Abstract
Purpose of review
To summarize recent advances in the discovery of chemical inhibitors targeting the HIV capsid and research on their mechanisms of action.
Recent findings
HIV infection is critically dependent on functions of the viral capsid. Numerous studies have reported the identification of a variety of compounds that bind to the capsid protein (CA); some of these inhibit reverse transcription and nuclear entry, steps required for infection. Other CA-targeting compounds appear to act by perturbing capsid assembly, resulting in noninfectious progeny virions. Inhibitors may bind to several different positions on the CA protein, including sites in both protein domains. However, the antiviral activity of many reported CA-targeting inhibitors has not been definitively linked to CA binding. Until recently, the low to moderate potency of reported CA-targeting inhibitors has precluded their further clinical development. In 2017, GS-CA1, a highly potent capsid inhibitor was described that holds promise for clinical development.
Summary
Small molecules that bind to the viral CA protein can be potent inhibitors of HIV infection.
Capsid-targeting drugs are predicted to exhibit high barriers to viral resistance, and ongoing work in this area is contributing to an understanding of the molecular biology of HIV uncoating and maturation.
Keywords: HIV, capsid, inhibitor, uncoating, maturation
Introduction
Infection by human immunodeficiency viruses remains a global public health threat. Although advances in antiretroviral therapy have been instrumental in reducing the spread and severity of HIV/AIDs, there remains no effective vaccine. Moreover, treatment is not curative; consequently patients must be treated for their entire lives. Poor adherence to therapy leads to viral resistance to existing drugs, resulting in a continuing need for new therapeutics, preferably against new drug targets. In this review, we highlight recent research efforts aimed at identifying inhibitors that directly target the HIV-1 capsid and determining their antiviral mechanisms. Although there are currently no CA-targeting compounds approved for clinical use, a potent capsid-targeting inhibitor recently reported holds promise for therapeutic development.
The HIV-1 capsid is a conical protein shell composed of a repeating hexameric lattice of the viral CA protein. Within the capsid are housed the viral RNA genome and associated proteins, including the enzymes reverse transcriptase and integrase. The capsid and its contents are collectively referred to as the viral core. CA itself consists of two domains (N-terminal and C-terminal domains, or NTD and CTD respectively) connected by a flexible linker (Fig. 1). The capsid is organized into hexameric and pentameric rings of CA, and the overall lattice is stabilized by several types of CA-CA interactions. The hexamers contain a central ring formed by CA-NTD interactions as well as an external ring formed by CA-CTD/CA-NTD interactions (Fig. 2). Hexamers are connected by CTD-CTD interactions at two- and three-fold symmetry axes in the lattice [reviewed in [5]]. Of the 1200 or so CA subunits in the capsid, the majority are hexamers, but the capsid also contains 12 CA pentamers (Fig. 2) that result in closure of the lattice. The presence of CA pentamers in the capsid of native HIV-1 particles has recently been confirmed by cryo-electron microscopy [4]. The placement of the pentamers determines the shape of the capsid, which is generally conical but can also be tubular or spherical.
Figure 1.
