Abstract
The present study aimed to explore the influence of sirtuin 1 (SIRT1) polymorphisms (rs12778366 and rs3758391) on diabetic foot (DF) susceptibility and severity in patients with type 2 diabetes mellitus (T2DM).
This case–control study recruited 142 patients with DF, 148 patients with T2DM, and 148 healthy controls. SIRT1 gene polymorphisms were sequenced by polymerase chain reaction (PCR) and direct sequencing method. The relative expression of SIRT1 mRNA was estimated using quantitative real-time PCR (qRT-PCR) assay. Odds ratio (OR) with 95% confidence interval (95% CI) were used to represent the association of SIRT1 polymorphisms with DF susceptibility and severity. The results were adjusted using logistic regression analysis.
C allele of rs12778366 polymorphism was significantly correlated with reduced DF susceptibility which deriving from healthy controls (adjusted OR = 0.364, 95% CI = 0.158–0.835) so was patients with T2DM (P = .047, OR = 0.591, 95%CI = 0.349–0.998), but the results became nonsignificant adjusted by clinical features (adjusted OR = 0.654, 95% CI = 0.391–1.094). We failed to find any significant association between rs3758391 polymorphisms and T2DM, DF susceptibility. No significant association has been discovered between SIRT1 polymorphisms and DF severity or characteristics. In addition, compared to healthy control and T2DM cases, patients with DF exhibited significant downregulation of SIRT1. The 2 studied polymorphisms had no effects on its gene expression (P > .05 for all).
SIRT1 rs12778366 polymorphism C allele might act as a protective factor for DF onset.
Keywords: diabetic foot, polymorphisms, SIRT1, type 2 diabetes mellitus
1. Introduction
Diabetes mellitus (DM) is a group of common metabolic disorders which is caused by the detect of insulin secretion (type 1 DM, T1DM) or insulin resistance (type 2 DM, T2DM).[1,2] High blood glucose is the main characteristics.[3] Long-term DM will lead to different complications.[4–6] Recent years, with the economic development and the alteration of life style, incidence of T2DM is increased.[7] As a common complication of DM, diabetic foot (DF) also had high prevalence and might lead to disability.[8] Combinations of neural and vascular lesions result in the DF development.[9,10] Infection, ulcer, and neuropathic osteoarthropathy are the common pathogenesis in patients with DF.[11–13] Previous studies suggest that oxidative stress,[14] inflammation,[15] angiogenesis,[16] and nervous lesions[17] are involving in the DF participant. These alterations will lead to the occurrence and difficult healing of foot ulcer. A variety of factors participate in the development of DM.[10,18] Individual response for risk factor is decided by genetic factors.
Sirtuin 1 (SIRT1), also known as NAD-dependent deacetylase sirtuin-1, it is encoded by the gene which is located at chromosome 10q21.3. High insulin resistance could reduce the SIRT1 expression, and then decrease the insulin sensitivity.[19]SIRT1 affects the function of neurons via limiting calorie intake.[20]SIRT1 is downregulated in patients with T2DM and associated with oxidative stress.[21] Suppressed SIRT1 expression promote the inflammation in patients with T2DM.[22] Besides, SIRT1 mediates the survival of endothelial cells.[23] Polymorphisms in SIRT1 gene might alter the expression or function of it, then contribute to different disorders, such as neural or vascular lesions. Glucose tolerance was elevated in C allele carriers of rs12778366.[24] Serum SIRT1 level is elevated in persons with rs3758391 and rs12778366 TT genotypes.[25] C allele of rs12778366 single-nucleotide polymorphism (SNP) was positively correlated with T2DM risk.[26]
Two SNPs, rs12778366 and rs3758391, are in promoter region of SIRT1 gene with the minor allele frequencies more than 0.1 in Chinese Han, Beijing population. Thus we speculated that these 2 SNPs might contribute to the development of DF. In present study, we selected rs12778366 and rs3758391 to detect the association between them and DF susceptibility.
2. Materials and methods
2.1. Subjects
Present study was approved by the ethic committee of The First Hospital of Zhangjiakou. Written informed consent was signed by each participant. Study process was conformed with the Declaration of Helsinki. Basic characteristics of participants were collected using questionnaire.
