Abstract
Increasing evidence supports the involvement of a catalytic subunit (PP2Ac) of protein phosphatase 2A (PP2A) in the mechanisms of systemic lupus erythematosus (SLE). This study was conducted to explore the association single nucleotide polymorphisms (SNPs) of PPP2CA with SLE susceptibility, serum cytokines levels, and clinical features in a Chinese Han population. A case–control association study was carried out in 1509 Chinese Han subjects (730 SLE patients and 779 healthy individuals). Genotyping for genetic variants of PPP2CA (rs10491322 and rs7704116) was performed using a polymerase chain reaction-high resolution melting (PCR-HRM) assay. In the cohort of SLE patients, we observed that rs10491322 and rs7704116 were positively increased SLE susceptibility (OR = 1.61, 95% CI = 1.13–2.31, P = .009; OR = 1.59, 95% CI = 1.17-2.15, P = .003, respectively). Interestingly, the AG genotype of rs10491322 carriers presented higher IL-6 (P < .001) and IL-17 (P < .001) than those with AA genotype carriers. Specifically, carriage of the rs10491322 G∗ allele led to a higher prevalence of arthritis in SLE patients (P = .01). This study demonstrated an association of PPP2CA (rs10491322 and rs7704116) with SLE susceptibility in a Chinese Han population. Furthermore, the minor allele of PPP2CA rs10491322 as a risk factor was correlated with immunologic disorders for SLE.
Keywords: polymorphism, PP2Ac, PPP2CA, systemic lupus erythematosus
1. Introduction
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disorder with a broad spectrum of clinical manifestations and multiple immunological abnormalities, which results from environmental factors interacting with genetic susceptibility variants.[1] T cells abnormalities in SLE patients are considered to have a pivotal role in the pathogenesis of SLE via contributing to the activation of autoreactive proinflammatory lymphocytes.[2]
Protein phosphatase 2A (PP2A) is a major serine/threonine phosphatase. It has been proved that PP2A negatively regulates nuclear factor-κB (NF-κB), mitogen-activated protein kinase (MAPK) and Wingless-type (Wnt) signal transduction pathways. These pathways are involved in many cellular processes, including cell cycle progression, DNA replication, gene transcription/translation, and cell differentiation.[3–5]
Considering that SLE-associated T cells abnormalities contribute to SLE by facilitating the loss of immune tolerance and activation of autoreactive proinflammatory lymphocytes,[2] PP2A has central roles in the pathogenesis of SLE via affecting the function of T cell. One of the SLE signaling defects is the Fc receptor γ chain (FcRγ) structurally and functionally replaces the T cell receptor (TCR)-associated cluster of differentiation 3ζ (CD3ζ) chain.[6] Another hallmark dysregulation of SLE T cells is a reduction in their ability to produce interleukin-2 (IL-2), an essential cytokine for T cell proliferation and effector functions.[7]
PP2A is composed with 3 subunits: a scaffold subunit A, a regulatory subunit B and a catalytic subunit C. The scaffold (subunit A) and catalytic (subunit C) proteins form a heterodimeric core that can associate with various regulatory (B) subunits, only the catalytic subunit C has serine/threonine phosphatase activity.[7] Gene PPP2CA encoded the catalytic subunit (PP2Acα) of PP2A. It has been confirmed that T cells in the SLE patients express abnormally high level of messenger RNA (mRNA), protein and enzymatic activity of a catalytic subunit of PP2Ac.[7] This enhanced expression of PP2Ac is one of the factors contributing to the molecular defects in SLE. PP2Ac facilitates the transcription of proinflammatory genes linked to T cell and promotes the hypomethylation of inflammatory cytokines DNA on the mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway, such as interleukin-6 (IL-6) and interleukin-17 (IL-17).[8–11] Elevated interleukins cause aberrant immunoregulation responses in SLE. Recently, a genome wide association study (GWAS) had revealed that disease-associated single nucleotide polymorphisms (SNPs) rs10491322 and rs7704116 in PPP2CA not only associated with susceptibility and renal disease of SLE but also had correlation with the expression of mRNA of PP2Ac in T cell from SLE patients.[12] However, there are few studies mentioning whether the genetic variants in PPP2CA had effect on the levels of cytokines and clinical manifestations.
