Abstract
PI3K/AKT and NOTCH1 signaling pathways are frequently dysregulated in T-cell acute lymphoblastic leukemias (T-ALL). Although we have shown that the combined activities of the class I PI3K isoforms p110γ and p110δ play a major role in the development and progression of PTEN null T-ALL, it has yet to be determined whether their contribution to leukemogenic programing is unique from that associated with NOTCH1 activation. Using a Lmo2-driven mouse model of T-ALL in which both the PI3K/AKT and NOTCH1 pathways are aberrantly upregulated, we now demonstrate that the combined activities of PI3Kγ/δ have both overlapping and distinct roles from NOTCH1 in generating T-ALL disease signature and in promoting tumor cell growth. Treatment of diseased animals with either a dual PI3Kγ/δ or a γ-secretase inhibitor (GSI) reduced tumor burden, prolonged survival, and induced proapoptotic pathways. Consistent with their similar biological effects, both inhibitors downregulated genes involved in cMYC-dependent metabolism in gene set enrichment analyses. Furthermore, overexpression of cMYC in mice or T-ALL cell lines conferred resistance to both inhibitors, suggesting a point of pathway convergence. Of note, interrogation of transcriptional regulators and analysis of mitochondrial function showed that PI3Kγ/δ activity played a greater role in supporting the disease signature and critical bioenergetic pathways. Results provide insight into the interrelationship between T-ALL oncogenic networks and the therapeutic efficacy of dual PI3Kγ/δ inhibition in the context of NOTCH1 and cMYC signaling.
Keywords: T-ALL, GSI, PI3Kγ/δ, cMYC, NOTCH1
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive disease characterized by frequent relapse, resistance to chemotherapy, and reduced survival as compared to B-precursor ALL (1). Consistent with most lymphoid malignancies, tumor development and disease progression depend upon deregulation of an oncogene(s) and/or tumor suppressor genes that permit unchecked proliferation and resistance to programmed cell death. Chromosomal translocations driven by T-cell receptor-regulatory elements occur frequently in T-ALL, resulting in aberrant expression of a select group of transcription factor oncogenes such as TAL1, LMO1, LMO2, TLX1, and TLX3 (2–4). These gene rearrangement events are typically accompanied by constitutive NOTCH1 signaling due to mutations within its heterodimerization (HD) and C-terminal proline-, glutamic acid-, serine-, and threonine-rich (PEST) domains, causing ligand-independent activation and increased stability of the γ-secretase cleaved intracellular domain (ICN1) (5, 6). Subsequent nuclear translocation of ICN1 stimulates transcription of downstream target genes such as HES1, DTX1, and MYC, the latter being an essential mediator of NOTCH1 signaling (7, 8). cMYC is a known master transcription factor that regulates genes essential for cell metabolism, proliferation and survival (9, 10).
Aberrant activation of the PI3K/AKT pathway has also been implicated in the oncogenic transformation and tumor progression of T-ALL, and is associated with aggressive biological features, drug resistance and poor prognosis (11–13). In the majority of cases this results from a loss or reduced function of the phosphatase and tensin homolog (PTEN) tumor suppressor, which limits levels of PIP3 generated by class I PI3Ks (14). Interestingly, NOTCH1 has also been reported to indirectly regulate this signaling pathway due to its ability to alter PTEN expression, which has been implicated in inducing GSI resistance in T-ALL by switching “oncogene addiction” (15). However, other studies have shown that PTEN status does not always correlate with GSI responsiveness, and that NOTCH1 and PI3K/AKT pathways may work in concert to promote tumor growth and survival (16, 17).
Previously, we have demonstrated the importance of the class I PI3K isoforms p110γ and p110δ in T cell development and their ability to cooperate as non-classical oncogenes in supporting leukemogenesis in the absence of negative regulation by PTEN (18, 19). This was further evidenced by our observations that a highly selective dual PI3Kγ/δ inhibitor CAL-130 significantly reduced disease burden, prolonged survival of mice with established PTEN null T-ALL, and induced apoptosis in human T-ALL tumor cells with aberrant PI3K/AKT signaling. Yet, it remains to be determined whether PI3Kγ/δ regulate transcriptional pathways typically associated with activated NOTCH1 (e.g. cMYC). Investigations along these lines are essential for establishing whether p110-selective small molecule inhibitors could synergize or substitute for GSIs in the treatment of T-ALL. Indeed, the variable antitumor effects of GSIs reported in phase I clinical trails would suggest that such an approach is warranted (20, 21). To this end, we evaluated the molecular and genetic interplay between these pathways using the Lmo2-driven mouse model of T-ALL where both PI3K/AKT and NOTCH1 are activated (22–23). Additionally, gene set enrichment and pathway analyses were used to determine if these disparate pro-oncogenic pathways controlled similar genes and transcriptional regulators that are determinants of the disease signature associated with T-ALL.
Materials and Methods
Mice
Cohorts of CD2-Lmo2 and Gt(ROSA)26Sortm13(CAG-MYC, -CD2*)Rsky/J;Lck-cre/Ptenfl/fl, both on C57BL/6J background, were monitored for the onset of leukemia (19, 22, 25). Experiments were performed in accordance with guidelines set forth by the Institutional Animal Care and Use Committee of Columbia University. Animals with established T-ALL received either the dual PI3Kγ/δ inhibitor CAL-130 (10 mg/kg every 8 hours; Calistoga Pharmaceuticals) (19) or the γ-secretase inhibitor dibenzazepine (DBZ; 10 μmol/kg IP daily; Tocris) (26) for a total of 7 days (27). Kaplan-Meier survival and statistical analyses were performed using GraphPad Prism Version 6.0 software. Values were considered significant at P < 0.05.
Primary leukemia samples and cell lines
Cryopreserved human T-ALL samples were provided by St. Jude Children’s Research Hospital and Vanderbilt University Medical Center after appropriate IRB review. All samples were collected with informed consent. Murine CD2-Lmo2 T-ALL cell lines 03007 and 03027 were generated as previously described (23). In brief, they were generated in the Davé lab (Vanderbilt University) from T-cell leukemia that arose in transgenic B6.CD2-Lmo2 mice. Once established in culture, aliquots of cells were banked in liquid nitrogen and samples obtained for this study in 2014. The cell lines were identified and then confirmed immediately before use by their immunophenotpye and by T-cell receptor Jβ rearrangement (23).
Retroviral transduction of murine cell line
Plasmid pMSCV-IRES-mCherry and pMSCV-cMyc-IRES-mCherry were kindly provided by the laboratory of Dr. Riccardo Dalla-Favera (Columbia University, NY, NY). Retroviruses were produced in ecotropic packaging cell line 293T Platinum-E according to manufacturer’s instructions (Cell Biolabs Inc). Viral transduction was performed using the RetroNectin® (Takara) and spinoculation method as previously described (28).
Mutation detection
Sequencing of the Notch1 and Pten genes was performed on primary mouse T-ALL cells by PCR using Pfu DNA polymerase (Stratagene) with primers specific for exon 34, and exons 3 through 7, respectively.
FACS
Preparation, staining, and detection of cell surface and cytoplasmic proteins in primary T-ALL cells and murine T-ALL cell lines were performed as previously described (19).
Western blot analysis
Cell lysates were prepared on ice in M-PER Mammalian Protein Extraction reagent (Pierce) containing a cocktail of protease and phosphatase inhibitors (19). Lysates were subjected to SDS-PAGE, transferred to PVDF membrane (Immobilon-P, Millipore), and membranes probed by overnight incubation (4°C) with appropriate primary antibodies. Bound antibodies were visualized with HRP-conjugated secondary antibodies and ECL chemistry (SuperPico West, Pierce).
Drug synergy and cell viability studies
Murine CD2-Lmo2 T-ALL cell lines 03007 and 03027 were plated at optimal density into 384-well tissue culture plates (Greiner 781080) employing the Cell::Explorer automation system (PerkinElmer). CAL-130 or the GSI CompE (ENZO Biochem) (29) (was added using HP D300 Digital Dispenser, and drug activity measured at 60 hours using Cell Titer Glo (Promega). The standard reference model of Bliss independence was employed for the results analysis (30).
