Abstract
Vaccinia virus (vv) early transcription can be reconstituted in vitro from purified virions; in this assay mRNAs are made inside the viral core and subsequently extruded. Although the in vitro process has been extensively characterized, relatively little is known about vv early transcription in vivo. In the present study the fate of vv early mRNAs in infected HeLa cells was followed by BrUTP transfection and confocal and electron microscopy. The extruded vv early mRNAs were found to be organized into unique granular cytoplasmic structures that reached a size up to 1 μm. By EM these structures appeared as amorphous electron-dense cytoplasmic aggregates that were surrounded by ribosomes. Confocal images showed that the RNA structures were located some distance away from intracellular cores and that both structures appeared to be aligned on microtubules (MTs), implying that MT tracks connected mRNAs and cores. Accordingly, intact MTs were found to be required for the typical punctate organization of viral mRNAs. Biochemical evidence supported the notion that vv mRNAs were MT associated and that MT depletion severely affected viral (but not cellular) mRNA synthesis and stability. By confocal microscopy the viral mRNA structures appeared to be surrounded by molecules of the translation machinery, showing that they were active in protein synthesis. Finally, our data suggest a role for a MT and RNA-binding viral protein of 25 kDa (gene L4R), in mRNA targeting away from intracellular cores to their sites of cytoplasmic accumulation.
INTRODUCTION
Vaccinia virus (vv) is the prototype member of the poxviruses containing a DNA genome of around 190 kb that encodes for >200 proteins. Vv DNA replication occurs in the cytoplasm of infected cells, rather than in the nucleus, and accordingly, the viral genome encodes for factors required for both cytoplasmic transcription as well as DNA replication (Moss, 1996, 1990).
The vv cytoplasmic life cycle is initiated upon virion entry, a process in which the particle loses its membranes at the plasma membrane (Krijnse Locker et al., 2000) and delivers the core containing ∼100 proteins into the cytoplasm. Within 20 min of infection from these cores a defined set of “early mRNAs” is made, in which about half the genome is transcribed. This expression of early genes is required to initiate the subsequent process of DNA replication, starting from ∼2 h postinfection. DNA replication triggers the transcription of late genes, late proteins being required for the assembly of new virions starting at ∼6 h postinfection. During virion assembly, a double-membraned cisterna derived from the smooth ER (sER) is wrapped around the viral genome to make the first infectious form of the virus, the brick-shaped intracellular mature virus (IMV; Sodeik et al., 1993). A second infectious form is generated when the IMV becomes enwrapped by membranes of the trans-Golgi network to form the extracellular enveloped virus (EEV; Schmelz et al., 1994)
During virion assembly, late in infection, the enzymes required for the process of cytoplasmic vv early transcription are packaged into the particle (Gershon and Moss, 1990). Although transcriptionally inactive during assembly, the enzymes become activated upon viral entry when the core ends up in the cytoplasm. One consequence of the fact that the enzymes required for vv early transcription are contained in the virions is that vv early mRNA synthesis can be reconstituted in vitro; incubation of purified virus with a mixture of detergent to disrupt the membrane, a reducing agent, Mg2+, and nucleotide triphosphates results in the production from vv cores of early mRNAs that are similar to those made early in infection (Kates and Beeson, 1970; Cooper and Moss, 1978; Pelham et al., 1978). This in vitro assay has enabled to study the molecular aspects of vv early transcription in detail (see e.g., Moss, 1990, 1991). Pioneering in vitro work by Kates and Beeson (1970) showed biochemically that mRNAs are first made inside the core and then released through (hypothetical) exit sites, extrusion from the core requiring the hydrolysis of ATP. It is generally assumed that in infected cells vv early mRNAs are also made inside the core from which they are subsequently extruded (see e.g., Metz et al., 1975). Despite the vast knowledge on the molecular requirements of vv transcription in vitro, relatively little is known about this process in the infected cell, in particular about the fate of the early mRNAs once extruded from the core. Biochemical studies have shown that viral early messengers are bound to polyribosomes in infected cells (Jeffert and Holowczak, 1971; Metz et al., 1975).
We have recently characterized the intracellular cores as they occur early in infection by analyzing their composition both biochemically and morphologically (Pedersen et al., 2000). In that study, intracellular cores accumulated in the presence or absence of the transcription inhibitor actinomycin D (act D) were compared. The data indicated that the outer surface of the core is composed of the proteins 4a/4b (genes A10L and A3L) as well as p39 (A4L), whereas the interior of the core contained the genome as well as the putative DNA-binding protein p25 (L4R). In the absence of act D, however, labeling for p25 as well as for the DNA was gradually lost from the remaining 4a/4b- and p39-containing core shell, before the latter was eventually degraded.
In mammalian cells, transcription as well as mRNA processing occur in specific regions of the nucleus (Lamond and Earnshaw, 1998; Lewis and Tollervey, 2000; Misteli, 2000). For translation, they leave the nucleus through the nuclear pores and associate with polyribosomes in the cytoplasm. These polysomes may associate with the rough ER for the cotranslational insertion of membrane proteins or for the translocation into the lumen of that organelle of secreted proteins. Alternatively, mRNAs encoding for cytoplasmic proteins may be bound to the cytoskeleton. The cytoskeletal element with which mRNAs associate depends on the species and the origin of the cell type. In Drosophila and Xenopus oocytes as well as in mammalian neuronal cells, microtubules (MTs) are generally required for the proper targeting of specific mRNAs. In contrast, in fibroblasts as well as in budding yeast, mRNAs appear to require actin for their localization (Wilhelm and Vale, 1993; Hesketh, 1996; for reviews see Hazelrigg, 1998; Oleynikov and Singer, 1998; Jansen, 1999).
Cytoskeletal-bound mRNAs may organize into supramolecular complexes, which by immunofluorescence (IF) microscopy appear as granular structures. The latter not only contain mRNAs but also RNA-binding proteins, and proteins involved in translation as well as proteins required for RNA targeting, which regulate cytoskeletal binding and/or transport (reviewed in Jansen, 1999). Studies on the localization of viral mRNAs have shown that these can also bind to the cytoskeleton (Lenk and Penman, 1979; van Venrooij et al., 1981; Bonneau et al., 1985). In some cases viral infection has been shown to lead to the release of cellular messengers from the cytoskeleton in favor of the binding of viral mRNAs. Because cytoskeletal association may be required for mRNA-stability as well as for their efficient translation (Jansen, 1999), release of cellular mRNAs from the cytoskeleton could then facilitate viral induced host protein synthesis shut-off.
In the present study we have followed the fate of vv early mRNAs in infected HeLa cells. We show that soon after synthesis, the viral messengers move away from the viral cores and organize into discrete granular structures. MTs are required for their granular organization as well as for efficient transcription and translation of the viral mRNAs. The current data suggest a likely role for the viral protein p25 (L4R) in targeting of the mRNAs to the granular structures.
MATERIALS AND METHODS
Chemicals
All chemicals were purchased from Sigma (St. Louis, MO) except for nocodazole (Calbiochem, San Diego, CA) and latrunculin A (Lat A; Molecular Probes, Eugene, OR).
Cell Culture and Virus Preparation
HeLa cells were grown essentially as described by Sodeik et al. (1993). The cells were treated with 10 μM nocodazole for 1 h or with 1 μM Lat A for 20 min at 37°C. Unless indicated differently, nocodazole was added 1 h before infection and left throughout infection. Puromycin (30 μg/ml) and cycloheximide (25 μg/ml) were added 30 min before fixation. The vaccinia virus strain WR was propagated in HeLa cells and semipurified as described (Pedersen et al., 2000).
Metabolic Labeling
Infected HeLa cells, nocodazole- or mock-treated, were labeled with 50 μCi/ml [35S]methionine or [3H]uridine (NEN-Du Pont, Boston, MA) in DMEM/5% FCS for 30 min at 37°C. After the incubation the cells were washed and collected in cold PBS in Eppendorf tubes. The cells were lysed in lysis buffer (50 mM HEPES, pH 6.9, 100 mM KCl, 1 mM DTT, and 0.5% NP-40), and the amount of protein was measured with the use of a Bio-Rad protein-assay (Bio-Rad, Hercules, CA). For the uridine incorporation, 100 U/ml RNase inhibitor (Promega, Madison, WI) was added to the lysis buffer. A similar amount protein contained in each sample corresponding to 0.5 OD595 was TCA precipitated on 3M paper (Minneapolis, MN), and the radioactivity was determined by liquid scintillation counting.
Light Microscopy Assay
Cells (1.5 × 105) were grown on 11-mm-diameter round coverslips in 24-multiwell dishes. After 16 h the cells, in exponential growth phase, were washed twice in serum-free DMEM and infected with WR for 15 min at 37°C at multiplicity of infection (MOI) of 50. After infection, the cells were washed and lipofected (Lipofectin reagent; Life Technologies-BRL, Gaithersburg, MD). Briefly, 10 mM of 5′-bromouridine 5′-triphosphate (BrUTP; Sigma) or 2 μg of an antisense oligonucleotide corresponding to the H5R gene (2′-O-methylated, 5′-GCCAUCUUUGUGAAACUAGUAUC-3′) coupled to four biotin moieties was preincubated for 30 min in 30 μl of transfection medium containing 3.7 μl of lipofectin. The samples were diluted in 300 μl of transfection medium and added to the cells in presence of 5 μg/ml act D. After 1-h incubation, the cells were extensively washed and incubated in complete culture medium containing 5 mM hydroxyurea (Sigma) and fixed at the indicated times post-act D washout. In one control experiment a biotinylated antisense oligo to snRNA U1 (a subunit of the spliceosome) identical to the one published previously (Carmo-Fonseca et al., 1991) was also lipofected into infected cells.
