Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a deadly disease and a major health problem in the United States. While the cytokine TGF-β has been implicated in PDAC development, it can exert both pro- and anti-tumorigenic effects that are highly context dependent and incompletely understood. Using three-dimensional (3D) cultures of KrasG12D-expressing mouse pancreatic epithelial cells we demonstrated that while exposure to exogenous TGF-β induced growth arrest of the KrasG12D cells, its subsequent removal allowed the cells to enter a hyper-proliferative, partially mesenchymal (PM) and progenitor-like state. This state was highly stable and was maintained by autocrine TGF-β signaling. While untreated KrasG12D cells formed cystic lesions in vivo, PM cells formed ductal structures resembling human PanINs, suggesting that they had attained increased oncogenic potential. Supporting this hypothesis, we determined that the PM cells share salient molecular and phenotypic features with the quasi-mesenchymal/squamous subtype of human PDAC, which has the worst prognosis of any of the recently identified subtypes. Transient pulses of TGF-β have been observed during pancreatitis, a major risk factor for PDAC. Our data suggest that transient TGF-β exposure is sufficient to induce the acquisition of stable PDAC-associated phenotypes in pre-neoplastic KrasG12D cells, providing novel molecular insight into the complex role of TGF-β in tumorigenesis.
Keywords: pancreatic cancer, TGF-β, partially mesenchymal, de-differentiation
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) has the worst prognosis of any human cancer with an all-stage 5-year survival of 8% (1). PDAC develops from precursor lesions called pancreatic intra-epithelial neoplasias (PanINs) that progress from low grade (PanIN-1 and 2) to high grade (PanIN-3) before becoming fully invasive PDAC (2). This histologically-defined progression is paralleled by the accumulation of genetic mutations that confer distinct transforming properties to the neoplastic cells. Early activating mutations in the proto-oncogene KRAS are followed by loss-of-function mutations in one or more tumor suppressors including CDKN2A, TP53, and SMAD4. Concurrent with progression of PanINs to PDAC is replacement of the surrounding pancreatic parenchyma with a desmoplastic tumor stroma consisting of a heterogeneous collection of cells, extracellular matrix proteins, and soluble factors thought to promote malignant progression.
One of the principal soluble factors implicated in pancreatic tumorigenesis is transforming growth factor β (TGF-β). TGF-β regulates a number of diverse processes including proliferation, pluripotency and differentiation (3). It induces cellular responses by binding to receptor serine/threonine kinases, leading to the activation of both canonical (SMAD4-dependent) and non-canonical (SMAD4-independent) signaling pathways. TGF-β levels are elevated in developing and established pancreatic tumors due to its secretion by both neoplastic (4, 5) and stromal (6) cells. Approximately 50% of PDACs harbor loss-of-function mutations in TGF-β signaling components, most commonly SMAD4, consistent with TGF-β having a tumor suppressive effect (7). This is supported by data from mouse models in which loss of Smad4 accelerates PDAC progression driven by mutant Kras (8–10). In these models, TGF-β suppresses PDAC development by inhibiting cell growth and inducing apoptosis. However, 50% of human PDACs harbor intact TGF-β signaling components, indicating that in many tumors the suppressive function of this signaling axis can be bypassed. Furthermore, SMAD4 loss occurs during the advanced stages of human PDAC development (11), suggesting that the tumor suppressive mechanisms of TGF-β signaling have a relatively small impact on the early stages of disease development. TGF-β has also been shown to induce pancreatic cancer cells to undergo epithelial-to-mesencymal transition (EMT)(12), a process that can promote invasion and metastasis (13). While heterozygous loss of Smad4 in Kras-driven mouse models indeed suppresses EMT and metastasis, homozygous loss paradoxically leads to increased metastasis (10). Collectively, these observations suggest that the phenotypic output of TGF-β signaling varies in a context and dose-dependent manner during PDAC development and progression.
In the present study, we exploited a three-dimensional (3D) culture system to characterize the responses of pancreatic epithelial cells harboring mutant Kras to TGF-β exposure. We demonstrated that transient exposure of KrasG12D cells to TGF-β was sufficient to induce their transition to a highly proliferative state that was associated with the acquisition of mesenchymal and progenitor-like properties. This phenotype was highly stable and was maintained by autocrine TGF-β signaling. Moreover, markers of this phenotype are expressed by human PDAC subtypes associated with more aggressive disease. These data uncover a previously unappreciated role of TGF-β signaling dynamics in PDAC development.
