SUMMARY
Since its discovery as a skeletal muscle-specific transcription factor able to reprogram somatic cells into differentiated myofibers, MyoD has provided an instructive model to understand how transcription factors regulate gene expression. Reciprocally, studies of other transcriptional regulators have provided testable hypotheses to further understand how MyoD activates transcription. Using MyoD as a reference, in this review, we discuss the similarities and differences in the regulatory mechanisms employed by tissue-specific transcription factors to access DNA and regulate gene expression by cooperatively shaping the chromatin landscape within the context of cellular differentiation.
Keywords: Gene expression, chromatin architecture, epigenetics, transcription factors, pioneer factors, MyoD, myogenesis
eTOC Blurb
In 1987, Davis, Weintraub, and Lassar reported that MyoD expression converted fibroblasts into myoblasts, opening the field of cell reprogramming. In this Review, we use the lessons learned from MyoD to discuss how transcription factors access DNA to regulate gene expression by cooperatively shaping the chromatin landscape.
Historical Perspective
Experiments performed by John B. Gurdon in the 1960s revealed that, when transplanted into an enucleated recipient egg, the nucleus derived from an intestinal cell of Xenopus laevis could sustain normal development to an adult fertile frog (Gurdon, 1962) (Gurdon and Uehlinger, 1966). An important corollary of these observations was that the somatic nucleus retains the ability to recapitulate all the information necessary to execute the multitude of genetic programs necessary for cell lineage specification and animal development. Genes repressed in intestinal cells could be reactivated by exposure to factors present in the enucleated egg. Using a different technique, heterokaryons generated by the fusion of human non-muscle cells with mouse muscle cells were shown to activate the transcription of human muscle genes. Human fibroblasts, which share a mesodermal origin with muscle cells, displayed faster kinetics and a higher frequency of muscle gene activation than other cells of different embryonic origin (e.g., keratinocytes and hepatocytes) (Blau et al., 1983) (Blau et al., 1985). Similar experiments performed with heterokaryons of chick skeletal myocytes and rat neuronal cells suggested the existence of putative chick skeletal myocyte-inducing factors with the ability to promote the expression of rat muscle genes (Wright, 1984). Finally, heterokaryons of human B lymphocytes and mouse C2C12 myotubes revealed that muscle gene expression program is dominant over that of B cells (Terranova et al. 2006). These findings suggested that a factor diffusing from muscle cells might be acting in trans on the silent muscle-specific genes of different non-muscle cell types to activate their expression.
Transfection of genomic DNA from either mouse fibroblasts treated with the DNA demethylating agent 5-azacytidine (Taylor and Jones, 1979) (Konieczny and Emerson, 1984) or mouse muscle cells into mouse embryonic fibroblasts resulted in the myogenic conversion of approximately 1 in 15,000 transfected colonies (Lassar et al., 1986). The low frequency of myogenic conversion was consistent with the probability of transfecting a single genetic determinant. In 1987, Davis, Weintraub, and Lassar reported that the expression of a single transfected cDNA (myoblast determination gene number 1, MyoD1) converted fibroblasts into myoblasts (Davis et al., 1987). This discovery was the de facto origin of the field of cell reprogramming, which culminated in the 2006 report that four defined factors could induce pluripotent stem cells from mouse embryonic and adult fibroblasts (Takahashi and Yamanaka, 2006).
While celebrating the 20th anniversary of Molecular Cell, this review also reflects on the 30th anniversary of the discovery of MyoD, as recently discussed by Andrew Lassar (Lassar, 2017). A study published in the first issue of Molecular Cell (Puri et al., 1997b) reported that MyoD functionally associates with the histone acetyltransferases (HATs) p300/CBP and PCAF/GCN5 at target genes. This and subsequent studies have provided functional and biochemical evidence that the ability of MyoD to activate transcription relies on the enzymatic activity of associated HATs (Sartorelli et al., 1999) (Polesskaya et al., 2000) (Dilworth et al., 2004) (Di Padova et al., 2007) and paved the way for understanding the epigenetic regulation of muscle gene expression, prompting the search for and identification of additional chromatin modifying complexes used by MyoD to activate transcription (Fong and Tapscott, 2013).
Accessing DNA Within Repressive Chromatin
A critical question related to gene transcription and cell reprogramming is how TFs gain access to their cognate DNA-binding motifs within condensed chromatin to activate lineage programs. MyoD has the unique ability to convert differentiated somatic cells of various embryonic origins into skeletal muscle - an outcome that is otherwise achieved by the cooperative activity of multiple tissue-specific transcription factors (TFs), as in the case of fibroblast trans-differentiation into neurons and cardiomyocytes (Caiazzo et al., 2011) (Pang et al., 2011) (Pfisterer et al., 2011) (Vierbuchen et al., 2010) (Ieda et al., 2010) (Qian et al., 2012). Likewise, somatic cell reprogramming into pluripotent cells requires the cooperative activity of multiple TFs (Takahashi and Yamanaka, 2006) (Takahashi et al., 2007). This distinctive feature of MyoD likely resides in the lack of a requirement for the co-expression of additional TFs, which typically synergize through cooperative binding to regulatory elements in the genome (Chronis et al., 2017) (Velasco et al., 2017), and raises the question of what molecular properties might license MyoD to activate the myogenic program once expressed in non-muscle cells. Clues were provided by experimental evidence of the radical acceleration of nuclear reprogramming when the acidic transcriptional activation domain of MyoD was fused to Oct4 (Hirai et al., 2013). The increased dynamics of DNA methylation, chromatin accessibility, histone modification and protein binding to pluripotent genes observed in these experiments suggests that at least one property that distinguishes MyoD from other lineage-specific TFs is accounted for by the unusual transcriptional potency of its activation domain. Interestingly, the activation domain does not overlap with the amino acidic sequences required for MyoD to promote chromatin accessibility (Gerber et al., 1997), further emphasizing the complexity of MyoD-mediated chromatin regulation and gene activation.
One of the proposed function of TF activation domains is to contact the Pre-Initiation Complex (PIC), which is typically nucleated by general transcription factors (GTFs). In this regard, it has been proposed that replacement of the canonical GTF, TFIID-TBP complex, with the alternative TRF3/TBP2, is required for activation of muscle-gene expression by MyoD (Deato and Tjian, 2007). However, further work established that TRF3/TBP2 is actually not required for skeletal myogeneiss (Gazdag et al., 2009) and it is not expressed in muscle cells (Malecova et al., 2016). Indeed, during myogenic differentiation TFIID-TBP continues to provide the general transcription factor (GTF) available for MyoD to activate gene expression. Of note, TAF3 appears a candidate GTF component that mediates interactions between MyoD, permissive chromatin (enriched in H3K4me3) and PIC, as also proposed by Deato and coworkers (Deato et al., 2008).
Reciprocal Cooperation between Pioneers and Followers
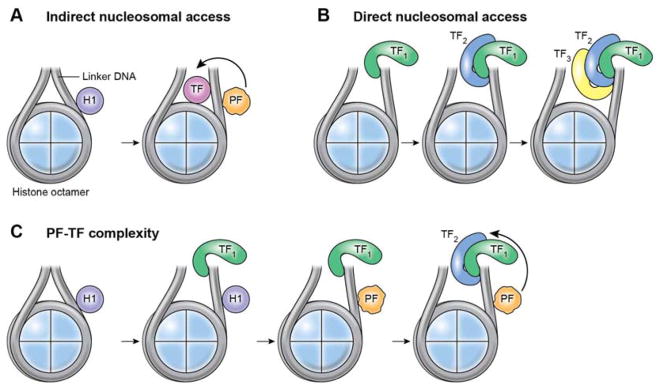
The transcription of developmentally or signal-regulated genomic region is generally associated with progressive chromatin unfolding (Stalder et al., 1980) (Wu et al., 1979). Chromatin decompaction can be initiated by a subset of TFs, called pioneer factors (PFs), that employ distinct molecular strategies to penetrate repressed chromatin to access their cognate DNA sequences complexed with histones in nucleosome structures (Iwafuchi-Doi and Zaret, 2014). Pioneer factors can scan nucleosomal DNA for their cognate DNA-binding motifs, reside for prolonged time, thus allowing for cooperative interactions with other TFs to occur for more stable DNA binding (Zaret and Mango, 2016). A prototypical TF with the properties of a PF is FoxA1 (HNF-3γ), whose “winged helix” DNA-binding domain structure shares structural similarities with the linker histone H1 (Clark et al., 1993). This feature allows FoxA to compete for and displace linker histones, thus permitting access to nucleosomes by other TFs, both at promoter and enhancer regions (Cirillo et al., 1998) (Iwafuchi-Doi et al., 2016) (Figure 1A). Similarly, Pax7 - a factor that specifies muscle stem (satellite) (Seale et al., 2000) and pituitary cell identity (Budry et al., 2012) - was shown to act as a PF for the Tbox TF Tpit (Lamolet et al., 2001) by recognizing its entire target sequence on nucleosomal DNA. In contrast, the pluripotency-reprogramming TFs Pou5fl (Oct4), Sox2, and Klf4 can bind nucleosomes in vitro and access silent sites enriched for nucleosomes by recognizing partial sequences of their respective DNA-binding motifs exposed on the nucleosome surface. C-Myc cooperatively facilitates the binding of pluripotent reprogramming factors (Soufi et al., 2015). In agreement with their genomic occupancy maps (Boyer et al., 2005), pluripotency reprogramming factors target nucleosomes synergistically (Soufi et al., 2015) (Figure 1). Such synergy is also exploited to silence active somatic enhancers during the early phases of cell reprogramming through recruitment of the histone deacetylase Hdac1 at somatic enhancers mediated by pluripotency TFs Oct4, Sox2, and Klf4 and decreased expression of the somatic TF Fra1 (Chronis et al., 2017). Human and mouse somatic cells may differ in this regard, with a more relevant role for stage-specific somatic TFs in modulating the binding of reprogramming factors in mouse cells (Chronis et al., 2017).
