Abstract
Background
Numerous studies have reported an association between cytotoxic T-lymphocyte associated antigen 4 gene (CTLA4) polymorphism and susceptibility to asthma, in different populations, but the results have been inconsistent. We performed a meta-analysis of 19 published case–control studies to obtain a reasonably accurate estimation of the relationship between CTLA4 polymorphism and asthma.
Methods
We searched the Pubmed, EMBASE, Chinese National Knowledge Infrastructure, and Wanfang databases and extracted data from 19 independent, eligible studies. Odds ratios (ORs) with 95% confidence intervals (CIs) and Egger test were separately used to assess the strength of associations and publication bias.
Results
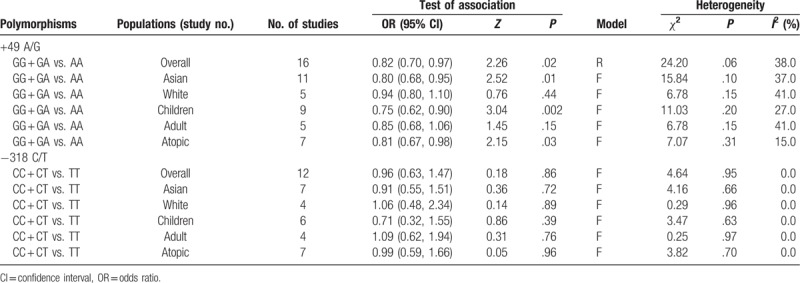
A total of 19 case–control studies involving 4831 cases and 4534 controls were identified. The combined results revealed that there was significant association between the +49A/G polymorphism and asthma (for GG + GA vs. AA: OR = 0.82, 95% CI = 0.70–0.97, P = .02). Stratification by race or age indicated a significant association between the CTLA-4 +49 GA+GG genotype and asthma in Asians (OR = 0.80, 95% CI = 0.68–0.95, P = .01) and children (OR = 0.75, 95% CI = 0.62–0.90, P = .002), but there was no association in whites (OR = 0.94, 95% CI = 0.80–1.10, P = .44) and adults (OR = 0.85, 95% CI = 0.68–1.06, P = .15). Additionally, there was a significant association with atopic asthma under the random-effects model (OR = 0.81, 95% CI = 0.67–0.98, P = .03). In addition, there was no significant association between the −318 C/T polymorphism and asthma risk.
Conclusions
Our meta-analysis results suggested that the +49A/G polymorphism in CTLA-4 was an important risk factor for asthma susceptibility, especially in Asian individuals, children, and atopic patients.
Keywords: asthma, cytotoxic T-lymphocyte associated antigen 4, gene polymorphism, meta-analysis
1. Introduction
Asthma is a chronic airway disease that is characterized by complex airway inflammation and variable airway obstruction.[1] The prevalence of asthma has been increasing in different countries around the world in recent decades, regardless of the level of development, and it poses a heavy financial burden to the family and country.[2] Numerous studies have suggested that the pathogenesis of asthma depends on the interaction between various susceptibility genes and environmental factors, including air pollution, allergens, and genetic variation.[3] T cells play a central role in asthma through the action of Th1-type cytokines generated in response to allergens.[4] A more precise understanding of the association between gene mutation and asthma can facilitate the development of novel therapeutic targets and prevention strategies that reduce the incidence of and mortality associated with asthma. Numerous studies have reported that CTLA4 plays an important role in the pathogenesis process of asthma.[5,6]
CTLA-4, a B7-binding protein expressed on activated T cells, and polymorphisms in CTLA4, a gene located on chromosome 2q33, have been proven to influence the development of asthma.[7] CTLA-4 is involved in the negative regulation of the immune response by blocking the CD28-mediated costimulatory molecules, and is associated with Th2 cell activation and differentiation.[8] Cumulative evidence suggests that the plasma CTLA-4 concentration is significantly higher in asthma patients than in healthy individuals, and CTLA-4-Ig effectively ameliorated airway hyper-responsiveness and reduced the level of serum IgE.[9]
Recent studies have identified several single nucleotide polymorphisms and have demonstrated a significant association between these polymorphisms and asthma.[10] CTLA4 + 49A/G and −318C/T polymorphisms were respectively associated with asthma severity and bronchial hyper-responsiveness by increasing the IgE responsiveness, which is a major factor in asthma, and increasing the sCTLA4 transcript, which may play a role in promoting bronchoconstriction.[11,12] Yao et al and Lee et al performed a meta-analyses to investigate the effect of CTLA4 genotype on the risk of asthma.[13,14] However, the results of the association between CTLA4 polymorphism and asthma susceptibility in different races have not been consistent. Therefore, we have here performed a meta-analysis of recent studies to gain further insight into this matter.
