Abstract
Exact coordination of growth plate chondrocyte proliferation is necessary for normal endochondral bone development and growth. Here we show that PTHrP and TGFβ control chondrocyte cell cycle progression and proliferation by stimulating signaling pathways that activate transcription from the cyclin D1 promoter. The TGFβ pathway activates the transcription factor ATF-2, whereas PTHrP uses the related transcription factor CREB, to stimulate cyclin D1 promoter activity via the CRE promoter element. Inhibition of cyclin D1 expression with antisense oligonucleotides causes a delay in progression of chondrocytes through the G1 phase of the cell cycle, reduced E2F activity, and decreased proliferation. Growth plates from cyclin D1–deficient mice display a smaller zone of proliferating chondrocytes, confirming the requirement for cyclin D1 in chondrocyte proliferation in vivo. These data identify the cyclin D1 gene as an essential component of chondrocyte proliferation as well as a fundamental target gene of TGFβ and PTHrP during skeletal growth.
INTRODUCTION
Endochondral bone growth is controlled by the coordinated proliferation and differentiation of growth plate chondrocytes (Cancedda et al., 1995). Numerous skeletal diseases (chondrodysplasias) are caused by genetic disturbances of these processes, resulting in skeletal deformities, dwarfism, and early onset osteoarthritis (Mundlos and Olsen, 1997a, 1997b). Despite the recent identification of many genes that control chondrocyte proliferation and differentiation (Mundlos and Olsen, 1997a, 1997b; Beier et al., 1999a), the intracellular signaling pathways and transcriptional mechanisms involved are not well defined. Transforming growth factor beta (TGFβ) and parathyroid hormone–related peptide (PTHrP) stimulate the proliferation of chondrocytes and chondrosarcoma cells in vitro (O'Keefe et al., 1988; Rosier et al., 1989; Guerne and Lotz, 1991; Loveys et al., 1993; Guerne et al., 1994). In addition, interruption of TGFβ or PTHrP signaling in vivo in mice causes a reduction in the number of proliferative chondrocytes as well as premature differentiation of these cells (Amizuka et al., 1994; Karaplis et al., 1994; Serra et al., 1997; Yang et al., 2001). These data suggest that both factors are required for normal chondrocyte proliferation in vivo and in vitro. However, the intracellular signaling pathways activated by PTHrP and TGFβ in chondrocytes as well as their target genes have yet to be identified.
Cell cycle genes appear to play an important role in the control of chondrocyte proliferation and differentiation (Beier et al., 1999a; LuValle and Beier, 2000). Progression through the eukaryotic cell cycle is controlled by the activity of a family of kinases called the cyclin-dependent kinases or CDKs (Weinberg, 1995). CDK activity is strictly controlled by a number of mechanisms, including phosphorylation status and the presence of inhibitory proteins such as p16Ink4 or p21Cip1/Waf1. An absolute requirement for the activity of a given CDK is the presence of its specific cyclin partner. The D-type cyclins (D1, D2, and D3) control progression through the G1 phase of the cell cycle in complexes with CDKs 4 and 6. Mice in which the cyclin D1 gene has been disrupted display severely reduced postnatal growth (Fantl et al., 1995; Sicinski et al., 1995), suggesting differences in skeletal growth and therefore in growth plate physiology. Cyclin D1 expression is induced by a variety of mitogenic stimuli in different cell types and is strictly controlled by transcriptional and posttranscriptional mechanisms. More recently, cyclin D1 expression in the growth plate has been shown to be specific for proliferating chondrocytes by in situ hybridization (Long et al., 2001). We have identified the cyclin D1 gene as a target of the transcription factors ATF-2 (activating transcription factor 2) and CREB (CRE-binding protein) in chondrocytes (Beier et al., 1999b). ATF-2–deficient mice display reduced chondrocyte proliferation resulting in dwarfism and skeletal deformities (Reimold et al., 1996). CREB-deficient mice show a similar growth phenotype, but their skeletons have not been analyzed in detail (Rudolph et al., 1998). However, overexpression of a dominant-negative form of CREB in chondrocytes of transgenic mice causes severe disturbances of growth plate architecture and growth (Long et al., 2001).
Because genetic interference with TGFβ and PTHrP signaling leads to growth phenotypes that are similar to those caused by interruption of the cyclin D1, ATF-2, or CREB genes, we hypothesized that transcriptional activation of the cyclin D1 gene through ATF-2 and/or CREB would be involved in the biological responses of growth plate chondrocytes to TGFβ and PTHrP. Here we demonstrate that TGFβ and PTHrP induce chondrocyte proliferation through induction of cyclin D1 transcription, mediated by the transcription factors ATF-2 and CREB. We also show that cyclin D1 is required for normal chondrocyte proliferation in vivo.
MATERIALS AND METHODS
Materials
The human cyclin D1 reporter plasmids −745 CD1Luc CREmut and −1745 CD1LUC CRE/AP-1mut have been generated through PCR-based mutagenesis (details available on request). The other cyclin D1 promoter plasmids (Albanese et al., 1995; Beier et al., 1999b) and the expression plasmids for dominant-negative ATF-2 (Beier et al., 1999b) and CREB (Ahn et al., 1998) have been described. All GAL4 components were purchased from Stratagene (La Jolla, CA). The E2F reporter plasmid pE2F-TA-Luc was purchased from Clontech (Palo Alto, CA), the rat cyclin A promoter plasmid pCyvAluc707 was a generous gift from K. Oda (Shimizu et al., 1998; Beier et al., 2000), and the human PTH/PTHrP receptor expression plasmids were provided by H. Jüppner (Schipani et al., 1999). TGFβ1 and PTHrP were from Calbiochem (La Jolla, CA).
Cell Culture, FACS Analyses, and Cell Counting
Primary mouse chondrocytes were isolated as described (Lefebvre et al., 1994; Beier et al., 1999c). Primary rat chondrocytes were isolated from the femoral heads of newborn rats. All primary chondrocytes and RCS (rat chondrosarcoma) cells (Mukhopadhyay et al., 1995) were cultured as described (Beier et al., 1999b, 2000). For FACS analyses of BrdU incorporation, RCS cells were serum-starved for 3 d and then stimulated with TGFβ (1 ng/ml), PTHrP (10−8 M), or both. Cells were labeled for 30 min with BrdU (10 μM) at 4-h intervals to obtain a time course, and the percentage of cells incorporating BrdU was determined by FACS. For proliferation assays, 1.5 × 105 cells/well were plated in 24-well plates and cultured for 3 d without growth factors, with TGFβ (1 ng/ml), PTHrP (10−8 M), or both. Cells were counted in a hemacytometer. The data shown represent the mean and SD from two independent experiments, each done in triplicate. Control (1 μM) or cyclin D1 antisense or control oligonucleotide (Ko et al., 1998) were used where indicated. Cell cycle distribution of cells was determined by measurement of DNA content with the use of FACS analysis of propidium iodine staining of DNA.