Figure 1.
Accessing DNA Within Repressive Chromatin. (A) Indirect TF nucleosomal access. Exploiting structural similarities with the linker histone H1, the pioneer factor (PF) FoXA1 competes for and displaces linker histones, thus permitting access to nucleosomes by other TFs. (B) Direct TF nucleosomal access. Pluripotency reprogramming factors target nucleosomes synergistically. (C) PF-TF cooperativity. Reciprocal cooperativity between PFs and TFs.
While PFs share the ability to penetrate repressed chromatin, the transcriptional outcome of their DNA binding is distinct. Reprogramming factors behave as both PFs and transcriptional activators. FoxA1 occupies the liver-specific albumin enhancer prior to gene activation (Gualdi et al., 1996). Similarly, Xenopus tropicalis Foxh1 binding precedes H3K4me1 deposition and p300 engagement (Charney et al., 2017), and Pax7 binding favors the subsequent engagement of MyoD, even after Pax7 removal (Lilja et al., 2017). Their function therefore seems to be to mark specific genomic regions to direct the recruitment of additional transcriptional activators and may not necessarily be to activate transcription. Beside its role in development, FoxA1 also exerts a PF function in promoting the signal-regulated recruitment of the estrogen receptor. FoxA1 silencing either abolishes or diminishes the recruitment of approximately 90% of estrogen receptor binding events (Carroll et al., 2005) (Laganiere et al., 2005). In other instances, FoXA1 binding might depend on the action of a steroid receptor (Rigaud et al., 1991) (Kong et al., 2011) (Caizzi et al., 2014). For instance, in breast cancer cells, a subset of FoxA1 sites located in close vicinity to estrogen or glucocorticoid receptor binding sites is occupied only after dexamethasone treatment (Swinstead et al., 2016). Under defined experimental conditions, residence times revealed by single-molecule tracking did not significantly differ between FoxA1 and steroid receptors, arguing against FoxA1 utilizing a long dwell time to create an open chromatin state (Swinstead et al., 2016). Whether FoxA1 exhibits longer residence times on chromatin in other cellular settings remains to be determined. In analogous cooperative examples, AP1 binding facilitates chromatin accessibility to the glucocorticoid receptor (Biddie et al., 2011). Furthermore, through colocalization with other TFs, NF-Y and related factors containing histone-fold domains structurally related to histone H2A and H2B promote accessibility to inactive or Polycomb-repressed genomic regions both in animals and plants (Fleming et al., 2013) (Oldfield et al., 2014) (Tao et al., 2017). Thus, as with reprogramming factors, there is a reciprocal, cooperative and site-specific requirement for the chromatin recruitment of FoxA1, AP1, steroid receptors, and other TFs (Figure 1).
Myogenesis: Even a Master Can Use Some Help
MyoD was the first TF shown to directly reprogram cell fate (Davis et al., 1987; Weintraub et al., 1989). MyoD interacts with a DNA consensus sequence (the E-box) through an evolutionarily conserved basic region shared with other members of the basic-helix-loop-helix (bHLH) family of TFs. MyoD and the related muscle-specific bHLH Myf5 access and remodel chromatin one order of magnitude more efficiently than Myogenin, a bHLH required for muscle differentiation whose basic region is identical to that of MyoD and Myf5. Two domains present in MyoD and Myf5 and absent in Myogenin mediate chromatin access and remodeling (Gerber et al., 1997). These two domains are necessary for the stable binding of MyoD to a non-canonical E-box at the Myogenin promoter occupied by an adjacent protein complex containing the homedomain TFs Pbx and Meis (Berkes et al., 2004). Pbx1 and MyoD physically interact (Knoepfler et al., 1999), and MyoD mutations disrupting cooperative binding to Pbx/Meis complexes prevent the stable recruitment of MyoD to the Myogenin promoter, subsequent chromatin remodeling, and transcriptional activation. Pbx binding to the Myogenin promoter occurs in undifferentiated myoblasts, when Myogenin is not yet transcribed, and precedes that of MyoD. Moreover, a Pbx-Meis complex is present at the Myogenin promoter in fibroblasts, prior to the activation of gene expression by exogenous MyoD, indicating that Pbx recruitment is independent of MyoD. In keeping with its role as a PF, mutating the Pbx binding site near the non-canonical E-box in the Myogenin promoter hinders MyoD binding (Berkes et al., 2004). In addition to serving as a modulator of muscle gene activation, Pbx is also part of a molecular switch involved in postnatal Myogenin repression, as revealed by its co-binding to the Myogenin promoter with the Y-box repressor MSY-3 in postnatal innervated muscle (Berghella et al., 2008). Notably, steroid hormone-induced FoxA1 binding occurs at target sites that are poorly enriched for canonical FoxA1 DNA motifs compared with the enrichment of hormone-independent FoxA1 binding sites (Swinstead et al., 2016). Thus, while a PF can help stabilize the binding of a TF to a non-canonical target site (e.g., Pbx1 and MyoD), the reverse can also occur when a steroid hormone facilitates the binding of FoxA1 to non-canonical sites. In addition to stabilizing MyoD at non-canonical E-box sequences, the requirement for Pbx to assist MyoD-dependent transcription might be related to the longer length of the basic-helix1 region of MyoD, which is predicted to result in suboptimal nucleosome binding compared with that of other bHLH neuronal TFs, such as Ascl1 (Soufi et al., 2015). Hence, Pbx may assist MyoD activity at both non-canonical and canonical E-boxes. Indeed, the initial observation of a cooperative role for Pbx/Meis and MyoD at the Myogenin gene has been confirmed and extended to a larger subset of genes regulated by MyoD, revealing the importance of homeodomain proteins in marking genes for subsequent activation (Maves et al., 2007) (Fong et al., 2015). A more general role of Pbx1 in regulating muscle gene expression is also consistent with the substantial genome-wide co-occupancy of MyoD and Pbx1 sites and the presence of Pbx1, along with the histone variant macroH2A1.2, at prospective muscle enhancer sites, allowing for transcriptional competency in differentiated muscle cells (Dell’Orso et al., 2016).
The general picture emerging from these studies suggests that, rather than operating according to a rigid division of labor, PFs and TFs cooperate in distinct biological processes with interchangeable roles and modalities to access and affect the activity of genomic regions controlling gene expression.