In the present study, we aimed to investigate the associations between the 2 SNPs in PPP2CA (rs10491322 at 3′-UTR and rs7704116 at intron 1) and the susceptibility of SLE, the levels of specific serum cytokines and certain clinical manifestations in a southwest Chinese SLE population.
2. Materials and methods
2.1. Patients and protocol
In total, 1509 Chinese Han subjects were recruited in the present study, including 730 SLE patients and 779 healthy controls from West China Hospital during 2013 to 2016. The SLE patients should meet the following criteria: diagnosed as SLE in compliance with the American College of Rheumatology (ACR) classification criteria for SLE revised in 1997.[13] The patients of SLE were all in the active stage with SLE disease activity index (SLEDAI) > 4. Clinical features of the patients could be collected completely at the same time. Clinical data on the cases were obtained from medical records. Hospitalized patients without drug-induced SLE. The healthy controls should meet these inclusion criteria: without any chronic, endemic infectious or any type of autoimmune disorders and with normal physical examination and blood tests.
This study was conducted according with the 1975 Declaration of Helsinki and was approved by the Ethics Committee of West China Hospital. All the patients have signed the informed consent to participate in this study and consented to sample collection.
2.2. Serological testing and clinical features collection
Laboratory assays of SLE were analyzed by the following methods: autoantibodies (including anti-ANA, -dsDNA, -Sm, -U1RNP, -SSA/Ro, -SSB/La, and -Rib-P antibodies) were tested by using ANA, anti-dsDNA, and ENA reagent kits from Euroimmun (Euroimmun, Lübeck, Germany), respectively. Complement 3 (C3) and complement 4 (C4) was tested in Beckman Coulter IMMAGE 800 immunoassay (Beckman Coulter, Inc. Brea). Serum urea, creatinine, cystatin C, and uric acid were tested in Roche cobas c702 system (Roche Diagnostics, Basel, Switzerland). All the tests were conducted according to manufacturers’ instruction. Clinical characteristics as disease parameters of the ACR criteria were collected and analyzed. Serum laboratory investigations of SLE patients were assessed at the same time during the clinical examination and assessment of SLE disease activity.
2.3. Polymorphism genotyping
Subjects were genotyped at the rs10491322 and rs7704116 SNPs in PPP2CA using the polymerase chain reaction-high resolution melting (PCR-HRM) methods. Light Cycler 480 (Roche Diagnostics, Bavaria, Germany) was used to perform the analysis. After the free circulating DNA was extracted from the peripheral blood by Genomic DNA kit (Biotake Corporation, Beijing, China), its concentration was measured by Nanodrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE). SNP genotyping was performed in a 20 μL reaction system. The whole genotyping process encompasses 4 programs, namely, predenaturation, amplification, high resolution melting, and cooling. When finished, the results were analyzed by the corresponding Gene Scanning Software v1.2 (Roche Diagnostics, Bavaria, Germany) primarily based on the shape of the melting curve.
2.4. Cytokine measurement
The serum concentrations of cytokines including IL-1β, IL-6, IL-10, IL-17, and IFN-γ were measured for randomly 161 SLE patients. The cytokines were quantitatively determined by using R&D Human Inflammation Assays. Procedures were performed according to the manufacturer's instructions. Blood sampling for assessing serum cytokines levels were performed at the same time of clinical examination and assessment of SLE disease activity.
2.5. Statistical analysis
Hardy–Weinberg equilibrium (HWE) was evaluated for each polymorphism independently. Statistical power was evaluated by a software “PS: Power and Sample size Calculation” (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize). Mean ± SD, median, and interquartile were used to describe the continuous variables with normal and skewed distribution respectively. Student t test or Mann–Whitney U test were used to compare demographic and clinical data between subgroups as appropriate. Allele case–control comparisons and qualitative data were analyzed by Pearson Chi-square test. Association of SNPs with susceptibility of SLE was calculated by figuring out the odds ratio (OR) and 95% confidence interval (95% CI). The association of SNPs with clinical manifestations was determined by χ2 test. All statistical analyses were used applying the Statistical Package for the Social Sciences (SPSS, SPSS, Inc., Chicago, IL), version 20.0. A 2-sided P-value < .05 was considered as statistically significant.
