representative male pupae hemizygous for the D. willistoni Ddc-MEE1-EGFP reporter transgene in an Abd-BMcp genetic background with ectopic Abd-B expression in the A4 segment. (A and A’) Regulatory activity measurements are represented as the % of the D. melanogaster Ddc-MEE1 mean A4 intensity plus the Standard Error of the Mean (SEM) in the wild type Abd-B genetic background. (B and B’) Regulatory activity measurements are represented as the % of the D. willistoni Ddc-MEE1 mean A4 intensity plus the Standard Error of the Mean (SEM) in the wild type Abd-B genetic background. Each activity measurement and SEM were derived using images for four biological replicates.
Purple background indicates the AscI (GGCGCGCC) and SbfI (CCTGCAGG) restriction enzymes sites that were added for cloning CRE versions into an EGFP reporter transgene vector. Sequences characteristic of binding sites for the AP-1+CREB, Grh, and EcRE transcription factors that were previously studied for the Ddc wound response enhancer activity are indicated by the yellow, teal, and blue background colors in the non-mutant sequence. The mutant versions for these sites are indicated by the black background with red letters, and the substituted nucleotides are shown in lowercase red letters.
Purple background indicates the AscI (GGCGCGCC) and SbfI (CCTGCAGG) restriction enzymes sites that were added for cloning into the EGFP reporter transgene vector. Gray background and white letters indicates sequences that comprise a scanning mutation of non-complementary transversions and for which there was not a conspicuous alteration in the pupal abdomen epidermis regulatory activity. Red background and white letters indicates sequences that comprise a scanning mutation of non-complementary transversions and for which the mutant CRE had a reduced regulatory activity in the abdomen. The lowercase nucleotide letters indicate the non-complementary transversions.
Reporter expression in the dorsal abdomens of (A and B) male and (A’ and B’) female P14-P15(i) stage transgenic D. melanogaster. (A and A’) Ddc-MEE1 reporter transgene inserted in the attP2 site in chromosome 3. (B and B’) Ddc-MEE1 reporter transgene inserted in the 51D site in chromosome 2.
(A–D) Regulatory activity of Ddc-MEE1 versions as seen by EGFP reporter transgene expression in the dorsal abdominal epidermis of P14-P15(i) stage transgenic D. melanogaster males. (A) Reporter expression driven by the wild type Ddc-MEE1. (B) Reporter expression driven by the Ddc-MEE1 with two mutant Grh binding sites was dramatically reduced. (C) Reporter expression was not noticeably altered when driven by a Ddc-MEE1 possessing mutations in two motifs characteristic of AP-1 binding sites and one motif characteristic of a CREB-A binding sites. (D) Reporter expression was not noticeably altered when driven by a Ddc-MEE1 with mutated version of a motif characteristic of the ecdysone receptor binding.
Highlights.
Drosophila melanogasterpale and Ddc expression are temporally and spatially dynamic
Ddc expression is augmented in species with derived melanic pigmentation patterns
Ddc expression is driven by an enhancer with an ancestral wound response activity
Augmented Ddc expression evolved through modifications in the wound response enhancer
A novel pigmentation trait required widespread enhancer evolution in realizator genes
Acknowledgments
Species stocks were purchased from the San Diego Drosophila Stock Center or provided by S.B. Carroll. Drosophila melanogaster UAS-RNAi lines were provided by the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The Grainy head antibody was generously provided by W. Mcginnis. SG and JH were supported by fellowships from the University of Dayton Graduate School. JH was additionally supported by a Graduate Research Fellowship from the National Science Foundation (DGE-1439647). TMW and MR were supported by a grant from the National Science Foundation (IOS-1555906). MR’s work on Drosophila pigmentation was supported by a grant from the National Institutes of Health (5R01GM114093-02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary Materials
Supplemental data associated with this article can be found in the online version at http://
References
- Abràmoff MD, Hospitals I, Magalhães PJ, Abràmoff M, 2004. Image Processing with ImageJ. Biophotonics Int 11, 36–42. [Google Scholar]