Engaging Histone Modifiers
As discussed above for PFs and TFs, there seems to be a reciprocal cooperation between TFs and chromatin accessibility in regulating gene transcription. The transcriptional coactivator p300 (Eckner et al., 1994) was initially shown to interact with numerous TFs, including MyoD and MEF2 (Eckner et al., 1996) (Yuan et al., 1996) (Puri et al., 1997a) (Sartorelli et al., 1997). Later studies revealed that p300 and the associated factor PCAF possess histone acetyltransferase (HAT) activity (Ogryzko et al., 1996) (Yang et al., 1996). PCAF and p300 were shown to interact with MyoD to perform different functions (Puri et al., 1997b) that have been subsequently molecularly clarified (Dilworth et al., 2004; Sartorelli et al., 1999). p300 and other HATs exert a central and genome-wide role in conferring H3K27 acetylation, a mark of transcriptional competency, at both promoter and enhancer regions (Calo and Wysocka, 2013). A case in point is the unusually high co-enrichment of p300, H3K27 acetylation and TFs at super-enhancers (Whyte et al., 2013). TFs mediate the recruitment of additional chromatin-modifying machines (CMMs) to direct them to specific genomic loci and, in doing so, guarantee the specificity of gene regulation. In addition to TFs, CMM components can mediate DNA sequence-specific recruitment. Recruitment modalities employed to position CMMs rely on specific DNA sequences, such as telomeric DNA (de Lange, 2005) (Erdel et al., 2017) or CpG islands (CGIs). The H3K4 methyltransferase complex Set1 targets non-methylated CGIs via its Cfp1 subunits (Lee and Skalnik, 2005) and Polycomb Repressive Complex 1.1 (PRC1.1) recognizes CGIs through the zinc-finger CxxC domain of KDM2B (Farcas et al., 2012). Analogously, Polycomb-like (PCL) proteins (PHF1, PHF19, and MTF2) mediate PRC2 recruitment to unmethylated CGIs (Li et al., 2017), and the propagation of PRC2 repression in Drosophila requires sequence-specific recruitment to DNA (Laprell et al., 2017). Thus, the entire collection of TFs and CMMs at regulatory elements in the genome (i.e., promoters and enhancers) appears to be directed by their combinatorial binding preference for distinct DNA sequences, thereby highlighting the importance of the integration between genetic and epigenetic determinants of gene expression.
Les Liaisons Dangereuses: Attracting and Repelling the Regulators
Heterokaryon experiments indicated that mouse muscle cells can reprogram the muscle gene expression of human non-muscle cells with different kinetics and efficiencies depending on their embryonic origin. Moreover, some cell types are refractory to MyoD-mediated conversion (Weintraub et al., 1989) (Schafer et al., 1990) (Albini et al., 2013). The pluripotent mouse cell line P19 can be converted into neurons via exogenous expression of the neurogenic bHLH NeuroD2 but not MyoD (Farah et al., 2000) (Fong et al., 2012). Conversely, MyoD can reprogram mouse embryonic fibroblasts and not P19 cells (Skerjanc et al., 1994) (Yoo et al., 2011). The lack of transcriptional activation of the neurogenic and myogenic programs by NeuroD2 and MyoD in non-competent cell types correlates well with decreased binding at the promoter-proximal sites of the corresponding genes. Moreover, studies by Fong et al. revealed that MyoD and NeuroD show a preference for a specific E-box motif, with the central dinucleotide as the genetic determinant of this preference (Fong et al., 2012). Overall, these findings demonstrate that the binding of bHLH TFs in different cell types is influenced by both epigenetic (e.g., chromatin accessibility to their DNA binding sites) and genetic (DNA sequence) determinants (Fong et al., 2012).
The functional interplay between genetic and epigenetic determinants is clearly demonstrated by the reciprocal interactions between TFs, CMM and histone modifications. The recruitment of CMMs to specific genomic locations occurs through DNA-binding proteins or long non-coding RNAs (lncRNAs) (Plath et al., 2003) (Rinn et al., 2007) (Zhao et al., 2008) (Lai et al., 2013). In turn, histone modifications introduced by CMMs regulate the access of TFs to chromatin. FoXA1 binding favors the deposition of H3K4me1/me2 (Taube et al., 2010) (Serandour et al., 2011) (Jozwik et al., 2016), and reciprocally, its engagement is favored by these two histone marks (Lupien et al., 2008). H3K4 and H3K79 methylation and H3 acetylation are a pre-requisite for Myc binding to either its E-box consensus or to alternative sequences (Guccione et al., 2006) (Figure 2). This Myc-chromatin signature might explain the behavior of Myc as a nonlinear amplifier of gene expression acting on already highly active genes (Nie et al., 2012) (Lin et al., 2012). As c-Myc assists the binding of Oct4, Sox2, and Klf4 during cell reprogramming (Soufi et al., 2012) (Soufi et al., 2015), it is relevant to understand how the Myc-chromatin signature is initially established. Sequential MyoD DNA binding, the recruitment of HATs and the further stabilization of the MyoD/HAT interaction with acetylated histones have been shown (Polesskaya et al., 2001) and might be implicated in the formation of active enhancers in muscle cells (Blum et al., 2012).
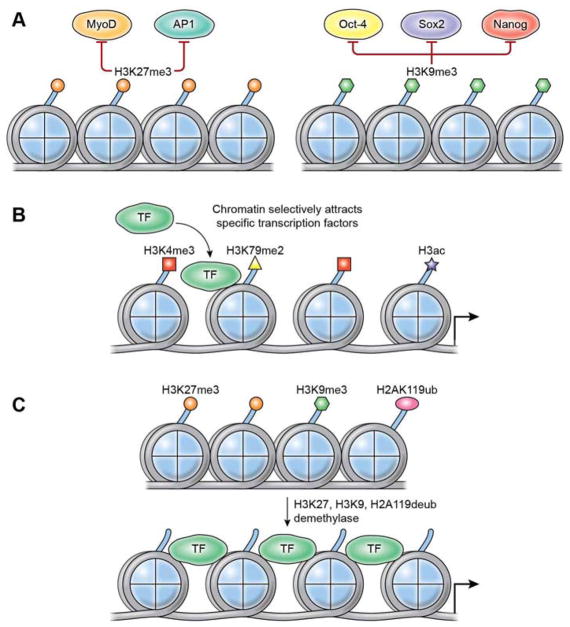
Figure 2.
Histone Marks Direct or Repel Transcription Factors. (A) Nucleosomes marked by H3K27me3 repel MyoD and AP1 binding (left panel) and those with H3K9me3 marks repel pluripotency factors Oct4, Sox2, and Nanog (right panel). (B) Myc target sites are enriched for H3K4me3 and H3K79me2 marks which precede and are independent of Myc binding. (C) Hundreds of genes resistant to nuclear reprogramming can be reactivated by the combinatorial overexpression of H3K9 and H3K27 demethylases, and the H2AK119 deubiquitylase.
Chromatin histone marks can also impede TF recruitment. PRC2 is involved in ensuring lineage fidelity by depositing H3K27 methylation (Schuettengruber et al., 2017). In undifferentiated muscle cells, Polycomb-dependent H3K27me3 opposes MyoD recruitment at repressed genes that become occupied by MyoD and activated when H3K27 is demethylated in differentiated cells (Caretti et al., 2004) (Figure 2). Accordingly, reduced H3K27me3 caused by interfering with either Polycomb Ezh2 or Suz12 component results in the premature expression of muscle-specific, as well as some non-myogenic bivalent genes (Juan et al., 2011) (Asp et al., 2011) (Liu et al., 2013) (Cesarini et al., 2015). This mechanism is evolutionarily conserved, as C. elegans PRC2 mes-2/E(Z) also counteracts the activity of hlh1/MyoD (Yuzyuk et al., 2009). Similarly, AP1 recruitment to genes in the epidermal differentiation locus is prohibited by H3K27me3 in epidermal basal progenitors (Figure 2). The transcription of these genes coincides with the loss of H3K27me and the differentiation of basal progenitors. Consistent with the regulatory role of H3K27me3, the expression of epidermal differentiation genes is precociously activated in basal cells derived from conditional Ezh2-null mice (Ezhkova et al., 2009). In line with a general role for H3K27me3 in regulating chromatin access to TFs, the delayed accumulation of H3K27me3 on nascent DNA during the early period of DNA replication offers a window of opportunity for the recruitment of several lineage-specific TFs during embryonic stem cell differentiation. The experimentally induced accumulation of H3K27me3 in nascent DNA blocks TF recruitment (Petruk et al., 2017). The inhibition of Polycomb proteins significantly decreases cell reprogramming. In contrast, SUV39H1 inhibition favors reprogramming (Onder et al., 2012). Polycomb and SUV39H1 deposit repressive histone marks. Thus, the unexpected finding that they have contrasting outcomes on cell reprogramming indicate that their combinatorial effects are complex. SUV39H1-mediated H3K9me3 repels binding by Oct4, Sox2, and Nanog (Figure 2), as indicated by the increased binding of these reprogramming factors in cells with reduced SUV39H1 and H3K9me3 and the accelerated appearance of reprogrammed colonies (Soufi et al., 2012) (Becker et al., 2017). As a given pre-existing chromatin signature can direct TF binding, as in the case of Myc (Guccione et al., 2006), combinations of different histone marks can also act as barriers to the recruitment of specific TFs (Figure 2). In certain biological processes, histone marks with opposing functions, as discussed for H3K27me3 and H3K9me3 in cell reprogramming, become functionally concordant. Mammalian nuclei transplanted into Xenopus oocytes switch from a somatic to an oocyte genetic program (Jullien et al., 2014). However, hundreds of genes are resistant to reprogramming. Most of these (~ 90%) can be reactivated by the combinatorial overexpression of chromatin modifiers such as H3K9 and H3K27 demethylases and H2AK119 deubiquitylase (Jullien et al., 2017) (Figure 2). Unexpectedly, these chromatin modifiers are less effective in reactivating resistant genes in the context of DNA hypomethylation (Jullien et al., 2017). Thus, while H3K27me3 and H3K9me3 have opposing effects during TF-mediated reprogramming (Onder et al., 2012), they act concordantly in repressing genes resistant to nuclear transfer reprogramming.