Cell lines were also grown in 6 well plates and individually treated with DMSO, CAL-130, IC87114 (Calistoga) (31) CompE for 72 hours. Proliferation and cell death were determined by cell counting with trypan blue and by staining with PI (BD Biosciences) followed by flow cytometric analysis, respectively. For primary murine and human T-ALL samples, cells were harvested from thymi of diseased animals and plated with MS5-DL1 stromal cells in the presence of recombinant IL-7 and FLT-3 (mouse) or recombinant IL-7, FLT-3 and SCF (human). T-ALL cell viability following 72 hours of drug treatment was assessed using the BD Cell Viability kit (BD Biosciences) coupled with fluorescent counting beads as previously described (19). After 72 hours of treatment, absolute number of cells in DMSO control was set to 100%.
Mitochondria isolation and respiration studies
Mitochondria were isolated from murine T-ALL cells treated with DMSO or CAL-130 (2.5 μM, 12 h) and respiration analyzed in TRIS-MOPS buffer using Clark’s type electrode (Hansatech, UK) as described (32).
OCR measurements
Murine CD2-Lmo2 T-ALL cell lines treated with CAL-130 (2.5μM, 12 h), CompE (1μM, 48 h) or vehicle (DMSO) were centrifuged and directly suspended in XF assay medium (Seahorse Biosciences) supplemented with 10 mM glucose (Sigma-Aldrich) and 10 mM sodium pyruvate (Invitrogen). For primary CD2-Lmo2 T-ALL tumors, cells were grown on MS5-DL1 stromal cells in the presence of recombinant IL-7 and FLT-3 (mouse) for 24 h before the addition of DMSO, CAL-130 (2.5 μM, 12 h) or CompE (1μM, 48 h). Tumor cells were then disrupted by pipetting, filtered through a 40 μm strainer to separate from the stromal cells, and centrifuged and suspended in XF assay medium (Seahorse Biosciences) supplemented with 10 mM glucose (Sigma-Aldrich) and 10 mM sodium pyruvate (Invitrogen). Primary tumors or tumor cell lines were added to a XF24 cell culture microplate coated with poly-L-Lysine (Sigma-Aldrich). After 30 minutes at 37°C, OCR was measured using a XFe24 Analyzer (Seahorse Biosciences) after serial injections of oligomycin (1μM), carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (0.75 μM), rotenone (1 μM) and antimycin A (1μM). Coupling efficiency, as a measure of the fraction of basal mitochondrial OCR used for ATP synthesis, was assessed by the decrease in respiration on inhibiting ATP synthase with oligomycin. Cell respiratory control ratio is the ratio of the uncoupled OCR (rate after FCCP injection) to OCR in the presence of oligomycin. Spare respiratory capacity was measured by the ratio of the uncoupled OCR to the basal rate. Experiments were performed using at least 3 biological replicates and all values were internally normalized (33).
Glucose Uptake Assessment
Murine CD2-Lmo2 T-ALL cell lines cells treated as described were resuspended in glucose-free RPMI-1640 medium containing 1% fetal calf serum (FCS) and 150 μM 2-NBDG (Life Technologies), incubated for 10 min at 37° C, washed with ice-cold PBS and analyzed using flow cytometry. Apigenin (100μM, 30 min treatment), an inhibitor of Glut1 expression, was used as a positive control.
Microarray procedure
A representative murine CD2-Lmo2 derived T-ALL cell line (03007) was treated with DMSO (0.6%) or CAL-130 (2.5 μM; 10 h) or CompE (1 μM; 48 h). Biological triplicates were performed with each experimental condition and gene expression profiling performed by Expression Analysis, Inc. using the Affymetrix Mouse Genome 430 array (#901570). Microarray data were then normalized using the Robust MultiChip Averaging (RMA) algorithm as implemented in the Bioconductor package Affy (34). A modified t-test was used in Bioconductor Limma package to identify differentially expressed probe sets between each drug treatment group and control group (35). The P-values from Limma analysis were adjusted by Benjamini and Hochberg method to control false discovery rate (FDR) and genes were ranked to identify top 50 up and 50 down regulated genes for each treatment condition (36).
Statistical analyses
Results are expressed as mean ± SEM for a minimum of 3 independent experiments. Statistical analysis was performed using either the Student unpaired t-test or two-way ANOVA followed by Bonferroni post-test. P values < 0.05 were considered significant. Further detailed statistical analysis of the microarray data and information on experimental methods can be found in supplemental materials available on the Molecular Cancer Therapeutics Web site.
Results
Lmo2-driven T-ALL is defined by constitutive activation of PI3K/AKT and NOTCH1 pathways
Disease penetrance was 100% with a median survival rate of 227 days in CD2-Lmo2 transgenic mice (Fig. 1A). Tumor cells displayed high levels of both phosphorylated AKT (Thr308 and Ser473) and intracellular NOTCH1 (ICN) with activation of downstream targets (Fig. 1B); all tumors expressed the p110γ and p110δ catalytic domains of class I PI3Ks (Fig. 1 C). Of note, the majority of Lmo2-driven T-ALL lacked expression of PTEN, acquired Notch1 mutations within the PEST domain (Supplementary Table S1) and had evidence of upregulation of the Notch1 target genes such as Deltex1 (~4-fold; Fig. 1D). Moreover, the majority of circulating blasts had an immunophenotype characteristic of NOTCH1-induced T-ALL in mice: CD8 single positive or CD4/CD8 double positive with high expression of CD25 (Fig. 2A and B). Thus, the CD2-Lmo2 transgenic animal model of T-ALL provided an ideal system to determine whether the combined activities of PI3Kγ/δ regulate similar or distinct oncogenic programs typically associated with NOTCH1 activation.
Figure 1.
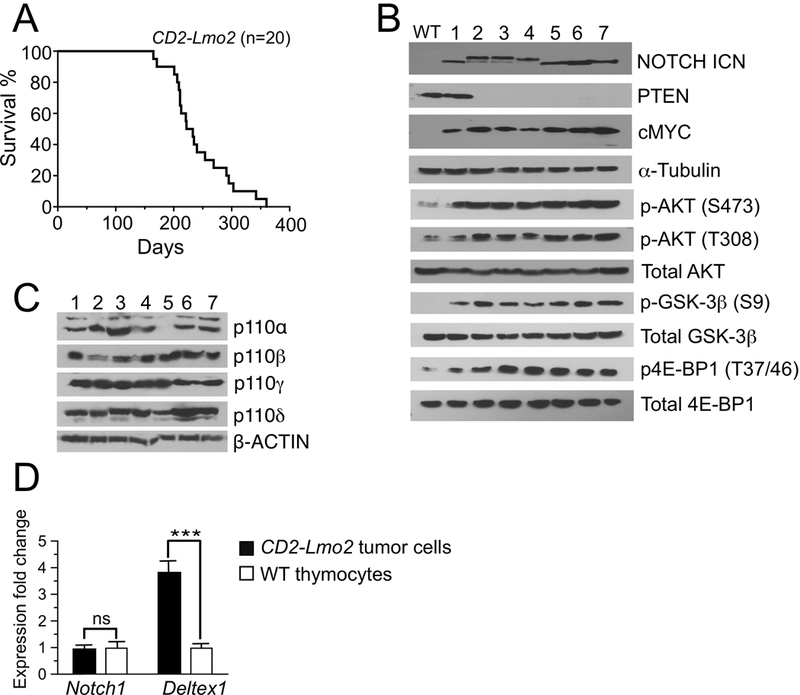
PI3K/AKT and NOTCH1 pathways are co-activated in Lmo2 driven T-ALL in mice. A, Kaplan-Meier survival curves for CD2-Lmo2 transgenic animals. Representative immunoblots of the primary T-ALL cells from diseased mice showing protein expression of PTEN and cMYC as well as AKT and NOTCH1 activation state (B), and expression of p110 catalytic subunits of class I PI3K (C). D, Expression levels of Notch1 and its target Deltex1 in primary murine tumors compared to wild-type (WT) DN3 thymocytes as determined by qRT-PCR (n = 7). ***, P < 0.0001; ns = not significant.
