Abstract
A comprehensive understanding of wheat responses to environmental stress will contribute to the long-term goal of feeding the planet. ALERNATIVE OXIDASE (AOX) genes encode proteins involved in a bypass of the electron transport chain and are also known to be involved in stress tolerance in multiple species. Here, we report the identification and characterization of the AOX gene family in diploid and hexaploid wheat. Four genes each were found in the diploid ancestors Triticum urartu, and Aegilops tauschii, and three in Aegilops speltoides. In hexaploid wheat (Triticum aestivum), 20 genes were identified, some with multiple splice variants, corresponding to a total of 24 proteins for those with observed transcription and translation. These proteins were classified as AOX1a, AOX1c, AOX1e or AOX1d via phylogenetic analysis. Proteins lacking most or all signature AOX motifs were assigned to putative regulatory roles. Analysis of protein-targeting sequences suggests mixed localization to the mitochondria and other organelles. In comparison to the most studied AOX from Trypanosoma brucei, there were amino acid substitutions at critical functional domains indicating possible role divergence in wheat or grasses in general. In hexaploid wheat, AOX genes were expressed at specific developmental stages as well as in response to both biotic and abiotic stresses such as fungal pathogens, heat and drought. These AOX expression patterns suggest a highly regulated and diverse transcription and expression system. The insights gained provide a framework for the continued and expanded study of AOX genes in wheat for stress tolerance through breeding new varieties, as well as resistance to AOX-targeted herbicides, all of which can ultimately be used synergistically to improve crop yield.
Introduction
Bread wheat (Triticum aestivum) feeds a significant portion of the world’s population and there has been substantial progress on boosting supply to meet the global increase in demand [1, 2]. While worldwide production and yield of wheat have gradually increased over the past decade (http://statistics.amis-outlook.org/data/index.html), these gains may be offset by predicted harvest losses to global climate change and a reduction in arable land [3, 4]. In addition to the decline in production quantity, the impending and growing environmental stress expected to cause a deterioration in wheat quality [5–7].
Stress response pathways in plants trigger changes in hormone biosynthesis, transcriptional activity and metabolic responses that are crucial for maintaining structural and functional integrity [8, 9]. One key biological component of plant metabolism and stress responses is the mitochondrion, which is the site of an ATP-generative electron shuffle involving multiple cytochrome oxidase and dehydrogenase complexes. Upon stress perception, electrons can also be shunted to an alternative oxidase, which is proposed to dissipate the energy as heat, reduces oxygen to water and limit reactive oxygen species (ROS) production [10–12]. In fact, thermogenic plants use ALTERNATIVE OXIDASE (AOX) to produce heat during respiration to facilitate pollen germination and to volatilize pollination attractants [13]. The AOX gene was first cloned from the thermogenic plant Sauromatum guttatum [14, 15]. Antibodies for this protein cross-reacted with similar proteins from non-thermogenic plants and this facilitated the study of these terminal oxidases in other species. With the availability of sequenced genomes and numerous molecular techniques, AOX genes have been identified and in some cases, functionally characterized in both dicots and monocots such as Arabidopsis thaliana, tobacco, carrot, mango, stone pine, cowpea, chickpea, barley, rice and maize [16–27]. AOX genes fall into two discrete subfamilies, Type 1 and Type 2. The former is present in both monocot and dicot species while the latter has so far only been found in dicots but is purported to have existed in ancient monocots [16, 17, 28]. AOX1 (Type 1 AOX) genes are very responsive to stresses as well as irregularities in respiratory metabolism [29–31]. The AOX2 (Type 2 AOX) genes control developmental processes such as germination, fertility and vegetative growth, but there is also some evidence for a role in stress response [32–35].
The initial cloning of two AOX genes from wheat [36] spurred a considerable amount of biochemical work and some expression studies indicating they are involved in numerous developmental processes as well as responses to stress [37–51]. On the genomic level, the number and spatiotemporal expression patterns of AOX genes has remained unclear in wheat. The availability of the wheat genome now makes it possible to conduct a genome-wide examination of the AOX family in the hexaploid and ancestral diploid species of this important monocot [52–54]. The current study investigated and identified the AOX gene family in hexaploid wheat and its diploid ancestors in the A (Triticum urartu), B (Aegilops speltoides) and D (Aegilops tauschii) subgenomes. Using multiple in silico resources and the latest transcriptome database [55], features such as phylogenetic evolutionary relationships, chromosomal locations, gene structures, promoter cis-elements, conserved motifs, subcellular localization and expression patterns were evaluated. Our findings provide a better understanding of the wheat AOX family members, promote our understanding of the regulation of this gene family and lay the groundwork for future study of AOX in wheat.
Materials and methods
Identification of the AOX gene family in wheat
The amino acid sequences of Waox1a and Waox1c (Genbank ID BAB88645.1 and BAB88646.1) were used in a BLASTP search (E-value threshold 1 e-1) on Ensembl Plants (http://plants.ensembl.org/index.html) and the International Wheat Genome Sequencing Consortium (IWGSC) URGI portal (https://urgi.versailles.inra.fr/blast/) in May 2018 [56, 57]. Concurrently the coding sequences of Waox1a and Waox1c (Genbank ID AB078882.1 and AB078883.1) were used in a BLASTN search (E-value threshold 10) on the aforementioned databases. Nucleotide sequences and unique protein IDs of matching sequences were obtained for T. aestivum as well as T. urartu (A subgenome), A. speltoides (B subgenome) and A. tauschii (D subgenome). The output from Ensembl Plants was classified by the databank as either high-confidence indicating that the data was fully supported by PacBio transcript sequencing as well as RNA-seq data, or low-confidence for sequences which had partial or no transcriptome data support (http://plants.ensembl.org/Triticum_aestivum/Info/Annotation/ - genebuild).
Phylogeny
The amino acid sequences of the AOX proteins from wheat, barley, Brachypodium distachyon, rice, and maize obtained from Ensembl Plants (http://plants.ensembl.org/index.html) [56], Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) [58] and the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) [59] were aligned with representative sequences from other monocots (Anasus comosus, Asparagus officinalis, Musa acuminata, Oropetalum thomaeum, Panicum virgatum, Spirodela polyrhiza, Symplocarpus renifolius, Zostera marina) using MUSCLE [60]. The amino acid alignment was analyzed using maximum likelihood (ML) with RAxML (7.7.1) [61] implementing the GTR-Gamma model and JTT substitution matrix with 100 bootstrap replicates. Bayesian inference (BI) analyses implementing a mixed AA model prior in MrBayes 3.2.2 [62] were run over 50 million generations with the first 25% removed for burn-in and assessed for convergence and stationarity using average standard deviation of split frequencies, potential scale reduction factor (PSRF) values approaching 1.0, and a large effective sample size assessed in Tracer v.1.4.1 [63].
Gene structure and protein analyses
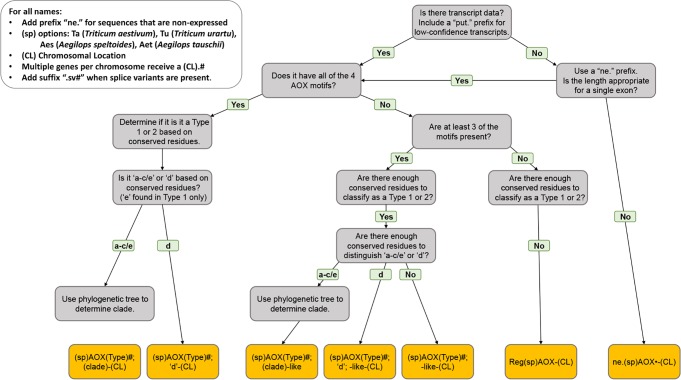
The coding sequence of each AOX gene was aligned with the genomic sequence in order to delineate the intron/exon boundaries using the Gene Structure Display Server program (http://gsds.cbi.pku.edu.cn/) [64]. Alignment of the protein sequences to search for relevant motifs and residues was done using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) [65] and the results used in subclassification via a protocol described by previous researchers [16, 66]. When all four motifs were present in high-confidence protein sequences, it was designated an AOX. Furthermore, when a motif was absent, the protein was given the suffix “-like”, and when all motifs were missing the corresponding gene was proposed to have a putative regulatory function and given the prefix “reg”. The low-confidence proteins with all motifs were given the prefix “put” (putative), those missing a motif were given the prefix “put” and the suffix “-like” and those with no motifs received the prefix “put.reg”. In some cases, the proteins were given the prefix “ne” for “non-expressed” to indicate a complete lack of transcript data but a similarity to a particular class of AOX proteins. Where the non-expressed protein bore no resemblance to a particular subclassification, the suffix symbol “•” was added as a stand-in for a future subclassification pending the availability of transcript data. For all hexaploid proteins, an indication was made of the chromosomal location of the corresponding gene provided by the Ensembl Plants database. The genes with splice variants were given the alphanumerical suffix “sv” followed by a number. In order to determine the orientation of genes on the same chromosomal arm, the sequences we obtained were aligned to the respective arms via SnapGene. A Needleman-Wunsch alignment was performed to determine transcript and protein percent identities using the Global Align program from NCBI with default parameters. The subcellular localization was predicted using TargetP (http://www.cbs.dtu.dk/services/TargetP/) [67] and putative protein modification sites were predicted using the Plant Protein Phosphorylation Database (http://www.p3db.org/index.php) and Musite (http://musite.net/) [68, 69] with a threshold score of 0.5. CpG islands in the gene body were determined using Cpgplot available in the European Molecular Biology Open Software Suite (EMBOSS) (http://www.bioinformatics.nl/cgi-bin/emboss/cpgplot) [70].
Promoter analyses
To identify cis-elements needed for various developmental cell functions as well as binding motifs for known regulators of AOX expression [71], 1500 bp upstream of the translation start site of the wheat AOX sequences was analyzed using plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), PlantPan2.0 (http://plantpan2.itps.ncku.edu.tw/index.html) and the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) [72–74]. CpG islands in the promoter region were found using Cpgplot available in EMBOSS (http://www.bioinformatics.nl/cgi-bin/emboss/cpgplot) [70].
Molecular modeling
The three-dimensional structures of the wheat AOX proteins were obtained via modeling to solved protein structures, using the Protein Homology/Analogy Recognition Engine version 2.0 server (Phyre2) [75]. This server was also used to predict the transmembrane topology of AOX proteins in the diploid and hexaploid and wheat species. Modeling and residues involved in the diiron center of the AOX proteins were visualized using Chimera (http://www.rbvi.ucsf.edu/chimera/) [76].
RNA expression analyses
The expression patterns of the hexaploid wheat AOX genes were obtained from the publicly available RNA-seq data from the wheat variety Chinese Spring on expVIP (http://www.wheat-expression.com/) [55]. The relative transcript abundance data from the seedling, vegetative and reproductive stages of development and over multiple tissue types were used to generate heat maps in order to visualize the similarities and differences in the TaAOX family. As previously described [77], the expression ratio for a given treatment compared to the control was used to generate heat maps for the transcripts under biotic and abiotic stress (https://www.rdocumentation.org/packages/gplots/versions/3.0.1/topics/heatmap.2).
Results and discussion
Identification and classification of the AOX gene family in wheat
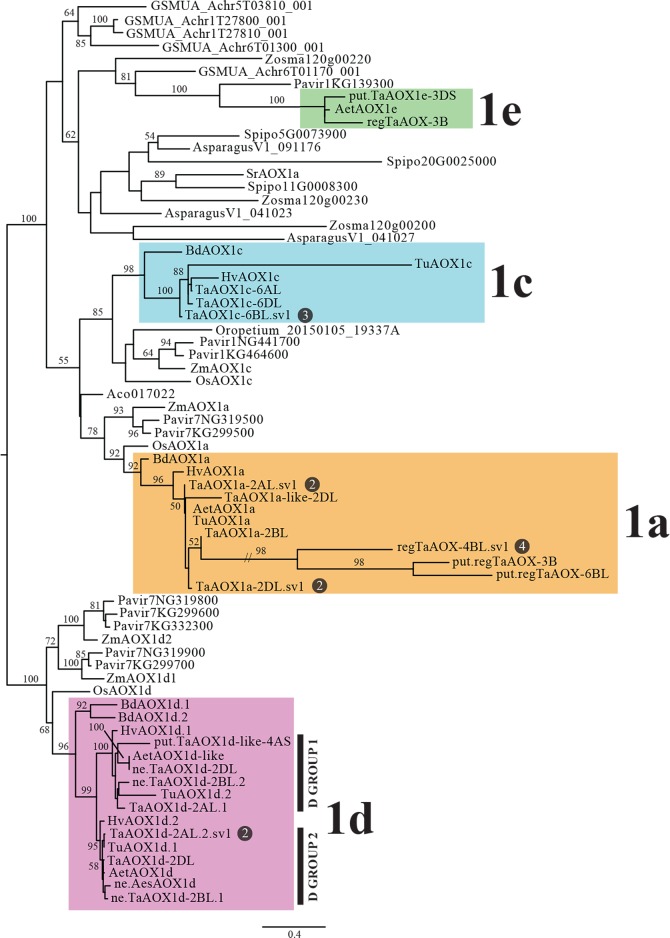
Two AOX coding sequences were previously cloned from wheat and named Waox1a and Waox1c [36]. These sequences as well as the Waox1a and Waox1c protein sequences were used in BLAST searches in order to identify additional wheat AOX genes and proteins. The obtained nucleotide sequences broadly fell into three groups, high-confidence where there was ample transcriptome and RNA-seq data, low-confidence where there was partial or no transcriptome data, and non-expressed where there was nucleotide similarity but no transcript data (Table 1). Based on previous work, all the corresponding proteins of AOX genes show a trend of unique motifs and residues known to dictate functionality [16, 66]. Therefore, in order to classify these genes and the corresponding proteins, the protein sequences were used in phylogenetic analysis resulting in the categorization into the clades AOX1a, AOX1c or AOX1e or AOX1d (Figs 1, S1 and S2; S1 Table). The outcomes were also supported by the specific AOX motifs and subclassification residues peculiar to each clade. Subsequently, the proteins and corresponding genes were named with additional indicators of chromosomal locations where necessary (Figs 1 and 2 and Tables 1 and 2 and S1 and S3 Figs and S2 Table). It must be noted that the residues of the wheat AOX proteins deviated in some cases from the observed residues thought to be highly conserved in the clades in other monocot species (Table 2) [16].
Table 1. Summary of all accession numbers of the AOX gene family in wheat done via a BLASTN search.
| Promoter | Classification | Ensembl Plants Gene ID |
| TaAOX1a-2AL | TaAOX1a-2AL.sv1 | TRIAE_CS42_2AL_TGACv1_093624_AA0283900.1 |
| TaAOX1a-2AL.sv2 | TRIAE_CS42_2AL_TGACv1_093624_AA0283900.2 | |
| TaAOX1a-2BL | TaAOX1a-2BL | TRIAE_CS42_2BL_TGACv1_132767_AA0439680.1 |
| TaAOX1a-2DL | TaAOX1a-2DL.sv1 | TRIAE_CS42_2DL_TGACv1_159044_AA0531270.1 |
| TaAOX1a-2DL.sv2 | TRIAE_CS42_2DL_TGACv1_159044_AA0531270.2 | |
| TaAOX1a-like-2DL | TaAOX1a-like-2DL | TRIAE_CS42_2DL_TGACv1_160367_AA0549780.1 |
| regTaAOX-4BL | regTaAOX-4BL.sv1 | TRIAE_CS42_4BL_TGACv1_321481_AA1061160.1 |
| regTaAOX-4BL.sv2 | TRIAE_CS42_4BL_TGACv1_321481_AA1061160.2 | |
| regTaAOX-4BL.sv3 | TRIAE_CS42_4BL_TGACv1_321481_AA1061160.3 | |
| regTaAOX-4BL.sv4 | TRIAE_CS42_4BL_TGACv1_321481_AA1061160.4 | |
| put.regTaAOX-3B | put.regTaAOX-3B | TRIAE_CS42_3B_TGACv1_221271_AA0735840 |
| put.regTaAOX-6BL | put.regTaAOX-6BL | TRIAE_CS42_6BL_TGACv1_499339_AA1578450 |
| TaAOX1c-6AL | TaAOX1c-6AL | TRIAE_CS42_6AL_TGACv1_471250_AA1505530.1 |
| TaAOX1c-6BL | TaAOX1c-6BL.sv1 | TRIAE_CS42_6BL_TGACv1_499881_AA1593950.1 |
| TaAOX1c-6BL.sv2 | TRIAE_CS42_6BL_TGACv1_499881_AA1593950.2 | |
| TaAOX1c-6BL.sv3 | TRIAE_CS42_6BL_TGACv1_499881_AA1593950.3 | |
| TaAOX1c-6DL | TaAOX1c-6DL | TRIAE_CS42_6DL_TGACv1_528632_AA1715280.1 |
| regTaAOX-3B | regTaAOX-3B | TRIAE_CS42_3B_TGACv1_221946_AA0753740.1 |
| put.TaAOX1e-3DS | put.TaAOX1e-3DS | TRIAE_CS42_3DS_TGACv1_271978_AA0912170 |
| TaAOX1d-2AL.1 | TaAOX1d-2AL.1 | TRIAE_CS42_2AL_TGACv1_094717_AA0302070.1 |
| TaAOX1d-2AL.2 | TaAOX1d-2AL.2.sv1 | TRIAE_CS42_2AL_TGACv1_093545_AA0282360.1 |
| TaAOX1d-2AL.2.sv2 | TRIAE_CS42_2AL_TGACv1_093545_AA0282360.2 | |
| TaAOX1d-2DL | TaAOX1d-2DL | TRIAE_CS42_2DL_TGACv1_162315_AA0562440.1 |
| put.TaAOX1d-like-4AS | put.TaAOX1d-like-4AS | TRIAE_CS42_4AS_TGACv1_308389_AA1027660 |
| TuAOX1d.1* | TuAOX1d.1* | TRIUR3_12374 |
| TuAOX1d.2* | TuAOX1d.2* | TRIUR3_19476 |
| TuAOX1c* | TuAOX1c* | TRIUR3_08189 |
| TuAOX1a* | TuAOX1a* | TRIUR3_10307 |
| AetAOX1d* | AetAOX1d* | F775_18387 |
| AetAOX1d-like* | AetAOX1d-like* | F775_43125 |
| AetAOX1e* | AetAOX1e* | F775_11948 |
| AetAOX1a* | AetAOX1a* | F775_17784 |
| Non-Expressed | ||
| Promoter | Classification | Ensembl Plants/IWGSC Fragment Location |
| N/A | ne.TaAOX1d-2BL.1 | RC.TGACv1_scaffold_129474_2BL:235,137–236,895 |
| N/A | ne.TaAOX1d-2BL.2 | RC.TGACv1_scaffold_129474_2BL:226867–227725 |
| N/A | ne.TaAOX1d-2DL | RC.TGACv1_scaffold_160654_2DL:15,057–16,369 |
| N/A | ne.TaAOX•-2AL | RC.TGACv1_scaffold_093545_2AL:13252–14775 |
| N/A | Ta.Fragment-7BL | TGACv1_scaffold_576971_7BL:58792–58921 |
| N/A | Tu.Fragment* | C163670370 1–226 |
| N/A | ne.AesAOX1d* | RC.TGAC_WGS_speltoides_v1_contig_403763 |
| N/A | ne.AesAOX•* | RC.TGAC_WGS_speltoides_v1_contig_239141 |
| N/A | ne.AesAOX•* | TGAC_WGS_speltoides_v1_contig_195745 |
| N/A | Fragment* | TGAC_WGS_speltoides_v1_contig_1601667 |
| N/A | Fragment* | TGAC_WGS_speltoides_v1_contig_1653744 |
| N/A | Fragment* | RC.TGAC_WGS_speltoides_v1_contig_2863348 |
| N/A | Aet.Fragment* | RC.C137891329 48–258 |
| N/A | Aet.Fragment* | RC.scaffold67708 39107–39328 |
| N/A | Aet.Fragment* | RC.scaffold94414 27790–27998 |
RC prefix designates sequences that were reverse-complemented in order to achieve AOX sequence identity.