RESULTS AND DISCUSSION
Transient TGF-β exposure alters cellular architecture and increases proliferation of KrasG12D cells
To investigate the response of pre-neoplastic pancreas cells to TGF-β exposure, LSL-KrasG12D;p48-Cre mice (KC)(14) were used in which KrasG12D is expressed within all pancreatic epithelial cells by p48 promoter-driven Cre recombinase-mediated excision of the LSL stop cassette. In KC mice, lesions resembling human PanINs develop with the potential to progress to PDAC. Pancreas explants were prepared from four-week-old KC mice (i.e., prior to PanIN formation) and subsequently used to generate a 3D cell culture by embedding the cells in Matrigel. Under these conditions, the KrasG12D cells (hereafter referred to as KC cells) formed hollow spheres that could be passaged weekly for at least one year (Figure 1A), in agreement with previous reports (15, 16). Analysis of the cultured cells for the expression of specific linage markers showed high levels of expression of the epithelial marker Krt8 (CK8) and negligible levels of immune, mesenchymal and islet cell markers (Figure S1A) confirming their epithelial identity.
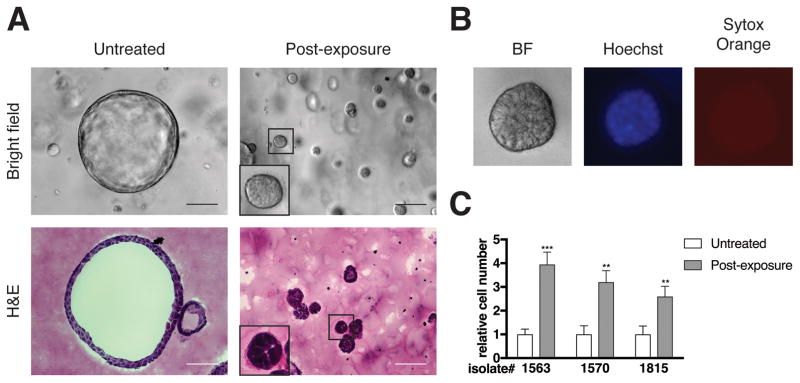
Figure 1. Transient TGF-β exposure alters the architecture and proliferation rate of KC cells.
Pancreas explants were harvested from LSL-KrasG12D;p48-Cre (KC, ref. 14) mice between 3.5 and 4.5 weeks of age as described previously (35), except that the cells were plated on Matrigel (354234, Corning; Corning, NY, USA). Following three days in culture at 37°C in 5% CO2 in air, the resulting ductal structures were removed from the Matrigel using Dispase (50 U/mL for 15 minutes at 37°C; 354235, Corning), trypsinized (0.05%, 2 × 3 minutes at 37°C, Gibco, Gaithersburg, MD, USA), and then embedded in Matrigel as described previously (16). Cells were treated without (untreated) or immediately with (post-exposure) human recombinant TGF-β1 (500 pg/mL; 240-B-002, R&D systems; Minneapolis, MN, USA) added to the culture medium. After two days, the TGF-β was removed and the cells were propagated for four weeks, after which experiments were performed. Media was replaced every two days. (A) Bright field images of live cells and hematoxylin and eosin (H&E; HHS32, HT110332 Sigma) stained sections are shown. Scale bars, 100 μm. (B) Bright field and fluorescent live images of TGF-β-treated cells stained with Hoechst 33342 (H1399, ThermoFisher; Waltham, MA, USA) and SYTOX Orange (S11368, ThermoFisher) according to the manufacturer’s protocols are shown. For (A) and (B), bright field and fluorescence images were obtained using a Zeiss Axiovert 200M microscope. Cells were prepared for cryosectioning (36) followed by H&E staining (16) as described previously with de-paraffinization and dehydration steps omitted, and imaged using a Nikon Eclipse 80i microscope. All images were processed using ImageJ and Adobe Photoshop software. Images are representative of five independent isolates. (C) Relative cell number of three independent isolates as determined by manual cell counting using a hemocytometer is shown. Error bars indicate mean +/− SD from independent experiments (n = 4–5). P values determined using a Student’s t test (unpaired, two tailed). **, P < 0.001. ***, p<0.0001. All animal care and procedures were approved by the Institutional Animal Care and Use Committee at NYU School of Medicine.