Figure 2.
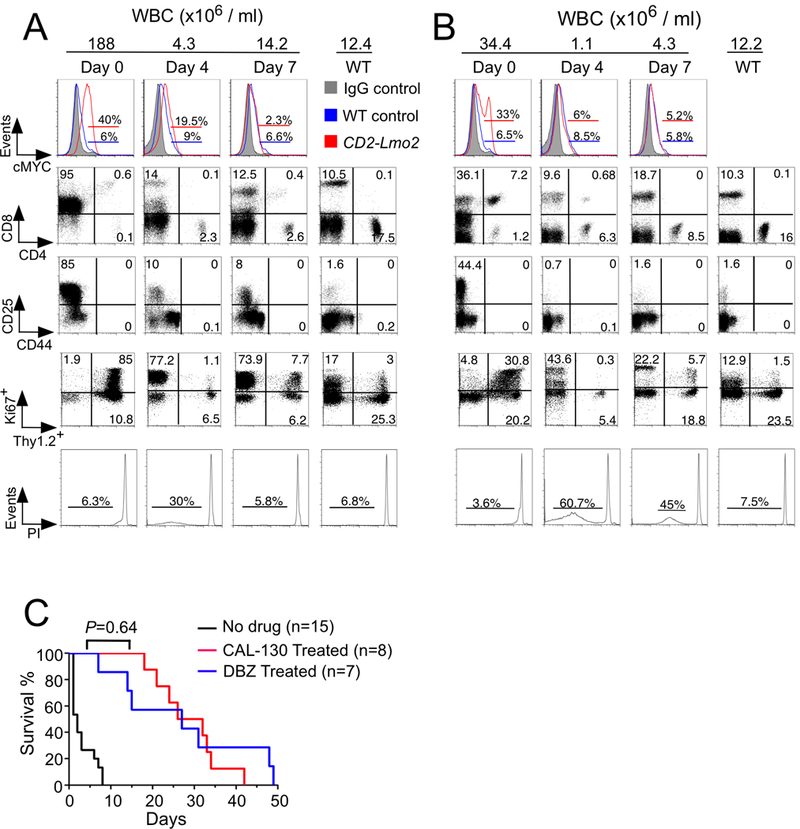
Combined PI3Kγ/δ blockade or GSI reduces tumor burden and prolongs the survival of mice with Lmo2-driven T-ALL. A and B, Flow cytometric analysis of disease status in CD2-Lmo2 transgenic mice immediately before and 4 days and 7 days after initiating treatment with either the dual PI3Kγ/δ inhibitor CAL-130 (A; 10 mg/kg every 8h) or the γ-secretase inhibitor DBZ (B; 10 μmol/kg daily) for 7 days. Tumor response to drug treatments was confirmed by changes in circulating blasts (Thy1.2+/Ki67+) and evidence of propidium iodide (PI)-staining. C, Kaplan-Meier survival curves for CD2-Lmo2 mice with T-ALL treated with either CAL-130 or DBZ for 7 days. The latter only included animals that received the full course of drug, P = 0.64 (log-rank test).
PI3Kγ/δ blockade or GSI reduces leukemia burden and prolongs survival
To determine the reliance of Lmo2-driven T-ALL on the PI3K/AKT versus NOTCH1 pathway, we treated diseased animals with either the dual PI3Kγ/δ inhibitor CAL-130 (10 mg/kg every 8 hours) or the potent γ-secretase inhibitor DBZ (10 μmol/kg per day) for 7 days. Previously we have shown that CAL-130 is highly selective for the p110γ and p110δ catalytic domains of class I PI3Ks by virtue of its ability to (1) yield an immunologic phenotype similar to that of mice lacking both p110 isoforms when administered to WT animals, and (2) significantly reduces tumor burden and prolong survival in mice that develop PTEN null T-ALL (19). Despite NOTCH1 activation, all mice had a rapid response to the dual PI3Kγ/δ inhibitor as evidenced by a reduction in circulating blasts and presence of apoptotic cells 4 days post treatment (detected sub-G0 population after propidium iodide (PI) staining) (Fig. 2A). Administration of DBZ also reduced disease burden (Fig. 2B) and resulted in a similar prolongation in survival: median of 27 days for DBZ-treated versus 29 days for CAL-130 treated animals (Fig. 2C, P = 0.64).
In order to investigate the direct effects of PI3K/AKT and NOTCH1 inhibition on tumor cell proliferation and survival, we performed in vitro co-culture of primary leukemic blasts from mice and human T-ALLs using MS5-DL1 stromal cells (19). Consistent with in vivo results, primary T-ALL cell viability was significantly reduced in the presence of either CAL-130 or the γ-secretase inhibitor CompE (Fig. 3A, Supplementary Fig. S1A and 1B, and Supplementary Table S1) while the viability of stromal cells was not affected by either compound.
Figure 3.
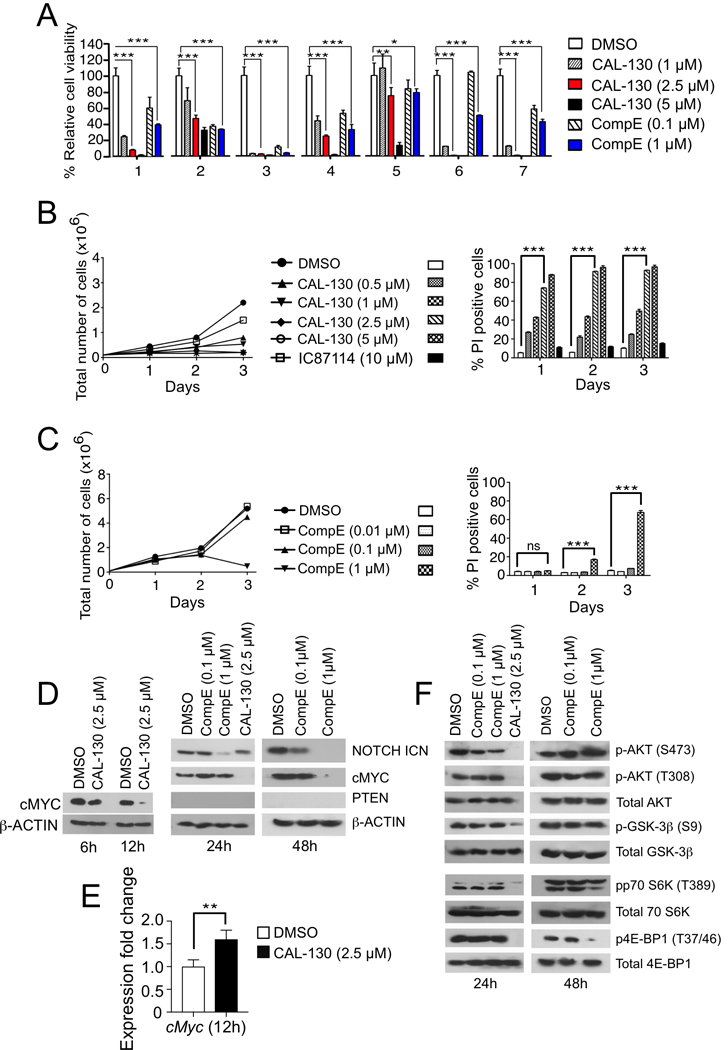
PI3Kγ/δ inhibition promotes tumor cell death irrespective of NOTCH1 activation status. A, Analyses of primary CD2-Lmo2 mouse T-ALL tumor cell viability in the presence of increasing concentrations of CAL-130 or CompE for 72 h. B-C, Proliferation and survival analysis of a representative Lmo2-driven T-ALL tumor cell line (03007) cultured in the presence of increasing concentrations of CAL-130 (B) or CompE (C) for 72 h. Data represent mean ± SEM (n = 3 in triplicate). Statistical significance was determined by Student’s t test for drug treated cells relative to control. *, P < 0.01; **, P < 0.001; ***, P < 0.0001; ns (not-significant). D, Immunoblots of PTEN and cMYC expression as well as NOTCH1 activation state in a representative CD2-Lmo2 T-ALL cell line (03007) cultured in the presence of CAL-130 (6, 12, and 24 hours) or CompE (24 and 48 hours). E, Expression levels of cMyc in the identical tumor cell line after 12 h of treatment with CAL-130 (2.5 μM) as determined by qRT-PCR (n = 3). F, Immunoblots depicting AKT and the activation state of its downstream effectors in the same cell line at the indicated duration of treatments.