Antibodies, Immunofluorescence, and Electron Microscopy
The following antibodies were obtained from commercial sources: anti-BrU (Harlan Sera-Lab, England), anti–β-tubulin (Amersham International, Amersham, UK), and anti-actin (Sigma). The antibodies against vaccinia proteins p25 (L4R) were raised in rabbit with the use of the corresponding GST-tagged proteins. The antibody to p39 (A4L; a kind gift of Mariano Esteban; Maa et al., 1990), anti-EF1α was a kind gift of George Janssen, IF4 E was a kind gift of Martina Muckenthaler. Cells were fixed for 15 min in 3% paraformaldeyde and washed three times in PBS containing 40 mM glycine. The cells were permeabilized in 0.1% Triton X-100 (TX-100) for 1 min, and the coverslips were blocked in a solution containing 2% FCS, 2% BSA in TBS (20 mM Tris-HCl, pH 7.5, 154 mM NaCl, 2 mM EGTA, 2 mM MgCl2) for 30 min. When indicated, the cells were Triton-extracted before fixation in 0.1% TX-100 in PHGM buffer (60 mM PIPES, pH 6.8, 25 mM HEPES, 2.5 mM Mg-acetate, 1 mM EGTA) for 2 min and fixed in 3% paraformaldeyde in PHGM buffer. Fixed cells were processed for immunofluorescence and analyzed by confocal microscopy (LSM 510; Zeiss, Jena, Germany). Image analysis of the total fluorescence was performed by NIH image 1.62. For electron microscopy (EM) localization of BrUTP-labeling, cells were grown overnight in a 6-cm dish. They were subsequently infected and lipofected with BrUTP as described for the light microscopy assay. Before fixation cells were TX-100 extracted for 2 min at room temperature. Fixed cells were prepared for cryosectioning as described (Griffiths, 1993). Epon embedding was according to Tilney et al. (1998). Briefly, uninfected cells and cells infected for 2 h in the presence of hydroxyurea were washed once with PBS and then fixed for 45 min at 4°C, in 1% glutaraldehyde and 1% osmium in phosphate buffer, pH 7.4. They were then washed extensively with water before dehydration in ethanol and overnight incubation at 4°C in the dark in uranyl acetate in 70% ethanol followed by Epon embedding as described in Griffiths (1993).
Cell Fractionating, RNA Isolation, and RNase Protection Assay
HeLa cells, infected or mock infected, were fractionated, at 2 h postinfection, as described (Cervera et al., 1981). Briefly, cells were washed with PBS and treated with extraction buffer (10 mM PIPES, pH 6.8, 100 mM KCl, 2.5 mM MgCl2, and 0.1% TX-100) at 4°C for 1 min and washed once with the same buffer without Triton. The material obtained under these condition was referred to as the soluble fraction. The cell remnants were scraped in cold cytoskeleton buffer (20 mM HEPES, pH 7.5, 0.5 M NaCl, 30 mM Mg-acetate, 0.5% deoxycholate, and 1% Tween-20), collected in Eppendorf tubes, and left for 5 min on ice. The suspension was passed through a low-gauge needle and centrifuged for 5 min at maximum speed. The recovered supernatants were referred to as the soluble fraction. Both fractions were treated with 200 μg/ml proteinase K in 0.5% SDS for 30 min at 37°C. The RNAs were purified by multiple round of phenol/chloroform (v/v 1:1) extractions and ethanol precipitated.
For the RNase protection assay 20 μg of total RNA was incubated in 20 μl of hybridization buffer (80% deionized formamide, 40 mM PIPES, pH 6.8, 400 mM NaCl, 5 mM EDTA, pH 8.0) containing 5000 cpm/ml [α-32P]UTP–labeled specific probe. The samples were incubated at 85°C for 5 min and the hybridized at 54°C for 5 h. Subsequently, the samples were treated with RNase A buffer (10 mM Tris, pH 7.5, 5 mM EDTA, 300 mM NaCl, and 10 mg/ml RNase A; Boehringer Mannheim, Mannheim, Germany) for 30 min at 30°C. After proteinase K treatment (5 μg/ml proteinase K, 0.5% SDS, 100 mM Tris, pH 7.5, for 30 min at 37°C) to inactivate RNase A activity, 5 μg of tRNA (Boehringer Mannheim) was added, and the samples were extracted twice with phenol/chloroform and ethanol precipitated. After centrifugation the pellet was resuspended in loading buffer (80% formamide, 0.5% bromophenol blue, 0.25% xylene cyanol, 10% glycerol), boiled, and loaded on a denaturing gel (7 M urea, acrylamide:bis acrylamide [30:1] in 0.5× Tris-Borate-EDTA). Gels were dried and autoradiographed. The evaluation of the protected RNA was performed by phosphoimager. The ribo-probe was prepared with the use of the “Riboprobe In Vitro Transcription System” (Promega), as indicated by the manufacturer.
Polyribosome Sucrose Gradients and Northern and Western Blotting
For the Northern blotting, 20 μg of total RNA, purified as described above, was separated by electrophoresis on 1.2% agarose gel containing 0.4 M MOPS, pH 7.0, and 0.66 M formaldehyde and transferred onto GeneScreen Plus membrane (NEN-Du Pont). The RNA blots were hybridized with H5R and β-actin probes [α-32P]ATP that were labeled by random priming (Promega) according to the manufacturer's instructions. The hybridization was performed in 1 M NaCl, 10% dextran sulfate, 1% SDS, 100 μg/ml sonicated salmon sperm DNA (Sigma), containing 1 × 106 cpm/ml probes at 60°C overnight. The blots were washed in 2× SSC and 1% SDS at the same temperature, followed autoradiography. The densitometric evaluation was performed by phosphoimager. For the Western blotting, 30 μg of total proteins was separated by SDS-PAGE with 12% resolution gel and 4% stacking gel. Samples were incubated in Laemmli sample buffer (1% SDS, 0.5% β-mercaptoethanol) at 95°C for 5 min. The gels were transferred to nitrocellulose membranes for immunoblotting with the use of a Bio-Rad semidry blotting system. The membranes were blocked in PBS, 0.2% Tween 20, and 5% milk powder for 2 h before incubation with anti–β-tubulin or anti–β-actin antibodies followed by horseradish peroxidase-tagged goat anti-mouse antibody (Bio-Rad). Proteins were detected by enhanced chemiluminescence (ECL; Amersham). The detection of mRNAs associated with polyribosomes was done as described (Korner et al., 1998). Briefly, 5 × 109 HeLa cells were infected for 90 min as described above. Ten minutes before harvesting, the cells were treated with 10 μg/ml cycloheximide (CX; Sigma). They were then washed with cold PBS containing CX and lysed for 10 min on ice in 0.5 ml of buffer (10 mM HEPES, pH 7.2, 0.15 M KCl, 10 mM MgCl2, 20 mM DTT, 0.5% NP-40, 150 μg/ml CX, and 100 U/ml RNasin; Promega). The lysates were centrifuged for 10 min in an Eppendorf centrifuge at 4°C. The supernatant was adjusted to 0.25 M KCl and layered on top of a 11-ml 10–40% sucrose gradient (in 20 mM HEPES, pH 7.2, 0.25 M KCl, 10 mM MgCl2, 20 mM DTT, 150 μg/ml CX) and centrifuged for 135 min at 32,000 rpm in a Beckman SW41 Ti rotor (Fullerton, CA) at 4°C. The gradient was fractionated in 24 fractions, and the polyribosomes profile was monitored by measuring the adsorbance at 254 nm. The RNA in each fraction was extracted as described for the Northern blotting procedure and then analyzed with a specific probe to H5R mRNA. To disintegrate polyribosomes, cells were either incubated with 100 μg/ml puromycin 1 h before lysis or the cell-lysate was treated with 25 mM EDTA before centrifugation. In the latter case the sucrose gradient also contained 25 mM EDTA.
RESULTS
After Synthesis, vv Early mRNAs Organize into Discrete Granular Structures
The following light microscopy assay was established to follow the fate of vv early mRNAs in infected HeLa cells. HeLa cells were infected with vv and subsequently lipofected with BrUTP. Because BrUTP-liposome uptake has been shown to peak within 30–60 min (Haukenes et al., 1997), infected cells were transfected for 1 h in the presence of the transcriptional inhibitor act D, after which the drug was washed out to allow a synchronized incorporation of BrU into (viral) mRNAs. On act D washout, an inhibitor of viral DNA replication was added, in order to restrict our observations to viral early transcription, independently of viral DNA synthesis and all subsequent stages of the vv life cycle. Infected and lipofected cells were fixed at various times after removal of act D. In all subsequent morphological experiments the time postinfection thus refers to the time after act D washout. BrU-labeled RNAs were visualized by confocal microscopy with the use of an antibody to BrU that also recognizes BrU.
