Abstract
As a dioxygenase, Ten-Eleven Translocation 2 (TET2) catalyzes subsequent steps of 5-methylcytosine (5mC) oxidation. TET2 plays a critical role in the self-renewal, proliferation, and differentiation of hematopoietic stem cells, but its impact on mature hematopoietic cells is not well-characterized. Here we show that Tet2 plays an essential role in osteoclastogenesis. Deletion of Tet2 impairs the differentiation of osteoclast precursor cells (macrophages) and their maturation into bone-resorbing osteoclasts in vitro. Furthermore, Tet2−/− mice exhibit mild osteopetrosis, accompanied by decreased number of osteoclasts in vivo. Tet2 loss in macrophages results in the altered expression of a set of genes implicated in osteoclast differentiation, such as Cebpa, Mafb, and Nfkbiz. Tet2 deletion also leads to a genome-wide alteration in the level of 5-hydroxymethylcytosine (5hmC) and altered expression of a specific subset of macrophage genes associated with osteoclast differentiation. Furthermore, Tet2 interacts with Runx1 and negatively modulates its transcriptional activity. Our studies demonstrate a novel molecular mechanism controlling osteoclast differentiation and function by Tet2, that is, through interactions with Runx1 and the maintenance of genomic 5hmC. Targeting Tet2 and its pathway could be a potential therapeutic strategy for the prevention and treatment of abnormal bone mass caused by the deregulation of osteoclast activities.
Keywords: Tet2, 5hmC, Macrophage, Osteoclast, Runx1
Introduction
Ten-Eleven Translocation 2 (TET2) is one of the most frequently mutated genes in adult myeloid malignancies, including myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN), chronic myelomonocytic leukemia (CMML) , and acute myeloid leukemia (AML) [1], [2], [3], [4]. TET2 is also found to be mutated in T–cell lymphoma (such as angioimmunoblastic T lymphomas) [5] and B-cell non-Hodgkin lymphomas (such as diffuse large B-cell lymphoma and mantle cell lymphoma) [6], [7]. Notably, TET2 mutations are prevalent in healthy elderly individuals aged >70 years (∼5%) and are associated with clonal hematopoiesis [8], [9], [10]. Tet2 deficiency in mice results in increased hematopoietic stem cell (HSC) self-renewal [11], [12], [13], [14]. Furthermore, Tet2 loss skews the differentiation of hematopoietic stem/progenitor cells (HSPCs) towards granulocytic/monocytic lineage, often resulting in monocytosis and accumulation of monocytes/macrophages in bone marrow (BM) and spleen of mice [11], [12], [13], [14]. Fusion of the monocyte-macrophages lead to the formation of multinucleated osteoclasts, the primary bone-resorbing cells in mammals [15]. Therefore, it would be inherently important to examine the impact of Tet2 loss on osteoclastogenesis and bone mass.
DNA methylation mediated by Dnmt3a, a de novo methyltransferase, plays a role in osteoclastogenesis [16]. Tet2 catalyzes the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) [17], [18], [19], and conceivably, Tet2 loss or loss-of-function mutations result in aberrant 5mC and 5hmC profiling of DNA [11], [20], raising the possibility that Tet2 may be involved in the epigenetic regulation of osteoclastogenesis. In this study, we investigated the role of Tet2 in osteoclastogenesis using previously established Tet2 knock-out (KO) mouse models. Our findings demonstrate that Tet2-deficient monocyte-macrophages do not differentiate efficiently into mature bone-resorbing osteoclasts. Additionally, Tet2−/− mice exhibit increased bone mass, likely due to fewer osteoclasts present in vivo. RNA-seq analysis and genome-wide profiling of 5hmC on wild type (WT) and Tet2−/− macrophages reveal that Tet2 loss leads to significant alterations in gene expression (such as Cebpa, Nfkbiz, Mafb, and Id2) and 5hmC profiling, with specific enrichment for genes related to osteoclast differentiation. Furthermore, Tet2 physically interacts with Runx1, and negatively modulates its transcriptional activity. This study reveals the critical role of Tet2 in osteoclast differentiation and function, implicating Tet2 in the regulation of bone remodeling. Thus, Tet2 is expected to be a potential therapeutic target in bone metabolic disorders with altered osteoclast activity such as osteopetrosis and osteoporosis.