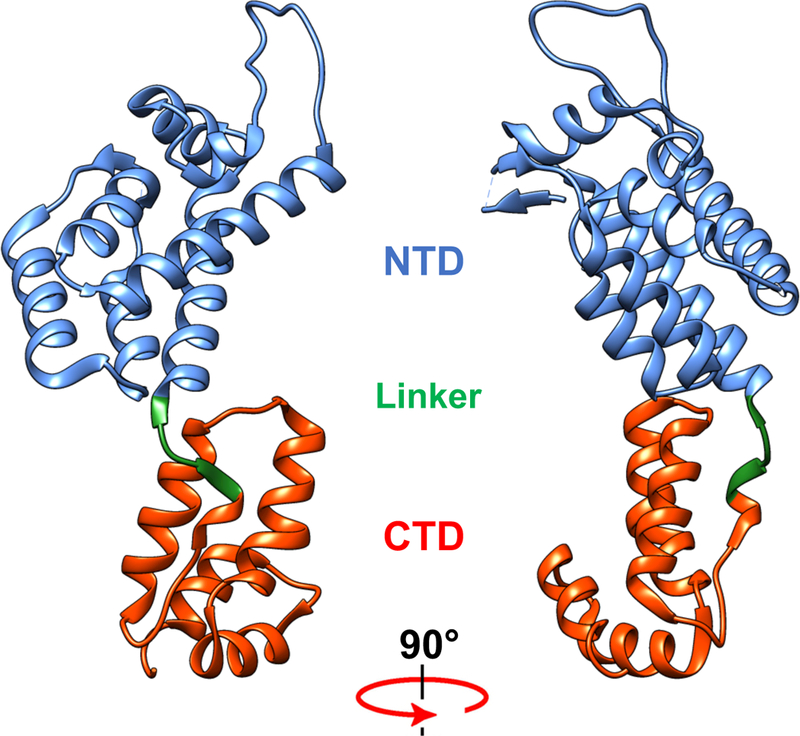
Structure of the HIV-1 CA protein. CA is shown as the conformation present within the X-ray crystal structure of the CA hexamer (4xfx.pdb)[1]. Shown are the N-terminal domain (NTD), the flexible linker, and the C-terminal domain (CTD). The structure was rendered with UCSF Chimera [2] as provided by SBGRID [3].
Figure 2.
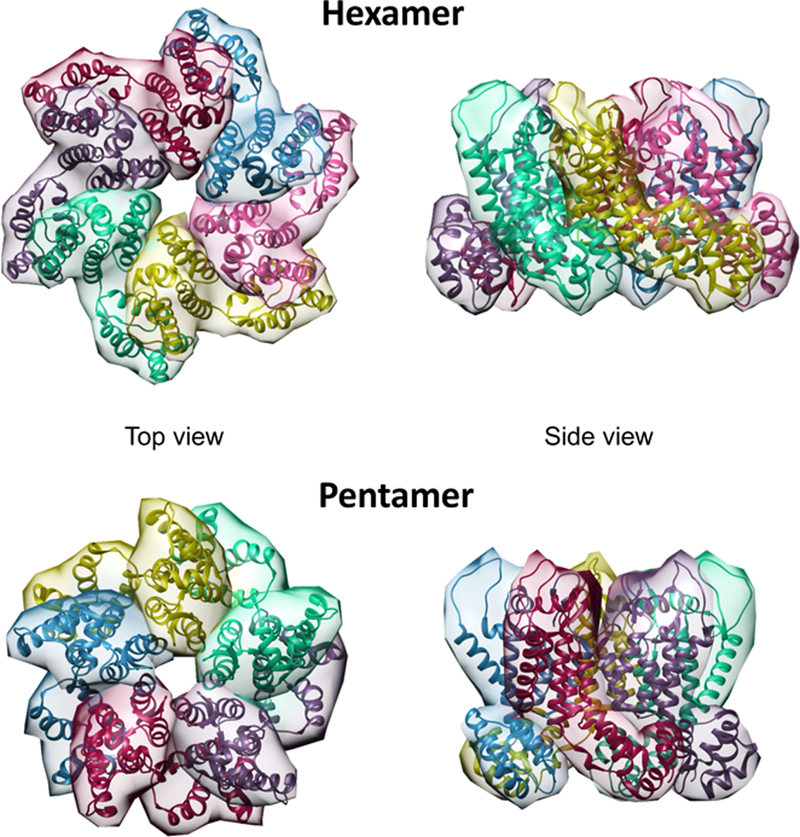
Structures of the HIV-1 CA hexamer and pentamer. The hexamer represents the X-ray crystal structure of the wild type CA hexamer (4xfx.pdb coordinates), and the pentamer was rendered from the electron tomographic structure of the native viral capsid (5mcy.pdb)[4]. The structures were rendered with Chimera’s Multiscale Models tool.