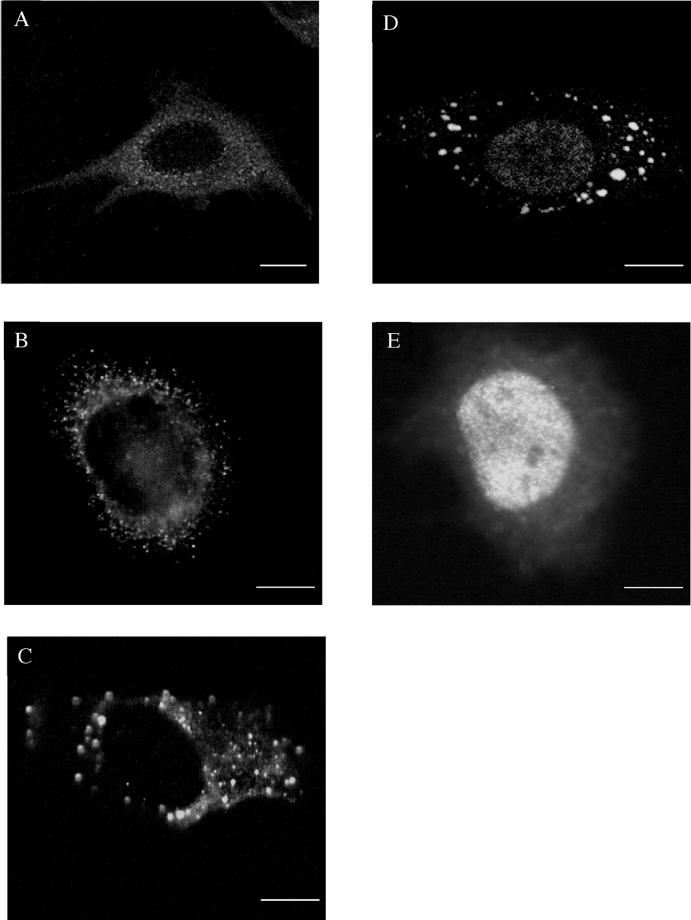
A representative time course is shown in Figure 1. Early in infection (30 min postinfection) a diffuse BrU pattern was seen in infected cells (Figure 1A), but from 45 min postinfection onward the labeling began to assemble into distinct punctate structures (Figure 1B). At 60 and 120 min postinfection (Figure 1, C and D) the labeling was predominantly associated with such granular structures, which reached a size up to ∼1 μm. No such cytoplasmic labeling was observed in uninfected BrUTP-transfected cells; instead, at 2 h after act D washout the nucleus was heavily labeled (Figure 1E). In contrast, in infected cells no nuclear BrU labeling was observed initially, but at later times a faint punctate nuclear labeling was consistently seen (Figure 1D). These data suggest that cellular transcription was mostly switched off, but some nuclear (cellular) transcription may still occur (see DISCUSSION).
Figure 1.
Over time vv early mRNAs become organized into discrete punctate structures. HeLa cells were infected and lipofected with BrUTP as described in MATERIALS AND METHODS. Viral mRNAs were detected by confocal microscopy with the use of a rat mAb against BrU- and FITC-conjugated anti-rat. In A at 30 min postinfection viral mRNAs are diffuse in the cytosol, but at later times progressively organize into punctate structures (B, 45 min; C, 60 min; D, 120 min postinfection). (E) Uninfected and BrUTP-transfected HeLa cell fixed at 2 h after act D washout, showing a strong nuclear staining. Scale bars, 10 μm.
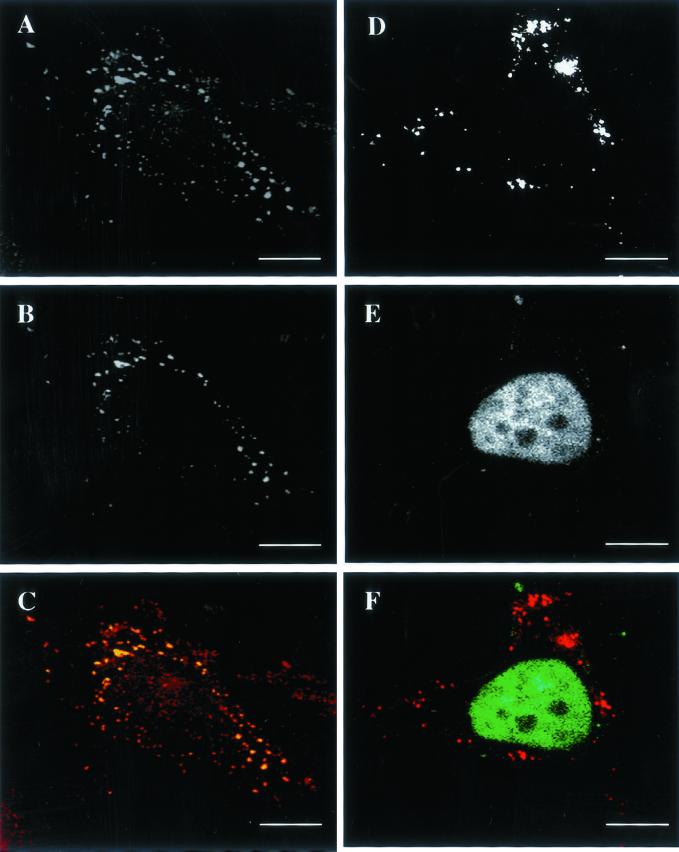
To demonstrate that the BrU labeling corresponded to viral mRNAs, the following control experiments were performed. First, infected cells were cotransfected with BrUTP and a biotinylated antisense RNA-oligonucleotide overlapping with the first 32 nucleotides of the 5′ end of H5R mRNA, an early/late vv gene. Infected and transfected cells were then double-labeled with anti-BrU and avidin-FITC. Figure 2, A–C, shows that the BrU- and the H5R oligonucleotide-labeling almost completely overlapped, confirming that the BrU pattern corresponded to viral mRNA localization in the cytoplasm of infected cells. Second, infected cells were cotransfected with BrUTP and an antisense oligo to a snRNA U1, a subunit of the spliceosome. When microinjected, the same antisense oligo has been shown before to localize throughout the nucleus, excluding the nucleoli (Carmo-Fonseca et al., 1991). On cotransfection of BrUTP and the U1 antisense oligo in vv-infected cells the former labeled typical cytoplasmic punctate structures, whereas the latter localized to the nucleus, seemingly excluding the nucleoli (Figure 2, D–F).
Figure 2.
The BrU labeling in infected cells colocalizes with vv H5R mRNA. Infected HeLa cells were cotransfected with BrUTP and with an antisense biotinylated oligonucleotide to H5R (A–C) or to snRNA U1 (D–F). The cells were fixed at 2 h postinfection and labeled with anti-BrU (A and D) or FITC-conjugated avidin (B and E). The merged images are shown in C and F. (C) Colocalization between the BrU-containing spots and the H5R pattern. (F) U1 snRNA localizes exclusively to the nucleus (apparently excluding the nucleoli) and does not colocalize with the BrU pattern in the cytoplasm of the infected cell. Scale bars, 10 μm.
These combined results made us confident that the BrU labeling in infected cells corresponded to the sites of viral early mRNA accumulation. Moreover, the time course of act D washout showed that after their synthesis, viral mRNAs become organized into distinct cytoplasmic structures that could reach 1 μm in size.
The Sites of mRNA Accumulation Are Distinct from Intracellular Cores
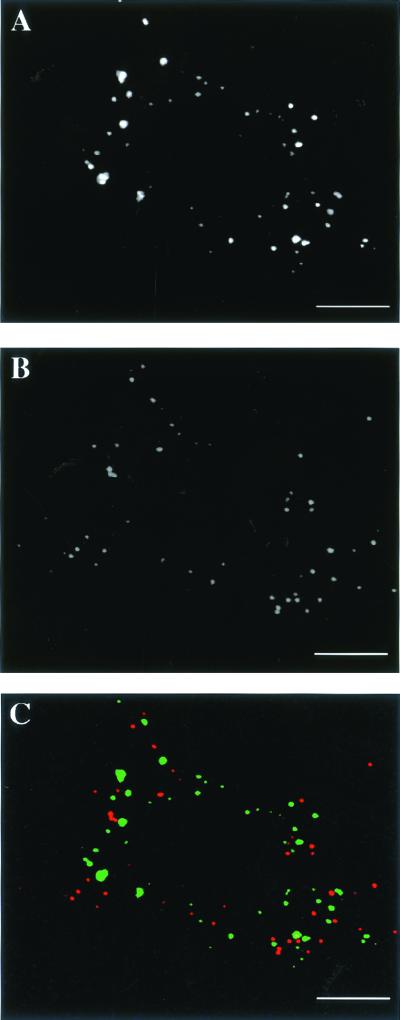
We next decided to compare the location of the RNA structures with the localization of the core, a rather robust brick-shaped structure that can survive in infected cells for many hours. For this we used an antibody to p39 (gene A4L), a protein that is associated with the surface of intracellular cores (Pedersen et al., 2000). By IF microscopy this antibody labeled intracellular cores only but not extracellular virions that remained attached to the plasma membrane (Krijnse Locker et al., 2000). On double-labeling, no colocalization between the BrU-labeled mRNAs and intracellular cores was observed by IF (Figure 3, A–C). Instead, the sites of vv mRNA accumulation appeared to be some distance away from the intracellular cores.
Figure 3.
The BrU pattern does not colocalize with intracellular cores by IF. Infected/transfected HeLa cells were fixed at 2 h postinfection and double-labeled with anti-BrU (A; green channel in C) and an antibody to p39 (A4L) that recognizes intracellular cores (B; red channel in C). The merged image in C shows that the RNA structures do not colocalize with the intracellular cores. Scale bars, 10 μm.