Results
Deletion of Tet2 impairs osteoclast differentiation and function
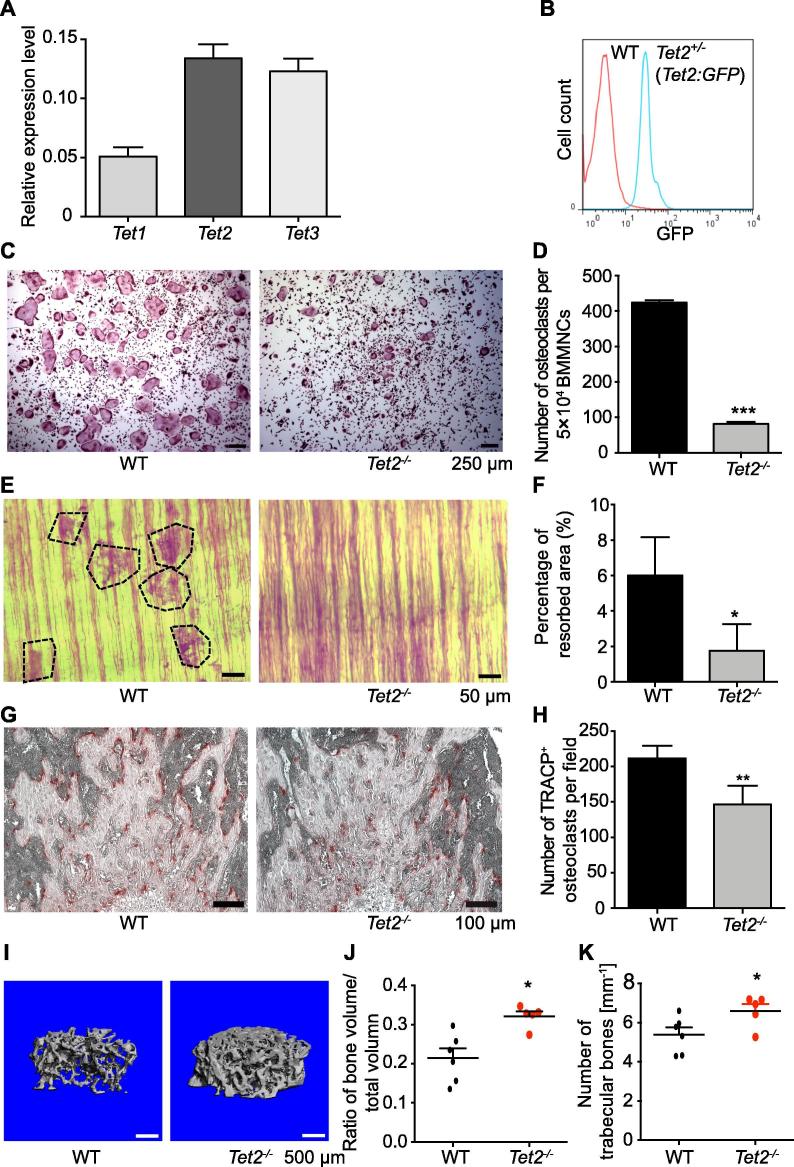
We first examined the mRNA expression levels of the genes encoding three Tet family proteins by qPCR in bone marrow derived macrophages in mice, and found that the expression level of both Tet2 and Tet3 was higher than that of Tet1 (Figure 1A), suggesting a potential role of Tet2 in osteoclastogenesis. Flow cytometric analyses on bone marrow derived macrophages (CD11b+) from heterozygous Tet2:nGFP knock-in mice [11] revealed that all macrophages expressed high level of GFP reporter, which correlates with Tet2 protein level (Figure 1B). We then examined the effects of Tet2 loss on osteoclast differentiation in vitro after the stimulation of macrophages with receptor activator for nuclear factor κ B ligand (RANKL) and macrophage colony-stimulating factor (M-CSF) and counted the tartrate-resistant acid phosphatase (TRACP) positive multinucleated osteoclasts [21]. Surprisingly, the Tet2−/− macrophage cultures contained significantly lower number of TRACP+ multinucleated osteoclasts compared to WT macrophage cultures (P < 0.001, Student’s t test) (Figure 1C and D). Most strikingly, there were few large-sized multinucleated osteoclasts in the Tet2−/− macrophage cultures (Figure 1C). To examine the resorptive activity by their osteoclast progenies, WT and Tet2−/− macrophages were cultured on whale dentin slides. It was found that osteoclasts formed in Tet2−/− macrophage cultures resorbed significantly less dentin surface compared to WT macrophage cultures (P < 0.05, Student’s t test) (Figure 1E and F). These data indicate that Tet2 plays a critical role in osteoclast differentiation and function in vitro.
Figure 1.
Deletion of Tet2 impairs osteoclast differentiation and function
A. Quantitative RT-PCR analysis of Tet1, Tet2, and Tet3 expression (relative to Actb) in the bone marrow-derived osteoclast precursor cells (macrophages, CD11b+ gated) of WT mice. B. Flow cytometric analysis for the GFP (Tet2) expression levels in bone marrow derived macrophages from a representative 8-week-old heterozygous Tet2:GFP knock-in mouse. C. Representative photomicrographs showing TRACP-staining of bone marrow-derived osteoclasts cultured in the presence of M-CSF and RANKL in WT and Tet2−/− mice, respectively. Scale bars, 250 μm. D. Quantitation of the number of TRACP+ osteoclasts derived from 5 × 104 BMMNCs. Data are presented as mean ± SEM, n = 5. E. Osteoclast cultures on dentine slices. Representative photomicrographs showing resorptive “pits” generated by osteoclast bone lytic activity. Scale bars, 50 μm. F. Quantitation of the area of the resorbed regions, referred to as ‘pits’. Data are presented as mean ± SEM, n = 3. G. Histological analysis using TRACP stain of the femur from 12-week-old WT and Tet2−/− male mice. Representative photographs (100×) of the trabecular bone following TRACP staining are shown. The red stained area indicates TRACP+ osteoclasts. Scale bars, 100 μm. H. Quantitation of the number of TRACP+ osteoclasts. Data are presented as mean ± SEM, n = 3. I. μCT analysis of the femur of 12-week-old WT and Tet2−/− male mice, respectively. Representative μCT imaging demonstrating the reconstructed 3D microstructure of femoral trabecular bones is shown. The cortical portion of each femur was removed to allow visualization of the metaphyseal architecture. J. Quantitation of the femoral bone volume/total volume assessed using μCT. K. Quantitation of the trabecular number assessed using μCT. *P < 0.05, **P < 0.01, ***P < 0.001. Student’s t test. TRACP, tartrate-resistant acid phosphatase; μCT, microcomputed tomography.
