Abstract
While advances in laboratory automation have dramatically increased throughput of compound screening efforts, development of robust cell-based assays in relevant disease models remain resource-intensive and time-consuming, presenting a bottleneck to drug discovery campaigns. To address this issue, we present a modified gene trap approach to efficiently generate pathway specific reporters that result in a robust “on” signal when the pathway of interest is inhibited. In this proof-of-concept study, we used vemurafenib and trametinib to identify traps that specifically detect inhibition of the mitogen-activated protein kinase (MAPK) pathway in a model of BRAFV600E driven human malignant melanoma. We demonstrate that insertion of our trap into particular loci results in remarkably specific detection of MAPK pathway inhibitors over compounds targeting any other pathway or cellular function. The accuracy of our approach was highlighted in a pilot screen of approximately 6000 compounds where 40 actives were detected including 18 MEK, 10 RAF, and 3 ERK inhibitors along with a few compounds representing previously under-characterized inhibitors of the MAPK pathway. One such compound, bafetinib, a second generation BCR/ABL inhibitor, reduced phosphorylation of ERK and when combined with trametinib, both in vitro and in vivo, reduced growth of vemurafenib resistant melanoma cells. While piloted in a model of BRAF-driven melanoma, our results set the stage for using this approach to rapidly generate reporters against any transcriptionally active pathway across a wide variety of disease-relevant cell-based models to expedite drug discovery efforts.
Keywords: Gene trap, high-throughput screening, B-Raf, MEK, MAP kinase, cancer
Introduction
Effective high-throughput screening (HTS) campaigns can establish a solid foundation upon which successful drug discovery efforts are built. Although automation platforms have advanced, and our collective understanding of molecular drivers of disease has greatly increased, converting such advances into therapies that induce durable patient responses has been challenging1. While there are many potential reasons for this, we viewed two as immediately addressable. First, cell-based assays, including reporter assays, are often developed in cell lines that are most amenable to genetic manipulation but not necessarily in models that best approximate the disease of interest. Second, the use of assays that report in the “down” or “off” direction following drug exposure often results in a limited signal-to-noise ratio leading to false positives, which can misdirect drug discovery resources1. To help address these issues, we set out to develop an improved reporter platform, initially focused on cancer drug development, with the following criteria in mind: First, the platform had to be HTS compatible, capable of testing thousands of samples in a reasonable time frame and cost-effective format. Second, assay development must be efficient, requiring no prior identification of promoter sites or site-specific targeting vector generation. Third, the readout must have a robust signal-to-noise ratio, ideally reporting an increase in signal upon pathway inhibition. Finally, the platform had to be flexible so that the technology can ultimately be employed to interrogate any transcriptionally active pathway in any cancer model.
The platform we devised was built on the foundation of two previous approaches: 1) gene expression profiling to derive transcriptional signatures of active oncogenic signaling cascades and 2) gene trap systems to identify and harness transcriptionally active genes as reporters of pathway activity. Changes in gene transcription can accurately capture the activation status of oncogenic signaling pathways2. This has been well demonstrated in studies that have shown that the RAS gene signature is a more comprehensive metric of RAS functional status than RAS mutation alone3. While gene expression signatures can accurately measure activity of oncogenic pathways, characterization can be labor-intensive, and multi-gene signatures are not easily amenable to HTS-based drug discovery. Therefore, we hypothesized that if single transcripts can be identified which accurately report on the activity state of an oncogenic pathway, these transcripts could be harnessed as robust reporters for use in comprehensive HTS campaigns.
Toward this end, we applied a modified gene trap approach. Gene traps are vectors containing a promoterless reporter cassette which is only expressed upon integration into an active gene locus4-6. While originally developed for insertional mutagenesis in the mouse, previous gene trap screens have been used to rapidly and efficiently identify transcriptionally responsive genes to exogenous stimuli such as Hepatocyte Growth Factor7. To test whether we could use a gene trap approach to isolate highly specific reporters of transcriptionally active oncogenic pathways for HTS, we established a scheme where random integration of a promoterless reporter into the host cell genome was followed by drug selection with a cancer pathway specific tool molecule. This approach allowed us to interrogate the genome in an unbiased manner and identify “trapped” genes where insertion of the promoterless reporter cassette highjacks the endogenous promoter and leads to increased signal, specifically in response to pathway inhibition. These reporters then provide the means to perform subsequent functional screens to identify novel oncogenic pathway inhibitors.
As proof of concept, we focused on the MAP kinase network in a model of BRAF mutant (V600E) melanoma. In addition to activating mutations of BRAF being clear drivers of melanoma and other malignancies, there are a number of tools available to test our hypothesis8. First, a number of highly characterized small molecule inhibitors of BRAF and downstream kinases, such as MEK, are available as inducing agents for our trapping studies9. Second, it is well-established that perturbation of the canonical MAP kinase cascade (BRAF/MEK/ERK) results in changes in the expression of distinct gene sets10-12. Additionally, many structurally diverse specific inhibitors of BRAF and MEK are incorporated into well-characterized annotated screening libraries, thus allowing us to assess the ability of our platform to accurately detect MAPK pathway inhibitors, out of large screening collections of compounds. Therefore, a gene trap screen was performed to identify traps that were responsive to both the BRAF inhibitor vemurafenib and the MEK inhibitor trametinib13-15. Identification of drug responsive traps was followed by a subsequent HTS pilot screen of ∼6000 diverse compounds to establish specificity of the approach and utility for future HTS campaigns designed to identify inhibitors of difficult-to-drug oncogenic targets.