The HIV capsid is a metastable structure: mutations that either stabilize or destabilize it generally result in low viral infectivity. Following penetration into target cells, HIV must undergo reverse transcription (copying the viral RNA into DNA, intracytoplasmic transport, nuclear entry, and integration.) All of these functions appear to depend on the viral capsid. During these early post-entry steps in infection, the capsid undergoes a stepwise disassembly process known as uncoating [6–14]. Perturbations in HIV capsid stability can result in premature uncoating; this frequently results in attenuated reverse transcription and a failure to establish productive infection. During its journey to the nucleus, the HIV capsid interacts with several host proteins that determine both the efficiency of nuclear entry and the distribution of integration sites in the host genome. Most of these interactions promote infection, but the noteworthy capsid-binding host proteins TRIM5α and MxB inhibit HIV infection, further demonstrating the critical role of the viral capsid during infection. Small molecule inhibitors that bind to the capsid can perturb the stability balance of the capsid and compete for host factor binding, thereby interfering with infection.
In the late phase of the HIV-1 life cycle, the capsid must assemble and mature into its final conical structure [15]. CA is formed by proteolytic cleavage of the viral Gag structural polyprotein during virus maturation. The CA region of Gag is critical for both HIV particle assembly and maturation (core formation); therefore, CA-targeting small molecule inhibitors may act at early as well as late stages of replication. The many critical functions of CA make it highly attractive as a pharmacologic target. CA is highly sensitive to mutations, thus explaining its high sequence conservation relative to other HIV proteins. [16, 17]. For these reasons, we expect that HIV resistance to CA-targeting drugs will require mutations that compromise the fitness of the virus, thus resulting in a high barrier to resistance and excellent therapeutic durability.
Old and New CA-targeting Inhibitors
CAP-1:
The first report of a CA-targeting HIV inhibitor appeared in 2003, when Summers and coworkers identified N-(3-chloro-4-methylphenyl)-N’-{2-[({5-[dimethylamino)-methyl]-2-furyl}-methyl)-sulfanyl]ethyl}urea) (CAP-1) via a computational search for small molecules that bound to the CA-NTD. CAP-1 reduced HIV-1 replication by 95% at a concentration of 100 μM. The compound binds to CA at the base of the NTD, near the linker region (Fig. 3). NMR spectroscopy and X-ray crystallography studies have shown that CAP-1 binding alters the conformation of the NTD; Phe32 is displaced from a buried position, opening a deep hydrophobic cavity where CAP-1 resides [18]. The compound inhibits the ability of CA to self-assemble in vitro, and results in virus particles of altered size and core morphology. CAP-1 does not inhibit infection at an early step, and does not appear to affect assembled capsid complexes in vitro [21]. Rather, it acts late during particle maturation, resulting in a defective core and thus noninfectious particles [22]. HIV resistance to CAP-1 has not been reported, leaving open the possibility of a functional target other than CA. Identification of more potent inhibitors targeting the CAP-1 binding site on CA would be helpful for further mechanistic studies.