These results prompted us to further investigate whether Tet2 is involved in the differentiation and function of osteoclasts in vivo using the Tet2 KO mouse models established in our laboratories [11], [14], [22]. We examined the long bone tissues of WT and Tet2−/− mice. Examination of the histological sections of the femur of Tet2−/− mice after TRACP staining revealed significantly reduced numbers of TRACP+ osteoclasts in trabecular bone tissues compared to age-matched WT mice (P < 0.01, Student’s t test) (Figure 1G and H). 3D micro-computed tomography (μCT) imaging showed that the femoral trabecular bone pattern of Tet2−/− mice appeared denser than that of WT mice (Figure 1I). Quantification of the 3D μCT data revealed that Tet2−/− mice had mild osteopetrosis with significantly increased femoral trabecular bone mass and trabecular number compared to those of WT mice (P < 0.05, Student’s t test) (Figure 1J and K). These results collectively indicate that Tet2 loss causes higher bone mass in mice, likely due to the impairment in osteoclast differentiation in vivo.
Deletion of Tet2 alters gene expression profiling in macrophages
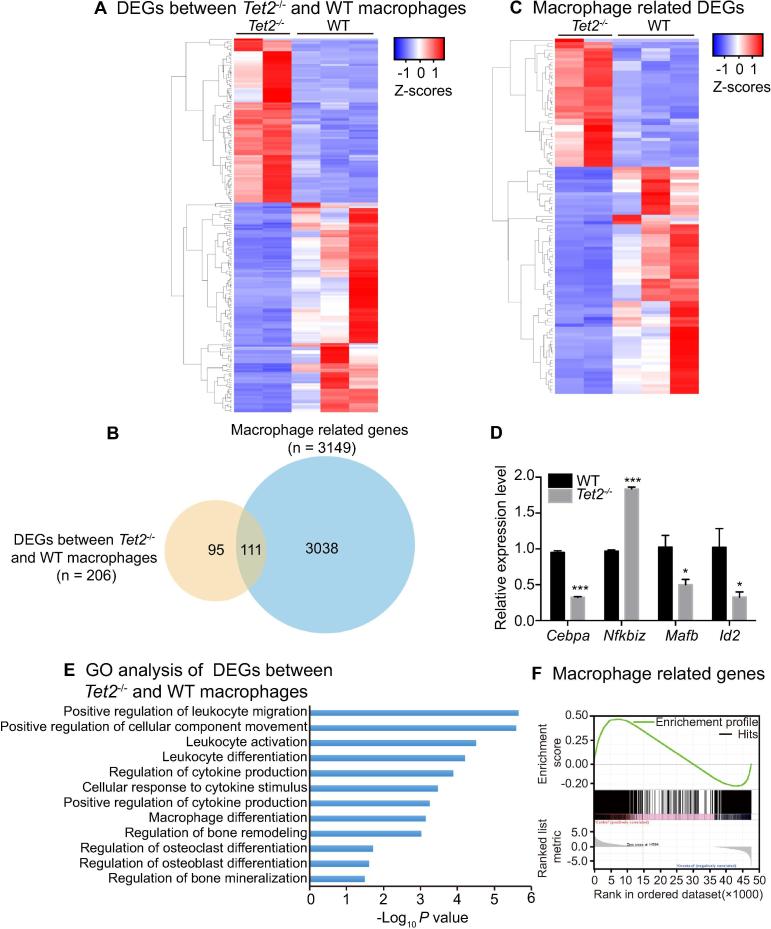
To explore the molecular mechanisms underlying the impaired osteoclast differentiation, mediated by Tet2 loss, RNA-seq was performed on macrophages derived from the bone marrow of WT and Tet2−/− mice. Analysis of the RNA-seq data revealed 206 differentially-expressed genes (DEGs) in macrophages from Tet2−/− mice in comparison to WT controls (Figure 2A, fold change (FC) ≥ 1.5 and FDR < 0.05; Table S1). Among these DEGs, 111 genes are related to macrophage development and/or differentiation according to the published gene list (3149 murine macrophage related genes [23]) (Figure 2B and C; Table S2). Interestingly, expression of Cebpa, which encodes CCAAT/enhancer-binding protein α, a critical transcription factor for osteoclast differentiation, is significantly down-regulated, while expression of Nfkbiz, which encodes nuclear factor kappa-B (NFκB) inhibitor zeta, another critical transcription factor for osteoclast differentiation, is up-regulated. These changes were further validated by quantitative RT-PCR (qPCR) (Figure 2D). Functional enrichment analysis using Ingenuity pathway analysis (IPA) indicated that the DEGs were enriched for macrophage differentiation and regulation of bone remodeling (Figure 2E). Consistently, the gene set enrichment analysis (GSEA) showed that the expression of macrophage-related genes was decreased in the Tet2−/− macrophages compared to WT macrophages (Figure 2F). These results are consistent with our observation that Tet2 loss impairs osteoclast differentiation and function.