- Arnone MI, Davidson EH, 1997. The hardwiring of development: organization and function of genomic regulatory systems. Development 124, 1851–64. [DOI] [PubMed] [Google Scholar]
- Arnoult L, Su K, Manoel D, Minervino C, Magrina J, Gompel N, Prud’homme B, 2013. Emergence and Diversification of Fly Pigmentation Through Evolution of a Gene Regulatory Module. Science 339, 1423–1426. 10.1126/science.1233749 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS, 2005. Drosophila: A laboratory handbook, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- Barolo S, Castro B, Posakony JW, 2004. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques 36, 436–40, 442. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K, 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U. S. A 104, 3312–7. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Furlong EEM, 2008a. cis-Regulatory networks during development: a view of Drosophila. Curr. Opin. Genet. Dev 18, 513–20. 10.1016/j.gde.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Bonn S, Furlong EEM, 2008b. cis-Regulatory networks during development : a view of Drosophila. Curr. Opin. Genet. Dev 513–520. 10.1016/j.gde.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–15. [DOI] [PubMed] [Google Scholar]
- Brudno M, Steinkamp R, Morgenstern B, 2004. The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Res 32, W41–4. 10.1093/nar/gkh361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja M, Herranz H, Estella C, Casal J, Lawrence P, Simpson P, Morata G, 2000. Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development 127, 3971–80. [DOI] [PubMed] [Google Scholar]
- Camino EM, Butts JC, Ordway A, Vellky JE, Rebeiz M, Williams TM, 2015. The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in cis and trans. PLOS Genet 11, e1005136 10.1371/journal.pgen.1005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Reece C, O’Keefe SL, Hawryluk GWL, Engstrom MM, Hodgetts RB, 2002. Induction of the early-late Ddc gene during Drosophila metamorphosis by the ecdysone receptor. Mech. Dev 114, 95–107. [DOI] [PubMed] [Google Scholar]
- Davidson EH, 2006. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. Elsevier Inc, Burlington, MA. [Google Scholar]
- Davis MM, O’Keefe SL, Primrose DA, Hodgetts RB, 2007. A neuropeptide hormone cascade controls the precise onset of post-eclosion cuticular tanning in Drosophila melanogaster. Development 134, 4395–4404. 10.1242/dev.009902 [DOI] [PubMed] [Google Scholar]
- Ferguson LC, Green J, Surridge A, Jiggins CD, 2011. Evolution of the insect yellow gene family. Mol. Biol. Evol 28, 257–272. 10.1093/molbev/msq192 [DOI] [PubMed] [Google Scholar]
- García-Bellido A, 1975. Genetic control of wing disc development in Drosophila. Ciba Found Symp 0, 161–82. [DOI] [PubMed] [Google Scholar]
- Glassford WJ, Johnson WC, Dall NR, Smith SJ, Liu Y, Boll W, Noll M, Rebeiz M, 2015. Co-option of an Ancestral Hox-Regulated Network Underlies a Recently Evolved Morphological Novelty. Dev. Cell 1–12. 10.1016/j.devcel.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB, 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–7. 10.1038/nature03235 [DOI] [PubMed] [Google Scholar]
- Gray S, Levine M, 1996. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev 10, 700–710. 10.1101/gad.10.6.700 [DOI] [PubMed] [Google Scholar]
- Groth AC, Calos MP, 2004. Phage Integrases: Biology and Applications. J. Mol. Biol 335, 667–678. 10.1016/j.jmb.2003.09.082 [DOI] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP, 2000. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. U. S. A 97, 5995–6000. 10.1073/pnas.090527097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinaux H, Bachem K, Battistara M, Rossi M, Xin Y, Jaenichen R, Le Poul, Y., Arnoult L, Kobler JM, Grunwald Kadow, I.C., Rodermund L, Prud’homme B, Gompel N, 2018. Revisiting the developmental and cellular role of the pigmentation gene yellow in Drosophila using a tagged allele. Dev. Biol 0–1. 10.1016/j.ydbio.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, Levine MS, 2008. Shadow enhancers as a source of evolutionary novelty. Science 321, 1314 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB, 2008. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 132, 783–93. 10.1016/j.cell.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB, 2006. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387–99. 10.1016/j.cell.2006.04.043 [DOI] [PubMed] [Google Scholar]