We next evaluated the effects of these inhibitors alone or in combination using two representative T-ALL cell lines, designated 03007 and 03027, which were derived from diseased CD2-Lmo2 transgenic animals (23). Consistent with results obtained with primary tumors, treatment of either cell line with CAL-130, but not the p110δ selective inhibitor IC87114, had a cytostatic effect and induced cell death within 24 hours based on PI staining (Fig. 3B and Supplementary Fig. S1C). The effects of CompE were similar albeit delayed (Fig. 3C and Supplementary Fig. S1D). To determine whether the combination of CAL-130 and CompE would be synergistic in nature, we used a high-throughput cell viability screen based on cellular ATP detection and the Bliss independence model to quantify the degree of synergy between the pathways (30). Excess over Bliss (EoB) was calculated as the difference between the observed effect and the predicted response; a 5–10% EoB response was considered as weak, 10–20% as moderate, and over 20% was considered as strong synergism. Results indicate that CAL-130 (2.5 μM) in combination with CompE did not yield a synergistic effect in either cell line, suggesting redundancy in the NOTCH1 and PI3K/AKT pathways in Lmo2-driven T-ALL (Supplementary Fig. S2).
PI3Kγ/δ regulate cMYC expression independently of NOTCH1
NOTCH1 directly regulates cMYC expression, which governs an oncogenic program supporting T-ALL cell growth and survival (7,8). Accordingly, CompE blocked NOTCH1 activation in Lmo2-driven T-ALL cell lines and also reduce protein levels of cMYC, albeit at distinct time points (24 hours and 48 hours, respectively) (Fig. 3D and Supplementary Fig. S1E). In contrast to the delayed effects associated with GSI, dual inhibition of PI3Kγ/δ activity abolished cMYC protein but not transcript expression within 12 hours of treatment (Fig. 3D and 3E). Importantly, CAL-130 was still able to inhibit activation of AKT (phosphorylation of Thr308 and Ser473) and its downstream targets such as glycogen synthase kinase-3β (GSK3β), the eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), and the ribosomal protein p70 S6 kinase (p70 S6K) (19), which play a major role in cell cycle progression and tumor cell proliferation (Fig. 3F and Supplementary Fig. S1F). Of note, blockade of PI3Kγ/δ activity or GSI abrogated the expression of the anti-apoptotic effectors MCL-1 and phospho-BAD (S136) and lead to upregulation of pro-apoptotic BIM (Supplementary Fig. S3A and S3B), with > 50% of CAL-130 treated cells becoming apoptotic by 24 hours in contrast to those treated with CompE as determined by flow cytometry (sub-G1) (Supplementary Figs. S3C-F). In contrast, the majority of effects of CompE were not observed until 48 hours.
In order to confirm that loss of cMYC protein expression is essential for CAL-130 or CompE-induced tumor cell death, we generated an Lmo2-driven murine T-ALL line with constitutive cMYC expression. Indeed, over expression of this oncogene permitted continued tumor cell growth despite incubation with either inhibitor. Moreover, cMYC protein levels remained constant in contrast to cells expressing vector alone (Fig. 4A-F). To further demonstrate that down regulation of cMYC is essential for the anti-tumorigenic effects CAL-130 and that it may exert this effect independent of NOTCH1 activation, we crossed Gt(ROSA)26Sortm13(CAG-MYC, -CD2*)Rsky/J mice that overexpress cMyc with Lck-cre/Pten(fl/fl) mice that lack Pten. Previously we have shown that T-ALL that develops in PTEN-null mice is exquisitely sensitive to the dual PI3Kγ/δ inhibition (19). In contrast, T-ALL with enforced cMYC expression was resistant to CAL-130 treatment (10 mg/kg every 8 hours for 7 days) and showed continued expansion of circulating blasts (Ki67+/Thy1.2+ population) and lack of PI-staining (Fig. 4G).
Figure 4.
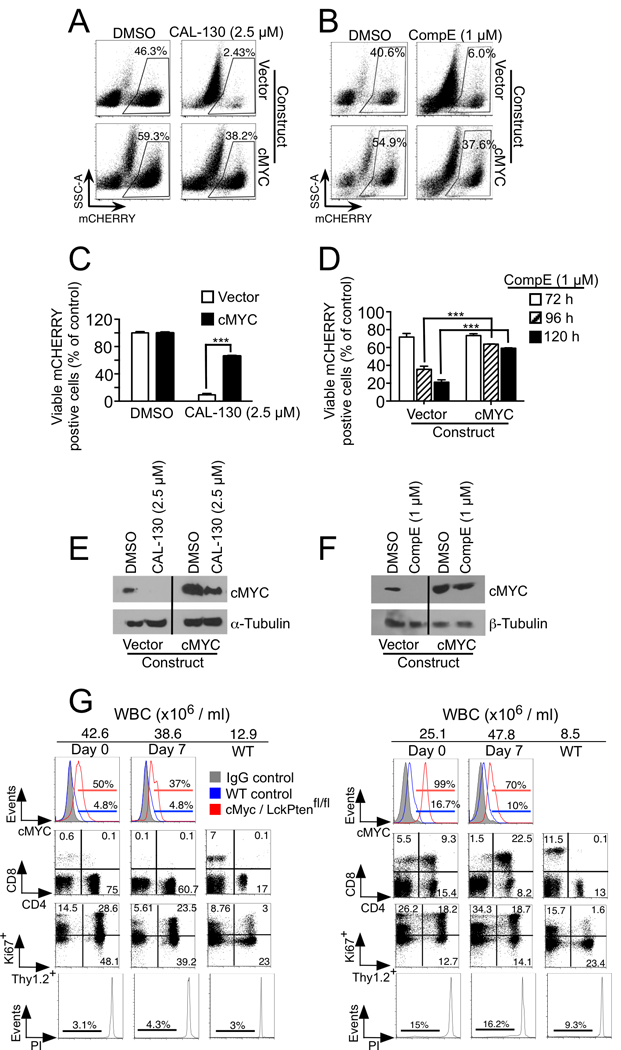
Overexpression of cMYC results in resistance to CAL-130 and GSI treatments. A-D, FACS dot plots (A and B) and viability bar graphs (C and D) of a representative CD2-Lmo2-driven T-ALL cell line (03007) transfected with mCherry-expressing empty or WT cMYC vector and treated with CAL-130 (72 hours), CompE (27, 96 and 120 hours) or DMSO control. Data represent mean ± SEM (***, P < 0.0001; n = 3, t test). cMYC protein expression in the same cells treated with CAL-130 (E) or CompE (F) for 24 and 48 hours, respectively. G, Flow cytometric analysis of disease status in two representative (n = 5) Gt(ROSA)26Sortm13(CAG-MYC, -CD2*)Rsky/J; Lck-cre/Pten(fl/fl) mice immediately before and 7 days after initiating treatment with dual PI3Kγ/δ inhibitor CAL-130 (10 mg/kg every 8h) for 7 days. Tumor drug response was determined by assessing for changes in circulating blasts (Thy1.2+/Ki67+) and propidium iodide (PI)-staining.