Results
Gene trap-based generation of BRAF/MAPK pathway-specific reporters
To test the hypothesis that high-specificity HTS-compatible reporters of oncogenic function can be rapidly generated without a priori identification of pathway-specific transcripts and mapping of promoter elements, we constructed a gene trap vector called SABRE (Splice Acceptor Bi-functional REporter). The vector construct pSABRE-1, consists of two non-destructive reporter elements (green fluorescent protein (GFP) and secreted, extracellular membrane-anchored Gaussia luciferase (mGLuc)) as well as a neomycin-resistance gene flanked by splice acceptor and SV40 polyadenylation sequences (Figure 1a). pSABRE-1 was packaged as lentiviral particles, transduced into the human BRAFV600E mutant malignant melanoma line A375, and transduced clones were selected in neomycin. Following initial drug selection, thousands of neomycin-resistant clones were identified. We then assessed whether we could employ a drug selection scheme following introduction of pSABRE-1 to identify and isolate A375 clones harboring traps that signal upon exposure to either the BRAF inhibitor vemurafenib or the MEK inhibitor trametinib. Two independent selection schemes, one based on detection of GFP, the other on luciferase signal were tested. Trametinib exposure for 24h followed by fluorescence-activated cell sorting (FACS) for GFP positive cells failed to reveal clones responsive to MEK inhibition. However, the non-destructive nature of the secreted membrane-anchored luciferase cassette provided an alternative selection scheme to identify vemurafenib and trametinib-responsive traps (Figure 1b). Plates containing cell colonies were exposed to luciferase substrate en-masse (105 clones/screen), imaged to generate a pre-treatment luciferase signal, treated with inhibitor for 24 hours, then reimaged to identify drug responsive cells. Clones exhibiting ≥3-fold changes in luciferase signal (in both the “up” and “down” direction) were isolated, expanded and retested. Eleven clones were identified in the initial screen (8 up, 3 down). Upon retesting, 3 “off-to-on” clones and 1 “on-to-off” clone exhibited luciferase signaling windows and dose dependent responses to trametinib indicating sufficiency for downstream HTS studies (Figures 1c and 2). These clones were sequenced to identify the integration sites of the SABRE reporters (Table 1). Further analysis by RNA-seq demonstrated changes in endogenous transcript levels in response to trametinib exposure. Interestingly, integration of the trapping cassette in the clone with the strongest off-to-on signal (SB01, Figure 2a) occurred within a pseudogene transcript called INTS4P2, which has never been previously linked to the canonical BRAF/MEK/ERK kinase cascade. Since SB01 exhibited the strongest signal in the “on” direction, suggesting excellent potential as a reporter for HTS, we focused on this clone for further analysis.
Figure 1. Vector design and screening approach.
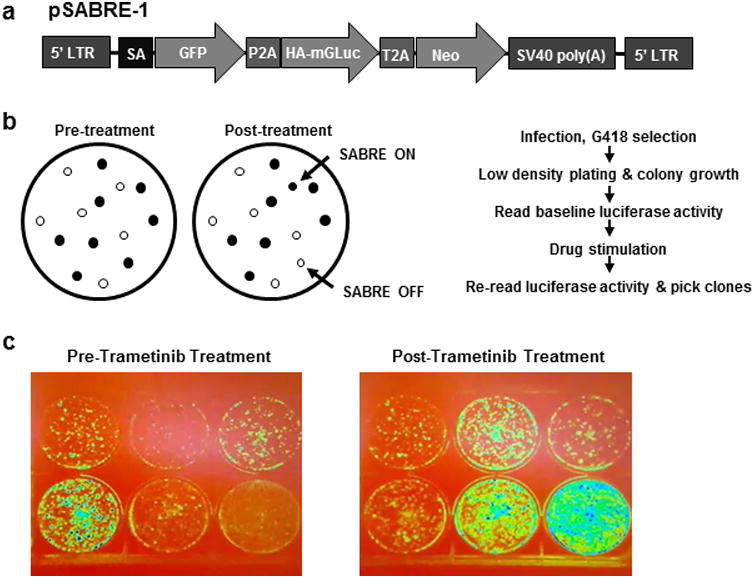
(a) Schematic of the SABRE gene trap vector, pSABRE-1. (b) Image-based screening approach using secreted, membrane anchored Gaussia luciferase (mGLuc). Infected cells are plated at a density where individual colonies can form. To assess basal expression levels, colonies are imaged pre-drug treatment. Pathway specific traps are identified by exposing all colonies to a pathway inhibitor for 24 hours followed by post-treatment imaging to identify both “ON” and “OFF” reporter lines. (c) Representative mGLuc image from the BRAF/MAPK inhibitor screening campaign. Isolated and expanded clones from the BRAF/MAPK pathway inhibitor screen visualized pre and post 24 hour exposure to trametinib. Note that 3 of the clones exhibit luciferase activation upon pathway inhibition (SABRE ON). In contrast, one clone exhibits decreased luciferase activity (SABRE OFF). Images are pseudo colored to better illustrate changes in luciferase expression
Figure 2. Identification of SABRE BRAF/MAPK pathway inhibitor gene trap clones.
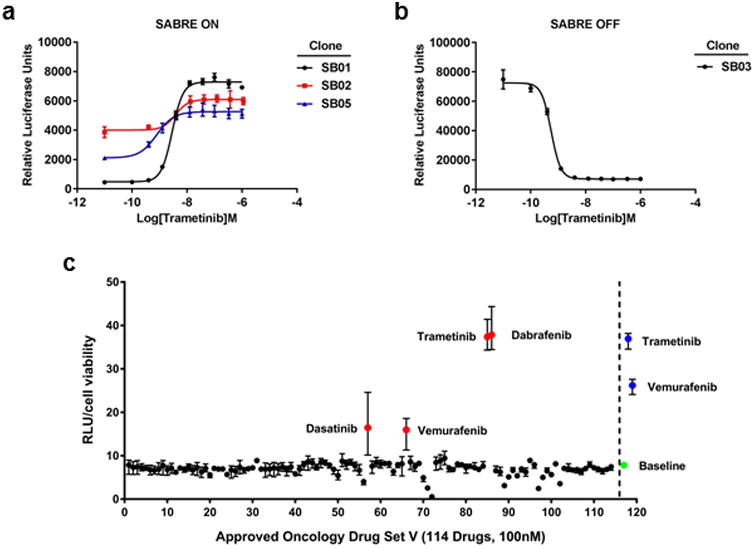
Dose response curves of luciferase activity in (a) 3 SABRE ON clones and (b) 1 SABRE OFF clone in 96-well plate assays. (c) Scatter plot of the luciferase results from the NCI DTP set screen. The red dots indicate drugs that were identified from the DTP set. The blue dots indicate control drugs added to the screen (100nM trametinib, 100nM vemurafenib). The green dot represents the baseline luciferase activity with DMSO control.
Table 1.
