Abstract
In mitosis, the anaphase-promoting complex (APC) regulates the onset of sister-chromatid separation and exit from mitosis by mediating the ubiquitination and degradation of the securin protein and mitotic cyclins. With the use of a baculoviral expression system, we have reconstituted the ubiquitin ligase activity of human APC. In combination with Ubc4 or UbcH10, a heterodimeric complex of APC2 and APC11 is sufficient to catalyze the ubiquitination of human securin and cyclin B1. However, the minimal APC2/11 ubiquitin ligase module does not possess substrate specificity, because it also ubiquitinates the destruction box deletion mutants of securin and cyclin B1. Both APC11 and UbcH10 bind to the C-terminal cullin homology domain of APC2, whereas Ubc4 interacts with APC11 directly. Zn2+-binding and mutagenesis experiments indicate that APC11 binds Zn2+ at a 1:3 M ratio. Unlike the two Zn2+ ions of the canonical RING-finger motif, the third Zn2+ ion of APC11 is not essential for its ligase activity. Surprisingly, with Ubc4 as the E2 enzyme, Zn2+ ions alone are sufficient to catalyze the ubiquitination of cyclin B1. Therefore, the Zn2+ ions of the RING finger family of ubiquitin ligases may be directly involved in catalysis.
INTRODUCTION
The orderly progression through mitosis relies on the sequential degradation of several cell cycle regulatory proteins mediated by the mitotic ubiquitination system (King et al., 1996a; Zachariae and Nasmyth, 1999; Nasmyth et al., 2000). This system first initiates the onset of sister-chromatid separation by ubiquitinating the anaphase inhibitor or securin (Nasmyth et al., 2000). Degradation of the ubiquitinated securin in turn activates the separase, which then cleaves a subunit of the cohesin complex, resulting in the loss of sister-chromatid cohesion and the onset of anaphase (Uhlmann et al., 1999, 2000; Waizenegger et al., 2000). The same machinery also mediates the ubiquitination and destruction of mitotic cyclins, leading to the inactivation of the cdc2 activity and the exit from mitosis (King et al., 1996a; Morgan, 1999; Zachariae and Nasmyth, 1999).
In this system, a large protein complex, called the anaphase-promoting complex (APC) or cyclosome, functions as the ubiquitin ligase (E3) (Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995; Tugendreich et al., 1995). In the presence of the ubiquitin-activating enzyme (E1) and certain ubiquitin-conjugating enzymes (E2s) such as Ubc4 and UbcH10 (called UbcX in Xenopus and E2-C in clams), APC catalyzes the attachment of ubiquitin to the lysine side chains of securin and mitotic cyclins (King et al., 1995; Aristarkhov et al., 1996; Yu et al., 1996). The polyubiquitinated APC substrates are then targeted to the 26S proteasome for degradation (Hershko and Ciechanover, 1998).
To ensure that APC substrates are degraded with proper timing, the ubiquitin ligase activity of APC is tightly regulated during the cell cycle (King et al., 1996a; Morgan, 1999). Significant progress has been made toward the understanding of the regulation of APC during the cell cycle. APC is turned on during mitosis, remains active through most of G1, and is rapidly inactivated at the G1/S transition (Fang et al., 1998; Kramer et al., 1998). The activity profile and substrate specificity of APC can be partially explained by the transient association of two related regulatory factors, Cdc20 and Cdh1, at specific stages of the cell cycle (Sigrist and Lehner, 1997; Schwab et al., 1997; Visintin et al., 1997; Fang et al., 1998; Kramer et al., 1998). Cdc20 binds to APC in mitosis, whereas Cdh1 interacts with APC both in mitosis and the G1 phase (Fang et al., 1998; Kramer et al., 1998). Binding of either Cdc20 or Cdh1 to APC increases the activity of APC drastically (Fang et al., 1998; Kramer et al., 1998). Although the exact mechanism by which Cdc20 and Cdh1 contribute to the ubiquitination reactions is unknown, several recent observations suggest that they might be directly involved in substrate recruitment (Schwab et al., 1997; Sigrist and Lehner, 1997; Visintin et al., 1997; Burton et al., 2001; Hilioti et al., 2001; Pfleger et al., 2001).
In contrast to the cell cycle regulation of APC, little is known about the mechanism by which APC catalyzes the ubiquitination reaction. This is partially due to the fact that the vertebrate APC contains at least 11 subunits (Yu et al., 1998; Grossberger et al., 1999; Gmachl et al., 2000). No clues about the biochemical functions of most APC subunits can be inferred from examining their amino acid sequences. However, sequence analysis of APC2 and APC11 has revealed homology with proteins involved in other ubiquitination systems, particularly the Skp1-Cullin-F-box (SCF) pathway (Yu et al., 1998; Zachariae et al., 1998; Deshaies, 1999; Ohta et al., 1999; Skowyra et al., 1999; Gmachl et al., 2000).
APC2 contains a region that is similar to a sequence in cullins, and thus is a distant member of the cullin family (Yu et al., 1998; Zachariae et al., 1998). The cullin family of proteins is essential for the ubiquitination of G1 cyclins, cyclin-dependent kinase inhibitors, and other important regulatory proteins in yeast and mammals (Kipreos et al., 1996; Willems et al., 1996; Feldman et al., 1997; Lyapina et al., 1998; Latres et al., 1999; Tan et al., 1999). A cullin protein in budding yeast, Cdc53p, is a part of the SCF ubiquitin ligase complex, which targets phosphorylated Sic1p and G1 cyclins for degradation in late G1 (Willems et al., 1996; Feldman et al., 1997). Similar SCF complexes containing cullin proteins have been found in mammals; they mediate the degradation of a variety of substrates, including IκB and E2F (Marti et al., 1999; Tan et al., 1999). APC11 contains a Zn2+-binding motif referred to as the RING-H2 finger and is homologous to Rbx1/Roc1/Hrt1 that physically interacts with Cul1 in mammals and Cdc53p in yeast (Zachariae et al., 1998; Kamura et al., 1999; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Gmachl et al., 2000). Because the Cul1/Rbx1 heterodimeric complex is the functional core of the SCF ubiquitin ligases, APC2 and APC11, by analogy, might also be directly involved in catalysis (Seol et al., 1999). In fact, it has recently been shown that APC11 alone is sufficient to ubiquitinate APC substrates in the presence of Ubc4 (Gmachl et al., 2000; Leverson et al., 2000). Surprisingly, UbcH10 does not support the ubiquitination reaction mediated by APC11 (Gmachl et al., 2000; Leverson et al., 2000).