Between January 2012 to June 2017, 537 patients with T2DM were diagnosed in The First Hospital of Zhangjiakou according to previous standards.[27] Among them, 142 patients with DF (DF group) and 148 patients without DF (DM group) were enrolled in this study. At the same time, 148 healthy individuals were recruited from the healthy checkup center (control group). Blood routine examinations and urine examinations demonstrated that they had normal glucose level, and they did not suffer from any inflammatory diseases, malignant diseases, or infectious diseases based on laboratory biochemical analyses. Age and gender frequencies were matched between patients and controls. Individuals with other DM complications, severe systemic diseases, tumors, neural diseases, vascular disease, inflammatory and immune diseases were excluded from this study. The disease severity of DF was estimated using Wagner staging system.[28]
2.2. Sample collection and genotyping method
After a 12-hour fasting, peripheral venous blood was collected from the participants in the early morning and stored at −80°C until used. GenElute Blood Genomic DNA kit (Sigma-Aldrich, Beijing, China) was used to extract genomic DNA from the blood samples.
Primer Premier 5.0 and Primer-BLAST were used to design the primer sequences of SIRT1 rs12778366 and rs3758391 SNPs based on GenBank database (NG_050664.1). Forward primer of rs12778366 was 5′-CTCCCTCCCCTTGACCCC-3′, reverse primer for rs12778366 was 5′-TCGCTAAGGTCCTATCTACATCCAAAA-3′. For rs3758391 SNP, the primers were 5′-GTTGGTTGCCTAAAGTCACGC-3′ (forward) and 5′-TTCCACTTTCCTCTCTCCCTGA-3′ (reverse). Polymerase chain reaction (PCR) reaction was as the following: predegeneration at 94°C for 8 minutes, then followed by 35 cycles of degeneration at 94°C for 50 seconds, annealing at 60°C for 40 seconds, extension at 72°C for 50 seconds, and the final extension at 70°C for 10 minutes. PCR products were sequenced by Sangon Biotech (Shanghai) Co, Ltd (Shanghai, China) using direct sequencing method.
2.3. RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was isolated from the collected blood samples using Trizol reagent (Invitrogen, Carlsbad, CA). Then the concentration and purity of the extracted RNA were estimated using NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington). The reverse transcription was carried out by PrimeScript 1st Strand cDNA synthesis kit (Takara, Dalian, China). The relative expression of SIRT1 mRNA was evaluated using quantitative real-time PCR (qRT-PCR) method which was performed by SYBR Premix Ex Taq (Takara) in the Applied Biosystems 7900 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). GAPDH served as the internal control. The specific primers were designed by Primer Premier 5.0 software, and their sequences were as follows: GAPDH forward, 5′-ATGGGGAAGGTGAAGGTCGG-3′; reverse, 5′-GACGGTGCCATGGAATTTGC-3′. SIRT1 forward, 5′-GCCTCATCTGCATTTTGATG-3′; reverse, 5′-TCTGGCATGTCCCACTATCA-3′. 2−ΔΔCt method was applied to calculate the relative expression of SIRT1 mRNA. Each test was repeated 3 times.
2.4. Statistical analysis
Hardy–Weinberg equilibrium (HWE) test was utilized to examine the genotype distributions of SIRT1 polymorphisms in control group. Genotype and allele frequencies were calculated by direct counting. The comparison of continuous variables between 2 groups was performed using Student t test. Association between SIRT1 polymorphisms and DF susceptibility were evaluated by Chi-squared test, and the results were estimated using odds ratio (OR) with 95% confidence interval (95% CI). The results were adjusted using logistic regression analysis. SPSS 18.0 was used to perform the calculations. Statistical significant level was set to .05.
3. Results
3.1. Basic characteristics
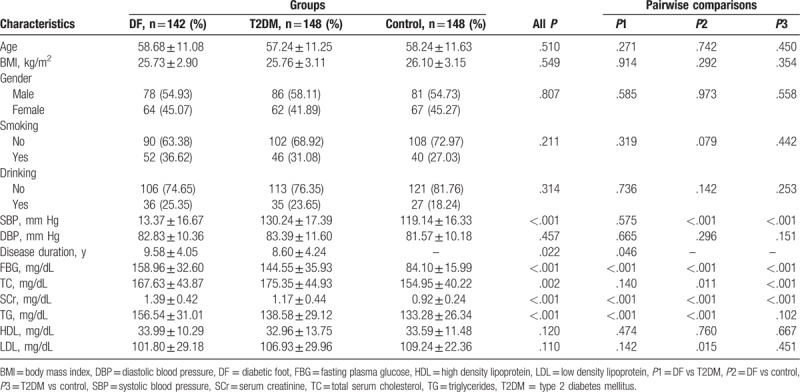
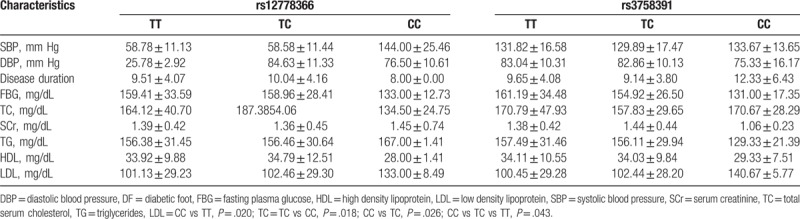
Numbers of males and females were 78 and 64 in DF group, 86 and 62 in T2DM group, and 81 and 67 in control group. Mean ages of these 3 groups were, respectively, 58.68 ± 11.08, 57.24 ± 11.25, and 58.24 ± 11.63 years (Table 1). There were no significant difference between DF group, T2DM group, and control group, indicating that the age and gender were well matched between groups. Systolic blood pressure (SBP), disease duration, fasting plasma glucose (FPG), serum creatinine (SCr), total serum cholesterol (TC), and triglyceride (TG) were significantly different between DF, T2DM, and control groups (Table 1, P < .05). However, other characteristics had no significant difference.