3. Results
3.1. Clinical features of recruited SLE patients
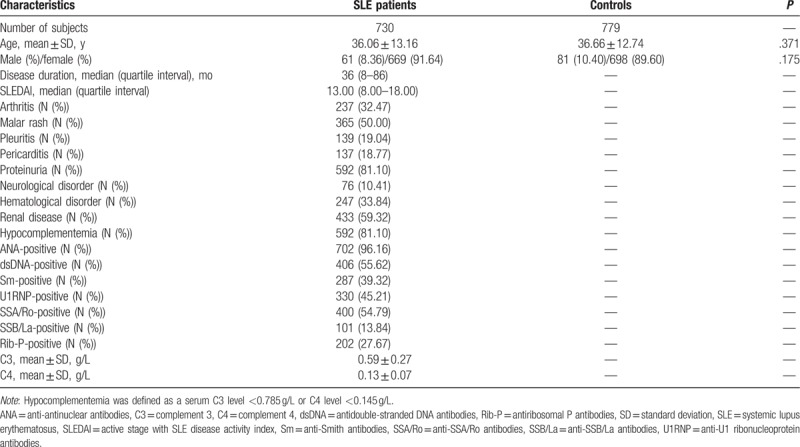
In this study, a total of 730 SLE patients (36.06 ± 13.16 years old) and 779 health controls (36.66 ± 12.74 years old) were recruited in this case–control study. The distribution of demographic features and clinical manifestations in the cases and controls are described in Table 1. The age and gender of participants from case–control groups were matched (P = .371 and P = .175, respectively). The median duration of SLE patients was 36 months (quartile interval: 8.00–86.00 months) and median SLEDAI score was 13.00 (quartile interval: 8.00–18.00). In the SLE group, the positive frequency of anti-ANA, -dsDNA, -Sm, -U1RNP, -SSA/Ro, -SSB/La, and -Rib-P antibodies were 96.16%, 55.62%, 39.32%, 45.21%, 54.79%, 13.84%, and 27.67%, respectively. In addition, the average serum C3 concentration was 0.59 ± 0.27 g/L, and average C4 was 0.13 ± 0.07 g/L. The average incidence of lupus nephritis (LN) was 59.32%.
Table 1.
The demographic and clinical characteristics of the subjects recruited in the study.
3.2. Genotyping and LD evaluation
All the included participants were genotyped by PCR-HRM methods for SNPs rs10491322 and rs7704116. To confirm the correctness of results, direct sequencing of PCR amplification products of randomly selected samples were applied and the results were consistent with all the corresponding genotypes results completely. All genotypes were distributed in concordance with the HWE, as determined at the 0.05 significance level.
Linkage disequilibrium evaluation was conducted by Haploview. As Fig. 1 shows rs10491322 and rs7704116 were not in strong linkage disequilibrium (r2 = 0.40) in a southwest Chinese population.
Figure 1.

Linkage disequilibrium for rs10491322 and rs7704116 of PPP2CA. The linkage disequilibrium plot shows r2 values between rs10491322 and rs7704116. There was not strong LD between these 2 polymorphisms (D′ = 0.64, r2 = 0.40).
3.3. Allele and genotype frequencies of SNPs in patients and controls in the study
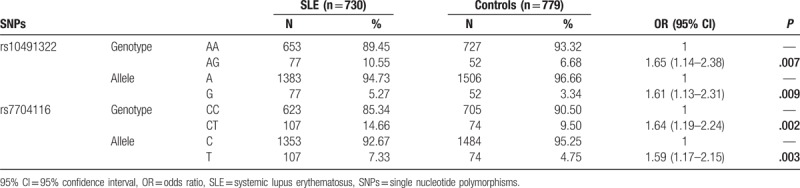
As Table 2 shows there were significant positive differences between the SLE patients group and the health controls. In this study, PPP2CA rs10491322 polymorphism was significantly associated with SLE susceptibility (OR = 1.61, 95% CI = 1.13–2.31, P = .009, statistical power = 0.899; AG-genotype vs AA-genotype, OR = 1.65, 95% CI = 1.14–2.38, P = .007). SNP rs7704116 was also significantly correlated with SLE susceptibility (OR = 1.59, 95% CI = 1.17–2.15, P = .003, statistical power = 0.954; CT-genotype vs CC-genotype, OR = 1.64, 95% CI = 1.19–2.24, P = .002).
