Abstract
Eukaryotic cells have the ability to degrade proteins and organelles by selective and nonselective modes of micro- and macroautophagy. In addition, there exist both constitutive and regulated forms of autophagy. For example, pexophagy is a selective process for the regulated degradation of peroxisomes by autophagy. Our studies have shown that the differing pathways of autophagy have many molecular events in common. In this article, we have identified a new member in the family of autophagy genes. GSA12 in Pichia pastoris and its Saccharomyces cerevisiae counterpart, CVT18, encode a soluble protein with two WD40 domains. We have shown that these proteins are required for pexophagy and autophagy in P. pastoris and the Cvt pathway, autophagy, and pexophagy in S. cerevisiae. In P. pastoris, Gsa12 appears to be required for an early event in pexophagy. That is, the involution of the vacuole or extension of vacuole arms to engulf the peroxisomes does not occur in the gsa12 mutant. Consistent with its role in vacuole engulfment, we have found that this cytosolic protein is also localized to the vacuole surface. Similarly, Cvt18 displays a subcellular localization that distinguishes it from the characterized proteins required for cytoplasm-to-vacuole delivery pathways.
INTRODUCTION
The yeast vacuole is one of the most versatile organelles in the cell (Klionsky et al., 1990). As the primary degradative compartment, the vacuole maintains an acidic lumen and imports various hydrolytic enzymes (Bryant and Stevens, 1998). For most of the nonvacuolar organelles and macromolecules, entry to the vacuole will lead to their disassembly or degradation. Therefore, the vacuolar targeting pathways for degradative substrates are highly regulated in response to environmental or physiological stimuli (Scott and Klionsky, 1998; Klionsky and Emr, 2000).
On nutrient-limiting conditions, the ability to degrade and reuse cellular components becomes essential for survival. In Saccharomyces cerevisiae, macroautophagy is induced by nitrogen starvation (Takeshige et al., 1992). During this process, cytoplasmic components are enwrapped within an enclosed double-membrane structure termed an autophagosome (Baba et al., 1994, 1995). The outer membrane of the autophagosome then fuses with the vacuolar membrane and delivers the cargo, confined by the inner membrane, to the vacuole lumen. The single-membrane delimiting the autophagic body is then degraded by vacuolar hydrolases to allow access to the cargo. A similar mechanism is used in the cytoplasm-to-vacuole targeting (Cvt) pathway that is used for the localization of the resident hydrolase aminopeptidase I (Ape1) (Harding et al., 1995). The newly synthesized precursor Ape1 (prApe1) undergoes dodecamerization (Kim et al., 1997) and further assembles into a large Cvt complex in the cytosol (Baba et al., 1997). Subsequently, the Cvt complex becomes enclosed within double-membrane Cvt vesicles that transport it to the vacuole (Scott et al., 1997). The mechanistic similarity is reflected by the genetic overlap between these two pathways (Harding et al., 1996; Scott et al., 1996). In fact, the Cvt complex goes to the vacuole via autophagosomes under nitrogen starvation conditions (Baba et al., 1997). The formation of the double-membrane autophagosomes or Cvt vesicles is a very unique and intrinsically complex process. Accordingly, most of the autophagy (apg and aut) and Cvt pathway (cvt) mutants are found blocked at the membrane sequestration stage (Klionsky and Ohsumi, 1999; Kim and Klionsky, 2000; Stromhaug and Klionsky, 2001).
Organelles are also selectively targeted to the vacuole when they are no longer needed. The autophagic turnover of peroxisomes has been well demonstrated in the methylotrophic yeast Pichia pastoris (Tuttle et al., 1993). When grown in methanol, peroxisome proliferation and the synthesis of methanol-utilizing enzymes such as alcohol oxidase are both greatly induced. After a shift to carbon sources such as glucose or ethanol, excess peroxisomes are selectively degraded either by micropexophagy or macropexophagy, respectively (Tuttle and Dunn, 1995). Macropexophagy involves sequestration of peroxisomes by multimembrane vesicles in the cytosol. This process also results in the uptake of cytosolic material and requires newly synthesized proteins. In contrast, during micropexophagy, only peroxisomes are sequestered by the arm-like protrusions of the vacuolar membrane, and the sequestration process is independent of protein synthesis. Corresponding pathway-specific mutants have been isolated (Sakai et al., 1998; Yuan et al., 1999). However, mutants that are defective in both pathways also exist (Sakai et al., 1998), suggesting a partial overlap in the molecular components between these two pathways.
Recently, sequence homology between S. cerevisiae APG/CVT genes and P. pastoris GSA genes has been identified (Kim et al., 1999; Yuan et al., 1999). In agreement with these findings, degradation of peroxisomes in S. cerevisiae has been shown to be a specific process. Excess peroxisomes are selectively degraded in a vacuolar protease-dependent manner, and this process is blocked in most of the characterized cvt/apg mutants (Hutchins et al., 1999). The observation that these pathways overlap indicates that these autophagic-like vacuolar import processes for various cargoes are closely related. The parallel studies in these two model systems have facilitated our understanding of the proteins that play a role in cytoplasm-to-vacuole transport in both organisms. Similarly, an increasing number of homologous Apg/Cvt proteins have been identified in higher eukaryotes, including mammalian cells (reviewed in Kim and Klionsky, 2000; Klionsky and Emr, 2000). Hence, the molecular mechanism for regulated autophagy revealed by studies in yeast is well conserved during evolution to fulfill important cellular functions in all eukaryotes.
In this article, we describe the identification and characterization of Cvt18/Gsa12, a protein required for the Cvt pathway, autophagy, and pexophagy in S. cerevisiae, and for autophagy and pexophagy in P. pastoris. Cvt18/Gsa12 has a cellular distribution that is distinct from other characterized Apg/Cvt/Gsa proteins and appears to function at a relatively early stage in these pathways.
MATERIALS AND METHODS
Materials
The S. cerevisiae strain S288C genomic DNA was from Research Genetics (Huntsville, AL). The pREMI vector was a gift from Dr. Benjamin S. Glick (University of Chicago, Chicago, IL). Expre35S35S Protein Labeling Mix was from PerkinElmer Life Sciences (Boston, MA). Rabbit antisera against Ape1, alkaline phosphatase (Pho8), Vma2, Pep12, Pgk1, and Fox3 and mouse monocolonal antibodies to Pho8 and Dpm1 have been previously described (Klionsky et al., 1992; Scott et al., 1996; Tomashek et al., 1996; Hutchins et al., 1999; Kim et al., 2001b). Mouse anti-GFP and rabbit anti-hemagglutinin (HA) antibodies were purchased from Covance Research Products (Richmond, CA). Yeast nitrogen base was from Difco (Detroit, MI). All other reagents were identical to those described previously (Kim et al., 2001a; Wang et al., 2001).
Strains and Media
The yeast strains used in this study are listed in Table 1. To disrupt the chromosomal PHO13 locus in DKY6281 (SEY6210 pho8Δ::TRP1), the pho13Δ::URA3 plasmid pPH13 (Kaneko et al., 1989) was digested with EcoRI and transformed into DKY6281 to generate strain JGY1. The plasmid pTS15 (pep4Δ::URA3) (Rothman et al., 1986) was digested with EcoRI and transformed into strain JGY3 to disrupt the PEP4 locus and generate the cvt18Δ pep4Δ strain JGY7.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| S. cerevisiae | ||
| SEY6210 | MATα leu2-3,112 ura3-51 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | (Robinson et al., 1988) |
| TVY1 | SEY6210 pep4Δ∷LEU2 | (Gerhardt et al., 1998) |
| MGY101 | SEY6210 apg5Δ∷LEU2 | (George et al., 2000) |
| DKY6281 | SEY6210 pho8Δ∷TRP1 | (Conradt et al., 1994) |
| JGY1 | DKY6281 pho13Δ∷URA3 | This study |
| JGY3 | SEY6210 cvt18Δ∷HIS5 | This study |
| JGY5 | JGY1 cvt18Δ∷HIS5 | This study |
| JGY7 | JGY3 pep4Δ∷URA3 | This study |
| WCG4a | MATα his3-11,15 leu2-3,112 ura3 | (Heinemeyer et al., 1993) |
| YMTA | WCG4a pep4Δ∷HIS3 | (Thumm et al., 1994) |
| JGY9 | WCG4a cvt18Δ∷HIS5 | This study |
| JGY11 | SEY6210 CVT18∷CVT18GFP(LEU2) | This study |
| MGY103 | WCG4a pex5Δ∷HIS5 | This study |
| P. pastoris | ||
| GS115 | his4 | (Cregg et al., 1985) |
| SMD1163 | his4 pep4 prb1 | (Tuttle and Dunn, 1995) |
| PPF1 | his4 arg4 | (Yuan et al., 1997) |
| WDK01 | PPF1 gsa1∷ARG4 | (Yuan et al., 1997) |
| WDK09 | PPF1 gsa9Δ∷ARG4 | (Kim et al., 2001b) |
| DMM1 | GS115∷pDM1 (PAOX1BFP-SKL, Zeocin) | (Kim et al., 2001b) |
| WDY7 | his4 gsa7 | (Yuan et al., 1999) |
| R2 | GS115 gsa12∷Zeocin | This study |
| R22 | GS115 gsa11-2∷Zeocin | (Stromhaug, et al., 2001) |
| ANB7 | R2 his4∷pPS64 (PGAPGFP/HA-GSA9, HIS4) | This study |
| ANB12 | WDK09 his4∷pPS64 (PGAPGFP/HA-GSA9, HIS4) | This study |
| WDY37 | R22 his4∷pPS69 (PGAPGFP/HA-GSA11, HIS4) | (Stromhaug, et al., 2001) |
| WDY46 | R2 his4∷pPS69 (PGAPGFP/HA-GSA11, HIS4) | This study |
| PHT12 | DMM1 his4∷pPHT-G12 (PGAPGFP/HA-GSA12, HIS4) | This study |
| PHT13 | R2 his4∷pPHT-G12 (PGAPGFP/HA-GSA12, HIS4) | This study |
S. cerevisiae and P. pastoris strains were grown or incubated in media as defined previously (Tuttle and Dunn, 1995; Scott et al., 2001). Synthetic minimal medium (SMD) contains glucose and nitrogen (0.67% yeast nitrogen base, 2% glucose, with auxotrophic amino acids and vitamins as needed), and SD-N is synthetic medium containing glucose, but limited for nitrogen (0.17% yeast nitrogen base without ammonium sulfate or amino acids, 2% glucose). Zeocin was added at 25 μg/ml when culturing Escherichia coli (DH5α) and 100 μg/ml when culturing P. pastoris.