To assess the effect of TGF-β on pre-neoplastic pancreas cells, KC cells were treated with recombinant TGF-β1 (hereafter referred to as TGF-β) and characterized morphologically. Within two days of exposure to TGF-β, the KC cells adopted a filled-sphere architecture. To test the reversibility of this response, the TGF-β-containing medium was replaced with normal growth medium (hereafter, transient TGF-β). The TGF-β-induced filled-sphere phenotype was maintained in subsequent passages even in the absence of exogenously added TGF-β (Figures 1A and 1B). These structures were composed exclusively of viable cells (Figure 1B). Filled-sphere architecture in 3D cultures is associated with de-differentiated or stem-like phenotypes (17). For example, whereas PanIN and primary PDAC cells form hollow spheres in 3D cultures, metastatic PDAC cells, which have stem-like properties (18), form filled spheres (15).
The KC cells also growth arrested prior to removal of exogenous TGF-β (data not shown) consistent with its well documented growth-suppressing effect (3). However, following removal of TGF-β, the cells regained a proliferative capacity that was significantly greater than baseline (Figure 1C). This post-exposure hyper-proliferative filled sphere-forming phenotype was maintained for at least 6 months following TGF-β removal. These results indicate that despite the transient nature of the exposure of KC cells to TGF-β, the transition that the cells undergo leads to a stable phenotype. By contrast, continuous TGF-β exposure resulted in filled sphere formation and stable growth arrest (Figures S1B and S1C), suggesting that transient TGF-β exposure is unique in its ability to induce a hyper-proliferative phenotype.
Transient TGF-β exposure causes KC cells to undergo a partial EMT and acquire progenitor-like properties
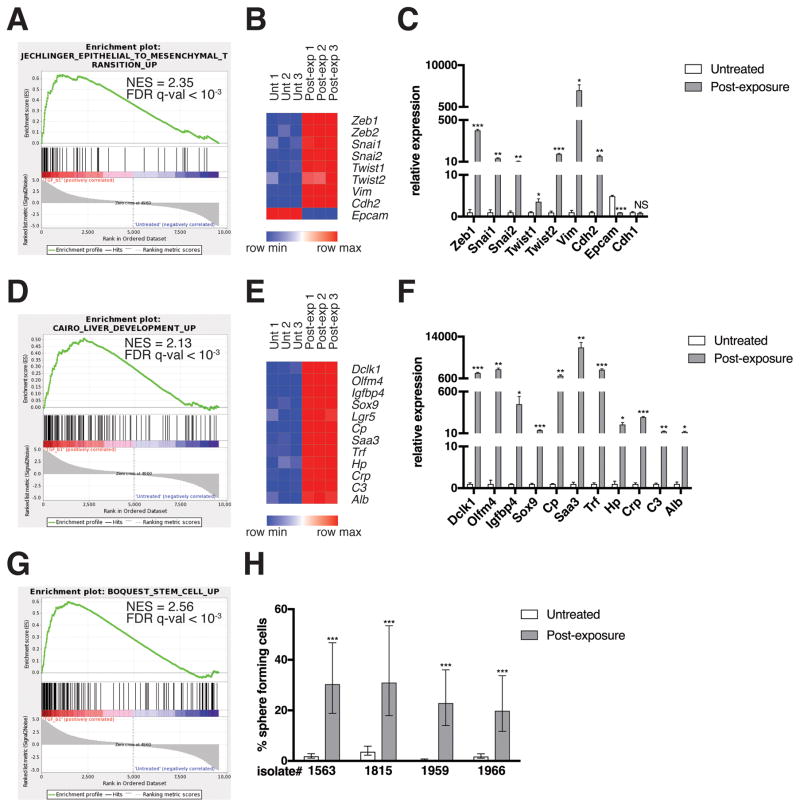
To understand the molecular changes associated with the filled sphere transition, the transcriptional profiles of untreated and transiently-exposed KC cells were assessed by RNA sequencing. Gene set enrichment analysis (GSEA) was performed using curated gene sets and oncogenic signatures in the Molecular Signatures Database (MSigDB)(19). Transient TGF-β exposure induced a number of gene sets related to EMT (Figure 2A) that included transcription factors (Zeb1, Zeb2, Snai1, Snai2, Twist1, Twist2), intermediate filaments (Vim), and cell adhesion molecules (Cdh2, Epcam) (Figure 2B). A subset of the most differentially expressed genes was validated by qRT-PCR (Figure 2C) and protein-based assays (Figures S2A and S2B). Notably, E-cadherin levels were unchanged in response to transient TGF-β treatment (Figures 2C and S2A) and TGF-β-treated cells displayed the normal junctional localization of E-cadherin (Figure S2C). As E-cadherin loss or mislocalization is the cardinal hallmark of EMT (13), we concluded that transient TGF-β exposure induced a partial EMT phenotype, which we will henceforth refer to as partially mesenchymal (PM).
