Abstract
Purpose
Our previous work evaluated individual prognostic and predictive roles of TP53, KRAS, and EGFR in non–small-cell lung cancer (NSCLC). In this analysis, we explore the prognostic and predictive roles of TP53/KRAS and TP53/EGFR comutations in randomized trials of adjuvant chemotherapy versus observation.
Patients and Methods
Mutation analyses (wild-type [WT] and mutant) for TP53, KRAS, and EGFR were determined in blinded fashion in multiple laboratories. Primary and secondary end points of pooled analysis were overall survival and disease-free survival. We evaluated the role of TP53/KRAS comutation in all patients and in the adenocarcinoma subgroup as well as the TP53/EGFR comutation in adenocarcinoma only through a multivariable Cox proportional hazards model stratified by trial.
Results
Of 3,533 patients with NSCLC, 1,181 (557 deaths) and 404 (170 deaths) were used for TP53/KRAS and TP53/EGFR analyses. For TP53/KRAS mutation status, no prognostic effect was observed (P = .61), whereas a borderline predictive effect (P = .04) was observed with a deleterious effect of chemotherapy with TP53/KRAS comutations versus WT/WT (hazard ratio, 2.49 [95% CI, 1.10 to 5.64]; P = .03). TP53/EGFR comutation in adenocarcinoma was neither prognostic (P = .83), nor significantly predictive (P = .86). Similar results were observed for both groups for disease-free survival.
Conclusion
We could identify no prognostic effect of the KRAS or EGFR driver and TP53 tumor suppressor comutation. Our observation of a potential negative predictive effect of TP53/KRAS comutation requires validation.
INTRODUCTION
Lung cancer is one of the most molecularly complex cancers, second only to malignant melanoma1-3; however, many of these genetic changes may not have functional importance, and it seems that a much smaller number of true driver mutations—with prognostic or predictive value—characterize this malignancy. KRAS, a member of the RAS family, was one of the first oncogenes to be identified in non–small-cell lung cancer (NSCLC).4-6 KRAS mutations occur most frequently in codons 12 and 13 and are usually found in nonsquamous histology and in cancers of smokers. Slebos et al7 were among the first to suggest that KRAS was prognostic of a poorer outcome. Since then, several meta-analyses have reported similar results, although there has been considerable variability among studies with respect to the magnitude of prognostic effect.8,9 In vitro experiments have suggested that RAS mutations might be associated with resistance to treatment10; however, clinical studies could not always confirm these preclinical effects.11
Mutations in the tumor suppressor gene, TP53, which encodes the p53 protein, are frequent in all subtypes of NSCLC, with reported mutation rates of approximately 39% to 46% in adenocarcinomas, 81% in squamous cell carcinomas, and 68% in large cell carcinomas.12,13 TP53 plays multiple roles in the prevention and suppression of abnormal cell growth through cell-cycle arrest, apoptosis, senescence, or control of metabolism and DNA repair. However, data on its prognostic or predictive effect in NSCLC are limited and inconclusive.14-16
Sensitizing epidermal growth factor receptor (EGFR) gene mutations were first reported in 200417,18 and, over the last decade, have become the most important molecular tool for treatment selection for advanced NSCLC.19 They occur almost exclusively in adenocarcinoma and non-neuroendocrine large cell carcinoma, significantly more frequently in patients who have never smoked, and they confer exquisite sensitivity to EGFR tyrosine kinase inhibitors.
Multiple driver mutations are less common in NSCLC, and their prognostic and predictive significance has not been well studied. In an attempt to clarify the impact of multiple TP53, KRAS, and EGFR mutations—the three most frequent driver oncogenes in NSCLC—and their interaction on survival benefit from platinum-based adjuvant chemotherapy (ACT), we performed this pooled analysis of four randomized trials of ACT or observation (OBS). We hypothesized that comutation of a driver oncogene and a tumor suppressor gene might be prognostic of poorer outcome in untreated patients and potentially predictive of lesser benefit from ACT.
PATIENTS AND METHODS
Clinical Trials
This study used the LACE (Lung Adjuvant Cisplatin Evaluation)20 database of 3,533 patients from three LACE cisplatin-based ACT trials—JBR.1021,22 IALT,23,24 and ANITA25—and CALGB-9633, which used carboplatin-based ACT.26 Scientists, clinicians, and statisticians from these trials represent the LACE-Bio Collaborative Group.