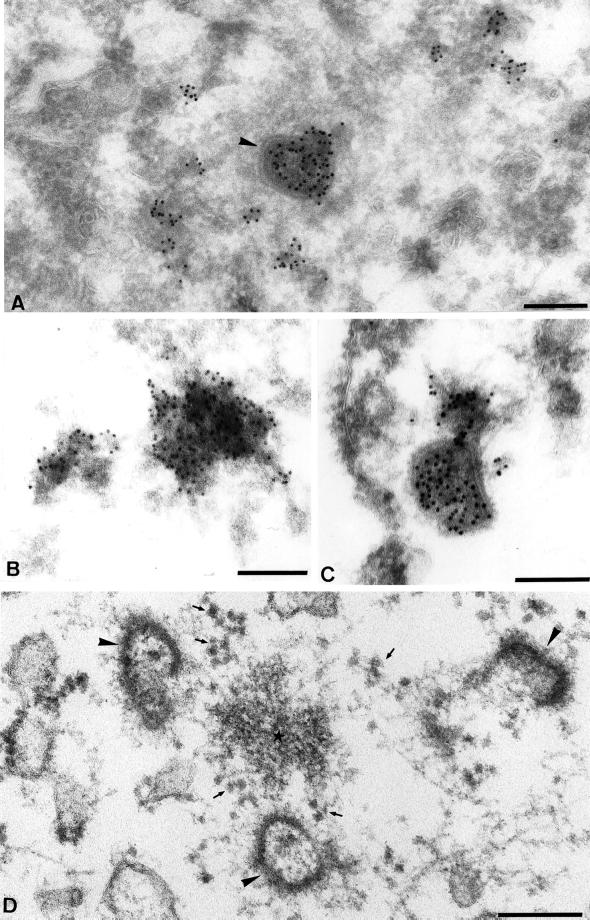
The absence of colocalization between the core and RNA labeling might indicate that in infected cells viral transcription happened (perhaps exclusively) outside the core or the amount of mRNAs accumulated inside the cores was too low to be detected. Alternatively, core-associated mRNAs were not accessible to antibody labeling by IF. The latter possibility was addressed by EM. Initially, cryosections were prepared from infected and BrUTP-transfected cells fixed and processed in a standard way, but no specific labeling could be detected on such sections. Significant labeling was, however, obtained when the cells were extracted with TX-100 before fixation. Abundant labeling was detected inside intracellular cores, on structures next to the core as well as some distance away from the core (Figure 4, A–C). They appeared as rather amorphous structures that on cryosections were difficult to recognize without antibody labeling, because their electron density was barely different from the surrounding cytoplasm (Figure 4, B and D). Furthermore, they did not appear to be associated with intracellular membranes, but because the cells were triton-extracted before fixation, this point could not be unequivocally addressed on cryosections. Therefore, cells infected in the same way but without BrUTP transfection were prepared for Epon embedding. A fixation protocol was chosen that highly extracts the cytosol (Tilney et al., 1998; see MATERIALS AND METHODS), because we expected that in conventionally fixed cells we would not be able to distinguish the mRNA structures from the surrounding cytoplasm. Epon embedding cannot usually be combined with antibody labeling, which made it difficult to unequivocally identify the viral mRNAs. Nevertheless, in infected cells but not in uninfected cells, we observed electron-dense cytoplasmic structures, apparently lacking a membrane, of ∼0.3–1 μm in diameter, close to or also some distance away from intracellular cores (Figure 4D). The occasional presence of cores close by and their size and structure as well as their absence in uninfected cells made us confident that they represented the mRNA structures labeled with anti-BrU on cryosections.
Figure 4.
EM localization of BrU in infected and transfected HeLa cells. (A–C) HeLa cells were infected, BrUTP-transfected, fixed after TX-100 extraction at 2 h postinfection and prepared for cryosectioning. In A a core is seen (arrowhead) that is heavily labeled for BrU. Some patches of BrU labeling are associated with the outside of this core. Strongly labeled BrU-positive patches, which seem associated with an electron-dense structure, are located some distance away from the core. (B) High-magnification view of a heavily labeled RNA-site. (C) A close-up of a heavily labeled core with BrU-positive material attached to it. (D) Cells were infected for 2 h as for the preparation of cryosections but without BrUTP transfection. They were fixed and prepared for Epon embedding. The image shows three cores (arrowheads) surrounding a putative site of viral mRNA accumulation (small star). The small arrows indicate ribosomes. Scale bars, 200 nm.
Moreover, they appeared to recruit ribosomes, indicating that they might be active in protein translation (see also below).
The combined data suggest that in infected cells vv transcription starts in the intracellular cores, from which the messengers are subsequently extruded and transported to discrete sites.
Microtubules Connect Cores and Viral mRNAs and Are Required for the Organization of the Latter
Cytoplasmic mRNAs are commonly bound to either the rough ER or to the cytoskeleton. It was therefore likely that the vv mRNA structures might also be associated with either membranes or with the cytoskeleton. When cells were extracted with TX-100 before fixation, the vv mRNA structures as well as intracellular cores appeared entirely unaffected, suggesting that both were associated with the cytoskeleton (see below).
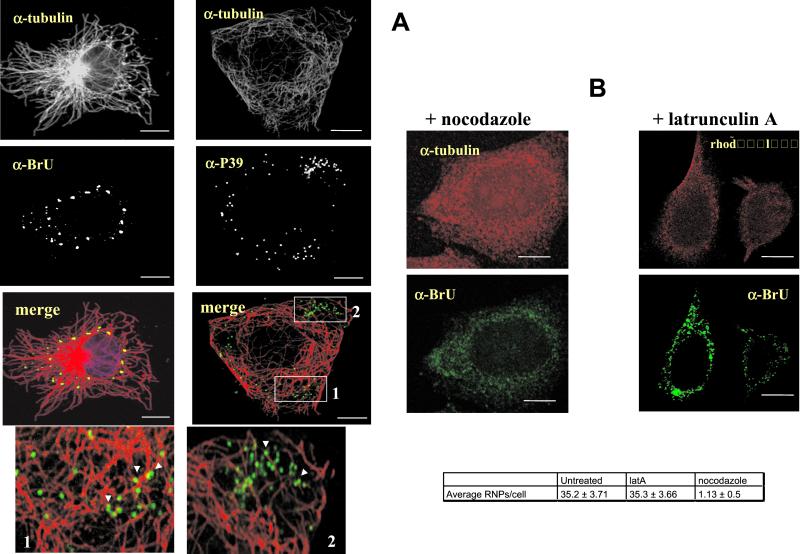
To address this issue in more detail, infected and BrUTP-transfected cells were fixed at 2 h postinfection and double-labeled with anti-BrU and either anti-tubulin (Figure 5A) or rhodamine-phalloidin (unpublished results). Although no colocalization was seen between actin and the vv mRNA labeling (unpublished results), the viral messengers did seem to follow MT tracks (Figure 5A; see also below). Because the above data suggested that viral mRNAs are made inside intracellular cores and then move to distinct sites where they accumulate, we asked whether cores might also be associated with MTs and that this cytoskeletal element thus connected both structures. Indeed, upon double-labeling with the core protein p39 and anti-tubulin, cores could be observed to be aligned on MT tracks (Figure 5A).
Figure 5.
vv early mRNAs and intracellular cores are aligned on microtubules by immunofluorescence. (A) Cells were infected and transfected with BrUTP for 2 h. Cells were extracted with TX-100 before fixation and double-labeling with either anti-tubulin and anti-BrU (left panel) to reveal the viral mRNA structures or anti-p39 (right panel) to label intracellular cores. The high-magnification view (indicated with 1 and 2) of the indicated areas 1 and 2 in the merge of anti-tubulin and p39 shows that intracellular cores are aligned on MT tracks (arrowheads). In B in the panel indicated with + nocodazole, cells were treated with 20 μM nocodazole 1 h before infection and BrUTP-transfection and fixed at 2 h postinfection in the continued presence of the drug. In the + latrunculin panel, cells were infected and transfected and treated 20 min before fixation with 1 μM Lat A and 2.5 μM taxol. Fixed cells were then double-labeled with anti-BrU and with either anti-tubulin or with rhodamine-phalloidin. Cells were fixed without prior TX-100 extraction. MTs (α-tubulin) as well as the viral mRNAs (α-BrU) show a diffuse pattern in the cytosol after nocodazole treatment. In the presence of Lat A, the phalloidin labeling is diffuse too, but the BrU pattern seems unaffected. Scale bars, 10 μm. In the table at the bottom of B, cells were infected and BrUTP-transfected for 2 h 20 min before fixation cells were mock treated or treated with either 40 μM of nocodazole or with 1 μM Lat A and 2.5 μM taxol. The cells were then extracted with TX-100, fixed, and labeled with anti-BrU. The amount of BrU-positive structure was counted in 30 infected/transfected cells. Because nocodazole-treated cells mostly lacked any BrU-positive structures, making it difficult to estimate which cells were transfected, cells that showed some residual BrU labeling were considered for counting. The values represent the average amount of BrU structures per cell and the SEM
We next tested whether MTs were required for the biogenesis of the distinct punctate vv mRNA structures seen by fluorescence at 60–120 min postinfection. Infected HeLa cells were treated with nocodazole before BrUTP transfection and fixation at 2 h postinfection. Whereas in control cells the viral mRNAs organized into the typical pattern shown in Figure 1D (unpublished results), in the presence of the drug a diffuse BrU pattern was observed, indicating that the mRNAs failed to organize in to the typical punctate structures (Figure 5B). As a control, the same experiment was also conducted in the presence of Lat A, a drug that binds to G-actin and appears to be a more potent actin-depolymerizing agent than cytochalasin D (Ayscough, 1998). Under these conditions the mRNA structures were not affected and developed in the same way as without drug treatment (Figure 5B).
To demonstrate in a more quantitative manner that the viral mRNA structures associated preferentially with MTs, the following experiment was performed. Cells were infected and transfected as usual, but before fixation the cells were incubated with TX-100 to extract molecules and subcellular components that are not bound to the cytoskeleton. Parallel sets of cells were treated with either nocodazole or with Lat A 20 min before TX-100 extraction to test whether the viral mRNA structures might be bound to MTs or to actin. After labeling with anti-BrU, the number of typical BrUTP-positive structures in transfected cells was subsequently counted in untreated or drug-treated cells. The table in Figure 5B shows that nocodazole treatment resulted in a dramatic decrease in the average number of structures per cell, whereas in the presence of Lat A this number was similar to the untreated control cells. Finally, the integrity of the newly formed mRNA structures was also dependent on intact MTs. When the mRNA structures were first allowed to organize and nocodazole was then added, the structures apparently disintegrated, because under these conditions the BrU labeling was diffuse rather than organized (unpublished results).