Table 1.
Characteristics of the participants.
3.2. The expression profiles of SIRT1 mRNA in the study population
The qRT-PCR was performed to investigate SIRT1 mRNA in the obtained blood samples. The results displayed in Figure 1 demonstrated that compared to healthy controls, SIRT1 expression was obviously downregulated in T2DM group (P = .021) and DF group (P < .001). Moreover, the mRNA levels of SIRT1 were significantly lower in DF group than that in T2DM group (P = .001).
Figure 1.
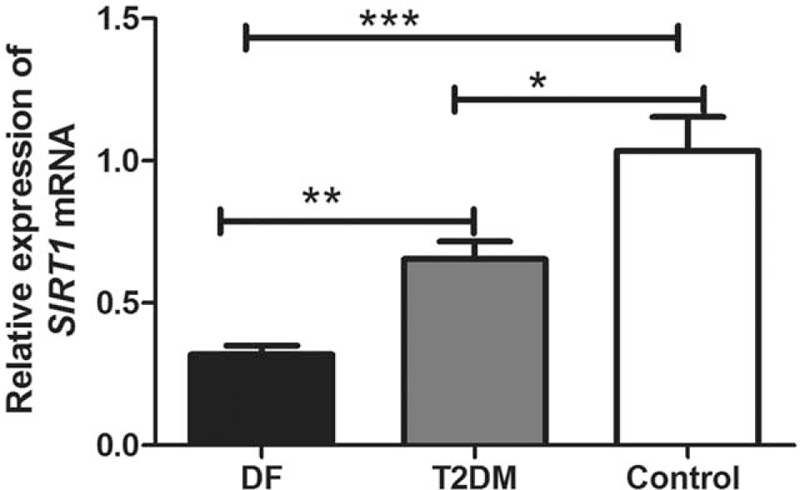
The expression patterns of sirtuin 1 (SIRT1) mRNA in the 3 studied groups. Compared to healthy controls, SIRT1 expression was obviously deceased in type 2 diabetes mellitus (T2DM) group (P = .021) and diabetic foot (DF) group (P < .001). Moreover, the mRNA levels of SIRT1 were significantly lower in DF group than that in T2DM group (P = .001). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
3.3. Association of SIRT1 polymorphisms with DF susceptibility
Genotype and allele distributions of SIRT1 gene rs12778366 and rs3758391 polymorphisms were conformed with HWE test in control group (Table 2, P > .05), suggesting that the study participants could represent the general population.
Table 2.
Genotype and allele distributions of SIRT1 polymorphisms.