PI3Kγ/δ and NOTCH1 have overlapping gene regulatory networks in Lmo2-driven T-ALL
To better understand the genetic basis by which PI3Kγ/δ and NOTCH1 regulate tumor progression in Lmo2-driven T-ALL, we generated gene expression signatures based on disease phenotype and drug treatment. Global gene expression profiles were obtained in triplicate from tumor cells cultured in the presence of CAL-130 (2.5 μM), CompE (1 μM), or DMSO. Drug treated cells were harvested at time points known to affect cMYC expression (12 hours and 48 hours, respectively). A Venn diagram was created to illustrate the overlap in genes altered by either PI3Kγ/δ blockade (red circle) or GSI (green circle) using a false discovery rate (FDR) of 0.0005 as cut off (Fig. 5A). Of note, ten-fold more genes were affected following PI3Kγ/δ blockade and included the majority (~62%) of genes altered by GSI monotherapy, a highly significant finding (Fisher’s exact test, P < 2.2×10−16). Heat maps were also generated from complete linkage hierarchical cluster analyses of statistically significant changes in gene expression associated with drug treatment versus vehicle control (Fig. 5B). In the case of PI3Kγ/δ blockade, down-regulated genes also included those involved in cMYC-regulated cellular metabolism and/or metabolic reprogramming such as the glycine/serine biosynthetic pathway (e.g. Phgdh, FDR = 6×10−8; Shmt1, FDR = 8.2×10−8; Shmt2, FDR = 6.2×10−8, folate metabolism (e.g. Mthfd1, FDR = 6.04×10−8), glycolysis (e.g. Hk-2, FDR = 5.4×10−8), nucleotide metabolism (Ak4, FDR = 2.3×10−8) and mitochondrial proline metabolism and synthesis (Pycr1, FDR = 3.1×10−7) (37–41). As expected, gene expression profiling of CompE-treated cells demonstrated significant downregulation of canonical NOTCH1 target genes including Hes1 (FDR = 1.1×10−7), Hey1 (FDR = 2.6×10−8), and Deltex1 (FDR = 4.4×10−6), as well as modulating the expression of genes involved in glycolysis (e.g. Slc2a3, FDR = 4.14×10−6; Hk2, FDR = 1.68×10−5) and nucleotide metabolism (e.g. Ak4, FDR=1.66×10−3) (42).
Figure 5.
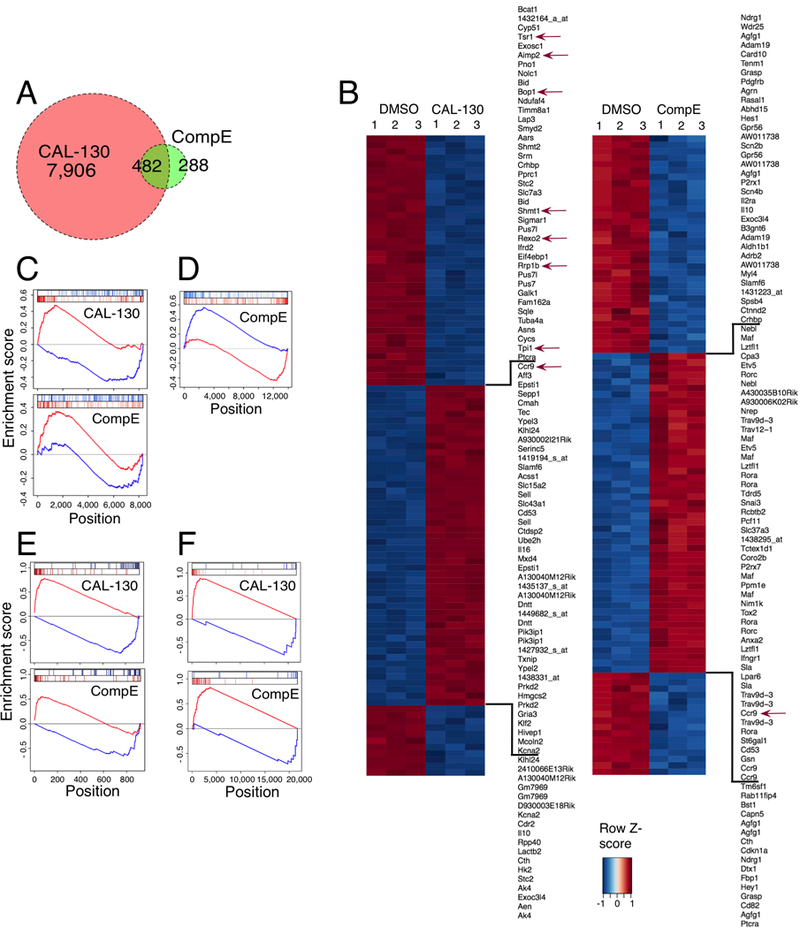
Blockade of PI3Kγ/δ activity reverses the oncogenic signature of Lmo2-driven T-ALL. A, Venn diagram comparing the number and overlap of genes altered by PI3Kγ/δ blockade (red circle) or GSI (green circle) using FDR < 0.0005 as cut off. Gene expression profiling was performed on triplicate samples of the CD2-Lmo2 T-ALL cell line (03007) treated with either CAL-130 (2.5μM, 10 hours) or CompE (1μM, 48 hours) as compared to DMSO control. B, Heat map representation of the top 100 differentially expressed genes (FDR < 0.00001) between DMSO and CAL-130 or CompE treated tumor cells. The scale bar shows color-coded differential expression, with red and blue indicating higher and lower levels of expression, respectively. Red arrows denote cMYC-regulated genes. C, Enrichment of top 100 activated and repressed genes from Lmo2-driven T-ALL disease signature (T-ALL versus WT mouse thymocytes) on the CAL-130 (top) and CompE (bottom) activity gene signature. Repressed genes are shown in blue, and over-expressed genes are shown in red. D, Comparison of GSI treatment signature for tumor cells (top 200 up and down regulated genes) on the consensus GSI perturbational signature obtained from the expression profiles (GSE5827) of seven human T-ALL cell lines treated with DMSO or CompE. E, Enrichment of top 40 activated and repressed T-ALL master regulator TFs (as inferred by ViPER based on the Lmo2-driven T-ALL versus WT murine thymocyte signature) on the CAL-130 (top) and CompE (bottom) activity signature, respectively. F, Enrichment of cMYC-regulated genes on the sorted CAL-130 (top) and CompE (bottom) treatment gene signature.
We next defined a murine T-ALL disease signature by ranking all genes based on their differential expression in T-ALL samples compared to wild type murine thymocytes. To determine the contribution of PI3Kγ/δ and NOTCH1 to the overall disease signature, we performed a gene set enrichment analysis (GSEA) of the 200 most transcriptionally activated and repressed genes following treatment with either CAL-130 or CompE in differentially expressed genes. Although both drugs significantly inverted the T-ALL disease signature, underscoring the importance of these signaling pathways in maintaining the leukemic phenotype (Fig. 5C), the enrichment was much more significant with CAL-130 (normalized enrichment score (NES) of −5.8 (P < 0.0001) for CAL-130 versus NES of −2.6 (P = 0.006) for CompE. To show that treatment of murine cells is highly consistent with treatment of human cells, we assessed whether the top 200 most differentially expressed genes following CompE treatment of murine T-ALL cells were significantly enriched in human-to-mouse mapped genes differentially expressed following CompE treatment of human cells (Fig. 5D). To accomplish this goal, we integrated data from seven distinct dataset representing human T-ALL cells treatment with CompE (7). Analysis confirms that results from drug treatment of Lmo2-driven murine T-ALL are relevant to human T-ALL.