- Johnson WC, Ordway AJ, Watada M, Pruitt JN, Williams TM, Rebeiz M, 2015. Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Drosophila Species. PLOS Genet 11, e1005279 10.1371/journal.pgen.1005279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Mcginnis W, 2010. Phosphorylation of Grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc Natl Acad Sci U S A 108, 650–655. 10.1073/pnas.1016386108/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1016386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Duncan I, 2002. Anteroposterior patterning in adult abdominal segments of Drosophila. Dev. Biol 242, 15–30. 10.1006/dbio.2001.0529 [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Levine M, 2012. Mechanisms of transcriptional precision in animal development. Trends Genet 28, 409–16. 10.1016/j.tig.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Davidson EH, 2005. Gene regulatory networks for development. Proc. Natl. Acad. Sci. U. S. A 102, 4936–42. 10.1073/pnas.0408031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace KA, Pearson JC, McGinnis W, 2005. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308, 381–5. 10.1126/science.1107573 [DOI] [PubMed] [Google Scholar]
- Monteiro A, Gupta MD, 2016. Identifying Coopted Networks and Causative Mutations in the Origin of Novel Complex Traits, 1st ed, Current Topics in Developmental Biology. Elsevier Inc; 10.1016/bs.ctdb.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Neckameyer WS, White K, 1993. Drosophila Tyrosine Hydroxylase is Encoded by the Pale Locus. J. Neurogenet 8, 189–199. [DOI] [PubMed] [Google Scholar]
- Ordway A, Hancuch KN, Johnson W, Wiliams TM, Rebeiz M, 2014. The expansion of body coloration involves coordinated evolution in cis and trans within the pigmentation regulatory network of drosophila prostipennis. Dev. Biol 1–10. 10.1016/j.ydbio.2014.05.023 [DOI] [PubMed] [Google Scholar]
- Pfeiffer B, Jenett A, Hammonds AS, Ngo Teri-T, B., Mirsa S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM, 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U. S. A 105, 9715–20. 10.1073/pnas.0803697105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Pirrotta V, 1995. Dosage Compensation of the Drosophila white Gene Requires Both the X Chromosome Environment and Multiple Intragenic Elements. Genetics 139 733–744. 10.1146/annurev-genet-110711-155454.Dosage [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Patel NH, Hinman VF, 2015. Unraveling the Tangled Skein: The Evolution of Transcriptional Regulatory Networks in Development. Annu. Rev. Genomics Hum. Genet 16 103–131. 10.1146/annurev-genom-091212-153423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB, 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663–7. 10.1126/science.1178357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Posakony JW, 2004. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev. Biol 271, 431–8. 10.1016/j.ydbio.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Tsiantis M, 2017. Enhancer evolution and the origins of morphological novelty. Curr. Opin. Genet. Dev 45, 115–123. 10.1016/j.gde.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Williams TM, 2017. Using Drosophila pigmentation traits to study the mechanisms of cis-regulatory evolution. Curr. Opin. Insect Sci 19, 1–7. 10.1016/j.cois.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Williams TM, 2011. Experimental approaches to evaluate the contributions of candidate cis-regulatory mutations to phenotypic evolution. Methods Mol. Biol 772. [DOI] [PubMed] [Google Scholar]
- Roeske MJ, Camino EM, Grover S, Rebeiz M, Williams TM, 2018. Cis-regulatory evolution integrated the bric-a-brac transcription factors into a novel fruit fly gene regulatory network. Elife 7, 1–28. 10.7554/eLife.32273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers WA, Grover S, Stringer SJ, Parks J, Rebeiz M, Williams TM, 2014. A survey of the trans-regulatory landscape for Drosophila melanogaster abdominal pigmentation. Dev. Biol 385, 417–432. 10.1016/j.ydbio.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Rogers WA, Salomone JR, Tacy DJ, Camino EM, Davis KA, Rebeiz M, Williams TM, 2013. Recurrent Modification of a Conserved Cis-Regulatory Element Underlies Fruit Fly Pigmentation Diversity. PLoS Genet 9, e1003740 10.1371/journal.pgen.1003740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers WA, Williams TM, 2011. Quantitative Comparison of cis-Regulatory Element (CRE) Activities in Transgenic Drosophila melanogaster. J. Vis. Exp 2–7. 10.3791/3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomone JR, Rogers WA, Rebeiz M, Williams TM, 2013. The evolution of Bab paralog expression and abdominal pigmentation among Sophophora fruit fly species. Evol. Dev 15, 442–57. 10.1111/ede.12053 [DOI] [PubMed] [Google Scholar]