These combined data strongly suggest that both intracellular cores and the sites of viral mRNA accumulation are associated with MTs, implying that both these structures are connected by this cytoskeletal element (see DISCUSSION).
Biochemical Evidence for the MT Association of Viral mRNAs
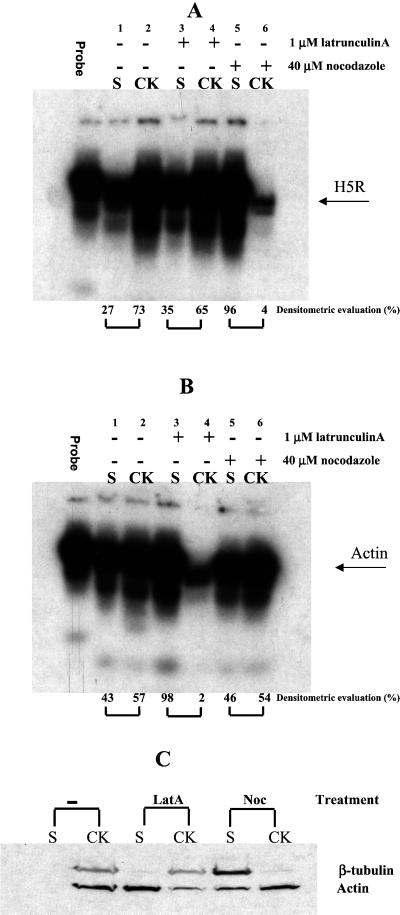
The putative association of viral mRNAs with MTs was further investigated with the use of an RNase protection assay. In this assay total mRNA is isolated from cells, the mRNAs of interest are hybridized to a 32P-labeled probe specific for these messengers, and unhybridized RNA is subsequently digested by RNase A treatment. The 32P-labeled protected fragment can then be used to quantify the amount of specific mRNAs contained in the sample. To determine whether viral mRNAs were bound to the cytoskeleton, infected cells were either mock-treated or treated with nocodazole or Lat A to depolymerize MTs or actin filaments, respectively. The cells were then extracted with detergent and divided into a detergent-resistant fraction, containing cytoskeletal elements, and a soluble fraction, containing mostly soluble proteins (see MATERIALS AND METHODS). However, initial Western blot analyses showed that Lat A treatment not only led to depolymerization of actin, but the drug also resulted in substantial amounts of tubulin shifting from the detergent-insoluble pool into the soluble fraction (unpublished results). Apparently, concentrations of Lat A that were required to depolymerize all F-actin also resulted in substantial depolymerization of MTs. To circumvent this problem, the Lat A treatment was subsequently done in the presence of a low concentrations of taxol to stabilize MTs. Under such conditions Western blot analysis using antibodies to actin or tubulin confirmed that both cytoskeletal elements were predominantly contained in the insoluble (detergent) fraction in untreated cells, whereas treatment of cells with Lat A (together with taxol) or nocodazole resulted in depolymerization of the majority of actin or MTs, respectively (Figure 6C).
Figure 6.
Biochemical evidence for MT association of viral mRNAs. (A–C) Cells were infected for 2 h. They were either left untreated or were treated with either 20 μM of nocodazole from 1 h before infection onward or with 1 μM Lat A together with 2.5 μM taxol 20 min before cell lysis. (A and B) Total RNA was extracted from infected cells from either the detergent-soluble (S) or -insoluble cytoskeletal fraction (CK), and the fractions were analyzed by an RNase protection assay with the use of probes to H5R and β-actin mRNA. The position of the H5R (A) and β-actin (B) protected mRNA is indicated on the right. The percentage of protected mRNAs in the cytoskeleton (CK) versus soluble (S) fraction of eachsample as assessed by phosphoimager analysis is indicated at the bottom of each panel. (C) Western blot analysis of actin and tubulin (the position of the respective protein is indicated) after nocodazole or Lat A treatment in the soluble (S) and cytoskeleton (CK) fraction. The proteins were detected with the use of antibodies to β-actin or β-tubulin followed by enhanced chemiluminescence.
To determine whether mRNAs were bound to the cytoskeleton, total RNA was subsequently extracted from the detergent-soluble and -insoluble pools, and specific mRNAs subjected to the RNase protection assay. As a representative probe for vv early mRNAs, we again used the H5R mRNA that, as shown above, localizes to the RNA-containing granular structures. A probe to β-actin mRNA, an mRNA that has been shown to be bound to actin in HeLa cells (Sundell and Singer, 1991), was used as a control.
Most of H5R mRNA (73%) appeared to be cytoskeletal-bound, and this pool was almost entirely released into the soluble fraction (96%) after treatment with nocodazole (Figure 6A), consistent with a association of this viral messenger with MTs. In contrast, the majority of the viral mRNAs remained bound to the cytoskeleton upon Lat A treatment (Figure 6A). Finally, when β-actin mRNA was subjected to the same analysis in infected cells it behaved as expected, because it was released into the soluble fraction after Lat A (but not nocodazole) treatment, confirming its association with actin filaments (Figure 6B).
In conclusion, both the immunofluorescence data as well as the RNase protection assay show that in infected cells the viral mRNAs are bound to MTs.
Microtubules Are Important for Viral but not for Cellular mRNA Metabolism
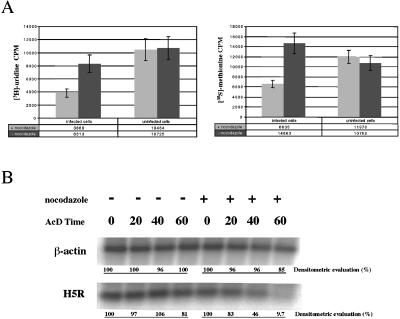
An alternative approach to address the role of MTs in viral mRNA organization was to test whether MT depolymerization had any effect on viral and cellular mRNA synthesis and on mRNA turnover. Indeed, nocodazole treatment affected RNA and protein synthesis in infected, but not in uninfected cells (Figure 7A) because in MT-depleted infected cells the incorporation of [3H]uridine was decreased by >50% and the rate of protein synthesis by more than twofold compared with untreated control cells (Figure 7A).
Figure 7.
Nocodazole treatment affects viral mRNA synthesis and stability. (A) HeLa cells were treated with nocodazole 1 h before infection in the presence of the drug. At 90 min postinfection the cells were labeled for 30 min with [3H]uridine (left) or [35S]methionine (right). The incorporation of uridine or methionine into RNAs and proteins, respectively, in total cell extracts (including cytoplasm and nuclei) was measured by liquid scintillation counting after TCA precipitation. The values represent the average and SDs of three experiments performed in duplicate. (B) The stability of H5R and β-actin (indicated) mRNAs was assessed in nocodazole-treated infected HeLa cells. Cells were nocodazole-treated 1 h before infection. At 1 h postinfection act D was added and mRNAs isolated from the infected cells at the indicated times after act D addition. The specific mRNAs were detected by Northern blotting, and the amount in each lane was determined by phospho-imager analysis (indicated at the bottom of each panel). The data show that the H5R mRNA is rapidly degraded in the absence of MTs.
MT depletion also affected viral (but not cellular) mRNA turnover. This was shown by isolating mRNAs from infected and nocodazole-treated cells at different times after act D addition to prevent further RNA synthesis, followed by Northern blots with the use of probes to H5R and to β-actin mRNA. Quantitation of such blots showed that after 1 h of chase >90% of the H5R mRNA was degraded, whereas β-actin mRNA was mostly unaffected (Figure 7B).
The collective data thus show an important role for MTs in the anchoring, organization and in the stability of vv early mRNAs. In contrast, as shown before, cellular mRNAs, represented by β-actin mRNA, likely require actin for anchoring and stability and remained actin-associated during infection. Consequently, the stability of this mRNA appeared unaffected in infected cells, both in the presence or absence of intact MTs (see DISCUSSION).
The Viral-induced Granular Structures Are Surrounded by Components of the Translation Machinery, and Viral mRNAs Associate with Polyribosomes
The typical organization as well as the association with microtubules of the viral mRNAs showed striking similarities to the mRNA localization in cultured oligodendrocytes. In these cells mRNAs also appear as granular structures that are bound to and move on MTs. Moreover, these mRNA-containing structures have been shown to contain components of the translation machinery (Barbarese et al., 1995).
We therefore investigated whether the viral mRNA structures might be organized in a similar way. For this, infected and BrUTP-transfected cells were double-labeled with antibodies to BrU and to proteins involved in translation.