Figure 2. EMT and de-differentiation signature genes are enriched in post-TGF-β-exposure KC cells.
(A, D, G) GSEA enrichment plots of the indicated gene sets amongst the genes upregulated in post-TGF-β exposure KC cells relative to untreated KC cells are shown. NES, normalized enrichment score. RNA was isolated using an RNeasy mini kit (74104, QIAGEN; Venlo, Netherlands) from cells generated from LSL-KrasG12D;Pdx1-Cre;LSL-YFP (KCY)(18) mice, which behaved identically to KC cells. Three technical replicates per experimental group from one isolate were analyzed by RNA sequencing. Genomic DNA was removed using an RNase-free DNase kit (79254, QIAGEN). RNA sequencing was performed using an Illumina HiSeq2500 instrument. To generate differential expression values, sequencing results were demultiplexed and converted to FASTQ format using Illumina Bcl2FastQ software. Paired-end reads were aligned to the mouse genome (build mm10/ GRCm38) using the splice-aware STAR aligner (37). PCR duplicates were removed using the Picard toolkit (38). The HTSeq package (39) was utilized to generate counts for each gene based on how many aligned reads overlap its exons. These counts were then normalized and used to test for differential expression using negative binomial generalized linear models implemented by the DESeq2 R package (40). P values were corrected for multiple hypothesis testing using the Benjamini-Hochberg method (41). Results were considered significant when p < 0.05. This data is publicly available via the Gene Expression Omnibus (42), accession #GSE101659. The Gene Set Enrichment Analysis (GSEA) software (19) was run using the curated gene sets and oncogenic signatures from the Molecular Signatures Database (MSigDB). (B, E) Heat maps of representative EMT and de-differentiation signature genes are shown. Expression levels shown are representative of log2(x + 1) transformed normalized read counts in each replicate. Expression relative to the maximum (red) and minimum (blue) level per gene across samples is shown. Heat maps were generated using GENE-E software (43). (C, F) mRNA levels of differentially expressed genes as measured by qRT-PCR are shown. Reverse transcription was performed using a QuantiTect reverse transcription kit (205311, QIAGEN). USB HotStart-IT SYBR Green qPCR Master Mix (75762, Affymetrix; Santa Clara, CA, USA) was used for amplification. See Table S1 for a list of primers used in this study. Samples were amplified on the Stratagene Mx 3005P. Differences in expression were determined using the 2−ΔΔCT method (44) using RPS29 as a housekeeping control. Expression levels from one isolate representative of the four individual isolates tested are shown. Three technical replicates were used per experimental group per isolate. Error bars indicate mean +/− SD from three technical replicates. P values determined using a Student’s t test (unpaired, two tailed). *, P < 0.05. **, P < 0.001. ***, P < 0.0001. NS, not significant. (H) Quantification of sphere formation capacity is shown. One sphere formation assay was performed per isolate for the four individual isolates tested. Cells were plated at densities ranging from 2000 cells/well-0.25 cells/well, 8 replicates per dilution, under conditions described previously to enrich for mammary stem cells (17). Following a 7-day incubation, the presence or absence of spheres was ascertained using bright field microscopy. Data is represented as the estimated percentage of cells able to form spheres, as calculated using ELDA statistical software (45), with error bars indicating the upper and lower limits of a 95% confidence interval. P values for differences between groups were generated by the software. ***, P < 0.0001.
While the completion of EMT is thought to be a critical step in tumor evolution and a prerequisite for invasion and metastasis, emerging data suggest that partial EMT may also contribute to tumor development. Forty-five percent of human PDACs contain vimentin-expressing tumor epithelial cells (20). Furthermore, transcriptomic analysis of human PDAC samples has identified a quasi-mesenchymal subtype with distinct biological properties and clinical behavior (21). In the mammary system, partial EMT has been associated with acquisition of stem-like properties (17). Interestingly, the PM cells that evolved following transient exposure to TGF-β were found to express gene sets associated with liver development (Figure 2D). A number of the differentially expressed liver genes were validated by qRT-PCR (Cp, Saa3, Trf, Hp, Crp, C3, Alb) (Figure 2F). As the liver is derived from foregut endoderm (22), which also gives rise to the pancreas, these data suggest that the PM cells have attained a progenitor-like state. Supporting this hypothesis, markers of several other foregut endoderm lineages, including intestinal stem cells (Olfm4, Igfbp4, Sox9, Lgr5) and intestinal tuft cells (Dclk1), were similarly upregulated in PM cells (Figures 2E and 2F). In addition, the PM cells expressed gene sets associated with stem cell phenotypes (Figure 2G) and had much higher sphere-forming potential than untreated KC cells in a sphere formation assay (Figure 2H), a well-established method to select for functional stem cells (17, 23), confirming that they had acquired stem-like properties.