Figure 3.
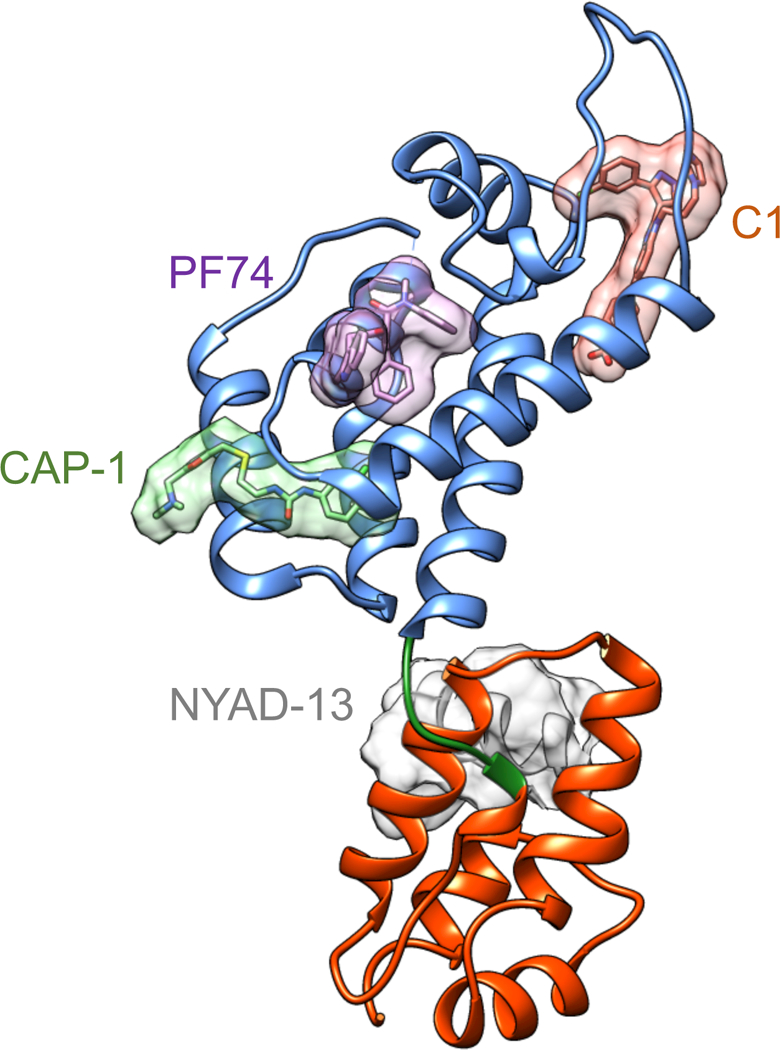
HIV-1 CA binding sites for three antiviral compounds and one peptide. C1, PF74, and CAP-1 occupy distinct sites on the NTD; the stapled peptide ligand NYAD-13, an analogue of CAI, binds to the CTD dimer interface. This figure illustrates several known binding sites for capsid-targeting HIV inhibitors and is a composite generated using the CA monomer from 4xfz.pdb [1] with the ligand coordinates aligned from 2jpr.pdb [18], 4e91.pdb [19], and 2l6e.pdb [20].
PF74:
PF-3450074 (PF74), first reported in 2010, exhibits broad spectrum inhibition of HIV isolates, with submicromolar potency (EC50 = 8–640 nM) [23–25]. High concentrations of PF74 interfere with early and late events in the virus lifecycle by destabilizing the HIV-1 capsid, resulting in premature uncoating and loss of reverse transcription and infection in early stages, and later, disrupting particle formation [25–27]. Interestingly, PF74 also stabilizes preassembled CA-NC tubes and stimulates the rate of CA self-assembly in vitro [21, 28]. This may be due to the dual effects exhibited by PF74; at concentrations lower than 2 μM, the compound competes for host protein binding to the capsid, but at 10 μM it induces premature uncoating, resulting in impaired reverse transcription [29*–31]. PF74 occupies a pocket at the NTD-CTD intersubunit interface, at which the host proteins CPSF6 and Nup153 also bind [32, 33] (Fig. 3). CPSF6 and Nup153 are two of many host proteins that enhance HIV infection by facilitating nuclear entry and/or integration [reviewed in [34]]. PF74’s antiviral activity is also influenced by another CA-binding host factor, cyclophilin A. Cyclophilin A binds the HIV-1 capsid and promotes HIV infection, but its precise mechanism is not well understood. Inhibition of CypA binding to CA in target cells reduces the antiviral activity of PF74, likely by modulating capsid structure and/or other host protein interactions with the capsid [29*]. The weight of evidence suggests that a major antiviral mechanism of PF74 is to perturb the binding of host factors to the incoming HIV capsid.
BI Compounds:
BI-1 and BI-2 are pyrrolopyrazolones that were discovered from a screen for HIV-1 replication inhibitors that stabilize capsid complexes [35]. While they both block replication in single and multiple round infections, BI-2 is more potent (EC50 values are 8.2 μM and 1.8 μM, respectively). As observed for PF74, BI-2 appears to destabilize the HIV-1 core, yet it promotes the self-assembly of capsid-like complexes of recombinant CA-NC protein in vitro [21, 27, 35]. BI-2 does not result in impaired HIV reverse transcription in target cells, but it inhibits HIV nuclear entry. BI-2 binds to the same site in the NTD as PF74; the compound is smaller, and does not make contacts with the CTD of the adjacent CA subunit [33]. Like PF74, BI-2 can influence the binding of CPSF6 and Nup153 to the viral capsid [27, 33]. Substitutions in the binding pocket (A105 and T107) conferred resistance to both BI-1 and BI-2 [35], confirming CA as the antiviral target. Overall, BI-2 appears to inhibit HIV infection by a mechanism similar to that exhibited at low concentrations of PF74.