Figure 2.
Deletion of Tet2 alters gene expression profiling in macrophages
A. Heatmap showing the relative abundance of DEGs between WT and Tet2−/− macrophages by RNA-seq analysis (FDR < 0.05 across all three algorithms, CuffDiff, edgeR, and DESeq2). Z-scores per transcript are shown and blue represents lower expression and red represents higher expression. B. Venn diagram showing the overlap between the macrophage related genes and DEGs between Tet2−/− and WT macrophages. C. Heatmap showing the relative abundance of the 111 overlapping genes in panel B using RNA-seq analysis (FDR < 0.05 across all three algorithms, CuffDiff, edgeR, and DESeq2). D. Quantitative RT-PCR analysis of phenotypic associated DEGs (relative to Actb) in Tet2−/−vs. WT macrophages that are important for osteoclast differentiation. Data are presented as mean ± SEM, n = 3. *P < 0.05, ***P < 0.001. Student’s t test. E. Ingenuity Pathway Analysis (IPA) of gene ontology biological processes enriched in pathways that are involved in hematopoietic cell differentiation. F. GSEA analysis plot shows decreased gene expression of the macrophage related genes in macrophages from Tet2−/− mice compared to those from WT mice (NES = 1.4399285 and FDR < 1×10−4). DEG, differentially-expressed gene; GSEA, gene set enrichment analysis; NES, nominal enrichment score; FDR, false discovery rate.
Tet2−/− macrophages present distinct DNA hydroxymethylation patterns
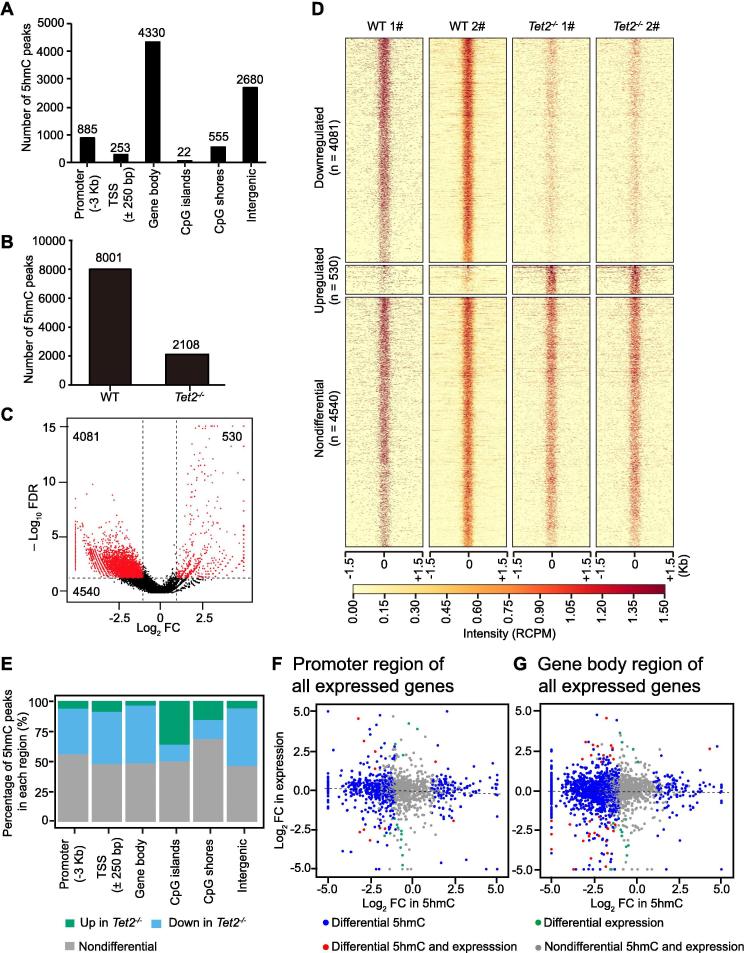
Considering of the enzymatic activity of Tet2 in converting 5mC into 5hmC, we performed 5hmC selective chemical labeling (hMe-Seal) high-throughput sequencing [24] to profile the whole-genome distribution of 5hmC in the macrophages from WT and Tet2−/− mice that we used for transcriptomic analyses (Table S3). Most of 5hmC modifications in the WT macrophages are located at gene body and intergenic regions (Figure 3A). Consistent with previous observations in mouse bone marrow Lin−c-Kit+ cells and embryonic stem cells [14], [25], Tet2 deletion led to a decrease in the number of 5hmC peaks in macrophages (Figure 3B). Tet2 deletion results in a significant reduction (downregulated) in 5hmC levels at 4081 genomic sites and a significant gain of 5hmC levels (upregulated) at 530 sites, while 5hmC levels at 4540 sites remained unchanged (nondifferential) (Figure 3C and D). The down-regulated 5hmC peaks upon Tet2 loss have a higher chance of being located in gene body, intergenic and transcriptional start site (TSS) regions, while the gained 5hmC peaks were more likely located at CpG islands/shores (Figure 3E). Linear correlation analysis of FC in 5hmC enrichment against the transcriptional changes showed that there appears to be no linear correlation between the changes in transcription and the changes in 5hmC in either the promoter or gene body regions (Figure 3F and G). However, when a chi-squared test was performed to examine the distribution of differential hydroxymethylated regions (DhMRs) in either the promoter or gene body regions of 206 DEG regions, it was found that DEGs contain significantly fewer 5hmC peaks in the gene body than nondifferential genes (P < 0.05 for upregulated expressed genes and P < 0.001 for downregulated genes, chi-squared test; Table S4). This observation indicates that a change in 5hmC levels may be associated with a change in gene expression levels, while the correlations could be either positive or negative.