To gain insights into the catalytic mechanism of APC, we have reconstituted the ubiquitin ligase activity of APC. The minimal ligase module of APC is the heterodimeric complex of the cullin-related protein APC2 and the RING finger protein APC11. Therefore, APC is a member of the expanding family of cullin-RING finger-based ubiquitin ligases. Although APC11 alone can ubiquitinate APC substrates with the use of Ubc4 as the E2 enzyme, the APC2/11 complex is required to catalyze the ubiquitination of APC substrates in the presence of UbcH10. Ubc4 interacts directly with APC11, whereas UbcH10 does not. Instead, UbcH10 binds to the C-terminal cullin domain of APC2. We further demonstrate that APC11 contains a third Zn2+-binding site, in addition to the two Zn2+ ions of the canonical RING-H2 finger. Interestingly, Zn2+ ions alone can catalyze the ubiquitination of APC substrates with the use of Ubc4 as the E2 enzyme, suggesting that Zn2+ ions of the RING finger-containing ubiquitin ligases might be directly involved in catalysis.
MATERIALS AND METHODS
Protein Expression with Use of Baculoviral System
In addition to the eight human APC subunits (APC1–8) reported previously (Yu et al., 1998), two more subunits were cloned with the use of polymerase chain reaction (PCR), APC10 and APC11, based on their sequence similarity to the yeast APC subunits. The recombinant baculoviruses were constructed with the Bac-to-Bac system (Invitrogen, Carlsbad, CA). Briefly, the human genes encoding APC subunits (APC1–11), Cdc20, and Cdh1 were amplified by PCR and cloned into the pFastBac, pFastBac-HT, or pFastBac-GST vectors with the use of suitable restriction sites for the production of untagged, N-terminal His6-tagged, or N-terminal GST-tagged proteins. These plasmids were then transformed into the Escherichia coli strain DH10Bac and the desired bacmids were isolated. These bacmids were transfected into adherent Sf9 cells for the packaging of baculoviruses. To limit the number of viruses used in the coinfections, four viruses each harboring two APC genes (APC1/8, APC2/7, APC3/6, and APC4/5) were also made with the use of the pFastBacDual vector that contains two independent promoters to transcribe both genes simultaneously. The initial viral stocks were then amplified with two successive rounds of infection of Sf9 cells in suspension. The titers of the final viral stocks were typically ∼1–5 × 108/ml, as measured with the Rapid Titer kit (CLONTECH, Palo Alto, CA). Small samples were taken during the second round of amplification and checked for protein production and solubility by immunoblotting with the relevant antibodies. The production of all 10 APC subunits, Cdc20, and Cdh1 proteins in soluble form was verified by either Coomassie staining or immunoblotting.
For the coinfection experiments, Sf9 or Hi5 insect cells were infected with a Multiplicity of infection of 5–10 for each virus for 40–50 h. Cells were harvested and lysed by Dounce homogenization. The lysates were cleared by centrifugation at 30,000 × g for 1 h. The human APC proteins was then purified via the appropriate tags and assayed for ubiquitin ligase activity or binding to other proteins. When necessary, the APC proteins were further purified by anion exchange and/or gel filtration chromatography.
Ubiquitination Assay
For the ubiquitination assays involving the intact APC, the α-APC3 (Cdc27) beads were incubated with 10 volumes of interphase Xenopus egg extracts for 2 h at 4°C and washed five times with XB (10 mM HEPES, pH 7.7, 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose) containing 500 mM KCl and 0.5% NP-40 and twice with XB. The interphase APC beads were then incubated for 1 h at room temperature with human Cdc20 or Cdh1 proteins. After incubation, the APC beads were again washed twice with XB and assayed for ubiquitin ligase activity. For the assays with the reconstituted APC, 5 μl of Ni2+-nitrilotriacetic acid (NTA) beads were incubated with 500 μl of the insect cell lysate containing the appropriate His6-tagged APC proteins, washed five times with XB, and assayed for ubiquitin ligase activity.
Each ubiquitination assay was performed in a volume of 5–10 μl. The reaction mixture contained ATP, 150 μM of bovine ubiquitin, 5 μM of the Myc-tagged human securin, or an N-terminal fragment of human cyclin B1, 5 μM of human E1, 2 μM of Ubc4 or UbcH10, and 2–5 μl of the APC beads. The reactions were incubated at room temperature for 1 h, quenched with SDS sample buffer, and analyzed by SDS-PAGE followed by immunoblotting with α-Myc.
Cyclin Degradation Assay
The Xenopus egg extracts were prepared as described previously (Murray, 1991). To assay cyclin degradation, the N-terminal fragment of human cyclin B1 (residues 1–102) with a Myc-tag and ubiquitin were added to the mitotic extracts at final concentrations of 100 nM and 150 μM, respectively. Aliquots of the reaction mixture were quenched by SDS sample buffer at the indicated time, separated by SDS-PAGE, and blotted with α-Myc.
Protein Binding Assay
To assay the binding among APC2, APC11, Ubc4, and UbcH10, one of the binding partners was expressed either in bacteria or insect cells as His6-tagged proteins. The other partner was in vitro translated in reticulocyte lysate in the presence of [35S]methionine. Purified His6-tagged proteins were bound to Ni2+-NTA beads, incubated with the 35S-labeled proteins, and washed three times with Tris-buffered saline containing 0.05% Tween. The 35S-labeled proteins retained on beads were analyzed by SDS-PAGE followed by autoradiography. The vectors encoding the APC2 fragments were constructed by PCR. The APC11 mutants were made with the use of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Zn2+-binding Assay
The purified APC2e/11 protein was dialyzed against TNG buffer (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 5% glycerol). The concentration of APC2e/11 was determined with the use of UV-VIS spectroscopy and Bradford assays (Bio-Rad, Hercules, CA). The Zn2+ ions bound by APC2e/11 were released by p-hydroxymercuriphenylsulfonic acid. The released Zn2+ was then coordinated by 4–2-pyridylazoresorcinol (PAR), and the resulting Zn2+-PAR2 complex absorbed light at 500 nm with an extinction coefficient of 6.6 × 104 M−1 cm−1. Specifically, aliquots of 1 mM p-hydroxymercuriphenylsulfonic acid were successively added to a mixture containing 1.5 μM of APC2e/11 and 100 μM of PAR, until a plateau of OD500 was reached. The maximum value of OD500 divided by the extinction coefficient then yielded the concentration of Zn2+.