Peptide Inhibitors:
CAI (CA inhibitor), first reported in 2005, is a CA-binding 12-mer peptide that was discovered in a phage display library screen [36]. CAI inhibits assembly of HIV-1 CA as well as a longer Gag fragment in vitro. It inhibits CA self-assembly in vitro by interacting with a conserved hydrophobic groove at the CA dimerization interface (Fig. 3), altering the structure of CA in this region. Substitutions in CA reduce the affinity for CAI and impair the maturation of particles, resulting in reduced HIV infectivity. Like many peptides, CAI cannot penetrate cell membranes [21, 36, 37], thus precluding its use as an antiviral agent. To improve its cell penetration, hydrocarbon stapling was utilized to convert CAI into a cell-penetrating peptide (named NYAD-1). NYAD-1 inhibits the assembly of immature and mature-like virus particles in vitro; it also reduces particle formation and inhibits HIV-1 maturation in cell culture. Remarkably, NYAD-1 can also target the incoming HIV capsid and inhibits infection at an early post-entry stage. NYAD-1 inhibits infection by a variety of HIV isolates with low micromolar potency (IC50=4–15 μM), making it attractive for therapeutic development [38].
C-A1:
The gyrase B inhibitor coumermycin A1 (C-A1) was observed to inhibit HIV-1 infection in a focused screen of known inhibitors targeting ATP-dependent DNA motors. CA-1 appears to have a dual antiviral effect: it reportedly inhibits viral gene expression by targeting the heat shock protein Hsp90, but it also inhibits HIV integration. C-A1 had no effect on nuclear entry or reverse transcription, indicating that it acts between nuclear entry and integration. C-A1 binds to the HIV capsid; resistance to the compound is conferred by a point mutation at CA 105, suggesting that C-A1-mediated inhibition may be partially due to a capsid-based mechanism. The CA mutant N74D is also resistant to C-A1; this mutant exhibits altered dependence on specific host factors involved in nuclear entry, suggesting a functional connection between CA-1 and HIV interactions with the nuclear pore complex. Interestingly, C-A1 also promotes the binding of CPSF6 to the capsid; CPSF6, normally a nuclear protein, blocks HIV-1 infection when it is mislocalized to the cytoplasm and may hyperstabilize the HIV-1 core, thus perturbing normal HIV uncoating. In the nucleus, however, CPSF6 promotes HIV-1 integration. These findings suggest a potential model in which C-A1 prevents CPSF6 dissociation from the viral capsid within the nucleus, potentially blocking integration [39, 40].
CK026, I-XW-053, compound 34:
CK026 was originally identified by in silico screening for ligands to the NTD-NTD intrahexamer interface [41]. CK-026 exhibited moderate antiviral potency. It is a relatively large molecule, and a smaller fragment of the compound, I-XW-053 also exhibited antiviral activity. Mechanistic studies showed that I-XW-053 interacts with CA in vitro and inhibits HIV reverse transcription in target cells. In a subsequent structure-activity relationship study, a derivative of I-XW-053 was reported to exhibit 11-fold improved antiviral activity. Howver, the actual CA binding site for these inhibitors has not been determined, and it is possible that they target additional viral proteins [42].
C1:
C1 was initially discovered as a novel inhibitor of CA assembly in vitro [43]. C1 binds to a unique site of the CA-NTD, near the base of the CypA-binding loop (Fig. 3). It inhibits HIV-1 replication by acting at a late step to disrupt proper assembly of the mature viral capsid, without altering Gag processing. C1 is inactive at the time of virus infection of target cells, indicating that the compound does not affect HIV-1 early events. C1 exhibits moderate antiviral activity (IC50 =57 μM); HIV-1 resistance can result from a single amino acid substitution within the compound binding site [19, 44*], thus confirming that CA is the bona fide antiviral target of the compound. Though the antiviral mechanism is not fully understood, C1 may act by perturbing the kinetics of CA assembly in the maturing particle, or may promote an off-target CA assembly pathway. Further exploration of this CA target should prove informative.