It is noteworthy that pancreatic progenitor markers Mnx1, Nkx2-2, Gata6, Hnf1b, and Pdx1 were downregulated in PM cells relative to untreated KC cells in the RNA sequencing dataset (Figure S2D). Furthermore, the SHH and Notch pathways did not appear markedly activated in the PM cells based on expression of downstream target genes (data not shown). Given that foregut endoderm development precedes pancreatic progenitor cell formation, these data are in line with the notion that the PM cells have acquired a pre-pancreatic progenitor-like state. These data reveal a previously unappreciated capacity for pancreas cells to activate genetic programs specific to multiple foregut endoderm lineages and introduce an additional level of context-dependent pancreas cell plasticity.
Markers of the PM state are expressed in human PDAC and are associated with aggressive disease
As some of the markers associated with the PM state, e.g., Olfm4, Cp, are among the most upregulated genes in human PDAC (24) and Olfm4 has been associated with worse prognosis (25), we sought to further investigate the potential functional relevance of the PM state to human PDAC. Recent tumor sequencing studies have described molecularly and clinically distinct PDAC subtypes (21, 26). We found the quasi-mesenchymal/squamous subtype of particular interest as this group of PDAC tumors is partly defined by expression of a gene program enriched for TGF-β and WNT signaling components and also features suppression of pancreatic progenitor genes by hyper-methylation. This subtype, like the PM cells described in this study, displays both epithelial and mesenchymal features. Notably, tumor subtypes with quasi-mesenchymal features are associated with the most aggressive disease in PDAC (21) and other organ systems (17, 27). By GSEA, the TGF-β and WNT signaling module from the quasi-mesenchymal/squamous subtype (GP3 as described in ref. 26) was significantly enriched in PM versus untreated KC cells (Figure S3A). In addition, a six-gene panel specific to a mouse model of quasi-mesenchymal/squamous PDAC (26) was upregulated in the PM cells relative to untreated KC cells (Figure S3B). These observations suggest that PM cells may represent a precursor population for the quasi-mesenchymal/squamous subtype of PDAC, though further studies are required to confirm this prediction.
The PM state is maintained by autocrine TGF-β signaling
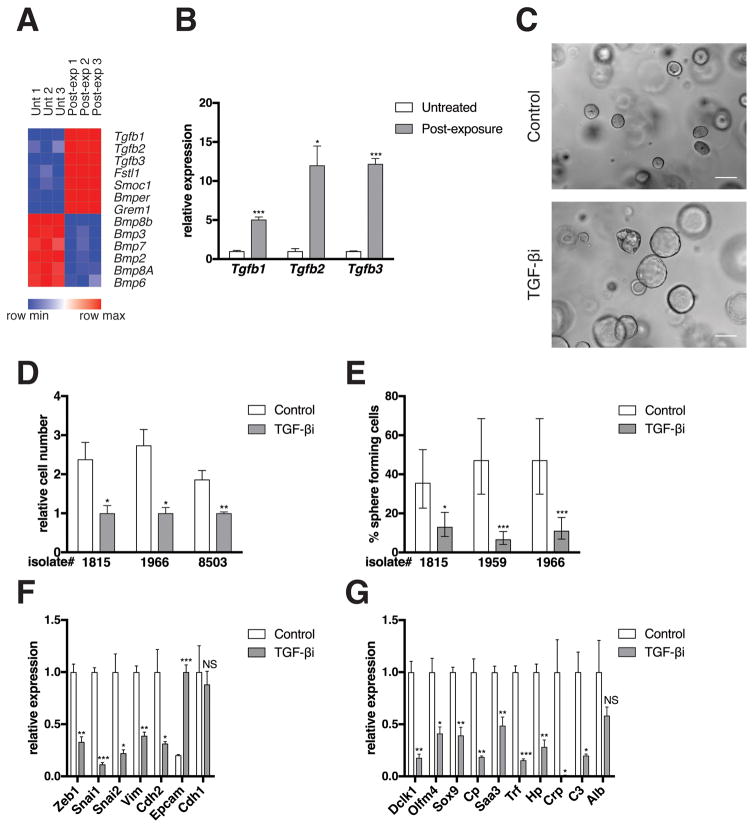
An intriguing aspect of the PM state is its persistence following removal of exogenous TGF-β. Given the documented capacity of TGF-β signaling to be maintained via an autocrine mechanism (32), we tested whether this could account for the sustained PM state. Indeed, all three isoforms of TGF-β were upregulated in PM cells (Figures 3A and 3B). In addition, PM cells expressed higher levels of BMP antagonists (e.g., Smoc1) and lower levels of BMPs (Figure 3A). As BMP signaling is antagonistic to TGF-β signaling (28), this trend is consistent with TGF-β pathway activation. Furthermore, while TGF-βR2 levels were stable or modestly suppressed in PM cells, TGF-βR1 was significantly upregulated in multiple isolates tested (Figure S4A) as were TGF-β downstream targets Skil and Smad7 (Figure S4B). To test the requirement of secreted TGF-β for maintaining the PM state, PM cells were