Another histone mark that has been shown to mediate transcriptional repression at promoters is H3K4 monomethylation (H3K4me1). MLL3/4-mediated H3K4me1 at the promoters of undifferentiated muscle cells, as well as in other cell types (such as embryonic fibroblasts, macrophages and embryonic stem cells), establishes boundaries that spatially demarcate the engagement of H3K4me3 readers. Upon activation of skeletal myogenesis, the conversion of H3K4me1 into the activation mark H3K4me3 by an alternative COMPASS methyltransferase leads to muscle gene promoter activation and transcription (Cheng et al., 2014). In another study, muscle-specific ablation of the MLL4 gene resulted in severe muscle defects, leading to breathing malfunction and death. Brown pre-adipocytes derived from MLL3/MLL4 knock-out mice and converted with MyoD failed to activate muscle gene expression, indicating a requirement of MLL3/MLL4 for enhancer activation and cell-type-specific expression (Lee et al., 2013).
Sculpting Chromatin by Remodelers
The ability of TFs to remodel chromatin generally depends on the recruitment of specialized chromatin remodeling complexes (CRCs) that alter the structure of the nucleosome. The chromatin unit comprises 147 base pairs of DNA wrapped around an octamer of core histones, namely, H2A, H2B, H3 and H4. Although several CRCs have been reported to assist TFs in altering nucleosomes, the SWItch/Sucrose Non-Fermentable (SWI/SNF) complex has been reported to be an essential effector of MyoD-mediated chromatin remodeling at previously silent loci (de la Serna et al., 2005) (Toto et al., 2016). The seminal finding that the enzymatic SWI/SNF sub-units Brg1 and Brm are necessary for MyoD-mediated conversion of mouse fibroblasts into skeletal muscle (de la Serna et al., 2001) was followed by several studies that emphasized the complexity of SWI/SNF usage by MyoD. These studies revealed how the dynamic composition of the SWI/SNF complex is exploited by MyoD to remodel the chromatin at specific loci (Puri and Mercola, 2012). SWI/SNF complexes typically contain one enzymatic subunit – either the ATPase Brahma (Brm) or Brg1 – and a collection of Brg1/Brm-associated factors (BAFs) that are conserved from yeast to humans, with an increased number of components and multiple variants for each subunit that are alternatively incorporated into specific SWI/SNF complexes with patterns of tissue-specific expression (Wang et al., 1996) (Ho and Crabtree, 2010). The evolutionarily expansion of SWI/SNF subunits indicates that dynamic combinatorial assembly can accommodate the demand for transcriptional control of more complex genomes. The multi-step SWI/SNF recruitment by MyoD to target loci has been shown, whereby initial association with the structural subunit BAF60C (encoded by SMARCD3) is followed by the recruitment of a functional complex that includes functional (ATPases) and structural “core” components - Baf47, Baf155, and Baf170 - required for chromatin remodeling activity (Forcales et al., 2012) (Figure 3). When extended to other models of terminal differentiation (e.g., neurogenesis and cardiogenesis), the TF preference for specific SWI/SNF variant sub-units appears to dictate localized chromatin remodeling throughout the genome for the tissue-specific control of gene expression (Takeuchi and Bruneau, 2009) (Lickert et al., 2004) (Lessard et al., 2007) (Yoo et al., 2009).
Figure 3.
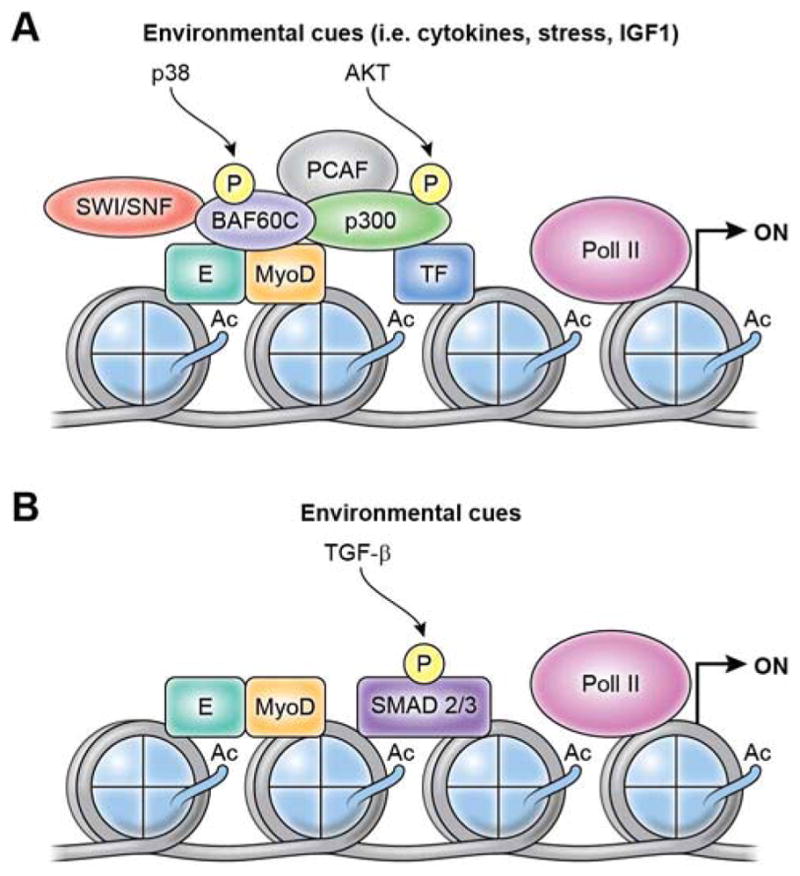
Two models of signal-regulated gene expression from MyoD-bound regulatory elements, either proximal promoters or distal enhancers. (A) Extracellular cues emanated from the regenerative muscle environment, such as cytokines, growth factors and stress, are converted into intracellular signaling leading to activation of effector kinases (e.g. p38 alpha/beta or AKT1-2) by targeting key components of CMM, including SWI/SNF chromatin remodeling complex (which is recruited via p38-mediated threonine 229 phosphorylation of murine BAF60C - encoded by SMARCD3) and the HAT p300 (which is recruited via AKT-mediated phosphorylation of serines 1734 and 1834) (Simone et al 2004; Serra et al. 2007; Forcales et al. 2012). While it is currently unknown whether these two events occur sequentially or simultaneously, they are interdependent, and the next effect of this interplay is histone hyperacetylation and chromatin remodeling. Genetic or pharmacological inhibition of either p38 or AKT signaling ex vivo generates two distinct populations of undifferentiated satellite cells with “intermediate phenotypes” that reflect the incomplete formation of chromatin modifications permissive for transcription at MyoD-bound regulatory elements. (B) Environmental activation of Smad2/3 DNA binding by TGF-β signaling is regulated by master transcription factors, such as MyoD, thereby enabling TGF-β-mediated regulation master transcription factor control of gene expression. For instance, MyoD expression in nonmuscle cells redirects Smad3 to MyoD-bound regulatory elements (Mullen et al. 2011). This is an example of TF cooperative loading to generate activated enhancers (or super-enhancers) for cell type-specific activation of gene expression. Given that the transcriptional outcome of TGF-β signaling largely depends on the intensity and/or duration of its activity, it is likely TGF-β signaling differentially impacts on MyoD-regulated gene expression during developmental myogenesis, as well as in post-natal life, during compensatory or maladaptive muscle regeneration.