Ebselen:
The small molecule ebselen was recently discovered by a novel screening approach employing a time-resolved fluorescence resonance energy transfer assay to identify inhibitors of CA dimerization [45]. This assay was used to screen a library of 1280 in vivo active drugs. Ebselen is an efficient inhibitor of CA dimerization in vitro and inhibits HIV infection with moderate potency (EC50 = 3.37 μM). Ebselen inhibits HIV reverse transcription in target cells; it also results in impaired uncoating based on a cell fractionation assay. The apparent capsid stabilization could be due to aggregation of the CTD in the presence of ebselen observed in vitro by nuclear magnetic resonance spectroscopy. Ebselen does not affect particle assembly and maturation, indicating that the inhibitor acts during early stages of infection. Further studies are needed to determine whether CA is the bona fide antiviral target of ebselen.
Antibodies-
Recently, monoclonal antibody research to inhibit viral infection within cells has flourished. Plagued by the same issue of poor cellular penetration that some peptide inhibitors face, monoclonal antibodies are now being conjugated to cell-penetrating peptides to generate cell-internalizing antibodies. A recent antibody against CA was discovered to reduce HIV-1 replication up to 73% and 49% in T lymphocytes and PBMCs, respectively, at a concentration of 10 ug/ml [46]. Pretreatment of cells with the antibody was more efficient at reducing HIV replication than adding antibody after viral incubation had begun, suggesting the antibody may be working at an early stage in infection. Together, the findings suggest that the approach holds promise for future therapeutic development].
GS-CA1:
A new CA-targeting HIV inhibitor was recently described at scientific meetings [47**, 48]. The compound, currently named GS-CA1, exhibits highly antiviral potency in human peripheral blood mononuclear cells (EC50 = 140 pM) with broad spectrum inhibition across all HIV clades. GS-CA1 binds to the same broadly conserved site occupied by with PF74, lying at the NTD-CTD intersubunit interface within CA hexamers. Like PF74, GS-CA1 exhibits a dual antiviral mechanism: it acts late to reduce HIV-1 particle infectivity (presumably via perturbation of maturation); it also targets the incoming mature viral capsid to inhibit infection. GS-CA1 also binds to the same location on CA as CPSF6 and NUP153. Overall, the current view of the compound’s mechanism indicates that it acts like PF74, but with much greater potency. In vitro resistance selection experiments identified 5 amino acid substitutions in the GS-CA1 binding pocket that independently conferred resistance. However, a recent study found that none of those mutations were actively circulating or found among 132 sample patients. Studies in rats indicate that a single subcutaneous injection maintains plasma concentrations of GS-CA1 well above the plasma-binding—adjusted effective concentration required to inhibit HIV replication by 95% for greater than 10 weeks, suggesting the possibility of long-acting treatment. Plans to begin Investigational New Drug-enabling toxicology studies and Phase 1 clinical trials in 2018 were recently announced, with the goal of developing a long-acting injectable formulation [47**, 48, 49**].
Conclusion
Efforts to identify capsid-targeting HIV inhibitors date back 15 years, with a variety of approaches and types of molecules described. In the past few years, this area has gained momentum, though with setbacks resulting in discontinuation of HIV antivirals research efforts by two major pharmaceutical companies. Renewed optimism in this area has been stimulated by structural and mechanistic information from basic research, and there is hope that a clinically useful HIV capsid inhibitor will be approved for therapy within the next several years.
Key Points.
The HIV capsid is an attractive therapeutic target owing to its high sequence conservation and multiple functions in replication.
The viral CA protein can be bound by small molecules at several positions in its structure, and CA-binding compounds can interfere with both early and late steps in the HIV replication cycle.
Although a variety of small molecule HIV inhibitors that bind to HIV-1 CA have been discovered, only for a few of these has CA been confirmed as the functional antiviral target.
The potent small molecule capsid-targeting inhibitor GS-CA1 recently described is a promising candidate for therapeutic development.
Acknowledgments
We thank the current and former members of the Aiken lab who have contributed to our understanding of the structure and function of the HIV capsid.