Previously, we have shown the existence of Master Regulator (MR) proteins responsible for causally (i.e., physically) regulating the transcriptional state of a cancer cell (43). As such, their activated and repressed targets are highly enriched in genes that are over- and under-expressed in tumor cells, respectively, and can thus be efficiently identified by analyzing a regulatory network that represents the transcriptional targets of every protein using the VIPER (Virtual Proteomics by Enriched Regulon) algorithm (44). VIPER is an extension of the Master Regulator Inference algorithm (MARINa) (45), which supports analysis of MR proteins on an individual sample basis. Such an analysis has helped elucidate novel mechanisms of tumorigenesis, progression, and drug sensitivity in glioma, leukemia, lymphoma, prostate and breast cancer, among others. As MR protein activity efficiently recapitulates overall tumor state and drug response, we first identified key T-ALL-specific MR proteins and then assessed whether compound treatment could effectively reverse their activity. MR proteins were inferred by VIPER analysis of a previously assembled, genome-wide T-ALL-specific regulatory network (interactome) using a signature of a Lmo2-driven murine T-ALL cell line (7) compared to wild type murine thymocytes. This interactome was previously used to successfully elucidate the Mechanism of Action (MoA) of small molecule compounds targeting human T-ALL pathways (7). The ViPER algorithm was then used to compute the activity of each of the top 40 T-ALL MR proteins following drug perturbation. This was accomplished by computing the enrichment of their regulons (i.e., the set of their transcriptional targets), as represented in the interactome, in genes differentially expressed following drug treatment (drug signature). Although the gene signatures for the two drugs overlapped significantly (P < 6 × 10−13, Fisher exact test), ~3-fold more T-ALL MRs were reverted by CAL-130 compared to GSI CompE (Fig. 5E and Supplementary Table S2; NES < −1.96 and P < 0.05). Of note, Lyl1, Tal1 and Hhex were not affected by CAL-130 according to our analysis. We also performed GSEA of ~4,500 annotated gene sets collected from Molecular Signatures Database (MSigDB) on the CAL-130 and CompE induced gene signatures to study the perturbed pathways (46). Although CAL-130 altered many of the same genes as compared to CompE, it had stronger effects on pathways regulated by PML (originally identified as a tumor suppressor in acute promyelocytic leukaemia) (NES 2.57 and P < 0.01 vs. NES 0.82 and P = 0.41), EDF1 (NES −3.58 and P < 0.005 vs. NES −0.53 and P = 0.6) and IRF9 (NES 3.18 and P < 0.005 vs. NES 0.07 and P = 0.944). Thus, CAL-130 treatment recapitulated majority of mechanism of action of CompE, while reversing the activity of a large number of additional master regulators and pathways associated with the T-ALL tumor cell state.
To access drug effects on cMYC activity, we analyzed published microarrays of human T-ALL (47) and used GSEA to compute the enrichment of cMYC-regulated genes differentially expressed following CAL-130 or CompE treatment (Fig. 5F and Supplementary Table S3). Although genes regulated by cMYC were observed in both drug treatment signatures (P < 10−3), CAL-130 treatment had a greater effect in terms of reversing the global expression of cMYC targets (NES of −6.23), compared to CompE (NES of −5.10). Of note, ~74% of genes regulated by cMYC were in the top 1,000 genes in the CAL-130 treated gene expression signature as compared to ~42% in the CompE treated gene signature (Supplementary Table S3).
PI3Kγ/δ and NOTCH1 differentially modulate metabolic and mitochondrial pathways in leukemic cells
To better assess the functional categories represented by the set of genes altered in response to the inhibitors, we performed pathway analysis using DAVID (48). A highly conservative threshold (P < 1×10−5) was used to identify the top down-regulated genes in CAL-130 or CompE treated Lmo2-driven T-ALL cells. In the case of PI3Kγ/δ blockade, there was a significant enrichment in functional categories related to cellular metabolism and most interestingly, pathways that regulate mitochondrial activity (Fig. 6A and 6B). CompE treatment not only altered similar metabolic pathways including those related to carbohydrate, nucleotide, pyruvate and TCA (Fig. 6C and Supplementary Fig. S4A and S4B), but also reduce glucose uptake in T-ALL tumor cells as observed for CAL-130 (Fig. 6D and Supplementary Fig. S5A); it did not, however, appear to affect genes that directly regulate mitochondrial function (7, 42, 49).
Figure 6.
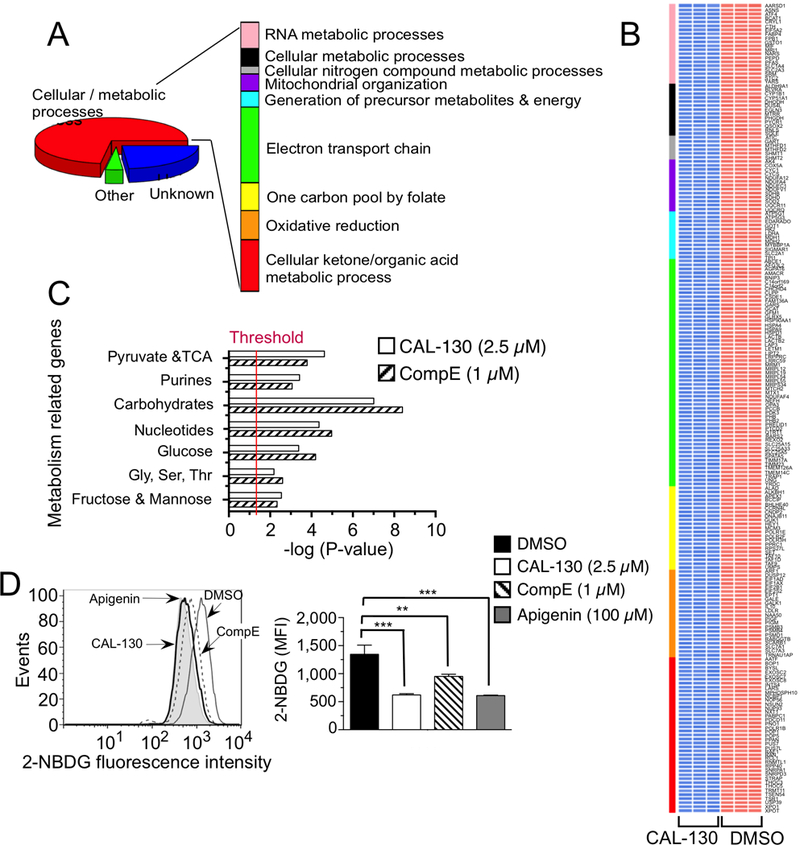
PI3Kγ/δ and NOTCH1 regulate metabolic pathways that contribute to the oncogenic programming of Lmo2-driven T-ALL. A, Functional annotation of top 297 genes down regulated (P < 0.00001) in a CAL-130 (2.5 μM, 10 h) treated CD2-Lmo2 T-ALL cell line (03007) using DAVID. Pathways with Benjamini Hochberg corrected P < 0.05 are shown. B, Heat map of metabolism-related gene expression profiles after CAL-130 or vehicle (DMSO) treatment (in triplicate). Colored bar denotes pathways annotated in A, and red and blue indicate high and low levels of gene expression, respectively. C, Summary of GSEA performed on the ranked genes according to the ratios of transcripts from DMSO, CAL-130 (2.5 μM, 10 h) or CompE (1μM, 48 h) treated cells. Six metabolic gene sets with P < 0.01 and a false discovery rate (FDR) of <0.05 (represents threshold) were considered significant. D, Inhibition of glucose uptake by PI3Kγ/δ blockade or NOTCH1 inhibition. Histograms (upper) and mean fluorescence intensity (MFI, lower) of 2-NBDG incorporation into tumor cells treated with DMSO, CAL-130 (2.5 μM, 14 h) or CompE (1μM, 72 h). Apigenin (100μM, 30 min treatment), an inhibitor of Glut1 expression, was used as a positive control. Data represent mean ± SEM (**, P < 0.01, ***, P < 0.001 relative to DMSO; n = 3, t test).
