Supplemental Digital Content is available in the text
Keywords: breast cancer, cell-free DNA, meta-analysis, plasma DNA, prognosis
Abstract
Purpose:
Circulating cell-free DNA (cfDNA) has been reported to predict outcomes in patients with various types of cancer. However, its prognostic value in patients with breast cancer is not well established still now. In this meta-analysis, we evaluated the prognostic role of cfDNA in breast cancer patients.
Methods:
We performed systematic searches in electronic databases to identify studies that evaluated the prognostic value of cfDNA in breast cancer patients. The end points were progression-free survival (PFS) and overall survival (OS). The hazard ratios (HRs) and their 95% confidence intervals (95% CIs) were extracted to assess the prognostic significance of cfDNA. Subgroup analyses were also conducted.
Results:
A total of 11 publications involving 1467 patients were included in this meta-analysis. cfDNA was shown to be significantly associated with PFS (HR 2.02, 95% CI 1.51–2.72, P < .001, I2 = 82%) and OS (HR 1.75, 95% CI 1.01–3.05, P < .001, I2 = 92%). The results of subgroup analyses also revealed that cfDNA was a good predictor of prognosis in breast cancer patients.
Conclusion:
Our meta-analysis indicated that cfDNA was associated with poor PFS and OS, thus it may help to predict outcomes of patients with breast cancer. However, further studies are needed to confirm our results.
1. Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-related death among women in the worldwide.[1] Although radical surgery, radiotherapy, and drug therapy have significantly reduced the risk of relapse and improved overall survival of breast cancer patients, a certain percentage of patients still develop early tumor recurrent or progression.[2,3] Thus, estimating relapse and monitoring metastases could contribute to a better outcome and improve quality of life for breast cancer patients.[4] In clinical practice, serum markers such as carbohydrate antigen (CA) 15-3, BR 27.29, mucin-like cancer-associated antigen, CA549, and carcinoembryonic antigen are the most common prognostic factors for monitoring patients and predicting their risk of relapse.[5] However, these serum biomarkers have limited usefulness due to their low sensitivity and specificity.[6] Therefore, new prognostic biomarkers are urgently needed to monitor the progression of breast cancer.
Circulating cell-free DNA (cfDNA), a type of cell-free nucleic acids, is defined as extracellular DNA in the blood.[7] The occurrence of circulating cfDNA is thought to be related to the apoptosis and necrosis of cells.[8] High concentrations and genetic alterations of cfDNA are revealed to be more likely present in cancer patients as compared to healthy controls.[9,10] Furthermore, many studies have reported the prognostic value of cfDNA in various kinds of solid neoplasm, including lung cancer, pancreatic cancer, prostate cancer, hepatocellular cancer, renal cell cancer, colorectal cancer, and breast cancer.[11–17] In addition, several studies have indicated that the levels of circulating cfDNA, methylation of cfDNA, loss of heterozygosity (LOH) of cfDNA, and cfDNA integrity are associated with disease outcome of breast cancer patients. However, such studies are conflicting in their results, and have not been systematically reviewed. The aim of this meta-analysis was to clarify the predictive and prognostic role of cfDNA in patients with breast cancer.
2. Methods
2.1. Search strategy and study selection
As this literature is a meta-analysis, ethical approval is not necessary. We searched relevant articles in PubMed, Embase, Cochrane, and the Ovid Medline database up to July 2017 without language or date restrictions. The search strategy included (“cfDNA” or “cell-free DNA” or “cell free DNA” or “plasma DNA” or “serum DNA” or “circulating DNA”) and (“breast cancer” or “breast neoplasm” or “breast tumor”) and (“prognosis” or “prognostic” or “predictive” or “prediction” or “outcome”). Moreover, related articles and reference lists of these articles were also reviewed to identify all available studies.
2.2. Inclusion and exclusion criteria
Two investigators independently evaluated the eligibility of studies (Jing Yang and Jing Zhang). A study would be included if it met the following inclusion criteria: All patients enrolled in the study were diagnosed with breast cancer; Studies investigated the prognostic value of cfDNA in breast cancer patients; Endpoints included progression-free survival (PFS) or overall survival (OS), and sufficient data were presented for extracting hazard ratio (HR) value; The latest or the most informative one was to include whether studies assessed the same patient population; and English language articles. The following exclusion criteria were applied: nonresearch publications such as case reports, editorials, reviews, letters, comments; and duplicate articles.