Disruption of Chromosomal Locus of CVT18 and Identification of GSA12
To knock out YFR021w (CVT18) in S. cerevisiae, the HIS5 gene from Schizosaccharomyces pombe was amplified with the use of plasmid pME3 (gift from Dr. Neta Dean, State University of New York, Stony Brook, NY) as template. The forward primer 5′ T T C T C T T-C G G C C T G A C A A T G T C T G A T T C A T C A C C C C C G G-G C T G C A G G A A T T C 3′ and the reverse primer 5′ C A A G A-T G G A A T A C T G T G A C A A T A T T A A G C A A T C G T C G- A C G G T A T C G A T A A G C 3′ contain 34 nucleotides of 5′ untranslated region (UTR) and 3′ UTR of the CVT18 gene, respectively. The resulting polymerase chain reaction (PCR) product contains the HIS5 gene flanked by the 5′ and 3′ UTR of CVT18, and was used to knock out the CVT18 gene via homologous recombination. The transformants were selected on SMD-His media. Replacement of the CVT18 coding region by the HIS5 gene was confirmed by a positive PCR reaction with the use of the genomic DNA from the transformants as template and the primer pair 5′ C A T A A G G C A-T C G T T A T T C C G 3′ (sequence of 5′ UTR of CVT18) and 5′ T C- A C A T G T A T C A T G C A C T G G 3′ (complementary sequence of part of the coding region of HIS5).
The P. pastoris gsa12 mutant was isolated following the restriction enzyme-mediated integration of a 2.0-kb pREMI plasmid that contained the ColE1 origin of replication and the Zeocin resistance gene under the control of the TEF1 promoter from S. cerevisiae and the EM7 promoter of E. coli. The pREMI plasmid was linearized with BamHI and used to transform GS115 cells. Transformation was done by electroporation with 1 μg of the linearized pREMI and 1 U of BamHI. Those cells that grew on Zeocin (100 μg/ml) plates were replica plated to YNMH (0.67% yeast nitrogen base, 0.4 mg/l biotin, 0.5% methanol, and 0.4 mg/l histidine) plates and allowed to grow for 2–3 d. The colonies were then transferred to nitrocellulose and placed onto YNDH (0.67% yeast nitrogen base, 0.4 mg/l biotin, 2% glucose, and 0.4 mg/l histidine) plates. After 12 h, those mutant colonies that retained alcohol oxidase (AOX) activity were identified by direct colony assays. This was done by lysing the cells in liquid nitrogen and then placing the nitrocellulose onto Whatman paper soaked with 33 mM potassium phosphate buffer, pH 7.5, containing 3.4 U/ml horseradish peroxidase, 0.53 mg/ml 2,2′-azino-bis(3-ethylbenz-thazoline-6-sulfonic acid), and 0.13% methanol at room temperature. The purple colonies that retained AOX activity were isolated and the gsa mutation verified by liquid assays (see below). The genomic DNA was then isolated, digested with EcoRI, religated, and the vector amplified in E. coli. On sequencing the genomic DNA fragment that was isolated along with the pREMI vector from the R2 mutant, we were able to completely assemble the GSA12 gene as well as 300 base pairs of the 5′ and 700 base pairs of the 3′ noncoding regions (National Center for Biotechnology Information accession number AF368421).
Antisera Preparation
To generate antiserum against Cvt18, we generated an MBP-CVT18 fusion construct. Part of the coding region of CVT18 was PCR amplified from genomic DNA with primers 5′ CCTTCGGTAGTCGACAGCTATTTAGTGTATCC 3′ and 5′ AGATGGAATACGTCGACAATATTAAGCAATCG 3′. The PCR product was digested with SalI and inserted into the SalI site of the pMAL-c2 vector. The MBP-Cvt18 fusion protein was expressed and purified from E. coli according to the manufacturer's instructions. Immunization of the rabbits and collection of the antiserum were carried out by the Comparative Animal Pathology Lab (University of California, Davis, Davis, CA) following the procedure described by Harlow and Lane (1999).
Plasmid Construction and Chromosomal Tagging
To clone CVT18 into pRS vectors, the forward primer 5′ CTTACATCTAGAGTAAGAAATACTTGC 3′ and the reverse primer 5′ TGCAAAAGTCTAGATTATACGCAGGAG 3′, both incorporating a XbaI site, were used to amplify CVT18 from S. cerevisiae genomic DNA. The PCR product was digested with XbaI and ligated to pRS415 linearized with XbaI to generate plasmid pCVT18(415). To make the Cvt18-GFP fusion, a two-step cloning approach was used. First, pCVT18(415) was digested with SpeI and BamHI to obtain the 5′ fragment of CVT18. This fragment was then ligated with the vector from pRS414-APG5GFP (George et al., 2000) that had been digested with the same enzymes. The 3′ fragment of CVT18 was amplified by PCR with the use of the forward primer 5′ AGGGATGATGCGGATCCAACAAGC 3′, which included the BamHI site within the CVT18 coding sequence, and the reverse primer 5′ CACTTCCGGGATCCATCCATCAAG 3′, which incorporated a BamHI site. The PCR product was digested with BamHI and inserted into the BamHI site in the above-mentioned construct behind the N-terminal region of CVT18 and in front of the green fluorescent protein (GFP) coding region. The resulting plasmid, pCVT18GFP(414), contains an in-frame fusion between CVT18 and GFP, with the 5′ UTR region of CVT18 and the actin termination sequence. The GFP gene codes for a modified GFP (S65T) from the jellyfish Aequorea victoria.
To integrate GFP at the chromosomal CVT18 locus, the plasmid pCVT18GFP(414) was digested with BglI. The fragments containing the CVT18GFP sequence were ligated into the BglI sites of plasmid vector pRS305. The pCVT18GFP(305) was subsequently digested with NsiI and used to transform wild-type (SEY6210) cells. Transformants were selected on −Leu plates and the chromosomal tagging of CVT18 with GFP was confirmed by Western blotting.
Gsa12 was tagged with the HA epitope at the N terminus by PCR amplification from genomic DNA with the use of a forward primer of 5′ GAATTCGAATTCATGTATCCATACGATGTTCCAGATTA-CGCGTCGCAACCTACAGATGAG 3′, which contained a start codon, an HA epitope, and an EcoRI site, and a reverse primer of 5′ CACACTTTGAATTCATAGGTGGGTA 3′, which contained an EcoRI site. HA-GSA12 was inserted into the EcoRI sites behind the GFP gene, which had been inserted into the EcoRI site of pIB2 (Sears et al., 1998). The resulting vector pPHT-G12 containing GFP/HA-GSA12 behind the constitutive and glucose-inducible GAPDH promoter was linearized by cutting within the HIS4 gene (e.g., with StuI) and used to transform by electroporation R2 (his4 gsa12::Zeocin) and DMM1 (his4:: pDM1 (PAOX1BFP-SKL, Zeocin)) cells. The plasmid encoding GFP/HA-GSA9, pPS64, was described previously (Kim et al., 2001b). A plasmid encoding GFP/HA-GSA11 will be described elsewhere (Stromhaug, et al., 2001)
An S. cerevisiae plasmid encoding GFP with a C-terminal serine-lysine-leucine (SKL) peroxisomal targeting signal was constructed. The sequence encoding the GFP-SKL fusion protein was amplified by PCR with the use of pCAPG5GFP (George et al., 2000) as a template and oligonucleotides that incorporated a BamHI restriction site into both the 5′ and 3′ ends of the DNA fragment (upstream: 5′ GCTGGATCCATGAGTAAAGGAGAAG 3′; downstream: 5′ CGAGGATCCTTATAATTTGGATAGTTCATCCATGCC 3′). The resulting PCR product was then gel purified and digested with BamHI to allow for cloning into the pCu416 plasmid (provided by Dr. Dennis Thiele, University of Michigan, Ann Arbor, MI) (Labbe and Thiele, 1999) to generate pCu416GFPSKL. The correct orientation of the GFP-SKL coding sequence was confirmed initially by restriction digest analysis and ultimately by fluorescence microscopy analysis of copper-induced expression.
Measurement of Peroxisome Degradation
The loss of peroxisomes by P. pastoris and S. cerevisiae was analyzed biochemically and morphologically. The degradation of peroxisomes in P. pastoris was evaluated by measuring the loss of AOX during glucose adaptation as described previously (Yuan et al., 1999). The degradation of peroxisomes in S. cerevisiae was determined by the loss of thiolase (Fox3) as described previously (Hutchins et al., 1999).
To monitor peroxisome levels in vivo in S. cerevisiae, wild-type (WCG4a and SEY6210), MGY103, TVY1, and JGY9 yeast strains harboring pCu416GFPSKL were grown to log phase in SMD before examination by scanning confocal fluorescence microscopy. Copper levels normally present in commercial synthetic minimal medium (Difco) were sufficient to induce observable GFP-SKL expression. The vital dye N-(triethlyammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4-64) was used for visualization of the vacuolar membranes as described previously (Noda et al., 2000) and was added at a 16 μM concentration to cultures 4 h before microscopy. Induction and degradation of peroxisomes was carried out essentially as described previously (Wang et al., 2001). Cells were examined for peroxisome levels on the Leica DM-IRB scanning confocal microscope (Leica, Deerfield, IL), with a 510–525-nm band pass filter for GFP in combination with a 585-nm filter to observe the FM 4-64. All samples were examined within 20 min after harvesting.
Fluorescence Microscopy
The cells of S. cerevisiae strain JGY11, having a chromosomally integrated CVT18GFP gene, were grown to mid log phase, labeled with FM 4-64 for 30 min, and chased in SMD or SD-N for 2 h. The GFP signals were visualized with the use of a Nikon E-800 fluorescent microscope (Mager Scientific, Dexter, MI). The images were captured with an ORCA II CCD camera (Hamamatsu, Bridgewater, NJ) with the use of Openlab software (Improvision, Lexington, MA). To examine the localization of various proteins, including Cvt9, Cvt19, Aut7, Apg5, and Apg2, the respective GFP fusion plasmids were transformed into the desired strains. The cells were grown to mid log phase or shifted to SD-N for 2 h, and the images were taken as described above.
The cellular distribution of Gsa12 during glucose-induced micropexophagy was examined by visualizing GFP-Gsa12 in cells coexpressing BFP-SKL as described previously (Kim et al., 2001b). To examine the localization of GFP-Gsa11, gsa11-2 (WDY37) and gsa12 (WDY46) cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) and shifted to YNM for 24 h. Cells were then shifted to YND for 3 h and examined by fluorescence microscopy as described above. To examine the localization of GFP-Gsa9, gsa9Δ (ANB12) and gsa12 (ANB7) cells were grown in minimal YND.
Electron Microscopy
P. pastoris strains were grown for 40 h in medium supplemented with 0.5% methanol and then transferred to medium containing 2% glucose or 0.5% ethanol for 3 h. The cells were washed with water and fixed in 1.5% KMnO4 in veronal-acetate buffer. The cells were then dehydrated, embedded in Epon 812, and sectioned for viewing on a JEOL 100CX transmission electron microscope as described previously (Tuttle and Dunn, 1995; Yuan et al., 1999).