Mutation Analyses
Mutations were analyzed by direct sequencing of PCR products of DNA that was extracted from formalin-fixed, paraffin-embedded sections. All personnel who conducted mutation analyses were blinded to study arm and outcome. All mutation analyses were performed in one laboratory for JBR.10 (Tsao) and IALT (Hainaut). For ANITA, KRAS and TP53 were analyzed in the Tsao and Hainaut laboratories, respectively. For CALGB-9633, KRAS and EGFR were analyzed in the Jänne laboratory and TP53 in the Hainaut laboratory. For technical reasons, EGFR test results are not available for the ANITA trial. Mutations are defined as wild-type (WT) or mutant (MUT), without further subtyping. EGFR mutation refers only to sensitizing mutations in exons 19 and 21. Full laboratory methods have been published previously.11,15,27
Statistical Methods
Overall survival (OS)—the primary end point—was defined as time from random assignment to death from any cause or last follow-up in surviving patients. Disease-free survival (DFS) was defined as time from random assignment to recurrence or death from any cause or last follow-up in surviving patients. We evaluated the prognostic and predictive value of TP53/KRAS comutation status and TP53/EGFR comutation status. For the latter, analysis was restricted to adenocarcinoma as EGFR mutation is uncommon in other histologic subtypes. Mutation status from these combinations was grouped into four classes (WT/WT, WT/MUT, MUT/WT, and MUT/MUT). Hazard ratios (HRs) and their CIs were estimated via a multivariable Cox proportional hazards model stratified by trial including the following core variables: treatment (ACT or OBS), sex, age (< 55, 55 to 64, ≥ 65 years), tumor stage (T1, T2, and T3/4), nodal stage (N0, N1, and N2), and histology (squamous, adenocarcinoma, or other), with minor modifications according to the type of combination (Appendix, online only). The predictive value of mutation status was assessed by a treatment-mutation status interaction. Prognostic analyses were performed in OBS patients. We assessed heterogeneity of prognostic and predictive values of mutation status across trials and according to histology (Appendix). Preplanned subgroup analyses were performed that assessed the prognostic effect of TP53/KRAS combination in adenocarcinoma, and KRAS (EGFR, respectively) in EGFR (KRAS, respectively) WT adenocarcinoma. Statistical significance was set at P < .01. Survival curves were based on Kaplan-Meier methods and presented with unadjusted HRs from the Cox model and P values using log rank statistic. Follow-up was estimated for both arms with the Schemper method.28 Statistical analyses were performed by using SAS (SAS/STAT User’s Guide, Version 9.3; SAS Institute, Cary, NC). All P values are two sided.
RESULTS
Patients
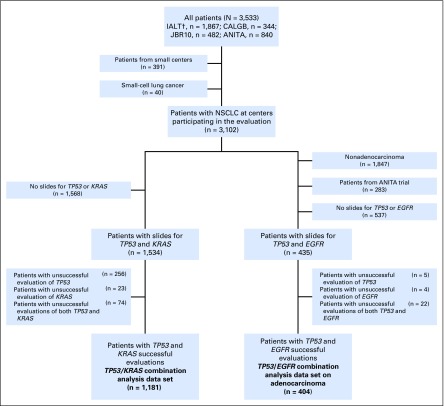
Of 3,533 patients who were randomly assigned (ANITA, n = 840; JBR.10, n = 482; IALT, n = 1,867; CALGB-9633, n = 344), 1,534 patients were eligible for TP53/KRAS combination analysis and 435 for TP53/EGFR combination analysis (Appendix Fig. A1, online only).
TP53/KRAS and TP53/EGFR Combinations
TP53/KRAS analysis included only patients for whom both TP53 and KRAS status were available (n = 1,181). Baseline demographics for patients with and without TP53/KRAS status known are shown in Appendix Table A1 (online only). Compared with patients with unknown status, there were more women (P = .02) with known status and more T2 tumors (P = .003)—stage was not significantly different between the two groups (P = .10). Among patients with known status, 888 (75%) were male and 569 (48%) were stage I, with mostly squamous cell carcinoma (43%) and adenocarcinoma (42%). Mutation distribution was as follows: 570 (48%) with no mutation, 186 (16%) with KRAS mutation, 376 (32%) with TP53 mutation, and 49 (4%) with TP53 and KRAS comutations (Table 1). Median follow-up was 5.5 years (range, 0.1 to 11.3 years) and 557 patients died.
Table 1.
Patient and Tumor Characteristics for Patients With Double Wild-Type, KRAS, or TP53 Mutated and Double-Mutant Tumors
TP53/EGFR analysis included only patients with adenocarcinoma with both TP53 and EGFR status available (n = 404). Baseline demographics for patients with and without known TP53/EGFR status are shown in Appendix Table A2 (online only). There were no significant differences between groups. Among patients with known status, 247 (61%) were male and 245 (61%) were stage I. Mutation distribution was as follows: 260 (64%) with no mutation, 31 (8%) with EGFR mutation, 95 (24%) with TP53 mutation, and only 18 (4%) with TP53 and EGFR comutations (Table 2). Median follow-up was 5.5 years (range, 0.2 to 11.1 years) and 170 patients died.
Table 2.