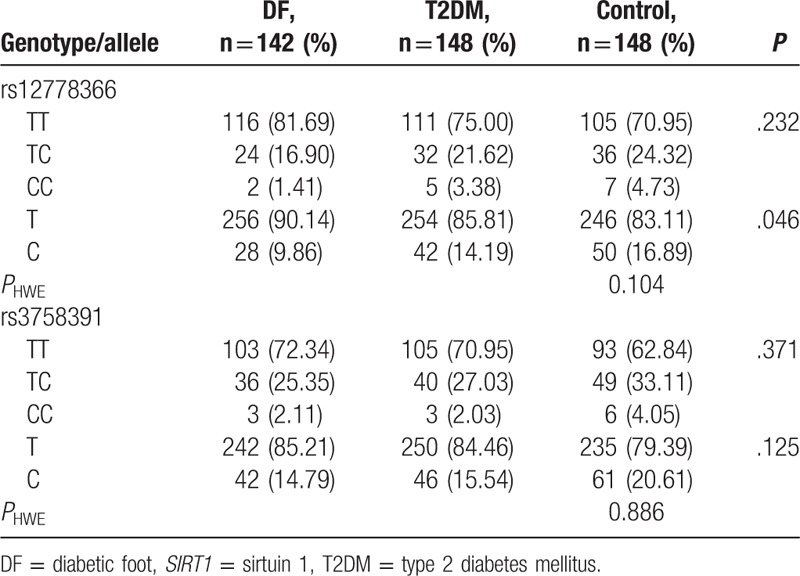
Genotype frequencies of rs12778366 SNP had no significant difference among groups (Table 2, P > .05). Association analysis suggested that no significant association has been discovered between rs12778366 genotypes and T2DM and DF susceptibility (Table 2, P > .05). Allele frequencies of rs12778366 SNP were distinctly different among DF, T2DM, and control groups (Table 2, P = .046). C allele of rs12778366 was more frequently discovered in controls than patients with T2DM and DF. When compared with rs12778366 T allele, C allele carriers had significantly higher risk of DF respectively originating from healthy individuals (Table 3; P = .013, OR = 0.328, 95% CI = 0.328–0.882) and the results were still significantly adjusted by environmental factors (P = .017, adjusted OR = 0.364, 95% CI = 0.158–0.835) so was patients with T2DM (P = .047, OR = 0.591, 95% CI = 0.349–0.998), but the difference was not significant adjusted by environmental factors (P = .106, adjusted OR = 0.654, 95% CI = 0.391–1.094).
Table 3.
Association of SIRT1 polymorphisms with DF susceptibility.
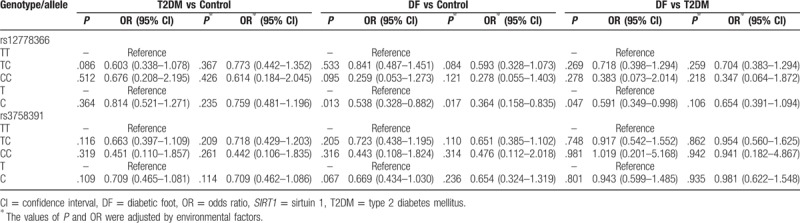
Genotype and allele distributions of rs3758391 SNP were not significantly different among 3 groups (P > .05). In the association analysis, we failed to find any significant association between rs3758391 SNP and T2DM, DF risk (Table 3, P > .05).
3.4. Effects of SIRT1 polymorphisms for DF severity
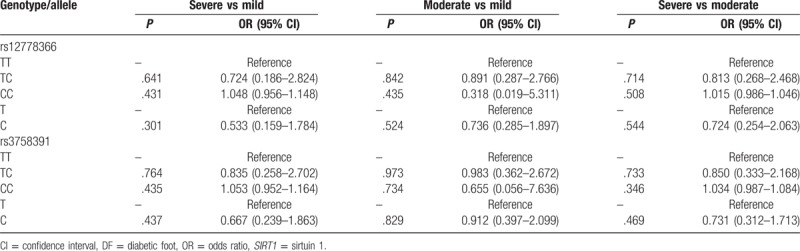
Based on Wagner staging system, 27 patients with DF were 0 to 1 stages (mild), 81 patients were in 2 to -3 stages (moderate), and 34 patients with DF were in 4 to 5 stages (severe). Genotype and allele distributions of rs12778366 and rs3758391 SNPs were detected in different stages (Table 4). However, no significant difference have been discovered in these stages (P > .05). Then we suggested that SIRT1 polymorphisms had no significant effects for DF severity (Table 4).
Table 4.
Effects of SIRT1 polymorphisms for DF severity.
3.5. Effects of SIRT1 polymorphisms for DF characteristics
We also explored the effects of SIRT1 polymorphisms for DF characteristics. However, no significant difference has been discovered between rs12778366 genotypes (Table 5). Similar results had also been discovered between rs3758391 genotypes (Table 5).
Table 5.
Effects of rs12778366 genotypes for DF characteristics.
3.6. The influences of SIRT1 polymorphisms on SIRT1 expression
Both of the 2 studied polymorphisms were located at the promoter region of SIRT1 gene. In our study, we estimated the genetic association of SIRT1 genotypes with its gene transfection. Analysis results suggested that there were no significant association between genotypes of rs12778366 and rs3758391 SNPs with expression levels of SIRT1 mRNA in any of the 3 studied groups (P > .05 for all) (Fig. 2A–F).