Membrane Association and Protease Sensitivity Analysis
For membrane flotation analysis, S. cerevisiae strains JGY3 and TVY1 were grown in SMD until log phase. Cells were converted to spheroplasts and osmotically lysed in lysis buffer PS200 (20 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 200 mM sorbitol) containing 5 mM MgCl2 as previously described (Scott and Klionsky, 1995). The pellet (P5) and supernatant (S5) fractions were separated by centrifugation at 5000 rpm for 5 min at 4°C. The P5 fraction from 5 O.D.600 units of cells was resuspended in 0.3 ml of 15% Ficoll and overlaid with 1 ml of 13% and then 0.2 ml of 2% Ficoll. All Ficoll solutions were dissolved in the above-mentioned osmotic lysis buffer. The gradient was centrifuged at 55,000 rpm (100,000 × g) in a TLS55 swinging bucket rotor (Beckman Coulter, Fullerton, CA) for 30 min at 4°C. The float fraction (0.5 ml) was collected from the top, and trichloroacetic acid (TCA) precipitated. For the protease sensitivity experiment, the strain JGY9 was treated essentially as described previously (Scott et al., 2001) with the use of 0.4% Triton X-100 and 40 μg/ml proteinase K where indicated. Identical results were obtained with the use of 5 μg/ml proteinase K. Ape1 and Pho8 were detected by immunoblotting.
Subcellular Fractionation and Membrane Biochemistry of Cvt18
Wild-type (SEY6210) cells were grown to mid log phase, harvested, converted to spheroplasts, and osmotically lysed in PS200 buffer containing 5 mM MgCl2. The total lysate was subjected to differential centrifugation as described (Wang et al., 2001). For the membrane biochemistry experiment, aliquots of 2 O.D.600 equivalent of the P13 pellet from the above-mentioned fractionation was resuspended in either lysis buffer, 1 M KCl, 0.1 M Na2CO3 (pH 11), 3 M urea, or 0.2% Triton X-100 in lysis buffer, and incubated at room temperature for 5 min. The reactions were stopped by adding TCA to a final concentration of 10%. Samples from the above-mentioned experiments were washed twice in acetone, resuspended in SDS-PAGE sample buffer, and samples of 0.2 O.D.600 equivalents were resolved by SDS-PAGE. Cvt18 was detected by Western blotting.
The OptiPrep density gradient analysis of Cvt18 was performed essentially as described (Kim et al., 2001) with the use of 15 O.D.600 units from the P13 fraction generated from S. cerevisiae wild-type strain SEY6210.
Other Procedures
Pulse/chase (Scott et al., 1996) and nitrogen starvation experiments were performed as described previously (Noda et al., 2000). The degradation of cellular proteins during nitrogen starvation was performed essentially as described previously (Tuttle and Dunn, 1995; Yuan et al., 1999).
RESULTS
Gsa12 Is Required for Autophagy and Both Micro- and Macropexophagy in Pichia pastoris
When fungal cells are exposed to specific environmental conditions such as methanol or oleic acid that require peroxisomal enzymes to assimilate these carbon sources for survival, they respond by synthesizing peroxisomes. Afterward, when the environmental conditions change to one than no longer requires peroxisomes for survival, these cells rapidly degrade the superfluous organelles. We have shown that in P. pastoris and S. cerevisiae the peroxisomes are degraded by the vacuole (Tuttle et al., 1993; Hutchins et al., 1999). In P. pastoris the sequestration events are selective and can proceed by one of two pathways. The metabolic shift from methanol to glucose induces micropexophagy, whereas the shift from methanol to ethanol induces macropexophagy (Tuttle and Dunn, 1995). To identify components required for these processes, we carried out a screen for mutants that are blocked in glucose-induced selective autophagy (gsa) of peroxisomes with the use of mutagenesis with the pREMI plasmid (see MATERIALS AND METHODS). After methanol induction, AOX is the primary matrix enzyme in the peroxisomes of methylotrophic yeasts, including P. pastoris, and serves as a convenient marker to monitor peroxisome levels.
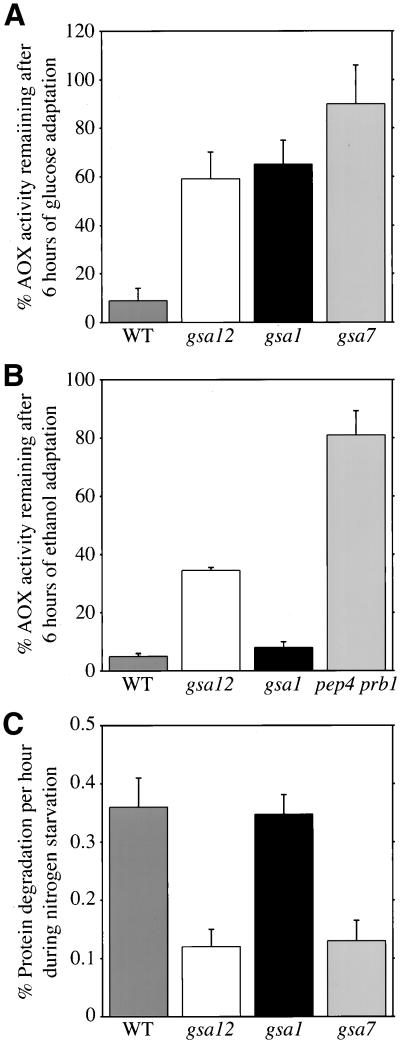
The degradation of peroxisomes was stimulated upon adaptation from growth on methanol medium to medium containing glucose as the sole carbon source (Figure 1A). After 6 h of glucose adaptation, as much as 80% of the AOX activity in wild-type GS115 cells was lost, indicating peroxisome degradation. In contrast, the glucose-induced loss of AOX activity was dramatically suppressed in gsa1 and gsa7 mutants. GSA1 encodes phosphofructokinase 1, which appears to be involved in the glucose-signaling pathway for pexophagy (Yuan et al., 1997). GSA7 is the gene that encodes the P. pastoris homolog of Apg7, an essential component for pexophagy as well as the Cvt pathway and autophagy (Kim et al., 1999; Yuan et al., 1999). Similar to these mutants, the degradative loss of AOX activity was dramatically impaired in the gsa12 mutants.
Figure 1.
gsa12 mutants are defective in pexophagy and autophagy. (A and B) Wild-type (WT, GS115), gsa12 (R2), gsa1 (WDKO1), gsa7 (WDY7), and pep4 prb1 (SMD1163) cells were grown in methanol for 36 h. At that time, cells were switched to medium containing either 2% glucose (A) or 0.5% ethanol (B). Aliquots were removed at 0 and 6 h of adaptation, the cells lysed, and AOX activities measured as described in MATERIALS AND METHODS. The data are expressed as a percentage of AOX activity remaining at 6 h relative to 0 h and represent the mean ± SD (n = 3–6). The gsa12 mutant was defective in peroxisome degradation by both micro- and macropexophagy. (C) Wild-type (WT, GS115), gsa12 (R2), gsa1 (WDKO1), and gsa7 (WDY7) cells were grown in minimal medium containing [14C]valine for 18 h. The cells were pelleted and resuspended in medium lacking amino acids and nitrogen and containing 10 mM cold valine chase. The production of TCA-soluble radioactivity was measured at 2, 5, 8, and 24 h of chase, and the rates calculated by linear regression of the slope of the line. The rates are presented as a mean ± SD (n = 3–5). The gsa12 mutant was defective in nonspecific protein degradation by autophagy.
The degradation of AOX was also enhanced when wild-type cells adapted from methanol to ethanol medium (Figure 1B). The loss of AOX activity was almost completely suppressed in the absence of the vacuolar endopeptidases Pep4 and Prb1. Ethanol-induced degradation of AOX was unaltered in gsa1 mutants, but suppressed in gsa12 mutants. This is consistent with a role for Gsa1 in a glucose-signaling event.
Nonselective autophagy accounts for the majority of the cellular degradative capacity during starvation conditions. Because many of our GSA and CVT genes have been found to be required for starvation-induced autophagy, we next examined the ability of the gsa12 mutant to degrade radiolabeled endogenous proteins in response to nitrogen starvation (Figure 1C). The endogenous proteins of wild-type, gsa1, gsa7, and gsa12 cells were labeled with [14C]valine for 16 h. Afterward, the cells were transferred to a nitrogen and amino acid starvation medium containing 10 mM cold valine. The rates of protein degradation were determined by measuring the production of TCA-soluble radioactivity over the course of 2–24 h of chase. Wild-type cells degraded endogenous proteins at a rate of 0.36% of total cellular protein per hour. This rate was unaffected in gsa1 cells. However, starvation-induced protein degradation was suppressed by as much as 70% in both gsa7 and gsa12 mutants.
GSA12 Encodes a Unique 60-kDa Protein
We next identified the gene that was disrupted by the insertion of the pREMI vector. The genomic DNA from R2 (his4, gsa12::Zeocin) cells was digested with EcoRI, religated, and the resulting vector containing the Zeocin-resistance gene amplified in E. coli. We then completely sequenced the genomic DNA that accompanied the pREMI vector. From this sequence, we were able to assemble the GSA12 gene in addition to 300 base pairs of the 5′- and 700 base pairs of the 3′-noncoding regions (National Center for Biotechnology Information accession number AF368421). We also sequenced into the beginning of a downstream putative open reading frame, which appears to encode a protein homologous to YFR022w in S. cerevisiae. With the use of appropriate primers, the entire GSA12 gene was amplified by PCR from the genomic DNA of wild-type cells and sequenced to verify that the pREMI disruption occurred between Y172 and K173. GSA12 encoded a 60-kDa protein of 543 amino acids that has high homology (51% identity and 68% similarity) with YFR021w that encodes a protein of unknown function in S. cerevisiae (Figure 2). There exist structural homologs of Gsa12 in S. pombe (CAC19764), Caenorhabditis elegans (T31883), Drosophila melanogaster (AAF50472), and Homo sapiens (T12539). Sequence analysis of GSA12 revealed no transmembrane domains, but a signature pattern for two WD40 domains between T216 and R300. Each homolog also has two WD40 domains (Figure 2). The GSA12 disruption in the R2 mutant occurs upstream of the WD40 repeat, suggesting that two-thirds of the protein is missing.
Figure 2.
Amino acid alignment of the Gsa12/Cvt18 proteins. The amino acid sequences of Gsa12 from P. pastoris, and Cvt18 from S. cerevisiae were aligned with the structural homologs from S. pombe (CAC19764) and H. sapiens (T12539). Amino acid identities (—) and gaps (… ) are indicated and the two WD40 domains that are found in all known homologs designated in bold.