Identification of pSABRE-1 integration sites.
| SABRE Clone | Chromosome/ Integration site | Nearest Gene | Nearest Gene Name | RNA-seq (log Fold Change) |
|---|---|---|---|---|
| SB01 | 7q11.21 | INTS4P2 | Integrator complex subunit 4 pseudogene 2 | 3.493 |
| SB02 | 17p13.1 | KDM6B | Lysine demethylase 6B | 0.639 |
| SB05 | 10q25.2 | ADD3 | Adducin 3 | 2.615 |
| SB03 | 12p13.1 | GPRC5A | G protein-coupled receptor class C group 5 member A | -3.225 |
HTS demonstrates BRAF/MAPK pathway specificity of SABRE reporter line
The utility of any drug discovery assay is highly dependent on its specificity. To determine if a single gene insertion generated by our traps could accurately report on specific inhibition of the MAPK pathway, we performed a pilot scale screen on the SABRE SB01 reporter line. We first tested the NCI DTP set (version V), composed of 114 approved anti-cancer agents including 3 inhibitors of the MAPK pathway that target BRAF (vemurafenib and dabrafenib) or MEK (trametinib)16. Consistent with the ability of our clones to accurately detect inhibitors within the MAPK pathway, out of the 114 drugs tested all 3 known MAPK pathway inhibitors resulted in significant activation of the luciferase reporter (Figure 2c). The only other agent exhibiting reporter activity above background was the multi-kinase inhibitor dasatinib, which has been previously shown to inhibit the MAPK pathway17. Based on the suggestive specificity exhibited in this small-scale assessment, we next tested whether this reporter could specifically and comprehensively identify other MAPK pathway inhibitors out of a library of thousands of compounds.
To prepare for HTS, the assay was miniaturized (1536-well format), adapted to automation and validated for screening performance (Figure 3a and b). The assay maintained dose-dependent luciferase signal and excellent performance in miniaturized and automated format (average Z′ values = 0.7). The miniaturized assay was used to screen an annotated library of approximately 6000 small molecules with known modes of action. The screening library was comprised of the well-characterized Library of Pharmacologically Active Compounds (LOPAC) set of 1280 compounds, ∼ 2,800 compounds from the NCGC Pharmaceutical Collection of clinically approved drugs, and 1,912 compounds from the NCATS Oncology Mechanism Interrogation PlatEs (MIPE) library composed of 745 approved drugs, 420 clinical trial-stage drugs, and 767 preclinical molecules18. The MIPE library was assembled with the intent to have diverse and redundant mechanisms of action represented to allow for target-based enrichment analysis of responses. The drugs were screened at either 4, 7, or 11 different concentrations, for the NPC, LOPAC and MIPE collections, respectively, to obtain dose response information for each compound, including potency and efficacy. Consistent with highly specific and comprehensive detection of RAF/MEK/MAPK pathway inhibitors, the screen yielded 40 actives (0.67% hit rate), of which 18 were MEK inhibitors, 10 were RAF inhibitors, and 3 were ERK inhibitors (Figure 3c and Table 2). Enrichment scores using Fisher's exact test were P=2.76E-11 and P=2.99E-20 for BRAF and MEK inhibitors respectively. Actives identified in the screen were re-tested in 9-point dilution series in the SB01 reporter assay as well as the on-to-off reporter line SB03. Interestingly, the dose response curves closely mirrored each other across the 2 lines and the rank order was maintained for all compounds tested (Figure 4, compare a to b and c to d). In addition to the RAF, MEK, and ERK inhibitors, the screen yielded several active compounds that had not been previously annotated as directly inhibiting MAPK pathway targets suggesting that our assay has potential to detect novel inhibitors of pathway signaling (Table 2). Three of these compounds (AMG-Tie2-1, AMG47a, and bafetinib) reconfirmed in our dose response assays in both reporter lines, and resulted in decreased ERK phosphorylation (Figure 4e-g). Taken together, these results demonstrate the exquisite accuracy of the SABRE reporters for detecting bona fide inhibitors of the canonical MAPK pathway.
Figure 3. HTS campaign.
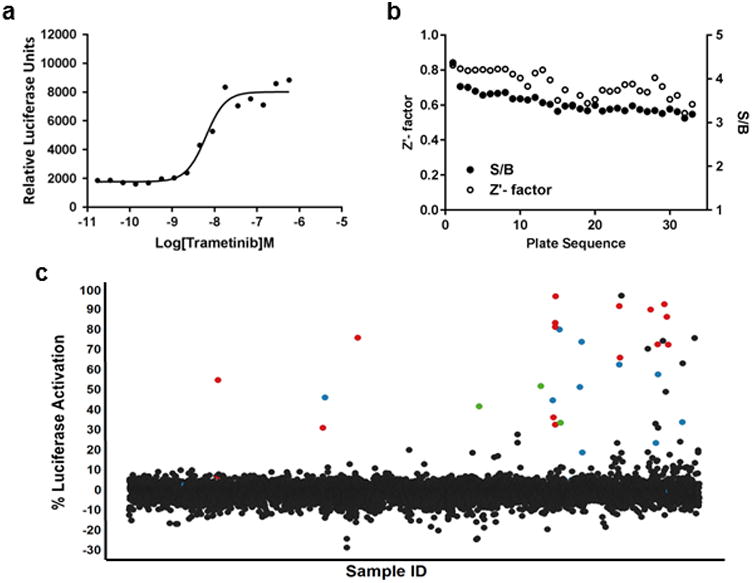
(a) The luciferase assay was miniaturized to a 1536-well format. (b) Clone SB01 was used to screen approximately 6000 compounds from several libraries, including the LOPAC, NPC and MIPE collections. The average Z′ factor for the screen was 0.7. (c) A scatter plot of the screening results demonstrates how well this assay performed. Blue dots represent BRAF inhibitors, red dots represent MEK inhibitors, and green dots represent ERK inhibitors.
Table 2.
Active compounds identified in the HTS.