Table 2.
Genotype distributions of PPP2CA in SLE patients and controls in Chinese Han population.
3.4. Association analysis of variants in PPP2CA with cytokines levels and clinical features
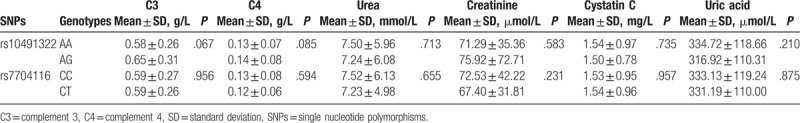
Analysis of the association of the SLE susceptibility loci with particular cytokines and disease manifestations are depicted in Tables 3 and 4. In this group of patients, PPP2CA rs10491322 was significantly associated with IL-6 (P < .001) and IL-17 (P < .001). Those heterozygous AG genotype carriers had higher serum IL-6 and IL-17 levels than those homozygous AA genotype carriers. In clinical manifestations, rs10491322 and rs7704116 were not associated with serum clinical parameters in Table 4.
Table 3.
Association between PPP2CA polymorphisms and cytokines in SLE patients.
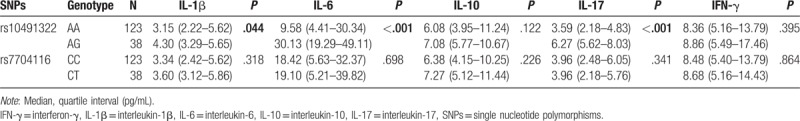
Table 4.
Associations between PPP2CA polymorphisms and clinical characteristics in SLE patients.
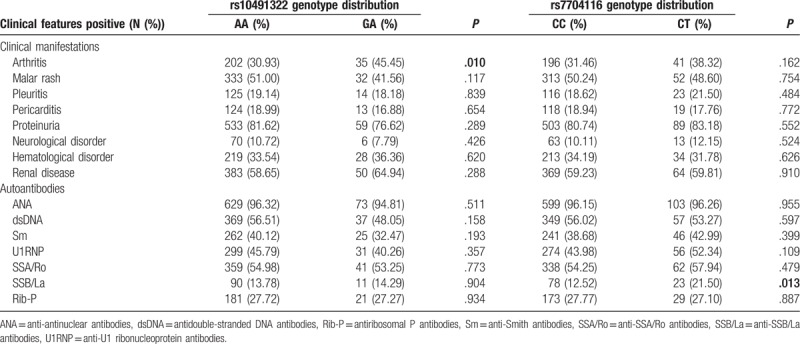
We next investigated whether these 2 genotyped SNPs were correlated with clinical manifestations of SLE. A significant difference could be observed between rs10491322 and arthritis (P = .01) (Table 5). The SLE patients carrying G allele of rs10491322 had higher frequency of arthritis compared with A allele carriers (AG: 45.45% vs AA: 30.93%). We also found rs7704116 was significantly associated with anti-SSB/La autoantibody in clinical ACR criteria (P = .013). Patients carrying T allele of rs7704116 had higher frequency of anti-SSB/La compared with C allele carriers (CT: 21.50% vs CC: 12.52%).
Table 5.
Frequencies of rs10491322 and rs7704116 genotypes with various clinical features.