RESULTS
Reconstitution of Ubiquitin Ligase Activity of Human APC
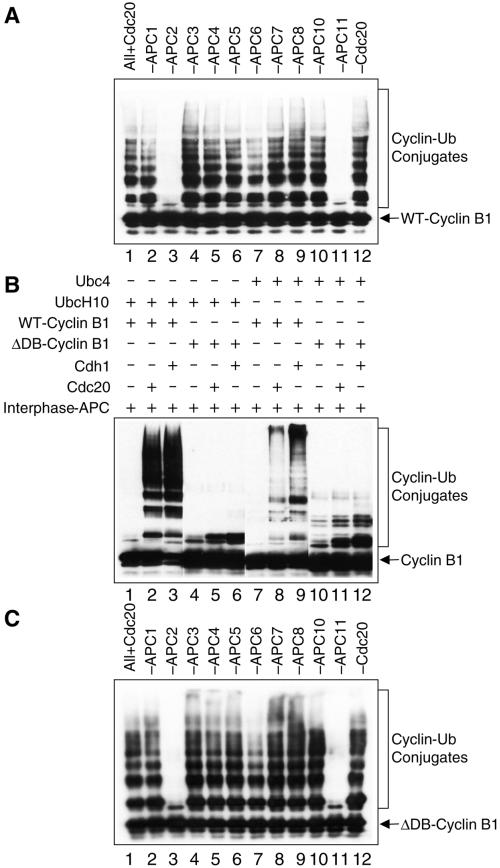
To investigate the mechanism of APC, we coinfected Hi5 insect cells with multiple recombinant baculoviruses harboring 10 APC genes and the cofactors Cdc20 or Cdh1. Because the APC proteins were His6-tagged at their N termini, they were purified from the insect cell lysate with the use of Ni2+-NTA beads and assayed for their ubiquitination activity. As shown in Figure 1A, when Hi5 cells were infected with viruses encoding 10 APC subunits (APC1–11) and Cdc20, the expressed APC proteins contained a ubiquitin ligase activity that, in combination with UbcH10, efficiently ubiquitinated human cyclin B1. This activity also supported ubiquitination of human securin (our unpublished data; see below). To determine which subunits were required for this activity, each virus was omitted individually from the coinfection. Only APC2 and APC11 were required for the ligase activity (Figure 1A). Similar results were obtained when the Cdh1 baculovirus was used in the coinfection instead of Cdc20 (our unpublished data). The fact that omission of APC2 or APC11 alone caused the loss of the reconstituted activity indicated that the purified ligase activity was not due to the presence of the endogenous APC from insect cells.
Figure 1.
Reconstitution of the ubiquitin ligase activity of APC. (A) Hi5 insect cells were coinfected with baculoviruses encoding the indicated APC subunits. The expressed His6-tagged APC proteins were isolated from the insect cell lysate with the use of the Ni2+-NTA beads and assayed for ubiquitin ligase activity in the presence of UbcH10. The reaction mixture was separated on SDS-PAGE and blotted with α-Myc to detect the C-terminally Myc-tagged cyclin B1 protein. Omission of the APC2 (lane 3) or APC11 (lane 11) viruses from the coinfection resulted in the loss of ubiquitin ligase activity. (B) Ubiquitin ligase activity of the intact APC. Either UbcH10 (lanes 1–6) or Ubc4 (lanes 7–12) was used as the E2 enzyme. To determine the D-box dependency of APCCdc20 and APCCdh1, either the wild-type (WT) cyclin B1 (lanes 1–3 and 7–9) or the D-box deletion mutant (ΔDB) of cyclin B1 (lanes 4–6 and 10–12) was used as substrates. (C) Same as A except that ΔDB-cyclin B1 was used as the substrate instead of the wild-type cyclin B1 protein.
Because Cdc20 and Cdh1 are positive regulators of APC, it was surprising to us that Cdc20 or Cdh1 were not required for the ubiquitin ligase activity of the reconstituted APC. We therefore compared the reconstituted APC activity with that of the intact APC from Xenopus egg extracts. Nearly all APC substrates contain the destruction box (D-box) or the KEN-box motifs, which are required for the efficient ubiquitination and degradation of these substrates (King et al., 1996b; Pfleger and Kirschner, 2000). Cdc20 and Cdh1 have been shown to confer the D-box and KEN-box specificity of APC (Burton et al., 2001; Hilioti et al., 2001; Pfleger et al., 2001). The activities of the intact APCCdc20 and APCCdh1 from Xenopus were assayed with cyclin B1 as the substrate. As shown in Figure 1B, purified Cdc20 and Cdh1 proteins greatly stimulated the ligase activity of the intact interphase Xenopus APC with UbcH10 as the E2 enzyme and wild-type cyclin B1 as substrate. However, neither the intact APCCdc20 nor APCCdh1 significantly ubiquitinated a D-box deletion mutant of cyclin B1 (ΔDB-cyclin B1). Similar results were obtained with Ubc4 as the E2 enzyme, although the patterns of cyclin B-ubiquitin conjugates formed by the two enzymes were different. Ubc4 appeared to be more processive than UbcH10 in the presence of either intact APCCdc20 or APCCdh1.
Because Cdc20 and Cdh1 were not essential for the reconstituted APC activity, we tested whether the ligase activity obtained with overexpressed APC proteins conferred D-box specificity, similar to the intact APC. Not surprisingly, the reconstituted human APC ubiquitinated ΔDB-cyclin B1 equally efficiently, indicating that the reconstituted activity did not possess substrate specificity (Figure 1C). This activity also required the presence of APC2 and APC11. Therefore, we reconstituted the minimal ligase activity of APC, which lacked D-box dependency. At present, we do not know the exact cause for the lack of D-box dependency of our reconstituted APC. However, several factors might contribute to this. First, the reconstituted APC might not have the correct quaternary arrangement of all the relevant subunits. Second, the set of subunits used to reconstitute the APC is not complete. Additional human APC subunits are required for the proper function of the reconstituted APC. Finally, it is also possible that the high concentrations of the reconstituted APC and the substrates in our in vitro reactions may have eliminated the need for high-affinity interactions between APC and substrates. We are currently investigating these possibilities.
Heterodimeric Complex of APC2 and APC11 Is the Minimal Ligase Module of APC
Because both APC2 and APC11 were required for the reconstituted APC activity, we tested whether they interacted with each other in the absence of the rest of the APC subunits. To characterize the immediate binding partners of APC11 in the APC complex, the GST-APC11 virus was coinfected with each of the other viruses in a pairwise manner. GST-APC11 and its associated subunits were then purified with glutathione-Sepharose beads and analyzed by SDS-PAGE followed by Coomassie staining and immunoblotting. As shown in Figure 2A, APC11 binds tightly to APC2 and weakly to APC6. The identities of APC2 and APC6 were verified by immunoblotting (our unpublished data). It was not apparent from the GST-APC11 pull-down experiment whether GST-APC11 interacts with APC10 because the His6-tagged APC10 (26 kDa) comigrated with proteolytic fragments of GST-APC11 on SDS-PAGE (our unpublished data). We therefore used a His6-tagged APC10 virus to infect Sf9 cells together with the GST-APC11 virus. The APC10 protein was then purified with Ni2+-NTA beads and analyzed by SDS-PAGE. APC10 binds tightly to GST-APC11 as revealed by Coomassie staining (Figure 2B) and immunoblotting with α-GST antibody (Figure 2C). Several large proteins contain the so-called DOC domains that are similar in sequence to APC10; some of these proteins also contain cullin homology domains or homologous to E6-AP C terminus (HECT) domains (Grossberger et al., 1999). E6-AP, the founding member of a family of proteins containing HECT domains, mediates the HPV E6-dependent degradation of p53 (Scheffner et al., 1995). It is likely that the DOC domains of these large multidomain proteins might also be involved in binding to yet unidentified RING-finger proteins.