Patient and Tumor Characteristics for Patients With Double Wild-Type, EGFR, or TP53 Mutated and Double-Mutant in Adenocarcinoma
Prognostic Effect of TP53 (WT and MUT) Combined With KRAS (WT and MUT)
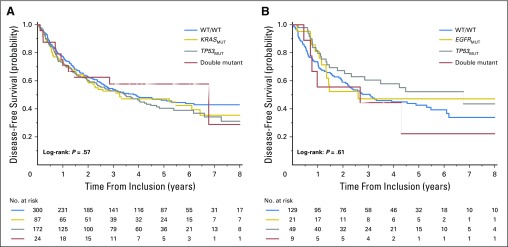
In the OBS arm, there was no significant difference in OS on the basis of mutation status (Fig 1A, Appendix Table A3, online only; multivariable Cox model, P = .61). No heterogeneity across trials (P = .66) or histology (P = .97) was observed. Results were similar for DFS (Appendix Table A4 and Appendix Fig A2A, online only).
Fig 1.
Overall survival. Unadjusted Kaplan-Meier curves of the prognostic effect in the observation arm. (A) TP53/KRAS combination. (B) TP53/EGFR combination. MUT, mutant; WT, wild type.
Predictive Effect of TP53 (WT and MUT) Combined With KRAS (WT and MUT)
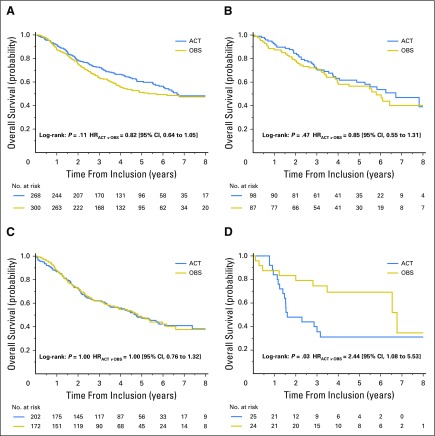
As shown in Table 3 and Figure 2, there was no significant difference in OS benefit from chemotherapy when patients with KRAS mutation (HR, 0.71 [95% CI, 0.45 to 1.10]; P = .13) were compared with those with double WT tumors (HR, 0.81 [95% CI, 0.63 to 1.04]; P = .09; interaction test, P = .63). Patients with TP53 mutant tumors did not seem to benefit from chemotherapy (HR, 0.99 [95% CI, 0.75 to 1.32]; P = .97). In contrast, patients with double-mutant tumors had significantly worse OS with chemotherapy (HR, 2.49 [95% CI, 1.10 to 5.64]; P = .03) compared with patients with double WT tumors (interaction test, P = .01). This latter result led to a chemotherapy effect on OS that was marginally heterogeneous across mutation status (P = .04). This marginal predictive effect was homogeneous across trials (P = .73) and histology (P = .85). For DFS, the same results were observed with an interaction (P = .04) as a result of a marginal effect of ACT in the KRAS mutant group (HR, 0.62 [95% CI, 0.41 to 0.92]; P = .02; Appendix Table A5 and Appendix Fig A3, online only).
Table 3.
Predictive Effect of TP53 and KRAS Combination on Overall Survival (n = 1,176)
Fig 2.
Overall survival. Unadjusted Kaplan-Meier curves of treatment effect according to TP53 and KRAS mutation status. (A) Double wild type. (B) TP53 wild-type, KRAS mutant. (C) TP53 mutant, KRAS wild type. (D) Double mutant. ACT, adjuvant chemotherapy; OBS, observation.
Prognostic Effect of TP53 (WT and MUT) Combined With EGFR (WT and MUT) in Adenocarcinoma
Trials that were included in this analysis include only IALT, JBR.10, and CALGB and are limited to patients with adenocarcinoma as few EGFR results were available for other histologies. In the OBS arm, there was no significant difference in OS on the basis of mutation status (Appendix Table A6, online only, and Fig 1B), EGFR, TP53, or double mutation compared with double WT subgroup (multivariable Cox proportional hazards model, P = .83). No heterogeneity across trials (P = .38) was observed. Results were similar for DFS (Appendix Table A7 and Appendix Fig A2B, online only).
Predictive Effect of TP53 (WT and MUT) Combined With EGFR (WT and MUT) in Adenocarcinoma
As shown in Table 4 and Figure 3, there was no significant difference in OS benefit from chemotherapy when patients with EGFR mutation (HR, 0.67 [95% CI, 0.20 to 2.21]; P = .51) were compared with those with double WT tumors (HR, 0.67 [95% CI, 0.46 to 0.99]; P = 0.05; interaction P = 1.00). Patients with TP53 mutant tumors did not seem to benefit from chemotherapy (HR, 0.89 [95% CI, 0.47 to 1.69]; P = .72), nor did patients with double-mutant tumors (HR, 1.06 [95% CI, 0.23 to 4.94]; P = 0.94). Overall, chemotherapy effect on OS was homogeneous across mutation status (P = .86). For DFS, there was a beneficial effect of ACT in double WT patients (HR, 0.63 [95% CI, 0.45 to 0.90]; P = .01) but significant interaction was not observed (P = .74; Appendix Table A8 and Appendix Fig A4, online only).
Table 4.
Predictive Effect of TP53 and EGFR Combination on Overall Survival in Adenocarcinoma (n = 399)
Fig 3.