4. Discussion
SLE is one of the most heterogeneous complex autoimmune diseases. It involves dysregulation of T and B cells, leading to overproduction of autoantibodies of self proteins and DNA, overproduction of inflammatory cytokines, formation of immune complexes, and infiltration of T cells into tissues.[14,15] These processes cause a variety of symptoms including arthritis, hematological disorder, proteinuria, and renal disease. In part because of the heterogeneity of the disease, SLE has been elusive. Most cases are caused by genetic susceptibility and environmental factors. Altered gene expression is usually accompanied by changes of SLE T cell functions.[16,17]
Indeed, some recent studies have shown that PP2Ac could operate as an essential regulatory mechanism factor to systemic inflammatory responses in T cells from SLE patients. SLE patients have higher levels of PP2Ac in T cells. This enhanced expression and activity translates into defects in T cell functions, such as reduced CD3ζ expression and decreased production of IL-2. Recent studies have shown that higher levels of PP2Ac may be an important factor causing hypomethylation of DNA in SLE patients.[7–11]
Moreover, in current years, several specific SNPs have been reported to be associated with susceptibility and pathogenesis of SLE in multiple populations.[18,19] A recent study reported that variant alleles of PPP2CA significantly associated with SLE susceptibility.[12] PPP2CA encoded the catalytic subunit (PP2Acα) of PP2A. PPP2CA is composed of 7 exons and 6 introns localized in chromosome 5q23–q31. The polymorphisms of PPP2CA (rs10491322 at 3′-UTR and rs7704116 at intron 1) were revealed as risk factors of SLE in multiple ethnic populations. SNP rs10491322 and rs7704116 showed strong association with SLE across Asian, European American, and Hispanic American populations (OR = 1.2, 95% CI = 1.1–1.3, meta-analysis P = 3.0 × 10−4 for rs10491322 and OR = 1.3, 95% CI = 1.1–1.3, meta-analysis P = 3.8 × 10−7 for rs7704116, respectively).[12,20] Although PP2Ac expression is clearly a significant aspect controlling the outcome of T cell signaling in SLE, the effects of such genetic variations on the immune responses including the serum cytokine levels and the clinical manifestations in SLE patients are currently unknown.
To our knowledge, it is this study that firstly analyzed the association between SLE susceptible gene PPP2CA and serum cytokines and clinical manifestations in southwest Chinese SLE patients.
The results of our present study indicated that PPP2CA rs10491322 and rs7704116 were significantly associated with higher SLE susceptibility (P = .009 and P = .003, respectively) in the Chinese population, which was consistent with the previous study. In this study, we found that rs10491322 and rs7704116 were not in strong linkage disequilibrium (r2 = 0.40) in southwest Chinese population.
We also found PPP2CA rs10491322 and rs7704116 were significant associated with serum cytokines and clinical manifestations in Chinese SLE population. Furthermore, we observed significant differences in serum IL-6 and IL-17 levels from SLE patients. In specially, SNP rs10491322 AG genotype carriers presented higher IL-6 (P < .001) and IL-17 (P < .001) levels than the patients who carried AA genotype. Moreover, significant positive association was observed between polymorphism rs10491322 and arthritis (P = .01). The patients who carried rs10491322 allele G tended to have higher risk of arthritis. In this study, we observed rs10491322 subgroups with an increase in IL-6, IL-17 expression and arthritis corresponding to the risk “G” alleles. However, the exact mechanisms how PPP2CA rs10491322 G minor allele affect predisposition to serum cytokines and arthritis are still not clear and functional studies did not show an effect of the rs10491322 genotypes on PP2Ac mRNA expression in SLE patients.
A previous functional study showed that individuals who carried the rs7704116 CT genotype had a higher production of PP2Ac mRNA and protein expression than those who carried CC genotype by T cells from SLE patients.[12] The expression of PP2Ac can be repressed by the binding of transcription factor Ikaros to rs7704116 site in the first intron.[20] Higher level of PP2Ac resulted in decreased amounts of IL-2 via dephosphorylation of cAMP-response element binding protein (CREB) and exerted its influence on disease susceptibility via a higher level of IL-6, IL-17 in T cells from SLE patients.[8–10,21] The signature of IL-6, a key cytokine in a proinflammatory immune response, has long been correlated with having detrimental effects in several autoimmune diseases including SLE.[22,23] IL-6 and its receptor might be as the potential therapeutic targets for the treatment of SLE.[24,25]
Our present study found that rs7704116 significantly associated with SLE susceptibility (P = .003), in aspect of autoantibodies, the present study showed that rs7704116 was correlated with anti-SSB/La autoantibody (P = .013). It is not consistent with the study of Tan et al,[12] who found that rs7704116 was associated with renal disease and anti-dsDNA in Asians only when SLE patients were compared to control subjects. Possible explanations for those above findings might be genetic heterogeneity, different sizes of the studied groups, variable clinical features of SLE or patient interaction with different environmental factors.