treated with the highly selective TGF-βR1 kinase inhibitor, LY364947 (29). LY36947 treatment effectively suppressed SMAD activation and the expression of canonical TGF-β target genes in PM cells (Figures S4C and S4D). TGF-βR1 inhibition reversed the changes in cellular architecture (Figure 3C), proliferation (Figure 3D), sphere forming efficiency (Figure 3E), and expression of EMT and de-differentiation genes (Figures 3F and 3G) induced by transient TGF-β exposure. Together, these data demonstrate that autocrine TGF-βR1-dependent signaling is required to maintain the PM state.
Figure 3. Autocrine TGF-β signaling maintains the PM phenotype.
(A) Heat map of TGF-β and BMP genes is shown. See Figure 2B for methods. (B) mRNA levels of differentially expressed genes as measured by qRT-PCR is shown. See Figure 2C for methods. (C) Bright field images are shown from a single isolate representative of three independent isolates tested. PM cells were either not treated (Control) or treated with the TGF-βR1 kinase inhibitor LY364947 (1 μg/mL; 616451, EMD Millipore; Mahopac, NY, USA) for six days (TGF-βi). Scale bars, 100 μm. See Figure 1A for image capture and processing methods. (D) Relative cell number of three independent isolates as determined by manual cell counting using a hemocytometer is shown (Cells from isolate 8503 were generated from a KCY mouse [see Figure 2A]). PM cells were treated with DMSO (Control) or LY364947 (TGF-βi) for six days. Error bars indicate mean +/− SD from three technical replicates. P values determined using a Student’s t test (unpaired, two tailed). *, P < 0.05. **, P < 0.001. (E) Quantification of sphere formation capacity is shown. PM cells were treated with DMSO (Control) or LY364947 (TGF-βi) for six days. One sphere formation assay was performed per isolate for the three isolates tested. See Figure 2H for methods. Data represented as described in Figure 2H. *, P < 0.05. ***, P < 0.0001. (F–G) mRNA levels of EMT (F) and de-differentiation signature (G) genes as measured by qRT-PCR is shown. PM cells were either not treated (Control) or exposed to LY364947 (TGF-βi) for six days. Expression levels from one isolate representative of the three individual isolates tested are shown. See Figure 2C for methods.
As exposure of treatment-naïve KC cells or PM cells to exogenous TGF-β1 resulted in growth arrest (data not shown), it was surprising that the hyper-proliferative phenotype of the PM cells was maintained by autocrine TGF-β signaling. Given the dose-dependent nature of TGF-β responses, differences in TGF-β levels between exogenous and autocrine conditions could explain this apparent contradiction. Furthermore, despite sharing a single receptor, TGF-β isoforms have discrete biological functions. For example, TGF-β2 elicits the upregulation of integrin β3 in mammary stem cells required for expansion of this cell population during pregnancy, whereas TGF-β1 causes this population to growth arrest (30). While all three TGF-β isoforms were able to induce the PM state (Figure S4E), it is unclear which isoform(s) are responsible for maintaining it as all three isoforms were upregulated in the PM cells and the TGF-βR1 inhibitor blocks the activity of all three. It is also possible that the PM state represents the combinatorial autocrine activity of all three isoforms.