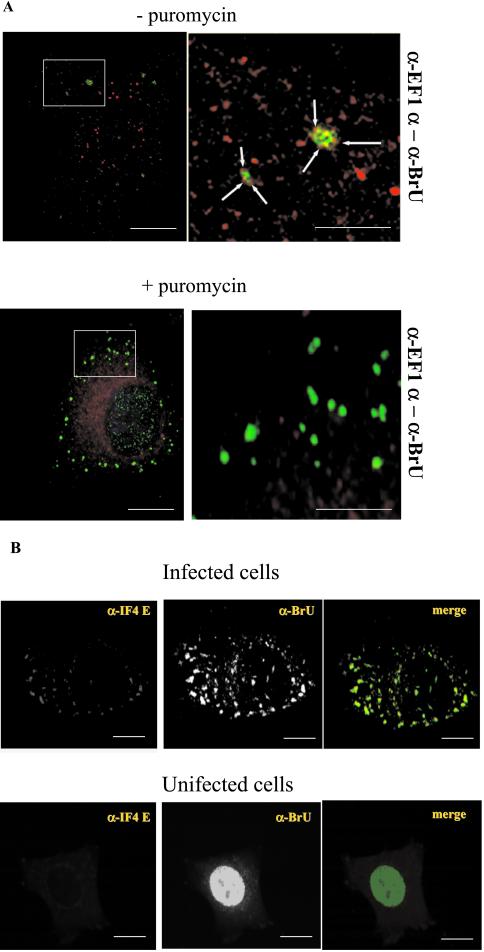
In uninfected cells antibodies to elongation factor 1α (EF1α) showed a punctate cytoplasmic pattern (unpublished results). In infected cells this pattern was mostly unchanged. However, we noticed that the protein was additionally recruited to the viral mRNA structures and seemed to surround the viral BrU-positive spots, while being absent from the central, RNA-containing part (Figure 8A). The labeling for EF1α around the viral mRNAs displayed a characteristic dotted pattern, typical of polyribosomes. To test whether this pattern represented ribosomes, cells were treated with puromycin before fixation (as well as with cycloheximide as control) to release ribosomes from the mRNAs. As expected, in puromycin- but not in cycloheximide-treated cells (unpublished results), the typical punctate pattern of EF1α around the mRNAs was lost, and the protein now displayed a diffuse rather than punctate cytoplasmic pattern (Figure 8A).
Figure 8.
vv early mRNAs are surrounded by EF1α, colocalize with IF4E, and associate with polyribosomes. (A) Infected and BrUTP-transfected HeLa cells were puromycin- or mock-treated for 30 min, fixed, and labeled with anti-EF1α (red channel) and anti-BrU (green channel) antibodies. The panels on the left side show the pattern at low magnification, whereas the right side panels show high-magnification views of the boxed areas indicated in the low-magnification image. In untreated cells, EF1α shows a punctate labeling in the cytoplasm as well as around the BrU-positive structures. In treated cells the EF1α pattern is more diffuse and does not surround the viral mRNAs. Bars, low-magnification images, 10 μm; insets, 2 μm. (B) Infected or uninfected cells were transfected with BrUTP, fixed at 2 h postinfection, and labeled with anti-BrU and anti-IF4 E antibodies. In infected cells the protein colocalizes with the BrU-positive structures of viral mRNAs. In uninfected cells the anti-BrU strongly labels the nucleus, whereas the antibodies to IF4E show a faint diffuse labeling in the cytoplasm. In the merge panel, the green channel is BrU labeling, and red is the antibody to IF4E. Bars, 10 μm.
A different labeling was observed with antibodies to initiation factor 4E, a protein that binds to the 5′ RNA cap and mediates ribosome binding (Sonenberg et al., 1978). As expected for a protein that is not abundantly expressed, the labeling for IF4E was barely detectable in uninfected cells and appeared as a faint cytosolic background labeling (Figure 8B). In infected cells, however, IF4E was apparently recruited to and concentrated in the viral mRNA structures because the antibody displayed distinct labeling that completely overlapped with the BrU-positive structures (Figure 8B).
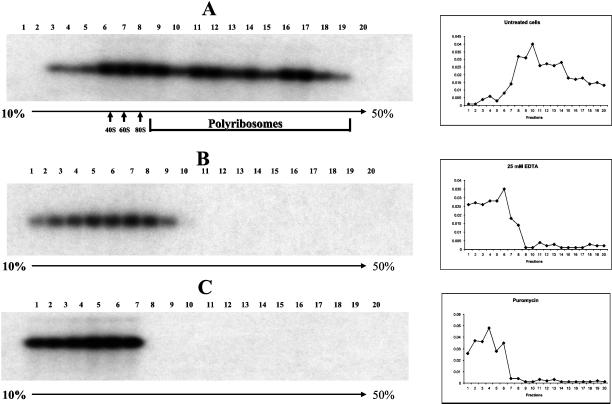
As an independent proof that the viral mRNAs indeed associated with polyribosomes, cell lysates of infected cells were subjected to polyribosome sucrose-gradients to separate (viral) mRNAs associated with monoribosomes and polyribosomes. After fractionation of such gradients, each fraction was subjected to Northern blots with the use of the H5R-specific probe. Figure 9 shows that a substantial amount of the H5R mRNAs were associated with fractions expected to contain polyribosomes. As an independent proof of polyribosomes association, cells were either treated with puromycin before lysis or the cell lysates were pretreated with EDTA before centrifugation and fractionation. Whereas the former treatment releases ribosomes from mRNAs, the latter is known to disintegrate polyribomes. As shown in Figure 9 under both conditions, the viral mRNAs could no longer be detected in the heavy sucrose fractions, indicating that these fractions represented H5R mRNAs associated with polyribosomes.
Figure 9.
H5R mRNAs associate with polyribosomes. Lysates of cells infected for 2 h in the presence of hydroxyurea were subjected to polyribosome gradients. Each fraction was subjected to Northern blotting with the use of a specific probe to H5R mRNA. The positions of the 40S, 60S, and 80S ribosomal subunits and the polyribosome containing fractions as well as the top (10% sucrose) and the bottom (50% sucrose) of the gradient are indicated. (A) The analysis of untreated cell lysates shows that a substantial amount of H5R mRNA associates with the heavy polyribosome-containing fractions. (B) The lysate was treated with 25 mM EDTA before centrifugation to disintegrate polyribosomes. (C) Infected cells were treated with 100 μg/ml puromycin before lysis to release ribosomes from mRNAs. In both B and C H5R mRNAs can no longer be detected in the fractions expected to contain polyribosomes. The panels on the right indicate the corresponding OD254 read-out of the different fractions.
The combined data thus demonstrate that the viral mRNA structures recruit polyribosomes as well as cellular components involved in translation. They therefore strongly suggest that the sites of viral mRNA accumulation observed by immunofluorescence are not transfection artifacts or “dead-end” structures but are likely active in viral early protein synthesis. Finally, the confocal images strongly suggested that the vv mRNAs were located in the center of the structures, whereas the polyribosomes (as detected by EF1α labeling) appear to be restricted to their periphery.
The Role of the Viral Protein p25
Having established that the sites of viral mRNA accumulation recruited cellular proteins, we next investigated whether any viral proteins were involved in the assembly of the mRNAs. To test for a potential role for newly synthesized viral proteins, a time-course experiment identical to the one in Figure 1 was repeated in the presence of cycloheximide to block viral and cellular protein synthesis. The viral mRNA sites organized in the same way as without this drug (unpublished results), indicating that newly synthesized viral (or cellular) proteins were not required for the typical organization of the viral mRNAs. We therefore tested whether viral proteins associated with the incoming cores were involved in this process. For this, infected and BrUTP-transfected cells were fixed at 2 h postinfection and double-labeled with anti-BrU and antibodies to viral core proteins. None of the core proteins tested (4a [A10L], 4b [A3L], p39 [A4L], and p11 [F17R]) appeared to colocalize with the site of mRNA accumulation (unpublished results).
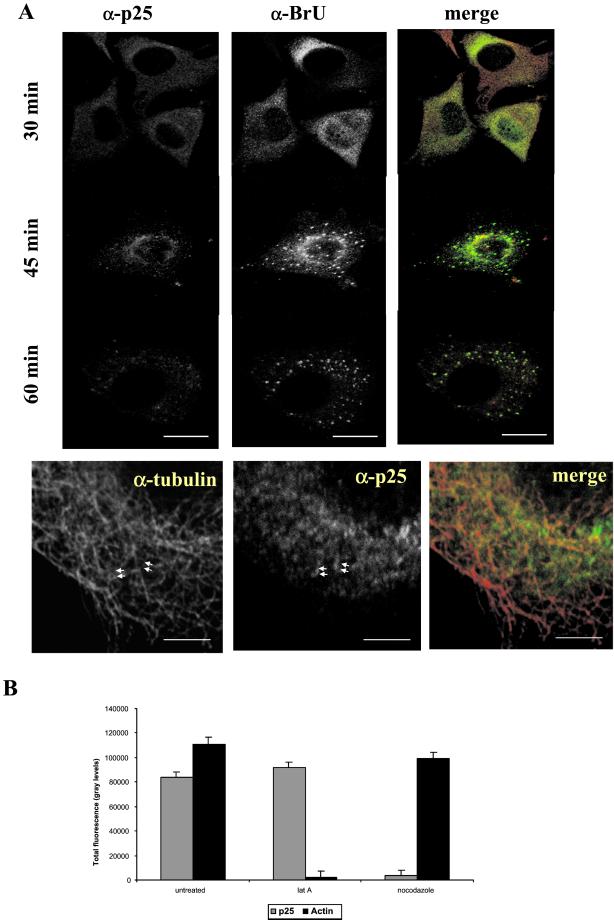
A different pattern was obtained when BrUTP-transfected infected cells were double-labeled with antibodies to p25 (L4R) and to BrU. A time course conducted as in Figure 1 but now double-labeled with anti-BrU and anti-p25 revealed that p25 was apparently able to leave the core, because at 30 min postinfection the p25 labeling displayed a diffuse cytoplasmic pattern similar to the one of BrU at this time of infection (Figure 10A). No such labeling for p25 was observed in act D–treated cells, suggesting that this pattern depended on active viral transcription (unpublished results). Although p25 did not localize to the typical punctate BrU structures seen at 60 min (nor at 120 min postinfection; unpublished results) postinfection, partial colocalization was observed at 30 and most convincingly at 45 min, a time when both the RNA and the protein showed a diffuse cytoplasmic pattern (Figure 10A).