Western Blot Analyses
Western blot analyses were performed as described (Beier et al., 1999b, 1999c) with antibodies against the following proteins: ATF-2 (SC-187; Santa Cruz Biotechnologies, Santa Cruz, CA), CREB (9192), phosphorylated ATF-2 (9221), and phosphorylated CREB (9191; all from New England Biolabs, Beverly, MA), cyclin D1 (MS-210-P; NeoMarkers, Fremont, CA), and actin (catalogue no. 1378 996; Boehringer Mannheim, Indianapolis, IN).
Transfections and Luciferase Assays
Transfections were done as described (Beier et al., 1999b, 1999c). Cotransfections were performed by transfecting cells with 1.0 μg of reporter plasmid (containing the firefly luciferase reporter gene), 0.1 μg of pRlSV40 (Promega, Madison, WI; encoding Renilla luciferase for standardization), and 0.1 μg of empty expression vector or expression vectors for dominant-negative forms of ATF-2 and CREB or for wild-type or mutant PTH/PTHrP receptors. For Gal4 assays, 1.0 μg of reporter construct (pFR-Luc) was cotransfected with 0.1 μg of Gal4 expression plasmid (pFA-ATF2 or pFA-CREB). The promoter data shown represent the mean and SD from two independent experiments, each performed in triplicate.
Histology
Knee joints from 6-week-old wild-type and cyclin D1 (−/−) mice were dissected, fixed in 10% formalin, decalcified in Cal-EX (Fisher Scientific, Pittsburgh, PA), and embedded in paraffin. Sections of 6-μm were stained with hematoxylin, Fast Green, and Saffranin O.
Mice
Mice carrying inactivating mutations of the cyclin D1 or ATF-2 genes were genotyped as described (Sicinski et al., 1995; Reimold et al., 1996). Animal experiments were performed in accordance with federal and institutional guidelines.
RESULTS
Mitogenic Stimulation of Chondrocytes Induces and Requires Cyclin D1 Expression
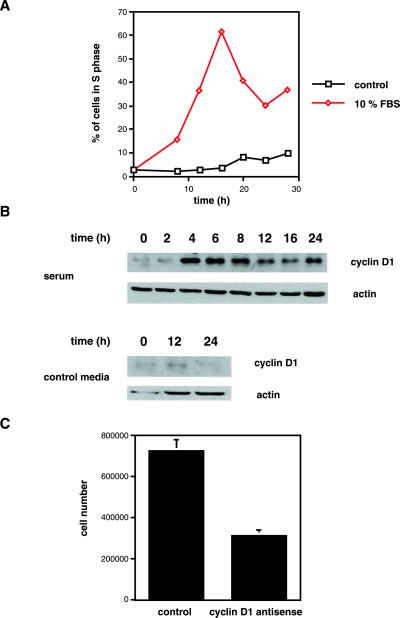
RCS cells (Mukhopadhyay et al., 1995) were serum-starved for 3 d to synchronize cells in the G0/1 phase of the cell cycle (Beier et al., 2000), followed by stimulation with 10% FBS. Serum stimulation induced progression of RCS cells into the S-phase of the cell cycle, as measured by FACS analyses of BrdU incorporation into replicating DNA (Figure 1A). Cells were harvested for protein analyses, and cyclin D1 protein expression was examined by Western blot to investigate whether cell cycle progression is accompanied by changes in cyclin D1 levels. Cyclin D1 is expressed a very low levels in the absence of serum, but expression was rapidly induced by serum stimulation, reaching maximal levels after 4 h of stimulation (Figure 1B).
Figure 1.
Cyclin D1 is required for serum-induced RCS proliferation. (A) RCS cells were serum-starved for 3 d, followed by stimulation with 10% FBS or control serum-free medium. Cells were labeled with BrdU in 4-h intervals. The percentage of cells incorporating BrdU was determined by FACS and is shown. (B) RCS cells were serum-starved for 3 d, followed by stimulation with 10% FBS or control medium for the indicated intervals, and harvested for the analyses of cyclin D1 protein expression by Western blot. Equal gel loading (5 × 105 cells/lane) was documented by probing for actin protein expression. (C) RCS cells (150,000/well) were plated, serum-starved for 3 d, and restimulated with 10% FBS in the presence of control or cyclin D1 antisense oligonucleotides. Cells were counted after 48 h of stimulation. The average and SD from two independent experiments (performed in triplicate wells each) are shown.
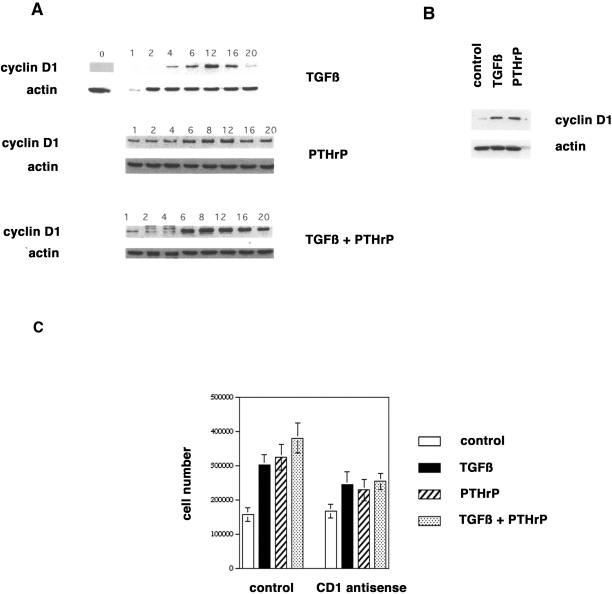
RCS cells were serum-starved as above and stimulated with 10% FBS in the presence of 10 μM control oligonucleotides or cyclin D1 antisense oligonucleotides (Ko et al., 1998) to determine whether cyclin D1 is required for the mitogenic activity of serum. Cells were counted after 2 d. Whereas the number of control cells increased by almost 500%, cyclin D1 antisense-treated cells displayed only a doubling in cell number (Figure 1C).
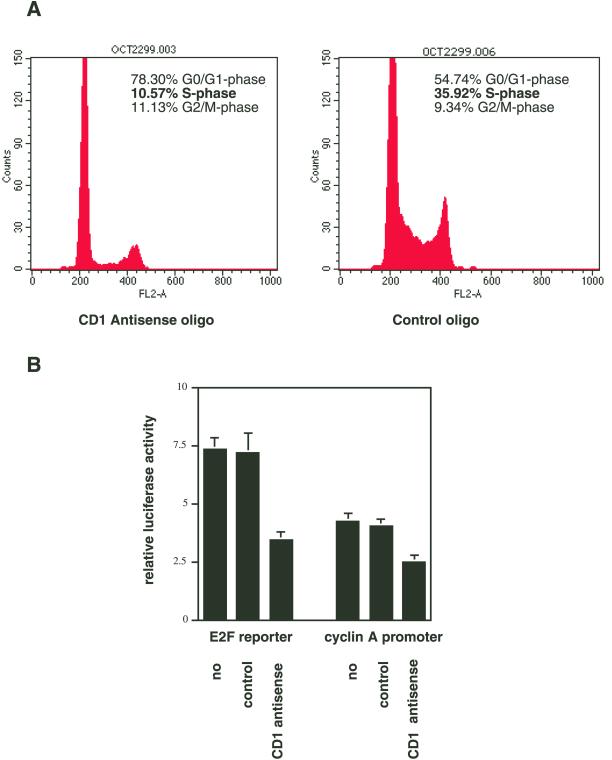
Cell cycle distributions of cyclin D1 antisense oligonucleotide–treated and control cells (in the presence of 10% FBS) were compared with the use of FACS analyses of propidium iodine–stained nuclear DNA. Only 10.6% of antisense-treated cells were in the S-phase of the cell cycle, whereas 78.3% of these cells were in the G0/1 phase. In contrast, 35.9% and 54.7% of control cells were in the S-phase and G0/1, respectively (Figure 2A). These data suggest that expression of cyclin D1 protein is required for normal progression through the G1 phase of the cell cycle in chondrocytes.