Other CRCs have been implicated in the MyoD-mediated control of chromatin remodeling for the regulation of target genes. Among them, the chromatin-remodeling complex SNF2-related CBP activator protein (SRCAP) and the SNF2 chromatin remodeling enzyme family member – chromodomain helicase DNA-binding domain 2 (Chd2) – has been proposed to link MyoD-mediated chromatin remodeling with histone exchange through the nucleosomal incorporation of specific histone variants H2A.Z and H3.3, respectively, which facilitate TF access and gene activation (Cuadrado et al., 2010) (Harada et al., 2012). Conversely, the forced overexpression of histone H3.1 to replace H3.3 suppressed the activation of skeletal myogenesis by favoring H3K27me3 accumulation and decreasing H3K4me3 levels at MyoD-bound promoters (Harada et al., 2015). Likewise, the incorporation of the histone variant mH2A1.2 is required for the activation of muscle gene expression by promoting H3K27 acetylation and the recruitment of the PF Pbx1 at prospective enhancers (Dell’Orso et al., 2016). A general interplay between lineage-specific TFs and histone variants regulates chromatin dynamics during development, nuclear reprogramming and cellular differentiation and is largely dependent on cell-cycle-regulated expression patterns and the activity of histone chaperones (Filipescu et al., 2014) (Buschbeck and Hake, 2017). Failure to activate the cell cycle, such as in a model of replicative senescence, likely limits the availability of chaperones for the deposition of histone variants that enable MyoD-chromatin remodeling, thereby accounting for the constitutive resistance of senescent cells to MyoD-mediated myogenic conversion (Latella et al., 2017). While this emphasizes the importance of cell-cycle progression in the MyoD-mediated activation of the myogenic program, the replication-independent H3.3 assembly into chromatin by the histone chaperones HIRA and Asf1a has been shown to generate permissive chromatin structures for polymerase II recruitment at MyoD-bound promoters, regardless of transcription-associated covalent histone modifications (Yang et al., 2011). Similarly, the histone chaperone and elongation factor Spt6 promotes skeletal myogenesis by favoring cooperation between Polymerase II and the histone demethylase KDM6A (UTX), although in this case, changes in histone modifications (e.g., reduced levels of H3K27me3) are involved (Wang et al., 2013). Thus, cell cycle-dependent and independent regulation of histone variant incorporation into nucleosomes likely reflects the existence of distinct regulatory processes involved in controlling gene expression in proliferating muscle progenitors and their post-mitotic differentiated progeny.
Finally, processive chromatin remodeling can be instigated by non-coding RNA generated from transcription within the MyoD super-enhancer region or other genomic regions, whereby these non-coding RNA activate MyoD and downstream myogenic genes (Cesana et al., 2011) (Mousavi et al., 2013) (Gong et al., 2015).
Signal-Mediated Regulation of TF Chromatin Accessibility
TF activity is typically regulated by environmental perturbations. In the case of lineage-specific TFs that control tissue progenitor identity and differentiation, environmental cues are important determinants of the temporal and spatial control of gene expression. How these cues are converted into regulatory signals that coordinate TF access to DNA and transcriptional activity to generate cell-type-specific responses continues to be investigated. During adult myogenesis (i.e., injury-induced muscle regeneration), the activation of muscle progenitor cells (otherwise referred to as satellite cells) coincides with both MyoD expression (Zammit et al., 2004) and exposure to multiple cues released by the cellular components of the regenerative environment (Brancaccio and Palacios, 2015). The direct regulation of MyoD by the environment has been proposed to occur through a variety of signals that can impact MyoD function both directly and indirectly. For instance, MyoD, which is a highly phosphorylated protein (Puri and Sartorelli, 2000), may be subject to regulatory post-translational control by specific kinases. However, in contrast to other signal-regulated TFs, a direct relationship between MyoD phosphorylation at specific residues and its ability to activate transcription has not been established. This might reflect differences in TF regulation by transient versus persistent stimuli. Indeed, TF-activation by transient phosphorylation, followed by de-phosphorylation and the termination of a transcription burst, typically defines a class of signal-responsive TFs implicated in the regulation of inflammatory or metabolic responses (Altarejos and Montminy, 2011) (Karin and Hunter, 1995). By contrast, serine-200 phosphorylation controls MyoD half-life during the cell cycle (Kitzmann et al., 1999), and tyrosine-30 phosphorylation regulates the MyoD response to DNA damage (Puri et al., 2002). Nonetheless, a direct link between phosphorylation and MyoD transcriptional activity has not been established. Rather, emerging evidence indicates that signaling to different components of the chromatin remodeling and transcriptional machineries converts environmental cues into epigenetic modifications that control gene expression in muscle progenitors (see below).
Adapting to Environmental Changes - The Signal-Dependent Control of Gene Expression
The activation of p38 kinases alpha and beta in MyoD-expressing satellite cells exposed to the milieu of cytokines and growth factors released within the regenerative environment promotes myogenic commitment through asymmetric division (Troy et al., 2012). p38 alpha and beta stimulate skeletal myogenesis through a variety of mechanisms, including the phosphorylation of MEF2 proteins (Zetser et al., 1999) (Wu et al., 2000), which mediate the recruitment of the Ash2L-containing H3K4me3 methyltransferase complex (Rampalli et al., 2007), as well as the phosphorylation of the SWI/SNF chromatin remodeling BAF60C subunit (Simone et al., 2004) (Forcales et al., 2012) and the SRCAP subunit ZNHIT1/p18 (Hamlet) (Cuadrado et al., 2010). The net effect is local H3K4me3 enrichment and chromatin remodeling at MyoD-bound loci, which ultimately facilitates Polymerase II access to target genes (Bergstrom et al., 2002) (Penn et al., 2004). Of note, upon stimulation, p38 alpha/beta kinases bind to chromatin at active promoters during myogenic differentiation (Simone et al., 2004) (Segales et al., 2016), which is consistent with a general model of signal-activated chromatin binding of p38 kinases leading to the phosphorylation of components of the chromatin remodeling and transcription machineries to enable TF-mediated activation of Pol II elongation and gene expression (Proft et al., 2006) (Ferreiro et al., 2010) (Pelet et al., 2011) (Silva et al., 2017). Interestingly, the p38-mediated formation of repressive chromatin has also been reported to coordinate the activation and repression of different subsets of genes implicated in the cell cycle, lineage determination and differentiation of satellite cells – a finding that has inspired a pharmacological strategy for satellite cell expansion via the intermittent pharmacological blockade of p38 kinases (or their downstream targets) (Palacios et al., 2010), which a few years later was repurposed to reactivate and expand aged satellite cells (Cosgrove et al., 2014) (Bernet et al., 2014).
An additional mechanism by which p38 signaling can control MyoD activity relies on the downstream kinase Msk-1, which phosphorylates KAP1 (KRAB [Kruppel-like associated box]-associated protein 1)/TRIM28, which functions as a scaffold to recruit co-activators (p300 and LSD1) or corepressors (G9a and HDAC1) at MyoD-bound promoters. Upon differentiation, the MSK1-mediated phosphorylation of KAP1 releases the corepressors and favors MyoD interactions with positive cofactors (Singh et al., 2015).
Changes in metabolic states during development or following muscle injury can also potentially affect myogenesis. MyoD and its associated HATs, p300 and PCAF, are deacetylated and inactivated by SIRT1 in an NAD+-dependent fashion (Fulco et al., 2003) (Motta et al., 2004). Satellite cells experience a metabolic switch from fatty-acid oxidation to glycolysis during activation (Ryall et al., 2015) (Machado et al., 2017). This metabolic change coincides with decreased NAD+ levels, reduced SIRT1 activity, and the promotion of a myogenic differentiation program (Ryall et al., 2015). In contrast, calorie restriction and exercise increase NAD+ production and lead to SIRT1 activation, resulting in the inhibition of skeletal muscle differentiation, increased myogenic colony formation, and the activation of oxidative metabolism (Fulco et al., 2008) (Canto et al., 2009) (Cerletti et al., 2012). In another example of signal-dependent metabolic modifications, IGF-1/PI3K/AKT-dependent activation of ATP citrate lyase (ACL) leads to the production of acetyl-CoA, which is utilized by HATs to promote histone H3 acetylation at the MyoD locus, followed by MyoD activation and cell differentiation (Das et al., 2017). While subject to metabolic regulation itself, MyoD can in turn influence the metabolic state of differentiated myotubes and myofibers by activating transcription of numerous metabolic genes including PGC-1β, a master regulator of mitochondrial biogenesis and oxidative metabolism (Shintaku et al., 2016). Thus, multiple signals from the surrounding environment converge on MyoD, its coregulators, and its genomic regulatory regions to determine the most appropriate transcriptional outcome.