2.3. Data extraction
Two investigators independently reviewed all eligible studies and extracted the data (Jing Yang and Linyan Chen). Information retrieved from these studies included author, year of publication, country, population source, number of enrolled patients, characteristics of patients (age, sex, stage), method of cfDNA assessment, origin of cfDNA (serum or plasma), follow-up, and survival data. We recorded all the data by using standard electronic tables.
2.4. Statistical analysis
HRs and its 95% confidence intervals (95% CIs) were adopted to clarify the prognostic value of cfDNA. HRs >1 indicated an elevated risk of disease progression or death. For studies in which HRs and 95%CIs were not available, we extracted survival rates from Kaplan–Meier curves by using Engauge Digitizer version 4.1.[18] To make sure the accuracy, 2 investigators (Jing Yang and Li Cheng) independently performed this process. In addition, data of multivariate analysis were preferable if both HRs of univariate and multivariate were retrievable. Log HR and its standard error for each study were retrieved following the method described by other authors.[18]
To assess the heterogeneity among studies, pooled HRs were initially calculated by using a fixed effects model. If there was significant heterogeneity among studies (P ≤ .1, I2 > 50%), random effects model was adopted.[19] We used Cochrane's Q statistics and I2 statistics to assess the homogeneity of the studies; I2 > 50% and P < .05 were considered significant for heterogeneity.
Subgroups were stratified by tumor-node-metastasis (TNM) stage (I-III, IV, I-IV), assay indicators (quantification or mutation), origin of cfDNA, sampling time (before treatment, after treatment, before and after treatment), and method of cfDNA assessment. We did not carry out the subgroup analysis if the number of study was not more than 1. We performed subgroup analysis to further investigate the potential factors associated with survival of breast cancer patients. Statistical analysis was conducted with Review Manager Version 5·2 (The Cochrane Collaboration). All P values are 2-tailed. The methods adopted in this study have been used in the previous study.[20]
2.5. Quality assessment
The quality of the included studies was assessed by using the Newcastle–Ottawa Quality Assessment Scale (NOS).[21] The NOS consists of 8 items, which were categorized into 3 main parts, including selection, comparability, and outcome. A star system is used to assess study quality. Studies with higher stars indicate higher quality. NOS 5 to 9 stars were considered to be high quality in this meta-analysis.[20]
2.6. Publication bias
Publication bias was evaluated by inspecting the symmetry of the funnel plot and tested with Begg and Egger adjusted rank correlation test.[22] Publication bias assessment was conducted with STATA version 12.1 (Stata Corporation, College Station, TX).
3. Results
3.1. Literature search results
We retrieved 7124 articles, of which 1744 were duplicates and 346 were non-English publication. By reviewing title and abstract, we found 2 review articles, 1 comments article, 1 editorials article, and 4989 articles out of the scope of our meta-analysis. Thirty-nine studies were recognized as potentially relevant publication, all of which were included in detailed assessment. After obtaining and reading the full articles, we excluded 18 articles because they did not estimate PFS or OS and without follow data; moreover, 9 articles were excluded because these studies did not provide sufficient data for extracting HRs value; 1 [23] of 2 studies from Takeshita et al[23,24] was excluded for overlapping of study population. Therefore, a total of 11 [24–34] studies met our inclusion criteria and were finally included in this meta-analysis. Figure 1 shows the study selection process of our meta-analysis.
Figure 1.

Literature search strategy and selection of study.
3.2. Study characteristics and quality
The basic characteristics of studies are summarized in Table 1. Eleven studies published between 2009 and 2017 were eligible for our meta-analysis. Study sample sizes ranged from 14 to 541 (median 100), with a total of 1467 patients. Among the 11 studies, 3 studies were from Germany (379 patients, 25.9%),[29,32,33] 3 were from America (273 patients, 18.6%),[28,30,34] 2 were from Japan (133 patients, 9.0%),[24,31] 1 was from India (86 patients, 5.9%),[27] and 1 was from Britain (55 patients, 3.8%)[26]; in addition, the samples of 1 study were obtained from participants of a phase3 clinical trial in which the patients were from 24 countries (541 patients, 36.8%).[25] Three studies (675 patients, 46.0%) collected samples before treatment,[25,30,32] 3 studies (291 patients, 19.8%) collected samples after treatment,[29,31,33] and 5 studies (501 patients, 34.2%) collected their samples before or after treatment.[24,26–28,34] HRs were extracted directly in 5 studies[24,25,27,30,32] and could be calculated in 6 studies.[26,28,29,31,33,34] The quality score ranged from 5 to 7. Six studies were evaluated with score 5,[27–30,32,34] 4 studies were evaluated with score 6,[24,25,31,33] and 1 study was evaluated with score 7.[26] All the studies were regarded as high quality. The characteristics and quality of studies enrolled in this meta-analysis are summarized in Table 1. S1 Table summarizes the NOS score of the enrolled studies.