*Indicates diploid promoters or protein isoforms.
Fig 1. Maximum likelihood (ML) phylogeny of AOX.
Numbers on branches are ML bootstrap percentages. The number of splice variant isomers for a protein are denoted in the dark gray circle when applicable. Colored boxes distinguish the different AOX clades.
Fig 2. Summary of protocol for AOX protein classification in wheat.
Table 2. Classification of wheat AOX proteins using Arabidopsis AOX1a as reference.
| Protein Name | Type 1 or Type 2 Residues | Type 1 (a-c/e) or Type 2(d) Residues | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 112 | 124 | 229 | 233 | 241 | 167 | 175 | 178 | 180 | 181 | 295 | |
| TaAOX1a-2AL.sv1 | |||||||||||
| TaAOX1a-2AL.sv2 | |||||||||||
| TaAOX1a-2BL | |||||||||||
| TaAOX1a-2DL.sv1 | |||||||||||
| TaAOX1a-2DL.sv2 | |||||||||||
| TaAOX1a-like-2DL | |||||||||||
| regTaAOX-4BL.sv1 | |||||||||||
| regTaAOX-4BL.sv2 | |||||||||||
| regTaAOX-4BL.sv3 | |||||||||||
| regTaAOX-4BL.sv4 | |||||||||||
| put.regTaAOX-3B | |||||||||||
| put.regTaAOX-6BL | |||||||||||
| TaAOX1c-6AL | |||||||||||
| TaAOX1c-6BL.sv1 | |||||||||||
| TaAOX1c-6BL.sv2 | |||||||||||
| TaAOX1c-6BL.sv3 | |||||||||||
| TaAOX1c-6DL | |||||||||||
| regTaAOX-3B | |||||||||||
| put.TaAOX1e-3DS | |||||||||||
| TaAOX1d-2AL.2.sv1 | |||||||||||
| TaAOX1d-2AL.2.sv2 | |||||||||||
| TaAOX1d-2AL.1 | |||||||||||
| TaAOX1d-2DL | |||||||||||
| put.TaAOX1d-like-4AS | |||||||||||
| TuAOX1a* | |||||||||||
| TuAOX1c* | |||||||||||
| TuAOX1d.1* | |||||||||||
| TuAOX1d.2* | |||||||||||
| AetAOX1a* | |||||||||||
| AetAOX1e* | |||||||||||
| AetAOX1d* | |||||||||||
| AetAOX1d-like* | |||||||||||
| ne.TaAOX1d-2BL.1 | |||||||||||
| ne.TaAOX1d-2BL.2 | |||||||||||
| ne.TaAOX1d-2DL | |||||||||||
| ne.AesAOX1d* | |||||||||||
Residues used are from Costa et al. 2014. Blue indicates presence of Type 1 residues. Red indicates a Type 2 residue. Green indicates residues for monocot Type 1(d). Yellow indicates Type 1(a-c/e). Purple represents amino acid residues that did not match either classification. Black represents residues that were absent.
*Denotes diploid wheat AOX proteins.
Consequently, 12 high-confidence AOX genes and four low-confidence genes were found in hexaploid wheat (Table 1 and S4 Fig). Four AOX genes were found in each of the two A and D subgenome diploid ancestors, T. urartu and A. tauschii. Four non-expressed hexaploid AOX genes and three non-expressed A. speltoides genes were also found. For all the genomes, gene fragments were found and these have been documented (Table 1). The genomes of the diploids are still being resolved and thus it is possible that with further work, some of these fragments discovered would be shown to be part of complete gene sequences. The current study focused on elucidating the information from both high- and low-confidence genes with the caveat that future work could resolve the low-confidence data provided. No analysis was done on the non-expressed genes beyond the phylogeny and protein classification as we have no experimental support for final transcript or translation features. These non-expressed sequences may be transcribed as given, or may undergo intronizations to give sequences which fall into the “-like” or regulatory categories. The artificial non-expressed protein sequences used in the phylogeny give an indication of evolutionary relationships and may indicate function if the protein form is maintained as we assume. However, it is possible that once transcribed and translated, putative intronizations may change the final structure. This simulated use of the non-expressed sequences was only possible for AOX1d as the “ne” nucleotide and protein sequences showed a strong alignment to the full coding regions and protein sequences of high-confidence TaAOX1d genes (S5 and S6 Figs). The assumption made was further validated by the observation that these AOX1d “ne” proteins grouped in the AOX1d clade in the phylogeny, an analysis also supported by all the required residues to meet this subclassification (Figs 1 and S1 and Table 2;) [16]. The other non-expressed genes in the AOX1a, AOX1c or AOX1e clades are multiexonic and therefore it was impossible to artificially determine intron exon boundaries and extrapolate the putative protein sequences.
With the exception of a few amino acid substitutions and an insertion, the proteins which were used in the initial search Waox1a (BAB88645.1) and Waox1c (BAB88646.1) were found to align most closely to TaAOX1a-2AL.sv1 and TaAOX1c-6AL respectively (S7 Fig). The substitutions may be due to a varietal difference since the Waox1a and Waox1c proteins were obtained from the wheat variety Mironovskaya 808 [36], whereas sequences from Chinese Spring have been used in this study. It must also be noted that the chromosomal locations noted by the previous researchers of the coding sequences Waox1a (AB078882.1) and Waox1c (AB078883.1) match that of TaAOX1a-2AL.sv1 and TaAOX1c-6AL respectively [36].
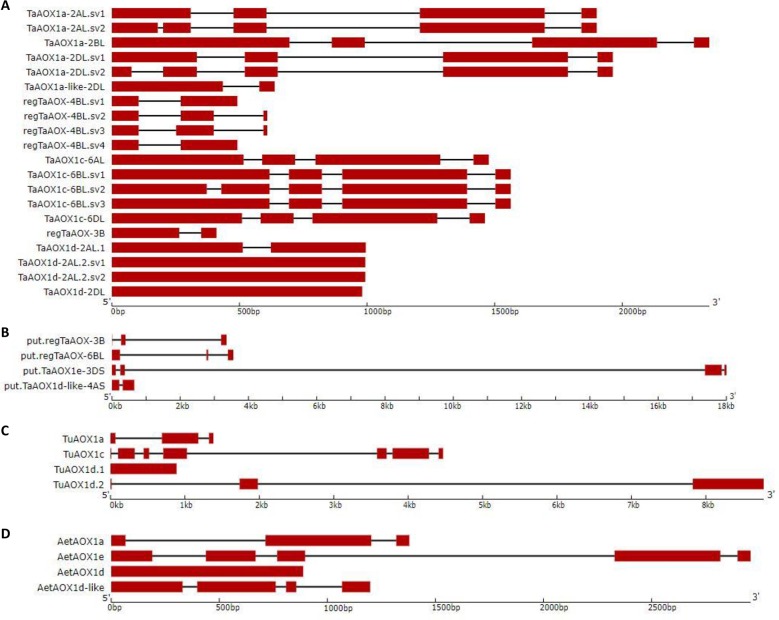
Hexaploid AOX transcripts ranged in size from 1180 bp to 3274 bp with coding regions between 249 bp and 1374 bp (Table 3 and S4 Fig). The notable exception was put.TaAOX1e-3DS where intron 2 was almost 17000 bp. There were splice variants in some AOX genes resulting from 5’ and 3’ alternative splice sites as well as intronizations (Table 4). Within the hexaploid coding regions, it was observed that the AOX1a clade generally had the longest introns while the AOX1d clade genes were intronless or single-intron (Fig 3). Given the few occurrences genes in the AOX1e clade, it was to conclude any gene structure patterns. The transcripts and coding sequences of the diploid AOX genes spanned 615 bp to 1305 bp (Table 3). There were one to six exons in the transcripts giving one to five exons within the start and stop codons (Fig 3 and Table 3). There was at least one gene from each of the diploid AOX1d clade which was also monoexonic within the coding regions, mirroring what was observed in the hexaploids (Fig 3 and Table 3). In contrast to the multi-exonic nature of most AOX genes described in the literature from a variety of species [27, 78, 79], it seems that hexaploid wheat shows clade-dependent differences in gene structures (Fig 3 and Table 3). In yeast, moss, A. thaliana, mice, rice and switchgrass it has been suggested that genes with fewer introns are rapidly activated or are highly responsive to environmental changes or stress [78, 80–82]. The simplicity of the gene structure denoted by the presence of few or no introns leads to faster processivity of the pre-mRNA, which in turn leads to faster accumulation of the protein. In contrast, other researchers have found that highly expressed genes had more complex gene organization as indicated by more and longer introns in both A. thaliana and rice [83, 84]. The expression strategies described above are both possible in the wheat AOX genes given the variation in gene structures between the clades and may be suggestive of a mosaic pattern of expression.
Table 3. Features of AOX genes in hexaploid and diploid wheat.
| Gene Name | Length of Gene (bp) | Transcript Length (bp) | Coding Sequence (bp) | # of Exons | # of Introns | |
|---|---|---|---|---|---|---|
| Hexaploid | TaAOX1a-2AL.sv1 | 2369 | 1456 | 987 | 4 | 3 |
| TaAOX1a-2AL.sv2 | 2369 | 1432 | 963 | 5 | 4 | |
| TaAOX1a-2BL | 6169 | 2102 | 1374 | 4 | 3 | |
| TaAOX1a-2DL.sv1 | 2419 | 1467 | 1011 | 4 | 3 | |
| TaAOX1a-2DL.sv2 | 2419 | 1341 | 885 | 5 | 4 | |
| TaAOX1a-like-2DL | 1515 | 1372 | 495 | 2 | 1 | |
| regTaAOX-4BL.sv1 | 4284 | 1571 | 327 | 5 | 4 | |
| regTaAOX-4BL.sv2 | 4284 | 3158 | 249 | 4 | 3 | |
| regTaAOX-4BL.sv3 | 4284 | 3176 | 267 | 4 | 3 | |
| regTaAOX-4BL.sv4 | 4284 | 3274 | 327 | 4 | 3 | |
| put.regTaAOX-3B | 3652 | 585 | 291 | 3 | 2 | |
| put.regTaAOX-6BL | 3833 | 718 | 441 | 3 | 2 | |
| TaAOX1c-6AL | 2019 | 1736 | 1194 | 4 | 3 | |
| TaAOX1c-6BL.sv1 | 2207 | 1940 | 1296 | 4 | 3 | |
| TaAOX1c-6BL.sv2 | 2207 | 1806 | 1239 | 6 | 5 | |
| TaAOX1c-6BL.sv3 | 2207 | 1863 | 1296 | 5 | 4 | |
| TaAOX1c-6DL | 2075 | 1801 | 1188 | 4 | 3 | |
| regTaAOX-3B | 1456 | 1370 | 324 | 2 | 1 | |
| put.TaAOX1e-3DS | 18352 | 1142 | 789 | 4 | 3 | |
| TaAOX1d-2AL.1 | 1290 | 1180 | 885 | 2 | 1 | |
| TaAOX1d-2AL.2.sv1 | 1423 | 1423 | 993 | 1 | 0 | |
| TaAOX1d-2AL.2.sv2 | 2395 | 1269 | 993 | 2 | 1 | |
| TaAOX1d-2DL | 1405 | 1405 | 981 | 1 | 0 | |
| put.TaAOX1d-like-4AS | 1065 | 963 | 552 | 2 | 1 | |
| A Genome | TuAOX1a* | 1383 | 615 | 615 | 3 | 2 |
| TuAOX1c* | 4469 | 1305 | 1305 | 7 | 6 | |
| TuAOX1d.1* | 888 | 888 | 888 | 1 | 0 | |
| TuAOX1d.2* | 8777 | 1212 | 1212 | 3 | 2 | |
| D Genome | AetAOX1a* | 1379 | 615 | 615 | 3 | 2 |
| AetAOX1e* | 2958 | 1098 | 1098 | 5 | 4 | |
| AetAOX1d* | 888 | 888 | 888 | 1 | 0 | |
| AetAOX1d-like* | 1198 | 870 | 870 | 4 | 3 |
*Denotes diploid wheat AOX genes.
Table 4. Splice variants of hexaploid wheat AOX genes.
| Gene Name | Status | Points of Difference |
|---|---|---|
| TaAOX1a-2AL.sv1 | Wildtype | Intron retention: Portion of exon 1 in wildtype is intron 1 in variant. |
| TaAOX1a-2AL.sv2 | Variant | |
| TaAOX1a-2BL | N/A | N/A |
| TaAOX1a-2DL.sv1 | Wildtype | Intron retention: Portion of exon 1 in wildtype is intron 1 in variant. |
| TaAOX1a-2DL.sv2 | Variant | |
| TaAOX1a-like-2DL | N/A | N/A |
| regTaAOX-4BL.sv1 | Wildtype | Intron retention: Portion of exon 2 in wildtype is part of intron 2 in variant. Portion of exon 4 in variant is intron 4 in wildtype. |
| regTaAOX-4BL.sv2 | Variant | |
| regTaAOX-4BL.sv3 | Variant | Intron retention: Portion of exon 2 in wildtype is a part of intron 2 in variant. Portion of exon 2 in variant is part of intron 1 in wildtype. Portion of exon 4 in variant is intron 4 in wildtype. |
| regTaAOX-4BL.sv4 | Variant | Intron retention: Portion of exon 4 in variant is intron 4 in wildtype. |
| put.regTaAOX-3B | N/A | N/A |
| put.regTaAOX-6BL | N/A | N/A |
| TaAOX1c-6AL | N/A | N/A |
| TaAOX1c-6BL.sv1 | Wildtype | Intron retention: Portion of exon 1 in wildtype is part of intron 1 in variant. Portion of exon 4 in wildtype is part of intron 5 in variant. |
| TaAOX1c-6BL.sv2 | Variant | |
| TaAOX1c-6BL.sv3 | Variant | Intron retention: Portion of exon 4 in wildtype is intron 4 in variant. |
| TaAOX1c-6DL | N/A | N/A |
| regTaAOX-3B | N/A | N/A |
| put.TaAOX1e-3DS | N/A | N/A |
| TaAOX1d-2AL.1 | N/A | N/A |
| TaAOX1d-2AL.2.sv1 | Wildtype | Intron retention: Portion of exon 1 in wildtype is part of intron 1 in variant. Alternative 3'UTR site: Portion of exon2 in variant is downstream of the gene sequence of the wildtype. |
| TaAOX1d-2AL.2.sv2 | Variant | Alternative 5'UTR site: part of the wildtype's 5'UTR is upstream of the gene sequence of the variant |
| TaAOX1d-2DL | N/A | N/A |
| put.TaAOX1d-like-4AS | N/A | N/A |
Fig 3. AOX gene structures of hexaploid and diploid wheat.
Exons are depicted in red, with introns being represented by black lines for (A) high-confidence T. aestivum AOX gene family, (B) low-confidence T. aestivum AOX gene family (C) T. urartu AOX gene family, and (D) A. tauschii AOX gene family.