Signal-Regulated Enhancer Activation
One of the most important aspects of lineage specification, namely, the determination of cell identity and cellular differentiation, is related to signal-directed genome usage by TFs, which typically refers to the activation of tissue-specific enhancers for lineage-specific gene expression (Heinz et al., 2015). Active enhancers are typically marked by H3K4me1, by the presence of HATs and relative enrichment in H3K27Ac, and by DNAse hypersensitivity, which reflects chromatin accessibility (Visel et al., 2009) (Krebs et al., 2011). Enhancers bound by unusually high concentrations of TFs, coactivators, and PolII to drive the transcription of genes involved in cell identity are referred as to super-enhancers (Whyte et al., 2013) (Parker et al., 2013) (Hnisz et al., 2013). While MyoD-bound enhancers have been identified in myoblasts and myotubes (Blum et al., 2012) (Mousavi et al., 2013), a current challenge is to understand how external signals direct the MyoD-mediated activation of enhancers during skeletal myogenesis as well as the formation of super-enhancers. In this regard, FoxO3 and MyoD may participate in establishing super-enhancers at hotspot regions (Peng et al., 2017). Models of enhancer activation in response to external cues can be extrapolated from several works.
For instance, the concomitant activation of the p38 and PI3K/AKT signaling cascades has been shown to promote chromatin accessibility, via SWI/SNF recruitment, and histone hyperacetylation, via p300 recruitment, at MyoD target genes (Serra et al., 2007) (Figure 3). While the presence of HATs and histone hyperacetylation are well established enhancer marks, the role of BAF components of SWI/SNF has emerged only recently (Shi et al., 2013) (Wang et al., 2017) (Mathur et al., 2017) (Nakayama et al., 2017) (Vierbuchen et al., 2017). A recent analysis of H3K4me1-associated proteins at mammalian enhancers revealed an enrichment of chromatin-remodeling complex BAF on enhancers in vivo, with H3K4me1-marked nucleosomes being more efficiently remodeled by the BAF complex in vitro (Local et al., 2018). A signal-mediated enhancer selection by the growth-factor-inducible TF FOS/JUN (AP-1) and SWI/SNF (BAF) chromatin remodeler has been recently described in fibroblasts, where FOS/JUN selects enhancers together with cell-type-specific TFs by cooperative binding to nucleosomal enhancers and SWI/SNF recruitment (Vierbuchen et al., 2017). By analogy, signal-mediated interactions between MyoD and the SWI/SNF complex (Simone et al., 2004) (Forcales et al., 2012) (Padilla-Benavides et al., 2017) potentially favor the cooperative binding of TFs to enhancers activated in response to environmental signals. A number of putative cooperating TFs (e.g., MEF2, Six, Runx, AP1, and Pbx1) have been found to occupy chromatin in close proximity to MyoD (Cao et al., 2010). Enhancer recruitment of multiple endogenous cooperating TFs and Pol II by MyoD can account for the distinctive ability of MyoD (even among myogenic bHLH TFs) (Conerly et al., 2016) (Gerber et al., 1997) to reprogram somatic cells into skeletal muscle without the co-expression of additional TFs. According to this model, MyoD nuclear reprogramming toward skeletal myogenesis requires synergistic activation of a feed-forward loop between MyoD, Six1 and Six4 proteins (Santolini et al., 2016) (Chakroun et al., 2015).
Another indirect signal-mediated regulator of MyoD activity that is related to cooperative TF occupancy in proximity to MyoD has been proposed based on the genome-wide binding of Smad3 in myotubes, which revealed MyoD-directed Smad3 binding to MyoD-bound sites in response to TGF-beta signaling as a general mechanism of master TF-directed occupancy of signal-activated TFs at cell-type-specific regions relatively devoid of nucleosomes (Mullen et al., 2011) (Ruetz et al., 2017) (Figure 3). This mechanism further reiterates the importance of the cooperative action of TF binding, which is reminiscent of the ligand-mediated activation of transcription typically exploited to control the activity of nuclear hormone receptors (e.g., androgens, estrogens or steroids).
Concluding Remarks and Future Perspectives
The fast pace of technological and conceptual advances in the study of gene expression challenges our understanding of the molecular and mechanistic basis of tissue-specific TF-regulated gene expression. For instance, the emerging interest in the relationship between the three-dimensional (3D) regulation of genome and TF activity is gaining increasing interest. Recent evidence has revealed that the genome is folded into a hierarchy of chromatin domains – including topologically associating domains (TADs) and insulated neighborhoods (INs) – that are generated by high-order chromatin interactions and facilitate or constrain interactions between regulatory elements and genes (Hnisz et al., 2016) (Dixon et al., 2012) (Phillips-Cremins et al., 2013) (Nora et al., 2012) (Sexton et al., 2012) (Dowen et al., 2014) (Dekker and Mirny, 2016). While these structures have the potential to spatially regulate TF activity, whether TFs are simply regulated by or may also actively contribute to 3D genome organization, perhaps by promoting looping between TF-bound genomic elements, remains to be established. The zinc-finger TF YY1 was recently reported to mediate enhancer-promoter loops in analogy to the CTCF-mediated structuring of TADs (Weintraub et al., 2017). A number of observations suggest that MyoD may also contribute to the 3D regulation of gene expression in muscle cells. First, ChIP-seq experiments revealed the pervasive genome binding of MyoD, with only a small percentage of bound DNA elements directly correlating with regional gene transcription (Cao et al., 2010). This observation suggests that MyoD can also globally regulate target genes expression from a distance, as already reported for the regulation of individual genes (Battistelli et al., 2014) (Busanello et al., 2012) (Harada et al., 2015). Moreover, MyoD binding events are often associated with histone acetylation, not necessarily in proximity to transcribed genes (Cao et al., 2010). Finally, physical and functional interactions have been reported between MyoD and the architectural protein CTCF (Delgado-Olguin et al., 2011), and the enrichment of MyoD binding was observed at cis-regulatory regions and at DNA elements bound by CTCF (Cao et al., 2010), often coinciding with IN boundaries (Dall’Agnese and Puri, unpublished results). Future work will determine whether MyoD directly regulates transcription by re-organizing interactions between functional and/or structural genomic elements during the activation of skeletal myogenesis.
Another emerging concept relates to the ability of TFs to cooperatively form super-enhancers in response to extracellular signals. The unusually high density of proteins (including TFs, Pol II, CMMs and components of the Mediator complex) interacting with nucleic acids may lead to the formation of non-membrane-bound organelles, or nuclear bodies, which compartmentalize specific biochemical processes, as in the case of P granules and nucleoli (Hyman et al., 2014) (Saha et al., 2016). Transcriptional regulatory events may also be compartmentalized. Nuclear bodies might be formed by phase separation through cooperative interactions between multivalent molecules (e.g., enhancer-bound TF and other proteins) undergoing reversible chemical modifications (such as phosphorylation and acetylation) (Hnisz et al., 2017). The phase separation model may well apply to the signal-mediated activation of MyoD-bound enhancers to activate the myogenic program in response to physiological cues (such as during developmental and adult myogenesis) but also under conditions of de-regulated activation of the myogenic program, such as during loss of satellite cell asymmetric division during aging and disease (Chakkalakal et al., 2012) (Dumont et al., 2015) and pathological myogenic conversion of non-muscle cells into rhabdomyosarcomas (RMS) (Tenente et al., 2017) (Drummond et al., 2018).