Figure 2.
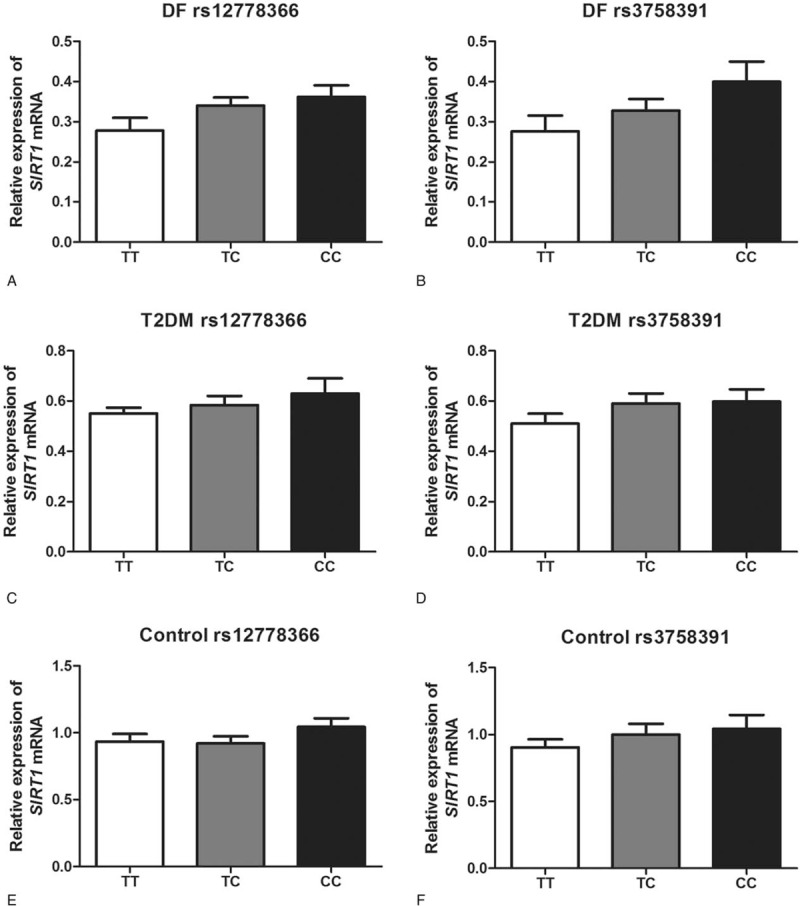
The genetic effects of SIRT1 polymorphisms on expression levels of SIRT1 in the study population. There were no significant association between genotypes of rs12778366 and rs3758391 SNPs with expression levels of SIRT1 mRNA in any of the three studied groups (P > .05 for all). DF = diabetic foot, SIRT1 = sirtuin 1, T2DM = type 2 diabetes mellitus.
4. Discussion
Neuronal and vascular disorders were participated in DF development. SIRT1 play protective effect for neuronal proliferation[29,30] and vascular cells.[31]SIRT1 could mediate the insulin resistance via inflammatory process, reactive oxygen species, gluconeogenesis, and adiponectin level.[24] In our study, we found that patients with T2DM and DF exhibited downregulation of SIRT1. The expression deficiency of SIRT1 gene might be involved in etiology of T2DM and its complications. Binding affinity between SIRT1 and p53 was reduced by rs3758391 C allele.[32] Hu and colleagues demonstrated that rs12778366 genotypes had no significant effects for mRNA expression level of SIRT1.[33] However, Rai et al suggested significantly higher expression level of SIRT1,[34] while rs3758391 TT genotype carriers had significantly higher mRNA expression level of SIRT1 than that in TC and CC genotype carriers.[33] Unfortunately, we did not observe any significant association between SIRT1 rs3758391 and rs12778366 polymorphisms in our study. The divergences might be attributed to the relatively small sample size.
Significantly higher glucose tolerance was discovered in rs12778366 C allele carriers.[24] Neurodevelopment is significantly affected by rs3758391 SNP.[35] These 2 SNPs have been explored in various diseases, such as pituitary adenoma, autoimmune thyroid disease, and breast cancer.[25,36,37] Acute coronary syndrome is protected by SIRT1 gene.[33] Breast cancer susceptibility was correlated with rs12778366 and rs3758391 polymorphisms.[25] Cardiovascular mortality risk was reduced by rs3758391 SNP.[38] These evidence suggested that SIRT1 gene rs12778366 and rs3758391 SNPs might play crucial role in T2DM development, then contributing to DF risk. Nevertheless, there was no previous study focused on the association in Chinese Han population. Thus we carried out this study.