Gsa12 Is Required for an Early Event in Micro- and Macropexophagy
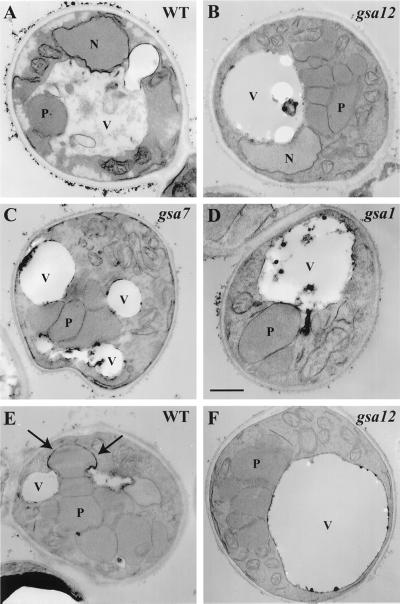
The process of glucose-induced micropexophagy can be categorized into distinct stages (Sakai et al., 1998). During the early steps of glucose adaptation, peroxisomes associate with the vacuolar membrane. Arm-like extensions of the vacuole initiate the peroxisome engulfment process. In the middle stage(s), vacuolar extensions become more pronounced, ultimately surrounding the peroxisomes. This is followed by the late stage(s), which involves a homotypic fusion event to fully enclose the peroxisome inside the vacuole. Subsequent breakdown of the peroxisome-containing inner vacuolar vesicle triggers the eventual degradation of the peroxisome. To determine the stage at which Gsa12 is required during micropexophagy, we compared the cellular morphology of the gsa12 mutant to wild-type, gsa1, and gsa7 strains at 3 h of glucose adaptation (Figure 3). In wild-type cells that have adapted to glucose for 3 h, few peroxisomes were present and many of those were observed in the vacuolar lumen (Figure 3A). In gsa1 mutants, the vacuole remains round with minimal interaction with peroxisomes, suggesting that Gsa1 is required for an early event of micropexophagy (Figure 3D). In contrast, in gsa7 mutants, the vacuole almost completely surrounds or engulfs a cluster of peroxisomes, suggesting that Gsa7 is required for a late event of micropexophagy (Figure 3C). Unlike wild-type cells, the gsa12 cells contained many peroxisomes at 3 h of glucose adaptation. In these mutants, micropexophagy is stalled at an early stage in which the vacuole is round with no apparent extensions engulfing the peroxisomes (Figure 3B). Even at 5 h of glucose adaptation the vacuole of the gsa12 mutants as visualized by electron microscopy was round (our unpublished results).
Figure 3.
gsa12 cells are defective in an early event of micro- and macropexophagy. Wild-type (WT, GS115), gsa12 (R2), gsa1 (WDKO1), and gsa7 (WDY7) cells were grown in methanol for 36 h. At that time, cells were switched to medium containing either 2% glucose (A–D) or 0.5% ethanol (E–F) for 3 h. Cells were then fixed in potassium permanganate, dehydrated, embedded, and sectioned for viewing on a JEOL 100CX transmission electron microscope (Tuttle and Dunn, 1995; Yuan et al., 1999). During glucose adaptation (A–D), peroxisomes can be seen in vacuoles of wild-type (WT) cells. The vacuoles in gsa12 cells remain round much like that observed in gsa1 mutants, but contrary to that seen in gsa7 mutants. In gsa7 cells, the vacuole can be seen to almost surround the peroxisomes. During ethanol adaptation (E and F), wrapping membranes around a single peroxisome (arrows) normally seen in wild-type cells (WT) were not observed in the gsa12 mutant. N, nucleus; P, peroxisome; V, vacuole. Bar, 0.5 μm.
Ethanol-induced macropexophagy is a process whereby individual peroxisomes are sequestered by two or more membranes of unknown origin. These autophagic vesicles then fuse with the vacuole thereby releasing “autophagic bodies” within the vacuole. On digestion of the limiting membranes of these autophagic bodies, the peroxisomes are then exposed to the degradative enzymes of the vacuole, resulting in the complete proteolysis of the peroxisomal enzymes. We next compared the cellular morphology of wild-type and gsa12 cells at 3 h of ethanol adaptation to evaluate the stage at which macropexophagy was blocked in gsa12 mutants (Figure 3). Multiple membranes were found to surround individual peroxisomes in wild-type cells (Figure 3E, arrows). In contrast, there was no evidence that more than a single membrane bound the peroxisomes in gsa12 cells (Figure 3F). Although double membranes were occasionally seen in the cytosol, these membranes did not appear to surround the peroxisomes. In addition, as was seen during glucose adaptation, the vacuole remained round in gsa12 cells. The morphological data suggest that Gsa12 is required for an early event of both micropexophagy (vacuolar engulfment) and macropexophagy (sequestration within double-membrane vesicles).
Cvt18 Is Required for the Cvt Pathway, Autophagy, and Pexophagy in S. cerevisiae
In S. cerevisiae, it has been demonstrated that the nitrogen starvation-induced degradative autophagy pathway and the biosynthetic Cvt pathway share much of the same molecular machinery (Harding et al., 1996; Scott et al., 1996; Klionsky and Ohsumi, 1999; Kim and Klionsky, 2000). In addition, peroxisome degradation under nitrogen-limiting conditions has also been shown to require most of the CVT/APG genes (Hutchins et al., 1999; Kim and Klionsky, 2000). Recently, homologs for the S. cerevisiae genes encoding an E1-like conjugating enzyme, Apg7/Cvt2 (Mizushima et al., 1998; Kim et al., 1999), and the Cvt pathway-specific protein Cvt9 have been found in P. pastoris and shown to be required for pexophagy (Yuan et al., 1999; Kim et al., 2001b). This observation suggests that conserved mechanisms are involved in the Cvt/autophagy pathway in S. cerevisiae and pexophagy in P. pastoris. Based on the above-mentioned phenomenon, we decided to examine whether the S. cerevisiae homolog of GSA12, the open reading frame YFR021w, could also be a component in the Cvt/autophagic pathway and pexophagy in this yeast. The chromosomal locus encoding YFR021w was replaced by the S. pombe HIS5 gene via homologous recombination. The resulting deletion mutant strain was subsequently examined for its phenotype in the Cvt, autophagy, and pexophagy pathways.
Ape1 is the best-characterized marker protein for transport through both the Cvt and autophagy pathways (Kim and Klionsky, 2000). Defects in either pathway can be observed by monitoring the processing of precursor Ape1 to the mature form, an event that is dependent on delivery to the vacuole. Both the wild-type and YFR021w deletion strain were subjected to Western blotting analysis with the use of antiserum to Ape1 (Figure 4A). The YFR021w knockout strain showed accumulation of prApe1 as opposed to the wild-type strain, where the majority of Ape1 was present as the mature form. Kinetic analyses confirmed that the Cvt pathway was blocked in the YFR021w deletion strain, whereas the processing of carboxypeptidase Y (Prc1) was only slightly delayed (our unpublished results). Prc1 is a resident hydrolase that is delivered to the vacuole through the Vps pathway (Bryant and Stevens, 1998). These results suggest that the Ape1 maturation defect in the YFR021w deletion strain is specific to the Cvt pathway but not due to a general block in vacuolar function. Therefore, we named YFR021w as CVT18 and the deletion strain as cvt18Δ.
Figure 4.
cvt18Δ strain is defective in the Cvt pathway, autophagy, and pexophagy in S. cerevisiae. (A) cvt18Δ strain accumulates precursor Ape1. Wild-type (WT, SEY6210), cvt18Δ (JGY3), and CVT18GFP (JGY11) strains were grown in SMD until mid log phase and then subjected to immunoblotting analysis for Ape1 and Cvt18 as described in MATERIALS AND METHODS. The position of prApe1, mature Ape1 (mApe1), and Cvt18 are as indicated. (B) Viability of the cvt18Δ strain is sensitive to nitrogen starvation. The Wild-type (WT, SEY6210), apg5Δ (MGY101), and cvt18Δ (JGY3) strains were grown in SMD until mid log phase and then shifted to SD-N. At the indicated time points, equal volumes of culture were withdrawn and plated onto YPD plates. After 3 d the number of colonies representing the viable cells in the culture were counted and plotted against time in starvation medium. (C) cvt18Δ strain does not have the ability to process the cytosolic marker protein Pho8Δ60 under nitrogen starvation conditions. WT (JGY1) and cvt18Δ (JGY5) cells expressing Pho8Δ60 were pulse-labeled in SMD for 10 min and chased in SD-N for the indicated times. The kinetics of Pho8Δ60 processing was analyzed by immunoprecipitation as described in MATERIALS AND METHODS. The precursor and mature forms of Pho8Δ60 are indicated. (D) cvt18Δ strain is defective in pexophagy. Wild-type (WT, WCG4a), pep4Δ (YMTA), and cvt18Δ (JGY7) strains were grown in YTO medium to allow the proliferation of peroxisomes and then were shifted to SD-N to induce pexophagy. Samples of equal volume were collected at the indicated time points and processed for Western blotting with the use of antibodies against the peroxisomal marker protein Fox3. The vacuolar protease-dependent degradation of peroxisomes is blocked in cvt18Δ.
Precursor Ape1 is delivered to the vacuole by both the Cvt and autophagy pathways, depending on the nutrient conditions (Scott et al., 1996; Baba et al., 1997; Klionsky and Ohsumi, 1999). The gsa12 mutant was shown to be defective for nonspecific protein degradation. Accordingly, we examined the cvt18Δ strain for its autophagic phenotype. Strains that are defective in autophagy display limited viability under starvation conditions (Tsukada and Ohsumi, 1993). The wild-type, apg5Δ (George et al., 2000), and cvt18Δ strains were grown to mid log phase, shifted to nitrogen starvation conditions, and their viability was determined over time. The wild-type strain survived nitrogen starvation >11 d without a significant decrease in viability (Figure 4B). In contrast, the number of viable cells decreased dramatically in the apg5Δ and cvt18Δ strains over the same time period. Different cvt/apg mutants show variable sensitivity to starvation (Scott et al., 1996). The rapid loss in viability of the cvt18Δ strain places it into the most sensitive group.
To provide a more quantitative analysis, we examined autophagy by following the vacuolar processing of the cytosolic marker protein Pho8Δ60 (Scott et al., 1996). CVT18 was knocked out in strain DKY6281, which lacks the chromosomal PHO8 gene, to generate strain JGY1. A centromeric plasmid carrying PHO8Δ60, pMUH8 (Hutchins et al., 1999), was transformed into the pho8Δ and pho8Δ cvt18Δ strains. Cells were pulse-labeled in SMD, washed, and subjected to a nonradioactive chase in SD-N. In the wild-type strain, a significant fraction of Pho8Δ60 was processed to its mature size due to its import into the vacuole via the autophagic pathway (Figure 4C). In cvt18Δ, however, Pho8Δ60 remained as the precursor form within the chase time of 8 h, indicating that its vacuolar import via autophagy was blocked.
The specific degradation of peroxisomes, pexophagy, induced by glucose adaptation or nitrogen starvation can be monitored in S. cerevisiae by following the vacuolar degradation of peroxisomal matrix protein Fox3 (Hutchins et al., 1999). Wild-type, pep4Δ, and cvt18Δ cells were subjected to pexophagy analysis. The proliferation of peroxisomes was induced by growing the cells in medium with oleic acid as the sole carbon source. Once shifted to SD-N, the excess peroxisomes were delivered to the vacuole and degraded in the wild-type strain as shown by the gradual decrease in the Fox3 level (Figure 4D). The degradation process is dependent on the vacuolar proteinase A (Hutchins et al., 1999), and the level of Fox3 remained the same even after 20 h in SD-N in the pep4Δ strain. The Fox3 level over time in the cvt18Δ strain also remained constant, suggesting that cvt18Δ cells are impaired for pexophagy in S. cerevisiae.