Table 1.
Characteristics of the included studies.

3.3. Prognostic value of cfDNA and heterogeneity
The results of meta-analysis revealed a significant association between cfDNA and PFS in patients with breast cancer, with a pooled HR of 2.02 (95% CI 1.51–2.72, n = 20). For OS, we found a pooled HR of 1.75 (95% CI 1.01–3.05, n = 12), which indicated that significant association could also be observed for OS. However, high heterogeneities were presented in the statistical tests among study group of PFS (I2 = 82%, P < .001) and OS (I2 = 92%, P < .001). Therefore, we adopted random effects model (Fig. 2).
Figure 2.

Forest plot of pooled hazard ratio (HR) for the impact of cfDNA on PFS (A) and OS (B) in breast cancer patients.
3.4. Subgroup analysis
Some factors are correlated with patient's prognosis and may bring heterogeneity to the overall analysis. As high heterogeneities were observed in the analysis of all studies, we performed a subgroup analysis stratified according to the TNM stage (I-III, IV, I-IV), region, method of cfDNA assessment, origin of cfDNA, cfDNA analysis (quantification or mutation), and number of patients. In the subgroup analysis stratified by TNM staging, the pooled HRs of stage I-III patients for PFS and OS were 7.62 (95% CI, 2.53- 22.97; P = .002; I2 = 74%) and 1.86 (95% CI, 0.87–3.98; P = .19; I2 = 41%), respectively; the pooled HRs of stage IV patients for PFS and OS were 1.22 (95% CI, 0.84–1.76; P < .001; I2 = 88%) and 1.59 (95% CI, 0.80–3.18; P < .001; I2 = 94%), respectively; the pooled HRs of stage I-IV patients for PFS and OS were 2.14 (95% CI, 1.67–2.73; P = .38; I2 = 6%) and 2.10 (95% CI, 1.03–4.29; P = .26; I2 = 20%), respectively. The presence of ESR1 mutation and high levels of ALU concentration were associated with a worse OS (HR 1.35, 95% CI 1.07–1.69, P = .60, I2 = 0; HR 3.97, 95% CI 2.69–5.87, P = .87, I2 = 0, respectively). In addition, our results indicated that PFS was poorer for patients with TP-53 mutation, and LOH (HR 2.57, 95% CI 1.73–3.83, P = .71, I2 = 0; HR 9.41, 95% CI 3.32–26.68, P = .14, I2 = 49%, respectively) than those without TP-53 mutation and LOH. The pooled HRs of patients with other mutations or high level of cfDNA for PFS and OS were 1.86 (95% CI, 1.18–2.94; P < .001; I2 = 81%) and 1.06 (95% CI, 0.40–2.79; P < .001; I2 = 96%), respectively. With respect to the sample, the majority of studies tested cfDNA in the plasma rather than in the serum of patients. The combined HRs of both groups were >1 (Fig. 3, S1–S9 Fig). Table 2 summarizes the results of subgroup analyses.
Figure 3.