In present study, we found that SBP, disease duration, FPG, SCr, TC, and TG levels were distinctly different between DF, T2DM, and control groups. T allele of rs12778366 SNP was more frequently discovered in healthy controls than that in patients with T2DM and DF. It suggested that rs12778366 T allele was obviously correlated with approximately 0.328 and 0.591 times reduced DF risk which developing from healthy persons and patients with T2DM. This was conformed with the result performed by Rai and colleagues which found that rs12778366 TT genotype carriers had high risk for T2DM.[34] On the contrary, Han et al indicated that C allele of rs12778366 SNP was positively correlated with increased T2DM susceptibility.[26]
We failed to find any significant association between rs3758391 and DF susceptibility. Study performed by Kovanen et al found that rs3758391 genotypes had no significant difference between patients with T2DM and controls.[39] However, Cruz and coworkers demonstrated that rs3758391 T allele is positively correlated with T2DM susceptibility.[40] Lv and colleagues suggested that rs3758391 had no significant association with susceptibility of different lung cancer subtype.[41] T allele of rs3758391 was positively correlated with nephritis in patients with systemic lupus erythematosus.[42]
To certify the mechanism of SIRT1 polymorphisms to DF development, we explored the effects of them for DF severity. Twenty-seven patients with DF were Wagner stages 0 to 1 (mild), 81 patients were Wagner stages 2 to 3 (moderate), and 34 patients with DF were Wagner stages 4 to 5 (severe). However, genotype and allele distributions of rs12778366 and rs3758391 SNPs had no significant difference between Wagner stages. It indicated that no significant effects have been caused by these 2 SNPs. At the same time, we failed to find any significant effects of rs12778366 and rs3758391 SNPs to DF characteristics, despite these characteristics were different between genotype carriers.
There were several limitations in present study. Firstly, statistical power was reduced by the relatively small sample size, particularly in severity analysis. Secondly, the development of DF and T2DM was regulated by the interactions between various genetic and environmental factors; however, these factors were not considered in this study. Thirdly, functions of SIRT1 polymorphisms in DF development were not certified in present study. In addition, to clearly state the genetic effects of SIRT1 polymorphisms on severity of DF, more staging system should be applied, such as DUSS system. Therefore, further study should be performed in the future to verify current result.
In conclusion, SIRT1 gene rs12778366 T allele might reduce individual susceptibility to DF in Chinese Han population. But SIRT1 rs12778366 and rs3758391 polymorphisms had no significant influence on SIRT1 expression, DF severity or clinical characteristics. The present study might be helpful for early screening and prevention of DF.
Author contributions
Conceptualization: Hongxia Tang.
Data curation: Guishan Zhang, Hongxia Tang.
Formal analysis: Guishan Zhang, Hongxia Tang, Luling Dong.
Funding acquisition: Guishan Zhang, Hongxia Tang, Luling Dong.
Investigation: Yi Peng, Luling Dong, Chunbin Gao.
Methodology: Yi Peng, Luling Dong, Chunbin Gao.
Project administration: Chunbin Gao.
Resources: Yi Peng, Chunbin Gao.
Software: Xiuhong Yang.
Supervision: Xiuhong Yang.
Validation: Xiuhong Yang, Ying Peng, Yanrong Xu.
Visualization: Xiuhong Yang, Ying Peng, Yanrong Xu.
Writing - original draft: Yi Peng, Guishan Zhang, Ying Peng, Yanrong Xu.
Writing - review & editing: Yi Peng, Guishan Zhang, Ying Peng, Yanrong Xu.
Footnotes
Abbreviations: 95% CI = 95% confidence interval, DBP = diastolic blood pressure, DF = diabetic foot, DM = diabetes mellitus, FPG = fasting plasma glucose, HWE = Hardy–Weinberg equilibrium, OR = odds ratio, PCR = polymerase chain reaction, qRT-PCR = quantitative real-time polymerase chain reaction, SBP = systolic blood pressure, SCr = serum creatinine, SIRT1 = sirtuin 1, SNPs = single-nucleotide polymorphisms, T2DM = type 2 diabetes mellitus, TC = total serum cholesterol, TG = triglycerides.
The authors have no funding and conflicts of interest to disclose.