Overall survival. Unadjusted Kaplan-Meier curves of treatment effect according to EGFR and TP53 mutation status, in adenocarcinoma. (A) Double wild type. (B) TP53 wild-type, EGFR mutant. (C) TP53 mutant, EGFR wild type. (D) Double mutant. ACT, adjuvant chemotherapy; OBS, observation.
Adenocarcinoma Subgroup Analyses
Because EGFR and KRAS mutations occur mainly in adenocarcinoma, this subgroup was analyzed separately. Patient selection is presented in Appendix Fig A5 (online only).
TP53 (WT and MUT) Combined With KRAS (WT and MUT)
The prognostic effect of TP53 and KRAS comutation was assessed in 377 patients with adenocarcinoma—257 patients and 120 deaths in the OBS arm—and no survival advantage was observed for any mutation subgroup (data not shown).
KRAS in EGFR WT
The prognostic effect of KRAS mutation was assessed further in the subgroup of EGFR WT adenocarcinoma—207 patients and 99 deaths in the OBS arm—and no OS difference was observed for patients with KRAS mutations (HR, 0.97 [95% CI, 0.62 to 1.53]; P = .91), and no heterogeneity among trials (P = .12).
EGFR in KRAS WT
The prognostic effect of EGFR mutation was assessed further in the subgroup of KRAS WT adenocarcinoma—160 patients and 77 deaths in the OBS arm—and no OS advantage was observed for patients with EGFR mutations (HR, 1.18 [95% CI, 0.66 to 2.12]; P = 0.57), and no heterogeneity among trials (P = .91).
DISCUSSION
In this study, we elected to focus on three oncogenes—KRAS, TP53, and EGFR—because they are the most frequently mutated in NSCLC and, importantly, the most frequently comutated genes.1-3
We previously reported that KRAS mutation was not a significant prognostic marker in resected early-stage NSCLC, whether analyzed collectively or by mutation subtype.11 More recently, we have reported similar results for TP53.16 In advanced disease, EGFR mutation is frequently associated with longer survival, although inconsistent results have been reported from surgical series.29,30 EGFR mutation occurs rarely in NSCLC subtypes other than adenocarcinoma. When we limited our analyses to adenocarcinoma, we could identify no prognostic effect for EGFR mutation even when patients with KRAS-mutated tumors were removed from the analysis (MUT v WT HR, 1.18 [95% CI, 0.66 to 2.12]; P = .57).
With the introduction of next-generation sequencing techniques, it is now recognized that NSCLC is a molecularly complex cancer1-3; however, most molecular changes are of little or unknown functional significance. Somatic driver mutations occur less frequently, with multiple driver mutations reported in approximately 3% to 44% of tumors studied, depending on platform used.31,32
TP53 mutation occurs in almost all small-cell lung cancers and is the most frequent driver mutation in NSCLC, occurring in approximately 50% of this malignancy.1-3 Our TP53 mutation rate in adenocarcinoma seems to be somewhat lower than that reported in some series but is consistent with a recent study from the Clinical Lung Cancer Genome Project that found TP53 mutations in 26.9% of cases,33 which is virtually identical to our result. The first multiplex platforms that were designed to study specific driver mutations did not include TP53 because of the multiplicity and complexity of genetic changes in this gene; therefore, early publications that have reported on mutation analyses in NSCLC undoubtedly underestimated the frequency of dual mutations. Kris et al31 reported only 3% of patients with dual mutations when up to 10 mutations were assessed. In contrast, Jao et al32 found dual mutations in 44% of tumors from a study by using next-generation sequencing platforms that included TP53. Furthermore, in studies that have included TP53 testing, this mutation is the most frequent comutation in all subtypes of NSCLC.1-3
With more than 1,000 patients, our study is the first and the largest, to our knowledge, to report on the prognostic and predictive effects of TP53 mutation in combination either with KRAS mutation or with EGFR mutation. Furthermore, our study examines a homogeneous group of surgically staged patients with complete follow-up of more than 5 years’ duration. After accounting for other prognostic variables in the OBS arm, we could demonstrate no prognostic effect of dual TP53/KRAS mutation compared with the WT/WT cohort or with cohorts with only one mutation. Results were similar when examining the adenocarcinoma subset that accounted for almost 70% of KRAS mutations. Because EGFR mutations, as expected, were virtually limited to adenocarcinoma, our analyses of the TP53/EGFR combination were limited to this NSCLC subgroup. Once again, we could demonstrate no prognostic effects of dual mutation. Similar results were reported recently by Kosaka et al,34 who identified no significant prognostic effect of TP53, KRAS, or EGFR mutations on multivariable analysis of 397 surgically resected adenocarcinomas. They did not report on the outcomes of patients with dual-mutant tumors. Cortot et al35 also reported a high frequency of dual EGFR or KRAS mutations with alterations in the p53/arf14 pathway, and emphasized the importance of further investigation to understand the effect of mutations in tumor suppressor genes in association with other driver mutations. Most recently, Reily et al36 reported a nonsignificant trend to shorter survival in 92 patients with co-occurring KRAS/TP53 mutated tumors compared with 147 with KRAS alone (HR, 1.25; P = .396). They also found that alterations of STK11, KEAP1, or NF2E2L with KRAS mutation were associated with significantly shorter survival on univariable analysis.