Figure 2.
APC11 interacts with APC2 and APC10. (A) GST-APC11 baculovirus was coinfected with APC1/8 (a single baculovirus encoding both APC1 and APC8), APC2/7, APC3/6, APC4/5, or Cdh1 viruses in a pairwise manner into Sf9 cells. GST-APC11 and its interacting proteins were purified on glutathione beads, separated on SDS-PAGE, and stained with Coomassie Blue. GST was added to the lysates of APC1/8, APC2/7, APC3/6, APC4/5, and Cdh1 and used as controls. APC11 interacted strongly with APC2 (lane 3) and weakly with APC6 (lane 5). As determined by mass spectrometry, the band at 55 kDa belonged to α/β-tubulin, which presumably associated nonspecifically with GST-APC11. (B) His6-tagged APC10 baculovirus was coinfected with the GST-APC11 virus into Sf9 cells. His6-APC10 and its interacting proteins were purified on Ni2+-NTA beads, separated on SDS-PAGE, and stained with Coomassie Blue. As controls, Ni2+-beads were added to lysates infected with GST-APC11 alone. The α/β-tubulin proteins again copurified with APC10 and APC11. (C) To verify the interaction between APC10 and APC11, the same samples from B were blotted with α-GST, confirming the identity of the GST-APC11 band.
To identify subunits that interact with APC2, the His6-tagged APC2 virus was used to infect Sf9 cells together with other viruses. The APC2 protein was then purified with Ni2+-NTA beads and analyzed by SDS-PAGE. We confirmed that APC2 interacts with APC11, and found no other strong interactions between APC2 and the rest of APC subunits (our unpublished data). Therefore, APC2 and APC11 formed a complex in the absence of the other APC subunits.
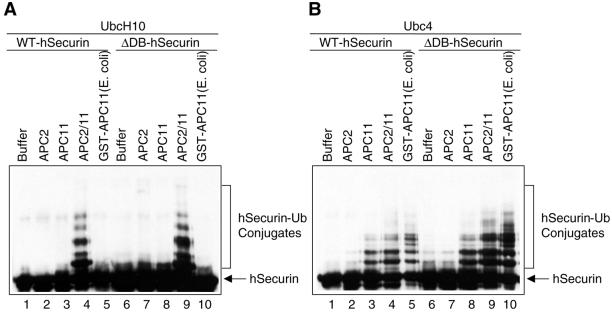
We next tested whether the subcomplex of APC2 and APC11 (APC2/11) was sufficient to support ubiquitination of APC substrates. With the use of UbcH10 as E2, APC2/11 catalyzed the ubiquitination of human securin, whereas either APC2 or APC11 alone had no activity (Figure 3A). Consistent with previous reports, APC11 alone expressed either in bacteria or insect cells was sufficient to ubiquitinate human securin in the presence of Ubc4 (Figure 3B). Similar data were obtained with the use of human cyclin B1 as the substrate (our unpublished data). Therefore, APC2/11 represents the minimal ligase module of APC, because it supports the ubiquitination of APC substrates with the use of either Ubc4 or UbcH10 as E2s.
Figure 3.
Heterodimeric complex of APC2 and APC11 possesses ubiquitin ligase activity. (A) Hi5 cells were either infected with His6-APC2 (lanes 2 and 7) and His6-APC11 (lanes 3 and 8) viruses individually, or coinfected with the His6-APC2 and His6-APC11 viruses (lanes 4 and 9). The APC2 and APC11 proteins were purified with Ni2+-NTA beads and assayed for ubiquitination activity with the use of UbcH10 as the E2 enzyme and wild-type human securin (lanes 1–5) or a D-box deletion mutant (ΔDB) of securin (lanes 6–10) as the substrates. Bacterial expressed GST-APC11 protein purified with glutathione-agarose beads was also tested for ligase activity (lanes 5 and 10). (B) Same as A except that Ubc4 was used as the E2 enzyme instead of UbcH10.
Ubc4 Interacts with APC11, whereas UbcH10 Binds to APC2
Both Ubc4 and UbcH10 support the ubiquitination reactions catalyzed by APC in an additive manner (Yu et al., 1996). It is unclear which enzyme is the physiological E2 of the APC pathway. Microinjection of a UbcH10 dominant-negative mutant protein into mammalian cells arrested cells in mitosis (Townsley et al., 1997). In addition, mutation of the Schizosaccharomyces pombe homolog of UbcH10, UbcP4, caused accumulation of cells in mitosis, similar to mutations of APC subunits (Osaka et al., 1997). These findings suggest that UbcH10 might be involved in the mitotic degradation system in living cells. However, in budding yeast, mutations of either the Ubc4/5 family E2s or the UbcH10 homolog Ubc11 did not cause obvious mitotic phenotype (Townsley and Ruderman, 1998). To further clarify this issue, we immunodepleted UbcX, the Xenopus homolog of UbcH10, from the mitotic Xenopus egg extract that contains active APC and degrades APC substrates with fast kinetics. As shown in Figure 4A, the α-UbcX antibody beads effectively depleted the UbcX protein from the mitotic extract. Although the mitotic extract depleted with a control antibody degraded cyclin B1 with a half-life of 5–10 min, cyclin B1 was stabilized in the UbcX-depleted extract. Because the concentration of UbcX in mitotic Xenopus extracts was estimated to be 50 nM by semiquantitative immunoblotting (our unpublished data), we added 50 nM of recombinant purified UbcX expressed in bacteria back to the UbcX-depleted extract. Addition of the bacterially expressed UbcX restored the ability of the mitotic extract to degrade cyclin B1. Taken together, UbcX is required for proper degradation of cyclin B1, and possibly other APC substrates, in Xenopus egg extracts. Unfortunately, we did not have access to an antibody that can immunodeplete Ubc4 from these extracts, and thus could not do similar experiments for Ubc4.
Figure 4.
Ubc4 binds to the ring protein APC11 whereas UbcH10 binds to the cullin protein APC2. (A) Xenopus homolog of UbcH10, UbcX, was immunodepleted from mitotic Xenopus egg extracts with the use of a polyclonal α-UbcX antibody coupled to Affiprep protein A beads (compare lanes 1 and 2). The preimmune serum was used as the control. The kinetics of cyclin B1 degradation was assayed in the control-depleted extract (top), the UbcX-depleted extract (middle), and the UbcX-depleted extract with physiological amount of purified UbcX added back (bottom). (B) Binding assays among Ubc4, UbcH10, APC2, and APC11. Purified His6-tagged Ubc4 and UbcH10 proteins were immobilized on Ni2+-NTA beads and incubated with 35S-labeled APC2 or APC11 proteins. After washing, the 35S-labeled proteins bound to beads were analyzed by SDS-PAGE followed by autoradiography. (C) Sequence alignment of the RING-binding loops of human Ubc4, UbcH7, and UbcH10.