Figure 10.
P25 (4RL) colocalizes with the viral mRNAs early in infection and may associate with MTs. Top panels in A: Infected and BrUTP-transfected HeLa cell were fixed at the indicated times postinfection and double-labeled with anti-BrU and anti-p25 antibodies. At 30 min postinfection the viral mRNAs partially colocalize with the diffuse p25 labeling, whereas at 45 min postinfection colocalization of the two markers is detected in the perinuclear region where the mRNAs appear still unorganized. At 60 min postinfection, when all of the mRNAs are organized into the typical punctate structures, no colocalization is detected. In the merge anti-BrU is shown in green and anti-p25 in red. Note that over time the p25 labeling decreases. Bottom panels in B: Cells were extracted with TX-100 before fixation at 30 min postinfection and double-labeling with antitubulin and anti-p25. Anti-p25 labeling appears to follow MT tracks (arrowheads). In the merge p25 is in the green and antitubulin in the red channel. Scale bars, top, 10 μm; bottom, 2 μm. (B) Infected HeLa cells were mock treated or treated with either nocodazole or Lat A (together with taxol) and Triton-extracted before fixation at 1 h postinfection. The cells were then labeled with antibodies to β-tubulin, p25, or rhodamine-phalloidin. Ten random pictures of both nocodazole- and mock-treated cells were taken with the use of the same parameters (the same amplitude, gain, and offset). The total fluorescence for p25 or actin was measured with the use of NIH image. The graph represents the average and SD of the total fluorescence of 10 cells for both treated and untreated cells.
Like the vv mRNAs, labeling for p25 appeared to be associated with MTs, confirming previous data (Ploubidou et al., 2000). When cells were double-labeled with anti-tubulin and p25, the latter labeling appeared to follow MT tracks (Figure 10A). To provide independent evidence that p25 might be bound to MTs, cells were extracted with TX-100 before fixation in the presence or absence of either nocodazole or Lat A. The p25 pattern was unaffected when cells were extracted with TX-100 before fixation, indicating that the protein was bound to the cytoskeleton (unpublished results). When cells were treated with nocodazole and subsequently extracted with TX-100 before fixation, most of the p25 labeling was lost (Figure 10B). The amount of p25 labeling was, however, unaffected when cells were treated with Lat A (Figure 10B). In contrast, labeling for actin was unaffected by nocodazole treatment but was entirely lost when cells were treated with Lat A (Figure 10B).
These combined data suggest first, that the sites of viral mRNA accumulation must be predominantly, if not exclusively, organized by cellular proteins, a subset of the candidates being described above. Nevertheless, it seems likely that the viral protein p25 plays a role in vv mRNA targeting because is was released from the core and interacted with MTs, concomittant with the synthesis of vv RNAs. Importantly, our observations are consistent with previous results on p25. The protein has been shown to bind DNA and RNA and to be involved in the process of early transcription (Yang and Bauer, 1988; Wilcock and Smith, 1996; Bayliss and Smith, 1997). Moreover a recent study showed this protein binds MTs both in vitro and in vivo (Ploubidou et al., 2000; see DISCUSSION).
DISCUSSION
Ever since the realization that the process of vv early transcription can be reconstituted in a test tube, the molecular details underlying this process have been extensively studied. However, relatively little was known about the fate of vv transcripts in infected cells and in particular about their subcellular localization. The present study shows that vv early mRNAs are not diffusely localized in the cytoplasm but soon after synthesis in the viral cores they are transported from their site of transcription in an MT-dependent manner to become organized into discrete structures. That MTs were the main cytoskeletal player in vv mRNA organization was shown by several lines of evidence; first, early transcripts were bound to MTs, second, their characteristic organization required intact MTs, and viral RNA and protein synthesis were significantly affected in the presence of nocodazole. The effects of nocodazole on mRNA and protein synthesis in infected cells (twofold decrease) are consistent with the data by Ploubidou et al. (2000), showing a threefold reduction in virus yields in nocodazole-treated cells compared with untreated controls.
vv-induced Host Protein Synthesis Shut-off
In nonneuronal cells mRNAs are generally assumed to be mostly bound to actin. The present study confirmed this view because β-actin mRNA was bound to actin and because this messenger was not significantly degraded upon MT depletion or viral infection. Furthermore, in uninfected cells (but not in infected cells) protein synthesis was not affected in the absence of MTs. One of the obvious mechanisms of viral-induced shut-off of host protein synthesis is the viral-induced release of cellular messengers from the cytoskeleton that results in inefficient translation and/or increased degradation of the cellular mRNAs. Because β-actin mRNA remained efficiently bound to the cytoskeleton and was not subjected to increased degradation in infected cells, these results suggest that vv host shut-off may act differently. The observed stability of β-actin mRNA in infected cells is consistent with at least two studies showing that cellular messenger do not undergo degradation upon vv infection (Rosemond-Hornbeak and Moss, 1975; Pedley and Cooper, 1984). Because vv infects a variety of cultured cells and because its genome encodes for >250 proteins, its mechanism of host shut-off is likely to be complex. Indeed, among the mechanisms suggested for this process are as follows: (1) a decrease in cellular RNA synthesis (Kit and Dubbs, 1962; Becker and Joklik, 1964; Pedley and Cooper, 1984), (2) release of cellular mRNAs from polyribosomes (Metz et al., 1975; Rosemond-Hornbeak and Moss, 1975), (3) inhibition of the translation of host messengers by the production from vv cores of small polyadenylated RNAs (Rosemond-Hornbeak and Moss, 1975; Cacoullos and Bablanian, 1993), and (4) the phosphorylation of ribosomal proteins (Buendia et al., 1987). The present study did not address the question of host shut-off directly, but it is interesting to note that upon vv infection BrUTP labeling in the nucleus was significantly decreased, although not completely absent. These results suggest that although cellular transcription is mostly switched off, some RNA synthesis (e.g., of rRNA and tRNA, RNAs likely to be vital for viral functions) does seem to occur.
The current results exemplify that in HeLa cells mRNAs as well as the factors required for binding of mRNAs to the cytoskeleton can associate both with actin and MTs, as suggested by several other studies (reviewed in Oleynikov and Singer, 1998).
Structural Dissection of the mRNAs
Our data show that vv mRNAs have an intrinsic property to organize into discrete structures that can reach a size up to 1 μm. Localization studies on poly(A) mRNAs in human diploid fibroblasts showed that these RNAs assembled into much smaller and more diffusely localized structures that are mostly bound to actin (Taneja et al., 1992; Bassell et al., 1994). Our (unpublished) observations confirm these studies, because the pattern for cellular cytoplasmic RNAs in HeLa cells (with the use of BrUTP transfection) appeared in small dots and not in the typical large granular vv-induced RNAs (unpublished results). The granular organization of the vv mRNAs shows similarities to mRNA structures in oligodendrocytes; in these cells myelin basic protein (MBP) mRNAs become organized into typical granular structures that can reach a size up to 0.8 μm and that like the viral mRNAs are associated with MTs (Ainger et al., 1993; Barbarese et al., 1995). As for MBP mRNAs in oligodendrocytes, the viral mRNAs also recruited components of the translation machinery, including EF1α (Barbarese et al., 1995). Our data show, in addition that polyribosomes and EF1α appeared to surround the viral mRNA structures and to be excluded from the central mRNA-containing part. Consequently, components associated with the mRNA-surrounding structure, which include ribosomes, ribosome-binding proteins such as EF1α are the most likely candidates to mediate MT binding. Our data furthermore suggest an exclusive role for cellular proteins in the organization of the typical granular vv mRNA structures because they appeared at 60–120 min postinfection. Cycloheximide did not affect their organization, showing that no newly synthesized viral (nor cellular) proteins are involved. Furthermore, none of the proteins of the incoming cores we tested localized to the mRNA-sites (except for the viral protein p25; see below). It is of interest to note that EF1α, an abundant cellular protein involved in protein translation, is also able to bind actin and MTs (Durso and Cyr, 1994) and has been shown to sever MTs (Shiina et al., 1994; reviewed in Condeelis, 1995). Whether EF1α mediates MT binding of the viral messengers or whether other cellular MAPs/motors are involved will require further studies. We tested for the presence of a number of obvious MT binding proteins, including CLIP-170 and Staufen, in the vv RNA structures, but none of these colocalized (unpublished results).
Why Do vv mRNAs Bind Microtubules Rather than Actin?
Although most cellular mRNAs in fibroblasts seem to prefer to bind to actin, there are a number of examples where mRNAs bind MTs. Evidence for mRNA movement along MTs comes from mRNA localization studies in neuronal cells as well as from mRNA localization and targeting during oocyte development of Drosophila (see INTRODUCTION). Much less evidence is available for a role of MTs in the movement of mRNAs in nonneuronal mammalian cells.
The present data suggest that in infected cells, as in vitro, mRNAs are made inside the core from which they are extruded and transported to sites away from the core. That mRNA synthesis may start inside the intracellular cores was shown by the extensive core-associated BrU labeling by EM. Additional evidence that vv early transcription in infected cells occurs inside the cores will be presented elsewhere (Mallardo et al., unpublished results).