Dual mutation status, however, does seem to be predictive and to influence the survival benefit that is derived from ACT. There were trends toward a beneficial effect from ACT in patients with WT/WT tumors and even in those with KRAS-mutated tumors. In contrast, patients with TP53-mutated tumors seemed to derive no benefit from ACT (HR, 0.99), and those with double-mutant tumors who were treated with ACT had significantly worse OS (HR, 2.49; P = .03; interaction P = .01). These trends also were observed in our analyses of TP53/EGFR in adenocarcinoma, although significant differences could not be demonstrated in this smaller subset. Patients with WT/WT tumors and those with EGFR mutant tumors seemed to benefit from ACT (HR, 0.67 for both groups), whereas there was no apparent benefit from ACT in those with double MUT TP53/EGFR tumors (HR, 1.06).
There are preclinical data that suggest that the silencing of p53 in A549 cell lines—that carry a KRAS mutation—enhances resistance of cells to cisplatin and paclitaxel, and that intact p53 is required for chemotherapy-induced apoptosis.37 Our findings are consistent with this hypothesis; however, we could not find any preclinical work that examined the effect of dual TP53/EGFR mutation with respect to response to chemotherapy.
Our work, to our knowledge, is the first to examine the negative predictive effect of dual mutations on survival benefit from ACT. Tomasini et al38 reported on a small group of patients who were treated with platinum-based chemotherapy for all stages of NSCLC who had undergone molecular profiling, including TP53 analysis. They observed a negative effect that was similar to ours in the adjuvant setting. Among 218 patients, 28 had tumors with dual TP53/KRAS mutations. Whereas there was no difference when comparing survival outcomes among the four groups of WT/WT, MUT/MUT, or single mutation, patients with dual TP53/KRAS mutations had significantly shorter overall survival than those with WT/WT tumors (HR, 2.06 [95% CI, 1.09 to 3.88] P = .02). Interaction could not be demonstrated as all patients received chemotherapy and there was no untreated control arm.
Dual TP53/EGFR mutations have been reported to occur frequently by other groups,39-42 and in one small study40 were associated with lower response to gefitinib and shorter survival. Labbe et al39 examined the effect of dual TP53/EGFR mutation in 103 patients with EGFR exons 19 or 21 mutations who received EGFR tyrosine kinase inhibitors for advanced disease. There was no difference in response rate for patients with comutated tumors, but progression-free survival was significantly shorter for patients who were treated with EGFR tyrosine kinase inhibitors (HR, 1.82; P = .039). Similar results were reported by Yu et al,40 who reported an even greater negative effect of dual mutation (HR, 2.7; P = .017).
This LACE-Bio report is the first to suggest that TP53/EGFR mutations also may have a negative effect on the benefit from chemotherapy. Patients with WT/WT adenocarcinoma seemed to benefit from platinum-based ACT, whereas those with TP53 or dual mutations did not. The mechanistic basis for this observation remains unclear. As these analyses were limited to the adenocarcinoma subset, our results lack statistical power and so they should be interpreted with caution and must be validated in other studies.
In summary, TP53 comutation with KRAS or EGFR is not a significant prognostic marker in patients with resected NSCLC. Dual TP53/KRAS mutation seems to be predictive of shortened survival in patients who are treated with platinum-based ACT, but the statistically significant result must be interpreted with caution in view of the small sample size, and validation studies are needed in both the adjuvant and advanced disease settings. Preclinical models are also warranted and may clarify the potential interaction of dual mutation and chemotherapy. Similar, although nonsignificant, trends toward a lack of survival benefit from ACT in patients with dual TP53/EGFR mutations as well as preclinical studies support our observation that tumor suppressor gene mutations combined with other genetic mutations may be associated with relative resistance to chemotherapy. With recent improvements in technology, next-generation sequencing is now cost effective and should be considered for all newly diagnosed patients with NSCLC to assess the mutational profile of each cancer. Although not a therapeutic target at this time, routine inclusion of TP53 mutation testing may clarify the role that this tumor suppressor gene plays, both alone and in combination with other driver mutations in lung cancer, and determine whether there is a negative interaction with treatment.
ACKNOWLEDGMENT
We thank Françoise Delassus for administrative support and Caroline Domerg and Abderrahmane Bourredjem for their help on data management and statistical analyses.