We next examined why APC2 was not required for the ubiquitination reactions of Ubc4. As shown in Figure 4B, UbcH10 interacted strongly with APC2, whereas it did not bind to APC11. In contrast, Ubc4 associated directly with APC11. It did not exhibit significant binding toward APC2. This finding explains why APC2 is only required for UbcH10-catalyzed reactions. The two E2s may recognize different binding determinants within the APC2/11 ligase module. The fact that UbcH10 did not bind APC11 is consistent with previous structural studies on the interactions between the Cbl RING domain and UbcH7, an E2 of the Ubc4 subfamily (Zheng et al., 2000). The two loops that are critical for binding to RING domains are conserved between Ubc4 and UbcH7 (Figure 4C). On the other hand, UbcH10 contains quite divergent amino acid sequences in these two loops. Therefore, Ubc4 and UbcH10 may be recruited to the intact APC with distinct mechanisms: Ubc4 recognize APC11 initially, whereas UbcH10 first interacts with APC2. However, it remains possible that, once they are bound to APC, Ubc4 and UbcH10 occupy a similar site on APC and use a similar mechanism for transferring ubiquitin.
Cullin Domain of APC2 Interacts with APC11 and UbcH10
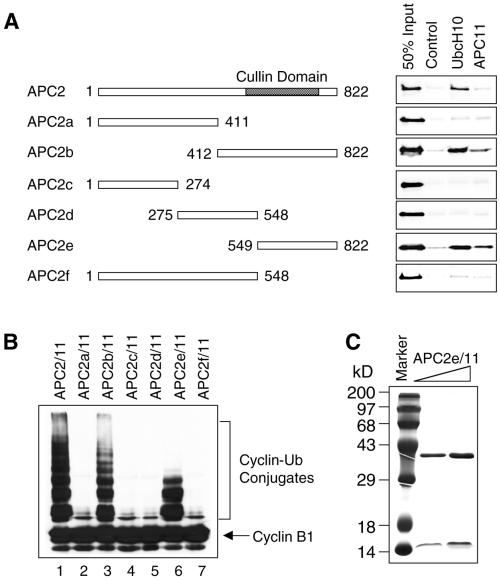
We next mapped the regions within APC2 that interact with APC11 and UbcH10. A series of truncation mutants of APC2 were constructed and tested for binding to APC11 and UbcH10. A C-terminal fragment of APC2, APC2e, spanning residues 549–822, was sufficient for binding to APC11 and UbcH10 (Figure 5A). This region almost coincides with the cullin homology region of APC2 that includes residues 512–750 (Yu et al., 1998). Interestingly, even although APC2 and APC11 formed an active complex when they were coexpressed in insect cells, the full-length APC2 protein did not bind to APC11 in this assay. This is consistent with the fact that, when APC2 and APC11 were expressed individually in insect cells and mixed after purification, no ubiquitin ligase activity was observed. Therefore, the full-length APC2 protein could not form a complex with APC11 post-translationally. However, smaller fragments of APC2 were able to bind to APC11 (Figure 5A).
Figure 5.
Cullin domain of APC2 is sufficient for binding to both APC11 and UbcH10. (A) Purified His6-tagged APC11 and UbcH10 proteins were immobilized on Ni2+-NTA beads and incubated with 35S-labeled APC2 or various APC2 truncation mutant proteins. After washing, the proteins retained on beads were analyzed by SDS-PAGE followed by autoradiography. (B) Complex of the cullin domain of APC2 and APC11 possesses ubiquitin ligase activity. A series of APC2 truncation mutants were expressed in insect cells together with APC11. APC proteins were purified with Ni2+-NTA beads and assayed for ubiquitination activity. (C) Purified APC2e/11 complex was analyzed by SDS-PAGE followed by Coomassie staining.
Because APC2e binds to both APC11 and UbcH10, we examined whether the APC2e/11 complex was an active ubiquitin ligase. The APC fragments were coexpressed with APC11 in insect cells, and assayed for their ability to ubiquitinate cyclin B1 in the presence of UbcH10. The APC2b and APC2e fragments ubiquitinated cyclin B1 efficiently (Figure 5B); both of these fragments retained the ability to bind to APC11 and UbcH10 (Figure 5A). Therefore, a complex of the cullin domain of APC2 and the RING finger protein APC11 is sufficient to catalyze ubiquitination of APC substrates, albeit with decreased efficiency.
One potential caveat of reconstituting the APC activity in insect cells is that certain insect APC proteins might associate with the expressed human APC proteins and contribute to the observed ligase activity. To rule out this possibility, we purified the APC2e/11 complex to homogeneity (Figure 5C) and determined the native size of the APC2e/11 complex by gel filtration chromatography and dynamic light scattering experiments. APC2e/11 cofractionated as a single species with an apparent molecular mass of 50 kDa on the gel filtration column (our unpublished data). Based on the intensity of staining on SDS-PAGE (Figure 5C), we estimated that APC2e and APC11 formed a complex of 1:1 stoichiometry. The calculated molecular mass of the complex is thus 50 kDa. Based on the light scattering experiment, the APC2e/11 complex was mono-dispersed with an apparent molecular mass of 46 kDa. Therefore, it is extremely unlikely that the APC2e/11 complex contains any insect proteins.
Zn2+-binding of APC11 Is Essential for Its Ubiquitin Ligase Activity
Because other RING finger proteins are known to coordinate Zn2+ ions, APC11 may also bind Zn2+. However, this has not been demonstrated experimentally. We thus performed a Zn2+-binding assay on the purified APC2e/11 complex (Yu and Schreiber, 1995). Surprisingly, based on four measurements, we found that APC2e/11 bound Zn2+ at a molar ratio of 3.2 ± 0.2. Similar results were obtained with purified GST-APC11 protein expressed in bacteria. Therefore, it appeared that, in addition to the two Zn2+ ions coordinated by the canonical RING finger motif, APC11 contained a third Zn2+-binding site (Figure 6A). Sequence alignment of the APC11 and Rbx1 proteins from various organisms reveals that three cysteines (Cys 34, Cys 37, and Cys 44 in human APC11) and one histidine (His 58 in human APC11) are conserved among these proteins (Figure 6A). These conserved residues do not belong to the canonical RING-H2 finger motif, and thus are good candidates for coordinating the third Zn2+ ion.
Figure 6.