Financial Support and Sponsorship
This work was supported in part by by NIH grants R01AI114339 (C.A.) and F31AI129747 (S.K.C.).
Footnotes
Conflicts of Interest
The authors have no conflicts of interest.
References
- 1.Gres AT, Kirby KA, KewalRamani VN, et al. STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349(6243):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. [DOI] [PubMed] [Google Scholar]
- 3.Morin A, Eisenbraun B, Key J, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattei S, Glass B, Hagen WJ, et al. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science. 2016;354(6318):1434–7. [DOI] [PubMed] [Google Scholar]
- 5.Byeon IJ, Meng X, Jung J, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139(4):780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamede JI, Cianci GC, Anderson MR, et al. Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc Natl Acad Sci U S A. 2017;114(34):E7169–E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis AC, Marin M, Shi J, et al. Time-Resolved Imaging of Single HIV-1 Uncoating In Vitro and in Living Cells. PLoS Pathog. 2016;12(6):e1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A. 2011;108(24):9975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankovic S, Varadarajan J, Ramalho R, et al. Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J Virol. 2017;91(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramalho R, Rankovic S, Zhou J, et al. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology. 2016;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arfi V, Lienard J, Nguyen XN, et al. Characterization of the behavior of functional viral genomes during the early steps of human immunodeficiency virus type 1 infection. J Virol. 2009;83(15):7524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosnefroy O, Murray PJ, Bishop KN. HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology. 2016;13(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da Silva Santos C, Tartour K, Cimarelli A. A Novel Entry/Uncoating Assay Reveals the Presence of at Least Two Species of Viral Capsids During Synchronized HIV-1 Infection. PLoS Pathog. 2016;12(9):e1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forshey BM, von Schwedler U, Sundquist WI, et al. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76(11):5667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol. 2015;13(8):484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rihn SJ, Wilson SJ, Loman NJ, et al. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013;9(6):e1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Verheyen J, Rhee SY, et al. Functional conservation of HIV-1 Gag: implications for rational drug design. Retrovirology. 2013;10:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly BN, Kyere S, Kinde I, et al. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373(2):355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemke CT, Titolo S, von Schwedler U, et al. Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein. J Virol. 2012;86(12):6643–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya S, Zhang H, Debnath AK, et al. Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid. J Biol Chem. 2008;283(24):16274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fricke T, Brandariz-Nunez A, Wang X, et al. Human cytosolic extracts stabilize the HIV-1 core. J Virol. 2013;87(19):10587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang C, Loeliger E, Kinde I, et al. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327(5):1013–20. [DOI] [PubMed] [Google Scholar]
- 23.Blair WS, Isaacson J, Li X, et al. A novel HIV-1 antiviral high throughput screening approach for the discovery of HIV-1 inhibitors. Antiviral Res. 2005;65(2):107–16. [DOI] [PubMed] [Google Scholar]
- 24.Rasaiyaah J, Tan CP, Fletcher AJ, et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503(7476):402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blair WS, Pickford C, Irving SL, et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010;6(12):e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J, Zhou J, Shah VB, et al. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol. 2011;85(1):542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricke T, Buffone C, Opp S, et al. BI-2 destabilizes HIV-1 cores during infection and Prevents Binding of CPSF6 to the HIV-1 Capsid. Retrovirology. 2014;11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lad L, Clancy S, Koditek D, et al. Functional label-free assays for characterizing the in vitro mechanism of action of small molecule modulators of capsid assembly. Biochemistry. 2015;54(13):2240–8. [DOI] [PubMed] [Google Scholar]
- 29.Saito A, Ferhadian D, Sowd GA, et al. Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74. J Virol. 2016;90(12):5808–23.* This study reported that the dose-response curve of PF74 consists of two distinct inhibitory phases that are regulated by CA-interacting host proteins. This paper provides novel insights into the mechanism of action of PF74 and the roles of host factors during HIV-1 early infection.