Appendix
Statistical Methods
To quantify the association between mutation status (both for TP53/KRAS and TP53/EGFR) and overall survival and disease-free survival, we first used the Cox proportional hazards regression model developed for TP53 analysis.16 This model (core model) was stratified by trial and included the following covariates: treatment (adjuvant chemotherapy [ACT] and observation [OBS]), sex, age (< 55, 55 to 64, > 64 years), tumor stage (T1, T2, T3/4), nodal stage (N0, N1, N2), and histology (squamous, adenocarcinoma, other). Histology was removed for TP53/EGFR analyses because they were limited to adenocarcinoma. We applied a process of several steps to develop a final model from this core model for both TP53/KRAS analysis and TP53/EGFR analysis. The first step was to study the correlation between KRAS and EGFR, respectively, and each covariate (age, sex, T stage, N stage, stage, histology only for KRAS, performance status, type of surgery, and lymphoid infiltration) via a univariable and multivariable logistic regression stratified by trial. Significant covariates (P < .20, univariable analysis) were included to a stratified logistic multivariable model. For KRAS, age, and histology remained statistically significant (P < .05). For EGFR, only sex remained statistically significant (P < .05). These covariates (age and histology for KRAS and sex for EGFR) were included in the core model. We added a composite marker (WT/WT, WT/MUT, MUT/WT, and MUT/MUT) that combined TP53 and KRAS (model 1) and combined TP53 and EGFR (model 2) to the core model. The next step was to check whether covariates not previously selected in each model may have an impact on the regression coefficients of the composite marker. If a variable caused a ≥ 20% change, the variable would be added to the model (Hosmer DW, et al: New York, NY, Wiley, 2008). For model 1, neither type of surgery, nor performance status changed the regression coefficients by more than 20%. For model 2, performance status was included in the model because it changed the regression coefficients by more than 20%. The last step was to check the proportional hazards assumption. This was performed by using the Schoenfeld residuals (survival R package), and no violation of this hypothesis was observed. Finally, model 1 included the following covariates: treatment (ACT and OBS), sex, age (< 55, 55-64, > 64 years), tumor stage (T1, T2, T3/4), nodal stage (N0, N1, N2), histology (squamous, adenocarcinoma, other), and composite marker TP53/KRAS. Model 2 included the following covariates: treatment (ACT and OBS), sex, age (< 55, 55-64, > 64 years), tumor stage (T1, T2, T3/4), nodal stage (N0, N1, N2), performance status (0, ≥ 1), and composite marker (TP53/EGFR). Univariable analyses showing relations between all analyzed covariates and overall survival are presented in Appendix Tables A9 and A10.
Heterogeneity of prognostic and predictive values of mutation status (TP53/KRAS and TP53/EGFR) across trials and according to histology was evaluated by introducing interaction terms in each model (two-order interaction for prognostic analyses and three-order interaction for predictive analyses).
Fig A1.
Flowchart of patients entered in the four trials and included in the (TP53, KRAS) and (TP53, EGFR) analyses. NSCLC, non–small-cell lung cancer.
Fig A2.
Disease-free survival. Unadjusted Kaplan-Meier curves of the prognostic effect in the observation arm. (A) TP53/KRAS combination. (B) TP53/EGFR combination. MUT, mutant; WT, wild type.
Fig A3.
Disease-free survival - Unadjusted Kaplan-Meier curves of treatment effect according to TP53 and KRAS mutation status. (A) Double wild type. (B) TP53 wild-type, KRAS mutant. (C) TP53 mutant, KRAS wild type. (D) Double mutant. ACT, adjuvant chemotherapy; OBS, observation.
Fig A4.
Disease-free survival - Unadjusted Kaplan-Meier curves of treatment effect according to TP53 and EGFR mutation status. (A) Double wild type. (B) TP53 wild-type, EGFR mutant. (C) TP53 mutant, EGFR wild type. (D) Double mutant. ACT, adjuvant chemotherapy; OBS, observation.
Fig A5.
Subgroup analyses. Flowchart of patients with adenocarcinoma entered in three trials and with KRAS and EGFR available mutation status.
Table A1.
Patient and Tumor Characteristics for Patients With and Without TP53/KRAS Mutation Status Results

Table A2.
Patient and Tumor Characteristics for Patients With and Without TP53/EGFR Mutation Status Results* in Adenocarcinoma

Table A3.
Prognostic Value of TP53 and KRAS Combination on Overall Survival in the Observation Arm

Table A4.
Prognostic Value of TP53 and KRAS Combination on Disease-Free Survival in the Observation Arm
Table A5.
Predictive Value of TP53 and KRAS Combination on Disease-Free Survival (n = 1,176)**
Table A6.
Prognostic Value of TP53 and EGFR Combination on Overall Survival in the Adenocarcinoma Observation Arm

Table A7.
Prognostic Value of TP53 and EGFR Combination on Disease-Free Survival in Adenocarcinoma and Observation Arm

Table A8.
Predictive Value of TP53 and EGFR Combination on Disease-Free Survival in Adenocarcinoma (n = 399)**
Table A9.
Univariable Cox Proportional Hazard Regression Model Presenting Relations Between Analyzed Covariates and Overall Survival for the TP53 and KRAS Combination

Table A10.