One APC11 protein molecule binds three Zn2+ ions. (A) Sequence alignment of APC11 and Rbx1 from various organisms (Hs, Homo sapiens; Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae; and Sp, S. pombe). The residues that coordinate the two Zn2+ ions of the canonical RING finger motif are labeled as open and closed circles, respectively. The residues for coordinating the third Zn2+ ion are labeled as open triangles. (B) Autoubiquitination assay of the APC11 mutants. 35S-labeled APC11 mutants were translated in vitro in reticulocyte lysate and incubated with a mixture of ubiquitin, ATP, and E1 in the presence and absence of Ubc4. The reaction mixture was separated on SDS-PAGE followed by autoradiography. The Zn2+-coordinating residues are labeled as in A. The mutations that affect Zn2+-binding are indicated by +, whereas the mutations that do not affect zinc binding are marked by −.
To determine the residues that coordinate Zn2+ ions, all cysteines and histidines of human APC11 were individually mutated to serines and alanines, respectively. Mutations of the Zn2+-binding ligands of the RING finger motif markedly reduced the expression levels of these proteins in bacteria (our unpublished data). Similar results were obtained when Cys 34, Cys 37, Cys 44, and His 58 were mutated. In contrast, mutations of Cys 7, Cys 33, Cys 54, His 65, and His 72 had no effect on the expression levels of the APC11 protein (our unpublished data). These findings are consistent with the notion that Cys 34, Cys 37, Cys 44, and His 58 coordinate the third Zn2+ ion. It is possible that mutations of the Zn2+-binding ligands destabilized the tertiary structure of APC11, resulting in the reduced level of expression of APC11. Obviously, loss of expression of APC11 mutant proteins in bacteria can be caused by factors other than protein folding. Because the APC11 mutant proteins that involve the putative Zn2+-binding ligands could not be obtained at sufficient purity and quantity, we could not determine whether mutations of these ligands actually caused the loss of Zn2+-binding.
When coexpressed with APC2, the APC11 mutants that did not affect Zn2+-binding still possessed ubiquitin ligase activity toward cyclin B1 in the presence of UbcH10 (our unpublished data). Because the APC11 mutants that perturbed Zn2+-binding were not expressed well, we could not compare the ubiquitin ligase activities of these mutants with those of the wild type or mutants that did not affect Zn2+-binding. To circumvent this problem, we obtained all APC11 mutant proteins with the use of the in vitro transcription and translation system in rabbit reticulocyte lysate. Many RING finger-based ubiquitin ligases also autoubiquitinate in the presence of the proper E2 enzyme. We therefore tested the autoubiquitination activity of the APC11 mutants in the presence of Ubc4. The wild-type APC11 protein and the APC11 mutants that did not affect Zn2+-binding were ubiquitinated efficiently when Ubc4 was added, based on the appearance of APC11-ubiquitin conjugates and the percentage of APC11 conjugated to ubiquitin (Figure 6B). Mutations of the eight Zn2+-binding ligands of the RING finger motif greatly reduced the autoubiquitination activity of APC11 (Figure 6B). In contrast, mutations of Cys 34, Cys 37, Cys 44, and His 58 that coordinated the third Zn2+ only slightly reduced the autoubiquitination activity of APC11 (Figure 6B). Thus, unlike the two Zn2+ ions of the RING-H2 finger motif, the third Zn2+ of APC11 might not be involved in catalysis (Figure 6B). This Zn2+ ion may only be important for maintaining the structural integrity of APC11. We also tested the binding of all APC11 mutants to APC2e. None of the mutations affected the binding of APC11 to APC2 (our unpublished data). Because the N-terminal region of APC11 and Rbx1 proteins is conserved and this region is not present in other RING-finger proteins, we speculate that the N-terminal 20 residues of APC11 and Rbx1 proteins are involved in binding to APC2 and Cul1, respectively.
Zn2+ Ions Alone Stimulate Activity of Ubc4 to Ubiquitinate Cyclin B1
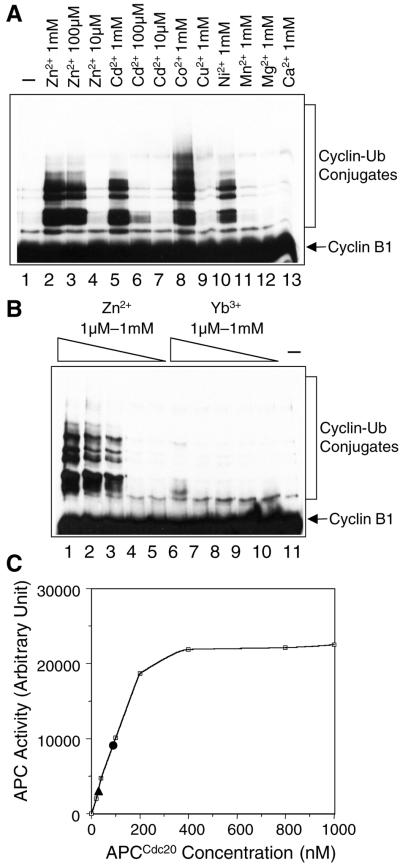
Recently, many RING finger proteins have been shown to possess ubiquitin ligase activities. Despite the relatively low sequence homology outside the RING finger motif, several RING finger proteins use the Ubc4/5 family of E2 enzymes in the ubiquitination reactions (Joazeiro et al., 1999; Lorick et al., 1999; Gmachl et al., 2000; Leverson et al., 2000). This suggested to us that the Zn2+ ions, the most obvious common feature of these RING finger proteins, might be directly involved in catalysis. Strikingly, we found that Zn2+ ions alone stimulated the ability of Ubc4 to ubiquitinate cyclin B1 (Figure 7A). Several other divalent cations, such as Cd2+, Co2+, and Ni2+, also enhanced the activity of Ubc4, whereas Mn2+, Mg2+, Ca2+, and Yb3+ had no effects (Figure 7, A and B). None of these cations stimulated the activity of UbcH10, which did not bind to RING proteins directly (our unpublished data). These findings further support the notion that Zn2+ may be directly responsible for the activity of the RING finger-containing ubiquitin ligases.
Figure 7.
Zn2+ ions alone stimulate the ubiquitination activity of Ubc4. (A) Various concentrations of Zn2+, Cd2+, and other divalent cations were added to a reaction mixture containing E1, ubiquitin, Ubc4, ATP, and cyclin B1. Ubiquitination of cyclin B1 was analyzed by immunoblotting with α-Myc. (B) Various concentrations of Zn2+ and Yb3+ were added to a reaction mixture containing E1, ubiquitin, Ubc4, ATP, and cyclin B1. Ubiquitination of cyclin B1 was analyzed by immunoblotting with α-Myc. (C) Comparison of the ubiquitin ligase activities of the intact APCCdc20, the reconstituted APC, and zinc ions alone. The ligase activity of APCCdc20 was plotted against the concentration used in the assay. The activities of the reconstituted APC at 5 μM and zinc ions alone at 100 μM were indicated by closed circle and triangle, respectively.