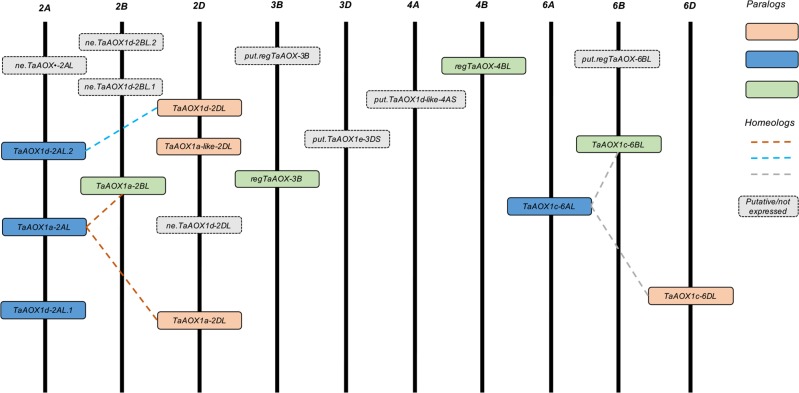
The paralogs and homeologs for the hexaploid AOXs were also identified by Ensembl Plants. There were paralogs in all the high-confidence hexaploid genes all located on long chromosomal arms. With the exception of TaAOX1a-like-2DL, regTaAOX-4BL, regTaAOX-3B and TaAOX1d-2AL.1 homeologs were found (Fig 4). The gene sequences were aligned with the draft physical genome sequences downloaded from Ensembl Plants (ftp://ftp.ensemblgenomes.org/pub/release-39/plants/fasta/triticum_aestivum/dna/). The higher resolution of the latest version of the genome facilitated the placement of the hexaploid genes on the chromosomal arms. It was clear that most of the AOX genes were on the long chromosomal arms with about half the total the long arms of the subgenomes of chromosome 2 (Fig 4). The two hexaploid AOX genes with the shortest distance on the same chromosome were ne.TaAOX1d-2BL.1 and ne.TaAOX1d-2BL.2 which were 8.6 kbp apart. TaAOX1a-2BL and ne.TaAOX1d-2BL.1 were the farthest apart with 301 Mbp between the two genes (S8 Fig). It is possible that with future work especially on the non-expressed and putative gene copies, the relationships between these genes and the high-confidence AOX gene copies will be better established. This will allow for a better study of the functionality of these paralogs within the hexaploid wheat genome.
Fig 4. Distribution of AOX homeologs and paralogs on hexaploid wheat chromosomes.
Boxes of the same color indicate paralogs, while lines of the same color indicate homeologous groups. Grey boxes with a dashed outline indicate putative or non-expressed genes.
Epigenetic gene regulation via CpG islands can lead to diversity and specificity in gene expression [85, 86]. The gene sequences discovered were input into the CpgPlot program in order to find putative CpG islands. The greatest CpG distribution was in the AOX1a clade followed by that of the AOX1c clade (Table 5). While no clear relationship could be established between these GC-rich regions and levels of transcription, it must be noted that these variations could become more relevant under other experimental conditions which were unexplored in the RNA-seq dataset used in the present study (exposure to phytohormones, salt stress, long-term heat and drought stress). CpG islands were not examined in the putative hexaploid genes and the diploids as significant gene regions were unresolved in the database.
Table 5. CpG islands in the promoters and gene bodies of the high-confidence TaAOX gene family.
| Gene Name | Promoter | Gene Body | ||||
|---|---|---|---|---|---|---|
| # of Islands | Regions | Length | # of Islands | Regions | Length | |
| TaAOX1a-2AL.sv1 | 1 | (1040..1444) | 405 | 2 | (47..845), (1192..1730) | 799, 539 |
| TaAOX1a-2AL.sv2 | ||||||
| TaAOX1a-2BL | 1 | (51..289) | 239 | 2 | (51..1249), (1622..2167) | 1199, 546 |
| TaAOX1a-2DL.sv1 | 1 | (1207..1444) | 238 | 2 | (47..910), (1273..1819) | 864, 547 |
| TaAOX1a-2DL.sv2 | ||||||
| TaAOX1a-like-2DL | 3 | (139..355), (554..1045), (1064..1444) | 217, 492, 381 | 1 | (47..464) | 418 |
| regTaAOX-4BL.sv1 | 3 | (427..690), (860..1203), (1208..1444) | 264, 344, 237 | 1 | (49..435) | 387 |
| regTaAOX-4BL.sv2 | 1 | (49..511) | 463 | |||
| regTaAOX-4BL.sv3 | 1 | (49..511) | 463 | |||
| regTaAOX-4BL.sv4 | 1 | (49..435) | 387 | |||
| TaAOX1c-6AL | 1 | (494..720) | 227 | 1 | (48..1356) | 1309 |
| TaAOX1c-6BL.sv1 | 0 | 2 | (49..522), (586..1431) | 474, 846 | ||
| TaAOX1c-6BL.sv2 | ||||||
| TaAOX1c-6BL.sv3 | ||||||
| TaAOX1c-6DL | 0 | 1 | (48..1327) | 1280 | ||
| regTaAOX-3B | 0 | 1 | (48..317) | 270 | ||
| TaAOX1d-2AL.1 | 0 | 1 | (48..938) | 891 | ||
| TaAOX1d-2AL.2.sv1 | 2 | (49..380), (768..1408) | 332, 641 | 1 | (47..937) | 891 |
| TaAOX1d-2AL.2.sv2 | ||||||
| TaAOX1d-2DL | 2 | (702..1064), (1079..1402) | 363, 324 | 1 | (47..925) | 879 |
AOX protein phylogeny reveals four lineages of wheat AOX genes, each with multiple copies
In order to hypothesize the major lineages of wheat AOX gene copies and infer the number and timing of duplication events, amino acid sequences of the diploid and hexaploid wheat AOX proteins in combination with other monocot sequences were used to generate a phylogenetic hypothesis of gene family evolution (Figs 1 and S1). All putative AOX copies in wheat and as many putative AOX1 paralogs as possible from other species were included in order to better assess the timing of duplication events. Four lineages of wheat AOX genes were found, corresponding to the AOX1a, AOX1c, AOX1e and AOX1d genes previously found [16, 17]. Wheat gene copies within each lineage are unevenly distributed: the AOX1a lineage includes seven T. aestivum copies (including regulatory, putative regulatory, and -like copies), but only one each from T. urartu and A. tauschii; the AOX1c lineage includes three T. aestivum copies and one from T. urartu; the AOX1d lineage includes seven T. aestivum copies, two T. urartu and three A. tauschii copies; and the AOX1e lineage includes two T. aestivum and one A. tauschii copies. Given the incomplete and only partially annotated nature of the T. urartu (A-subgenome) and A. tauschii (D-subgenome) genomes and the lack of availability of the A. speltoides (B-subgenome) genome, it is unclear how to interpret the lack of copies or variable number of copies of some of the duplication types. Some gene types (e.g., AOX1c) fit what we would expect for a hexaploid (three copies), while other types have more variable copy number in wheat (AOX1a, seven copies). AOX1d appears to be a triticoid-specific duplication with what appears to be two clades of AOX1d genes in wheat and relatives, with the duplication after the divergence of triticoid grasses from Brachypodium (Figs 1 and S1). It is important to note that while Brachypodium also has two AOX1d gene copies, those are from a different duplication event than the triticoid duplication.
In addition to the variation in copy number among AOX1 clades, the variation in the number of splice variants is unevenly distributed. As with the number of gene copies, the AOX1a clade also has the most splice variants with eight splice variants found from three genomic copies, whereas only three splice variants from one gene copy are found in the AOX1c lineage and two splice variants from one gene copy in the AOX1d lineage (Fig 3 and Table 1). No splice variants were found for the AOX1e genes. The pattern of duplication and divergence in the AOX1a gene lineage needs further study, but suggests that these genes are undergoing rapid duplication and divergence in sequence characteristics and, presumably, function. Whether the co-occurrence of multiple splice variants and duplication of genomic copies are connected will require further evaluation, but it is plausible that splice variant reintroduction to the genome as a duplication mechanism could drive the gene proliferation that we document here [87, 88]
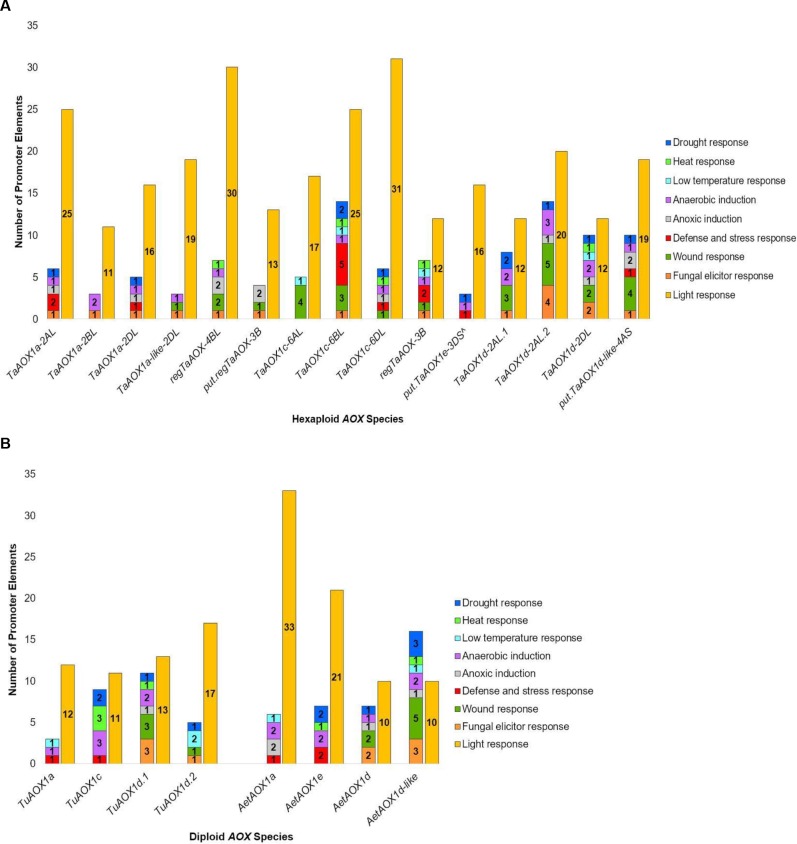
Promoter analyses reveal regulatory motifs in wheat AOX gene family
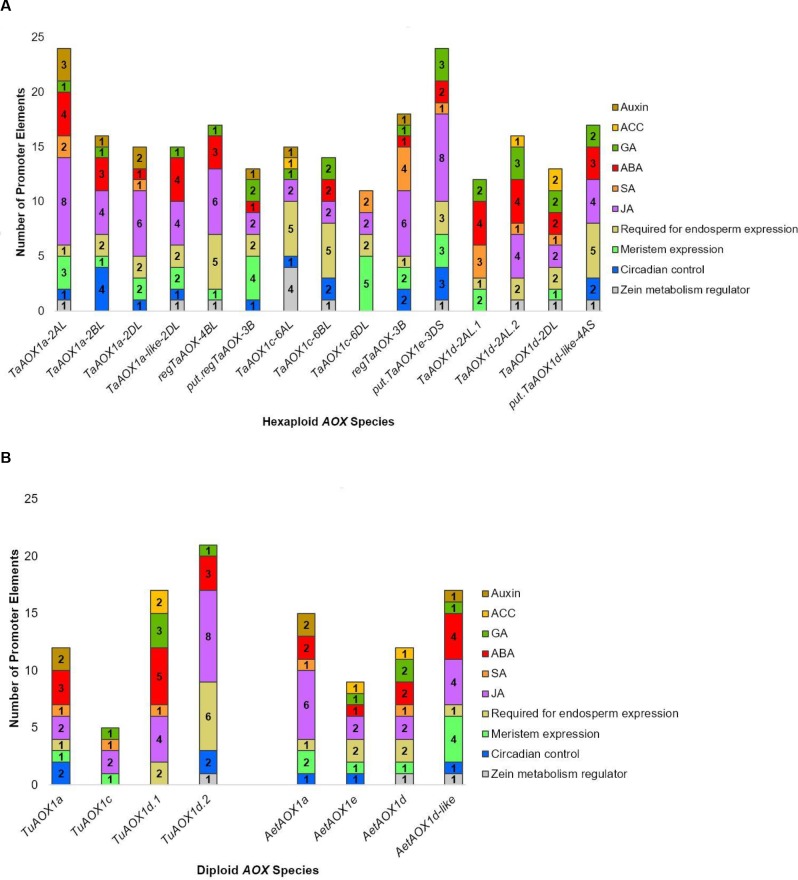
To identify putative regulatory elements of the wheat AOX genes, promoter elements were identified in the 1500 bp sequence upstream of the translation start site (S4 Fig). In the hexaploid and diploid species, the greatest proportion of elements was for light response (Fig 5). It was also apparent that there was diversity between family members with regards to elements involved in hormonal, developmental, biotic and abiotic environmental responses (Figs 5 and 6). Cumulatively, the greatest numbers of these hexaploid response elements were found in TaAOX1a-2AL, regTaAOX-4BL and TaAOX1c-6BL. The smallest numbers were observed in TaAOX1a-2BL, put.regTaAOX3B, TaAOX1d-2AL.1 and TaAOX1d-2DL (Figs 5 and 6). Out of these the highest numbers of environmental response elements were found in regTaAOX-4BL, TaAOX1c-6BL, TaAOX1c-6DL, and TaAOX1d-2AL.2 (Fig 5A). The highest numbers of hexaploid hormonal and developmental response elements were found in TaAOX1a-2AL, regTaAOX-3B and put.TaAOX1e-3DS (Fig 6A). The jasmonic acid (JA) and abscisic acid (ABA) response elements were also common across many family members in the hexaploid and diploid species (Fig 6). There was a large proportion of specific response elements in some promoters, e.g. TaAOX1a-2AL (JA), TaAOX1a-2BL (circadian control) and TaAOX1d-2AL.2 (fungal elicitor response) (Figs 5A and 6A). While the low temperature response elements were absent in the hexaploid AOX1a promoters, they were present in the diploid AOX1a promoters and may indicate levels of control peculiar to the diploid species (Fig 5). Many of these factors such as light, heat, drought, ABA, and SA have already been shown to cause induction of AOX in wheat and other plants [10, 37–39, 42]. However, the presence of motifs for gibberellic acid (GA), jasmonic acid (JA), ethylene (ACC) and others (Fig 6) suggest that there is still a lot of work to be done in terms of how these phytohormones and developmental factors are integrated into the framework of AOX expression and regulation. The commonality of certain elements across many family members could also give ways of inducing the expression of some or all of the low-confidence and non-expressed AOX genes that have been reported in the current study. It must be noted that some regions of the promoters were unresolved and therefore additional motifs may be found that could be specific to the diploids or show similar elements as in the hexaploid counterparts.
Fig 5. Putative cis-elements for abiotic response in promoter regions.
(A) TaAOX gene family and (B) diploid AOX gene families. The promoter for put.regTaAOX-6BL was not analyzed due to the majority of the promoter sequence being unresolved.
Fig 6. Putative cis-elements for hormonal and developmental responses in promoter regions.
(A) TaAOX gene family and (B) diploid AOX gene families. The promoter for put.regTaAOX-6BL was not analyzed due to the majority of the promoter sequence being unresolved.
Previous research has revealed a number of positive and negative regulators of AOX (S3 Table). Generally, regulator motifs were common in the AOX1a and AOX1d clades and absent in the AOX1c and AOX1e clades. Motifs were found for known positive regulators of AOX (Tables 6, 7, S3 and S4) in all the promoters except those of put.regTaAOX-3B, put.regTaAOX-6BL, TaAOX1c-6AL, TaAOX1c-6BL, TaAOX1c-6DL, regTaAOX-3B, put.TaAOX1e-3DS, put.TaAOX1d-like-4AS, TuAOX1d.2, TuAOX1c and AetAOX1e. The NAC Domain Containing Protein 17 (ANAC017) (At1g34190) is considered to be a critical positive regulator of AOX [89, 90] and motifs for this protein were well-represented in the AOX1a and AOX1d clades (TaAOX1a-2AL, TaAOX1a-2BL, TaAOX1a-2DL, TaAOX1a-like-2DL, TaAOX1d-2AL.2, TaAOX1d-2DL, TuAOX1d.1, AetAOX1a, AetAOX1d, and AetAOX1d-like) (Tables 6 and 7). Another positive regulator WRKY DNA-Binding Protein 63 (AtWRKY63, At1g66600) [91] was more common in the AOX1d clade (TaAOX1a-like-2DL, TaAOX1d-2AL.2, TaAOX1d-2DL, TuAOX1d.1 and AetAOX1d) (Tables 6 and 7). Motifs for the negative regulator ABA Insensitive 4 (ABI4, At2g40220) [92] were found in all the promoters except TaAOX1c-6AL, TaAOX1c-6BL and AetAOX1e (Tables 6, 7, S3 and S4). The binding sites for another negative regulator WRKY DNA-Binding Protein 40 (ATWRKY40, At1g80840) [90] existed only in the TaAOX1c-6BL promoter (Tables 6, 7, S3 and S4).
Table 6. Occurrence of conserved motifs for known positive and negative AOX regulators in the promoters of the TaAOX gene family.
| Locus | TaAOX1a-2AL | TaAOX1a-2BL | TaAOX1a-2DL | TaAOX1a-like-2DL | regTaAOX-4BL | put.regTaAOX-3B | TaAOX1c-6AL | TaAOX1c-6BL | TaAOX1c-6DL | regTaAOX-3B | put.TaAOX1e-3DS | TaAOX1d-2AL.1 | TaAOX1d-2AL.2 | TaAOX1d-2DL | put.TaAOX1d-like-4AS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At1g32870 | 3 | 2 | 3 | 1 | 1 | 1 | |||||||||
| At1g34190 | 2 | 2 | 2 | 1 | 1 | 1 | |||||||||
| At1g66600 | 1 | 3 | 1 | ||||||||||||
| At3g10500 | 2 | 2 | 2 | 1 | 1 | 1 | |||||||||
| At5g04410 | 1 | 2 | 1 | 1 | 1 | ||||||||||
| At1g80840* | 1 | ||||||||||||||
| At2g40220* | 4 | 2 | 4 | 6 | 8 | 10 | 1 | 1 | 6 | 2 | 2 | 4 | 2 | ||
| CTTGNNNNNCAMG | 2 | 2 | 2 | 2 | 2 | 2 | |||||||||
| YTTGNNNNNVAMV | 8 | 5 | 6 | 4 | 2 | 3 | 4 | 1 | 1 | 1 | 4 | 3 |
The promoter for put.regTaAOX-6BL was not analyzed due to the majority of the promoter sequence being unresolved.