Cvt Pathway Is Blocked at Membrane Sequestration/Enclosure Stage in cvt18Δ Strain
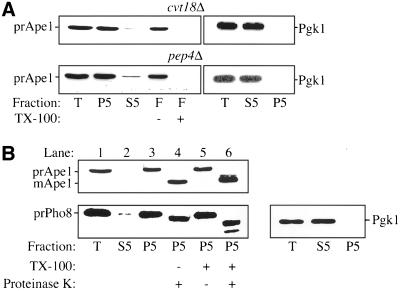
The Cvt pathway, autophagy, and pexophagy all involve delivery of cargo from the cytoplasm into the vacuole via a vesicular mechanism involving membrane fusion (Kim et al., 2000; Stromhaug and Klionsky, 2001). Accordingly, it is required that the cargo be sequestered within membranes before it can enter the vacuolar lumen. Most of the cvt/apg mutants have been shown to be required for different stages of Cvt vesicle/autophagosome formation (Klionsky and Ohsumi, 1999; Kim and Klionsky, 2000). To further investigate the role of Cvt18 in these pathways, we examined the state of prApe1 in the cvt18Δ strain. The first step of sequestration involves binding of prApe1 to the enwrapping membrane. We carried out a membrane flotation experiment to determine whether prApe1 is membrane associated. Due to the relatively low buoyant density of the lipid components, proteins associated with the membrane will appear in the float fraction at the top of a density gradient. Addition of detergent will disrupt the lipid bilayer and therefore prevent the flotation of the membrane. As described in MATERIALS AND METHODS, the P5 fractions from osmotically lysed spheroplasts of the pep4Δ and cvt18Δ strains were loaded at the bottom of a Ficoll step gradient, and subjected to centrifugation of 100,000 × g for 30 min. It has been shown previously that prApe1 accumulates in the vacuole in a pep4Δ strain (Harding et al., 1995). As a result, prApe1 inside the vacuole in pep4Δ cells floats to the top of the Ficoll gradient (Figure 5). Addition of detergent disrupts the appearance of prApe1 in the float fraction, indicating that its location in this fraction is membrane or lipid dependent. The cvt18Δ strain also showed a detergent-sensitive flotation of prApe1, indicating that the prApe1 is associated with certain membrane structures in this strain. In this experiment, the cytosolic marker Pgk1 was located exclusively in the supernatant fraction, indicating that prApe1 was not in the float fraction due to its presence in unlysed spheroplasts.
Figure 5.
Precursor Ape1 in the cvt18Δ strain is membrane associated and protease accessible. For the membrane flotation experiment, both the cvt18Δ (JGY3) and pep4Δ (TVY1) strains were grown in SMD until mid log phase. The cells were converted to spheroplasts and lysed as described in MATERIALS AND METHODS. The total cell lysates were centrifuged at 5000 rpm for 5 min to separate the pellet (P5) and supernatant (S5) fractions. The majority of prApe1 was recovered in the P5 fraction in both strains. (A) P5 fraction was resuspended in 15% Ficoll either in the absence or presence of 0.2% TX-100 and then overlaid with 13 and 2% Ficoll solution. The gradients were subsequently centrifuged at 100,000 × g for 30 min at 4°C. The float fraction was collected from the top of the gradient. The prApe1 was recovered in the float fraction in both strains in the absence of detergent, and the addition of TX-100 to the P5 fraction disrupted the membrane association. (B) For the protease-protection experiment, the P5 fraction was obtained as described above from the cvt18Δ pep4Δ (JGY9) strain. The P5 fraction was resuspended in lysis buffer and then mixed with equal volumes of lysis buffer, 40 μg/ml proteinase K, 0.4% TX-100, or proteinase K plus 0.4% TX-100, and incubated on ice for 30 min. The reactions were stopped by mixing with 20% TCA. Samples were resolved by SDS-PAGE and examined by immunoblot with serum to Ape1 and Pho8 as indicated. Even in the absence of detergent, the prApe1 in cvt18Δ was digested to its mature form by proteinase K. Protection of the Pho8 propeptide in the absence of detergent indicates integrity of the vacuole. After lysis, the T, S5, and P5 fractions in both the flotation and protease-protection experiments were also probed for the cytosolic marker Pgk1. Recovery of Pgk1 in the S5 fractions indicates efficient lysis of spheroplasts.
To examine whether the prApe1 has been completely enclosed within a membrane compartment, we subsequently analyzed its sensitivity to protease digestion. A cvt18Δ pep4Δ strain was generated for this purpose to allow the use of Pho8 as an internal control. Due to the absence of proteinase A, the precursor Pho8 (prPho8) delivered to the vacuole is not processed properly and retains its C-terminal propeptide region. Precursor Pho8 also has a cytosolic tail derived from the N terminus (Klionsky and Emr, 1989). Spheroplasts were osmotically lysed under conditions that disrupt the plasma membrane but retain the integrity of subcellular compartments. The vacuole is relatively fragile and serves as a convenient measure of compartment integrity. Under our lysis conditions the cytosolic tail of the prPho8 was accessible to exogenous protease digestion without the addition of detergent (Figure 5B, bottom, lane 4), indicating that the spheroplasts had been lysed. Similarly, recovery of Pgk1 exclusively in the supernatant fraction indicated efficient lysis of spheroplasts. The cleavage of the prPho8 cytosolic tail can be seen as a small molecular mass shift by SDS-PAGE. In contrast, the lumenal propeptide of prPho8 was protected from protease digestion by the vacuolar membrane, confirming the integrity of this organelle, and by extension other membrane-enclosed compartments. When detergent was added along with proteinase K, both the cytosolic tail and propeptide were cleaved, resulting in a larger shift in the molecular mass (Figure 5B, bottom, lane 6). The lower band in lane 6 was a specific digestion product of prPho8 by proteinase K that is seen in the presence of detergent (Klionsky and Emr, 1989). The analysis of Pho8 confirms that subcellular compartments were not ruptured during the osmotic lysis procedure.
Precursor Ape1 in the cvt18Δ strain was sensitive to proteinase K treatment even in the absence of detergent (Figure 5B, lane 4), suggesting that prApe1 in cvt18Δ cells was not completely enclosed within a membrane bound vesicle. The intermediate band that appears following proteinase K digestion in the presence of detergent (Figure 5B, top, lane 6) is thought to be due to an effect of detergent on the prApe1 conformation (Oda et al., 1996). From the above-mentioned observations, we concluded that Cvt18 is not required for the initial membrane association of prApe1, but it is involved in the membrane sequestration or enclosure stage of Cvt vesicle formation.
Pexophagy Is Blocked at Prevacuolar Stage in cvt18Δ Strain
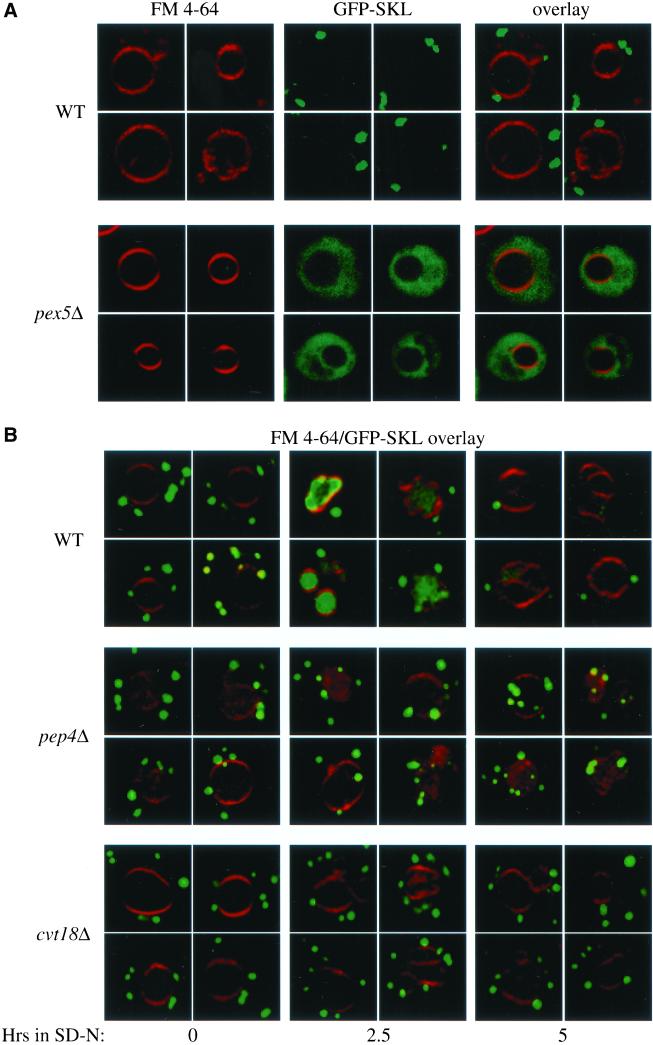
To extend our analysis on the site of action of Cvt18 we decided to examine the fate of peroxisomes in vivo after the induction of pexophagy. The analysis of prApe1 suggested that the defect in this strain was at a stage before sequestration within a completed vesicle. We also showed that peroxisome degradation as monitored by following degradation of Fox3 was blocked in the cvt18Δ strain (Figure 4D). Coupling these observations allowed us to predict that peroxisomes in the cvt18Δ strain would accumulate outside the vacuole in a nonsequestered state. To follow the state of the peroxisomes during pexophagy, we constructed a fusion protein consisting of GFP modified by a C-terminal extension with the type I peroxisomal targeting signal SKL. Vacuoles labeled with the dye FM 4-64, and peroxisomes were visualized by confocal fluorescence microscopy. In wild-type cells in nutrient-rich conditions, GFP-SKL was targeted to the peroxisomes and could be detected as large punctate structures outside of the vacuole (Figure 6A). As a control, we examined the distribution of GFP-SKL in the peroxisome biogenesis mutant pex5Δ (Veenhuis et al., 2000). The Pex5 protein is required for import of SKL-containing peroxisomal proteins. In contrast to the wild-type strain, pex5Δ cells displayed a diffuse fluorescent staining pattern, indicating that the punctate structures detected in the wild-type strain were dependent on a characterized component of the peroxisomal import machinery and hence corresponded to authentic peroxisomes.
Figure 6.
Pexophagy is blocked at a prevacuolar stage in the cvt18Δ mutant. (A) GFP-SKL chimera is localized to peroxisomes. Cells from Wild-type (WT, WCG4a) and pex5Δ (MGY103) strains expressing GFP-SKL from a CEN plasmid were grown to mid log phase in SMD as described in MATERIALS AND METHODS, and observed on a scanning confocal microscope. The vital dye FM 4-64 was added to cultures to allow visualization of the vacuolar membrane. (B) Peroxisomes accumulate outside of the vacuole during pexophagy in the cvt18Δ mutant. Cells from WT (WCG4a), pep4Δ (YMTA), and cvt18Δ (JGY9) strains were grown to mid log phase in SMD and transferred to YTO to induce peroxisome proliferation, and subsequently transferred to SD-N to induce degradation of the excess peroxisomes as described in MATERIALS AND METHODS. Cells were examined by scanning confocal microscopy after incubation in SD-N at the indicated times.