2. Methods
2.1. Search strategy
We searched the Pubmed, EMBASE, Chinese National Knowledge Infrastructure (CNKI), and Wanfang databases for the period of January 2000 to July 2017. The following MeSH terms were searched: “Asthma or Asthmatic” and “CTLA-4 or CTLA-4 + 49 A/G or CTLA-4 -318 C/T or cytotoxic T-lymphocyte associated antigen 4 or cytotoxic T-lymphocyte associated antigen 4 + 49 A/G or cytotoxic T-lymphocyte associated antigen 4 -318 C/T or CD152,” and “polymorphism or variant or mutation.” Furthermore, all references cited in the identified publications and review articles were searched manually to identify additional articles. No ethics committee approvement was necessary for this meta-analysis, which does not contain any studies with human participants or animals performed by any of the authors.
2.2. Inclusion criteria and excluded criteria
Published studies that met the following criteria were included in this meta-analysis: a case–control study on CTLA4 polymorphism and asthma susceptibility, published before July 2017; asthma diagnosed according to the Global Initiative for Asthma guidelines or the diagnostic criteria for asthma established by the Respiratory Society, Chinese Medical Association; the frequency distribution of the corresponding genotype was available for estimating an odds ratio (OR) with a 95% confidence interval (CI); the genotype distribution of the control population had to be in Hardy–Weinberg equilibrium (HWE). The exclusion criteria were as follows: reviews, meta-analysis, or animal studies; and lack of genotype data.
2.3. Data extraction and qualitative assessment
Two reviewers independently reviewed the full text of the eligible studies, and the following data were extracted into the predesigned data collection database. To ensure the accuracy of the data, we carried out multiple validations of the data by different individuals. The following information was extracted from each study: first author's name, year of publication, original country, ethnicity, age group, atopic status, sample size, asthma definition, genotyping method, CTLA4 polymorphism, and genotype number in cases and control. The quality of each selected study was also independently assessed by 2 reviewers who used the Newcastle–Ottawa Scale.[15,16] Any potential disagreement was adjudicated by discussion.
2.4. Statistical analysis
Pooled ORs and 95% CIs were applied to evaluate the strength of association between the +49 A/G and -318 C/T CTLA4 polymorphisms and asthma risk. The statistical significance of the OR was determined by the Z test. Q and I2 statistics were used to assess the heterogeneity among the eligible studies.[17] Heterogeneity was considered significant for P < .10. A random-effects model (P < .10) or a fixed-effect model (P ≥ .10) was used to sum up the pooled OR. The P value of HWE among the control groups within each study was checked by the exact test using an HWE calculator; P values >.05 indicated that the population was in genetic equilibrium. Furthermore, publication bias was examined visually by means of a funnel plot of log OR against its standard error. Publication bias was checked statistically by the Egger test. P value <.05 means that evidence of potential publication bias.[18]
All analyses were performed using the Revman 5.3 (Nordic Cochrane Center, Copenhagen, Denmark), STATA 12.0 software (Stata Corporation, College Station, TX), and Microsoft Office Excel 2003 (Microsoft Corporation, Redmond, WA). P values < .05 were considered statistically significant, whereas the test of heterogeneity used a level of 0.10.
3. Results
3.1. Literature selection and subject characteristics
A total of 102 relevant articles were identified after an initial search, and 38 articles were removed because of duplication. After reviewing the titles and abstracts, 38 articles were excluded because they were reviews, not clinical studies, or were irrelevant to the CTLA4 + 49 A/G and -318 C/T polymorphisms. After 26 full-text articles were assessed for eligibility, a further 8 articles that were not case–control studies or did not present usable data were excluded. When reviewing the full-text of the selected studies, we found one article that contained 2 cohort studies, and each was considered as a separate case–control study.[19] Thus, a total of 19 case–control studies in 18 articles were identified,[1,11,12,19–33] including 4831 cases and 4534 controls. The selection process is shown in Figure 1. There were 16 studies on the +49 A/G and 12 studies on the -318 C/T SNPs. Among the eligible studies, there were 12 studies[1,11,12,19–21,23,24,27,30–32] involving Asians, and 5 studies[22,25,28,29,33] involving whites. Six studies[11,12,19,24,29,33] were performed in adults, 11 studies[1,19–21,23,26–28,30–32] were performed in children, and 2 studies[22,25] included both adults and juveniles. Five studies[19,21,22,24,26] included atopic asthma patients, 3 studies[11,19,20] included both atopic and non-atopic asthma patients, and the data for these patients could be separately extracted. Ten studies[12,23,25,27–33] did not offer detailed information about patients’ atopic status. The definition of asthma in these eligible studies included different criteria (physician-diagnosed, American Thoracic Society diagnostic criteria, National Heart, Lung, and Blood Institute/World Health Organization guideline, Chinese Asthma Diagnosis criteria for children, questionnaire survey, National Institutes of Health criteria). Genotypes were determined by various methods (polymerase chain reaction-restriction fragment length polymorphism, Illumina Bead Array System, Taqman-ASA, Illumina BeadXpress platform, Sequenom platform, Affymetrix Genome-Wide Human SNP Array 5.0). Selected characteristics from each study are shown in Table 1. Genotype frequencies and HWE examination results are presented in Tables 2 and 3.