Figure 3.
Tet2−/− macrophages display distinct DNA hydroxymethylation signatures
A. Distribution of 5hmC-enriched regions with genomic features in WT macrophages. B. Bar plot of total 5hmC peaks detected in WT and Tet2−/− macrophages, respectively. C. Volcano plot of the whole-genome 5hmC profile. Red dots represent DhMRs with a log2 FC > 1 or < −1 and an FDR < 0.05, whereas black dots represent peaks of significant enrichment with no difference between macrophages from WT and Tet2−/− mice. D. Heatmap showing enrichment in RCPM in the 5hmC peaks that are downregulated, upregulated, or nondifferential in macrophages from Tet2−/− mice in comparison to WT mice. E. Bar charts showing the changed proportion of peaks in major genomic regions. F. FC of expression of all expressed genes between Tet2−/− and WT plotted against FC of normalized 5hmC read counts in promoter regions (TSS ± 3 kb) between Tet2−/− and WT. G. FC of expression of all expressed genes between Tet2−/− and WT plotted against FC of normalized 5hmC read counts in gene body regions between Tet2−/− and WT. DhmR, differential hydroxymethylated region; FC, fold change; RCPM, read counts per million; TSS, transcriptional start site.
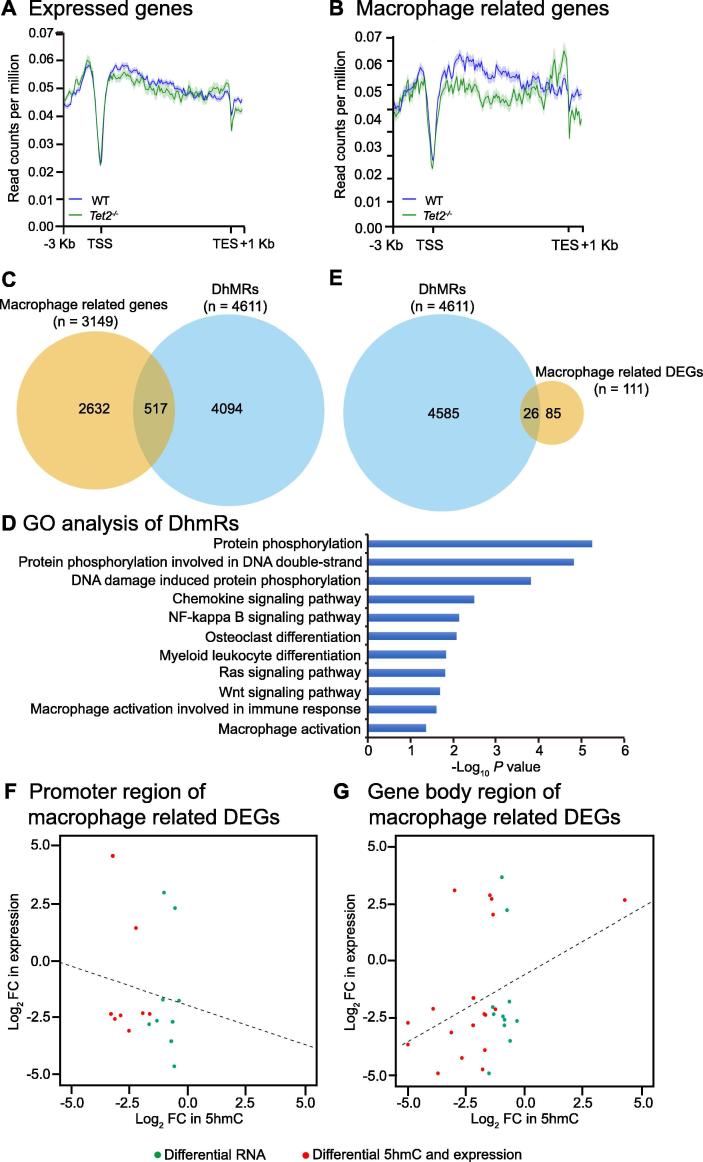
Tet2 deletion only led to a slight decrease of the 5hmC levels on all expressed genes in macrophages (Figure 4A), a greater degree of 5hmC loss in the gene body and a tendency of 5hmC gain at the transcriptional end sites (TES) were observed for the macrophage related genes (Figure 4B). Indeed, 517 of the 3149 macrophage related genes are marked with DhMRs (Figure 4C). Functional enrichment and pathway analysis showed that DhMRs were enriched in genes related to NFκB signaling pathway and osteoclast differentiation (Figure 4D). Integration of DhMRs and macrophage related DEGs from Tet2−/− in comparison to WT mice revealed that 26 of the 111 macrophage related DEGs are marked with DhMRs (Figure 4D). However, we did not observe a significant linear correlation between DhMRs and expression changes of these 111 macrophage related DEGs, regardless of the DhMR genomic locations at either promoter or gene body regions (Figure 4F and G). These observations suggest a role of 5hmC in the marking of specific genes, which might enable cells to alter gene expression in response to additional stimuli.