*Denotes negative regulators.
Table 7. Occurrence of conserved motifs for known positive and negative AOX regulators in the promoters of the diploid AOX gene families.
| Locus | TuAOX1a | TuAOX1c | TuAOX1d.1 | TuAOX1d.2 | AetAOX1a | AetAOX1e | AetAOX1d | AetAOX1d-like |
|---|---|---|---|---|---|---|---|---|
| At1g32870 | 1 | 1 | 3 | 1 | 2 | |||
| At1g34190 | 1 | 2 | 1 | 2 | ||||
| At1g66600 | 2 | 1 | ||||||
| At3g10500 | 1 | 2 | 1 | 1 | ||||
| At5g04410 | 1 | |||||||
| At2g40220* | 2 | 1 | 2 | 1 | 5 | 3 | 4 | |
| CTTGNNNNNCAMG | 2 | 2 | 2 | |||||
| YTTGNNNNNVAMV | 2 | 3 | 3 | 4 | 3 | 2 | 3 | 6 |
*Denotes negative regulators.
Previous researchers analyzed A. thaliana transcriptome data obtained via induction with mitochondrial regulation perturbation reagents. Further analysis on the promoters of the highly responsive and upregulated genes showed the presence of the cis-regulatory element CTTGNNNNNCAMG [93] labeled the mitochondrial dysfunction motif (MDM). All the genes with the MDM motif which were upregulated during disruption of mitochondrial retrograde regulation were thereafter referred to as the MITOCHONDRIAL DYSFUNCTION STIMULON (MDS) genes which include AOX. The protein ANAC013 (At1g32870) controls the MDS genes by direct interaction with the MDM motif CTTGNNNNNCAMG or the alternative YTTGNNNNNVAMV (sequence variation in orthologs) (Tables 6, 7, S3 and S4). [93]. In the current study, a search was conducted for the MDM motif in the wheat AOX gene family promoter regions. The stringent motif, CTTGNNNNNCAMG, was found in all promoters except regTaAOX-4BL, put.regTaAOX-3B, put.regTaAOX-6BL, TaAOX1c-6AL, TaAOX1c-6BL, regTaAOX-3B, put.TaAOX1e-3DS, TaAOX1d-2AL.1, put.TaAOX1d-like-4AS, TuAOX1a, TuAOX1c, TuAOX1d.2, AetAOX1d and AetAOX1e. The alternative MDM motif (YTTGNNNNNVAMV) was found in all promoters except regTaAOX-3B and put.regTaAOX-6BL (Tables 6, 7, S3 and S4). Furthermore, the promoters for TaAOX1a-2AL, TaAOX1a-2DL, TaAOX1a-like-2DL, TaAOX1c-6AL, TaAOX1d-2AL.2, TuAOX1a, TuAOX1d.2, and AetAOX1e contained the YTTGNNNNNVAMV motif but with only one nucleotide deviation from the stringent motif (S4 Table). This single nucleotide deviation was also found amongst some of the 24 MDM motif genes identified in a previous study [93]. This could indicate that for TaAOX1c-6AL and TuAOX1a, which lacked the stringent motif, they could still be controlled by ANAC013. The results found using the YTTGNNNNNVAMV motif could therefore be false positives as in A. thaliana there are other genes containing this MDM motif that have regulators other than ANAC013. They could also indicate that although not directly controlled by ANAC013, they may still be involved in the network of mitochondrial retrograde regulation [93]. If valid, this motif distribution allows for greater levels of control in how this gene family is expressed and may provide clues as to how to induce the expression of some or all of the non-expressed wheat AOX genes.
It must also be noted that the pattern or mode of expression could differ between the hexaploids and the diploids. Given that the promoter regions of the diploid species are still being sequenced, it is entirely possible that additional motifs of other aforementioned positive regulators could be discovered. This would further our knowledge of the regulation of AOX in the wild ancestors of bread wheat. Motifs for the hypoxia responsive promoter element [94] as well as known positive and negative regulators of A. thaliana AOX, At5g13610, At5g63610, At5g12290, At5g07690 and At1g32230 [95–99] were missing from both the hexaploid and diploid promoters (Tables 6, 7, S3 and S4). It is possible that these factors are restricted to dicots and have evolved a different form of control or have significantly diverged in grasses or monocots.
We also examined the CpG islands in the AOX promoters (S4 Fig). In the promoter regions, the largest distribution of CpG islands was in the genes of the AOX1a clade followed by the AOX1d clade. The lowest CpG distribution was in the AOX1c and AOX1e clades (Table 5). Similar to the AOX gene sequences, there was no obvious relationship between the number or length of CpG islands and the regulation of the transcripts from these promoters. Again, it is entirely possible that variations may emerge under new experimental conditions such as longer-term abiotic stresses and exposure to phytohormones.
AOX expression is diverse among all family members over tissue types and developmental stages
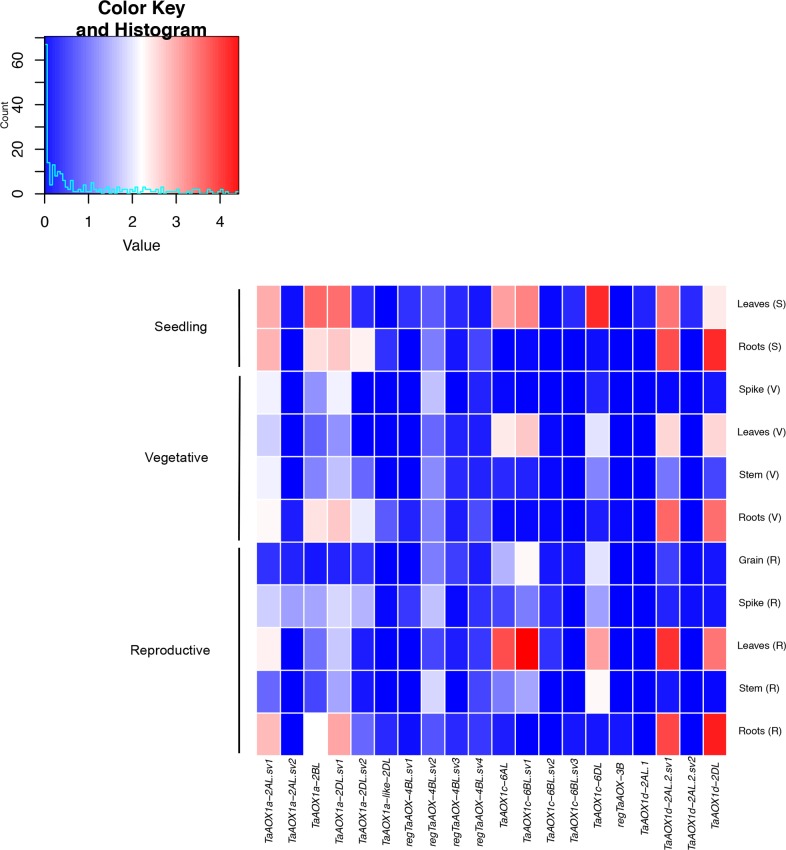
To provide further insight into wheat AOX function, RNA-seq data were obtained from studies deposited into the wheat database expVIP [55]. Data for the high-confidence AOX genes was obtained and expression was found in multiple tissues at various developmental stages as well as over various environmental conditions. Ten out of 20 possible transcripts were expressed in all tissues examined at all three developmental stages (seedling, vegetative and reproductive) (Fig 7 and S5 Table). These were, TaAOX1a-2AL.sv1, TaAOX1a-2BL, TaAOX1a-2DL.sv1, regTaAOX-4BL.sv2, regTaAOX-4BL.sv4, TaAOX1c-6AL, TaAOX1c-6BL.sv1, TaAOX1c-6DL, TaAOX1d-2AL.2.sv1 and TaAOX1d-2DL. There was a low-level of expression for TaAOX1a-2AL.sv2, TaAOX1a-like-2DL, regTaAOX-4BL.sv1, regTaAOX-4BL.sv3, regTaAOX-4BL.sv4, TaAOX1c-6BL.sv2, TaAOX1c-6BL.sv3, regTaAOX-3B, TaAOX1d-2AL.1 and TaAOX1d-2AL.2sv2 in all tissue and developmental stages tested. Of note were TaAOX1a-2AL.sv1, TaAOX1a-2BL, TaAOX1a-2DL.sv1, TaAOX1d-2AL.2.sv1 and TaAOX1d-2DL which had higher expression in the root at all three developmental stages (Fig 7 and S5 Table). There were a few transcripts with high root expression in particular stages. TaAOX1a-2DL.sv2 and had higher root expression at the seedling and vegetative stages (Fig 7 and S5 Table).
Fig 7. Heat map of expression profiles for high-confidence TaAOX genes at different developmental stages.
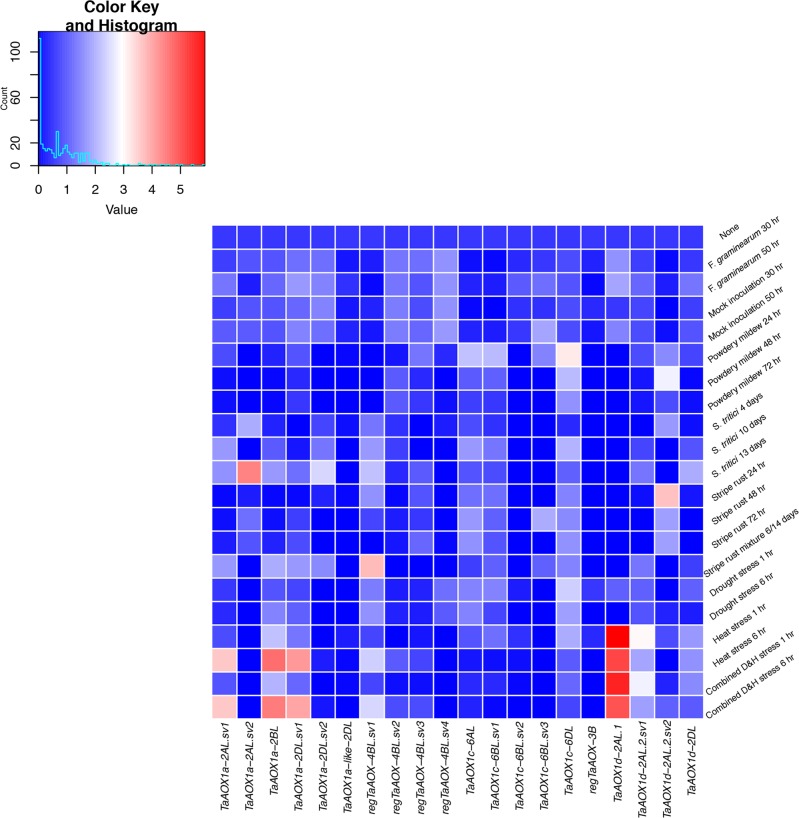
TaAOX1a-2BL, TaAOX1a-2DL.sv1, regTaAOX-4BL.sv1, TaAOX1c-6AL, TaAOX1c-6BL.sv1, TaAOX1c-6DL, TaAOX1d-2AL.1, TaAOX1d-2AL.2.sv1 and TaAOX1d-2DL were upregulated under drought stress (Fig 8 and S5 Table). Under heat stress, the highest level of transcript expression was notably TaAOX1a-2BL, TaAOX1a-2DL.sv1 and TaAOX1d-2AL.1. In addition, TaAOX1a-2AL.sv1, regTaAOX-4BL.sv1, regTaAOX-4BL.sv2, TaAOX1c-6AL, TaAOX1c-6BL.sv1, TaAOX1c-6DL, TaAOX1d-2AL.2.sv1, TaAOX1d-2AL.2.sv2 and TaAOX1d-2DL were expressed under heat but to a lesser extent. In contrast, TaAOX1a-2AL.sv2 and TaAOX1c-6BL.sv3 showed no expression under heat or drought stress (Fig 8 and S5 Table). Overall, heat stress had a higher impact on expression levels than drought stress. Under various forms of biotic stress (Fusarium graminearum, powdery mildew, stripe rust, Septoria tritici), TaAOX1a-2AL.sv2, TaAOX1a-2DL.sv2, regTaAOX-4BL.sv1, TaAOX1c-6DL and TaAOX1d-2AL.2.sv2 showed high expression levels (Fig 8 and S5 Table). TaAOX1a-2AL.sv1, TaAOX1a-2BL, TaAOX1a-2DL.sv1, TaAOX1a-2DL.sv2, regTaAOX-4BL.sv3, TaAOX1c-6AL, TaAOX1c-6BL.sv1, TaAOX1c-6BL.sv3, TaAOX1d-2AL.2.sv1 and TaAOX1d-2DL were also upregulated during biotic stress (Fig 8 and S5 Table). RegTaAOX-3B was mostly dormant during biotic stress. The mosaic pattern of results indicates diversity in the level of expression between gene family members and between splice variants of the same gene. This allows for nuance and complexity in function over numerous environmental and physiological conditions. In contrast to previous research, there was no clear relationship between the number of exons and the level of transcript expression [81–84]. It may be that unique physiology, polyploidization and the alternative splicing machinery have given rise to alternate forms of transcriptional regulation in wheat. There was no expression data available for the diploid ancestors. However, polyploidization can lead to neofunctionalization [100, 101] and therefore it is possible that the expression patterns and subsequent protein activities may differ between the hexaploid and diploid wheat species. This highlights the need for expression data in the diploid progenitor species as well.
Fig 8. Heat map of expression profiles for high-confidence TaAOX genes under biotic and abiotic stresses.
With regard to the Chinese Spring reference transcriptome, the expVIP database which integrates the most reliable gene models from Ensembl Plants, the most consistent and reliable information that has been supported by expression data from other species [102]. The transcriptome data generated by this database relied on genomic resources such as nullitetrasomic lines as well as the latest RNA-seq software in the analysis [55] to pinpoint the chromosomal localizations and ensure accurate placement on the physical map. In addition, this database contains information that has been shown by other researchers to be validated by alternate sources [77, 103]. A comparison of the transcript sequences shows that there are enough differences for further validation via future qPCR experiments (S6 Table). The current study provides information which can be used in future biological experiments over the same or different conditions used in this study. Different varieties may express these AOX genes differently and therefore drawing broad conclusions may not apply to other wheat cultivars or landraces or diploid species. It is critical that elite cultivars being used by researchers be utilized in further studies to further validate the sequences and gene structures shown in the present study. Future experiments may lead to the discovery of new alleles which may show different expression patterns than those revealed in the present study. Subsequent research may also show the occurrence of gene copy number variations, a phenomenon known to occur in the polyploid wheat and which has been shown to cause a spectrum of phenotypic differences that may depend on the geographic region of the cultivars in question [104, 105]. Researchers may therefore find that their respective cultivars have more AOX copies which may provide adaptation advantages nonexistent the Chinese Spring cultivar. The possibilities described may require alternate ways of triggering the induction of the newly discovered gene copies as well as the low-confidence and the non-expressed AOX genes in order to validate the transcripts and ensure that qPCR primers designed would be gene-specific. For example, hormone elicitors such as jasmonic acid (JA) could trigger expression as JA-responsive elements are found in some wheat AOX promoters (Fig 6). Alternatively, reagents that disrupt mitochondrial regulation could be used as in previous studies [93]. Regardless, these data lay the groundwork and generates new hypotheses concerning AOX gene expression and function given the isoforms discovered.
Determining expression profiles of AOX genes in the diploid ancestors could lead to a better understanding of how these genes evolved in the polyploid species as has been shown in wild relatives of rice [106, 107]. This process could also lead to potential germplasm sources which can be introgressed to improve marketable wheat varieties [108]. The results for short term abiotic stress (1 hour and 6 hours) indicate expression levels under temporary heat and drought stress, but more experimentation is need to determine expression under more sustained levels of stress. Variance in AOX expression and or copy number in wheat varieties with contrasting levels of resilience or tolerance to biotic or abiotic stress in a general or tissue-specific manner will facilitate the discovery of germplasm to create more marketable varieties. Importantly, AOX can have a strong effect on root morphology and may be upregulated roots under stress [109, 110]. Investigating the level of expression under infection by root pathogens and symbiotic soil microbes may aid in the elucidation of mechanism of susceptibility or resistance as well as symbiosis.