Figure 2.
Cyclin D1 is required for G1 progression and E2F-dependent gene expression in RCS cells. (A) Subconfluent RCS cells were incubated with cyclin D1 antisense or control oligo nucleotides for 24 h and harvested for FACS analysis. Cell cycle distribution was determined by propidium iodine staining of nuclear DNA. (B) Plasmids encoding the firefly luciferase gene under control of an E2F-responsive promoter or the cyclin A promoter were transfected into RCS cells, together with the plasmid pRlSV40. Cells were incubated without oligonucleotides (no), control oligonucleotides, or cyclin D1 antisense oligonucleotides for 48 h. Cells were harvested, and firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity.
Cyclin D1 is thought to exert its regulation of cell cycle progression through the regulation of the activity of E2F transcription factors, which in turn control the transcription of downstream target genes, such as the cyclin A gene, involved in cell cycle progression (Weinberg, 1995). RCS cells were transfected with the E2F reporter plasmid pE2F-TA-Luc (encoding the firefly luciferase gene under the control of a promoter containing several E2F sites) and incubated for 48 h with either no oligonucleotides, control oligonucleotides, or cyclin D1 antisense oligonucleotides (10 μM each). Whereas control cells displayed luciferase activity similar to untreated cells, treatment with cyclin D1 antisense oligonucleotides caused a 53% reduction in promoter activity. Similarly, cyclin D1 antisense treatment led to a 41% reduction in the activity of a cyclin A promoter reporter (Figure 2B).
Cyclin D1 Is Required for Normal Chondrocyte Proliferation In Vivo
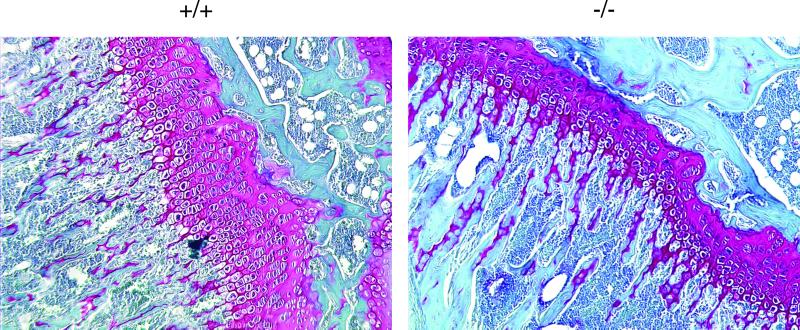
Cyclin D1–deficient mice show reduced postnatal growth (Fantl et al., 1995; Sicinski et al., 1995). To verify the importance of cyclin D1 in chondrocyte proliferation in vivo, we compared the histology of growth plates from 6-week-old, wild-type and cyclin D1–deficient (−/−) mice. Growth plates from −/− mice have a 50% reduction in the size of the proliferative zone of the growth plate when compared with wild-type littermates (Figure 3).
Figure 3.
Cyclin D1 is required for chondrocyte proliferation in vivo. Growth plates (stained red) from tibias of 6-week-old wild-type (+/+) or cyclin D1–deficient (−/−) mice were analyzed by histology with the use of hematoxylin/Fast Green/Saffronin O staining. The zone of proliferative chondrocytes (small, flattened chondrocytes, arranged in columns, on the right side of each growth plate) is reduced by 50% in −/− mice.
TGFβ and PTHrP Induce Cell Cycle Progression and Proliferation in Chondrocytes
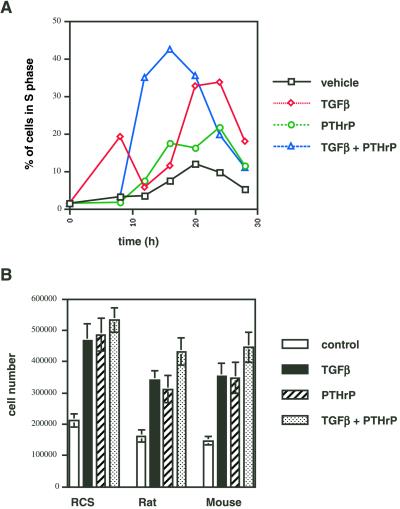
We investigated the effects of TGFβ and PTHrP on cell cycle progression by FACS analyses of BrdU incorporation into replicating DNA. Serum starvation of RCS cells for 3 d results in accumulation of >90% of the cells in the G0/1 phase of the cell cycle (Beier et al., 2000). Addition of TGFβ (1 ng/ml) or PTHrP (10−8 M) to serum-starved cells caused reentry into the cell cycle and stimulation of DNA synthesis, beginning at 8–12 h and reaching its maximum at 20–24 h (Figure 4A). Simultaneous stimulation with both growth factors accelerated and enhanced this induction of replication.
Figure 4.
Induction of chondrocyte proliferation by TGFβ and PTHrP. (A) RCS cells were serum-starved for 3 d, stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both, and labeled with BrdU in 4-h intervals. The percentage of cells incorporating BrdU was determined by FACS and is shown. (B) RCS cells and primary rat and mouse chondrocytes were plated in 24-well plates (105 cells/well), stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both for 3 d, and then counted with the use of a hemacytometer.
Direct cell counting of chondrocytes incubated either without growth factors or with TGFβ, PTHrP, or both for 3 d revealed that proliferation was strongly increased in the presence of growth factors in both RCS cells and primary chondrocytes. RCS cells proliferated even in the absence of growth factors, but primary cells (rat and mouse) did not multiply without exogenous growth factors. The presence of either growth factor resulted in a doubling of cell number in primary chondrocytes, and the combination of both factors had a slightly stronger effect for all three cell types (Figure 4B).
TGFβ and PTHrP Induce Cyclin D1 Expression
Next we examined the effect of TGFβ and PTHrP on cyclin D1 expression in RCS cells. RCS cells were serum-starved for 3 d, followed by stimulation with serum-free control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or a combination of both growth factors. Either growth factor induced expression of cyclin D1 protein within 4–6 h, with maximal levels reached at 8–12 h. The combination of both factors caused accelerated induction and higher maximal levels of cyclin D1 protein (Figure 5A). In contrast, neither factor had an influence on the protein levels of the cyclin D1–associated kinases CDK4 and CDK6 (our unpublished results). To verify these data in primary cells, we serum-starved primary rat chondrocytes for 2 d and stimulated them with the growth factors as above for 8 h. Both TGFβ and PTHrP induced cyclin D1 expression (Figure 5B).
Figure 5.