The GFP-SKL chimera was introduced into wild-type, pep4Δ, and cvt18Δ strains. The cells were incubated in medium containing oleic acid as the sole carbon source to induce peroxisome proliferation and then shifted to nitrogen starvation conditions for the indicated times. In wild-type cells, incubation in SD-N led to the appearance of the GFP signal inside the vacuolar lumen and a gradual decrease of the cytosolic punctate structures (Figure 6B). Eventually, the vacuolar GFP signal also decreased, presumably due to the degradation of the peroxisomes inside the vacuole. In the pep4Δ strain shifted to SD-N, the GFP signal also appeared in the vacuolar lumen, but in contrast to the wild-type strain the peroxisomes accumulated there instead of being degraded. In the cvt18Δ strain the peroxisomes did not appear in the vacuolar lumen but instead accumulated in the cytoplasm after shifting to nitrogen starvation conditions. These data, coupled with the analysis of the block in prApe1 import, further suggest that cvt18Δ cells were blocked at a stage before sequestration of cytoplasmic cargo.
Subcellular Localization of GFP-Gsa12 and Cvt18-GFP
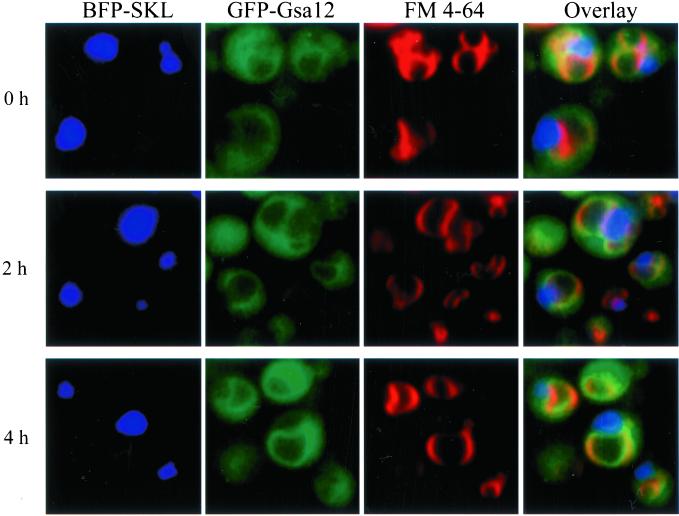
We have found that Gsa12 is required for the involution of the vacuole that occurs during micropexophagy. To better define the role of Gsa12 in micropexophagy, we examined its localization by fluorescence microscopy with the use of a functional GFP/HA-Gsa12 fusion protein. This construct was able to complement the gsa12 phenotype of the PHT13 strain (our unpublished results). We compared the localization of GFP/HA-Gsa12 to that of the vacuole and peroxisomes during glucose adaptation. We used the red dye FM 4-64 to label the vacuole membrane and a blue fluorescent protein that was targeted to peroxisomes (BFP-SKL) (Kim et al., 2001b). Cells expressing BFP-SKL behind the methanol-inducible AOX promoter and GFP/HA-Gsa12 behind the constitutive and glucose-inducible GAPDH promoter were grown in minimal methanol medium. FM 4-64 was added at 6 h, and after an additional 14 h the cells were switched to minimal glucose medium. At 0, 2, and 4 h of glucose adaptation, the triple-labeled cells were examined by fluorescence microscopy (Figure 7). At all time points, GFP/HA-Gsa12 was present within the cytosol and at the vacuole surface. The vacuole labeling colocalized with the vacuole membrane dye FM 4-64. GFP/HA-Gsa12 appeared to distribute evenly along the entire vacuole surface, including the sequestering arms that were engulfing the BFP-SKL-labeled peroxisomes. GFP/HA-Gsa12 was predominantly absent from the vacuole lumen. The GFP/HA-Gsa12 examined in these experiments was under control of the glucose-inducible GAPDH promoter. The localization pattern of GFP/HA-Gsa12 was essentially the same under both methanol (noninducing) and glucose conditions (Figure 7), indicating that the fluorescent pattern was not the result of overexpression.
Figure 7.
Gsa12 localizes to the vacuole membrane. Wild-type (PHT12) cells expressing both BFP-SKL and GFP-Gsa12 were grown in methanol medium in the presence of FM 4-64 and then adapted to glucose medium for 0, 2, or 4 h as indicated. The cells were visualized in situ by fluorescence microscopy. Peroxisomes were identified by the presence of BFP, which was targeted by its SKL signal, and the vacuole by FM 4-64. GFP-Gsa12 was found in the cytosol and at the vacuole membrane. Colocalization of GFP-Gsa12 with the FM 4-64 marker can be visualized by yellow-orange in the overlayed image. GFP-Gsa12 was also found associated with those vacuole arms that were sequestering the peroxisomes.
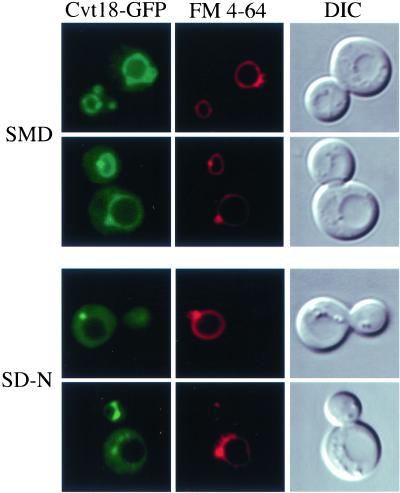
To localize Cvt18 in vivo, the chromosomal CVT18 locus was tagged with GFP. The resulting strain was normal for prApe1 maturation, suggesting that the Cvt18-GFP fusion protein is fully functional (Figure 4A). The Cvt18-GFP signal was observed by fluorescence microscopy. At the same time, the vacuole was visualized by FM 4-64 staining. In both vegetative growth and starvation conditions, Cvt18-GFP showed a vacuolar rim signal with bright spots, plus a significant level of cytosolic staining (Figure 8). The noncytosolic GFP signal overlapped with FM 4-64 staining. Under nitrogen starvation conditions, the punctate dots appeared to be stronger relative to the vacuolar rim staining. A similar localization pattern was seen under nitrogen starvation-induced pexophagy conditions (shifting from oleic acid to SD-N; our unpublished results). The distribution pattern of Cvt18 was also examined in a number of cvt/apg mutant strains. In all of the mutants examined, the fluorescent staining pattern remained unchanged (our unpublished results).
Figure 8.
Localization of Cvt18-GFP in S. cerevisiae under exponential growth (SMD) and nitrogen starvation (SD-N) conditions. The CVT18GFP (JGY11) strain was grown in SMD to mid log phase. The vacuolar membrane was labeled with FM 4-64, and the cells were incubated in either SMD or SD-N for 2 h before being observed by fluorescence microscopy as described in MATERIALS AND METHODS. The Cvt18-GFP signal can be detected in both the cytosol and on the vacuolar membrane. The noncytosolic Cvt18-GFP signal largely overlaps with FM 4-64 and also is concentrated at certain spots on or close to the vacuole.
Cvt18 Is Peripherally Associated with a Novel Compartment
To identify Cvt18 in cell lysates, polycolonal antiserum to Cvt18 was raised against the MBP-Cvt18 fusion protein as described in MATERIALS AND METHODS. Protein extracts were prepared and examined by immunoblotting (Figure 4A). In wild-type cells, the serum detected a predominant band that migrated at 55 kDa that was not present in the cvt18Δ strain. This size fits well with the predicted molecular mass of 54.9 kDa based on the deduced amino acid sequence of Cvt18. In the Cvt18-GFP strain, a band at ∼85 kDa corresponding to the Cvt18-GFP fusion protein was detected. These data indicated that the anti-Cvt18 serum recognized the Cvt18 protein. Pulse/chase analysis of Cvt18 revealed that it is a very stable protein in vivo under both vegetative growth and starvation conditions in either wild-type or pep4Δ strains (our unpublished results).
To examine the subcellular localization of Cvt18 by a biochemical approach, differential centrifugation was used to separate the total cell lysate into low-speed pellet (P13), high-speed pellet (P100), and soluble (S100) fractions as described in MATERIALS AND METHODS. Most of the Cvt18 protein was found to be soluble, and only a small percentage of protein was recovered from the low-speed pellet fraction (Figure 9A). The fractionation pattern of the cytosolic marker protein Pgk1 (S100) and the vacuolar membrane protein Pho8 (P13) were both as expected.
Figure 9.
Subcellular fractionation of Cvt18 and characterization of its membrane association. (A) Cvt18 is pelletable. The wild-type (SEY6210) strain was grown in SMD to mid log phase, converted to spheroplasts, and osmotically lysed in buffer (PS200 containing 5 mM MgCl2) as described in MATERIALS AND METHODS. The total cell lysate (T) was centrifuged at 13,000 × g for 10 min at 4°C to separate the low-speed pellet (P13) and supernatant (S13) fractions. The S13 fraction was then subjected to an additional centrifugation at 100,000 × g for 30 min at 4°C to separate the high-speed pellet (P100) and supernatant (S100) fractions. A small population of Cvt18 was recovered from the P13 fraction, whereas the majority of it was in the soluble S100 fraction. The fractionation patterns of the vacuolar membrane protein Pho8 (P13) and the cytosolic protein Pgk1 (S100) were both as expected. (B) Biochemical characterization of Cvt18 membrane association. Aliquots of the P13 fraction from the above-mentioned experiment were resuspended in lysis buffer, 1 M KCl, 0.1 M Na2CO3 (pH 11), 3 M urea, or 0.2% TX-100, and incubated at room temperature for 5 min as described in MATERIALS AND METHODS. The samples were centrifuged again at 13,000 × g for 10 min at 4°C to separate pellet and supernatant fractions. Cvt18, the integral membrane protein Pep12 and the peripheral membrane protein Vma2 were detected by immunoblotting. Cvt18 behaves as a peripheral membrane protein. (C) Equilibrium density gradient analysis of Cvt18. The spheroplasts from a mid log phase culture of wild-type (SEY6210) cells were lysed in gradient buffer (PS200 containing 1 mM MgCl2, 1 mM EDTA and 1 mM dithiothreitol). The P13 fraction was resuspended in the gradient buffer and loaded on top of a 0–66% OptiPrep linear gradient as described in MATERIALS AND METHODS. The gradient was centrifuged at 100,000 × g for 14 h at 4°C. Thirteen fractions of equal volume were collected from the top to the bottom and analyzed by Western blotting. Cvt18 peaks at fraction 3. It does not appear to cofractionate with the vacuolar marker Pho8, the endosomal protein Pep12, or the endoplasmic reticulum marker Dpm1.
The membrane association of the pelletable Cvt18 was subsequently characterized by treatment with various reagents. The P13 fraction was resuspended and then incubated in lysis buffer, 1 M KCl, 0.1 M Na2CO3 (pH 11), 3 M urea, or 0.2% Triton X-100. The membrane association of Cvt18 was completely disrupted by high pH or nonionic detergent, and partially disrupted by urea (Figure 9B). Buffer alone or high salt treatment did not affect its membrane association. Therefore, Cvt18 is peripherally associated with membranes via protein–protein interactions. Under the same conditions, the integral membrane protein Pep12 could only be stripped off the membrane by Triton X-100, whereas the peripheral membrane protein Vma2 was stripped from the pellet fraction by both detergent and high pH.