PM cells form ductal structures resembling PanINs in vivo
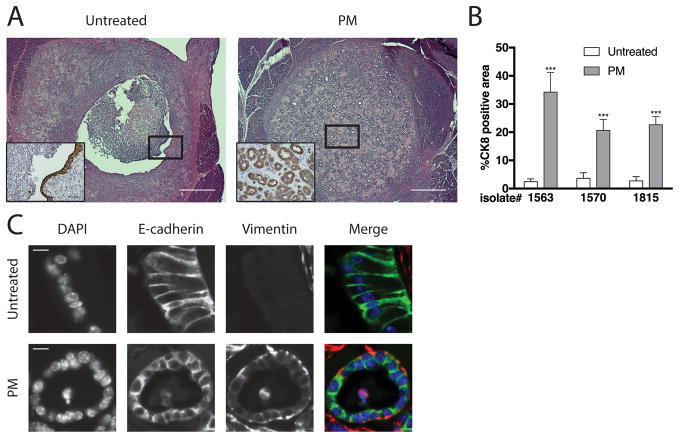
To investigate the functional consequences of the transition to the PM state in vivo, untreated KC or PM cells were orthotopically transplanted into the pancreata of syngeneic C57Bl/6 mice. A higher percentage of Ki67+ epithelial cells was observed in lesions derived from PM cells relative to lesions derived from untreated KC cells at one week (Figure S5), consistent with the proliferative advantage of PM cells in vitro. Indeed, two weeks following injection, PM lesions harbored significantly more epithelial cells than untreated lesions (Figures 4A and 4B). In addition, the untreated cells formed cyst-like structures lined by a single layer of epithelial cells while the PM cells formed ductal structures resembling early PanINs (Figure 4A). Furthermore, the transplanted PM cells expressed both E-cadherin and vimentin (Figure 4C), providing evidence that the PM state was maintained in vivo. These data imply that acquisition of the PM state may contribute to an increase in the oncogenic potential of KC cells.
Figure 4. PM cells form ductal lesions resembling PanINs in vivo.
Untreated or PM cells were implanted into pancreata of WT syngeneic C57/Bl6 mice (027, Charles River Laboratories, Wilmington, MA, USA) as described previously (16). Two weeks after implantation, the mice were sacrificed and the pancreata harvested. Tissue processing, sectioning, histology, immunohistochemistry, immunostaining, and image acquisition, processing, and analysis were performed as described previously (16). (A) Sections stained with H&E or immunostained for CK8 (insets) are shown. Scale bars, 500 μm or 50μm (insets). Images shown are representative of three independent experiments performed with separate isolates (n = 4–6 mice per group). Sample size was based on previously published studies (16). No randomization or blinding was performed. CK8 antibody (1:200, TROMA-Ic, DSHB, University of Iowa; Iowa City, IA, USA). (B) Quantification of %CK8 positive area within the lesion as determined using ImageJ software is shown. Error bars indicate mean +/− SD. P values determined using a Student’s t test (unpaired, two tailed). ***, P < 0.0001. (C) Representative sections immunostained for E-cadherin and vimentin are shown. Antibodies used were E-cadherin (1:200; 610181, BD, Franklin Lakes, NJ, USA). Vimentin (1:200; 5741, Cell Signaling Technology; Danvers, MA, USA). Scale bars, 10μm.
Conclusions
Activation of KRAS is thought to be required for PanIN formation, as nearly all PanIN-1 lesions in humans harbor oncogenic KRAS mutations (2). However, pancreatic epithelial cells in adult mice are resistant to transformation by oncogenic Kras, suggesting that oncogenic KRAS is not sufficient for PanIN formation but requires cooperation with other factors, the best characterized of which is pancreatitis (31). Pancreatitis, a fibroinflammatory condition of the pancreas, is a significant risk factor for the development of PDAC (32, 33). While mouse studies have established that pancreatitis cooperates with oncogenic Kras to drive PanIN formation (31), the mechanisms underlying this synergism are unclear.
TGF-β levels transiently rise following induction of pancreatitis in rats (34). Similarly, using publicly available microarray data, we found that all three TGF-β isoforms are transiently elevated following induction of pancreatitis in mice (Figure S6). We propose that the pulses of TGF-β observed during pancreatitis may serve to convert oncogenic KRAS-expressing cells into a partially mesenchymal population that can drive PanIN initiation and progression. Transient TGF-β exposure could therefore serve as a mechanism through which the oncogenic potential of mutant KRAS-expressing pancreatic cells is unmasked.