Figure 4.
Correlation of DhMRs with gene expression in Tet2−/− vs. WT macrophages
A. Distributions of average 5hmC enrichment in all expressed genes (>1 RCPM) in WT and Tet2−/− macrophages. B. Distributions of average 5hmC enrichment in macrophage related genes in WT and Tet2−/− macrophages. C. Venn diagram showing the overlap between macrophage related genes and DhMRs identified between Tet2−/− and WT macrophages. 517 out of 3149 macrophage related genes contain DhMRs in either their promoter or gene body genomic regions. D. Gene ontology analysis of genes with differential 5hmC peaks between WT and Tet2−/− macrophages. E. Venn diagram showing the overlap between the macrophage related DEGs and DhMRs identified between Tet2−/− and WT macrophages. 26 out of 111 macrophage related DEGs contain DhMRs in either their promoter or gene body regions. F. and G. FC of expression of the macrophage related DEGs between Tet2−/− and WT plotted against FC of normalized 5hmC read counts in promoter regions (TSS ± 3 kb) (F) and gene body region (G) between Tet2−/− and WT. TSS, transcriptional start site; TES, transcriptional end site; DhmR, differential hydroxymethylated region; GO, gene ontology; DEG, differentially expressed gene; FC, fold change.
Tet2 associates with Runx1 to regulate its transcriptional activity
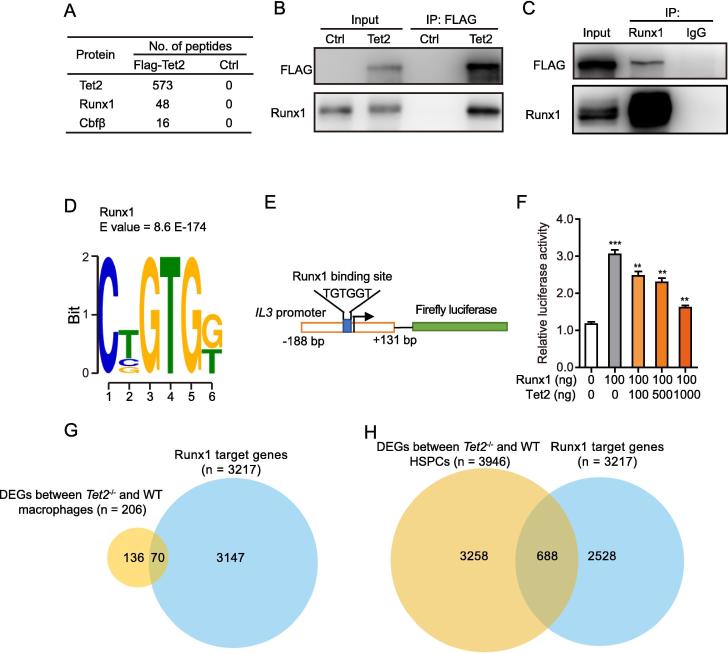
Tet2 deletion leads to expression changes of a specific set of genes in macrophages. Located in the nucleus and act as a DNA-modifying enzyme, Tet2 interacts with numerous nuclear proteins to exert its function [26]. It is therefore possible that Tet2 interacts with specific transcription factors to modulate their transcriptional activities, thus Tet2 loss may alter the expression of common target genes for Tet2 and its interactors. To test this hypothesis, we performed anti-FLAG affinity purifications for MEL cells stably expressing FLAG-Tet2 (∼4 fold of endogenous Tet2 expression [27]), to survey potential Tet2 binding partners. LC-MS/MS analysis revealed about 66 of high-confidence candidate Tet2-associating proteins, including some known Tet2-interactors such as O-linked N-acetylglucosamine transferase (Ogt) and non-POU-domain-containing, octamer binding protein (Nono) [28], [29], [30], [31] and several unknown candidates such as topoisomerase (DNA) II alpha (Top2α), YY1 transcription factor (Yy1), Runt-related transcription factor 1 (Runx1), and core binding factor beta (Cbfβ) (Figure 5A and Table S5). Among them, Runx1, which forms a heterodimeric complex with Cbfβ, is a transcription factor that plays a critical role in regulating HSPC proliferation and differentiation [32], as well as osteoclastogenesis [33]. We, therefore, focused on Runx1 in this study. The interaction of Tet2 with Runx1 was confirmed by reciprocal co-immunoprecipitation using HEK293T cells overexpressing FLAG-Tet2 (Figure 5B and C, and Figure S1). We then performed de novo motif enrichment analysis using our recently published genome-wide data for Tet2 binding sites identified in MEL cells overexpressing FLAG-tagged Tet2 [27]. Such comparative analysis could potentially confirm and even identify the regulatory partners/competitors of Tet2. Interestingly, we found that Runx1-binding DNA motifs are relatively enriched in the FLAG-Tet2 ChIP-seq peak regions (Figure 5D and Table S6), providing additional evidence for their interactions with Tet2.
Figure 5.