| Compound | Primary MOA | Max Response | IC50(uM) |
|---|---|---|---|
| Dabrafenib | RAF kinase B inhibitor | 33.987 | 0.0432 |
| Takeda-6d | RAF kinase B/VEGFR2 inhibitor | 33.192 | 0.1367 |
| AZ-628 | RAF kinase B/C inhibitor | 51.409 | 0.8627 |
| Vemurafenib | RAF kinase B inhibitor | 73.922 | 1.2186 |
| PLX-4720 | RAF kinase B inhibitor | 44.904 | 1.3673 |
| AR-00341677 | RAF kinase B inhibitor | 62.686 | 3.4346 |
| L-779450 | RAF kinase C inhibitor | 23.652 | 3.8537 |
| SB-590885 | RAF kinase B inhibitor | 80.14 | 7.6892 |
| MLN-2480 | RAF Kinase inhibitor | 57.695 | 7.6892 |
| ZM-336372 | RAF Kinase inhibitor | 46.306 | 17.2140 |
| TAK-733 | MEK1 inhibitor | 66.085 | 0.0216 |
| AZD-8330 | MEK inhibitor | 72.517 | 0.0385 |
| Trametinib | MEK1/2 inhibitor | 91.863 | 0.0544 |
| PD-0325901 | MEK inhibitor | 96.663 | 0.1218 |
| Pimasertib | MEK1/2 inhibitor | 92.778 | 0.1367 |
| PD-318088 | MEK1/2 inhibitor | 86.414 | 0.1721 |
| Cobimetinib | MEK1 inhibitor | 72.788 | 0.2728 |
| Selumetinib | MEK1/2 inhibitor | 81.37 | 0.3548 |
| ARRY-162 | MEK1/2 inhibitor | 90.085 | 0.6107 |
| Selumetinib | MEK1/2 inhibitor | 83.503 | 0.7689 |
| U0126 | MEK1/2 inhibitor | 10.921 | 1.5101 |
| C.I. 1040 | MEK inhibitor | 32.77 | 1.5342 |
| SL-327 | MEK1/2 inhibitor | 75.98 | 2.4315 |
| C.I. 1040 | MEK inhibitor | 1.594 | 2.5118 |
| Hypothemycin | MEK1/2 inhibitor | -5.244 | 7.6892 |
| RDEA-119 | MEK1/2 inhibitor | 36.386 | 17.2140 |
| PD-098059 | MEK1/2 inhibitor | 54.967 | 19.0115 |
| U0126 | MEK1/2 inhibitor | 31.118 | 24.3154 |
| Pluripotin | Dual RASGAP/ERK inhibitor | 41.839 | 7.6892 |
| NCGC00242487 | ERK inhibitor | 33.695 | 7.6892 |
| FR-180204 | ERK1/2 inhibitor | 51.997 | 19.3144 |
| AMG-47a | LCK Kinase inhibitor | 6.002 | 3.4346 |
| AMG-Tie2-1 | TIE-2 inhibitor | 97.001 | 6.1077 |
| Bafetinib | BCR-ABL Kinase inhibitor | 74.545 | 6.8530 |
| 3-Methyladenine | PI3K inhibitor | 70.604 | 6.8530 |
| VU0482089-2 | MCL inhibitor | 75.893 | 7.6892 |
| LY-2811376 | Beta-Secretase inhibitor | 63.305 | 21.6712 |
| BIX 02188 | MEK 5 inhibitor | 49.118 | 21.6712 |
| Dasatinib | BCR-ABL inhibitor | 27.776 | 21.6712 |
| BMS-536924 | IGF-1R inhibitor | 31.152 | 24.3154 |
Figure 4. Confirmation of HTS hits.
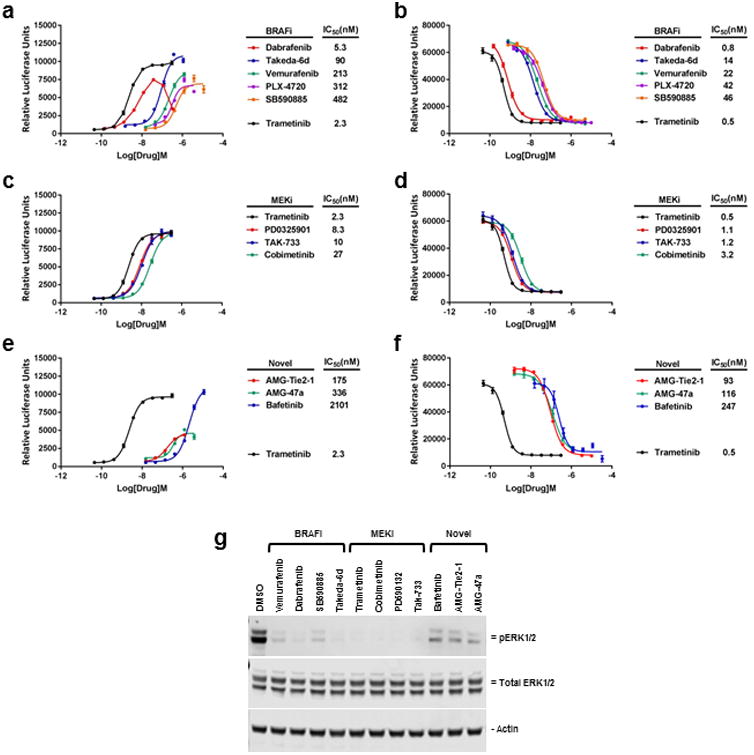
The ability of some of the compounds identified in the HTS were confirmed by 96-well plate assay to both (a, c, e) activate reporter activity in clone SB01 (SABRE ON) as well as (b, d, f) suppress reporter activity in clone SB03 (SABRE OFF). Error bars ± SD. (g) Treatment of SB01 cells for 24 hours with 1uM of each indicated drug results in decreased ERK1/2 phosphorylation as determined by Western blot analysis (20ug per lane).
Identification of a potential novel drug combination to address BRAF resistant melanoma
Acquired drug resistance to BRAF inhibitors is a common occurrence in melanoma. We next investigated whether we could use SABRE to identify compounds that retain inhibitory activity against the MAPK pathway following emergence of BRAF inhibitor resistance. To do so we generated a vemurafenib resistant A375 SABRE reporter line (SB01-VR) through continued exposure to increasing concentrations of vemurafenib (Figure 5a and b). We then tested the ability of some of the top drug candidates identified in the screen to induce luciferase expression in the SB01-VR line. As expected, the resistant SABRE line did not respond to BRAFV600E specific inhibitors (vemurafenib, PLX-4720 and dabrafenib), but maintained responsiveness to MEK inhibitors (trametinib, cobimetinib, and PD0325901) and BRAF inhibitors (SB590885 and Takeda-6d) (Figure 5c and d). Interestingly, although less potent than the MEK inhibitor trametinib, several of the novel pathway inhibitors including bafetinib, AMG-47a, and AMG-Tie2-1 all retained activity in the SB01-VR line (Figure 5e). This suggests that these compounds may have utility for treating BRAF inhibitor-resistant disease, either as single agents or in combination with MEK inhibitors.
Figure 5. Characterization of vemurafenib resistant SABRE line.