Figure 1.
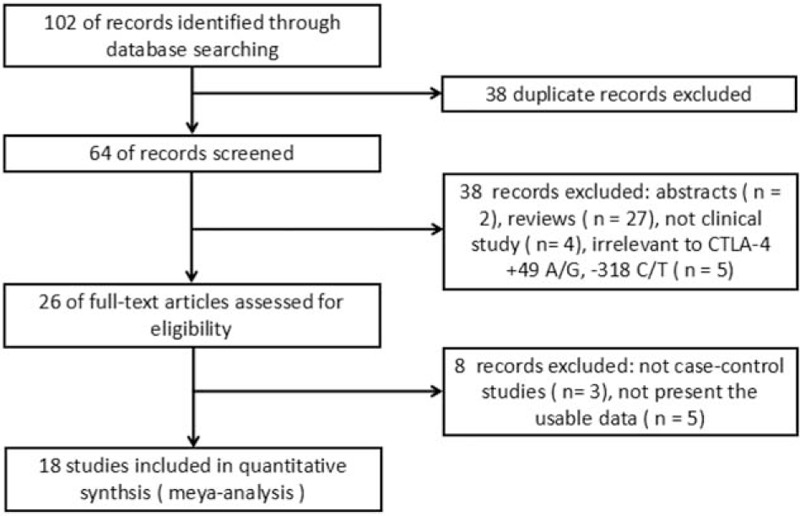
Flow of study identification, inclusion, and exclusion.
Table 1.
Characteristics of the studies included in the meta analysis.
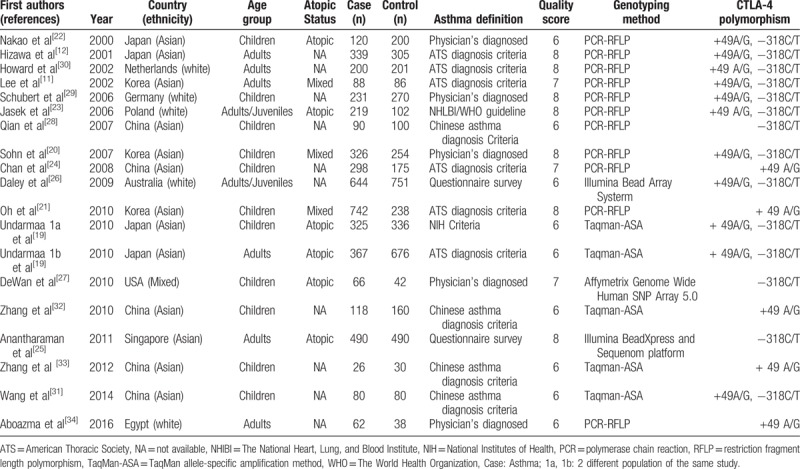
Table 2.
The distribution of the +49A/G genetic polymorphism included in meta analysis.
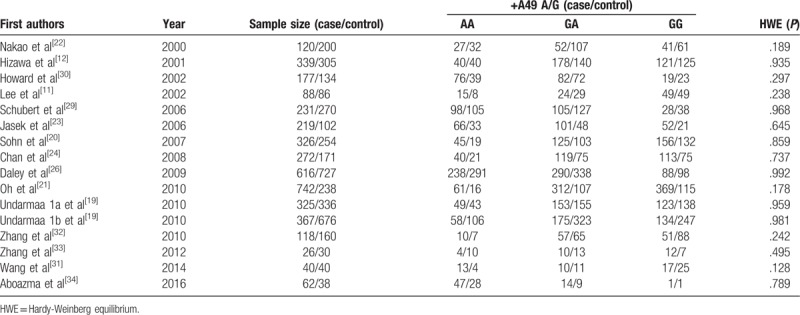
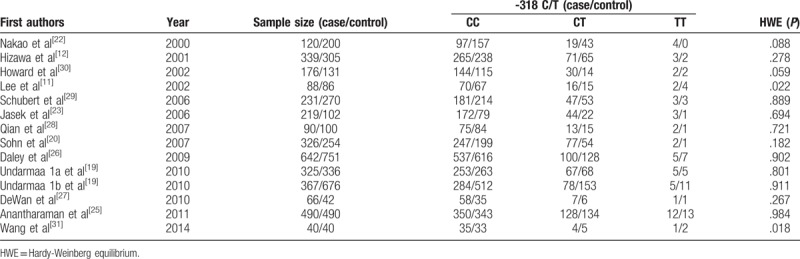
Table 3.