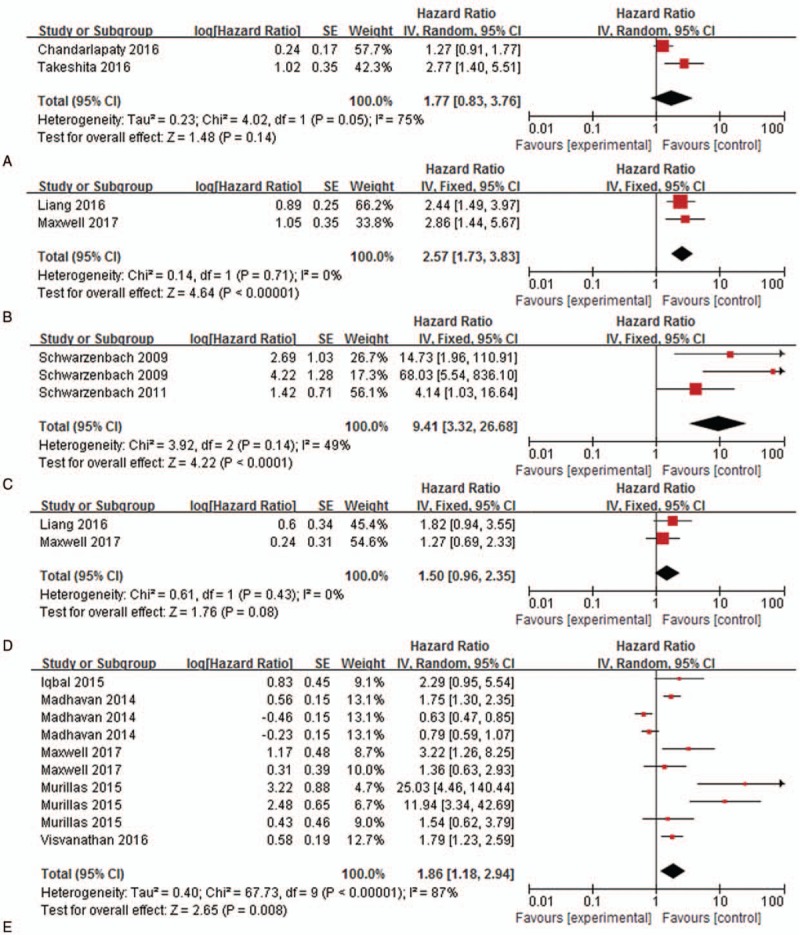
Subgroup analysis of PFS studies stratified according to the assay indicators. (A) ESR1, (B) TP-53, (C) LOH, (D) PIK3CA, (E) Other assay indicators.
Table 2.
Subgroup analysis of included studies.

3.5. Publication bias
Begg test and Eegg test were performed to assess the publication bias in our meta-analysis. The results revealed no evidence of significant publication bias in eligible studies of OS (Begg test, P = .447) and PFS (Eegg test, P = .106). The P values for publication bias of each subgroup are summarized in Table 2.
4. Discussion
As a noninvasive approach, detection of cfDNA has been reported to be a potential method to predict survival in patients with breast cancer.[35] Furthermore, the development of more effective methods such as quantitative polymerase chain reaction (qPCR) and droplet digital polymerase chain reaction (ddPCR) has allowed both screening and validation of genomic alteration in cfDNA, thus ensuring the availability of cfDNA detection.[36] To the best of our knowledge, this is the first meta-analysis to systematically estimate the association between cfDNA and prognosis of breast cancer patients. This meta-analysis enrolled 11 publications including 1336 patients with breast cancer. The results revealed that concentration, mutation of cfDNA, and cfDNA integrity were significantly associated with outcome of breast cancer patients. Compared with patients with mutation or high levels of cfDNA, patients without mutation or with low levels of cfDNA tend to have a favorable PFS.
Previous studies have reported that cfDNA could be observed in plasma of healthy individuals but was increased in patients with cancer.[37] The main source of circulating cfDNA is apoptosis of cells in healthy controls, whereas both apoptosis and necrosis contribute to elevated levels of cfDNA in cancer patients.[38] The result of our subgroup analysis stratified by assay indicators suggested that elevated ALU and presence of LOH were significantly correlated with patients’ poor PFS and OS; TP-53 mutation was shown to be significantly associated with worse PFS; ESR1 mutation was suggested to be significantly associated with poor OS. In addition, our results showed a tendency of poor outcome among breast cancer patients with PIK3CA and some other mutations, although without statistical significance. As a predictive HR value of more than 2.0 was considered to be statistically strong,[39] ALU concentration, TP-53 mutation, and presence of LOH were proved to be good predictors for survival. Moreover, LOH was shown to be the most effective factor that was associated with shorter PFS and OS. It might be because loss of tumor suppress gene and cyclin-dependent kinase inhibitor could promote cell proliferation.[33]
Subgroup analysis based on patients’ TNM staging demonstrated that cfDNA was a good prognostic maker for PFS in stage I-III and stage I-IV breast cancer patients, while the prognostic value of cfDNA for PFS among stage IV patients remained to be validated. However, cfDNA was shown to be a reliable prognostic marker in patients with stage I-IV. As to the subgroups stratified by sampling time, the results indicated that cfDNA could be a good predictor for PFS among patients before treatment and after treatment, but its prognostic value for OS among this patient population should be further interpreted. Furthermore, in the population mixed with patients before and after treatment, cfDNA was proved to be a good predictor. Regarding origin of sample, both serum and plasma were having good origin in the detection of cfDNA.