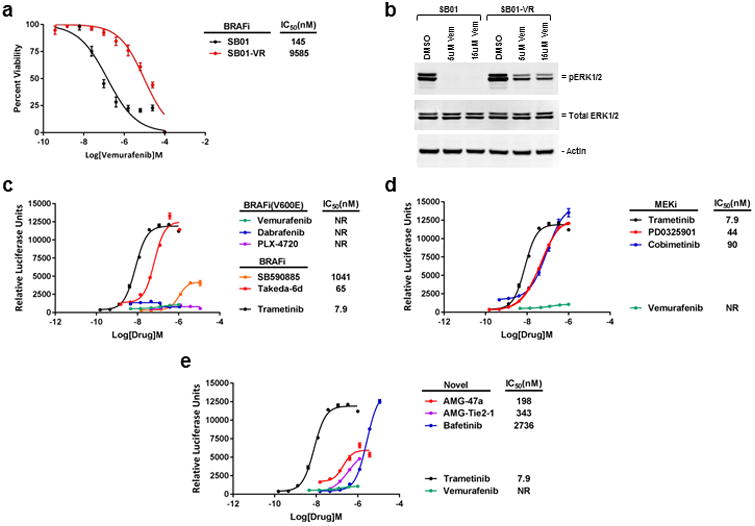
(a) Comparison of the vemurafenib resistant SB01-VR cells (red line) to the SB01 parental cells (black line). Viability curves show a 60-fold increase in IC50 value in the SB01-VR cells compared to the SB01 cells after 72 hours vemurafenib treatment. (b) Following 2 hours incubation with the indicated concentrations of vemurafenib, significant levels of ERK1/2 phosphorylation can still be detected by Western blot analysis in the SB01-VR line, as compared to the parental vemurafenib sensitive SB01 line (20ug per lane). (c-e) 96-well plate assays to assess luciferase activity in the SB01-VR line in response to various drug treatments. NR, no response. Error bars ± SD.
We focused further studies on the second generation BCR-ABL inhibitor bafetinib, since it has an established safety profile in human cancer patients and was progressed to phase II trials19. We assessed whether the combination of bafetinib and trametinib could outperform either drug alone in assays reflecting inhibition of MAPK signaling (pERK Western blot analysis) and tumor cell proliferation. Consistent with potential as an effective novel drug combination to investigate in BRAF inhibitor-resistant disease, the combination of the two drugs resulted in substantial decreases in ERK phosphorylation and A375 SB01-VR colony growth in vitro, as compared to either agent alone (Figure 6a-c). We also assessed the activity of the drug combination on SB01-VR xenografts in vivo (Figure 6d). Because trametinib has potent activity in this line, we first identified a dosing regimen of trametinib that reduced tumor growth rates yet enabled identification of drugs that improved efficacy when used in combination. Consistent with the results of our cell-based assays, treatment with the combination of bafetinib and trametinib, but not with either drug alone, significantly reduced tumor growth (between days 7-16 of the 21 day dosing period) compared to control mice (P=0.0159, Mann-Whitney test) (Figure 6d). In addition to demonstrating that SABRE lines can correctly identify known pathway inhibitors and identify new candidates, the in vivo studies reveal the ability to identify drug candidates that act in tumor cells rendered resistant to the drug used to first establish the SABRE line.
Figure 6. Combination drug treatment of SB01-VR cells in vitro and in vivo.
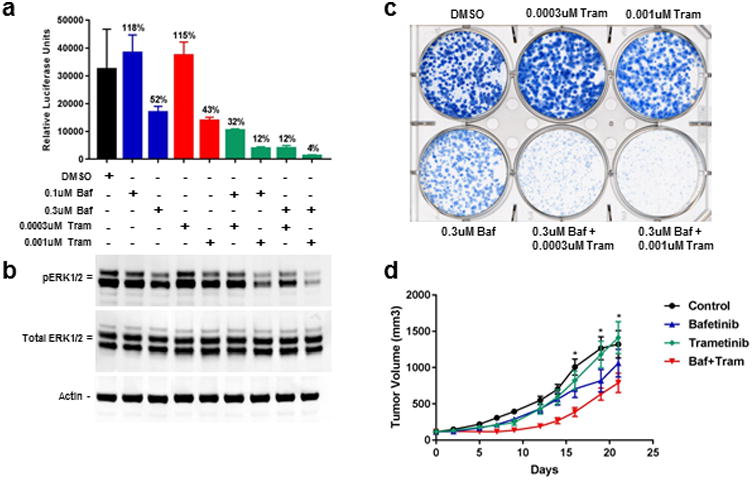
(a) CellTiter-Glo viability assay of SB01-VR cells treated with the indicated drug combinations for 14 days. (b) Western blot analysis of ERK1/2 phosphorylation in SB01-VR cells incubated for 24 hours with the indicated drug treatments (20ug per lane). (c) Colony formation assay with the SB01-VR line. SB01-VR cells treated with single agent (trametinib or bafetinib) or in combination at two different concentrations for 14 days. (d) In vivo PO dosing for 21 days of mice bearing SB01-VR xenografts ± SEM. No blinding of the treatment groups was done. Asterisks indicate control and trametinib treated mice that had to be euthanized early due to ulcerations.
Discussion
The data presented demonstrate that a gene trap approach can be used to generate robust and highly specific reporters that accurately detect inhibitors of oncogenic pathways. The gene trap reporter we used for our screen identified nearly all the known BRAF and MEK inhibitors out of a library of ∼6000 compounds as well as several compounds that had not been primarily associated with the canonical MAP kinase pathway. The “novel” pathway inhibitors detected in our screen validated as inhibiting ERK phosphorylation in subsequent experiments, emphasizing the utility of our approach to identify and repurpose existing compounds for use in new disease areas. Importantly, unlike the conventional approach to reporter generation, ours does not require identification of pathway specific response elements prior to construct generation, assay development, and validation, dramatically increasing the efficiency of reporter generation. Moreover, a single SABRE vector can theoretically be used to generate reporters for any transcriptionally active pathway of interest. Given that oncogenic signaling networks have built in redundancies allowing for adaptive bypass of primary target inhibition, the SABRE platform has the potential to identify and develop sets of novel pathway inhibitors to cut off multiple routes leading to tumor resistance to targeted therapies.
Our screen and subsequent hit validation studies revealed several compounds that could potentially address acquired resistance to BRAF inhibitors such as vemurafenib. The identification of bafetinib provides an example of how our approach can be used to repurpose a compound, originally developed as a second generation BCR/ABL inhibitor. Furthermore, our approach highlighted BRAF inhibitors such as SB590885 and Takeda-6d, that retained activity in our vemurafenib resistant model. Interestingly the chemical structure of Takeda-6d is similar to that of Pan-RAF inhibitor TAK-632 which demonstrated potent anti-tumor effects in BRAF-mutant melanoma cells with acquired resistance to BRAF inhibitors20-22.