The distribution of the -318 C/T genetic polymorphism included in meta analysis.
3.2. CTLA-4 + 49 A/G polymorphism and asthma susceptibility
Fifteen studies in HWE were pooled, and the total sample sizes for asthma and control groups were 4068 and 3767, respectively.[1,11,12,19–23,25,28–33] The results of this meta-analysis are presented in a forest plot (Fig. 2). Heterogeneity among individual estimates of the ORs was observed (I2 = 38%, P = .06) and the sample size data of the case and control groups were combined by means of a random effects model. The pooled OR was 0.82 (95% CI = 0.70–0.97, P = .02). This result suggested that the CTLA4 + 49 A/G polymorphism caused an increased risk of asthma in a worldwide population. In subgroup analysis by ethnicity, the CTLA4 + 49 A/G polymorphism was associated with asthma risk in the Asian population (OR = 0.80, 95% CI = 0.68–0.95, P = .01), but not in the white population (OR = 0.94, 95% CI = 0.80–1.10, P = .44).
Figure 2.
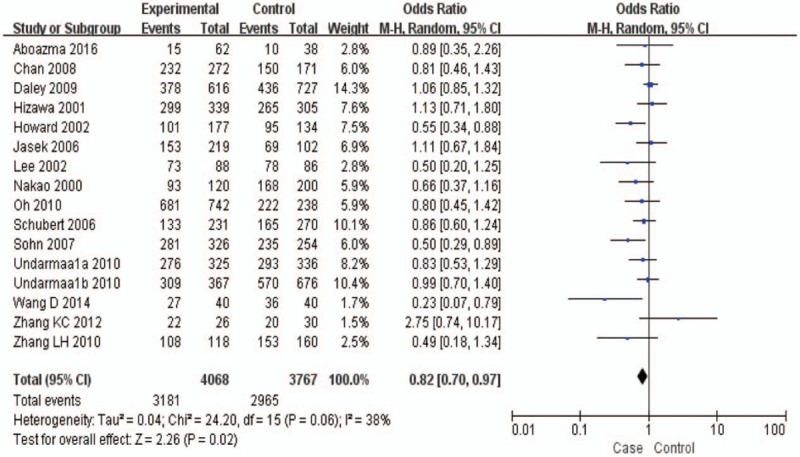
Meta-analysis for the association between asthma and the CTLA-4 + 49 A/G polymorphism (GG + GA vs. AA) with a random-effects model.
Stratifying subjects by age yielded a significant association between the CTLA4 + 49 A/G polymorphism and asthma risk in children (OR = 0.75, 95% CI = 0.62–0.90, P = .002), but no association was found in adults (OR = 0.85, 95% CI = 0.68–1.06, P = .15). Moreover, significant association was also observed in atopic asthma (OR = 0.81, 95% CI = 0.67–0.98, P = .03). All data are summarized in Table 4.
Table 4.
Results of the meta-analysis of CTLA-4 polymorphism on asthma and sensitivity analyses.
3.3. CTLA-4 -318 C/T polymorphism and asthma susceptibility
Twelve studies determined the association between the -318 C/T polymorphism and asthma risk.[12,19,20,21–29] All but 2 studies were in HWE[11,30]; those in HWE were pooled. The total sample sizes for asthma and control groups were respectively, 3391 and 3657. The sample size data of the case and control groups were combined by means of a random-effects model. The pooled OR was 0.96 (95% CI = 0.63–1.47, P = .86) (Fig. 3). Thus, there was no significant association between the CTLA4 -318 C/T polymorphism and asthma.
Figure 3.