- 30.Peng K, Muranyi W, Glass B, et al. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife. 2014;3:e04114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matreyek KA, Yucel SS, Li X, et al. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 2013;9(10):e1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharya A, Alam SL, Fricke T, et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A. 2014;111(52):18625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AJ, Jacques DA, McEwan WA, et al. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog. 2014;10(10):e1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita M, Engelman AN. Capsid-Dependent Host Factors in HIV-1 Infection. Trends Microbiol. 2017;25(9):741–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamorte L, Titolo S, Lemke CT, et al. Discovery of novel small-molecule HIV-1 replication inhibitors that stabilize capsid complexes. Antimicrob Agents Chemother. 2013;57(10):4622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sticht J, Humbert M, Findlow S, et al. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12(8):671–7. [DOI] [PubMed] [Google Scholar]
- 37.Barklis E, Alfadhli A, McQuaw C, et al. Characterization of the in vitro HIV-1 capsid assembly pathway. J Mol Biol. 2009;387(2):376–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Zhao Q, Bhattacharya S, et al. A cell-penetrating helical peptide as a potential HIV-1 inhibitor. J Mol Biol. 2008;378(3):565–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen NY, Zhou L, Gane PJ, et al. HIV-1 capsid is involved in post-nuclear entry steps. Retrovirology. 2016;13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vozzolo L, Loh B, Gane PJ, et al. Gyrase B inhibitor impairs HIV-1 replication by targeting Hsp90 and the capsid protein. J Biol Chem. 2010;285(50):39314–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kortagere S, Madani N, Mankowski MK, et al. Inhibiting early-stage events in HIV-1 replication by small-molecule targeting of the HIV-1 capsid. J Virol. 2012;86(16):8472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kortagere S, Xu JP, Mankowski MK, et al. Structure-activity relationships of a novel capsid targeted inhibitor of HIV-1 replication. J Chem Inf Model. 2014;54(11):3080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemke CT, Titolo S, Goudreau N, et al. A novel inhibitor-binding site on the HIV-1 capsid N-terminal domain leads to improved crystallization via compound-mediated dimerization. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 6):1115–23. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Zhou J, Halambage UD, et al. Inhibition of HIV-1 Maturation via Small-Molecule Targeting of the Amino-Terminal Domain in the Viral Capsid Protein. J Virol. 2017;91(9).* The authors reported that the small molecule inhibitor Compound 1 interferes with capsid assembly during virion maturation, and that a single amino acid mutation in CA confers resistance. This study describes a novel mechanism by which a capsid-targeting small molecule can inhibit HIV-1 replication.
- 45.Thenin-Houssier S, de Vera IM, Pedro-Rosa L, et al. Ebselen, a Small-Molecule Capsid Inhibitor of HIV-1 Replication. Antimicrob Agents Chemother. 2016;60(4):2195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali SA, Teow SY, Omar TC, et al. A Cell Internalizing Antibody Targeting Capsid Protein (p24) Inhibits the Replication of HIV-1 in T Cells Lines and PBMCs: A Proof of Concept Study. PLoS One. 2016;11(1):e0145986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tse W, Link J, Mulato A, et al. Discovery of Novel Potent HIV Capsid Inhibitors with Long-acting Potential. Conference on Retroviruses and Opportunistic Infections. 2017. Abstract 38- New HIV Drugs, Formulations, Combinations, and Resistance.** This presentation described GS-CA1, a highly potent HIV inhibitor that binds to the same site as PF74 and acts by a similar mechanism. Gilead Sciences will undertake clinical trials of the GS-CA1 in 2018.
- 48.Jarvis L Conquering HIV’s capsid. Chemical and Engineering News. 2017; 95(31): 23–25. [Google Scholar]
- 49.Perrier M, Bertine M, Le Hingrat Q, et al. Prevalence of gag mutations associated with in vitro resistance to capsid inhibitor GS-CA1 in HIV-1 antiretroviral-naive patients. J Antimicrob Chemother. 2017;72(10):2954–5.** This paper outlines the GS-CA1 resistance mutants in CA. The authors examined the occurrence of those mutations in HIV-infected individuals, and none of the HIV mutations conferring resistance to GS-CA1 were observed. The absence of mutations in circulating HIV strains suggests that the efficacy of GS-CA1 will not be limited by pre-existing viral polymorphisms.