Tet2 associates with Runx1
A. Proteins identified by mass spectrometry from MEL cells stably expressing FLAG-Tet2 after purification of Tet2-associated proteins. Spectral counts for each interacting protein are shown. B. and C. Reciprocal co-immunoprecipitations and Western blots confirming the interaction between Tet2 and Runx1 in HEK293T cells overexpressing FLAG-tagged Tet2. Flag (B) or Runx1 immunoprecipitation was performed for nuclear lysates of HEK293T cells overexpressing FLAG-vector (Ctrl) or FLAG-Tet2 (Tet2) using (C) before immunoblotting using the indicated antibodies. The isotype IgG was used as negative control in panel C. D. Motif enrichment analysis identifying the significant enrichment of Runx1 DNA binding motifs in the Tet2-binding sites. Tet2 ChIP-seq peaks were submitted to the MEME suite. E. Diagram of IL3-promoter-luciferase construct showing the Runx1 binding consensus site. F. HEK293T cells cotransfected with IL3-promoter luciferase reporter plasmid, 100 ng Runx1 and increasing concentrations of Tet2 expression plasmids. Renilla luciferase plasmid was used as an internal transfection control, and FC of reporter activity induced by Runx1/Tet2 expression relative to that of the Renilla luciferase control were plotted. Data are presented as mean ± SEM, n = 3. **P < 0.01, ***P < 0.001. G. Venn diagram showing the overlap between Runx1 target genes with DEGs between Tet2−/− and WT macrophages. H. Venn diagram showing the overlap between the Runx1 target genes with DEGs between Tet2−/− and WT HSPCs (lin−c-Kit+ gated). FC, fold change; HSPC, hematopoietic stem/progenitor cell.
Given the physical association between Tet2 and Runx1, we wondered whether Tet2 is capable of modulating the transcriptional activity of Runx1. To answer this question, we carried out luciferase reporter assays using an IL-3 promoter construct containing a Runx1 consensus binding site (Figure 5E). Transfection of Runx1 into HEK293T cells increased the luciferase activity by two-fold over the baseline level (Figure 5F), demonstrating that Runx1 mediates robust transcription activity. Interestingly, expression of Tet2 significantly reduced Runx1-mediated transcriptional activity in a dosage-dependent manner (Figure 5F), suggesting that Tet2 negatively regulates Runx1 transcriptional activity. In addition, by analyzing the RNA-seq data on osteoclast precursor and lin−c-Kit+ (LK, hematopoietic stem cells enriched group) cells from WT and Tet2−/− mice, we identified a significant portion of DEGs in macrophages (70 out of 206 genes, 34%) and LK cells (688 out of 3946, 17.5%) to be the Runx1 target genes (Figure 5G and H, Tables S7 and S9).
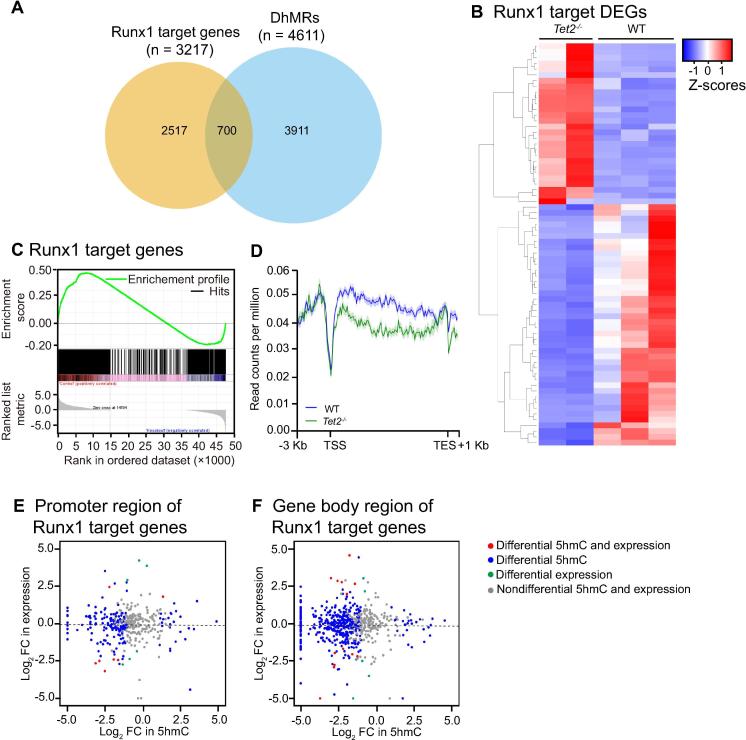
To further examine whether Tet2 modulates the expression of Runx1 target genes by its enzymatic activity, we then overlapped the DhMRs with Runx1 target genes [34] in macrophages, and found that a significant portion of the Runx1 target genes (700 out of 3217 genes; 22%) were marked by DhMRs (Figure 6A and Table S9). Among these 700 Runx1 target genes marked by DhMRs, 70 genes were differentially expressed in macrophages (Figure 6B). Additionally, GSEA confirmed a decrease in Runx1 target genes in Tet2−/− cells in comparison to WT cells (Figure 6C). Furthermore, a greater degree of 5hmC loss was observed in the gene body regions for Runx1 target genes than for all expressed genes (Figure 4A, Figure 6D). Besides Runx1 target genes, Ep300 target genes [35], [36] and Pu.1 target genes [34] were also decreased in Tet2−/− cells as shown by GSEA (Figure S2A and B), and the 5hmC in the gene body regions of these target genes were also decreased compared to all expressed genes (Figure 4A, Figure S2C and D). Similar to the correlation performed in macrophage related DEGs, in the 3217 genes known to be targeted by Runx1, no linear correlations were found between changes in gene expression and changes in 5hmC (promoter or gene body regions; Figure 6E and F). These data collectively indicate that Tet2 physically interacts with Runx1, regulating Runx1 transcriptional activity and expression of Runx1/Tet2 common target genes.