Induction of cyclin D1 expression by TGFβ and PTHrP. (A) RCS cells were serum-starved for 3 d, stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both for the indicated intervals, and harvested for the analyses of cyclin D1 protein expression by Western blot. Equal gel loading (5 × 105 cells/lane) was documented by probing for actin protein expression. (B) Primary rat chondrocytes were serum-starved for 1 d, stimulated with control medium, TGFβ (1 ng/ml), or PTHrP (10−8 M) for 8 h, and harvested for the analyses of cyclin D1 protein expression by Western blot. Equal gel loading (5 × 105 cells/lane) was documented by probing for actin protein expression. (C) Primary rat chondrocytes were plated in 24-well plates (105 cells/well). The cells were incubated with control or cyclin D1 antisense oligonucleotides and stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both for 3 d, and then counted with the use of a hemacytometer.
Primary rat chondrocytes were incubated with the growth factors in the presence of either control or cyclin D1 antisense oligonucleotides. Cells were counted after 3 d of incubation. Incubation with control oligonucleotides allowed a doubling of cell number within 3 d, similar to untreated cells (compare to Figure 4B). In contrast, cyclin D1 antisense oligonucleotides inhibited the mitogenic effects of TGFβ and PTHrP by ∼40 and 55%, respectively (Figure 5C).
Transcriptional Regulation of Cyclin D1 Expression by PTHrP and TGFβ
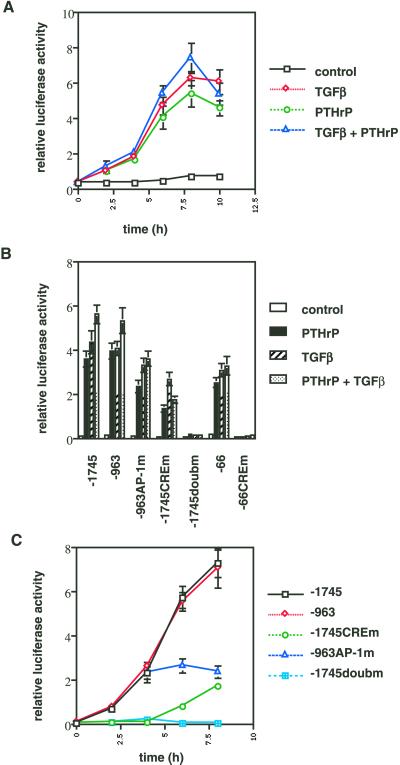
We transfected the plasmid −1745 CD1LUC (Figure 6), containing 1745 nucleotides of the human cyclin D1 promoter (Albanese et al., 1995), into RCS cells and stimulated the transfected cells (after 3 d of serum-starvation) with TGFβ, PTHrP, or both to determine the effects of these growth factors on cyclin D1 promoter activity. All three stimulations caused a strong increase in promoter activity with a time course similar to that observed at the cyclin D1 protein level, reaching maximal activity at 8 h (Figure 7A).
Figure 6.
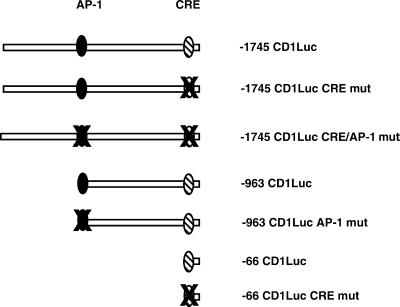
Cyclin D1 promoter constructs. Overview of the human cyclin D1 promoter constructs used in this study. The described AP-1 and CRE elements are indicated.
Figure 7.
Cyclin D1 promoter induction by PTHrP and TGFβ requires the AP-1 and CRE sites. (A) The plasmid −1745 CD1Luc was transfected into RCS cells, together with the plasmid pRlSV40. After transfection, cells were serum-starved for 3 d and stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both for 0, 2, 4, 6, 8, and 10 h. Cells were harvested at these time points, and firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity. (B) The cyclin D1 promoter constructs −1745 CD1Luc (−1745), −963 CD1Luc (−963), −963 CD1Luc AP-1mut (−963AP-1 m), −1745 CD1Luc CREmut (−1745CREm), −1745 CD1LucCRE/AP-1mut (−1745doubm), −66 CD1Luc (−66), and −66 CD1Luc CREmut (−66CREm) were transfected into RCS cells, together with pRlSV40. After transfection, cells were serum-starved for 3 d and stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both for 8 h. Cells were harvested, and firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity. (C) The cyclin D1 promoter constructs −1745 CD1Luc (−1745), −963 CD1Luc (−963), −963 CD1Luc AP-1mut (−963AP-1 m), −1745 CD1Luc CREmut (−1745CREm), and −1745 CD1LucCRE/AP-1mut (−1745doubm) were transfected into RCS cells, together with pRlSV40. After transfection, cells were serum-starved for 3 d and stimulated with control medium or a combination of TGFβ (1 ng/ml) and PTHrP (10−8 M) for 0, 2, 4, 6, or 8 h. Cells were harvested at the indicated time points, and firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity
Next we transfected several point and deletion mutants of the cyclin D1 promoter (Figure 6) into RCS cells in order to identify the cis-active sites that confer the transcriptional responses to TGFβ and PTHrP. Cells were stimulated with TGFβ, PTHrP, or both for 8 h, after 3 d of serum-starvation. Deletion of the sequences between −1745 and −963 of the promoter did not affect promoter induction under any experimental conditions (Figure 7B). However, a mutation in the AP-1 site at position −953 reduced the response to PTHrP by 34%, to TGFβ by 24%, and to the combination by 36%. Deletion of further sequences in the promoter up to position −66 did not cause significant changes in the responses to either growth factor (relative to the −963 AP-1 mutant). Mutation in the CRE (cyclic AMP responsive element) in the −66 promoter fragment abolished the response to both factors individually and in combination. Mutation of the CRE in the context of the 1745-base pair promoter caused a reduction of 62% in the response to PTHrP, 55% in the response to TGFβ, and 69% in the response to the combination of both factors. Simultaneous mutation of both the AP-1 site and the CRE element in the full-length promoter completely abolished the response to the growth factors (Figure 7B). These data identify the CRE and AP-1 sites in the 1745-base pair promoter as the major determinants in the transcriptional response to TGFβ and PTHrP.
We next examined the temporal induction of the cyclin D1 promoter by these growth factors. Because both factors seem to utilize the same DNA elements for promoter induction, only the combination of PTHrP and TGFβ was used in these experiments. Cells were transfected, serum starved for 3 d, and stimulated with growth factors. Cells were harvested for determination of luciferase activity at 2-h intervals. The wild-type −1745 and −963 promoter constructs displayed induction similar to that seen in Figure 7A. Mutation of the AP-1 site did not affect the early induction of the promoter up to 4 h but abolished further increases in promoter activity in the second 4 h. In contrast, mutation of the CRE (in the full-length promoter) inhibited the initial induction of promoter activity but still allowed some promoter activation at 6 and 8 h. Mutation of both sites completely abolished promoter responsiveness (Figure 7C).