A reasonable question regarding our approach is “why not just use expression profiling data followed by CRISPR to clone into the loci of highly drug-regulated transcripts?” While this sounds efficient, there are several key problems with this strategy. First, there is no way of knowing, a priori, whether reporter insertion into a regulated transcript site will lead to deleterious tumor cell effects that could decrease cell viability or signal strength of the reporter. With SABRE, insertion into genes required for cell viability or reporter signal are automatically eliminated since we screen for increased reporter function. Only the most robust traps are selected. Second, although CRISPR is relatively efficient, novel targeting vector generation is required for every targeted insertion23. Therefore, the gene trap approach provides an even greater level of efficiency over a CRISPR-based approach.
The insertion of our trap into the pseudogene INTS4P2 is interesting in that to our knowledge, INTS4P2 has never been reported to be associated with the MAP kinase pathway. Given that tumor cell types exhibit unique transcriptional profiles to external stimuli, the high specificity and responsiveness of the pSABRE-1 reporters, such as SB01, may be context specific to the cell type in which they are created. Early experiments performed in our lab to assess transcriptional regulation of INTS4P2 in response to trametinib across multiple cell lines, suggests that this is indeed the case. This suggests that the one-size-fits-all approach for reporter generation where a common promoter element is identified, linked to a reporter, and then artificially overexpressed will likely be limited for most drug discovery efforts due to a low signal to noise ratio. The gene trap approach described here has the potential to solve this issue.
Beyond establishing proof-of-concept, this study highlighted potential ways to improve the pSABRE-1 vector and its use in isolating traps with high signal-to-noise in the “on” direction. One potential issue is inclusion of all reporter elements including the neomycin-resistance gene in the trapping cassette. Since expression of neomycin currently relies on insertion into a gene with a constitutively active promoter, this design has the potential to bias for traps that report in the “off” direction (SB03) or for traps with low signal-to-noise in the “on” direction (SB02 and SB05). The identification of a high signal-to-noise clone like SB01 used here, suggests that the level of neomycin expression required for sufficient drug resistance is relatively low compared to the level of expression required for observable luciferase expression. To increase the efficiency of identification of clones such as SB01 in future studies, we are currently exploring vector designs that separate expression of the neomycin-resistance gene from the rest of the reporter cassette.
While the well-defined, highly “druggable” BRAF/MEK/ERK pathway was used here to demonstrate proof of concept for the approach, the technology may be best applied to interrogate transcriptionally active but notoriously difficult to drug targets such as RAS and MYC. RAS and MYC proteins have often been labeled “undruggable” due to the relatively smooth surface structure, devoid of obvious deep pockets, which serve as targets for binding of conventional small drug-like molecules24. Approximately one third of all cancers are driven by activating mutations of RAS and nearly 50% of cancers display increased MYC expression25, 26. Despite decades of research, drugs that safely curb RAS or MYC activity have yet to be discovered. Therefore, we believe that the SABRE platform provides a uniquely elegant opportunity to generate specific reporters for these more challenging oncogenic pathways and thus provide a system that can be used to more efficiently screen small molecule and/or peptide libraries.
Materials and Methods
Generation of SABRE cell lines
A375 cells (American Type Culture Collection, Manassas VA, USA) were RADIL tested (IDEXX Laboratories, Westbrook ME, USA) and maintained in Dulbecco's Modified Eagle's Medium (Gibco, Life Technologies, Carlsbad, CA, USA) with 10% FBS and penicillin-streptomycin. The pSABRE-1 vector was purchased from Xactagen, LLC (Shoreline, WA, USA). Lentivirus was generated by transfecting HEK293T cells with pSABRE-1, pMD2.G, and psPAX2 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Lentiviral supernatant with 5ug/ml polybrene was used to infect A375 cells. Infected cells were selected with 1mg/ml Geneticin for up to 10 days. To establish individual cell colonies for screening, cells were plated at low density in 150mm dishes and allowed to grow for two weeks. Colonies were exposed to luciferase substrate en-masse (10ug/ml Coelenterazine, NanoLight Technologies, Pinetop, AZ, USA) and imaged with a Kodak-4000R image station to generate pre-treatment luciferase signals. Plates were treated with inhibitor (10uM vemurafenib or 0.1-10uM trametinib; SelleckChem, Houston, TX, USA) for 6-24 hours prior to repeating the image-based luciferase assay. Clonal cell lines were established by serial dilution in 96-well plates. Vemurafenib-resistant SB01 cells (SB01-VR) were generated by growing cells in increasing concentrations of vemurafenib (up to 2uM).
Gaussia Luciferase Assay
Cells were plated (10,000 – 20,000 cells/90ul/well) in white 96-well plates. The next day, 10ul of 10× drug solution was added to each well in a 9-point dilution series in triplicate. Following 24 hour drug exposure, wells were rinsed with Opti-MEM I Reduced Serum Medium without phenol red (Gibco) and 100ul of Gluc Assay Buffer (10ug/ml Coelenterazine in Opti-MEM I Reduced Serum Medium without phenol red) was added. The luciferase signal was quantitated on a BioTek Synergy microplate reader (Winooski, VT, USA) using Gen5 v1.11 software. Viability was determined by adding 10ul PrestoBlue Cell Viability Reagent (Invitrogen) to the wells. Compounds: trametinib, vemurafenib, dabrafenib, PLX-4720, SB590885, PD0325901, TAK-733, cobimetinib, bafetinib (SelleckChem); AMG-47a (MedChem Express, Monmouth Junction, NJ, USA); AMG-Tie2-1, Takeda-6d (AdipoGen Life Sciences, San Diego, CA, USA). IC50 values were calculated using GraphPad Prism v7.03. (La Jolla, CA, USA)
Lentiviral Integration Site Analysis
Lentiviral insertion sites were determined using the Lenti-X Integration Site Analysis Kit and the SMARTer RACE 5′/3′ Kit following the manufacturer's instructions (Clontech, Mountain View, CA, USA). PCR products were sequenced (Genewiz, South Plainfield, NJ, USA) and analyzed using NCBI BLAST27.