To further define the subcellular localization of the membrane-associated Cvt18, the low-speed pellet fraction (P13) was applied on top of a linear OptiPrep density gradient and subjected to equilibrium centrifugation. Cvt18 peaked at fraction 3 (Figure 9C). The vacuolar membrane protein Pho8 was mainly localized at the top of the gradient in fraction 1. The endosomal marker Pep12 was collected from fraction 7–9, whereas Dpm1, the endoplasmic reticulum marker, peaked at fraction 9. We detected a second peak for Cvt18 also at fraction 9. However, the appearance of several marker proteins at this position suggests that this population may not represent a distinct compartment. In addition, overexpression of Cvt18 results in its migration at the denser part of the gradient (our unpublished results), further suggesting that this population may not be physiologically relevant. Surprisingly, the gradient analysis did not show a cofractionation of Cvt18 with the vacuolar membrane, seemingly in contradiction with the distribution pattern we observed with the GFP signal in vivo. However, we found that the vacuolar rim staining of Cvt18-GFP was immediately lost upon osmotic lysis of spheroplasts (our unpublished results), suggesting a weak association of the protein with the vacuole membrane. Consequently, this population of Cvt18 was most likely not included in the P13 fraction that was loaded onto the gradient. A number of other Cvt/Apg proteins, including Apg9, Cvt9, and Apg2, cofractionate with each other at denser fractions in similar gradients (Kim et al., 2001; Wang et al., 2001). The Cvt18 in fraction 3 clearly did not colocalize with these proteins.
Absence of Gsa12/Cvt18 Affects Other Cvt/Apg or Gsa Proteins
Since the initial genetic analysis of the Cvt and autophagy pathway in S. cerevisiae, a number of proteins involved in these pathways have been studied in great detail (Klionsky and Ohsumi, 1999; Kim and Klionsky, 2000; Stromhaug and Klionsky, 2001). Determining interactions among these proteins has greatly facilitated our understanding of their functions at the molecular level. We sought to further narrow down the possible role of Cvt18 in these pathways by examining the localization of various Cvt/Apg proteins in the cvt18Δ mutant compared with other strains defective in cytoplasm-to-vacuole transport.
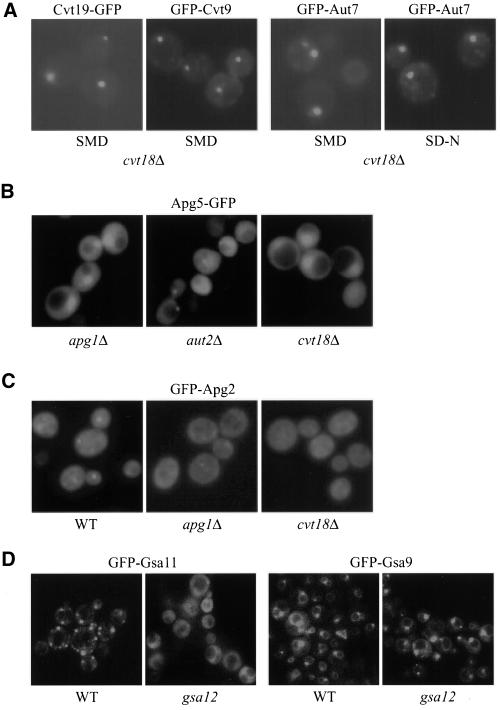
Cvt19 is a newly identified receptor for prApe1 in both the Cvt pathway and autophagy (Scott et al., 2001), and it has been shown to be a peripheral membrane protein. Presumably, Cvt19 requires another protein(s) for its membrane binding. We examined the localization of Cvt19-GFP in the cvt18Δ strain and found that it was not altered (Figure 10A). Cvt9 is involved in selective vacuolar import processes such as the Cvt pathway and pexophagy but not for nonselective autophagy (Kim et al., 2001b). The GFP-Cvt9 fusion displayed a perivacuolar punctate localization. This pattern was also observed in the cvt18Δ strain (Figure 10A). Cvt9 may be part of a multicomponent complex composed of a number of phosphorylated proteins, including the Apg1 kinase, Apg13, Apg17, and Vac8 (Kamada et al., 2000; Scott et al., 2000). The phosphorylation state of Apg13 in both rich and starvation conditions in the cvt18Δ strain was the same as that in a wild-type strain (our unpublished results). These data suggest that Cvt18 does not affect either the putative Cvt19 receptor complex required for import of resident hydrolases, or the Apg1 kinase complex that appears to control the conversion between the Cvt and autophagy pathways.
Figure 10.
Localization of Cvt/Apg/Gsa proteins in cvt18Δ/gsa12. (A) Cvt19-GFP, GFP-Cvt9, and GFP-Aut7 localization in cvt18Δ is normal. The cvt18Δ cells transformed with plasmids expressing the indicated proteins were grown in SMD to mid log phase or shifted to SD-N for 2 h as indicated. (B) Apg5-GFP was expressed in the indicated strains and the cells were grown to mid log phase in SMD. Apg5-GFP can be seen as a faint punctate dot in cells defective in conjugation of Apg12 to Apg5 or lipidation of Aut7, such as aut2Δ, but not in other strains, including apg1Δ. Apg5-GFP did not display a punctate dot in the cvt18Δ strain. The large fluorescent structure seen in the apg1Δ and cvt18Δ strains corresponds to the nucleus and results from overexpression of Apg5-GFP. (C) cvt18Δ mutant is defective in localization of GFP-Apg2. GFP-Apg2 was introduced into the indicated strains and the cells were grown to mid log phase in SMD and shifted to SD-N for 2 h. All samples were examined by fluorescence microscopy as described in MATERIALS AND METHODS. A faint punctate dot corresponding to GFP-Apg2 can be seen in the wild-type (WT) strain. A weaker punctate dot can occasionally be seen in strains such as apg1Δ and cvt18Δ. (D) gsa12 mutant displays a defect in localization of GFP-Gsa11. GFP-Gsa11 and GFP-Gsa9 were expressed in gsa11 (WDY37) and gsa9Δ (ANB12) strains, respectively, and in gsa12 (WDY46 and ANB7) cells. The GFP-Gsa11 and GFP-Gsa9 proteins complement the respective gsa11 and gsa9Δ mutations and are labeled as wild-type (WT) for simplicity. Cultures were analyzed by fluorescence microscopy as described in MATERIALS AND METHODS. When wild-type cells were adapted from methanol to glucose medium for 3 h, GFP-Gsa11 localized to one or more structures juxtaposed to the vacuole. In the gsa12 mutant, GFP-Gsa11 was not present in these structures but was predominantly cytosolic. GFP-Gsa9 localized to the vacuole surface and a structure adjacent to the vacuole when cells were grown in minimal glucose medium. This distribution was unaffected in the gsa12 mutant.
Aut7 marks the limiting membrane of the forming Cvt vesicles/autophagosomes and travels into the vacuole via the Cvt/Apg pathway (Kirisako et al., 1999; Huang et al., 2000). Aut7 is likely to be involved in the membrane expansion of the autophagosome (Abeliovich et al., 2000). Lipidation of Aut7 is required for its membrane association (Ichimura et al., 2000; Kirisako et al., 2000). In addition, defects in the ubiquitin-like Apg12-Apg5 conjugation system have also been shown to abolish Aut7 membrane binding (Kim et al., 2001a). We examined GFP-Aut7 localization in the cvt18Δ strain in both exponential growth and starvation conditions (Figure 10A) and found no difference compared with the wild-type strain (Shintani et al., 2001).
Apg5 is one component of the conserved conjugation system essential for the Cvt/Apg pathways (Mizushima et al., 1998a,b; George et al., 2000). The mammalian Apg5 protein has been shown to transiently bind to the forming autophagosomes and detach from the membrane at or before the vesicle enclosure step (Mizushima et al., 2001). In S. cerevisiae, Apg5-GFP displayed a predominantly cytosolic distribution and was observed as punctate dots only in certain mutant strains such as apg7Δ that are defective in the conjugation of Apg12 to Apg5 and the membrane recruitment of Aut7 (George et al., 2000; Kim et al., 2001a). We examined Apg5-GFP in a number of mutant strains, including those affecting conjugation, Aut7 lipidation, and the Apg1 kinase complex, and compared them with the cvt18Δ strain (Figure 10B). Consistent with the previous results, a bright punctate Apg5-GFP signal was detected in the conjugation mutants (our unpublished results). However, a smaller or dimmer Apg5-GFP dot was observed in the Aut7 lipidation mutants such as aut2Δ. These bright or dim dots were rarely seen in other strains examined, including cvt18Δ. The above-mentioned GFP-Aut7 and Apg5-GFP localization data suggested that Cvt18 may not be involved in the same steps as the conjugation machinery or the lipidation system that affects membrane recruitment of Aut7.
It has recently been reported that Apg2/Gsa11 is required for the Cvt pathway, autophagy, and pexophagy in S. cerevisiae and pexophagy in P. pastoris (Wang et al., 2001; Stromhaug, et al., 2001). GFP-Apg2 displayed a punctate structure in certain mutant strains but not in others, including apg1Δ and apg9Δ. We compared the fluorescent pattern of GFP-Apg2 in the cvt18Δ strain with that in the apg1Δ and apg9Δ strains. In the majority of the cvt18Δ cells no punctate GFP-Apg2 signal was observed (Figure 10C). A few cells with GFP dots could occasionally be observed, in which the dots were relatively weak. This phenotype was similar to what was observed in the apg1Δ strain (Shintani et al., 2001). When wild-type P. pastoris cells expressing GFP-Gsa11 were grown to saturation or shifted from methanol to glucose, a punctate GFP signal was observed (Figure 10D). This distribution pattern was not seen in the gsa12 strain. We also examined the influence of Gsa12 on the cellular localization of GFP-Gsa9. GFP-Gsa9 resides at a region juxtaposed to the vacuolar surface in wild-type cells that are not undergoing pexophagy (Kim et al., 2001b; Figure 10D). In contrast, with the altered localization observed for GFP-Gsa11 in the gsa12 mutant, the cellular distribution of GFP-Gsa9 was not affected by the absence of Gsa12. The loss of GFP-Apg2/GFP-Gsa11 punctate staining in cvt18Δ or gsa12 mutant strains in both yeasts suggested that the association of Apg2/Gsa11 with this punctate dot structure has been disrupted in the cvt18Δ/gsa12 mutant.
DISCUSSION
Maintaining a proper balance between biogenesis and degradation is important for the normal function of cells. The mechanism of macromolecular and organelle turnover has been studied in different model systems. Similar to the mammalian lysosome, the yeast vacuole is the major site for organelle degradation and for cytosolic turnover under starvation conditions. Multiple pathways are responsible for packaging cargoes and delivering them into the vacuole. Here we describe a protein, Cvt18/Gsa12, which is required for the Cvt pathway, autophagy, and pexophagy in S. cerevisiae, and for both macro- and micropexophagy as well as autophagy in P. pastoris.