Acknowledgments
We thank the “muscle community” for their support, helpful guidance and suggestions, and apologize in advance if references to their work have been inadvertently omitted. Work in the Sartorelli laboratory is supported by the NIAMS IRP through NIH grants AR041126 and AR041164. Work in the Puri laboratory is supported by NIH grants R01AR056712, R01AR052779, P30 AR061303, and MDA AFM and EPIGEN grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albini S, Coutinho P, Malecova B, Giordani L, Savchenko A, Forcales SV, Puri PL. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell reports. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E149–158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistelli C, Busanello A, Maione R. Functional interplay between MyoD and CTCF in regulating long-range chromatin interactions during differentiation. J Cell Sci. 2014;127:3757–3767. doi: 10.1242/jcs.149427. [DOI] [PubMed] [Google Scholar]
- Becker JS, McCarthy RL, Sidoli S, Donahue G, Kaeding KE, He Z, Lin S, Garcia BA, Zaret KS. Genomic and Proteomic Resolution of Heterochromatin and Its Restriction of Alternate Fate Genes. Molecular cell. 2017;68:1023–1037. e1015. doi: 10.1016/j.molcel.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghella L, De Angelis L, De Buysscher T, Mortazavi A, Biressi S, Forcales SV, Sirabella D, Cossu G, Wold BJ. A highly conserved molecular switch binds MSY-3 to regulate myogenin repression in postnatal muscle. Genes & development. 2008;22:2125–2138. doi: 10.1101/gad.468508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Molecular cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Molecular cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Molecular cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes & development. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio A, Palacios D. Chromatin signaling in muscle stem cells: interpreting the regenerative microenvironment. Front Aging Neurosci. 2015;7:36. doi: 10.3389/fnagi.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L’Honore A, Vallette S, Brue T, Figarella-Branger D, Meij B, Drouin J. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes & development. 2012;26:2299–2310. doi: 10.1101/gad.200436.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busanello A, Battistelli C, Carbone M, Mostocotto C, Maione R. MyoD regulates p57kip2 expression by interacting with a distant cis-element and modifying a higher order chromatin structure. Nucleic Acids Res. 2012;40:8266–8275. doi: 10.1093/nar/gks619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Hake SB. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat Rev Mol Cell Biol. 2017;18:299–314. doi: 10.1038/nrm.2016.166. [DOI] [PubMed] [Google Scholar]
- Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Caizzi L, Ferrero G, Cutrupi S, Cordero F, Ballare C, Miano V, Reineri S, Ricci L, Friard O, Testori A, et al. Genome-wide activity of unliganded estrogen receptor-alpha in breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4892–4897. doi: 10.1073/pnas.1315445111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Molecular cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Developmental cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes & development. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell stem cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarini E, Mozzetta C, Marullo F, Gregoretti F, Gargiulo A, Columbaro M, Cortesi A, Antonelli L, Di Pelino S, Squarzoni S, et al. Lamin A/C sustains PcG protein architecture, maintaining transcriptional repression at target genes. The Journal of cell biology. 2015;211:533–551. doi: 10.1083/jcb.201504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakroun I, Yang D, Girgis J, Gunasekharan A, Phenix H, Kaern M, Blais A. Genome-wide association between Six4, MyoD, and the histone demethylase Utx during myogenesis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015;29:4738–4755. doi: 10.1096/fj.15-277053. [DOI] [PubMed] [Google Scholar]
- Charney RM, Forouzmand E, Cho JS, Cheung J, Paraiso KD, Yasuoka Y, Takahashi S, Taira M, Blitz IL, Xie X, et al. Foxh1 Occupies cis-Regulatory Modules Prior to Dynamic Transcription Factor Interactions Controlling the Mesendoderm Gene Program. Developmental cell. 2017;40:595–607. e594. doi: 10.1016/j.devcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Molecular cell. 2014;53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017;168:442–459. e420. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. The EMBO journal. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. Distinct Activities of Myf5 and MyoD Indicate Separate Roles in Skeletal Muscle Lineage Specification and Differentiation. Developmental cell. 2016;36:375–385. doi: 10.1016/j.devcel.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Corrado N, Perdiguero E, Lafarga V, Munoz-Canoves P, Nebreda AR. Essential role of p18Hamlet/SRCAP-mediated histone H2A.Z chromatin incorporation in muscle differentiation. The EMBO journal. 2010;29:2014–2025. doi: 10.1038/emboj.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Morvan F, Morozzi G, Jourde B, Minetti GC, Kahle P, Rivet H, Brebbia P, Toussaint G, Glass DJ, et al. ATP Citrate Lyase Regulates Myofiber Differentiation and Increases Regeneration by Altering Histone Acetylation. Cell reports. 2017;21:3003–3011. doi: 10.1016/j.celrep.2017.11.038. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nature genetics. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes & development. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Molecular cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes & development. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Olguin P, Brand-Arzamendi K, Scott IC, Jungblut B, Stainier DY, Bruneau BG, Recillas-Targa F. CTCF promotes muscle differentiation by modulating the activity of myogenic regulatory factors. The Journal of biological chemistry. 2011;286:12483–12494. doi: 10.1074/jbc.M110.164574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orso S, Wang AH, Shih HY, Saso K, Berghella L, Gutierrez-Cruz G, Ladurner AG, O’Shea JJ, Sartorelli V, Zare H. The Histone Variant MacroH2A1.2 Is Necessary for the Activation of Muscle Enhancers and Recruitment of the Transcription Factor Pbx1. Cell reports. 2016;14:1156–1168. doi: 10.1016/j.celrep.2015.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Padova M, Caretti G, Zhao P, Hoffman EP, Sartorelli V. MyoD acetylation influences temporal patterns of skeletal muscle gene expression. The Journal of biological chemistry. 2007;282:37650–37659. doi: 10.1074/jbc.M707309200. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11593–11598. doi: 10.1073/pnas.0404192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schujiers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond CJ, Hanna JA, Garcia MR, Devine DJ, Heyrana AJ, Finkelstein D, Rehg JE, Hatley ME. Hedgehog Pathway Drives Fusion-Negative Rhabdomyosarcoma Initiated From Non-myogenic Endothelial Progenitors. Cancer Cell. 2018;33:108–124. e105. doi: 10.1016/j.ccell.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, Rudnicki MA. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes & development. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes & development. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Erdel F, Kratz K, Willcox S, Griffith JD, Greene EC, de Lange T. Telomere Recognition and Assembly Mechanism of Mammalian Shelterin. Cell reports. 2017;18:41–53. doi: 10.1016/j.celrep.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro I, Barragan M, Gubern A, Ballestar E, Joaquin M, Posas F. The p38 SAPK is recruited to chromatin via its interaction with transcription factors. The Journal of biological chemistry. 2010;285:31819–31828. doi: 10.1074/jbc.M110.155846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipescu D, Muller S, Almouzni G. Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu Rev Cell Dev Biol. 2014;30:615–646. doi: 10.1146/annurev-cellbio-100913-013311. [DOI] [PubMed] [Google Scholar]
- Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome research. 2013;23:1195–1209. doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Tapscott SJ. Skeletal muscle programming and re-programming. Curr Opin Genet Dev. 2013;23:568–573. doi: 10.1016/j.gde.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. Genetic and epigenetic determinants of neurogenesis and myogenesis. Developmental cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Johnson NM, Farr GH, 3rd, Maves L, Tapscott SJ. Conversion of MyoD to a Neurogenic Factor: Binding Site Specificity Determines Lineage. Cell reports. 2015;10:1937–1946. doi: 10.1016/j.celrep.2015.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, et al. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. The EMBO journal. 2012;31:301–316. doi: 10.1038/emboj.2011.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Developmental cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Molecular cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L, Torres-Padilla ME. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes & development. 2009;23:2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes & development. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Developmental cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes & development. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nature cell biology. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Gurdon JB, Uehlinger V. “Fertile” intestine nuclei. Nature. 1966;210:1240–1241. doi: 10.1038/2101240a0. [DOI] [PubMed] [Google Scholar]
- Harada A, Mallappa C, Okada S, Butler JT, Baker SP, Lawrence JB, Ohkawa Y, Imbalzano AN. Spatial re-organization of myogenic regulatory sequences temporally controls gene expression. Nucleic Acids Res. 2015;43:2008–2021. doi: 10.1093/nar/gkv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S, Kumamaru H, Saiwai H, Tsubota T, Kurumizaka H, et al. Chd2 interacts with H3.3 to determine myogenic cell fate. The EMBO journal. 2012;31:2994–3007. doi: 10.1038/emboj.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. 2013;100:105–113. doi: 10.1093/cvr/cvt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Molecular cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes & development. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell reports. 2016;17:2715–2723. doi: 10.1016/j.celrep.2016.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes & development. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J, Miyamoto K, Pasque V, Allen GE, Bradshaw CR, Garrett NJ, Halley-Stott RP, Kimura H, Ohsumi K, Gurdon JB. Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming. Molecular cell. 2014;55:524–536. doi: 10.1016/j.molcel.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J, Vodnala M, Pasque V, Oikawa M, Miyamoto K, Allen G, David SA, Brochard V, Wang S, Bradshaw C, et al. Gene Resistance to Transcriptional Reprogramming following Nuclear Transfer Is Directly Mediated by Multiple Chromatin-Repressive Pathways. Molecular cell. 2017;65:873–884. e878. doi: 10.1016/j.molcel.2017.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- Kitzmann M, Vandromme M, Schaeffer V, Carnac G, Labbe JC, Lamb N, Fernandez A. cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life and myogenic activity. Mol Cell Biol. 1999;19:3167–3176. doi: 10.1128/mcb.19.4.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Research. 1999;27:3752–3761. doi: 10.1093/nar/27.18.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SL, Li G, Loh SL, Sung WK, Liu ET. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol. 2011;7:526. doi: 10.1038/msb.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny SF, Emerson CP., Jr 5-Azacytidine induction of stable mesodermal stem cell lineages from 10T1/2 cells: evidence for regulatory genes controlling determination. Cell. 1984;38:791–800. doi: 10.1016/0092-8674(84)90274-5. [DOI] [PubMed] [Google Scholar]
- Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Molecular cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamolet B, Pulichino AM, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–859. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Laprell F, Finkl K, Muller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- Lassar AB. Finding MyoD and lessons learned along the way. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- Latella L, Dall’Agnese A, Boscolo FS, Nardoni C, Cosentino M, Lahm A, Sacco A, Puri PL. DNA damage signaling mediates the functional antagonism between replicative senescence and terminal muscle differentiation. Genes & development. 2017;31:648–659. doi: 10.1101/gad.293266.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. The Journal of biological chemistry. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lilja KC, Zhang N, Magli A, Gunduz V, Bowman CJ, Arpke RW, Darabi R, Kyba M, Perlingeiro R, Dynlacht BD. Pax7 remodels the chromatin landscape in skeletal muscle stem cells. PLoS One. 2017;12:e0176190. doi: 10.1371/journal.pone.0176190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell reports. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Local A, Huang H, Albuquerque CP, Singh N, Lee AY, Wang W, Wang C, Hsia JE, Shiau AK, Ge K, et al. Identification of H3K4me1-associated proteins at mammalian enhancers. Nature genetics. 2018;50:73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado L, Esteves de Lima J, Fabre O, Proux C, Legendre R, Szegedi A, Varet H, Ingerslev LR, Barres R, Relaix F, et al. In Situ Fixation Redefines Quiescence and Early Activation of Skeletal Muscle Stem Cells. Cell reports. 2017;21:1982–1993. doi: 10.1016/j.celrep.2017.10.080. [DOI] [PubMed] [Google Scholar]
- Malecova B, Dall’Agnese A, Madaro L, Gatto S, Coutinho Toto P, Albini S, Ryan T, Tora L, Puri PL. TBP/TFIID-dependent activation of MyoD target genes in skeletal muscle cells. eLife. 2016;5 doi: 10.7554/eLife.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CW. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nature genetics. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 Represses Forkhead Transcription Factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nature genetics. 2017;49:1613–1623. doi: 10.1038/ng.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, Jothi R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Molecular cell. 2014;55:708–722. doi: 10.1016/j.molcel.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Benavides T, Nasipak BT, Paskavitz AL, Haokip DT, Schnabl JM, Nickerson JA, Imbalzano AN. Casein kinase 2-mediated phosphorylation of Brahma-related gene 1 controls myoblast proliferation and contributes to SWI/SNF complex composition. The Journal of biological chemistry. 2017;292:18592–18607. doi: 10.1074/jbc.M117.799676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell stem cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Program NCS, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelet S, Rudolf F, Nadal-Ribelles M, de Nadal E, Posas F, Peter M. Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science. 2011;332:732–735. doi: 10.1126/science.1198851. [DOI] [PubMed] [Google Scholar]
- Peng XL, So KK, He L, Zhao Y, Zhou J, Li Y, Yao M, Xu B, Zhang S, Yao H, et al. MyoD- and FoxO3-mediated hotspot interaction orchestrates super-enhancer activity during myogenic differentiation. Nucleic Acids Res. 2017;45:8785–8805. doi: 10.1093/nar/gkx488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes & development. 2004;18:2348–2353. doi: 10.1101/gad.1234304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Cai J, Sussman R, Sun G, Kovermann SK, Mariani SA, Calabretta B, McMahon SB, Brock HW, Iacovitti L, et al. Delayed Accumulation of H3K27me3 on Nascent DNA Is Essential for Recruitment of Transcription Factors at Early Stages of Stem Cell Differentiation. Molecular cell. 2017;66:247–257. e245. doi: 10.1016/j.molcel.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E, Hucho F, Robin P, Harel-Bellan A. CREB-binding Protein/p300 activates MyoD by acetylation. The Journal of biological chemistry. 2000;275:34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol Cell Biol. 2001;21:5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Molecular cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO Journal. 1997a;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nature genetics. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- Puri PL, Mercola M. BAF60 A, B, and Cs of muscle determination and renewal. Genes & development. 2012;26:2673–2683. doi: 10.1101/gad.207415.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Molecular cell. 1997b;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nature structural & molecular biology. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruetz T, Pfisterer U, Di Stefano B, Ashmore J, Beniazza M, Tian TV, Kaemena DF, Tosti L, Tan W, Manning JR, et al. Constitutively Active SMAD2/3 Are Broad-Scope Potentiators of Transcription-Factor-Mediated Cellular Reprogramming. Cell stem cell. 2017;21:791–805. e799. doi: 10.1016/j.stem.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall JG, Dell’Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, et al. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell stem cell. 2015;16:171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, Osborne-Nishimura E, Mahamid J, Jahnel M, Jawerth L, et al. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell. 2016;166:1572–1584. e1516. doi: 10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini M, Sakakibara I, Gauthier M, Ribas-Aulinas F, Takahashi H, Sawasaki T, Mouly V, Concordet JP, Defossez PA, Hakim V, et al. MyoD reprogramming requires Six1 and Six4 homeoproteins: genome-wide cis-regulatory module analysis. Nucleic Acids Res. 2016;44:8621–8640. doi: 10.1093/nar/gkw512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Molecular & Cellular Biology. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Molecular cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- Schafer BW, Blakely BT, Darlington GJ, Blau HM. Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature. 1990;344:454–458. doi: 10.1038/344454a0. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Segales J, Islam AB, Kumar R, Liu QC, Sousa-Victor P, Dilworth FJ, Ballestar E, Perdiguero E, Munoz-Canoves P. Chromatin-wide and transcriptome profiling integration uncovers p38alpha MAPK as a global regulator of skeletal muscle differentiation. Skelet Muscle. 2016;6:9. doi: 10.1186/s13395-016-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome research. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, Jones DR, Du K, Jhala US, Simone C, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Molecular cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes & development. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintaku J, Peterson JM, Talbert EE, Gu JM, Ladner KJ, Williams DR, Mousavi K, Wang R, Sartorelli V, Guttridge DC. MyoD Regulates Skeletal Muscle Oxidative Metabolism Cooperatively with Alternative NF-kappaB. Cell reports. 2016;17:514–526. doi: 10.1016/j.celrep.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Cavero S, Begley V, Sole C, Bottcher R, Chavez S, Posas F, de Nadal E. Regulation of transcription elongation in response to osmostress. PLoS genetics. 2017;13:e1007090. doi: 10.1371/journal.pgen.1007090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nature genetics. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, Faralli H, Choi J, Youn HD, Dilworth FJ, et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes & development. 2015;29:513–525. doi: 10.1101/gad.254532.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerjanc IS, Slack RS, McBurney MW. Cellular aggregation enhances MyoD-directed skeletal myogenesis in embryonal carcinoma cells. Mol Cell Biol. 1994;14:8451–8459. doi: 10.1128/mcb.14.12.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 2016;165:593–605. doi: 10.1016/j.cell.2016.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y. Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature. 2017 doi: 10.1038/nature24300. [DOI] [PubMed] [Google Scholar]
- Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. The Journal of biological chemistry. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Tenente IM, Hayes MN, Ignatius MS, McCarthy K, Yohe M, Sindiri S, Gryder B, Oliveira ML, Ramakrishnan A, Tang Q, et al. Myogenic regulatory transcription factors regulate growth in rhabdomyosarcoma. eLife. 2017;6 doi: 10.7554/eLife.19214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto PC, Puri PL, Albini S. SWI/SNF-directed stem cell lineage specification: dynamic composition regulates specific stages of skeletal myogenesis. Cell Mol Life Sci. 2016;73:3887–3896. doi: 10.1007/s00018-016-2273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy A, Cadwallader AB, Fedorov Y, Tyner K, Tanaka KK, Olwin BB. Coordination of satellite cell activation and self-renewal by Par-complex-dependent asymmetric activation of p38alpha/beta MAPK. Cell stem cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S, Ibrahim MM, Kakumanu A, Garipler G, Aydin B, Al-Sayegh MA, Hirsekorn A, Abdul-Rahman F, Satija R, Ohler U, et al. A Multi-step Transcriptional and Chromatin State Cascade Underlies Motor Neuron Programming from Embryonic Stem Cells. Cell stem cell. 2017;20:205–217. e208. doi: 10.1016/j.stem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, Greenberg ME. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Molecular cell. 2017;68:1067–1082. e1012. doi: 10.1016/j.molcel.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez-Cruz G, Sartorelli V. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. The EMBO journal. 2013;32:1075–1086. doi: 10.1038/emboj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & development. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nature genetics. 2017;49:289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 2017 doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE. Induction of muscle genes in neural cells. The Journal of cell biology. 1984;98:427–435. doi: 10.1083/jcb.98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. The EMBO journal. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:85–90. doi: 10.1073/pnas.1009830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. Journal of Biological Chemistry. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Developmental cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? The Journal of cell biology. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. The Journal of biological chemistry. 1999;274:5193–5200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]