Comparative analysis of wheat AOX proteins shows potential role of protein properties in functional diversity
The corresponding proteins obtained from the AOX transcripts in both the hexaploid and diploid wheat species were used in phylogenetic analysis in order to determine their classification to the clades AOX1a, AOX1c, AOX1e and AOX1d. Ten high-confidence hexaploid isoforms were classified as AOX1a, five as AOX1c, one as AOX1e and four as AOX1d. An additional two low-confidence hexaploid isoforms were in the AOX1a clade and one each in the AOX1e and AOX1d clades respectively (Figs 1 and S1). For the diploid T. urartu, one protein each was identified in the AOX1a and AOX1c clades and two in the AOX1d clades. In A. tauschii, one protein each was identified in the AOX1a and AOX1e clades and two in the AOX1d clades (Figs 1 and S1). Overall, the protein lengths ranged from 82 amino acids to 457 amino acids (Table 8). Generally, the theoretical isoelectric points of the AOX1d and AOX1e proteins were the lowest while that of the AOX1c proteins was the highest. In some cases, splice variant isoforms were shown to have distinct isoelectric points (TaAOX1a-2AL.sv1 and TaAOX1a-2AL.sv2; TaAOX1a-2DL.sv1 and TaAOX1a-2DL.sv2; regTaAOX-4BL.sv1, regTaAOX-4BL.sv2 and regTaAOX-4BL.sv3) (Table 8). Plants are able to alter their gene expression in response to external pH [111, 112]. The presence of protein isoforms with varying isoelectric points may therefore help with adaptability to external acidity or alkalinity. It has been experimentally shown that AOX may functionally substitute for the plastid terminal oxidase (PTOX) [113] and it is possible that in the polyploid and ancestral wheat genomes, some of the AOX proteins could perform non-canonical functions or play support roles for other organellar proteins. Different compartments have different acidic or basic environments levels [114] and if this flexibility in functionality works in the AOX proteins in wheat, protein isoforms with efficiencies in a pH range will aid in functionality in multiple cell organelles or compartments. In addition, the isoelectric point can affect protein localization [115, 116]. It is therefore plausible that the range of isoelectric points play a role in the determination of subcellular localization and functionality.
Table 8. Features of AOX proteins in the wheat genomes.
| Protein Name | Protein Length (Amino Acids) | Molecular Weight (KDa) | Theoretical Isoelectric Point | Export Probability to Mitochondria |
|---|---|---|---|---|
| TaAOX1a-2AL.sv1 | 328 | 36.7 | 7.90 | 0.97 |
| TaAOX1a-2AL.sv2 | 320 | 36.0 | 7.51 | 0.97 |
| TaAOX1a-2BL | 457 | 50.5 | 9.60 | 0.12 |
| TaAOX1a-2DL.sv1 | 336 | 37.6 | 8.40 | 0.94 |
| TaAOX1a-2DL.sv2 | 294 | 33.4 | 7.30 | 0.82 |
| TaAOX1a-like-2DL | 164 | 18.8 | 6.50 | 0.76 |
| regTaAOX-4BL.sv1 | 108 | 12.7 | 10.90 | 0.93 |
| regTaAOX-4BL.sv2 | 82 | 9.9 | 9.90 | 0.88 |
| regTaAOX-4BL.sv3 | 88 | 10.6 | 10.10 | 0.91 |
| regTaAOX-4BL.sv4 | 108 | 12.7 | 10.90 | 0.93 |
| put.regTaAOX-3B | 96 | 11.1 | 4.66 | 0.20 |
| put.regTaAOX-6BL | 146 | 15.5 | 8.58 | 0.79 |
| TaAOX1c-6AL | 397 | 43.7 | 9.80 | 0.64 |
| TaAOX1c-6BL.sv1 | 431 | 47.5 | 10.20 | 0.29 |
| TaAOX1c-6BL.sv2 | 412 | 45.5 | 10.30 | 0.28 |
| TaAOX1c-6BL.sv3 | 431 | 47.6 | 10.20 | 0.29 |
| TaAOX1c-6DL | 395 | 43.7 | 9.70 | 0.57 |
| regTaAOX-3B | 107 | 12.1 | 6.70 | 0.77 |
| put.TaAOX1e-3DS | 262 | 30.0 | 6.97 | 0.08 |
| TaAOX1d-2AL.1 | 294 | 33.5 | 7.20 | 0.17 |
| TaAOX1d-2AL.2.sv1 | 330 | 37.1 | 7.20 | 0.81 |
| TaAOX1d-2AL.2.sv2 | 330 | 37.1 | 7.20 | 0.81 |
| TaAOX1d-2DL | 326 | 36.7 | 7.20 | 0.85 |
| put.TaAOX1d-like-4AS | 183 | 21.0 | 7.79 | 0.25 |
| TuAOX1a* | 204 | 23.5 | 6.90 | 0.49 |
| TuAOX1c* | 434 | 49.8 | 5.90 | 0.15 |
| TuAOX1d.1* | 295 | 33.6 | 6.80 | 0.24 |
| TuAOX1d.2* | 403 | 44.6 | 9.90 | 0.21 |
| AetAOX1a* | 204 | 23.5 | 6.90 | 0.49 |
| AetAOX1e* | 365 | 40.8 | 7.40 | 0.49 |
| AetAOX1d* | 295 | 33.6 | 6.80 | 0.24 |
| AetAOX1d-like* | 289 | 32.6 | 8.60 | 0.58 |
The theoretical isoelectric points were obtained with the SnapGene Program and the mitochondrial localization probabilities were obtained with TargetP. A high value in the last column indicates a greater likelihood of localization to the mitochondria.
*Denotes diploid wheat AOX proteins.
Using the subcellular localization program TargetP, it was observed that there was a range of mitochondrial localization probabilities for AOX proteins. Generally, the hexaploid AOX1c clade had very low probability of export to the mitochondria and this trend continued with all the diploid isoforms (Table 8). Most prediction software focus on the N-terminal region of the protein in order to determine the subcellular localization. However an internal localization signal may exist as in the parasite Trypanosoma brucei (TbAOX) and in other nuclear-encoded plant proteins with no clear N-terminal signals [117, 118]. The region in TbAOX that contains this internal signal (residues 115 to 146) has some sequence similarity to the wheat isoforms but there are also clear differences, making it impossible to extrapolate outcomes from one species to another (S9 Fig). The charge of the amino acids in the sequence can also indicate the final protein destination and this may be further complicated by various protein modifications in vivo [119–121]. Given all the alternatives, it is clear that there is potential for a substantial amount of functional diversity and complexity, which could manifest as tissue or subcellular specificity as well as functional redundancy, some of which may require reporter fusions to dissect [122, 123]. This is plausible as it has been shown that AOX has both developmental and physiological functionality some of which may suggest species-specific or clade-specific functionalities [10, 31, 124].
Protein modifications such as phosphorylation, acetylation and glycosylation have been shown to be critical for multiple cellular processes in plants and it is possible that this may be the case with AOX as well [12, 71, 125–130]. Using the Plant Protein Phosphorylation Database and the Musite prediction program with a cut-off score of 0.5 as a baseline [68, 69], we found 14 predicted phosphorylation sites in TuAOX1a and two predicted phosphorylation sites in TuAOX1c (Table 9 and S7). There were two and one predicted acetylation sites in TaAOX1a-2DL.sv1 and TaAOX1d-2AL.1 respectively. There was one predicted glycosylation site each in TaAOX1a-2AL.sv1, TaAOX1a-2AL.sv2, TaAOX1a-2DL.sv1, put.regTaAOX-3B, TaAOX1c-6DL and TaAOX1d-2DL, and 28 sites in TaAOX1a-2DL.sv2. The dramatic difference in the number of predicted glycosylation sites between two splice variant isoforms (TaAOX1a-2DL.sv1 and TaAOX1a-2DL.sv2) (Tables 9 and S7) introduces the possibility of variable regulation and functionality which needs to be studied further.
Table 9. Putative post-translational modification sites in wheat AOX proteins.
| Protein | Phosphorylation Sites |
| TuAOX1a* | T9, T17, T34, T78, T131, T162, T168, T197, S12, S51, S58, S111, S129, S185, |
| TuAOX1c* | Y145, Y214 |
| Acetylation Sites | |
| TaAOX1d-2AL.1 | K222 |
| TaAOX1a-2DL.sv1 | K59, K67 |
| TuAOX1d.2* | K4 |
| AetAOX1d-like* | K110 |
| Glycosylation | |
| TaAOX1a-2AL.sv1 | S113 |
| TaAOX1a-2AL.sv2 | S105 |
| TaAOX1a-2DL.sv1 | S121 |
| TaAOX1a-2DL.sv2 | T24, T25, T61, T73, T75, T78, T99, T107, T124, T168, T221, T252, T258, T287, S2, S8, S19, S27, S44, S45, S52, S66, S79, S102, S141, S148, S201, S219 |
| put.regTaAOX-3B | S11 |
| TaAOX1c-6DL | T73 |
| TaAOX1d-2DL | S111 |
| TuAOX1d.2* | S188 |
| AetAOX1d* | S80 |
The predictions were obtained using Musite from the Plant Protein Phosphorylation Database.
*Indicates diploid isoforms.
Molecular modeling depicts conservation of diiron center residues and isoform variance in transmembrane topology
In order to determine the three-dimensional structure of the wheat AOX isomers, homology models of wheat AOX proteins were made using the crystal structure of TbAOX as a reference (model 3vvaD in Phyre2). With the exception of the low-confidence protein put.regTaAOX-6BL, all other proteins modeled to the TbAOX with over 95% confidence, sequence identity between 34 to 46% and coverage ranging from 20% to 96% depending on the isoform (range: 82 to 457 amino acids). These results offer a preliminary understanding of the structure of these proteins in wheat (Fig 9 and S8 and S9 Tables and S1 Appendix). The proteins which had most or all the motifs required for the diiron center were modeled with a similar global conformation and active site configuration indicating a likely similarity in three-dimensional structural conformation (Fig 9 and S10 Table and S1 Appendix).
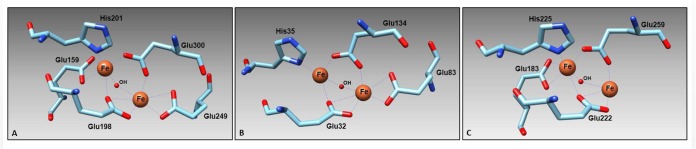
Fig 9. Proposed representative 3-D structure of the residues in the active site diiron center.
(A) 3-D structural representation of TaAOX diiron center residues. (B) 3-D structural representation of TaAOX1a-like-2DL diiron center residues. (C) 3-D structural representation of AetAOX1d-like diiron center residues.
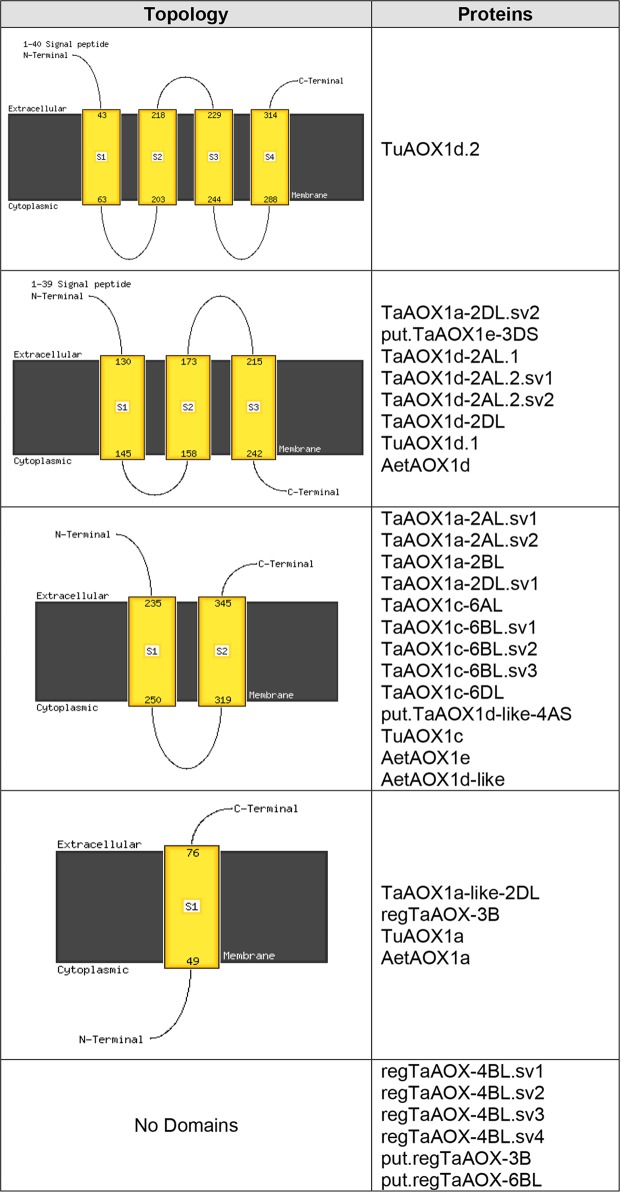
Regardless of clade, most of the modeled wheat AOX proteins had one to three transmembrane domains except regTaAOX-4BL.sv1 to sv4, put.regTaAOX-3B and put.regTaAOX-6BL which had none. Notably, TuAOX1d.2 had four transmembrane domains, the highest of all the proteins analyzed (Fig 10 and S8 and S9 Tables). TaAOX1a-like-2DL, regTaAOX-3B and TuAOX1a had the smallest number of transmembrane domains (Fig 10 and S8 and S9 Tables). Mirroring the observation of heterogeneity in protein properties between splice variant isoforms earlier observed, TaAOX1a-2DL.sv1 and TaAOX1a-2DL.sv2 had two and three transmembrane domains respectively, an example of the phenotypic diversity resulting from alternative splicing which could suggest functional diversification. Transmembrane domains have been shown to be key in the determination of protein localization in plants [131, 132] and the differences observed in wheat may facilitate the functional characterization of these proteins in the future.
Fig 10. Transmembrane topologies of wheat AOX proteins predicted by Phyre2.
Comparative analysis of wheat AOX proteins and identification of key functional residues in critical domains
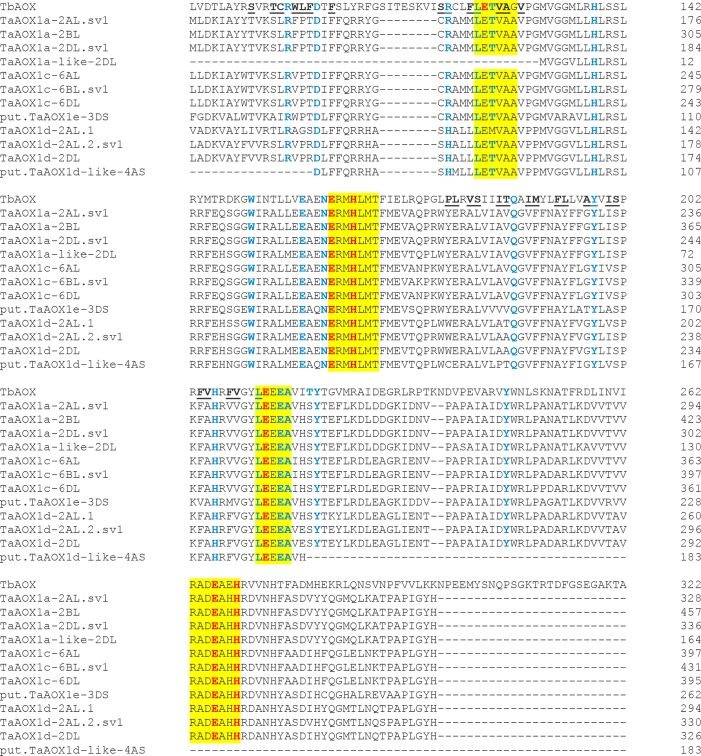
To identify conserved amino acid residues required for AOX function, the corresponding proteins of all wheat AOX transcripts and splice variants were aligned with the AOX sequence from the parasite Trypanosoma brucei (TbAOX) [133] (Figs 11–13; S2 Table; S2 Fig). Signature motifs required for AOX functionality via the diiron center, LETVAA, ERMHLMT, LEEEA and RADEAHH, were found in 22 out of 32 protein sequences (S2 Table). The protein TaAOX1a-like-2DL, lacked the first motif LETVAA, another put.TaAOX1d-like-4AS lacked the motif RADEAHH and a third AetAOX1d-like lacked the motif LEEEA. The protein TaAOX1d-2AL.1, had a slight modification LEMVAA which nevertheless conserved the glutamate critical for the diiron center. All the regulatory proteins lacked all four of these trademark motifs (Figs 11–13; S2 Table; S2 Fig). Previous research shows that mutations in any of the residues needed for coordinating the diiron center in the active site cause a partial or complete attenuation of AOX activity (Figs 11–13 and S11 Table) [12, 133–136]. This could suggest low or abolished activity for TaAOX1a-like-2DL, put.TaAOX1d-like-4AS and AetAOX1d-like which lack the motifs LETVAA, RADEAHH and LEEEA respectively, and consequently the critical glutamate residues needed to coordinate the diiron center (Figs 11 and 13; S11 and S12 Tables). The highly conserved threonine residue on other AOX proteins is a methionine in TaAOX1d-2AL.1 (TbAOX number scheme T124) (Fig 11). The difference in polarity between threonine (polar) and methionine (nonpolar) may have implications for enzyme activity and functionality. In the recombinant S. guttatum AOX protein (rSgAOX) that was tested, T179A substituted mutant (TbAOX number scheme T124) had severely reduced activity [134]. It is plausible that the same reduced enzyme activity could be observed in TaAOX1d-2AL.1. However, it must be noted that even though the hydrophobicity of the substitution in wheat mirrors that of rSgAOX T179A, the effect of the conformational change on AOX efficiency needs to be experimentally established in order to confirm an identical reduction in function.
Fig 11. Alignment of select TaAOX (hexaploid wheat) proteins with TbAOX (T. brucei).
Yellow highlights indicate conserved motifs. Red font indicates residues proposed to coordinate the diiron center of the active site. Blue font indicates residues experimentally tested for loss of activity by previous researchers. Underlined residues are involved in the TbAOX hydrophobic cavity. Splice variants were identical for the protein region analyzed. The “reg” proteins were not analyzed due to the absence of the conserved motifs.
Fig 13. Alignment of AetAOX (A. tauschii) proteins with TbAOX (T. brucei).
Yellow highlights indicate conserved motifs. Red font indicates residues proposed to coordinate the diiron center of the active site. Blue font indicates residues experimentally tested for loss of activity by previous researchers. Underlined residues are involved in the TbAOX hydrophobic cavity.