Univariable Cox Proportional Hazards Regression Model Presenting Relations Between Analyzed Covariates and Overall Survival for the TP53 and EGFR Combination

Footnotes
Supported by the Canadian Cancer Society Research Institute (Canada), la Ligue Nationale Contre le Cancer (France), le Programme National d’Excellence Spécialisé Cancer du poumon de l’Institut National du Cancer (France), the National Cancer Institute (United States), and an unrestricted grant from Sanofi.
Clinical trial information: NCT00002583, NCT00002823, NCT00002852.
AUTHOR CONTRIBUTIONS
Conception and design: Frances A. Shepherd, Gwénaël Le Teuff, Pierre Hainaut, Jean-Pierre Pignon, Thierry Le Chevalier, Lesley Seymour, Jean-Charles Soria, Ming-Sound Tsao
Administrative support: Jean-Pierre Pignon, Lesley Seymour
Provision of study materials or patients: Frances A. Shepherd, Thierry Le Chevalier, Lesley Seymour, Jean-Yves Douillard, Stephen Graziano, Robert Pirker, Martin Filipits, Robert Kratzke, Jean-Charles Soria
Collection and assembly of data: Benjamin Lacas, Gwénaël Le Teuff, Pierre Hainaut, Pasi A. Jänne, Jean-Pierre Pignon, Thierry Le Chevalier, Lesley Seymour, Stephen Graziano, Elizabeth Brambilla, Robert Pirker, Martin Filipits, Ming-Sound Tsao
Data analysis and interpretation: Benjamin Lacas, Gwénaël Le Teuff, Pierre Hainaut, Pasi A. Jänne, Jean-Pierre Pignon, Thierry Le Chevalier, Lesley Seymour, Jean-Yves Douillard, Elizabeth Brambilla, Robert Pirker, Martin Filipits, Robert Kratzke, Jean-Charles Soria, Ming-Sound Tsao
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non–Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Frances A. Shepherd
No relationship to disclose
Benjamin Lacas
No relationship to disclose
Gwénaël Le Teuff
No relationship to disclose
Pierre Hainaut
No relationship to disclose
Pasi A. Jänne
Stock or Other Ownership: Gatekeeper Pharmaceuticals
Consulting or Advisory Role: AstraZeneca, Boehringer Ingelheim, Pfizer, Chugai Pharma, Genentech, ARIAD Pharmaceuticals, Ignyta, Loxo, ACEA Biosciences
Research Funding: Astellas Pharma, AstraZeneca, Puma Biotechnology, Daiichi Sankyo
Patents, Royalties, Other Intellectual Property: Postmarketing royalties from DFCI owned intellectual proerty on EGFR mutations licensed to Laboratory Corp
Jean-Pierre Pignon
Research Funding: Bayer (Inst)
Thierry Le Chevalier
No relationship to disclose
Lesley Seymour
Stock or Other Ownership: AstraZeneca
Consulting or Advisory Role: Boehringer Ingelheim
Research Funding: Merck (Inst), AstraZeneca (Inst), Novartis (Inst), Innate Pharma (Inst)
Jean-Yves Douillard
Honoraria: Amgen, Bayer, Roche, Sanofi, AstraZeneca, Sirtex
Research Funding: Merck Serono (Inst)
Stephen Graziano
No relationship to disclose
Elizabeth Brambilla
No relationship to disclose
Robert Pirker
Consulting or Advisory Role: Merck Sharp & Dohme, Synta
Speakers' Bureau: Boehringer Ingelheim, Eli Lilly, Roche, Pierre Fabre
Martin Filipits
Consulting or Advisory Role: AstraZeneca, Boehringer Ingelheim, Merck Sharp & Dohme, Eli Lilly, Ratiopharm, Novartis, Roche, Pfizer
Robert Kratzke
No relationship to disclose
Jean-Charles Soria
No relationship to disclose
Ming-Sound Tsao
Consulting or Advisory Role: AstraZeneca, Boehringer Ingelheim
Research Funding: AstraZeneca (Inst)
Patents, Royalties, Other Intellectual Property: Patent on prognostic gene signature in patients with early-stage non–small-cell lung cancer
REFERENCES
- 1.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma Nature 511543–550.2014. [Erratum: Nature 514 2622014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers Nature 489519–525.2012. [Erratum: Nature 491 2882012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey JJ. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 5.Rodenhuis S, van de Wetering ML, Mooi WJ, et al. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–935. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 6.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slebos RJC, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–565. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 8.Huncharek M, Muscat J, Geschwind JF. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: A combined analysis of 881 cases. Carcinogenesis. 1999;20:1507–1510. doi: 10.1093/carcin/20.8.1507. [DOI] [PubMed] [Google Scholar]
- 9.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodenhuis S, Boerrigter L, Top B, et al. Mutational activation of the K-ras oncogene and the effect of chemotherapy in advanced adenocarcinoma of the lung: A prospective study. J Clin Oncol. 1997;15:285–291. doi: 10.1200/JCO.1997.15.1.285. [DOI] [PubMed] [Google Scholar]
- 11.Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato S, Han SY, Liu W, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang CL, Taki T, Adachi M, et al. Mutations of p53 and K-ras genes as prognostic factors for non-small cell lung cancer. Int J Oncol. 1998;12:553–563. doi: 10.3892/ijo.12.3.553. [DOI] [PubMed] [Google Scholar]