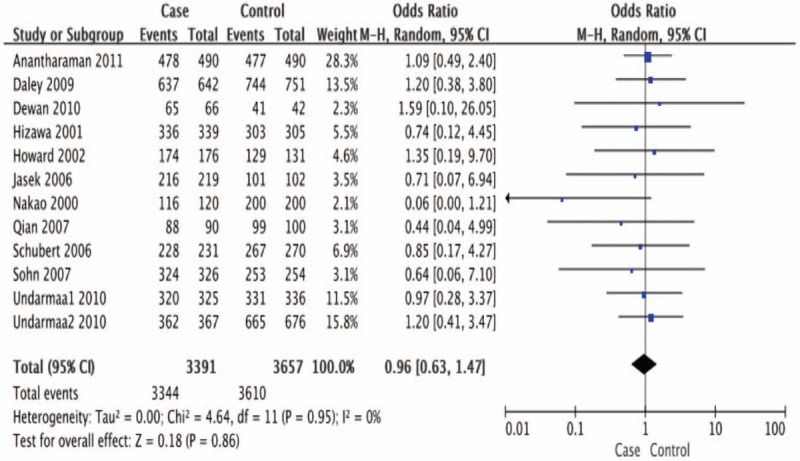
Meta-analysis for the association between asthma and the CTLA-4 -318 C/T polymorphism (CC + CT vs. TT) with a fixed-effects model.
Sensitivity analyses were performed according to atopic status, but no association between the CTLA4 -318 C/T polymorphism and asthma risk was found in the atopic population (OR = 0.99, 95% CI = 0.59–1.66, P = .96). Furthermore, stratifying subjects by age and ethnicity also showed no significant genetic effect (Table 4).
3.4. Sensitivity analysis
We performed a sensitivity analysis to assess the stability of the results by sequentially removing each eligible study. The results showed no statistical significance in terms of the pooled ORs or 95% CIs, indicating that our results were robust and reliable (Fig. 4). Additionally, Egger test indicated that there was no significant publication bias (t = −1.38, P = .189, and t = −0.86, P = .410) (Fig. 5).
Figure 4.
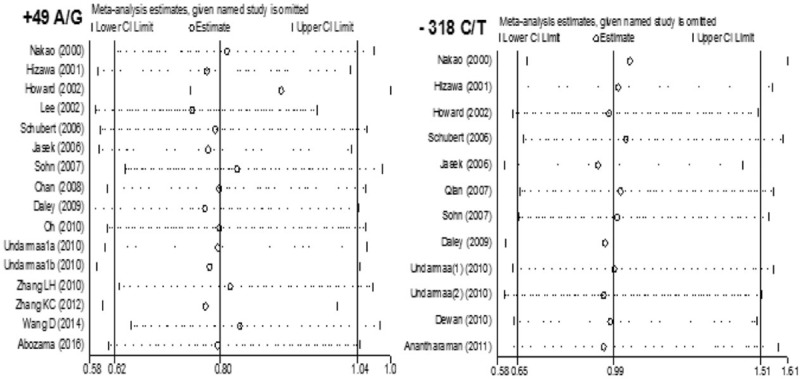
Sensitivity analysis on the associations between CYLA-4 +49A/G and -318 C/T polymorphism and asthma risk.
Figure 5.
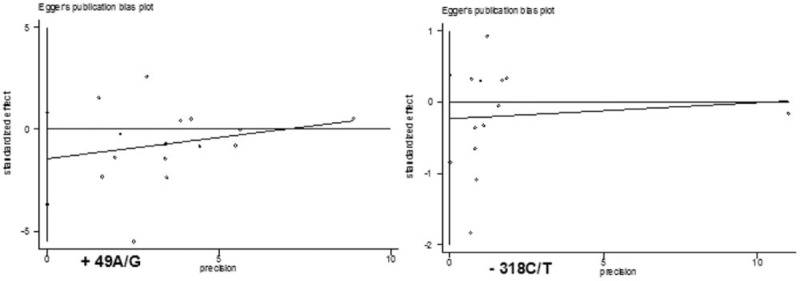
Funnel plot for publication bias in selection of studies on the CTLA-4 +49A/G and -318 C/T polymorphism.
4. Discussion
Asthma is a complicated inflammatory airway disease, and its pathogenesis is mainly related to environmental pollution and genetic inheritance[34]. Although the etiology of asthma has not been fully determined, genetic studies have established that asthma susceptibility involves a genetically determined Th cell differentiation,[8] and some studies have confirmed a significant association between CTLA4 genetic polymorphisms and asthma. To illustrate the effect of CTLA4 genetic polymorphism on the risk of asthma, Yao et al[13] and Lee et al[14] performed meta-analyses. Lee et al revealed a significant association between the +49A/G polymorphism and asthma risk in Asian populations, but Yao et al suggested that there was a significant association between the CTLA-4 +49A/G polymorphism and asthma only in white populations, but not in Asian populations. We therefore performed a meta-analysis including new data, to assess this conclusion.
In the present meta-analysis, a total of 4769 asthma cases and 4496 controls were used to evaluate the relationship between +49A/G and -318C/T polymorphisms in CTLA4 and the risk of asthma. We show that the +49A/G polymorphism was a moderate risk factor of asthma in the overall study population. In the subgroup meta-analysis, we found that there was a significant association between the CTLA4 +49A/G polymorphism and asthma in Asian populations, but not in white populations. Interestingly, our meta-analysis result of association between the +49A/G polymorphism and asthma in Asian populations was in accordance with the findings of Lee et al.