- Scholnick SB, Bray SJ, Morgan BA, Mccormick CA, 1993. CNS and Hypoderm Regulatory Elements of the Drosophila melanogaster Dopa Decarboxylase Gene. Science (80-.). 234, 998–1002. [DOI] [PubMed] [Google Scholar]
- Smith AF, Posakony JW, Rebeiz M, 2017. Automated tools for comparative sequence analysis of genic regions using the GenePalette application. Dev. Biol 429, 158–164. 10.1016/j.ydbio.2017.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M, 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science (80-.). 254, 1385–1387. [DOI] [PubMed] [Google Scholar]
- True JR, Yeh S-D, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou S-R, Han Q, Li J, 2005. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet 1, e63 10.1371/journal.pgen.0010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoder JH, 2012. Hox-mediated regulation of doublesex sculpts sex-specific abdomen morphology in Drosophila. Dev. Dyn 241, 1076–90. 10.1002/dvdy.23791 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A, 2003. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet 19, 495–504. 10.1016/S0168-9525(03)00194-X [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Vaccaro K, Carroll SB, 2002. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol 12, 1547–56. [DOI] [PubMed] [Google Scholar]
- Wright TRF, 1987. The Genetics of Biogenic Amine Metabolism, Sclerotization, and Melanization in Drosophila melanogaster, in: Scandalios JG, Caspari EW (Eds.), Advances in Genetics. Harcourt Brace Jovanovich, pp. 127–222. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Milestones during Drosophila pupal development can be identified by the presence of conspicuous morphological markers. The earliest stage analyzed in this study was P10, which can be identified by the appearance of red-colored eyes, and anteriorly-positioned malpighian tubes on the abdomen. Pupae at the P11 stage can be distinguished by the addition of light gray wing pigmentation, and thin head and thorax bristles. Pupae at the slightly more advanced P12 stage were identified by grey wing pigmentation, and gray bristles on the dorsal head, abdomen and thorax. Pupae at the P13 stage can be identified by the appearance of black wing and bristle pigmentation. At the even more advanced P14-15(i) stage, pupae were identified by black wing pigmentation, and black bristles on the abdomen and thorax, disappearance of the malpighian tubes, and meconium visible at the posterior-tip of the abdomen. Stage P15(ii) is not represented here, but these samples were identified as flies that recently eclosed from their puparium.
(A) Key detailing the names, locations, and effects of Ddc-MEE1 scanning mutations (SM) in transgenic male D. melanogaster pupae. Scanning mutations are indicated as gray blocks when abdomen activity appeared unaltered, and as red blocks when activity was conspicuously reduced. (B–M) The EGFP expression pattern in the male abdomen at the P14-P15(i) developmental stage driven by the Ddc-MEE1 scan mutant sequences that had no apparent effect on abdomen CRE activity. Representative results for SM11 and SM12 appear in Figure 7.
Puple background indicates the AscI (GGCGCGCC) and SbfI (CCTGCAGG) restriction enzymes sites that were added for cloning CRE versions into an EGFP reporter transgene vector. Sequences characteristic of binding sites for the AP-1+CREB, Grh, and EcRE transcription factors that were previously studied for the Ddc wound response enhancer activity are indicated by the yellow, teal, and blue background colors. The orthologous sequence from D. virilis, MEE1(vir), was included as an outgroup non-Sophophora species. The D. virilis sequence possesses wound response CRE activity (Mace et al. 2005).