As subgroup analysis classified by analytical methods, our results revealed that ddPCR and some other methods were more effective than qPCR in detection of cfDNA. The HR value of ddPCR group in prediction of PFS was 3.44. As reported in previous studies, a prognostic parameter with HR>2 is considered to be useful, which indicated that ddPCR was a good method to detect cfDNA.[39] This might be the reason that ddPCR technology has an excellent precision in quantification of sample.[40] In this way, ddPCR should consider to be adopted in the detection of cfDNA.
There were several limitations in our meta-analysis. First, the number of included studies is relatively small, which is partially due to the fact that articles in other languages were not included in our meta-analysis or that we excluded several studies for that HRs cannot be extracted. Second, heterogeneity was observed in this meta-analysis, so we performed subgroup analyses. Third, only published studies were included in this meta-analysis.
Despite these limitations, our meta-analysis is very valuable and crucial. First of all, we searched the relevant studies by using searching words; meanwhile, related articles and reference lists of these articles were also reviewed to identify all available studies. Second, 2 investigators independently examined the eligibility of studies and performed the quality assessment; all the eligible studies were demonstrated as having high quality. Third, patients of different ethnicities were included in our meta-analysis, which increased the generalizability of our results. Last but not least, we performed subgroup analyses to explore whether our results were influenced by other confounding factors, which demonstrated that our findings were reliable.
5. Conclusion
Our meta-analysis revealed that cfDNA could serve as a good prognostic factor for patients with breast cancer, which may help clinicians identify patients at a high risk of relapse or progression. Elevated circulating cfDNA, TP-53 mutation, and ESR1 mutation were significantly associated with worse survival of patients. However, the results of our meta-analysis need to be validated by further research in this field.
Author contributions
Conceptualization: Jing Yang, Li Cheng, Jing Zhang, Xuelei Ma.
Data curation: Jing Yang, Jing Zhang, Linyan Chen.
Formal analysis: Jing Yang.
Methodology: Jing Yang, Li Cheng, Jing Zhang, Denian Wang, xinli Guo, Xuelei Ma.
Software: Jing Yang, Li Cheng, Linyan Chen, Denian Wang, xinli Guo.
Supervision: Xuelei Ma.
Validation: Jing Yang, Jing Zhang.
Writing – original draft: Jing Yang, Li Cheng, Denian Wang.
Writing – review & editing: Jing Yang, Li Cheng, Jing Zhang, Linyan Chen, Denian Wang, xinli Guo, Xuelei Ma.
Supplementary Material
Footnotes
Abbreviations: CA = carbohydrate antigen, cfDNA = cell-free DNA, CIs = confidence intervals, HR = hazard ratio, HRs = hazard ratios, LOH = loss of heterozygosity, NOS = Newcastle–Ottawa Quality Assessment Scale, OS = overall survival, PFS = progression-free survival, TNM stage = tumor-node-metastasis stage.
JY, LC, and XM contributed equally to this work.
The authors declare no conflicts of interest.