However, we could not deduce that the +49A/G polymorphism plays no role in asthma in white populations based on 5 studies. Because such few studies might lead to unstable results or may be affected by a combination of environmental exposures and different genetic backgrounds, further studies on the effect of the +49A/G polymorphism on asthma is needed for verification.
Furthermore, we also observed an association between the +49A/G polymorphism and asthma in children (P = .002), but found no association in adults (P = .15).
Stratifying subjects by atopic status indicated a significantly increased risk of asthma in patients with atopy (P = .03). Thus, the +49A/G polymorphism may play a role in the pathophysiological process of allergic asthma. Some studies have also shown that serum sCTLA-4 concentrations are increased in patients with allergic asthma and after allergen inhalation and ingestion in sensitized asthmatic patients.[35,36]
We further found a lack of association between the -318C/T polymorphism and asthma susceptibility. However, Hizawa et al[12] reported that patients with asthma who were homozygous for the -318C allele in the CTLA4 promoter region had higher levels of total serum IgE than patients with asthma who carried the -318T allele. Moreover, the -318T allele was also associated with increased asthma severity and might be a candidate gene marker for severe asthma.[11,27] We could not conclude that there was no association between the -318C/T polymorphism and asthma risk because asthma is a complex genetic disease, and multiple genes are involved in its pathophysiology. Furthermore, the limited total sample size of the studies included in this meta-analysis may have influenced our findings. Larger sample size studies are required to assess the association between -318 C/T polymorphism and asthma risk more comprehensively.
Asthma is characterized by hyperresponsiveness, eosinophilic inflammation and elevated IgE levels, and is thought to be mediated by CD4+ T lymphocytes.[37] The morbidity and mortality associated with asthma have increased because of environmental aggravation in recent years, particularly in children. Therefore, effective early screening and treatment are important. Our meta-analysis found that the +49 A/G polymorphism in CTLA4 might be a risk factor for asthma susceptibility, especially in Asian individuals, children, and patients with atopy. Hence, clinicians could provide early intervention for asthma patients by promoting early screening for this CTLA4 polymorphism. Childhood is a sensitive stage for asthmatic attack and it has been reported that 80% of children older than 3 years are allergic to indoor conditions.[38] For these childhood asthma cases, especially those with atopy, CTLA4 SNP genotyping could help clinicians to implement rational therapy and reduce the impact of the disease on these children's development and growth. Clinicians should therefore consider implementing this genotyping as a routine test for hospitalized asthma patients, as would be done for cancer-related markers.
Publication bias and heterogeneity can influence the results of meta-analyses, and could result in potential overestimation of effect sizes. In this meta-analysis, publication bias was checked statistically using Egger test; no significant publication bias was found for studies of the +49 A/G and -318 C/T polymorphisms. In addition, no significant heterogeneity was found in most of the overall comparisons for these 2 polymorphisms. Therefore, heterogeneity did not seem to influence the results of the evaluation, suggesting the reliability of our results.
Meta-analysis involves the integration of all comparative studies to increase the sample size, to achieve more accurate results. However, this meta-analysis had some limitations. First, all studies included in this meta-analysis were identified from selected databases, and a publication bias may have occurred. Second, only 5 of the 17 studies were conducted in non-Asian populations. Third, publication bias may have been present because all studies included in this meta-analysis were published only in Chinese and English languages; some relevant studies in other languages may not have been identified. Finally, data were not stratified by sex, lifestyle, or environmental variables.
5. Conclusion
Our meta-analysis suggested that the +49 A/G polymorphism in CTLA4 might be a risk factor for asthma susceptibility, especially in Asian individuals, children, and patients with atopy. We found no evidence for association of the -318 C/T polymorphism with asthma susceptibility. However, further large-scale studies should be carried out to validate this conclusion.
Author contributions
Data curation: Yan Zheng. Formal analysis: Yan Zheng, Hongluan Wang. Investigation: Yan Zheng, Liyang Liao, Luxia You. Methodology: Hongluan Wang, Linlin Luo. Software: Yan Zheng. Supervision: Jun Wang, Qiugen Li. Validation: Hongluan Wang. Writing – original draft: Yan Zheng. Writing – review and editing: Yan Zheng, Hongluan Wang.
Data curation: Yan Zheng, Hongluan Wang.
Formal analysis: Hongluan Wang, Linlin Luo, Liyang Liao.
Resources: Luxia You.
Supervision: Jun Wang, Qiugen Li.
Writing – review & editing: Yan Zheng.