In conclusion, to our knowledge, it is our present study that firstly explores the effect of functional genetic variants in PPP2CA in a southwest Chinese population. We found that allele G of SNP rs10491322 and allele T of SNP rs7704116 in PPP2CA were risk factors for SLE. Patients who carried rs7704116 allele T significantly associated with risk SLE susceptibility and anti-SSB/La autoantibody. Specifically, the rs10491322 allele G carriers associated with SLE susceptibility, presented higher serum cytokine levels (IL-6 and IL-17) and high risk of arthritis than patient carriers of the wild genotype. However, it is worthwhile to mention that there are some limitations in our present study. The subject of patients in our case–control study is relatively small and only Han Chinese individuals in southwest China were included. Therefore, it is important to collaborate gene–gene interactions in larger ethnic populations SLE genetic studies in the future.
Author contributions
Junlong Zhang, Bin Yang, and Lanlan Wang designed the study; Junlong Zhang and Hengxu Wu were responsible for recruitment of subjects; Junlong Zhang, Hengxu Wu, and Bin Yang performed experiments and conducted data management; Junlong Zhang, Yongkang Wu, Bin Yang, and Lanlan Wang performed statistical analyses and interpreted results; Junlong Zhang and Yanming Meng wrote the manuscript. All authors reviewed the manuscript.
Conceptualization: Junlong Zhang, Lanlan Wang.
Data curation: Junlong Zhang, Bin Yang, Lanlan Wang.
Formal analysis: Junlong Zhang.
Funding acquisition: Junlong Zhang, Bin Yang.
Investigation: Junlong Zhang.
Methodology: Junlong Zhang, Hengxu Wu.
Project administration: Junlong Zhang, Hengxu Wu.
Resources: Junlong Zhang.
Software: Junlong Zhang.
Supervision: Yongkang Wu, Bin Yang, Lanlan Wang.
Validation: Junlong Zhang, Yongkang Wu, Bin Yang, Lanlan Wang.
Visualization: Yongkang Wu, Bin Yang, Lanlan Wang.
Writing – original draft: Junlong Zhang, Yanming Meng.
Writing – review & editing: Junlong Zhang, Bin Yang, Lanlan Wang.
Footnotes
Abbreviations: 95% CI = 95% confidence interval, ACR = American College of Rheumatology, ANA = anti-antinuclear antibodies, C3 = complement 3, C4 = complement 4, CD3ζ = cluster of differentiation 3ζ, CREB = cAMP-response element binding protein, dsDNA = antidouble-stranded DNA antibodies, ERK = extracellular signal-regulated kinase, FcRγ = Fc receptor γ chain, GWAS = genome wide association study, HWE = Hardy–Weinberg equilibrium, IFN-γ = interferon-γ, IL-1β = interleukin-1β, IL-2 = interleukin-2, IL-6 = interleukin-6, IL-10 = interleukin-10, IL-17 = interleukin-17, MAPK = mitogen-activated protein kinase, MEK = mitogen-activated protein kinase kinase, mRNA = messenger RNA, NF-κB = nuclear factor-κB, OR = odds ratio, PCR-HRM = polymerase chain reaction-high resolution melting, PP2A = protein phosphatase 2A, PP2Ac = protein phosphatase 2A catalytic subunit, Rib-P = antiribosomal P antibodies, SLE = systemic lupus erythematosus, SLEDAI = active stage with SLE disease activity index, Sm = anti-Smith antibodies, SNPs = single nucleotide polymorphisms, SSA/Ro = anti-SSA/Ro antibodies, SSB/La = anti-SSB/La antibodies, TCR = T cell receptor, U1RNP = anti-U1 ribonucleoprotein antibodies, Wnt = Wingless-type.
Funding: This work was supported by the National Natural Science Foundation of China (no. 81601830 and no. 81772258). These funders supported our work including study design, decision to publish and preparation of the manuscript.
The authors have no conflicts of interest to disclose.