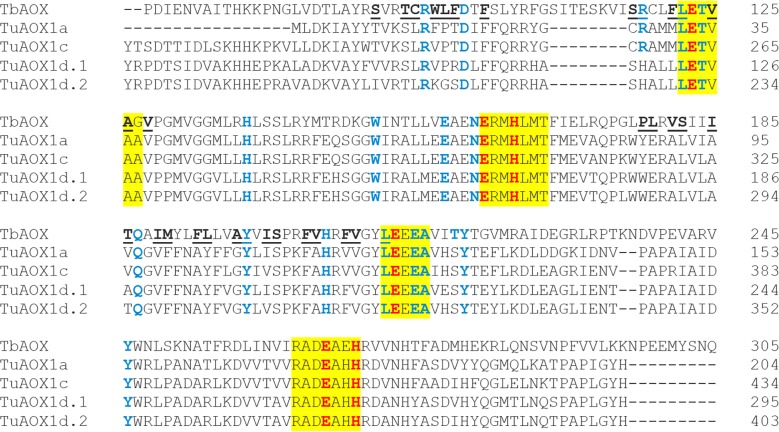
Fig 12. Alignment of TuAOX (T. urartu) proteins with TbAOX (T. brucei).
Yellow highlights indicate conserved motifs. Red font indicates residues proposed to coordinate the diiron center of the active site. Blue font indicates residues experimentally tested for loss of activity by previous researchers. Underlined residues are involved in the TbAOX hydrophobic cavity.
A T219V mutation, which leads to a significant change in side chain chemistry and configuration, causes an almost complete loss of function in recombinant T. brucei AOX (rTbAOX) (S11 Table) [133]. In wheat, there is a T219S (TbAOX number scheme) substitution conserved in all the diploid and hexaploid AOX except regTaAOX-4BL, regTaAOX-3B, put.TaAOX1d-like-4AS and AetAOX1d-like where this residue is nonexistent (Figs 11 and 13; S12 Table). The substitution maintains side chain properties but the effect of the lost methyl group on the enzymatic outcome remains to be determined. The proteins TaAOX1a-like-2DL, put.TaAOX1d-like-4AS and AetAOX1d-like are missing residues which have been experimentally shown to greatly reduce or abolish activity (S12 Table). Where nonexistent, it may imply an alternate protein configuration in that region which may change the enzyme efficiency or allow for high efficiency in a distinct role.
The crystal structure of TbAOX shows that this protein exists as a homodimer. At the dimer interface in TbAOX, there are six completely conserved residues and 12 highly conserved residues [133] some of which show significant loss of activity when mutated (H138, Q187) (S11 Table). Excluding AetAOX1d-like, these six residues are completely conserved in wheat AOX proteins as well (Table 10). With regards to the 12 highly conserved residues for the dimer interface, six are identical to TbAOX (M131, L139, S141, A159, M167, R180) except in the case of AetAOX1d-like (Tables 10 and S13). This high level of conservation across species emphasizes the importance of these residues in this functional capacity. There were three substitutions that were peculiar to the AOX1d clade and the AOX1a-like proteins (M135V, R147H, L156M). There were also one substitutions that were conserved in all wheat clades (M145F) (Tables 10 and S13). Two other substitutions (I183V, D148S) are also conserved in all the wheat clades with the exception of put.TaAOX1d-like-4AS (D148N) and AetAOX1d-like (I183V is nonexistent) (Tables 10 and S13). None of these conserved substitutions match any of the substitutions thought to support an AOX Type AOX1d classification done by other researchers [16] and thus may indicate a divergence peculiar to this AOX clade in wheat which could inform function. These substitutions need further characterization in order to test their effect on AOX dimerization and efficiency in wheat.
Table 10. Comparison of residues at dimerization interface between the TbAOX and wheat AOX proteins.
| TbAOX Residue Numbers | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Completely Conserved in TbAOX | Highly Conserved in TbAOX | |||||||||||||||||
| H138◆ | L142 | R143 | R163 | L166 | Q187◆ | M131 | M135 | L139 | S141 | M145 | R147 | D148 | L156 | A159 | M167 | R180 | I183 | |
| TaAOX1a-2AL.sv1 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1a-2AL.sv2 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1a-2BL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1a-2DL.sv1 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1a-2DL.sv2 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1a-like-2DL | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TaAOX1c-6AL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1c-6BL.sv1 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1c-6BL.sv2 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1c-6BL.sv3 | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1c-6DL | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| put.TaAOX1e-3DS | H | L | R | R | L | Q | M | A | L | S | F | Q | S | L | A | M | R | V |
| TaAOX1d-2AL.1 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TaAOX1d-2AL.2.sv1 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TaAOX1d-2AL.2.sv2 | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TaAOX1d-2DL | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| put.TaAOX1d-like-4AS | H | L | R | R | L | Q | M | V | L | S | F | H | N | M | A | M | R | V |
| TuAOX1a* | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TuAOX1c* | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| TuAOX1d.1* | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| TuAOX1d.2* | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| AetAOX1a* | H | L | R | R | L | Q | M | M | L | S | F | Q | S | L | A | M | R | V |
| AetAOX1e* | H | L | R | R | L | Q | M | A | L | S | F | Q | S | L | A | M | R | V |
| AetAOX1d* | H | L | R | R | L | Q | M | V | L | S | F | H | S | M | A | M | R | V |
| AetAOX1d-like* | H | L | R | R | L | - | M | V | L | S | F | H | S | M | A | M | - | - |
Completely conserved residues (yellow), highly conserved (green), and semi-conserved (white) in the wheat AOX family are shown. The “reg” proteins were not analyzed due to the absence of the functionally necessary motifs.
*Denotes diploid wheat AOX proteins.
◆Denotes residues which have been experimentally tested.
Another highly essential domain is the hydrophobic cavity consisting of 33 residues known to facilitate quinol-binding in the TbAOX active site (Table 11) [135]. Except in the cases of TaAOX1a-like-2DL, put.TaAOX1d-like-4AS and AetAOX1d-like, nine out of the 33 residues show complete conservation with TbAOX (F102, L122, V125, A126, V128, Y198, S201, V209, L212) emphasizing the crucial nature of these residues in active site efficiency (Tables 11 and S14). With the exception of put.TaAOX1d-like-4AS and AetAOX1d-like, there are eight substitutions which are conserved in all the wheat residues in this hydrophobic cavity (L179E, V181A, S182L, I189V, M190F, F193A, L194Y, V205A) (Tables 11 and S14). The AOX1e clade also has distinct residues or substitutions (T94A, C95M or C95I, W97, A197T, I200A, F204V, F208M) (Tables 11 and S14). With the exception of AetAOX1d-like, there are five residues or substitutions conserved in all wheat AOX1d proteins (S117, R118H, F121L, P178W, F208) (Tables 11 and S14). In addition, there are residues or substitutions peculiar to AOX1d group 1 (S91I, T94, L98G, F99S, T186) or AOX1d group 2 (S91V, F99R, T186A) efficiency (Fig 1; Tables 11 and S14). Substitutions of some of these residues have been shown to cause partial or complete attenuation of AOX activity (R118, L122, Y198 and L212) (Tables 11, S11 and S12). In rTbAOX, R118A and R118Q abolish nearly all AOX function even though some side chain chemistry is conserved for the latter mutation (S11 Table) [133]. The observation that R118H is conserved in one wheat AOX clade suggests that this substitution may be important for how the Type AOX1d isoforms function in wheat and the R118H mutation should be studied in this context.
Table 11. Comparison of residues in the hydrophobic cavity of the TbAOX and wheat AOX proteins.
| TbAOX Residue Numbers in Hydrophobic Cavity | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S91 | T94 | C95 | W97 | L98 | F99 | F102 | S117 | R118◆ | F121 | L122◆ | V125 | A126 | V128 | P178 | L179 | V181 | S182 | I185 | T186 | I189 | M190 | F193 | L194 | A197 | Y198◆ | I200 | S201 | F204 | V205 | F208 | V209 | L212◆ | |
| TaAOX1a-2AL.sv1 | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TaAOX1a-2AL.sv2 | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TaAOX1a-2BL | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TaAOX1a-2DL.sv1 | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TaAOX1a-2DL.sv2 | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TaAOX1a-like-2DL | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | E | A | L | A | V | V | F | A | Y | G | Y | V | S | F | A | V | V | L |
| TaAOX1c-6AL | T | S | L | V | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TaAOX1c-6BL.sv1 | T | S | L | V | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | V | S | F | A | V | V | L |
| TaAOX1c-6BL.sv2 | T | S | L | V | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | V | S | F | A | V | V | L |
| TaAOX1c-6BL.sv3 | T | S | L | V | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | V | S | F | A | V | V | L |
| TaAOX1c-6DL | T | S | L | V | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | V | S | F | A | V | V | L |
| Put.TaAOX1e-3DS | T | A | I | W | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | V | V | V | F | A | Y | T | Y | A | S | V | A | M | V | L |
| TaAOX1d-2AL.1 | I | T | L | A | G | S | F | S | H | L | L | V | A | V | W | E | A | L | A | T | V | F | A | Y | G | Y | V | S | F | A | F | V | L |
| TaAOX1d-2AL.2.sv1 | V | S | L | V | P | R | F | S | H | L | L | V | A | V | W | E | A | L | A | A | V | F | A | Y | G | Y | I | S | F | A | F | V | L |
| TaAOX1d-2AL.2.sv2 | V | S | L | V | P | R | F | S | H | L | L | V | A | V | W | E | A | L | A | A | V | F | A | Y | G | Y | I | S | F | A | F | V | L |
| TaAOX1d-2DL | V | S | L | V | P | R | F | S | H | L | L | V | A | V | W | E | A | L | A | A | V | F | A | Y | G | Y | I | S | F | A | F | V | L |
| put.TaAOX1d-like-4AS | - | - | - | - | - | - | F | S | H | L | L | V | A | V | C | E | A | L | P | T | V | F | A | Y | G | Y | V | S | F | A | F | V | L |
| TuAOX1a* | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| TuAOX1c* | T | S | L | V | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | V | S | F | A | V | V | L |
| TuAOX1d.1* | V | S | L | V | P | R | F | S | H | L | L | V | A | V | W | E | A | L | A | A | V | F | A | Y | G | Y | I | S | F | A | F | V | L |
| TuAOX1d.2* | I | T | L | K | G | S | F | S | H | L | L | V | A | V | W | E | A | L | A | T | V | F | A | Y | G | Y | V | S | F | A | F | V | L |
| AetAOX1a* | T | S | L | F | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | G | Y | I | S | F | A | V | V | L |
| AetAOX1e* | T | A | M | W | P | T | F | C | R | M | L | V | A | V | Y | E | A | L | A | V | V | F | A | Y | T | Y | A | S | V | A | M | V | L |
| AetAOX1d* | V | S | L | V | P | R | F | S | H | L | L | V | A | V | W | E | A | L | A | A | V | F | A | Y | G | Y | I | S | F | A | F | V | L |
| AetAOX1d-like* | I | T | L | A | G | S | F | S | H | L | L | V | A | V | - | - | - | - | - | - | V | F | A | Y | G | Y | I | S | L | - | - | - | - |
Green represents polar residues while red represents hydrophobic residues. Yellow represents residues with cyclic side-chains, and gray represent glycine. The “reg” proteins were not analyzed due to the absence of the functionally necessary motifs.
*Denotes diploid wheat AOX proteins.
◆Denotes residues which have been experimentally tested.
The ratios of hydrophobic to polar residues have been shown to be critical in how this cavity binds to substrates in the active site. With the exception of AOX1d group 1 and TaAOX1a-like-2DL, it was very clear that the ratios of sidechain chemistries were conserved in the clades (S15 Table). Functionally, these permutations in the wheat AOX amino acid sequences may suggest a gradient of enzyme function and activity which can be explored in the future. The absence of a significant number of these conserved hydrophobic-cavity residues in the TaAOX1a-like-2DL and AetAOX1d-like proteins (Table 11) may suggest distinct physiological roles for these isoforms such as interaction with other isoforms or proteins to exert control of subcellular localization or processes [137–139]. The situation in wheat may be similar to what has been suggested in other organisms with unique environments and physiologies, where residues critical for the configuration and stability of the active site or hydrophobic cavity in one species may be negligible in another [135, 140].
The information garnered could provide clues on how these proteins function in a polyploid monocot or grass species in general and highlights the urgent need for the biochemical elucidation of more AOX isoforms in diverse plant species. Given the similarities that exist within the wheat AOX isoforms especially those obtained via alternative splicing, it may be helpful to try to dissect how they function in different biological contexts (S16 Table). This can be done by mutation studies to study the effect of the substitutions or deletions, and biochemical methods which detect isoforms as well as posttranslational modifications which may change how and where these isomers function [141, 142].
Conclusions
Exploiting the structure of AOX holds potential for treating both human and plant diseases [135, 143, 144]. For plants, this is of paramount importance as there is the need to maintain or improve yield of important food and cash crops [145]. Elucidating the structure of AOX proteins in wheat may allow for many possibilities in addition to those already stated in this study. In A. thaliana, AOX induction has been shown to be a consequence of herbicide toxicity rather than evidence for tolerance [146]; however, the effect may be different in monocots. One could potentially exploit differences in the active site or hydrophobic substrate-binding cavities between hexaploid wheat isoforms and diploid wheat isoforms in order to more effectively design herbicides which affect diploid, wild and weedy ancestral wheat while allowing optimal growth of domesticated wheat which is used for food. The same approach can be used for designing better herbicides against other monocot and dicot weeds which can have a devastating effect on crop yield [147, 148]. On the other hand, the domestication of wheat has caused a loss in alleles which may be beneficial for yield. It may be advantageous to explore AOX structures in wild relatives and select those with the dual advantages of high expression and efficiency [149, 150]. The alleles for these isoforms that then correlate with resistance or tolerance to various forms of biotic and abiotic stress could then be introgressed into marketable wheat varieties or used as a template in order to effect gene-editing in highly profitable hexaploid wheat [151, 152]. In general, the TbAOX has been shown to have the best efficiency [135] but this may change as more plant AOX structures from organisms such as extremophiles and other monocots and dicots with variant ploidy are genetically and biochemically studied. The identification of the AOX gene family in wheat will contribute towards this process.
Supporting information
The number of splice variant isomers for a protein are denoted in the dark gray circle when applicable. Colored boxes distinguish the different AOX clades.
(PDF)
(PDF)
Blue indicates presence of Type 1 residues. Red indicates a Type 2 residue. Green indicates residues for monocot Type 1(d). Yellow indicates Type 1(a-c/e). Purple represents amino acid residues that did not match either classification. Black represents residues that were absent.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Diagram not to scale.
(PDF)
The “reg” proteins were not analyzed due to absence of the amino acids within this region.
(PDF)
RC in the wheat sequences refers to sequences which had high sequence similarity in the reverse complement from which the protein was translated.
(XLSX)
The highlighted residues in the motif are critical in the active site diiron center. X indicates presence of motifs. *Indicates diploid wheat isoforms.
(PDF)
*Denotes negative regulators of AOX expression.
(PDF)
The nucleotide sequence in bold indicates cases where the motif YTTGNNNNNVAMV has a single nucleotide deviation (site of change underlined) from the MDM motif CTTGNNNNNCAMG. *Denotes negative regulators of AOX expression.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The model used was c3vvaD. *Models to c3rylB.
(PDF)
The model used was c3vvaD.
(PDF)
*Denotes diploid AOX proteins.
(PDF)
The residues that are not highlighted are from Moore et al. 2013; red highlights are residues from Shiba et al. 2013; green highlights are residues from Young et al. 2014 and Crichton et al. 2010.
(PDF)
The residues that are not highlighted are from Moore et al. 2013; red highlights are residues from Shiba et al. 2013; green highlights are residues from Young et al. 2014 and Crichton et al. 2010.
(XLSX)
(PDF)
(PDF)
(XLSX)
(XLSX)
(ZIP)
Acknowledgments
We thank Dr. Chelsea K. Thornburg for advice on protein analysis and for critical reading of the manuscript. We also thank Kathryn J. Fitzgerald for laboratory assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Orville A. Vogel Wheat Research Fund in a grant awarded to K.A.S. http://cahnrs.wsu.edu/research/grant-resources/internal-competitive-grants/vogel-rfp/.