Footnotes
Abbreviations: CI = confidence interval, CTLA-4 = cytotoxic T-lymphocyte associated antigen 4, HWE = Hardy-Weinberg equilibrium, OR = odds ratio, SNP = single nucleotide polymorphism.
YZ and HW equally contributed to the manuscript, and should be considered as the co-frist author. JW and QGL supervised the study.
Funing/Support: This study was supported by the National Natural Sciences Foundation of China (30960143) and the Major Projects of Jiangxi Provincial Science and Technology Department (2015BBB70267).
The authors have no conflicts of interest to disclose.
References
- [1].Oh KY, Kang MJ, Choi WA, et al. Association between serum IgE levels and the CTLA4 +49A/G and FCER1B -654C/T polymorphisms in Korean children with asthma. Allergy Asthma Immunolo Res 2010;2:127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev 2011;242:10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bouzigon E, Nadif R, Le MN, et al. Genetic and environmental factors of asthma and allergy: Results of the EGEA study. Rev Mal Respir 2015;32:822–40. [DOI] [PubMed] [Google Scholar]
- [4].Cao J, Zhang L, Huang S, et al. Aberrant production of soluble co-stimulatory molecules CTLA-4 and CD28 in patients with chronic hepatitis B. Microb Pathog 2011;51:262–7. [DOI] [PubMed] [Google Scholar]
- [5].Munthe-Kaas MC, Carlsen KH, Helms PJ, et al. CTLA-4 polymorphisms in allergy and asthma and the TH1/TH2 paradigm. J Allergy Clin Immunol 2004;114:280–7. [DOI] [PubMed] [Google Scholar]
- [6].Botturi K, Lacoeuille Y, Cavaillès A, et al. Differences in allergen-induced T cell activation between allergic asthma and rhinitis: Role of CD28, ICOS and CTLA-4. Respir Res 2011;12:1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kawayama T, Matsunaga K, Kaku Y, et al. Decreased CTLA4(+) and Foxp3(+) CD25(high)CD4(+) cells in induced sputum from patients with mild atopic asthma. Allergol Int 2013;62:203–13. [DOI] [PubMed] [Google Scholar]
- [8].Wang CJ, Heuts F, Ovcinnikovs V, et al. CTLA-4 controls follicular helper T-cell differentiation by regulating the strength of CD28 engagement. Proc Natl Acad Sci U S A 2015;112:524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ying L, Fu Z, Luo J, et al. Cytotoxic T lymphocyte antigen 4 immunoglobulin modified dendritic cells attenuate allergic airway inflammation and hyperresponsiveness by regulating the development of T helper type 1 (Th1)/Th2 and Th2/regulatory T cell subsets in a murine model of asthma. Clin Exp Immunol 2011;165:130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang H, Shin J, Kim K, et al. CTLA-4 exon 1 and promoter gene polymorphisms in childhood asthma. J Allergy Clin Immunol 2006;117:S324–1324. [Google Scholar]
- [11].Lee SY, Lee YH, Shin C, et al. Association of asthma severity and bronchial hyperresponsiveness with a polymorphism in the cytotoxic T-lymphocyte antigen-4 gene. Chest 2002;122:171–6. [DOI] [PubMed] [Google Scholar]
- [12].Hizawa N, Yamaguchi E, Jinushi E, et al. Increased total serum IgE levels in patients with asthma and promoter polymorphisms at CTLA4 and FCER1B. J Allergy Clin Immunol 2001;108:74–9. [DOI] [PubMed] [Google Scholar]
- [13].Yao YS, Wang LH, Chang WW, et al. Association between CTLA-4 exon-1 +49A/G polymorphism and asthma: an updated meta-analysis. Int J Clin Exp Med 2015;8:3107–13. [PMC free article] [PubMed] [Google Scholar]
- [14].Lee YH, Choi SJ, Ji JD, et al. The CTLA-4 +49 A/G and −318 C/T polymorphisms and susceptibility to asthma: a meta-analysis. Mol Biol Rep 2012;39:8525–32. [DOI] [PubMed] [Google Scholar]
- [15].Zhao J, Xu W, Zhang Z, et al. Prognostic role of HER2 expression in bladder cancer: a systematic review and meta-analysis. Int Urol Nephrol 2015;47:87–94. [DOI] [PubMed] [Google Scholar]
- [16].Zeng YJ, Lai W, Liu L, et al. Prognostic significance of neuroendocrine differentiation in colorectal adenocarcinoma after radical operation: a meta-analysis. J Gastrointest Surg 2014;18:968–76. [DOI] [PubMed] [Google Scholar]