As aforementioned, blockade of PI3Kγ/δ activity preferentially altered genes that affect mitochondrial activity, such as those involved in the electron transport chain and oxidative phosphorylation (Fig. 7A and Supplementary Fig. S4C). Consistent with this observation, CAL-130 but not CompE significantly reduced in situ oxygen consumption rates (OCRs), ATP levels and spare respiratory capacity, the latter indicating that PI3Kγ/δ inhibition limits the ability of tumor cell lines and primary tumor cells to respond to stress or metabolic challenge (Fig. 7B-E and Supplementary Fig. S5B-H). Further evidence to support these observations was provided by directly studying mitochondria isolated from CAL-130 treated tumor cells. Indeed, a robust inhibition of resting and phosphorylating respirations rates in purified mitochondria were observed suggesting that the alterations in genes encoding the respiratory chain complexes I (e.g. Ndufa12), II (e.g. Sdhb), and III (e.g. Cyc1) were functionally significant (Fig. 6B and 7F). Our experiments also suggested dysregulation of complex IV as evidenced by the marked depression of mitochondrial respiration with TMPD-ascorbate as substrate (50). Of note, CAL-130 treatment affected the expression of mitochondrial protein serine hydroxymethyl transferase (SHMT2; Figure 7F, insert). SHMT2 is a key enzyme in the serine / glycine synthesis pathway that plays a critical role in the metabolic reprogramming in cancer and is a transcriptional target of cMYC (51). Thus, PI3Kγ/δ inhibition not only affects major metabolic pathways that support tumor homeostasis but also affects the bioenergetics of T-ALL cells.
Figure 7.
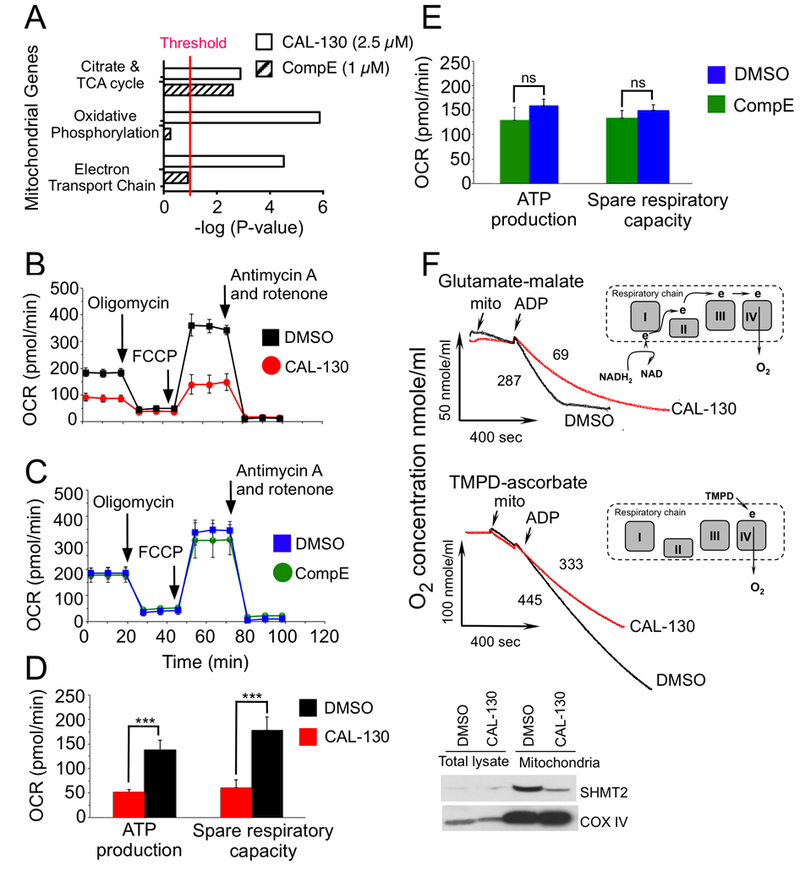
PI3Kγ/δ regulate distinct bioenergetic pathways that contribute to the oncogenic programming of Lmo2-driven T-ALL. A, Summary of mitochondrial GSEA analysis performed on the ranked genes according to the ratios of transcripts from DMSO, CAL-130 (2.5 μM, 10 h) or CompE (1μM, 48 h) treated cells. Gene sets with a P < 0.01 and a false discovery rate (FDR) of < 0.05 (represents threshold) were considered significant. B and C, Oxygen consumption rates (OCR) in the same cell line treated with DMSO vs. CAL-130 (2.5μM) for 12 hours (B) or DMSO vs. CompE (1μM) for 48 hours (C) under basal conditions and in response to the indicated mitochondrial inhibitors. Changes after oligomycin and FCCP application are indicative for respiration linked to ATP synthesis and the maximal respiratory capacity, respectively. D and E, ATP production and spare respiratory capacity in CAL-130 (D) or CompE (E) treated tumor cells. Data represent mean ± SEM (***, P < 0.0001, n = 3, t test). F, Respiration of mitochondria isolated from a CD2-Lmo2 T-ALL cell line (03007) following exposure to CAL-130 (2.5 μM, 12 hours) or DMSO and supported with two different substrates: complex-I dependent glutamate-malate (upper panel) or complex-IV linked substrate, TMPD-ascorbate (lower panel). Red and black tracings denote drug and DMSO treated cells, respectively. The points of initiation of resting, phosphorylating and uncoupled respirations are indicated (arrows). The values of the phosphorylating respiration rates (PRR) are provided. Schematic substrate-dependent changes in electron supply for mitochondrial respiratory chain is presented above each tracing. Insert shows western blot analysis for the mitochondria expressed SHMT2 protein in treated tumor cells.
Discussion
Cumulative evidence indicates that the PI3K/AKT and NOTCH1 pathways not only contribute to the malignant transformation of T cell progenitors but also play a major role in regulating the growth, metabolism, and survival of T-ALL. Here, we explored the interdependence of these oncogenic pathways in promoting tumor cell maintenance, which has profound implications for the development of a rational therapeutic strategy to treat this aggressive hematologic malignancy. It is for this reason that we chose the Lmo2-driven transgenic mouse model of T-ALL that is characterized by the spontaneous accumulation of several oncogenic mutations in T cell precursors, resulting in the co-activation of PI3K/AKT and NOTCH pathways. In the context of this model, we now provide evidence that (1) although there is no preferential addiction of tumor cells to either pathway, PI3Kγ/δ blockade appears to exert a more rapid and potent effect on tumor cells than GSI, and (2) there is a lack of synergy between PI3K/AKT and NOTCH1 in supporting tumor cell proliferation and survival.
Previous studies have demonstrated that NOTCH1 can indirectly impact on the activity of the PI3K/AKT pathway by controlling a transcriptional network that regulates PTEN expression in leukemic T cells (15). Indeed, this mechanism is believed to be responsible for bypassing the requirement for aberrant NOTCH1 signaling that normally supports tumor cell growth and metabolism. In contrast, our studies indicate that NOTCH1 does not regulate PI3Kγ/δ activity or PTEN expression, suggesting that these pathways can function independently of one another. This is exemplified by the lack of correlation between activation status of either pathway and the ability of primary mouse or human T-ALL cells to respond to treatment with CompE or CAL-130. That said, they do appear to control similar oncogenic networks that are indispensible for tumor cell maintenance. Results indicate a genetic underpinning for these observations as evidenced by the considerable overlap in genes regulated by both signaling pathways, including cMYC. The latter was confirmed by the ability of constitutively expressed cMYC to reverse the effects of either inhibitor. Collectively, these data demonstrate that PI3Kγ/δ can support a NOTCH1-independent pathway of cMYC activation in T-ALL, which may account for similar reduction in cell growth and survival but lack of synergistic effect observed in response to CAL-130 and/or CompE (52).