ATF-2 and CREB Mediate Transcriptional Activation of the Cyclin D1 CRE by TGFβ and PTHrP
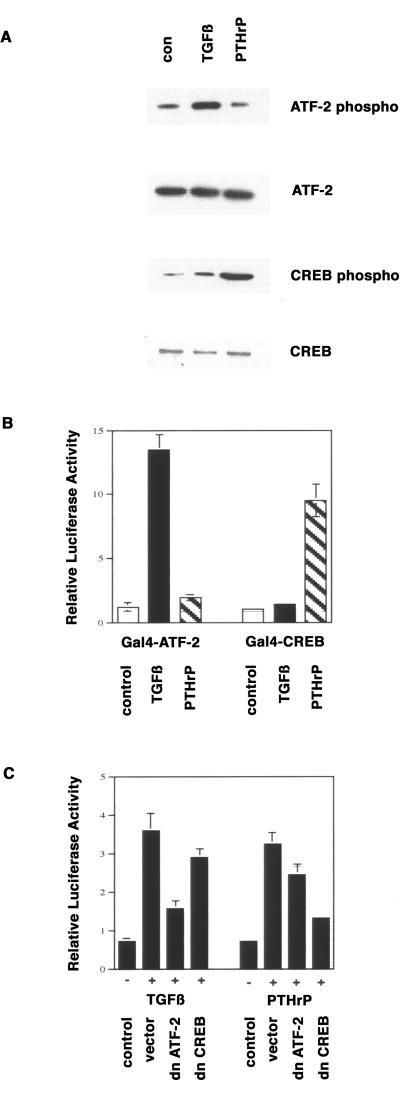
Because mutation in the CRE had the largest quantitative effect on the promoter response to both factors, we focused on the analyses of this site in response to TGFβ and PTHrP. We had previously shown that the transcription factors CREB and ATF-2 bind to the cyclin D1 CRE in chondrocytes (Beier et al., 1999b). Activity of both transcription factors is regulated by phosphorylation. We examined the phosphorylation of ATF-2 and CREB by pathways initiated by TGFβ and PTHrP with the use of phospho-specific antibodies. TGFβ treatment led to an increase in ATF-2 phosphorylation, without affecting CREB phosphorylation. In contrast, PTHrP caused a clear increase in the phosphorylation of CREB but not of ATF-2. The total levels of CREB and ATF-2 proteins did not change significantly during this treatment (Figure 8A).
Figure 8.
Activation of the cyclin D1 CRE by TGFβ and PTHrP is mediated by ATF-2 and CREB. (A) RCS cells were serum-starved for 3 d and stimulated for 8 h with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both. Cells were harvested for the analyses of protein expression of ATF-2 and of phosphorylated forms of ATF-2 and CREB. (B) The plasmids pFA-ATF-2 and pFA-CREB (encoding Gal4-ATF-2 and Gal4-CREB fusion proteins, respectively) were transfected into RCS cells together with the Gal4 reporter plasmid pFRluc and pRlSV40. After transfection, cells were serum-starved for 3 d and stimulated with control medium, TGFβ (1 ng/ml), or PTHrP (10−8 M) for 8 h. Cells were harvested, firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity. (C) The plasmid −1745 CD1Luc was transfected into RCS cells together with pRlSV40 and empty expression vector or expression vectors for dominant-negative forms of ATF-2 and CREB. After transfection, cells were serum-starved for 3 d and stimulated with control medium, TGFβ (1 ng/ml), PTHrP (10−8 M), or both for 8 h. Cells were harvested, and firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity.
We measured the transcriptional activation of a reporter gene under the control of Gal4-binding sites with the use of fusion proteins containing the Gal4 DNA-binding domain and transcriptional activation domains of either ATF-2 or CREB in order to correlate growth factor–induced phosphorylation of these transcription factors with their activity. TGFβ treatment of RCS cells for 8 h enhanced ATF-2 activity 11-fold, with very little activation of CREB (Figure 8B). In contrast, PTHrP caused a 9.5-fold increase in CREB activity with only marginal effects on ATF-2.
We also showed that dominant-negative forms of ATF-2 and CREB could block the activation of the cyclin D1 promoter by TGFβ and PTHrP. Expression vectors for dominant-negative ATF-2 (Beier et al., 1999b) and CREB (Ahn et al., 1998) were cotransfected with −1745 CD1Luc into RCS cells, followed by 3 d of serum starvation before stimulation with TGFβ and PTHrP for 8 h. Dominant-negative ATF-2 caused a 72% reduction in promoter activation by TGFβ and a 28% reduction in activation by PTHrP (Figure 8C). In contrast, dominant-negative CREB caused an 84% reduction in promoter activation by PTHrP and a 17% reduction in activation by TGFβ. The observed mild cross-inhibition by the dominant-negative constructs might reflect a true but minor requirement for ATF-2 in PTHrP signaling and for CREB in TGFβ signaling. However, these effects may also be due to slight nonspecific actions caused by ectopic expression of these dominant-negative plasmids.
ATF-2–deficient Chondrocytes Display a Reduced Proliferative Response to TGFβ
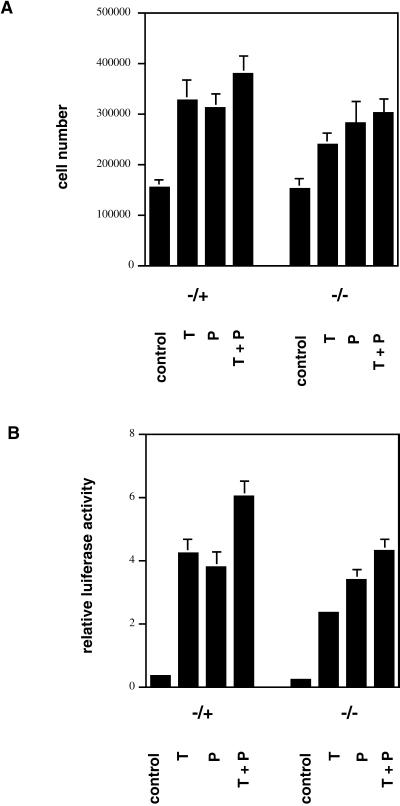
We made use of chondrocytes isolated from ATF-2–deficient mice (Reimold et al., 1996) to investigate the role of this transcription factor in the proliferative response to TGFβ and PTHrP. Primary chondrocytes from heterozygous (−/+) and homozygous (−/−) ATF-2–deficient mice were incubated for 3 d in the absence of serum with or without TGFβ, PTHrP, or both. Both growth factors caused proliferative responses in (−/+) cells that were similar to those observed in wild-type chondrocytes (compare to Figure 1B), whereas the response to TGFβ and the combination was clearly reduced in (−/−) cells. In contrast, the response to PTHrP was nearly normal in (−/−) cells (Figure 9A).
Figure 9.
ATF-2 is necessary for the proliferative response of chondrocytes to TGFβ. (A) Chondrocytes from heterozygous (−/+) or homozygous (−/−) ATF-2–deficient mice were plated in 24-well plates (105 cells/well), stimulated with control medium, TGFβ (1 ng/ml; T), PTHrP (10−8 M; P), or both (T+P) for 3 d, and counted with the use of a hemacytometer. (B) The plasmid −1745 CD1Luc was transfected into primary chondrocytes from heterozygous (−/+) or homozygous (−/−) ATF-2–deficient mice, together with pRlSV40. After transfection, cells were serum-starved for 3 d and stimulated with control medium, TGFβ (1 ng/ml; T), PTHrP (10−8 M; P), or both (T+P) for 8 h. Cells were harvested, firefly luciferase activity was measured and standardized to Renilla luciferase activity to yield the relative luciferase activity.