- [17].Chen B, Benedetti A. Quantifying heterogeneity in individual participant data meta-analysis with binary outcomes. Syst Rev 2017;6:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou HM, Liu AL, Zhou BZ, et al. Interleukin-10 gene rs1800896 polymorphism increases risk of acute pancreatitis. Medicine 2017;96:e9006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Undarmaa S, Mashimo Y, Hattori S, et al. Replication of genetic association studies in asthma and related phenotypes. J Hum Genet 2010;55:342–9. [DOI] [PubMed] [Google Scholar]
- [20].Sohn MH, Kim SH, Song TW, et al. Cytotoxic T lymphocyte-associated antigen-4 gene polymorphisms confer susceptibility to atopic asthma in Korean children. Pediatr Pulmonol 2007;42:542–7. [DOI] [PubMed] [Google Scholar]
- [21].Nakao F, Ihara K, Ahmed S, et al. Lack of association between CD28/CTLA-4 gene polymorphisms and atopic asthma in the Japanese population. Exp Clin Immunogenet 2000;17:179–84. [DOI] [PubMed] [Google Scholar]
- [22].Jasek M, Luszczek W, Obojski A, et al. Distribution of CTLA-4 polymorphisms in allergic asthma. Int Arch Allergy Immunol 2006;141:223–9. [DOI] [PubMed] [Google Scholar]
- [23].Chan IH, Tang NL, Leung TF, et al. Study of gene-gene interactions for endophenotypic quantitative traits in Chinese asthmatic children. Allergy 2008;63:1031–9. [DOI] [PubMed] [Google Scholar]
- [24].Anantharaman R, Andiaappan AK, Nilkanth PP, et al. Genome-wode association study identifies PERLD1 as asthma candidate gene. BMS Med Genet 2011;12:170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Daley D, Lemire M, Akhabir L, et al. Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet 2009;125:445–59. [DOI] [PubMed] [Google Scholar]
- [26].DeWan AT, Triche EW, Xu X, et al. PDE11A associations with asthma: results of a genome-wide association scan. J Allergy Clin Immunol 2010;126:871–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qian L, LU Jin, JR, YL Relationship between polymorphism of cytotoxic T lymphocyte associated antigen gene promoter-318 C/T and serum total IgE level. J Clin Pediatr 2007;8:660–3. [Google Scholar]
- [28].Schubert K, von Bonnsdorf H, Burke M, et al. A comprehensive candidate gene study on bronchial asthma and juvenile idiopathic arthritis. Dis Markers 2006;22:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Howard TD, Postma DS, Hawkins GA, et al. Fine mapping of an IgE-controlling gene on chromosome 2q: analysis of CTLA4 and CD28. J Allergy Clin Immunol 2002;110:743–51. [DOI] [PubMed] [Google Scholar]
- [30].Wang D, LIDF, Yuan Y, et al. Study on the association between CTLA4 genes SNPS and pediactric asthma susceptibility in Shenzhen. Chin J Health Lab Technol 2014;24:549–11. [Google Scholar]
- [31].Zhang LH, Miu XP, Wang HP. The relationship of cytotoxic T-lymphocyte antigen-4 functional genetic polymorphism and the risk for the occurrence of children asthma. Chin J Birth Health Heredity 2010;18:135–6. [Google Scholar]
- [32].Zhang KC, Li WC, Chen F. CTLA-4 gene polymorphisms and its impact on the expression of cytokines in children with bronchial asthma. Shandong Med J 2012;3:55–7. [Google Scholar]
- [33].Aboazma SM, Elsamanoudy A, Mokhtar N, et al. Cytotoxic T-lymphocyte antigen 4 (CTLA-4) gene polymorphisms in bronchial asthma. Int J Adv Res 2016;4:35–42. [Google Scholar]
- [34].Li X, Howard TD, Zheng SL, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 2010;125:328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Qin XJ, Shi HZ, Qin SM, et al. Effects of allergen inhalation and oral glucocorticoid on serum soluble CTLA-4 in allergic asthmatics. Allergy 2005;60:774–9. [DOI] [PubMed] [Google Scholar]
- [36].Shi HZ, Mo XY, Zhong XN. Soluble CTLA-4 in sera of patients with bronchial asthma. J Asthma 2005;42:133–9. [PubMed] [Google Scholar]
- [37].Kavvoura FK, Akamizu T, Awata T, et al. Cytotoxic T-lymphocyte associated antigen 4 gene polymorphisms and autoimmune thyroid disease: a meta-analysis. J Clin Endocrinol Metab 2007;8:3162–70. [DOI] [PubMed] [Google Scholar]
- [38].Kim J, Lee S, Woo SY, et al. The indoor level of house dust mite allergen is associated with severity of atopic dermatitis in children. J Korean Med Sci 2013;1:74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