We next quantitatively compared the activities of the intact APCCdc20, the reconstituted APC, and the Zn2+ ions alone. Various concentrations of APCCdc20 were used in the in vitro ubiquitination assay with the use of cyclin B1 as the substrate. The ligase activity of APC was measured by the intensities of the cyclin-ubiquitin conjugates, which were normalized by the number of ubiquitin molecules in the conjugates (Figure 7C). The activity of the reconstituted APC at 5 μM was similar to that of the intact APCCdc20 at 90 nM, indicating that the intact APCCdc20 was 55 times more active than the reconstituted APC. Zinc ions at 100 μM exhibited ligase activity comparable to 25 nM of APCCdc20. Thus, the activity of zinc ions alone was 4000 weaker than that of the intact APC.
DISCUSSION
APC as a Member of Cullin-RING Finger Family of Ubiquitin Ligases
Among the three classes of enzymes of the ubiquitin pathway, the ubiquitin ligases are the most divergent, in terms of both composition and function (Hershko and Ciechanover, 1998). All ubiquitination systems use a single ubiquitin-activating enzyme (E1) for the first step of the reaction (Hershko and Ciechanover, 1998). Although certain ubiquitin-conjugating enzymes (E2s) are used in different ubiquitination systems, all E2 enzymes are homologous in sequence and contain the ubiquitin-conjugation (UBC) domain (Hershko and Ciechanover, 1998). In contrast, the ubiquitin ligases (E3s) are loosely defined as entities that collaborate with E1 and E2 to ligate ubiquitin chains on substrates. They can be unrelated in amino acid sequence and use distinct biochemical mechanisms for catalysis.
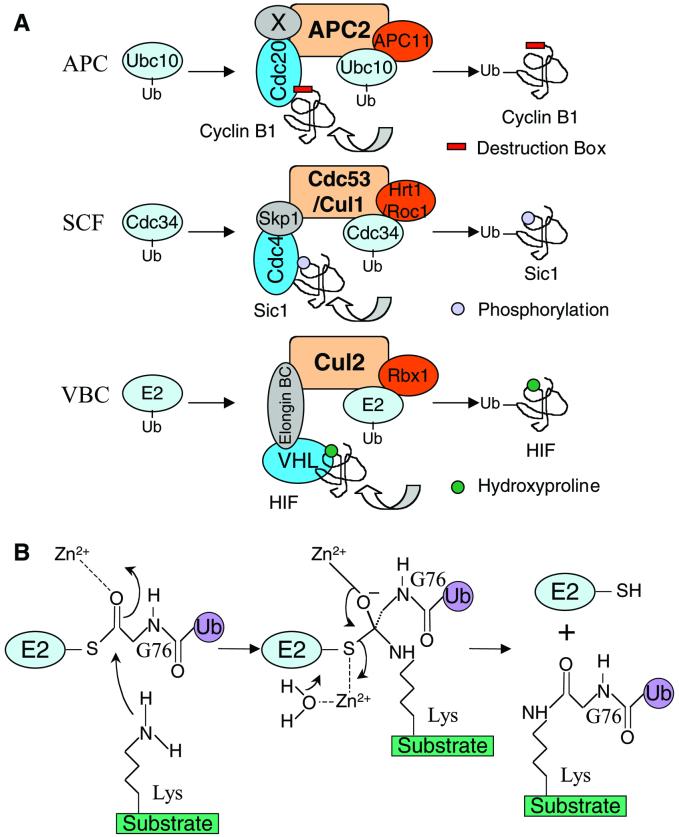
The E3s identified so far can be divided into three major families. The first family consists of HECT domain-containing proteins (Scheffner et al., 1995). The second family of E3s includes Ubr1, c-Cbl, Mdm2, IAPs, and other RING finger-containing proteins, which appear to contain both the E2-binding domain and the substrate recognition motif in a single polypeptide chain (Joazeiro et al., 1999; Xie and Varshavsky, 1999; Fang et al., 2000; Yang et al., 2000). In contrast, the third group of E3s consists of large protein complexes that contain a minimal ligase core of a cullin and RING-H2 heterodimer, such as the SCF and the VBC complexes (Deshaies, 1999; Kamura et al., 1999). Based on the data presented herein, the minimal ligase module of APC is comprised of APC2 (a distant member of the cullin family) and APC11 (a RING-H2 finger protein). Therefore, APC belongs to the third group of ubiquitin ligases (Figure 8A).
Figure 8.
(A) APC belongs to the cullin-RING family of ubiquitin ligases. (B) Proposed role of Zn2+ in Ubc4-catalyzed ubiquitination reactions. See DISCUSSION for details.
Substrate Recognition by APC and Its Regulation during Cell Cycle
In addition to the fact that APC, SCF, and VBC ligase complexes all contain a cullin-RING finger heterodimeric ligase core, there is another analogy between APC and the other two systems (Figure 8A). The minimal ligase modules of these complexes are connected to various adaptor proteins that serve to recruit substrates. In the case of SCF complex, the substrate-binding proteins, such as Cdc4, Grr1, Skp2, and β-TRCP, contain the F-box motif and interact with Skp1, which in turn associates with the cullin protein Cdc53 or hCul1 (Deshaies, 1999). Likewise, VHL, the substrate receptor of the VBC ligase complex, is tethered to the hCul2/Rbx1 ligase core through the elongin C protein, which shares structural similarity with Skp1 (Kamura et al., 1999). The association between VHL and the elongin BC complex is mediated by the SOCS-box motif of VHL (Kamura et al., 1998). The substrate recognition factors of APC are the WD40 repeat-containing proteins Cdc20 and Cdh1 (Burton et al., 2001; Hilioti et al., 2001; Pfleger et al., 2001). We have observed direct binding between Cdc20 or Cdh1 and the APC2/11 ubiquitin ligase core (our unpublished data). However, binding of Cdc20 or Cdh1 to APC2/11 does not stimulate the ligase activity of APC2/11. Thus, it is unclear which subunit(s) of APC is required to anchor Cdc20 or Cdh1 to the ligase core in a functional way. It is also unknown what structural feature of Cdc20 and Cdh1 is recognized by APC.
The minimal ligase modules of APC and other cullin-RING type of E3s appear to be constitutively active, at least in vitro (Seol et al., 1999). The regulation of the ubiquitination processes involving these enzymes resides in substrate binding. However, APC differs significantly from the SCF and VBC complexes in the specific mode of regulating substrate binding. Both SCF and VBC complexes recognize post-translationally modified substrates: the F-box proteins of SCF complexes bind phosphorylated protein substrates, whereas the VHL protein of the VBC complex recognizes a novel hydroxyproline moiety of the HIF substrate (Skowyra et al., 1997; Ivan et al., 2001; Jaakkola et al., 2001). Therefore, the critical regulation of SCF and VBC pathways is at the level of the substrates. In contrast, although all APC substrates contain specific sequence motifs, such as the D-box and KEN-box, post-translational modifications of the APC substrates are not required for their efficient ubiquitination. Instead, it is the association of Cdc20 or Cdh1 to APC that is tightly regulated during the cell cycle. Dissociation of Cdc20 or Cdh1 from APC during the S, G2, and prophase of the cell cycle, in essence, serves to inhibit the substrate binding ability and thus the activity of APC (Fang et al., 1998).