References
- [1].Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet 2015;47:1457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lisnevskaia L, Murphy G, Isenberg D. Systemic lupus erythematosus. Lancet 2014;384:1878–88. [DOI] [PubMed] [Google Scholar]
- [3].Zhang T, Park KA, Li Y, et al. PHF20 regulates NF-kappaB signalling by disrupting recruitment of PP2A to p65. Nat Commun 2013;4:2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rahman MM, Rumzhum NN, Morris JC, et al. Basal protein phosphatase 2A activity restrains cytokine expression: role for MAPKs and tristetraprolin. Sci Rep 2015;5:10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kumar A, Pandurangan AK, Lu F, et al. Chemopreventive sphingadienes downregulate Wnt signaling via a PP2A/Akt/GSK3beta pathway in colon cancer. Carcinogenesis 2012;33:1726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Juang YT, Wang Y, Jiang G, et al. PP2A dephosphorylates Elf-1 and determines the expression of CD3zeta and FcRgamma in human systemic lupus erythematosus T cells. J Immunol 2008;181:3658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Katsiari CG, Kyttaris VC, Juang YT, et al. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J Clin Invest 2005;115:3193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Crispin JC, Apostolidis SA, Rosetti F, et al. Cutting edge: protein phosphatase 2A confers susceptibility to autoimmune disease through an IL-17-dependent mechanism. J Immunol 2012;188:3567–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sunahori K, Juang YT, Kyttaris VC, et al. Promoter hypomethylation results in increased expression of protein phosphatase 2A in T cells from patients with systemic lupus erythematosus. J Immunol 2011;186:4508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Apostolidis SA, Rauen T, Hedrich CM, et al. Protein phosphatase 2A enables expression of interleukin 17 (IL-17) through chromatin remodeling. J Biol Chem 2013;288:26775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sunahori K, Nagpal K, Hedrich CM, et al. The catalytic subunit of protein phosphatase 2A (PP2Ac) promotes DNA hypomethylation by suppressing the phosphorylated mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/phosphorylated ERK/DNMT1 protein pathway in T-cells from controls and systemic lupus erythematosus patients. J Biol Chem 2013;288:21936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tan W, Sunahori K, Zhao J, et al. Association of PPP2CA polymorphisms with systemic lupus erythematosus susceptibility in multiple ethnic groups. Arthritis Rheum 2011;63:2755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. [DOI] [PubMed] [Google Scholar]
- [14].Wahren-Herlenius M, Dorner T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet 2013;382:819–31. [DOI] [PubMed] [Google Scholar]
- [15].Murphy G, Lisnevskaia L, Isenberg D. Systemic lupus erythematosus and other autoimmune rheumatic diseases: challenges to treatment. Lancet 2013;382:809–18. [DOI] [PubMed] [Google Scholar]
- [16].Bradley SJ, Suarez-Fueyo A, Moss DR, et al. T cell transcriptomes describe patient subtypes in systemic lupus erythematosus. PLoS ONE 2015;10:e0141171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chung SA, Nititham J, Elboudwarej E, et al. Genome-wide assessment of differential dna methylation associated with autoantibody production in systemic lupus erythematosus. PLoS ONE 2015;10:e0129813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sun C, Molineros JE, Looger LL, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet 2016;48:323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Morris DL, Sheng Y, Zhang Y, et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet 2016;48:940–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nagpal K, Watanabe KS, Tsao BP, et al. Transcription factor Ikaros represses protein phosphatase 2A (PP2A) expression through an intronic binding site. J Biol Chem 2014;289:13751–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sunahori K, Juang YT, Tsokos GC. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J Immunol 2009;182:1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Morris G, Berk M, Galecki P, et al. The neuro-immune pathophysiology of central and peripheral fatigue in systemic immune-inflammatory and neuro-immune diseases. Mol Neurobiol 2016;53:1195–219. [DOI] [PubMed] [Google Scholar]
- [23].Tan W, Gu Z, Shen B, et al. PTEN/Akt-p27(kip1) signaling promote the BM-MSCs senescence and apoptosis in SLE patients. J Cell Biochem 2015;116:1583–94. [DOI] [PubMed] [Google Scholar]
- [24].Stohl W. Future prospects in biologic therapy for systemic lupus erythematosus. Nat Rev Rheumatol 2013;9:705–20. [DOI] [PubMed] [Google Scholar]
- [25].Wallace DJ, Strand V, Merrill JT, et al. Efficacy and safety of an interleukin 6 monoclonal antibody for the treatment of systemic lupus erythematosus: a phase II dose-ranging randomised controlled trial. Ann Rheum Dis 2017;76:534–42. [DOI] [PMC free article] [PubMed] [Google Scholar]