References
- [1].Lindfors E, Gopalacharyulu PV, Halperin E, et al. Detection of molecular paths associated with insulitis and type 1 diabetes in non-obese diabetic mouse. PLoS One 2009;4:e7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xu H, Wang Q, Sun Q, et al. In type 2 diabetes induced by cigarette smoking, activation of p38 MAPK is involved in pancreatic beta-cell apoptosis. Environ Sci Pollut Res Int 2018;25:9817–27. [DOI] [PubMed] [Google Scholar]
- [3].Miyoshi Y, Ogawa O, Oyama Y. Nivolumab, an anti-programmed cell death-1 antibody, induces fulminant type 1 diabetes. Tohoku J Exp Med 2016;239:155–8. [DOI] [PubMed] [Google Scholar]
- [4].Umemura T, Kawamura T, Hotta N. Pathogenesis and neuroimaging of cerebral large and small vessel disease in type 2 diabetes: a possible link between cerebral and retinal microvascular abnormalities. J Diabetes Investig 2017;8:134–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Duarte JS, Bello C, Ferreira R, et al. Atherogenic dislipidemia in patients with type 2 diabetes linked to microvascular complications and heart disease. Atherosclerosis 2017;263:e265. [Google Scholar]
- [6].Yamazaki D, Hitomi H, Nishiyama A. Hypertension with diabetes mellitus complications. Hypertens Res 2018;41:147–56. [DOI] [PubMed] [Google Scholar]
- [7].Mayoral Gonzalez B, Riano Galan I, Rodriguez Dehli C, et al. Epidemiology of type 1 diabetes in Asturias: 2002-2011. Endocrinol Diabetes Nutr 2018;65:68–73. [DOI] [PubMed] [Google Scholar]
- [8].Khan A, Junaid N. Prevalence of diabetic foot syndrome amongst population with type 2 diabetes in Pakistan in primary care settings. J Pak Med Assoc 2017;67:1818–24. [PubMed] [Google Scholar]
- [9].Noor S, Ahmad J, Parwez I, et al. Culture-based screening of aerobic microbiome in diabetic foot subjects and developing non-healing ulcers. Front Microbiol 2016;7:1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qiu H, Schooling CM, Sun S, et al. Long-term exposure to fine particulate matter air pollution and type 2 diabetes mellitus in elderly: a cohort study in Hong Kong. Environ Int 2018;113:350–6. [DOI] [PubMed] [Google Scholar]
- [11].Chin YF, Yeh JT, Yu HY, et al. Knowledge of the warning signs of foot ulcer deterioration among patients with diabetes. J Nurs Res 2017. [DOI] [PubMed] [Google Scholar]
- [12].Suryaletha K, John J, Radhakrishnan MP, et al. Metataxonomic approach to decipher the polymicrobial burden in diabetic foot ulcer and its biofilm mode of infection. Int Wound J 2018;15:473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Babamahmoodi F, Shokohi T, Ahangarkani F, et al. Rare case of Aspergillus ochraceus osteomyelitis of calcaneus bone in a patient with diabetic foot ulcers. Case Rep Med 2015;2015:509827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bolajoko EB, Akinosun OM, Anetor J, et al. Relationship between selected micronutrient deficiencies and oxidative stress biomarkers in diabetes mellitus patients with foot ulcers in Ibadan, Nigeria. Turk J Med Sci 2017;47:1117–23. [DOI] [PubMed] [Google Scholar]
- [15].Davis FM, Kimball A, Boniakowski A, et al. Dysfunctional wound healing in diabetic foot ulcers: new crossroads. Curr Diab Rep 2018;18:2. [DOI] [PubMed] [Google Scholar]
- [16].Kim S, Kim J, Choi J, et al. Polydeoxyribonucleotide improves peripheral tissue oxygenation and accelerates angiogenesis in diabetic foot ulcers. Arch Plast Surg 2017;44:482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Almuhannadi H, Ponirakis G, Khan A, et al. Diabetic neuropathy and painful diabetic neuropathy: Cinderella complications in South East Asia. J Pak Med Assoc 2018;68:85–9. [PubMed] [Google Scholar]
- [18].Lefrancois T, Mehta K, Sullivan V, et al. Evidence based review of literature on detriments to healing of diabetic foot ulcers. Foot Ankle Surg 2017;23:215–24. [DOI] [PubMed] [Google Scholar]
- [19].Sun C, Zhang F, Ge X, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab 2007;6:307–19. [DOI] [PubMed] [Google Scholar]
- [20].Srivastava S, Haigis MC. Role of sirtuins and calorie restriction in neuroprotection: implications in Alzheimer's and Parkinson's diseases. Current Pharm Des 2011;17:3418–33. [DOI] [PubMed] [Google Scholar]