Why then does PI3Kγ/δ blockade appear to exert a more rapid and potent anti-leukemic effect than inhibition of NOTCH1 by GSI? Gene expression profiling and pathway analysis based on DNA microarrays of drug treated Lmo2-driven T-ALL provided possible answers. On a global scale, CAL-130 treatment had a greater effect on altering the disease signature compared to CompE, as demonstrated by the reversal of top master regulators (e.g. HNRNPAB, Tp53, PA2G4, ASH2L and SNAPC5) and affects on pathways (e.g. PML, EDF1, and IRF9) associated with the CD2-Lmo2 T-ALL tumor cell state. This was consistent with the ability of CAL-130 to down regulate a 10-fold greater number of genes at an early time point. Interestingly, the majority of altered genes are known to support cellular metabolism and/or the metabolic reprogramming implicated in tumor progression, including those involved in serine/glycine metabolism, polyamine synthesis, and glycolysis. Indeed, the ability of cancer cells to alter their metabolism is essential to support the increased energy demands associated with rapid tumor growth. Germane to this study is the ability of the PI3K/AKT pathway to act as a master regulator of aerobic glycolysis and cellular biosynthesis that critically relies on mitochondrial bioenergetics (53, 54). In fact, the shift from oxidative to more glycolytic metabolism is one of the most common changes found in tumor cells. Although our results clearly demonstrated that PI3Kγ/δ blockade or GSI significantly reduce glucose uptake in leukemic cells, only the former was able to rapidly alter mitochondrial function. Indeed, we noted that genes encoding proteins involved in oxidative phosphorylation were uniquely susceptible to CAL-130, including those essential for the mitochondrial respiratory chain complex formation (FDR < 0.005). Consistent with this observation was the robust inhibition of mitochondrial resting and phosphorylating respirations rates. This finding is of particular interest as enhancement in the bioenergetics properties of tumor cells is thought to be a prerequisite for survival.
Overall, our results suggest the existence of a regulatory circuit linking NOTCH1 and PI3K/AKT signaling, with cMYC serving as a crucial node of convergence that intertwines but does preferentially support the activity of either pathway. Moreover, we provide new insights into the mechanism(s) by which the combined activities of PI3Kγ/δ contribute to the maintenance of T-ALL and how pharmacological blockade of these two isoforms exerts a more rapid and pronounced effect on tumor cell proliferation than GSI, yet can yield similar rates of survival of diseased animals. This involves regulating transcriptional pathways and genes typically associated with NOTCH1, as well as distinct metabolic and bioenergetic pathways essential for tumor progression and survival. These results provide a rationale for the combined targeting of p110γ and p110δ catalytic domains of class I PI3Ks in T-ALL where NOTCH and/or PI3K/AKT pathways are dysregulated.
Supplementary Material
Acknowledgements
We thank Juan Carlos Zuniga-Pflucker for providing the MS5-DL1 cells, Andrew Nicholson, Anastasiya Egorova, and Anna Binyamin for genotyping and animal husbandry, and St. Jude Children’s Research Hospital for providing primary T-ALL cells from patients.
Financial support
This research is supported by National Cancer Institute grant R01CA169162–01 (T.G. Diacovo), Department of Veterans Affairs (I01BX001799, U.P.D.), the Vanderbilt Ingram Cancer Center (P30 CA68485) (U.P. Davé).
Footnotes
Conflict of Interest Disclosure. The authors declare no potential conflicts of interest
References
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med 2006; 354:166–78. [DOI] [PubMed] [Google Scholar]
- 2.Teitell MA, Pandolfi PP. Molecular genetics of acute lymphoblastic leukemia. Annu Rev Pathol 2009;4:175–98. [DOI] [PubMed] [Google Scholar]
- 3.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest 2012;122:3398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer 2006;6:347–59. [DOI] [PubMed] [Google Scholar]
- 5.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol 2008;3:587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004;306:269–71. [DOI] [PubMed] [Google Scholar]
- 7.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 2006;103:18261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 2006;20:2096–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang CV. MYC on the path to cancer. Cell 2012;149:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer N, Penn LZ. Reflecting on 25 years with MYC Nat Rev Cancer 2008;8:976–90. [DOI] [PubMed] [Google Scholar]
- 11.Blackburn JS, Liu S, Wilder JL, Dobrinski KP, Lobbardi R, Moore FE, et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 2014; 25: 366–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jotta PY, Ganazza MA, Silva A, Viana MB, da Silva MJ, Zambaldi LJ, et al. Negative prognostic impact of PTEN mutation in pediatric T-cell acute lymphoblastic leukemia. Leukemia 2010;24:239–42. [DOI] [PubMed] [Google Scholar]
- 13.Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengline E, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol 2013;31:4333–42. [DOI] [PubMed] [Google Scholar]
- 14.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest 2008;118:3762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007;13:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood 2009; 113:6172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medyouf H, Gao X, Armstrong F, Gusscott S, Liu Q, Gedman AL, et al. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood 2010;115:1175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swat W, Montgrain V, Doggett TA, Douangpanya J, Puri K, Vermi W, et al. Essential role of PI3Kdelta and PI3Kgamma in thymocyte survival. Blood 2006;107:2415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, De Keersmaecker K, et al. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell 2012;21:459–72. [DOI] [PubMed] [Google Scholar]
- 20.Deangelo DJ, Stone RM, Silverman LB, Stock W, Attar EC, Fearen I, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. J Clin Oncol. 2006;24(18S):6585. [Google Scholar]
- 21.Papayannidis C, DeAngelo DJ ,Stock W, Huang B, Shaik MN, Cesari R et al. A Phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer Journal. 2015;5:e350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleveland SM, Smith S, Tripathi R, Mathias EM, Goodings C, Elliott N, et al. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells 2013;31:882–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleveland SM, Goodings C, Tripathi RM, Elliott N, Thompson MA, Guo Y, et al. LMO2 induces T-cell leukemia with epigenetic deregulation of CD4. Exp Hematol 2014; 42:581–93 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 2010;327:879–83. [DOI] [PubMed] [Google Scholar]
- 25.Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol 2012;13:1092–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milano J, McKay J, Dagenais C, Foster-Brown L, Pognan F, Gadient R, et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004; 82:341–58. [DOI] [PubMed] [Google Scholar]
- 27.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med 2009;15:50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Sadelain M, Brentjens R. Retroviral transduction of murine primary T lymphocytes. Methods Mol Biol 2009;506:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doerfler P, Shearman MS, Perlmutter RM. Presenilin-dependent gamma-secretase activity modulates thymocyte development. Proc Natl Acad Sci U S A. 2001;98:9312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A 2003;100:7977–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential role of phosphoinositide 3-kinase delta in neutrophil directional movement. J Immunol. 2003;170:2647–54. [DOI] [PubMed] [Google Scholar]
- 32.Niatsetskaya ZV, Sosunov SA, Matsiukevich D, et al. The oxygen free radicals originating from mitochondrial complex I contribute to oxidative brain injury following hypoxia-ischemia in neonatal mice. J Neurosci 2012;32:3235–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J 2011;435:297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300. [Google Scholar]
- 37.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol 2012;30:671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanning NJ, Looyenga BD, Kauffman AL, Niemi NM, Sudderth J, DeBerardinis RJ, et al. A mitochondrial RNAi screen defines cellular bioenergetic determinants and identifies an adenylate kinase as a key regulator of ATP levels. Cell Rep 2014;7:907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A 2012;109:8983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 2013;13:572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013;24:213–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herranz D, Ambesi-Impiombato A, Sudderth J, Sanchez-Martin M, Belver L, Tosello V, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med 2015;21:1182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer 2017;17:116–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 2016;48:838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 2013;24:766–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis:a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia 2006;20:1279–87. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, and Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 49.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol 2005;6:881–8. [DOI] [PubMed] [Google Scholar]
- 50.Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat Protoc 2014;9:421–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikiforov MA A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 2002;22:5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnet M, Loosveld M, Montpellier B, Navarro JM, Quilichini B, Picard C, et al. Posttranscriptional deregulation of MYC via PTEN constitutes a major alternative pathway of MYC activation in T-cell acute lymphoblastic leukemia. Blood 2011;117:6650–9. [DOI] [PubMed] [Google Scholar]
- 53.DeBerardinis RJ1, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11–20. [DOI] [PubMed] [Google Scholar]
- 54.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