The recent finding that vv cores bind MTs in vitro (Ploubidou et al., 2000) is consistent with our observations showing that intracellular cores are aligned on MT tracks by IF. Because the mRNA structures also bound to MTs and because MTs appear to be required for the typical granular organization away from the intracellular core, these combined data imply that core-synthesized viral mRNA move along MTs to the site of their accumulation and subsequent translation. Our data furthermore argue for a role for the vv core-associated protein p25 (L4R) in this process. In addition to its ability to bind both DNA as well as RNA in vitro (Yang and Bauer, 1988; Bayliss and Smith, 1997), this protein is also able to bind MTs (Ploubidou et al., 2000). Importantly, p25 has been implicated in the process of early transcription, because virions lacking p25 were defective in the latter process (Wilcock and Smith, 1994, 1996). Our data show that p25 leaves the core at the same time as the mRNAs, and after this event it then transiently colocalizes with the mRNAs before these become organized into granular structures. These data suggest that p25, by binding both to MTs and to the viral mRNAs, may play an important role in the structural organization of the viral messengers, perhaps by assisting their transport/binding along MTs. We have attempted to address some of those open questions, with the use of a recombinant virus in which the synthesis of p25 can be regulated (Wilcock and Smith, 1994). Unfortunately the phenotype of this virus was too leaky to make any firm conclusions about the role of p25 in early transcription in vivo.
In conclusion, we would like to propose that the process of vv early transcription offers a well-defined synchronized system for a more detailed characterization of mRNA transport and anchoring on MTs in nonneuronal cells as well as the biogenesis of mRNA/protein complexes involved in translation.
ACKNOWLEDGMENTS
We thank Martina Muckenthaler for antibodies and expert advice; Jens Rietdorf for help with confocal microscopy; and Gareth Griffiths, Michael Kiebler, Anne Ephrussi, Michael Way, Aspasia Ploubidou, and Martina Muckenthaler for critically reading the manuscript. This work was supported by an EU TMR and a Biotech grant.
REFERENCES
- Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. Use of latrunculin-A, an actin monomer-binding drug. Methods Enzymol. 1998;298:18–25. doi: 10.1016/s0076-6879(98)98004-1. [DOI] [PubMed] [Google Scholar]
- Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Powers CM, Taneja KL, Singer RH. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J Cell Biol. 1994;126:863–876. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss CD, Smith GL. Vaccinia virion protein VP8, the 25 kDa product of the L4R gene, binds single-stranded DNA and RNA with similar affinity. Nucleic Acid Res. 1997;25:3984–3990. doi: 10.1093/nar/25.20.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker Y, Joklik WK. Messenger RNA in cells infected with vaccinia virus. Proc Natl Acad Sci USA. 1964;51:577–585. doi: 10.1073/pnas.51.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau A-M, Darveau A, Sonenberg N. Effect of viral infection on host protein synthesis and mRNA association with cytoplasmic cytoskeletal structure. J Cell Biol. 1985;100:1209–1218. doi: 10.1083/jcb.100.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia B, Person-Fernandez A, Beaud G, Madjar J-J. Ribosomal protein phosphorylation in vivo and in vitro by vaccinia virus. Eur J Biochem. 1987;162:95–103. doi: 10.1111/j.1432-1033.1987.tb10547.x. [DOI] [PubMed] [Google Scholar]
- Cacoullos C, Bablanian R. Role of polyadenylated RNA sequences (POLADS) in vaccinia virus infection: correlation between accumulation of Polads and extent of shut-off in infected cells. Cell Mol Biol Res. 1993;39:657–664. [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI. In vivo detection of snRNA-rich organelles in the nuclei of mammalian cells. EMBO J. 1991;10:1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera M, Dreyfuss G, Penman S. Messenger RNA is translated when associated with the cytoskeletal framework in normal and VSV-infected HeLa cells. Cell. 1981;23:113–120. doi: 10.1016/0092-8674(81)90276-2. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Moss B. Transcription of vaccinia virus mRNA coupled to translation in vitro. Virology. 1978;88:149–165. doi: 10.1016/0042-6822(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Durso NA, Cyr RJ. A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1 alpha. Plant Cell. 1994;6:893–905. doi: 10.1105/tpc.6.6.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon PD, Moss B. Early transcription factor subunits are encoded by vaccinia virus late genes. Proc Natl Acad Sci USA. 1990;87:4401–4405. doi: 10.1073/pnas.87.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G. Fine structure immunocytochemistry. Heidelberg, Germany: Springer-Verlag; 1993. [Google Scholar]
- Haukenes G, Szilvay AM, Brokstad KA, Kanestrom A, Kalland KH. Labeling of RNA transcripts of eukaryotic cells in culture with BrUTP using liposome transfection reagent (DOTAP) Biotechniques. 1997;22:308–312. doi: 10.2144/97222st03. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T. The destinies and destinations of RNAs. Cell. 1998;95:451–460. doi: 10.1016/s0092-8674(00)81613-x. [DOI] [PubMed] [Google Scholar]
- Hesketh JE. Sorting of messenger RNAs in the cytoplasm: mRNA localization and the cytoskeleton. Exp Cell Res. 1996;225:219–236. doi: 10.1006/excr.1996.0172. [DOI] [PubMed] [Google Scholar]
- Jansen R-P. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- Jeffert ER, Holowczak JA. RNA synthesis in vaccinia-infected L cells: inhibition of ribosome formation and maturation. Virology. 1971;46:730–744. doi: 10.1016/0042-6822(71)90075-4. [DOI] [PubMed] [Google Scholar]
- Kates J, Beeson J. Ribonucleic acid synthesis in vaccinia virus. I. The mechanism of synthesis and release of RNA in vaccinia cores. J Mol Biol. 1970;50:1–18. doi: 10.1016/0022-2836(70)90100-2. [DOI] [PubMed] [Google Scholar]
- Kit S, Dubbs DR. Biochemistry of vaccinia-infected mouse fibroblasts (strain L-M). I. Effects on nucleic acid and protein synthesis. Virology. 1962;18:274–285. doi: 10.1016/0042-6822(62)90014-4. [DOI] [PubMed] [Google Scholar]
- Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meoitic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse Locker J, Kuehn A, Rutter G, Hohenberg H, Wepf R, Griffiths G. Vaccinia virus entry at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol Biol Cell. 2000;11:2497–2511. doi: 10.1091/mbc.11.7.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Sci. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lenk R, Penman S. The cytoskeletal framework and poliovirus metabolism. Cell. 1979;16:289–301. doi: 10.1016/0092-8674(79)90006-0. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Maa J-S, Rodriguez JF, Esteban M. Structural and functional characterization of a cell surface binding protein of vaccinia virus. J Biol Chem. 1990;265:1569–1577. [PubMed] [Google Scholar]
- Metz DH, Esteban M, Danielescu G. The formation of virus polysomes in L cells infected with vaccinia virus. J Gen Virol. 1975;27:181–195. doi: 10.1099/0022-1317-27-2-181. [DOI] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Moss B. Poxviridae the viruses and their replication. In: Fields BN, Knipe DM, Chanock RM, Hirsch MS, Melnick JL, Monath TP, Roizman B, editors. Fields Virology. New York: Raven Press.; 1996. pp. 2637–2671. [Google Scholar]
- Moss B. Regulation of vaccinia virus transcription. Annu Rev Biochem. 1990;59:661–688. doi: 10.1146/annurev.bi.59.070190.003305. [DOI] [PubMed] [Google Scholar]
- Moss B, Ahn B-Y, Amegadzie B, Gershon PD, Keck JG. Cytoplasmic transcription system encoded by vaccinia virus. J Biol Chem. 1991;266:1355–1358. [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Snijder EJ, Schleich S, Roos N, Griffiths G, Krijnse Locker J. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J Virol. 2000;74:3525–3536. doi: 10.1128/jvi.74.8.3525-3536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley S, Cooper RJ. The inhibition of HeLa cell RNA synthesis following infection with vaccinia virus. J Gen Virol. 1984;65:1687–1697. doi: 10.1099/0022-1317-65-10-1687. [DOI] [PubMed] [Google Scholar]
- Pelham HRB, Sykes JMM, Hunt T. Characteristics of a coupled cell-free transcription and translation system directed by vaccinia cores. Eur J Biochem. 1978;82:199–209. doi: 10.1111/j.1432-1033.1978.tb12012.x. [DOI] [PubMed] [Google Scholar]
- Ploubidou A, Moreau V, Ashman K, Reckman I, Gonzalez C, Way M. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemond-Hornbeak H, Moss B. Inhibition of host protein synthesis by vaccinia virus: fate of cell mRNA and synthesis of small poly(A)-rich polyribonucleotides in the presence of actinomycin D. J Virol. 1975;16:34–42. doi: 10.1128/jvi.16.1.34-42.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz M, Sodeik B, Ericsson M, Wolffe E, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1 alpha. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- Sodeik B, Doms RW, Ericsson M, Hiller G, Machamer CE, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci USA. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell C, Singer R. Requirements of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Taneja KL, Lifshits LM, Fay FS, Singer RH. Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Why are different cross-linkers necessary for actin bundle formation in vivo and what does each cross-link contribute? J Cell Biol. 1998;143:121–133. doi: 10.1083/jcb.143.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Venrooij WJ, Sillekens PTG, van Eekelen CAG, Reinders RJ. On the association of mRNA with the cytoskeleton in uninfected and adenovirus-infected human KB cells. Exp Cell Res. 1981;135:79–91. doi: 10.1016/0014-4827(81)90301-3. [DOI] [PubMed] [Google Scholar]
- Wilcock D, Smith GL. Vaccinia virus core protein VP8 is required for virus infectivity, but not for core protein processing or for INV and EEV formation. Virology. 1994;202:294–304. doi: 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- Wilcock D, Smith GL. Vaccinia virions lacking core protein VP8 are deficient in early transcription. J Virol. 1996;70:934–943. doi: 10.1128/jvi.70.2.934-943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, P, Bauer WR. Purification and characterization of vaccinia virus structural protein VP8. Virology. 1988;167:578–584. [PubMed] [Google Scholar]