RNA-seq
A375 cells were treated for 24 hours with 100nM trametinib or DMSO. RNA-seq libraries were prepared from total RNA using the TruSeq RNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA) and a Sciclone NGSx Workstation (PerkinElmer, Waltham, MA, USA). Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies). Sequencing was performed using an Illumina HiSeq 2500 employing a paired-end, 50 base read length (PE50) approach. Image analysis and base calling were performed using Illumina's Real Time Analysis v1.18 software, followed by ‘demultiplexing’ of indexed reads and generation of FASTQ files, using Illumina's bcl2fastq Conversion Software v1.8.4 (http://support.illumina.com/downloads/bcl2fastq_conversion_software_184.html). Low quality reads were filtered prior to alignment to the reference genome (UCSC hg38 assembly) using TopHat v2.1.028. Counts were generated from TopHat alignments for each gene using the Python package HTSeq v0.6.129. Genes which did not have at least 1 CPM in at least 1 sample were discarded, followed by TMM normalization using the Bioconductor package edgeR v3.12.0.30. LogFC and logCPM values were calculated from the normalized results.
HTS
SABRE reporter assays were conducted in 1536-well white solid bottom plates. 1000 cells/well were seeded in 4ul Opti-MEM I Reduced Serum Medium without phenol red, using a Multidrop Combi Reagent dispenser and small pin cassette (Thermo Scientific, Fisher Scientific, Fair Lawn, NJ, USA). After overnight incubation, 23nl of compound solution in DMSO was transferred using a Kalypsys pintool. Plates were covered with stainless steel Kalypsys lids and incubated at 37°C for 24 hours. Then 4ul of GLuc Assay Buffer (final concentration 40ug/ml) was added using a Multidrop Combi Reagent dispenser and small pin cassette. Relative luciferase units (RLU) were quantified using ViewLux (PerkinElmer).
For library screening, compounds were transferred to columns 5-48, and controls were added in columns 1-4 of the 1536-well plate. Column 1 contained media only; column 2 contained cells with DMSO, while columns 3 and 4 contained a dose response of trametinib (starting at a final concentration of 500nM, with 2-fold dilutions) and a fixed concentration of trametinib at 100nM, respectively. Compounds were tested as dose responses starting at a stock concentration of 10mM (final concentration of 57uM) in DMSO, and diluted 3-fold, also with DMSO. RLUs for each well were normalized to the median RLUs from the DMSO control wells as 0% signal, and median RLUs from 100nM trametinib wells as 100% signal. Dose response data was analyzed as described previously31-33. Briefly, activity of the compounds from the dose response qHTS screen was determined based on two parameters: i) % activity at the maximum concentration of compound tested (MAXR); and ii) logIC50 for those compounds with a dose response, as determined by the Curve Response Class (CRC) classification from dose response HTS, in which normalized data is fitted to a 4-parameter dose response curve using a custom grid-based algorithm to generate a CRC score for each compound dose response. CRC values of 1.1, 1.2, 2.1, 2.2 are considered highest quality hits; CRC values of 1.3, 1.4, 2.3, 2.4 and 3 are inconclusive hits; and a CRC value of 4 are inactive compounds. See Supplemental Data for list of MAXR, CRC and logIC50 for the compounds screened.
We identified the annotated targets for these compounds and computed the enrichment for each target, compared to background, using Fisher's exact test. The background was defined as all targets annotated in the MIPE collection. The P-value was adjusted for multiple hypothesis testing using the Benjamini-Hochberg method.
Western Blot
Protein lysates were prepared in MPER (ThermoScientific, Waltham, MA, USA) containing protease (Roche, Indianapolis, IN, USA) and phosphatase (Sigma-Aldrich, St. Louis, MO, USA) inhibitors. Lysates were resolved by 4-12% NuPage Bis-Tris SDS-PAGE (Invitrogen) and transferred to nitrocellulose. Antibodies: Rabbit anti-p44/42 MAPK (Erk1/2) (1:1,000, #4695 Cell Signaling Technology, Danvers, MA, USA), Rabbit anti-Phospho-p44/42 MAPK (Erk1/2), Thr202/Tyr204 (1:2,000, #4370, Cell Signaling Technology), Mouse anti-β-actin (1:3,000, #926-42212, LI-COR, Lincoln, NE, USA), IRDye 680RD Goat anti-Rabbit (1:15,000, 925-68071, LI-COR) and IRDye 800CW Donkey anti-Mouse (1:15,000, 92532212, LI-COR). Blots were developed using the LI-COR Odyssey CLx Imaging System.
Colony and Cell Proliferation Assays
For colony formation assays, cells (1,000/well) were seeded in 6-well plates and allowed to adhere overnight. Bafetinib, trametinib, combinations or 0.5% DMSO was added to the media. Cells were treated for 14 days, with media and drugs replaced after 7 days. Colonies were fixed and stained with 0.5% methylene blue (Sigma-Aldrich) in 50% EtOH. For short-term proliferation assays, cells (8,000/well) were seeded in 96-well plates and allowed to adhere overnight. Drugs were added to each well and incubated for 72 hours. For long-term cell proliferation assays, cells (60/well) were seeded in 96-well plates and allowed to adhere overnight. Drugs were added to each well and cells were treated for 14 days, with media and drugs replaced after 7 days. Viability was assessed using CellTiter-Glo (Promega, Madison, WI, USA) or PrestoBlue (Invitrogen).
In Vivo
All animal procedures were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee. Nu/Nu female mice 4 weeks old were purchased from Envigo (East Millstone, NJ, USA). SB01-VR xenografts were established by subcutaneous injection of 5×10ˆ6 cells into the right flank. Five days after injection, mice were randomized into groups of 10 to obtain groups with similar starting average tumor size. Groups of 10 mice allowed us to observe tumor size differences of 15% or more that are 1.1 standard deviation units with 80% power in a 2-sided test with 0.05 level of significance. Bafetininb, trametinib, combination bafetinib and trametinib, or vehicle (0.5% methylcellulose, 5% DMSO, 0.2% Tween 80) was administered by oral gavage for 21 consecutive days. Bafetinib was dosed at 200mg/kg/d (100mg/kg twice daily); trametinib was dosed at 0.1mg/kg once a day34, 35. Mice treated with drug combinations were dosed with 100mg/kg bafetinib plus 0.1mg/kg trametinib in the morning and only 100mg/kg bafetinib at night. Control mice were also dosed twice daily. Tumor sizes were assessed three times a week by caliper measurement and volumes calculated from the formula, tumor size (cm3)=(L×Wˆ2×3.14159/6) using Study Advantage Tumor Tracker software (Jenkintown, PA, USA).