References
- 1.Reynolds M, Bonnett D, Chapman SC, Furbank RT, Manes Y, Mather DE, et al. Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. Journal of Experimental Botany. 2011;62(2):439–52. 10.1093/jxb/erq311 [DOI] [PubMed] [Google Scholar]
- 2.Longin CFH, Reif JC. Redesigning the exploitation of wheat genetic resources. Trends in Plant Science. 2014;19(10):631–6. 10.1016/j.tplants.2014.06.012 [DOI] [PubMed] [Google Scholar]
- 3.Zhao C, Liu B, Piao SL, Wang XH, Lobell DB, Huang Y, et al. Temperature increase reduces global yields of major crops in four independent estimates. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(35):9326–31. 10.1073/pnas.1701762114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, et al. Food Security: The Challenge of Feeding 9 Billion People. Science. 2010;327(5967):812–8. 10.1126/science.1185383 [DOI] [PubMed] [Google Scholar]
- 5.Balla K, Rakszegi M, Li ZG, Bekes F, Bencze S, Veisz O. Quality of Winter Wheat in Relation to Heat and Drought Shock after Anthesis. Czech Journal of Food Sciences. 2011;29(2):117–28. [Google Scholar]
- 6.Ashraf M. Stress-Induced Changes in Wheat Grain Composition and Quality. Critical Reviews in Food Science and Nutrition. 2014;54(12):1576–83. 10.1080/10408398.2011.644354 [DOI] [PubMed] [Google Scholar]
- 7.Rakszegi M, Lovegrove A, Balla K, Lang L, Bedo Z, Veisz O, et al. Effect of heat and drought stress on the structure and composition of arabinoxylan and beta-glucan in wheat grain. Carbohydrate Polymers. 2014;102:557–65. 10.1016/j.carbpol.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 8.Wang WX, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218(1):1–14. 10.1007/s00425-003-1105-5 [DOI] [PubMed] [Google Scholar]
- 9.dos Reis SP, Lima AM, de Souza CRB. Recent Molecular Advances on Downstream Plant Responses to Abiotic Stress. International Journal of Molecular Sciences. 2012;13(7):8628–47. 10.3390/ijms13078628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanlerberghe GC. Alternative Oxidase: A Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis during Abiotic and Biotic Stress in Plants. International Journal of Molecular Sciences. 2013;14(4):6805–47. 10.3390/ijms14046805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore AL, Albury MS. Further insights into the structure of the alternative oxidase: from plants to parasites. Biochemical Society Transactions. 2008;36:1022–6. 10.1042/BST0361022 [DOI] [PubMed] [Google Scholar]
- 12.Moore AL, Shiba T, Young L, Harada S, Kita K, Ito K. Unraveling the Heater: New Insights into the Structure of the Alternative Oxidase. Annual Review of Plant Biology, Vol 64. 2013;64:637–63. 10.1146/annurev-arplant-042811-105432 [DOI] [PubMed] [Google Scholar]
- 13.Ito K, Seymour RS. Expression of uncoupling protein and alternative oxidase depends on lipid or carbohydrate substrates in thermogenic plants. Biology Letters. 2005;1(4):427–30. 10.1098/rsbl.2005.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhoads DM, McIntosh L. Isolation and characterization of a cDNA clone encoding an alternative oxidase protein of Sauromatum guttatum (Schott). Proceedings of the National Academy of Sciences of the United States of America. 1991;88(6):2122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elthon TE, Nickels RL, McIntosh L. Monoclonal-antibodies to the alternative oxidase of higher-plant mitochondria. Plant Physiology. 1989;89(4):1311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa JH, McDonald AE, Arnholdt-Schmitt B, Fernandes de Melo D. A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion. 2014;19 Pt B:172–83. [DOI] [PubMed] [Google Scholar]
- 17.Costa JH, dos Santos CP, Lima BDE, Netto ANM, Saraiva K, Arnholdt-Schmitt B. In silico identification of alternative oxidase 2 (AOX2) in monocots: A new evolutionary scenario. Journal of Plant Physiology. 2017;210:58–63. 10.1016/j.jplph.2016.12.009 [DOI] [PubMed] [Google Scholar]
- 18.Borecky J, Nogueira FTS, de Oliveira KAP, Maia IG, Vercesi AE, Arruda P. The plant energy-dissipating mitochondrial systems: depicting the genomic structure and the expression profiles of the gene families of uncoupling protein and alternative oxidase in monocots and dicots. Journal of Experimental Botany. 2006;57(4):849–64. 10.1093/jxb/erj070 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Xi DM, Wang J, Zhu DF, Guo XQ. Functional analysis reveals effects of tobacco alternative oxidase gene (NtAOX1a) on regulation of defence responses against abiotic and biotic stresses. Bioscience Reports. 2009;29(6):375–83. 10.1042/BSR20080133 [DOI] [PubMed] [Google Scholar]
- 20.Campos MD, Cardoso HG, Linke B, Costa JH, de Melo DF, Justo L, et al. Differential expression and co-regulation of carrot AOX genes (Daucus carota). Physiologia Plantarum. 2009;137(4):578–91. 10.1111/j.1399-3054.2009.01282.x [DOI] [PubMed] [Google Scholar]
- 21.Considine MJ, Daley DO, Whelan J. The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiology. 2001;126(4):1619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frederico AM, Zavattieri MA, Campos MD, Cardoso HG, McDonald AE, Arnholdt-Schmitt B. The gymnosperm Pinus pinea contains both AOX gene subfamilies, AOX1 and AOX2. Physiologia Plantarum. 2009;137(4):566–77. 10.1111/j.1399-3054.2009.01279.x [DOI] [PubMed] [Google Scholar]
- 23.Costa JH, Mota EF, Virginia Cambursano M, Alexander Lauxmann M, Nogueira de Oliveira LM, Silva Lima MdG, et al. Stress-induced co-expression of two alternative oxidase (VuAox1 and 2b) genes in Vigna unguiculata. Journal of Plant Physiology. 2010;167(7):561–70. 10.1016/j.jplph.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 24.Sweetman C, Soole KL, Jenkins CLD, Day DA. Genomic structure and expression of alternative oxidase genes in legumes. Plant, Cell & Environment. 2018; 10.1111/pce.13161 [DOI] [PubMed] [Google Scholar]
- 25.Li F, Zhang Y, Wang MM, Wu XL, Guo XQ. Molecular cloning and expression characteristics of alternative oxidase gene of cotton (Gossypium hirsutum). Molecular Biology Reports. 2008;35(2):97–105. 10.1007/s11033-007-9058-6 [DOI] [PubMed] [Google Scholar]
- 26.Wanniarachchi VR, Dametto L, Sweetman C, Shavrukov Y, Day DA, Jenkins CLD, et al. Alternative Respiratory Pathway Component Genes (AOX and ND) in Rice and Barley and Their Response to Stress. International Journal of Molecular Sciences. 2018;19(3):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polidoros AN, Mylona PV, Pasentsis K, Scandalios JG, Tsaftaris AS. The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress. Redox Report. 2005;10(2):71–8. 10.1179/135100005X21688 [DOI] [PubMed] [Google Scholar]
- 28.Considine MJ, Holtzapffel RC, Day DA, Whelan J, Millar AH. Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiology. 2002;129(3):949–53. 10.1104/pp.004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpova OV, Kuzmin EV, Elthon TE, Newton KJ. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell. 2002;14(12):3271–84. 10.1105/tpc.005603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinakar C, Vishwakarma A, Raghayendra AS, Padmasree K. Alternative Oxidase Pathway Optimizes Photosynthesis During Osmotic and Temperature Stress by Regulating Cellular ROS, Malate Valve and Antioxidative Systems. Frontiers in Plant Science. 2016;7:17 10.3389/fpls.2016.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha B, Borovskii G, Panda SK. Alternative oxidase and plant stress tolerance. Plant Signaling & Behavior. 2016;11(12):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saisho D, Nakazono M, Lee KH, Tsutsumi N, Akita S, Hirai A. The gene for alternative oxidase-2 (AOX2) from Arabidopsis thaliana consists of five exons unlike other AOX genes and is transcribed at an early stage during germination. Genes & Genetic Systems. 2001;76(2):89–97. [DOI] [PubMed] [Google Scholar]
- 33.Chai TT, Simmonds D, Day DA, Colmer TD, Finnegan PM. Photosynthetic Performance and Fertility Are Repressed in GmAOX2b Antisense Soybean. Plant Physiology. 2010;152(3):1638–49. 10.1104/pp.109.149294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chai TT, Simmonds D, Day DA, Colmer TD, Finnegan PM. A GmAOX2b antisense gene compromises vegetative growth and seed production in soybean. Planta. 2012;236(1):199–207. 10.1007/s00425-012-1601-6 [DOI] [PubMed] [Google Scholar]
- 35.Costa JH, Jolivet Y, Hasenfratz-Sauder M-P, Orellano EG, Lima MdGS, Dizengremel P, et al. Alternative oxidase regulation in roots of Vigna unguiculata cultivars differing in drought/salt tolerance. Journal of Plant Physiology. 2007;164(6):718–27. 10.1016/j.jplph.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 36.Takumi S, Tomioka M, Eto K, Naydenov N, Nakamura C. Characterization of two non-homoeologous nuclear genes encoding mitochondrial alternative oxidase in common wheat. Genes & Genetic Systems. 2002;77(2):81–8. [DOI] [PubMed] [Google Scholar]
- 37.Bartoli CG, Gomez F, Gergoff G, Guiamet JJ, Puntarulo S. Up-regulation of the mitochondrial alternative oxidase pathway enhances photosynthetic electron transport under drought conditions. Journal of Experimental Botany. 2005;56(415):1269–76. 10.1093/jxb/eri111 [DOI] [PubMed] [Google Scholar]
- 38.Feng H, Duan J, Li H, Liang H, Li X, Han N. Alternative respiratory pathway under drought is partially mediated by hydrogen peroxide and contributes to antioxidant protection in wheat leaves. Plant Production Science. 2008;11(1):59–66. [Google Scholar]
- 39.Feng H, Li X, Duan J, Li H, Liang H. Chilling tolerance of wheat seedlings is related to an enhanced alternative respiratory pathway. Crop Science. 2008;48(6):2381–8. [Google Scholar]
- 40.Garmash EV, Dymova OV, Malyshev RV, Plyusnina SN, Golovko TK. Developmental changes in energy dissipation in etiolated wheat seedlings during the greening process. Photosynthetica. 2013;51(4):497–508. [Google Scholar]
- 41.Garmash EV, Malyshev RV, Shelyakin MA, Golovko TK. Activities of respiratory pathways and the pool of nonstructural carbohydrates in greening leaves of spring wheat seedlings. Russian Journal of Plant Physiology. 2014;61(2):160–8. [Google Scholar]
- 42.Garmash EV, Grabelnych OI, Velegzhaninov IO, Borovik OA, Dalke IV, Voinikov VK, et al. Light regulation of mitochondrial alternative oxidase pathway during greening of etiolated wheat seedlings. Journal of plant physiology. 2015;174:75–84. 10.1016/j.jplph.2014.09.016 [DOI] [PubMed] [Google Scholar]
- 43.Garmash EV, Velegzhaninov IO, Grabelnych OI, Borovik OA, Silina EV, Voinikov VK, et al. Expression profiles of genes for mitochondrial respiratory energy-dissipating systems and antioxidant enzymes in wheat leaves during de-etiolation. Journal of Plant Physiology. 2017;215:110–21. 10.1016/j.jplph.2017.05.023 [DOI] [PubMed] [Google Scholar]
- 44.Grabel'nykh OI, Pobezhimova TP, Pavlovskaya NS, Koroleva NA, Borovik OA, Lyubushkina IV, et al. Antioxidant function of alternative oxidase in mitochondria of winter wheat during cold hardening. Biologicheskie Membrany. 2011;28(4):274–83. [Google Scholar]
- 45.Grabelnych OI, Borovik OA, Tauson EL, Pobezhimova TP, Katyshev AI, Pavlovskaya NS, et al. Mitochondrial energy-dissipating systems (alternative oxidase, uncoupling proteins, and external NADH dehydrogenase) are involved in development of frost-resistance of winter wheat seedlings. Biochemistry-Moscow. 2014;79(6):506–19. 10.1134/S0006297914060030 [DOI] [PubMed] [Google Scholar]
- 46.Jacoby RP, Millar AH, Taylor NL. Wheat Mitochondrial Proteomes Provide New Links between Antioxidant Defense and Plant Salinity Tolerance. Journal of Proteome Research. 2010;9(12):6595–604. 10.1021/pr1007834 [DOI] [PubMed] [Google Scholar]
- 47.Naydenov NG, Khanam S, Siniauskaya M, Nakamura C. Profiling of mitochondrial transcriptome in germinating wheat embryos and seedlings subjected to cold, salinity and osmotic stresses. Genes & Genetic Systems. 2010;85(1):31–42. [DOI] [PubMed] [Google Scholar]
- 48.Vassileva V, Simova-Stoilova L, Demirevska K, Feller U. Variety-specific response of wheat (Triticum aestivum L.) leaf mitochondria to drought stress. Journal of Plant Research. 2009;122(4):445–54. 10.1007/s10265-009-0225-9 [DOI] [PubMed] [Google Scholar]
- 49.Sugie A, Murai K, Takumi S. Alteration of respiration capacity and transcript accumulation level of alternative oxidase genes in necrosis lines of common wheat. Genes & Genetic Systems. 2007;82(3):231–9. [DOI] [PubMed] [Google Scholar]
- 50.Mizuno N, Sugie A, Kobayashi F, Takumi S. Mitochondrial alternative pathway is associated with development of freezing tolerance in common wheat. Journal of Plant Physiology. 2008;165(4):462–7. 10.1016/j.jplph.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 51.Naydenov NG, Khanam SM, Atanassov A, Nakamura C. Expression profiles of respiratory components associated with mitochondrial biogenesis during germination and seedling growth under normal and restricted conditions in wheat. Genes & Genetic Systems. 2008;83(1):31–41. [DOI] [PubMed] [Google Scholar]
- 52.Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Research. 2017;27(5):885–96. 10.1101/gr.217117.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia JZ, Zhao SC, Kong XY, Li YR, Zhao GY, He WM, et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496(7443):91–5. 10.1038/nature12028 [DOI] [PubMed] [Google Scholar]
- 54.Ling HQ, Zhao SC, Liu DC, Wang JY, Sun H, Zhang C, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496(7443):87–90. 10.1038/nature11997 [DOI] [PubMed] [Google Scholar]
- 55.Borrill P, Ramirez-Gonzalez R, Uauy C. expVIP: a Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiology. 2016;170(4):2172–86. 10.1104/pp.15.01667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolser D, Staines DM, Pritchard E, Kersey P. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data In: Edwards D, editor. Plant Bioinformatics: Methods and Protocols, 2nd Edition. Methods in Molecular Biology. 1374. Totowa: Humana Press Inc; 2016. p. 115–40. [DOI] [PubMed] [Google Scholar]
- 57.Mayer KFX, Rogers J, Dolezel J, Pozniak C, Eversole K, Feuillet C, et al. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345(6194):11. [DOI] [PubMed] [Google Scholar]
- 58.Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research. 2012;40(D1):D1178–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, et al. Database Resources of the National Center for Biotechnology. Nucleic Acids Research. 2017;45(D1):D12–D7. 10.1093/nar/gkw1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–7. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Systematic Biology. 2008;57(5):758–71. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 62.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology. 2012;61(3):539–42. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v. 1.6. http://beastbioedacuk/Tracer. 2014.