The role of ATF-2 in transcriptional activation of the cyclin D1 promoter by TGFβ and PTHrP was determined by transfecting the plasmid −1745 CD1Luc into primary chondrocytes isolated from heterozygous and homozygous ATF-2–deficient mice. The response of −1745 CD1Luc to TGFβ was reduced by 55% in ATF-2 null chondrocytes (Figure 9B). The response to PTHrP was not changed significantly, whereas transcriptional activation by the combination of growth factors was reduced by 25%. These data demonstrate the importance of ATF-2 in the TGFβ response but not in the PTHrP response.
DISCUSSION
The growth plate phenotype observed in cyclin D1–deficient mice resembles that of PTHrP-deficient mice (Amizuka et al., 1994; Karaplis et al., 1994), of mice deficient for the Smad3 gene, which is involved in TGFβ signal (Yang et al., 2001) and of transgenic mice overexpressing a dominant-negative TGFβ-receptor (Serra et al., 1997). We therefore postulated that the cyclin D1 gene might be a target of these growth factors in skeletal growth. In vivo and in vitro data suggest that both PTHrP and TGFβ are necessary (although probably not sufficient) for normal proliferation of chondrocytes. Evidence for antiproliferative action of TGFβ on chondrocytes in transgenic mice has been presented by Serra et al. (1999), but this group has recently shown that the antiproliferative effect of TGFβ on chondrocytes is likely indirect, mediated by the perichondrium, whereas the direct (paracrine or autocrine) effects of TGFβ on chondrocytes are mitogenic (R. Serra, personal communication). More recently, Yang and coworkers (2001) have shown that interruption of TGFβ signaling through targeted mutagenesis of the Smad3 gene leads to a reduction in the number of proliferating chondrocytes in the growth plate. In summary, the bulk of published evidence as well as the data presented here support a role for TGFβ as a stimulator of chondrocyte proliferation and inhibitor of differentiation (O'Keefe et al., 1988; Battegay et al., 1990; Ballock et al., 1993; Guerne et al., 1994; Bohme et al., 1995). It therefore appears that TGFβ has very complex effects on chondrocyte physiology, depending, for example, on the concentration of growth factor, site of action, differentiation status of chondrocytes, or interplay with other signaling molecules. Further in vivo and in vitro studies will be necessary for the complete elucidation of these complex actions of TGFβ in chondrocytes.
It has recently been proposed that some of the actions of TGFβ may be achieved through the control of PTHrP expression (Serra et al., 1999). Because both growth factors activate cyclin D1 expression with similar kinetics, this scenario is not very likely in this case; it appears rather that both signals converge on the cyclin D1 promoter.
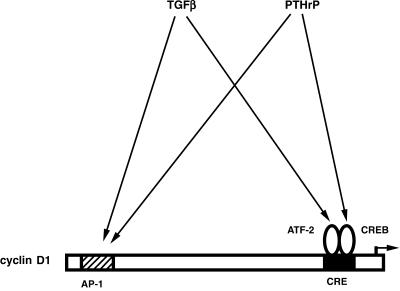
Because the time course of cyclin D1 protein and promoter induction are very similar, it is likely that cyclin D1 induction by TGFβ and PTHrP occurs primarily at the level of transcription. Our data demonstrate that the AP-1 and CRE sites are required for induction of the cyclin D1 promoter by TGFβ and PTHrP (summarized in Figure 10). Kinetic analyses of promoter induction suggests that activation of the cyclin D1 promoter by these growth factors is a biphasic process, with the CRE conferring the first phase of induction and the AP-1 site responsible for the second phase. This is consistent with the presence of ATF-2 and CREB proteins in unstimulated cells; activation of these transcription factors by phosphorylation can cause a relatively rapid induction of CRE activity. In contrast, AP-1 proteins are present only at very low levels in unstimulated cells (Ionescu et al., 2001; Beier and LuValle, unpublished observations). Activation of AP-1 activity is therefore likely to require synthesis of AP-1 proteins and occurs later than activation of the CRE. For example, both TGFβ and signaling through the PTH/PTHrP receptor have been shown to induce the expression of c-Fos in chondrocytes (Lee et al., 1994; Osaki et al., 1999). In addition, TGFβ is able to induce AP-1 activity directly through the stimulation of the MAP kinase JNK (c-Jun-N-terminal kinase) in other cell types, such as fibrosarcoma cells (Hocevar et al., 1999).
Figure 10.
Model of regulation of the cyclin D1 promoter by PTHrP and TGFβ. Both growth factors induce cyclin D1 promoter activity through the AP-1 and CRE elements. TGFβ effects require the transcription factor ATF-2, whereas the effects of PTHrP are mediated by the related transcriptional regulator CREB.
We conclude that induction of the cyclin D1 CRE by TGFβ and PTHrP is mediated by the transcription factors ATF-2 and CREB, respectively. In vivo models clearly demonstrate the importance of both of these transcription factors in endochondral ossification (Reimold et al., 1996; Long et al., 2001). Our data clearly show that the response to TGFβ is mediated primarily by the transcription factor ATF-2, whereas the related transcription factor CREB confers promoter stimulation largely in response to PTHrP. Transcription factors of the ATF/CREB families are generally regulated at the level of phosphorylation, rather than through DNA-binding (Montminy, 1997). In addition, it has been shown recently that PTHrP does not affect DNA-binding of CREB in chondrocytes. Rather, PTHrP stimulates the activity of CREB that is already bound to its DNA response elements, thereby inducing transcriptional activation (Ionescu et al., 2001). We therefore focused our analyses of ATF-2 and CREB regulation by PTHrP and TGFβ on their phosphorylation status and activity. ATF-2 activity is reported to be controlled by the MAP kinases JNK and p38 (Gupta et al., 1995, 1996; Livingstone et al., 1995; Raingeaud et al., 1996). Both of these kinases have recently been shown to be regulated by TGFβ, in both Smad-dependent and -independent pathways (Hanafusa et al., 1999; Hocevar et al., 1999; Sano et al., 1999). We are in the process of analyzing the signaling mechanism(s) connecting TGFβ and ATF-2. The effects of PTHrP on chondrocytes are thought to be mediated by protein kinase A (Schipani et al., 1995; Schipani et al., 1997). CREB can be a direct substrate of protein kinase A (Gonzalez and Montminy, 1989), but it can also be targeted by other kinases such as Akt/protein kinase B (Du and Montminy, 1998) or p38-dependent pathways (Tan et al., 1996; Iordanov et al., 1997; Pierrat et al., 1998). We are currently examining these possibilities.