Mechanism of Ubiquitin Transfer of APC-catalyzed Reactions
E3-catalyzed ubiquitination reactions may use two distinct mechanisms for conjugating ubiquitin to substrates. In the case of E6-AP-mediated ubiquitination of p53, Ubc4 (the E2 in the system) first transfers ubiquitin to E6-AP to form an E3-ubiquitin thioester, which then attaches the ubiquitin to the lysine residues of p53 (Scheffner et al., 1995). However, this does not seem to be the prevailing mechanism for all ubiquitination reactions. For the SCF complexes, it appears that the ubiquitination reactions do not involve the formation of an E3-ubiquitin thioester (Seol et al., 1999). Instead, the SCF complexes serve as a scaffold to bring together the E2 enzyme Cdc34 and the substrates. The RING protein Rbx1 of SCF enhances the ubiquitin transfer activity of Cdc34, and ubiquitin is then transferred directly from the Cdc34 thioester to substrates.
Aside from the analogy between APC and SCF, there is additional evidence to suggest that APC might also use the latter scaffolding mechanism for ubiquitin transfer. First, we and others were unable to detect a ubiquitin thioester of any APC subunits so far. Second, in the presence of active APCCdc20 or APCCdh1, Ubc4 seems to generate cyclin-ubiquitin conjugates of higher molecular weight than those generated by UbcH10 (Figure 1B) (Yu et al., 1996). The E3-thioester model would predict that, no matter which E2 is used, the same APC-ubiquitin thioester acts as the intermediate for relaying the ubiquitin to cyclin. The two E2s would then have little influence over the pattern of cyclin-ubiquitin conjugates. Therefore, the fact that Ubc4 and UbcH10 support the formation of cyclin-ubiquitin conjugates with different patterns favors the scaffolding model. One possible explanation for the higher molecular weight conjugates catalyzed by Ubc4 might be that Ubc4 transfers ubiquitin faster than UbcH10. Thus, during the residence time of substrate binding to APC, Ubc4 might recharge and transfer ubiquitin more frequently than UbcH10, thus forming higher molecular weight conjugates than UbcH10. Finally, all cysteines of APC11 have been mutated. Except the cysteines coordinating Zn2+ ions, mutation of other cysteines does not abolish the ubiquitin ligase activity of APC11 with Ubc4 as the E2. Therefore, it is unlikely that an APC-ubiquitin thioester is involved in these ubiquitination reactions.
Role of Zn2+ Ions in Catalysis
The Zn2+ ions of the RING finger E3 ligases are clearly essential for maintaining the structural integrity of these proteins. Besides the structural role, are the Zn2+ ions of the RING finger proteins also directly involved in catalysis, similar to many Zn2+-binding metalloenzymes? Two lines of evidence presented herein suggest that this might be the case. First, we found that APC11 binds a third Zn2+ ion, aside from the two Zn2+ ions that form the RING finger motif. Mutations of the residues that coordinate the third Zn2+ appear to destabilize the structure of APC11. Yet, these APC11 mutants still possess E3 ligase activity. In contrast, mutations of the Zn2+-binding residues of the canonical RING motif not only disrupt protein structure but also abrogate the ligase activity of APC11. This suggests that, in addition to the structural role, the two Zn2+ ions of the RING motif might be directly involved in catalysis. This notion is further strengthened by the unexpected finding that Zn2+ ions alone can stimulate the ability of Ubc4 to ligate ubiquitin to cyclin B1. Zn2+ might facilitate two steps in the ubiquitin ligation reaction (Figure 8B). It might stabilize the oxyanion of the putative tetrahedral intermediate of the reaction. Along this line, it is worth noting that polycations can stimulate the autoubiquitination activity of Cdc34, and the polycations have been proposed to stabilize the oxyanion intermediate (Seol et al., 1999; Zachariae and Nasmyth, 1999). Zn2+ might also promote the release of the active thiol group of the E2 from the tetrahedral intermediate. Consistent with this notion, several cations that can potentially coordinate oxyanions, such as Cu2+ and Yb3+, do not have the same effect as Zn2+. Obviously, the mechanism by which Zn2+ ions at high concentrations catalyze the ubiquitination reaction may not mimic the mode of action of the RING-containing ubiquitin ligases. Further experiments are required to clarify these issues.
ACKNOWLEDGMENTS
We thank Marc Kirschner for encouragement at the early stages of this project. We also thank Hui Zou for providing the human securin plasmids. This work is supported by the Damon Runyon-Walter Winchell Foundation, the Robert A. Welch Foundation, the Burroughs Wellcome Fund, the Packard Foundation, and the National Institutes of Health grant GM-61542.
REFERENCES
- Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman JV. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast HSl1p are required APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Feldman R, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters J-M. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci USA. 2000;97:8973–8978. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossberger R, Gieffers C, Zachariae W, Podtelejnikov AV, Schleiffer A, Nasmyth K, Mann M, Peters J-M. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J Biol Chem. 1999;274:14500–14507. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Chung Y, Mochizuki Y, Hardy CF, Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, et al. Targeting of HIFα to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Jr, Conaway RC, Conaway JW. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters J-M, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996a;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996b;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. Cul-1 is required for cell cycle exit in c-elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters J-M. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Latres E, Chiaur DS, Pagano M. The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- Leverson JD, Joazeiro CA, Page AM, Huang H, Hieter P, Hunter T. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina SA, Correll CC, Kipreos ET, Deshaies RJ. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc Natl Acad Sci USA. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Osaka F, Seino H, Seno T, Yamao F. A ubiquitin-conjugating enzyme in fission yeast that is essential for the onset of anaphase in mitosis. Mol Cell Biol. 1997;17:3388–3397. doi: 10.1128/mcb.17.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Seol JH, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Aristarkhov A, Beck S, Hershko A, Ruderman JV. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Ruderman JV. Functional analysis of the Saccharomyces cerevisiae UBC11 gene. Yeast. 1998;14:747–757. doi: 10.1002/(SICI)1097-0061(19980615)14:8<747::AID-YEA271>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. The CDC27HS protein co-localizes with the CDC16HS protein to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1—A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Yu H, King RW, Peters J-M, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- Yu H, Peters J-M, King RW, Page A, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1223. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- Yu H, Schreiber SL. Cloning, Zn2+ binding, and structural characterization of the guanine nucleotide exchange factor human Mss4. Biochemistry. 1995;34:9103–9110. doi: 10.1021/bi00028a020. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews P, Galova M, Stark M, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from budding yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]