Supplementary Material
Acknowledgments
We would like to thank Jason Frazier, Robert Finney, Emily J. Girard, Madison W. Nakamoto and the staff of the Genomics Shared Resource and the Comparative Medicine Shared Resource at the Fred Hutchinson Cancer Research Center. This research was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers R43CA171366 (R.A.K.), RO1CA193841 (A.J.M. and J.M.O.), RO1CA155360 and the Seattle Children's Brain Tumor Research Endowment (J.M.O.).
Footnotes
Conflict of Interest: Shelli Morris and Jim Olson are shareholders in Presage Biosciences and Jim Olson serves on the Presage Board. Richard Klinghoffer and Connor Burns are employees and shareholders of Presage Biosciences.
References
- 1.Kaelin WG., Jr Common pitfalls in preclinical cancer target validation. Nature reviews Cancer. 2017;17(7):425–40. doi: 10.1038/nrc.2017.32. [DOI] [PubMed] [Google Scholar]
- 2.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 3.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC medical genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes & development. 1991;5(9):1513–23. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 5.Friedrich G, Soriano P. Insertional mutagenesis by retroviruses and promoter traps in embryonic stem cells. Methods in enzymology. 1993;225:681–701. doi: 10.1016/0076-6879(93)25044-3. [DOI] [PubMed] [Google Scholar]
- 6.Friedel RH, Soriano P. Gene trap mutagenesis in the mouse. Methods in enzymology. 2010;477:243–69. doi: 10.1016/S0076-6879(10)77013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medico E, Gambarotta G, Gentile A, Comoglio PM, Soriano P. A gene trap vector system for identifying transcriptionally responsive genes. Nature biotechnology. 2001;19(6):579–82. doi: 10.1038/89343. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nature reviews Clinical oncology. 2017;14(8):463–82. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 10.Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment cell research. 2007;20(3):216–21. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 11.Pavey S, Johansson P, Packer L, Taylor J, Stark M, Pollock PM, et al. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23(23):4060–7. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- 12.Kannengiesser C, Spatz A, Michiels S, Eychene A, Dessen P, Lazar V, et al. Gene expression signature associated with BRAF mutations in human primary cutaneous melanomas. Molecular oncology. 2008;1(4):425–30. doi: 10.1016/j.molonc.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi T, Kakefuda R, Tajima N, Sowa Y, Sakai T. Antitumor activities of JTP-74057 (GSK1120212), a novel MEK1/2 inhibitor, on colorectal cancer cell lines in vitro and in vivo. International journal of oncology. 2011;39(1):23–31. doi: 10.3892/ijo.2011.1015. [DOI] [PubMed] [Google Scholar]
- 15.Abe H, Kikuchi S, Hayakawa K, Iida T, Nagahashi N, Maeda K, et al. Discovery of a Highly Potent and Selective MEK Inhibitor: GSK1120212 (JTP-74057 DMSO Solvate) ACS medicinal chemistry letters. 2011;2(4):320–4. doi: 10.1021/ml200004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monga M, Sausville EA. Developmental therapeutics program at the NCI: molecular target and drug discovery process. Leukemia. 2002;16(4):520–6. doi: 10.1038/sj.leu.2402464. [DOI] [PubMed] [Google Scholar]
- 17.Li YJ, He YF, Han XH, Hu B. Dasatinib suppresses invasion and induces apoptosis in nasopharyngeal carcinoma. International journal of clinical and experimental pathology. 2015;8(7):7818–24. [PMC free article] [PubMed] [Google Scholar]
- 18.Mathews Griner LA, Zhang X, Guha R, McKnight C, Goldlust IS, Lal-Nag M, et al. Large-scale pharmacological profiling of 3D tumor models of cancer cells. Cell death & disease. 2016;7(12):e2492. doi: 10.1038/cddis.2016.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos FP, Kantarjian H, Cortes J, Quintas-Cardama A. Bafetinib, a dual Bcr-Abl/Lyn tyrosine kinase inhibitor for the potential treatment of leukemia. Current opinion in investigational drugs. 2010;11(12):1450–65. [PubMed] [Google Scholar]
- 20.Okaniwa M, Hirose M, Arita T, Yabuki M, Nakamura A, Takagi T, et al. Discovery of a selective kinase inhibitor (TAK-632) targeting pan-RAF inhibition: design, synthesis, and biological evaluation of C-7-substituted 1,3-benzothiazole derivatives. Journal of medicinal chemistry. 2013;56(16):6478–94. doi: 10.1021/jm400778d. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura A, Arita T, Tsuchiya S, Donelan J, Chouitar J, Carideo E, et al. Antitumor activity of the selective pan-RAF inhibitor TAK-632 in BRAF inhibitor-resistant melanoma. Cancer research. 2013;73(23):7043–55. doi: 10.1158/0008-5472.CAN-13-1825. [DOI] [PubMed] [Google Scholar]
- 22.Okaniwa M, Hirose M, Imada T, Ohashi T, Hayashi Y, Miyazaki T, et al. Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds. Journal of medicinal chemistry. 2012;55(7):3452–78. doi: 10.1021/jm300126x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Human molecular genetics. 2014;23(R1):R40–6. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 24.Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nature reviews Cancer. 2017;17(8):502–8. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nature reviews Drug discovery. 2014;13(11):828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D, et al. MYC Deregulation in Primary Human Cancers. Genes. 2017;8(6) doi: 10.3390/genes8060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Jadhav A, Southal N, Huang R, Nguyen DT. A grid algorithm for high throughput fitting of dose-response curve data. Current chemical genomics. 2010;4:57–66. doi: 10.2174/1875397301004010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(31):11473–8. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southall N, Jadhav A, Huang R, Nguyen T, Wang Y. Enabling the large scale analysis of quantitative high throughput screening data. In: Seethala R, Zhang L, editors. Handbook of Drug Screening. Second. Boca Raton, FL, USA: CRC Press; 2009. pp. 442–64. [Google Scholar]
- 34.Kimura S, Naito H, Segawa H, Kuroda J, Yuasa T, Sato K, et al. NS-187, a potent and selective dual Bcr-Abl/Lyn tyrosine kinase inhibitor, is a novel agent for imatinib-resistant leukemia. Blood. 2005;106(12):3948–54. doi: 10.1182/blood-2005-06-2209. [DOI] [PubMed] [Google Scholar]
- 35.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(5):989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