Long et al. (2001) have shown that overexpression of a dominant-negative form of CREB in chondrocytes of transgenic mice causes skeletal deformities, perinatal death, and reduced proliferation of chondrocytes, consistent with the role of CREB in chondrocyte proliferation as suggested here. These authors did not find a change in cyclin D1 expression in the growth plates of transgenic animals by in situ hybridization. However, we suggest that CREB is only one of a number of transcription factors controlling cyclin D1 expression in chondrocytes and that disruption of CREB function would lead to a decrease in but not a complete block from cyclin D1 expression in chondrocytes. Such quantitative changes cannot be shown clearly by in situ hybridization. Quantitative comparison of cyclin D1 expression between chondrocytes from wild-type and dominant-negative CREB-expressing mice, as done by us with ATF-2–deficient chondrocytes (Beier et al., 1999b), will be necessary to resolve this issue.
Our data demonstrate that cyclin D1 is necessary for normal chondrocyte proliferation in vivo and in vitro. Inhibition of cyclin D1 expression in vitro reduces the proliferative response of chondrocytes to mitogenic stimulation through a delay in the G1 phase of the cell cycle and reduced E2F activity. These findings are supported in vivo by the dwarfism and the reduced thickness of the proliferative zone of the growth plate in cyclin D1–deficient mice. These results suggest that the function of cyclin D1 in chondrocytes is similar to that in most other cell types, the stimulation of G1 progression and proliferation. However, nonproliferative roles of cyclin D1 and other D-type cyclins have been reported in a number of systems, for example, during PC12 cell differentiation (Yan and Ziff, 1995). Therefore it was necessary to establish the exact function of cyclin D1 in chondrocytes. It should be noted, however, that proliferation of chondrocytes is not completely blocked in both models, suggesting redundant mechanisms. In agreement with this, we have observed expression of both cyclin D2 and cyclin D3 in primary chondrocytes (Ali, Beier, and LuValle, unpublished observations). Similarly, the remaining activity of the cyclin A promoter in the absence of cyclin D1 expression is likely caused by both parallel pathways (e.g., cyclin D2- or D3-dependent) and by direct, E2F-independent promoter activation, for example, through the cyclin A CRE (Beier et al., 2000).
Our results suggest that the cyclin D1 gene acts as an integrator of different mitogenic stimuli in chondrocytes. It will be of great interest to determine whether additional positive or negative regulators of chondrocyte proliferation will also control the expression of cyclin D1, given that our data, while supporting our conclusions that activation of cyclin D1 by TGFβ and PTHrP is necessary for chondrocyte proliferation, do not demonstrate that it is sufficient. In addition, the elucidation of the complete signaling pathways from receptors to transcription factors and the identification of other targets of TGFβ and PTHrP in chondrocytes will significantly enhance our understanding of skeletal development and diseases.
ACKNOWLEDGMENTS
We are grateful to B. de Crombrugghe and V. Lefebvre for RCS cells, C. Vinson for the dominant-negative CREB expression vector, P. Sicinski for cyclin D1–deficient mice, and L. Glimcher and A. Reimold for ATF-2–deficient mice. We also thank Ruth Seerattan for technical support and Todd Barnash for digital artwork. This work was supported by grants from the Medical Research Council of Canada, the Alberta Cancer Foundation, the Alberta Heritage Foundation for Medical Research, and the Arthritis Society to P.L., and by R01CA70897, R01CA75503, and 5-P30-CA13330–26 (to R.G.P.). F.B. was supported by postdoctoral fellowships from Deutscher Akademischer Austauschdienst, Deutsche Forschungsgemeinschaft, and the Alberta Heritage Foundation for Medical Research.
Abbreviations used:
- AP-1
activator protein 1
- ATF-2
activating transcription factor 2
- CDK
cyclin-dependent kinase
- CRE
cyclic AMP response element
- CREB
CRE-binding protein
- PTHrP
parathyroid hormone–related peptide
- RCS
rat chondrosarcoma
- TGFβ
transforming growth factor beta
REFERENCES
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Amizuka N, Warshawsky H, Henderson JE, Goltzman D, Karaplis AC. Parathyroid hormone-related peptide-depleted mice show abnormal epiphyseal cartilage development and altered endochondral bone formation. J Cell Biol. 1994;126:1611–1623. doi: 10.1083/jcb.126.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballock RT, Heydemann A, Wakefield LM, Flanders KC, Roberts AB, Sporn MB. TGF-beta 1 prevents hypertrophy of epiphyseal chondrocytes: regulation of gene expression for cartilage matrix proteins and metalloproteases. Dev Biol. 1993;158:414–429. doi: 10.1006/dbio.1993.1200. [DOI] [PubMed] [Google Scholar]
- Battegay EJ, Raines EW, Seifert RA, Bowen-Pope DF, Ross R. TGF-beta induces bimodal proliferation of connective tissue cells via complex control of an autocrine PDGF loop. Cell. 1990;63:515–524. doi: 10.1016/0092-8674(90)90448-n. [DOI] [PubMed] [Google Scholar]
- Beier F, Leask TA, Haque S, Chow C, Taylor AC, Lee RJ, Pestell RG, Ballock RT, LuValle P. Cell cycle genes in chondrocyte proliferation and differentiation. Matrix Biol. 1999a;18:109–120. doi: 10.1016/s0945-053x(99)00009-8. [DOI] [PubMed] [Google Scholar]
- Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc Natl Acad Sci USA. 1999b;96:1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier F, Taylor AC, LuValle P. The Raf-1/MEK/ERK pathway regulates the expression of the p21(Cip1/Waf1) gene in chondrocytes. J Biol Chem. 1999c;274:30273–30279. doi: 10.1074/jbc.274.42.30273. [DOI] [PubMed] [Google Scholar]
- Beier F, Taylor AC, LuValle P. Activating transcription factor 2 is necessary for maximal activity and serum induction of the cyclin A promoter in chondrocytes. J Biol Chem. 2000;275:12948–12953. doi: 10.1074/jbc.275.17.12948. [DOI] [PubMed] [Google Scholar]
- Bohme K, Winterhalter KH, Bruckner P. Terminal differentiation of chondrocytes in culture is a spontaneous process and is arrested by transforming growth factor-beta 2 and basic fibroblast growth factor in synergy. Exp Cell Res. 1995;216:191–198. doi: 10.1006/excr.1995.1024. [DOI] [PubMed] [Google Scholar]
- Cancedda R, Descalzi Cancedda F, Castagnola P. Chondrocyte differentiation. Int Rev Cytol. 1995;159:265–358. doi: 10.1016/s0074-7696(08)62109-9. [DOI] [PubMed] [Google Scholar]
- Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Guerne PA, Lotz M. Interleukin-6 and transforming growth factor-beta synergistically stimulate chondrosarcoma cell proliferation. J Cell Physiol. 1991;149:117–124. doi: 10.1002/jcp.1041490115. [DOI] [PubMed] [Google Scholar]
- Guerne PA, Sublet A, Lotz M. Growth factor responsiveness of human articular chondrocytes: distinct profiles in primary chondrocytes, subcultured chondrocytes, and fibroblasts. J Cell Physiol. 1994;158:476–484. doi: 10.1002/jcp.1041580312. [DOI] [PubMed] [Google Scholar]
- Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Campbell D, Derijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Ninomiya-Tsuji J, Masuyama N, Nishita M, Fujisawa J, Shibuya H, Matsumoto K, Nishida E. Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-beta-induced gene expression. J Biol Chem. 1999;274:27161–27167. doi: 10.1074/jbc.274.38.27161. [DOI] [PubMed] [Google Scholar]
- Hocevar BA, Brown TL, Howe PH. TGF-beta induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. EMBO J. 1999;18:1345–1356. doi: 10.1093/emboj/18.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu AM, Schwarz EM, Vinson C, Puzas JE, Rosier RN, Reynolds PR, O'Keefe RJ. PTHrP modulates chondrocyte differentiation through AP-1 and CREB signaling. J Biol Chem. 2001;2:2. doi: 10.1074/jbc.M006564200. [DOI] [PubMed] [Google Scholar]
- Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf HJ, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, Mulligan RC. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994;8:277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- Ko TC, Yu W, Sakai T, Sheng H, Shao J, Beauchamp RD, Thompson EA. TGF-beta1 effects on proliferation of rat intestinal epithelial cells are due to inhibition of cyclin D1 expression. Oncogene. 1998;16:3445–3454. doi: 10.1038/sj.onc.1201902. [DOI] [PubMed] [Google Scholar]
- Lee K, Deeds JD, Chiba S, Un-No M, Bond AT, Segre GV. Parathyroid hormone induces sequential c-fos expression in bone cells in vivo: in situ localization of its receptor and c-fos messenger ribonucleic acids. Endocrinology. 1994;134:441–450. doi: 10.1210/endo.134.1.8275957. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, De Crombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 1994;14:329–335. doi: 10.1016/0945-053x(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Schipani E, Asahara H, Kronenberg H, Montminy M. The CREB family of activators is required for endochondral bone development. Development. 2001;128:541–550. doi: 10.1242/dev.128.4.541. [DOI] [PubMed] [Google Scholar]
- Loveys LS, Gelb D, Hurwitz SR, Puzas JE, Rosier RN. Effects of parathyroid hormone-related peptide on chick growth plate chondrocytes. J Orthop Res. 1993;11:884–891. doi: 10.1002/jor.1100110615. [DOI] [PubMed] [Google Scholar]
- LuValle P, Beier F. Cell cycle control in growth plate chondrocytes. Front Biosci. 2000;5:D493–D503. doi: 10.2741/luvalle. [DOI] [PubMed] [Google Scholar]
- Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay K, Lefebvre V, Zhou G, Garofalo S, Kimura JH, de Crombrugghe B. Use of a new rat chondrosarcoma cell line to delineate a 119-base pair chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-alpha 1(II) collagen gene. J Biol Chem. 1995;270:27711–27719. doi: 10.1074/jbc.270.46.27711. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Olsen BR. Heritable diseases of the skeleton. Part I: Molecular insights into skeletal development-transcription factors and signaling pathways. FASEB J. 1997a;11:125–132. doi: 10.1096/fasebj.11.2.9039954. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Olsen BR. Heritable diseases of the skeleton. Part II: Molecular insights into skeletal development-matrix components and their homeostasis. FASEB J. 1997b;11:227–233. [PubMed] [Google Scholar]
- O'Keefe RJ, Puzas JE, Brand JS, Rosier RN. Effect of transforming growth factor-beta on DNA synthesis by growth plate chondrocytes: modulation by factors present in serum. Calcif Tissue Int. 1988;43:352–358. doi: 10.1007/BF02553278. [DOI] [PubMed] [Google Scholar]
- Osaki M, Tsukazaki T, Yonekura A, Miyazaki Y, Iwasaki K, Shindo H, Yamashita S. Regulation of c-fos gene induction and mitogenic effect of transforming growth factor-beta1 in rat articular chondrocyte. Endocr J. 1999;46:253–261. doi: 10.1507/endocrj.46.253. [DOI] [PubMed] [Google Scholar]
- Pierrat B, Correia JS, Mary JL, Tomas-Zuber M, Lesslauer W. RSK-B, a novel ribosomal S6 kinase family member, is a CREB kinase under dominant control of p38alpha mitogen-activated protein kinase (p38alphaMAPK) J Biol Chem. 1998;273:29661–29671. doi: 10.1074/jbc.273.45.29661. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Grusby MJ, Kosaras B, Fries JW, Mori R, Maniwa S, Clauss IM, Collins T, Sidman RL, Glimcher MJ, Glimcher LH. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature. 1996;379:262–265. doi: 10.1038/379262a0. [DOI] [PubMed] [Google Scholar]
- Rosier RN, O'Keefe RJ, Crabb ID, Puzas JE. Transforming growth factor beta: an autocrine regulator of chondrocytes. Connect Tissue Res. 1989;20:295–301. doi: 10.3109/03008208909023900. [DOI] [PubMed] [Google Scholar]
- Rudolph D, Tafuri A, Gass P, Hammerling GJ, Arnold B, Schutz G. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc Natl Acad Sci USA. 1998;95:4481–4486. doi: 10.1073/pnas.95.8.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y, Harada J, Tashiro S, Gotoh-Mandeville R, Maekawa T, Ishii S. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-beta signaling. J Biol Chem. 1999;274:8949–8957. doi: 10.1074/jbc.274.13.8949. [DOI] [PubMed] [Google Scholar]
- Schipani E, Jensen GS, Pincus J, Nissenson RA, Gardella TJ, Juppner H. Constitutive activation of the cyclic adenosine 3′,5′-monophosphate signaling pathway by parathyroid hormone (PTH)/PTH-related peptide receptors mutated at the two loci for Jansen's metaphyseal chondrodysplasia. Mol Endocrinol. 1997;11:851–858. doi: 10.1210/mend.11.7.9934. [DOI] [PubMed] [Google Scholar]
- Schipani E, Kruse K, Juppner H. A constitutively active mutant PTH-PTHrP receptor in Jansen-type metaphyseal chondrodysplasia. Science. 1995;268:98–100. doi: 10.1126/science.7701349. [DOI] [PubMed] [Google Scholar]
- Schipani E, Langman C, Hunzelman J, Le Merrer M, Loke KY, Dillon MJ, Silve C, Juppner H. A novel parathyroid hormone (PTH)/PTH-related peptide receptor mutation in Jansen's metaphyseal chondrodysplasia. J Clin Endocrinol Metab. 1999;84:3052–3057. doi: 10.1210/jcem.84.9.6000. [DOI] [PubMed] [Google Scholar]
- Serra R, Johnson M, Filvaroff EH, LaBorde J, Sheehan DM, Derynck R, Moses HL. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. J Cell Biol. 1997;139:541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra R, Karaplis A, Sohn P. Parathyroid hormone-related peptide (PTHrP)-dependent and -independent effects of transforming growth factor beta (TGF-beta) on endochondral bone formation. J Cell Biol. 1999;145:783–794. doi: 10.1083/jcb.145.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Nomura Y, Suzuki H, Ichikawa E, Takeuchi A, Suzuki M, Nakamura T, Nakajima T, Oda K. Activation of the rat cyclin A promoter by ATF2 and Jun family members and its suppression by ATF4. Exp Cell Res. 1998;239:93–103. doi: 10.1006/excr.1997.3884. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ, Weinberg RA. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb MJ. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Yan GZ, Ziff EB. NGF regulates the PC12 cell cycle machinery through specific inhibition of the Cdk kinases and induction of cyclin D1. J Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]