Supplementary Material
Acknowledgments
The authors thank L.J Taylor for discussions and help with article preparation and the members of the Bar-Sagi laboratory for comments. The authors also thank the New York University School of Medicine Genome Technology Center (GTC), especially Adriana Heguy and Igor Dolgalev, for help designing, performing, and analyzing RNA sequencing experiments. The GTC is partially supported by the Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center. This work was further supported by NIH/NCI grant CA210263 (D. Bar-Sagi). J. Handler was supported by NIH grant T32GM007308. J. Cullis was supported by NIH grants 5-T32 CA 009161-39 and 5-T32AI100853-04. E.A. Vucic is supported by a Canadian Institutes of Health Research postdoctoral fellowship (146792).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
Supplementary Information is available at Oncogene’s website.
References
- 1.American Cancer Society. Cancer Facts & Figures 2017. Atlanta: American Cancer Society; 2017. [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–49. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, et al. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105:1846–56. doi: 10.1016/0016-5085(93)91084-u. [DOI] [PubMed] [Google Scholar]
- 5.Hezel AF, Deshpande V, Zimmerman SM, Contino G, Alagesan B, O’Dell MR, et al. TGF-beta and alphavbeta6 integrin act in a common pathway to suppress pancreatic cancer progression. Cancer Res. 2012;72:4840–5. doi: 10.1158/0008-5472.CAN-12-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Principe DR, DeCant B, Mascarinas E, Wayne EA, Diaz AM, Akagi N, et al. TGFbeta Signaling in the Pancreatic Tumor Microenvironment Promotes Fibrosis and Immune Evasion to Facilitate Tumorigenesis. Cancer Res. 2016;76:2525–39. doi: 10.1158/0008-5472.CAN-15-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–43. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell. 2015;161:1345–60. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–6. [PubMed] [Google Scholar]
- 12.Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, Ruhland C, et al. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–8. [PubMed] [Google Scholar]
- 13.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 15.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–47. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handra-Luca A, Hong SM, Walter K, Wolfgang C, Hruban R, Goggins M. Tumour epithelial vimentin expression and outcome of pancreatic ductal adenocarcinomas. Br J Cancer. 2011;104:1296–302. doi: 10.1038/bjc.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadler TW. Langman’s medical embryology. 13. Wolters Kluwer; Philadelphia, USA: 2015. [Google Scholar]
- 23.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–10. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 24.Goonesekere NC, Wang X, Ludwig L, Guda C. A meta analysis of pancreatic microarray datasets yields new targets as cancer genes and biomarkers. PLoS One. 2014;9:e93046. doi: 10.1371/journal.pone.0093046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takadate T, Onogawa T, Fukuda T, Motoi F, Suzuki T, Fujii K, et al. Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int J Cancer. 2013;132:1368–82. doi: 10.1002/ijc.27797. [DOI] [PubMed] [Google Scholar]
- 26.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 27.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, et al. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016 doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–40. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawyer JS, Anderson BD, Beight DW, Campbell RM, Jones ML, Herron DK, et al. Synthesis and activity of new aryl- and heteroaryl-substituted pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain. J Med Chem. 2003;46:3953–6. doi: 10.1021/jm0205705. [DOI] [PubMed] [Google Scholar]
- 30.Desgrosellier JS, Lesperance J, Seguin L, Gozo M, Kato S, Franovic A, et al. Integrin alphavbeta3 drives slug activation and stemness in the pregnant and neoplastic mammary gland. Dev Cell. 2014;30:295–308. doi: 10.1016/j.devcel.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernandez-Porras I, Canamero M, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–39. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, et al. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med. 1993;328:1433–7. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 33.Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, et al. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849–52. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riesle E, Friess H, Zhao L, Wagner M, Uhl W, Baczako K, et al. Increased expression of transforming growth factor beta s after acute oedematous pancreatitis in rats suggests a role in pancreatic repair. Gut. 1997;40:73–9. doi: 10.1136/gut.40.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Means AL, Meszoely IM, Suzuki K, Miyamoto Y, Rustgi AK, Coffey RJ, Jr, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132:3767–76. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 36.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–40. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.http://broadinstitute.github.io/picard
- 39.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 42.https://www.ncbi.nlm.nih.gov/geo/
- 43.https://software.broadinstitute.org/GENE-E/
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Cullis J, Siolas D, Avanzi A, Barui S, Maitra A, Bar-Sagi D. Macropinocytosis of Nab-paclitaxel Drives Macrophage Activation in Pancreatic Cancer. Cancer Immunol Res. 2017;5:182–90. doi: 10.1158/2326-6066.CIR-16-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.