In the methylotrophic yeast P. pastoris, shifting the carbon source from methanol to either ethanol or glucose induces macro- or micropexophagy, respectively. These two processes are mechanistically different, which is reflected by mutants and inhibitors that are specific to one pathway but not the other. However, mutants have been isolated that are impaired in both pathways, suggesting that common molecular components are involved in both. Gsa12 is one of the proteins that is required for both pathways and also for autophagy. A morphological analysis of the gsa12 mutant under conditions that induce pexophagy suggests that it may be blocked in a relatively early stage of the micropexophagy process. This is most easily seen by comparing the state of the vacuole in the gsa12 mutant to other mutants that are also defective in micropexophagy. For example, both Gsa1 (Yuan et al., 1997) and Gsa12 are required for early events of micropexophagy. However, ethanol-induced macropexophagy and starvation-induced autophagy proceed normally in gsa1 mutants, suggesting that Gsa1 is required for an event unique to glucose-induced micropexophagy such as glucose signaling. Indeed, GSA1 encodes phosphofructokinase, an enzyme that regulates glucose metabolism (Yuan et al., 1997). In contrast, Gsa12 is involved in macropexophagy and autophagy, presumably fulfilling a common function in these pathways. Gsa7 is a protein that is required for both micropexophagy and autophagy (Yuan et al., 1999). The engulfment of peroxisomes by the vacuole during glucose adaptation proceeds almost to completion in the gsa7 mutant (Yuan et al., 1999; Figure 3). In contrast, vacuole involution and the formation of sequestering arms that engulf the peroxisomes during micropexophagy, and that define the middle and late stages of the process, do not occur in gsa12 mutants. That is, in the absence of Gsa12, the vacuole structure remains relatively static (Figure 3).
At present, the precise function of Gsa12 in micro- and macropexophagy and autophagy is not known. It could be a signaling component that acts downstream of Gsa1 but is required for the induction of both forms of pexophagy and autophagy. A second possibility is that it may be involved in the early membrane events common to vacuole and autophagosome sequestration. It is not known whether the vacuolar membrane has any specific role in the early stages of macropexophagy or macroautophagy. Therefore, the lack of vacuolar rearrangements in gsa12 could be a result of the failure to form a sequestering autophagosome and the subsequent absence of contact and fusion between these vesicles and the vacuole. Last, there exists a nonvacuolar pool of Gsa12 that may associate with the sequestering membranes, thereby assisting in the rearrangements of these membranes to engulf cellular organelles. However, the role of Gsa12 in membrane rearrangements necessary for autophagic sequestration remains unknown.
In S. cerevisiae, the cvt18 mutant is defective in the Cvt pathway, autophagy, and pexophagy. Overlapping of these pathways has been well documented. As seen with most of the other apg/cvt mutants, prApe1 in the cvt18 mutant is also blocked at a membrane-associated but protease-accessible state, indicating that Cvt18 is required for membrane sequestration (Figure 5). This finding is consistent with the concept that the vesicle formation process is the most demanding step of cytoplasm-to-vacuole transport and may require the coordinated work of a number of proteins and lipids. The process of sequestering vesicle formation is also a unique feature that distinguishes these pathways from most of the other intracellular trafficking pathways. On the other hand, the membrane-associated, protease-sensitive state of prApe1 may represent a broad stage along its way into the vacuole. In different cvt/apg mutants that are blocked at this step, prApe1 showed a difference in magnesium concentration-dependent membrane binding (Kim et al., 2001b; Scott et al., 2001). This may indicate that the nature or the extent of membrane association is different when the Cvt pathway is blocked in those mutants. The membrane binding of prApe1 in cvt18Δ at different magnesium concentrations was comparable with that in most of the mutants such as apg1Δ and apg12Δ but unlike what was observed in the cvt19Δ and cvt9Δ mutants that displayed a weakened binding interaction (our unpublished results). Further analysis is required to resolve the detailed steps in the vesicle formation process revealed by prApe1 membrane binding.
Alternatively, a number of proteins involved in these pathways have been shown to belong to several functional groups. For newly identified mutants, studying the interaction of the corresponding protein with these previously characterized components may help reveal its potential function. Along these lines, we examined the localization of various proteins in the cvt18Δ and gsa12 strains. In both S. cerevisiae and P. pastoris, the localization of Apg2/Gsa11 was altered in cvt18Δ/gsa12 mutant strains (Figure 10). It has been shown that Apg2 colocalizes with a population of Aut7 in S. cerevisiae (Shintani et al., 2001). Because Aut7 localizes normally in cvt18Δ, it is likely that the absence of Cvt18/Gsa12 actually prevents the association of Apg2/Gsa11 with the punctate structure instead of the formation of such a structure. Alternatively, the inability of GFP-Apg2/GFP-Gsa11 to associate with this structure in cvt18/gsa12 mutants could be an indirect effect because this punctate localization is also abolished in a number of other mutants. Additional experiments will be needed to determine the precise role of Cvt18/Gsa12 in the localization of Apg2/Gsa11.
We have found that Cvt18 is localized on the vacuolar membrane as well as a perivacuolar punctate structure in vivo (Figure 8). This distribution pattern is unique among all the characterized Apg/Cvt proteins. To date, a number of proteins involved in these pathways have been localized by fluorescence microscopy. Several Cvt/Apg proteins, including Apg2, Cvt9, Cvt19, and Apg9 have also been shown to reside at punctate dots near the vacuole (Noda et al., 2000; Kim et al., 2001b; Scott et al., 2001; Wang et al., 2001). Clearly, these dots are functionally important for the vesicle formation process. The identity of those structures and their relationship with the perivacuolar Cvt18-GFP dots as well as with the process of cytoplasm-to-vacuole transport are currently under further study.
Interestingly, the association of Cvt18 with the vacuolar membrane is easily disrupted by osmotic lysis, but its binding to the perivacuolar punctate structure(s) is retained under the same conditions, suggesting that the nature of the association might be different. We speculate that this perivacuolar structure corresponds to certain membranes that migrated mainly at fraction 3 in the OptiPrep gradient analysis. The localization is not dependent on a functional Cvt/Apg pathway (our unpublished results), arguing against the possibility that Cvt18 reaches the vacuole via Cvt vesicles or autophagosomes.
Further insight can be gained by comparing Cvt18 and Gsa12 in these two systems. Both proteins showed a vacuolar membrane association, suggesting that this localization is important for their function. In addition, there is a cytosolic population of both proteins, and the interaction of Cvt18 with the vacuolar membrane is clearly weak. Accordingly, we can speculate that there is a dynamic equilibrium between the cytosolic and membrane-associated pools of these proteins. The molecular nature of the membrane attachment is not yet known. Both proteins, as well as the human and S. pombe homologs, contain WD40 repeats (Figure 2). This pattern is generally found two or more times (up to 8 times in the G protein β-subunit) in WD-repeat proteins (PROSITE), where the WD-repeats are thought to be involved in mediating protein–protein interactions. At present, two-hybrid and affinity chromatography analyses have not identified other proteins that interact with Cvt18 or Gsa12.
In the P. pastoris gsa12 mutant, micropexophagy is blocked at an early stage with minimum changes of the vacuolar morphology. There is also a strong block in nonspecific autophagic protein degradation. In S. cerevisiae, the deletion of CVT18 causes a block in pexophagy and the Cvt pathway. There is also a severe defect in survival during nitrogen starvation, suggesting that it is difficult to bypass the requirement for the Cvt18 protein under these conditions. Our current understanding of these various pathways in the above-mentioned two different model systems, however, is still too limited to allow a more detailed comparison between them. Isolation and characterization of additional components that are shared by these overlapping but distinct pathways will shed light on the common mechanisms involved in cytoplasm-to-vacuole transport.
ACKNOWLEDGMENTS
We thank Drs. Sidney Scott and Maria Hutchins for helpful discussions. This work was supported by National Institutes of Health Public Health Service grant GM-53396 (to D.J.K), a grant from The Norwegian Cancer Society (to P.E.S.), and National Science Foundation grant MCB-9817002 (to W.A.D.).
Abbreviations used:
- AOX
alcohol oxidase
- Ape1
aminopeptidase I
- Apg
autophagy
- Cvt
Cytoplasm to vacuole targeting
- FM 4-64
N-(triethlyammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide
- Pgk1
cytosolic phosphoglycerate kinase
- Pho8
vacuolar alkaline phosphatase
- PVC
prevacuolar compartment
- UTR
untranslated region
REFERENCES
- Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B, Haas A, Wickner W. Determination of four biochemically distinct, sequential stages during vacuole inheritance in vitro. J Cell Biol. 1994;126:99–110. doi: 10.1083/jcb.126.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Barringer KJ, Hessler AY, Madden KR. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J Biol Chem. 1998;273:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 61–99. [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Toh-e A, Banno I, Oshima Y. Molecular characterization of a specific p-nitrophenylphosphatase gene, PHO13, and its mapping by chromosome fragmentation in Saccharomyces cerevisiae. Mol Gen Genet. 1989;220:133–139. doi: 10.1007/BF00260867. [DOI] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Kim J, Scott SV, Klionsky DJ. Alternative protein sorting pathways. Int Rev Cytol. 2000;198:153–201. doi: 10.1016/s0074-7696(00)98005-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Labbé S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Hunter CP, Valls LA, Stevens TH. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci USA. 1986;83:3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm to vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. In vitro reconstitution of cytoplasm to vacuole protein targeting in yeast. J Cell Biol. 1995;131:1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. Delivery of proteins and organelles to the vacuole from the cytoplasm. Curr Opin Cell Biol. 1998;10:523–529. doi: 10.1016/s0955-0674(98)80068-9. [DOI] [PubMed] [Google Scholar]
- Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Sears IB, O'Connor J, Rossanese OW, Glick BS. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast. 1998;14:783–790. doi: 10.1002/(SICI)1097-0061(19980615)14:8<783::AID-YEA272>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2 functions in autophagosome formation on the perivacuolar structure. J Biol Chem. 2001;276:30452–30460. doi: 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]
- Stromhaug, P.E., Bevan, A., and Dunn, Jr., W.A. (2001). Gsa11 encodes a unique 208 kDa protein required for pexophagy and autophagy in P. pastoris. J. Biol. Chem. (in press). [DOI] [PubMed]
- Stromhaug PE, Klionsky DJ. Approaching the molecular mechanism of autophagy. Traffic. 2001;2:524–531. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tomashek JJ, Sonnenburg JL, Artimovich JM, Klionsky DJ. Resolution of subunit interactions and cytoplasmic subcomplexes of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1996;271:10397–10404. doi: 10.1074/jbc.271.17.10397. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WA., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- Veenhuis M, Salomons FA, Van Der Klei IJ. Peroxisome biogenesis and degradation in yeast: a structure/function analysis. Microsc Res Tech. 2000;51:584–600. doi: 10.1002/1097-0029(20001215)51:6<584::AID-JEMT8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn WA, Jr, Klionsky DJ. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Stromhaug PE, Dunn WA., Jr Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA., Jr Glucose-induced microautophagy in Pichia pastoris requires the α-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]