- 14.Scoccianti C, Vesin A, Martel G, et al. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: The EUELC cohort. Eur Respir J. 2012;40:177–184. doi: 10.1183/09031936.00097311. [DOI] [PubMed] [Google Scholar]
- 15.Ma X, Rousseau V, Sun H, et al. Significance of TP53 mutations as predictive markers of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer. Mol Oncol. 2014;8:555–564. doi: 10.1016/j.molonc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Le Teuff G, Lacas B, et al. Prognostic and predictive effect of TP53 mutations in non-small cell lung cancer patients from adjuvant cisplatin-based therapy randomized trials: A LACE-Bio pooled analysis. J Thorac Oncol. 2016;11:850–861. doi: 10.1016/j.jtho.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 18.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 19.Russo A, Franchina T, Ricciardi GR, et al. A decade of EGFR inhibition in EGFR-mutated non small cell lung cancer (NSCLC): Old successes and future perspectives. Oncotarget. 2015;6:26814–26825. doi: 10.18632/oncotarget.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pignon J-P, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 21.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 22.Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: Updated survival analysis of JBR-10. J Clin Oncol. 2010;28:29–34. doi: 10.1200/JCO.2009.24.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 24.Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the International Adjuvant Lung Cancer Trial evaluating adjuvant cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol. 2010;28:35–42. doi: 10.1200/JCO.2009.23.2272. [DOI] [PubMed] [Google Scholar]
- 25.Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): A randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 26.Strauss GM, Herndon JE, II, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jänne PA, Wang X, Socinski M.A, et al. Randomized phase II trial of erlotinib alone or with carboplatin and paclitaxel in patients who were never or light former smokers with advanced lung adenocarcinoma: CALGB 30406 trial. J Clin Oncol. 2012;30:2063–2069. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17:343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 29.Fang S, Wang Z. EGFR mutations as a prognostic and predictive marker in non-small-cell lung cancer. Drug Des Devel Ther. 2014;8:1595–1611. doi: 10.2147/DDDT.S69690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Wang T, Zhang J, et al. Prognostic value of epidermal growth factor receptor mutations in resected non-small cell lung cancer: a systemic review with meta-analysis. PLoS One. 2014;9:e106053.0. doi: 10.1371/journal.pone.0106053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jao K., Tomasini P., Kamel-Reid S., et al. Prognostic effect of single versus multiple somatic mutations in non-small cell lung cancer (NSCLC) J Clin Oncol 33, 2015. (abstr 7521) [Google Scholar]
- 33.Clinical Lung Cancer Genome Project (CLCGP) Network Genomic Medicine (NGM) A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4:22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 35.Cortot AB, Younes M, Martel-Planche G, et al. Mutation of TP53 and alteration of p14(arf) expression in EGFR- and KRAS-mutated lung adenocarcinomas. Clin Lung Cancer. 2014;15:124–130. doi: 10.1016/j.cllc.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Reily GJ, Jordan E, Kim HR, et al. Association of outcomes and co-occurring genomic alterations in patients with KRAS-mutant non-small cell lung cancer J Clin Oncol 34, 2016. (abstr 9019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Zhou Y, Li Y, et al. Mutations of p53 and KRAS activate NF-κB to promote chemoresistance and tumorigenesis via dysregulation of cell cycle and suppression of apoptosis in lung cancer cells. Cancer Lett. 2015;357:520–526. doi: 10.1016/j.canlet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Tomasini P, Jao K, Kamel-Reid S, et al. Predictive value of co-existing KRAS and TP53 mutations on response to chemotherapy in non-small cell lung cancer (NSCLC) J Clin Oncol 33, 2015. (abstr 11060) [Google Scholar]
- 39.Labbe C, Korpanty G, Tomasini P, et al. Prognostic and predictive effects of p53 mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC) J Clin Oncol 34, 2016. (abstr 11585) [DOI] [PubMed] [Google Scholar]
- 40.Yu HA, Jordan E, Ni A, et al. Concurrent genetic alterations identified by next-generation sequencing in untreated, metastatic EGFR-mutant lung cancers J Clin Oncol 34, 2016. (abstr 9053) [Google Scholar]
- 41.Lim SM, Kim HR, Cho EK, et al. Targeted sequencing identifies genetic alterations that confer primary resistance to EGFR tyrosine kinase inhibitor (Korean Lung Cancer Consortium) Oncotarget. 2016;7:36311–36320. doi: 10.18632/oncotarget.8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bria E, Pilotto S, Amato E, et al. Molecular heterogeneity assessment by next-generation sequencing and response to gefitinib of EGFR mutant advanced lung adenocarcinoma. Oncotarget. 2015;6:12783–12795. doi: 10.18632/oncotarget.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]