(A) Reporter expression driven by the Ddc-MEE1 CRE in the 51D attP insertion site. (B and C) Reporter expression driven respectively by the truncated Ddc-MEE364 and Ddc-MEE130 CREs in the 51D attP site. Compared to the larger CRE, the truncated forms drove a similar pattern of expression, yet modestly-reduced activities. Regulatory activity measurements are represented as the % of the wild type D. melanogaster Ddc-MEE1 mean A5 segment intensity plus the Standard Error of the Mean (SEM). Each activity measurement and SEM were derived using images for five biological replicates.
(A) EGFP reporter expression for a representative male pupae hemizygous for the D. melanogaster Ddc-MEE1-EGFP reporter transgene in a wild type Abd-B expressing genetic background. (A’) EGFP reporter expression for a representative male pupae hemizygous for the D. melanogaster Ddc-MEE1-EGFP reporter transgene in an Abd-BMcp genetic background with ectopic Abd-B expression in the A4 segment. (B) EGFP reporter expression for a representative male pupae hemizygous for the D. willistoni Ddc-MEE1-EGFP reporter transgene in a wild type Abd-B expressing genetic background. (B’) EGFP reporter expression for a representative male pupae hemizygous for the D. willistoni Ddc-MEE1-EGFP reporter transgene in an Abd-BMcp genetic background with ectopic Abd-B expression in the A4 segment. (A and A’) Regulatory activity measurements are represented as the % of the D. melanogaster Ddc-MEE1 mean A4 intensity plus the Standard Error of the Mean (SEM) in the wild type Abd-B genetic background. (B and B’) Regulatory activity measurements are represented as the % of the D. willistoni Ddc-MEE1 mean A4 intensity plus the Standard Error of the Mean (SEM) in the wild type Abd-B genetic background. Each activity measurement and SEM were derived using images for four biological replicates.
Purple background indicates the AscI (GGCGCGCC) and SbfI (CCTGCAGG) restriction enzymes sites that were added for cloning CRE versions into an EGFP reporter transgene vector. Sequences characteristic of binding sites for the AP-1+CREB, Grh, and EcRE transcription factors that were previously studied for the Ddc wound response enhancer activity are indicated by the yellow, teal, and blue background colors in the non-mutant sequence. The mutant versions for these sites are indicated by the black background with red letters, and the substituted nucleotides are shown in lowercase red letters.
Purple background indicates the AscI (GGCGCGCC) and SbfI (CCTGCAGG) restriction enzymes sites that were added for cloning into the EGFP reporter transgene vector. Gray background and white letters indicates sequences that comprise a scanning mutation of non-complementary transversions and for which there was not a conspicuous alteration in the pupal abdomen epidermis regulatory activity. Red background and white letters indicates sequences that comprise a scanning mutation of non-complementary transversions and for which the mutant CRE had a reduced regulatory activity in the abdomen. The lowercase nucleotide letters indicate the non-complementary transversions.
Reporter expression in the dorsal abdomens of (A and B) male and (A’ and B’) female P14-P15(i) stage transgenic D. melanogaster. (A and A’) Ddc-MEE1 reporter transgene inserted in the attP2 site in chromosome 3. (B and B’) Ddc-MEE1 reporter transgene inserted in the 51D site in chromosome 2.
(A–D) Regulatory activity of Ddc-MEE1 versions as seen by EGFP reporter transgene expression in the dorsal abdominal epidermis of P14-P15(i) stage transgenic D. melanogaster males. (A) Reporter expression driven by the wild type Ddc-MEE1. (B) Reporter expression driven by the Ddc-MEE1 with two mutant Grh binding sites was dramatically reduced. (C) Reporter expression was not noticeably altered when driven by a Ddc-MEE1 possessing mutations in two motifs characteristic of AP-1 binding sites and one motif characteristic of a CREB-A binding sites. (D) Reporter expression was not noticeably altered when driven by a Ddc-MEE1 with mutated version of a motif characteristic of the ecdysone receptor binding.