- 64.Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nobre T, Campos MD, Lucic-Mercy E, Arnholdt-Schmitt B. Misannotation Awareness: A Tale of Two Gene-Groups. Frontiers in Plant Science. 2016;7:7 10.3389/fpls.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300(4):1005–16. 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- 68.Yao QM, Ge HY, Wu SQ, Zhang N, Chen W, Xu CH, et al. (PDB)-D-3 3.0: From plant phosphorylation sites to protein networks. Nucleic Acids Research. 2014;42(D1):D1206–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao JJ, Thelen JJ, Dunker AK, Xu D. Musite, a Tool for Global Prediction of General and Kinase-specific Phosphorylation Sites. Molecular & Cellular Proteomics. 2010;9(12):2586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends in Genetics. 2000;16(6):276–7. [DOI] [PubMed] [Google Scholar]
- 71.Wagner S, Van Aken O, Elsässer M, Schwarzländer M. Mitochondrial energy signaling and its role in the low oxygen stress response of plants. Plant Physiology. 2018;176(2):1156–70. 10.1104/pp.17.01387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research. 2002;30(1):325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin JP, Tian F, Yang DC, Meng YQ, Kong L, Luo JC, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research. 2017;45(D1):D1040–D5. 10.1093/nar/gkw982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, et al. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research. 2016;44(D1):D1154–D60. 10.1093/nar/gkv1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 2015;10(6):845–58. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25(13):1605–12. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 77.Ma J, Yang YJ, Luo W, Yang CC, Ding PY, Liu YX, et al. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). Plos One. 2017;12(7):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castro JA, Ferreira MDG, Silva RJS, Andrade BS, Micheli F. Alternative oxidase (AOX) constitutes a small family of proteins in Citrus clementina and Citrus sinensis L. Osb. Plos One. 2017;12(5):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Velada I, Cardoso HG, Ragonezi C, Nogales A, Ferreira A, Valadas V, et al. Alternative Oxidase Gene Family in Hypericum perforatum L.: Characterization and Expression at the Post-germinative Phase. Frontiers in Plant Science. 2016;7:16 10.3389/fpls.2016.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan HD, Zhang AL, Chen J, He XY, Xu B, Xie GQ, et al. Genome-Wide Analysis of the PvHsp20 Family in Switchgrass: Motif, Genomic Organization, and Identification of Stress or Developmental-Related Hsp20s. Frontiers in Plant Science. 2017;8:15 10.3389/fpls.2017.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stenoien HK. Compact genes are highly expressed in the moss Physcomitrella patens. Journal of Evolutionary Biology. 2007;20(3):1223–9. 10.1111/j.1420-9101.2007.01301.x [DOI] [PubMed] [Google Scholar]
- 82.Mukhopadhyay P, Ghosh TC. Relationship between Gene Compactness and Base Composition in Rice and Human Genome. Journal of Biomolecular Structure & Dynamics. 2010;27(4):477–88. [DOI] [PubMed] [Google Scholar]
- 83.Ren XY, Vorst O, Fiers M, Stiekema WJ, Nap JP. In plants, highly expressed genes are the least compact. Trends in Genetics. 2006;22(10):528–32. 10.1016/j.tig.2006.08.008 [DOI] [PubMed] [Google Scholar]
- 84.Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP. Effect of 5 ' UTR introns on gene expression in Arabidopsis thaliana. Bmc Genomics. 2006;7:13 10.1186/1471-2164-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wen SS, Wen NA, Pang JS, Langen G, Brew-Appiah RAT, Mejias JH, et al. Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(50):20543–8. 10.1073/pnas.1217927109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei XJ, Song XW, Wei LY, Tang SQ, Sun J, Hu PS, et al. An epiallele of rice AK1 affects photosynthetic capacity. Journal of Integrative Plant Biology. 2017;59(3):158–63. 10.1111/jipb.12518 [DOI] [PubMed] [Google Scholar]
- 87.Iniguez LP, Hernandez G. The Evolutionary Relationship between Alternative Splicing and Gene Duplication. Frontiers in Genetics. 2017;8:7 10.3389/fgene.2017.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of Gene Duplication in Plants. Plant Physiology. 2016;171(4):2294–316. 10.1104/pp.16.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, et al. A Membrane-Bound NAC Transcription Factor, ANAC017, Mediates Mitochondrial Retrograde Signaling in Arabidopsis. Plant Cell. 2013;25(9):3450–71. 10.1105/tpc.113.113985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH, et al. Mitochondrial and Chloroplast Stress Responses Are Modulated in Distinct Touch and Chemical Inhibition Phases. Plant Physiology. 2016;171(3):2150–65. 10.1104/pp.16.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Aken O, Zhang BT, Law S, Narsai R, Whelan J. AtWRKY40 and AtWRKY63 Modulate the Expression of Stress-Responsive Nuclear Genes Encoding Mitochondrial and Chloroplast Proteins. Plant Physiology. 2013;162(1):254–71. 10.1104/pp.113.215996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giraud E, Van Aken O, Ho LHM, Whelan J. The Transcription Factor ABI4 Is a Regulator of Mitochondrial Retrograde Expression of ALTERNATIVE OXIDASE1a. Plant Physiology. 2009;150(3):1286–96. 10.1104/pp.109.139782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, et al. The Membrane-Bound NAC Transcription Factor ANAC013 Functions in Mitochondrial Retrograde Regulation of the Oxidative Stress Response in Arabidopsis. Plant Cell. 2013;25(9):3472–90. 10.1105/tpc.113.117168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gasch P, Fundinger M, Muller JT, Lee T, Bailey-Serres J, Mustroph A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell. 2016;28(1):160–80. 10.1105/tpc.15.00866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yao X, Li JJ, Liu JP, Liu KD. An Arabidopsis mitochondria-localized RRL protein mediates abscisic acid signal transduction through mitochondrial retrograde regulation involving ABI4. Journal of Experimental Botany. 2015;66(20):6431–45. 10.1093/jxb/erv356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, et al. Cyclin-dependent Kinase E1 (CDKE1) Provides a Cellular Switch in Plants between Growth and Stress Responses. Journal of Biological Chemistry. 2013;288(5):3449–59. 10.1074/jbc.M112.416727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moellering ER, Benning C. Phosphate Regulation of Lipid Biosynthesis in Arabidopsis Is Independent of the Mitochondrial Outer Membrane DGS1 Complex. Plant Physiology. 2010;152(4):1951–9. 10.1104/pp.110.153262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang XH, Ivanova A, Vandepoele K, Radomiljac J, Van de Velde J, Berkowitz O, et al. The Transcription Factor MYB29 Is a Regulator of ALTERNATIVE OXIDASE1a(1). Plant Physiology. 2017;173(3):1824–43. 10.1104/pp.16.01494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brosche M, Blomster T, Salojarvi J, Cui FQ, Sipari N, Leppala J, et al. Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by RADICAL-INDUCED CELL DEATH1. Plos Genetics. 2014;10(2):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, et al. The fate of duplicated genes in a polyploid plant genome. Plant Journal. 2013;73(1):143–53. 10.1111/tpj.12026 [DOI] [PubMed] [Google Scholar]
- 101.Hughes TE, Langdale JA, Kelly S. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Research. 2014;24(8):1348–55. 10.1101/gr.172684.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brinton J, Simmonds J, Uauy C. Ubiquitin-related genes are differentially expressed in isogenic lines contrasting for pericarp cell size and grain weight in hexaploid wheat. Bmc Plant Biology. 2018;18:17 10.1186/s12870-018-1233-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pearce S, Vazquez-Gross H, Herin SY, Hane D, Wang Y, Gu YQ, et al. WheatExp: an RNA-seq expression database for polyploid wheat. Bmc Plant Biology. 2015;15:8 10.1186/s12870-014-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wurschum T, Boeven PHG, Langer SM, Longin CFH, Leiser WL. Multiply to conquer: Copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. Bmc Genetics. 2015;16:8 10.1186/s12863-015-0167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wurschum T, Longin CFH, Hahn V, Tucker MR, Leiser WL. Copy number variations of CBF genes at the Fr-A2 locus are essential components of winter hardiness in wheat. Plant Journal. 2017;89(4):764–73. 10.1111/tpj.13424 [DOI] [PubMed] [Google Scholar]
- 106.Scafaro AP, Galle A, Van Rie J, Carmo-Silva E, Salvucci ME, Atwell BJ. Heat tolerance in a wild Oryza species is attributed to maintenance of Rubisco activation by a thermally stable Rubisco activase ortholog. New Phytologist. 2016;211(3):899–911. 10.1111/nph.13963 [DOI] [PubMed] [Google Scholar]
- 107.Covshoff S, Szecowka M, Hughes TE, Smith-Unna R, Kelly S, Bailey KJ, et al. C-4 Photosynthesis in the Rice Paddy: Insights from the Noxious Weed Echinochloa glabrescens. Plant Physiology. 2016;170(1):57–73. 10.1104/pp.15.00889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Placido DF, Campbell MT, Folsom JJ, Cui XP, Kruger GR, Baenziger PS, et al. Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiology. 2013;161(4):1806–19. 10.1104/pp.113.214262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hedayati V, Mousavi A, Razavi K, Cultrera N, Alagna F, Mariotti R, et al. Polymorphisms in the AOX2 gene are associated with the rooting ability of olive cuttings. Plant Cell Reports. 2015;34(7):1151–64. 10.1007/s00299-015-1774-0 [DOI] [PubMed] [Google Scholar]
- 110.Feng HQ, Guan DD, Sun K, Wang YF, Zhang TG, Wang RF. Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochimica Et Biophysica Sinica. 2013;45(12):985–94. 10.1093/abbs/gmt094 [DOI] [PubMed] [Google Scholar]
- 111.Lager I, Andreasson O, Dunbar TL, Andreasson E, Escobar MA, Rasmusson AG. Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell and Environment. 2010;33(9):1513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kosova K, Vitamvas P, Urban MO, Prasil IT, Renaut J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Frontiers in Plant Science. 2018;9:22 10.3389/fpls.2018.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fu AG, Liu HY, Yu F, Kambakam S, Luan S, Rodermel S. Alternative Oxidases (AOX1a and AOX2) Can Functionally Substitute for Plastid Terminal Oxidase in Arabidopsis Chloroplasts. Plant Cell. 2012;24(4):1579–95. 10.1105/tpc.112.096701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shen JB, Zeng YL, Zhuang XH, Sun L, Yao XQ, Pimpl P, et al. Organelle pH in the Arabidopsis Endomembrane System. Molecular Plant. 2013;6(5):1419–37. 10.1093/mp/sst079 [DOI] [PubMed] [Google Scholar]
- 115.Kiraga J, Mackiewicz P, Mackiewicz D, Kowalczuk M, Biecek P, Polak N, et al. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. Bmc Genomics. 2007;8:16 10.1186/1471-2164-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schwartz R, Ting CS, King J. Whole proteome pl values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Research. 2001;11(5):703–9. 10.1101/gr.158701 [DOI] [PubMed] [Google Scholar]
- 117.Hamilton V, Singha UK, Smith JT, Weems E, Chaudhuri M. Trypanosome alternative oxidase possesses both an N-Terminal and internal mitochondrial targeting signal. Eukaryotic Cell. 2014;13(4):539–47. 10.1128/EC.00312-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murcha MW, Kmiec B, Kubiszewski-Jakubiak S, Teixeira PF, Glaser E, Whelan J. Protein import into plant mitochondria: signals, machinery, processing, and regulation. Journal of Experimental Botany. 2014;65(22):6301–35. 10.1093/jxb/eru399 [DOI] [PubMed] [Google Scholar]
- 119.Garg SG, Gould SB. The Role of Charge in Protein Targeting Evolution. Trends in Cell Biology. 2016;26(12):894–905. 10.1016/j.tcb.2016.07.001 [DOI] [PubMed] [Google Scholar]
- 120.Selinski J, Hartmann A, Kordes A, Deckers-Hebestreit G, Whelan J, Scheibe R. Analysis of posttranslational activation of alternative oxidase Isoforms. Plant Physiology. 2017;174(4):2113–27. 10.1104/pp.17.00681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kumar K, Muthamilarasan M, Bonthala VS, Roy R, Prasad M. Unraveling 14-3-3 proteins in C-4 panicoids with emphasis on model plant Setaria italica reveals phosphorylation-dependent subcellular localization of RS splicing factor. Plos One. 2015;10(4):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee A, Lee SS, Jung WY, Park HJ, Lim BR, Kim HS, et al. The OsCYP19-4 gene is expressed as multiple alternatively spliced transcripts encoding isoforms with distinct cellular localizations and PPIase activities under cold stress. International Journal of Molecular Sciences. 2016;17(7):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koo SC, Yoon HW, Kim CY, Moon BC, Cheong YH, Han HJ, et al. Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochemical and Biophysical Research Communications. 2007;360(1):188–93. 10.1016/j.bbrc.2007.06.052 [DOI] [PubMed] [Google Scholar]
- 124.Del-Saz NF, Ribas-Carbo M, McDonald AE, Lambers H, Fernie AR, Florez-Sarasa I. An in vivo perspective of the role(s) of the alternative oxidase pathway. Trends in Plant Science. 2018;23(3):206–19. 10.1016/j.tplants.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 125.Hartl M, Finkemeier I. Plant mitochondrial retrograde signaling: post-translational modifications enter the stage. Frontiers in Plant Science. 2012;3:7 10.3389/fpls.2012.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hosp F, Lassowskat I, Santoro V, De Vleesschauwer D, Fliegner D, Redestig H, et al. Lysine acetylation in mitochondria: From inventory to function. Mitochondrion. 2017;33:58–71. 10.1016/j.mito.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 127.Gibbs DJ. Emerging functions for N-terminal protein acetylation in plants. Trends in Plant Science. 2015;20(10):599–601. 10.1016/j.tplants.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blanco-Herrera F, Moreno AA, Tapia R, Reyes F, Araya M, D'Alessio C, et al. The UDP-glucose: glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. Bmc Plant Biology. 2015;15:12 10.1186/s12870-014-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takano S, Matsuda S, Funabiki A, Furukawa J, Yamauchi T, Tokuji Y, et al. The rice RCN11 gene encodes beta 1,2-xylosyltransferase and is required for plant responses to abiotic stresses and phytohormones. Plant Science. 2015;236:75–88. 10.1016/j.plantsci.2015.03.022 [DOI] [PubMed] [Google Scholar]
- 130.Zhang M, Henquet M, Chen ZZ, Zhang HR, Zhang Y, Ren XZ, et al. LEW3, encoding a putative alpha-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant Journal. 2009;60(6):983–99. 10.1111/j.1365-313X.2009.04013.x [DOI] [PubMed] [Google Scholar]
- 131.Jiang N, Wiemels RE, Soya A, Whitley R, Held M, Faik A. Composition, Assembly, and Trafficking of a Wheat Xylan Synthase Complex. Plant Physiology. 2016;170(4):1999–2023. 10.1104/pp.15.01777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yao DS, Qi WW, Li X, Yang Q, Yan SM, Ling HL, et al. Maize opaque10 encodes a cereal-specific protein that is essential for the proper distribution of zeins in endosperm protein bodies. Plos Genetics. 2016;12(8):25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 133.Shiba T, Kido Y, Sakamoto K, Inaoka DK, Tsuge C, Tatsumi R, et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(12):4580–5. 10.1073/pnas.1218386110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Young L, May B, Pendlebury-Watt A, Shearman J, Elliott C, Albury MS, et al. Probing the ubiquinol-binding site of recombinant Sauromatum guttatum alternative oxidase expressed in E. coli membranes through site-directed mutagenesis. Biochimica Et Biophysica Acta-Bioenergetics. 2014;1837(7):1219–25. [DOI] [PubMed] [Google Scholar]
- 135.May B, Young L, Moore AL. Structural insights into the alternative oxidases: are all oxidases made equal? Biochemical Society Transactions. 2017;45:731–40. 10.1042/BST20160178 [DOI] [PubMed] [Google Scholar]
- 136.Crichton PG, Albury MS, Affourtit C, Moore AL. Mutagenesis of the Sauromatum guttatum alternative oxidase reveals features important for oxygen binding and catalysis. Biochimica Et Biophysica Acta-Bioenergetics. 2010;1797(6–7):732–7. [DOI] [PubMed] [Google Scholar]
- 137.Nicolas M, Rodriguez-Buey ML, Franco-Zorrilla JM, Cubas P. A Recently Evolved Alternative Splice Site in the BRANCHED1a Gene Controls Potato Plant Architecture. Current Biology. 2015;25(14):1799–809. 10.1016/j.cub.2015.05.053 [DOI] [PubMed] [Google Scholar]
- 138.Pirrello J, Leclercq J, Dessailly F, Rio M, Piyatrakul P, Kuswanhadi K, et al. Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. Bmc Plant Biology. 2014;14:17 10.1186/1471-2229-14-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Haak DC, Fukao T, Grene R, Hua ZH, Ivanov R, Perrella G, et al. Multilevel Regulation of Abiotic Stress Responses in Plants. Frontiers in Plant Science. 2017;8:24 10.3389/fpls.2017.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pennisi R, Salvi D, Brandi V, Angelini R, Ascenzi P, Polticelli F. Molecular Evolution of Alternative Oxidase Proteins: A Phylogenetic and Structure Modeling Approach. Journal of Molecular Evolution. 2016;82(4–5):207–18. 10.1007/s00239-016-9738-8 [DOI] [PubMed] [Google Scholar]
- 141.Stastna M, Van Eyk JE. Analysis of protein isoforms: Can we do it better? Proteomics. 2012;12(19–20):2937–48. 10.1002/pmic.201200161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Doll S, Burlingame AL. Mass Spectrometry-Based Detection and Assignment of Protein Posttranslational Modifications. Acs Chemical Biology. 2015;10(1):63–71. 10.1021/cb500904b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miguez M, Reeve C, Wood PM, Hollomon DW. Alternative oxidase reduces the sensitivity of Mycosphaerella graminicola to QOI fungicides. Pest Management Science. 2004;60(1):3–7. 10.1002/ps.837 [DOI] [PubMed] [Google Scholar]
- 144.Xu T, Wang YT, Liang WS, Yao F, Li YH, Li DR, et al. Involvement of alternative oxidase in the regulation of sensitivity of Sclerotinia sclerotiorum to the fungicides azoxystrobin and procymidone. Journal of Microbiology. 2013;51(3):352–8. [DOI] [PubMed] [Google Scholar]
- 145.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484(7393):186–94. 10.1038/nature10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zulet A, Gil-Monreal M, Zabalza A, van Dongen JT, Royuela M. Fermentation and alternative oxidase contribute to the action of amino acid biosynthesis-inhibiting herbicides. Journal of Plant Physiology. 2015;175:102–12. 10.1016/j.jplph.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 147.Umm EK, Khan MA. Prediction of grain yield losses in wheat (Triticum aestivum L.) under different densities of wild oat (Avena fatua L.). Pakistan Journal of Botany. 2015;47:239–42. [Google Scholar]
- 148.Khan MA, Wahid F, Umme K, Marwat KB, Gul B, Inayatullah M, et al. Evaluating the threshold levels of Neslia apiculata in wheat and its effects on crop yield losses in swat valley of Khyber Pakhtunkhwa. Pakistan Journal of Botany. 2015;47:87–91. [Google Scholar]
- 149.Nobre T, Oliveira M, Arnholdt-Schmitt B. Wild Carrot Differentiation in Europe and Selection at DcAOX1 Gene? Plos One. 2016;11(10):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brozynska M, Furtado A, Henry RJ. Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant Biotechnology Journal. 2016;14(4):1070–85. 10.1111/pbi.12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim D, Alptekin B, Budak H. CRISPR/Cas9 genome editing in wheat. Functional & Integrative Genomics. 2018;18(1):31–41. [DOI] [PubMed] [Google Scholar]
- 152.Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances. 2015;33(1):41–52. 10.1016/j.biotechadv.2014.12.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number of splice variant isomers for a protein are denoted in the dark gray circle when applicable. Colored boxes distinguish the different AOX clades.
(PDF)
(PDF)
Blue indicates presence of Type 1 residues. Red indicates a Type 2 residue. Green indicates residues for monocot Type 1(d). Yellow indicates Type 1(a-c/e). Purple represents amino acid residues that did not match either classification. Black represents residues that were absent.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Diagram not to scale.
(PDF)
The “reg” proteins were not analyzed due to absence of the amino acids within this region.
(PDF)
RC in the wheat sequences refers to sequences which had high sequence similarity in the reverse complement from which the protein was translated.
(XLSX)
The highlighted residues in the motif are critical in the active site diiron center. X indicates presence of motifs. *Indicates diploid wheat isoforms.
(PDF)
*Denotes negative regulators of AOX expression.
(PDF)
The nucleotide sequence in bold indicates cases where the motif YTTGNNNNNVAMV has a single nucleotide deviation (site of change underlined) from the MDM motif CTTGNNNNNCAMG. *Denotes negative regulators of AOX expression.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The model used was c3vvaD. *Models to c3rylB.
(PDF)
The model used was c3vvaD.
(PDF)
*Denotes diploid AOX proteins.
(PDF)
The residues that are not highlighted are from Moore et al. 2013; red highlights are residues from Shiba et al. 2013; green highlights are residues from Young et al. 2014 and Crichton et al. 2010.
(PDF)
The residues that are not highlighted are from Moore et al. 2013; red highlights are residues from Shiba et al. 2013; green highlights are residues from Young et al. 2014 and Crichton et al. 2010.
(XLSX)
(PDF)
(PDF)
(XLSX)
(XLSX)
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.