- [21].Bo S, Togliatto G, Gambino R, et al. Impact of sirtuin-1 expression on H3K56 acetylation and oxidative stress: a double-blind randomized controlled trial with resveratrol supplementation. Acta Diabetol 2018;55:331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Peng J, Zhou Y, Deng Z, et al. miR-221 negatively regulates inflammation and insulin sensitivity in white adipose tissue by repression of sirtuin-1 (SIRT1). J Cell Biochem 2018;119:6418–28. [DOI] [PubMed] [Google Scholar]
- [23].Jiang B, Jen M, Perrin L, et al. SIRT1 overexpression maintains cell phenotype and function of endothelial cells derived from induced pluripotent stem cells. Stem Cells Dev 2015;24:2740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Figarska SM, Vonk JM, Boezen HM. SIRT1 polymorphism, long-term survival and glucose tolerance in the general population. PLoS One 2013;8:e58636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rizk SM, Shahin NN, Shaker OG. Association between SIRT1 gene polymorphisms and breast cancer in Egyptians. PLoS One 2016;11:e0151901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Han J, Wei M, Wang Q, et al. Association of genetic variants of SIRT1 with type 2 diabetes mellitus. Gene expression 2015;16:177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care 2008;31(Suppl 1):S55–60. [DOI] [PubMed] [Google Scholar]
- [28].Wagner FW., Jr The dysvascular foot: a system for diagnosis and treatment. Foot Ankle 1981;2:64–122. [DOI] [PubMed] [Google Scholar]
- [29].Li H, Wang R. Blocking SIRT1 inhibits cell proliferation and promotes aging through the PI3K/AKT pathway. Life Sci 2017;190:84–90. [DOI] [PubMed] [Google Scholar]
- [30].Koronowski KB, Khoury N, Saul I, et al. Neuronal SIRT1 (silent information regulator 2 homologue 1) regulates glycolysis and mediates resveratrol-induced ischemic tolerance. Stroke 2017;48:3117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lin Y, Zhao Y, Liu E. High glucose upregulates endothelin type B receptors in vascular smooth muscle cells via the downregulation of Sirt1. Int J Mol Med 2018;41:439–45. [DOI] [PubMed] [Google Scholar]
- [32].Dong Y, Guo T, Traurig M, et al. SIRT1 is associated with a decrease in acute insulin secretion and a sex specific increase in risk for type 2 diabetes in Pima Indians. Mol Genet Metab 2011;104:661–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hu Y, Wang L, Chen S, et al. Association between the SIRT1 mRNA expression and acute coronary syndrome. J Atheroscler Thromb 2015;22:165–82. [DOI] [PubMed] [Google Scholar]
- [34].Rai E, Sharma S, Kaul S, et al. The interactive effect of SIRT1 promoter region polymorphism on type 2 diabetes susceptibility in the North Indian population. PLoS One 2012;7:e48621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brett ZH, Sheridan M, Humphreys K, et al. A neurogenetics approach to defining differential susceptibility to institutional care. Int J Behav Dev 2015;39:150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Glebauskiene B, Vilkeviciute A, Liutkeviciene R, et al. Association of FGFR2 rs2981582, SIRT1 rs12778366, STAT3 rs744166 gene polymorphisms with pituitary adenoma. Oncol Lett 2017;13:3087–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sarumaru M, Watanabe M, Inoue N, et al. Association between functional SIRT1 polymorphisms and the clinical characteristics of patients with autoimmune thyroid disease. Autoimmunity 2016;49:329–37. [DOI] [PubMed] [Google Scholar]
- [38].Zillikens MC, van Meurs JB, Sijbrands EJ, et al. SIRT1 genetic variation and mortality in type 2 diabetes: interaction with smoking and dietary niacin. Free Radic Biol Med 2009;46:836–41. [DOI] [PubMed] [Google Scholar]
- [39].Kovanen L, Donner K, Partonen T. SIRT1 polymorphisms associate with seasonal weight variation, depressive disorders, and diastolic blood pressure in the general population. PLoS One 2015;10:e0141001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cruz M, Valladares-Salgado A, Garcia-Mena J, et al. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev 2010;26:261–70. [DOI] [PubMed] [Google Scholar]
- [41].Lv Y, Lin S, Peng F. SIRT1 gene polymorphisms and risk of lung cancer. Cancer Manag Res 2017;9:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Consiglio CR, Juliana da Silveira S, Monticielo OA, et al. SIRT1 promoter polymorphisms as clinical modifiers on systemic lupus erythematosus. Mol Biol Rep 2014;41:4233–9. [DOI] [PubMed] [Google Scholar]