Supplemental Digital Content is available for this article.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 2005; 365(9472):1687–1717. [DOI] [PubMed] [Google Scholar]
- [3].Coughlin SS, Ekwueme DU. Breast cancer as a global health concern. Cancer Epidemiol 2009;33:315–8. [DOI] [PubMed] [Google Scholar]
- [4].Cardoso F, Castiglione M. Locally recurrent or metastatic breast cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol 2009;20(suppl 4):15–8. [DOI] [PubMed] [Google Scholar]
- [5].Donepudi MS, Kondapalli K, Amos SJ, et al. Breast cancer statistics and markers. J Cancer Res Ther 2014;10:506–11. [DOI] [PubMed] [Google Scholar]
- [6].Mirabelli P, Incoronato M. Usefulness of traditional serum biomarkers for management of breast cancer patients. BioMed Res Int 2013;2013:685641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker: a critical appraisal of the literature. Clin Chim Acta 2010;411:1611–24. [DOI] [PubMed] [Google Scholar]
- [8].Schwarzenbach H, Stoehlmacher J, Pantel K, et al. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci 2008;1137:190–6. [DOI] [PubMed] [Google Scholar]
- [9].Goebel G, Zitt M, Zitt M, et al. Circulating nucleic acids in plasma or serum (CNAPS) as prognostic and predictive markers in patients with solid neoplasias. Dis Markers 2005;21:105–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426–37. [DOI] [PubMed] [Google Scholar]
- [11].Ellinger J, Muller SC, Stadler TC, et al. The role of cell-free circulating DNA in the diagnosis and prognosis of prostate cancer. Urol Oncol 2011;29:124–9. [DOI] [PubMed] [Google Scholar]
- [12].Hauser S, Zahalka T, Ellinger J, et al. Cell-free circulating DNA: diagnostic value in patients with renal cell cancer. Anticancer Res 2010; 30:2785–2789. [PubMed] [Google Scholar]
- [13].Moynahan ME, Chen D, He W, et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR+, HER2- advanced breast cancer: results from BOLERO-2. Br J Cancer 2017;116:726–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Piciocchi M, Cardin R, Vitale A, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int 2013;7:1050–7. [DOI] [PubMed] [Google Scholar]
- [15].Ryan BM, Lefort F, McManus R, et al. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut 2003;52:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh N, Gupta S, Pandey RM, et al. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest 2015;33:78–85. [DOI] [PubMed] [Google Scholar]
- [17].Sirera R, Bremnes RM, Cabrera A, et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011;6:286–90. [DOI] [PubMed] [Google Scholar]
- [18].Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- [19].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [20].Basnet S, Zhang ZY, Liao WQ, et al. The prognostic value of circulating cell-free DNA in colorectal cancer: a meta-analysis. J Cancer 2016;7:1105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [22].Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Stat Med 2001;20:641–54. [DOI] [PubMed] [Google Scholar]
- [23].Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Prognostic role of PIK3CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci 2015;106:1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget 2016;7:32504–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol 2016;2:1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [DOI] [PubMed] [Google Scholar]
- [27].Iqbal S, Vishnubhatla S, Raina V, et al. Circulating cell-free DNA and its integrity as a prognostic marker for breast cancer. Springerplus 2015;4:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liang DH, Ensor JE, Liu ZB, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 2016;155:139–49. [DOI] [PubMed] [Google Scholar]
- [29].Madhavan D, Wallwiener M, Bents K, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat 2014;146:163–74. [DOI] [PubMed] [Google Scholar]
- [30].Maxwell KN, Soucier-Ernst D, Tahirovic E, et al. Comparative clinical utility of tumor genomic testing and cell-free DNA in metastatic breast cancer. Breast Cancer Res Treat 2017;164:627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nakauchi C, Kagara N, Shimazu K, et al. Detection of TP53/PIK3CA mutations in cell-free plasma DNA from metastatic breast cancer patients using next generation sequencing. Clin Breast Cancer 2016;16:418–23. [DOI] [PubMed] [Google Scholar]
- [32].Schwarzenbach H, Muller V, Milde-Langosch K, et al. Evaluation of cell-free tumour DNA and RNA in patients with breast cancer and benign breast disease. Mol BioSyst 2011;7:2848–54. [DOI] [PubMed] [Google Scholar]
- [33].Schwarzenbach H, Pantel K, Kemper B, et al. Comparative evaluation of cell-free tumor DNA in blood and disseminated tumor cells in bone marrow of patients with primary breast cancer. Breast Cancer Res 2009;11:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Visvanathan K, Fackler MS, Zhang Z, et al. Monitoring of serum dna methylation as an early independent marker of response and survival in metastatic breast cancer: TBCRC 005 prospective biomarker study. J Clin Oncol 2017;35:751–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Canzoniero JV, Park BH. Use of cell free DNA in breast oncology. Biochim Biophys Acta 2016;1865:266–74. [DOI] [PubMed] [Google Scholar]
- [36].Casciano I, Vinci AD, Banelli B, et al. Circulating tumor nucleic acids: perspective in breast cancer. Breast Care (Basel) 2010;5:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646–50. [PubMed] [Google Scholar]
- [38].Pinzani P, Salvianti F, Orlando C, et al. Circulating cell-free DNA in cancer. Methods Mol Biol (Clifton, NJ) 2014;1160:133–45. [DOI] [PubMed] [Google Scholar]
- [39].Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia 2001;6:375–92. [DOI] [PubMed] [Google Scholar]
- [40].Taylor SC, Laperriere G, Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci Rep 2017;7:2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