Figure 6.
Enrichment of Runx1 target genes with DhMRs
A. Venn diagram showing the overlap between Runx1 target genes with DhMRs identified between Tet2−/− and WT macrophages. 700 out of 3217 Runx1 target genes overlap with DhMRs. B. Heatmap showing the relative abundance of Runx1 target DEGs (70 out of 206 DEGs) between WT and Tet2−/− macrophages by RNA-seq analysis (FDR < 0.05 across all three algorithms CuffDiff, edgeR, and DESeq2). Z-scores per transcript are shown with blue representing lower expression and red representing higher expression in Tet2−/−. C. GSEA analysis plot showing decreased gene expression of the Runx1 signature in Tet2−/− macrophages compared with WT macrophages (NES = 1.411 and FDR < 1 × 10−4). D. Distributions of average 5hmC enrichment at Runx1 target genes in WT and Tet2−/− macrophages. E. and F. FC of expression of the Runx1 target genes between Tet2−/− and WT plotted against FC of normalized 5hmC read counts in promoter regions (TSS ± 3 kb) (E) and gene body regions (F) between Tet2−/− and WT. DhmR, differential hydroxymethylated region; DEG, differentially expressed gene; GSEA, gene set enrichment analysis; NES, nominal enrichment score; FDR, false discovery rate.
Discussion
Osteoclasts are primary cells for bone resorption, and their differentiation is tightly regulated. Recent studies have uncovered that epigenetic regulatory mechanisms, such as DNA methylation [16], play a critical role in determining osteoclast differentiation. However, little is known about the regulation of osteoclastogenesis by TET family demethylation enzymes. In this study, we use Tet2 knock-out mouse models to illustrate the importance of Tet2 and Tet2-mediated 5hmC maintenance in osteoclast differentiation and function in vitro and in vivo. Deletion of Tet2 impairs osteoclast differentiation from their monocyte/macrophage precursors in vitro in the presence of M-CSF and RANKL. More importantly, Tet2−/− mice have mild osteopetrosis with significantly increased bone mass, suggesting a functional defect of osteoclasts in vivo.
Loss-of-function mutations in TET2 are common in both lymphoid and myeloid malignancies [1], [2], [3], [4], [5], [6], [13]. TET2 mutations are also detected in elderly individuals with clonal hematopoiesis [8], [9], [10]. Both Tet2−/− mice and patients with TET2 mutations often have increased number of monocyte/macrophage osteoclast precursors, due to a skewed differentiation of HSPCs to monocytic lineage. Despite an increased population of precursor cells, the number of osteoclasts is significantly decreased in Tet2−/− mice, likely due to the impairment of osteoclast differentiation. Given the high incidence of TET2 mutations in patients with myeloid malignancies and elderly individuals, it would be of importance to monitor the bone mass in these populations harboring TET2 mutations, and also to screen for potential presence of TET2 mutations in individuals with osteopetrosis.
Tet2 loss in macrophages dysregulates a set of genes implicated in osteoclast differentiation, including Cebpa and Nfkbiz. NFκB is one of the most important transcription factors for osteoclast differentiation regulated by RANKL. Inactivation of NFκB pathway by interruption of either inhibitor of NFκB kinase subunit α (IKKα) or β (IKKβ) results in the disruption of osteoclast differentiation [37]. It has been shown that Cebpa−/− mice develop a severe osteopetrotic phenotype because of defective osteoclastogenesis [38]. Therefore, the down-regulation of Cebpa and up-regulation of Nfkbiz (through inhibition of NFκB activity) in Tet2−/− macrophages may contribute to the impaired osteoclast differentiation and the osteopetrotic bone phenotype in Tet2-deficient mice. In addition, the genes differentially expressed in macrophages upon Tet2 loss are enriched with Runx1 target genes. Our proteomic studies identifies Runx1 as a binding partner of Tet2. Moreover, luciferase reporter assays show that Tet2 negatively modulates Runx1 transcriptional activity. It has been shown that the deletion of Runx1 in adult hematopoietic stem cells in adult mice produces a myeloproliferative phenotype [32] and that Runx1 loss accelerates osteoclastogenesis [33]. In addition, during the preparation of our manuscript, a study reported a physical interaction of RUNX1 and TET2, and a colocalization of RUNX1 and TET2 in the genomic regions in a human T-lymphocyte line [39]. It is possible that Tet2 loss activates Runx1 transcriptional activity, contributing to the impaired osteoclast differentiation.
The genomic distribution of 5mC demethylation is regulated in a complicated manner. 5hmC profiling on Tet2−/− vs. WT macrophages reveals that Tet2 deletion leads to a genome-wide alteration in the number and intensity of 5hmC peaks. Interestingly, DhMRs are enriched in genes implicated in osteoclast differentiation, suggesting a role of 5hmC deregulation in the impaired osteoclast differentiation observed in Tet2−/− macrophages. Despite DhMR marking in specific genes in Tet2−/− macrophages, no overall linear correlation between alterations in 5hmC levels and gene expression changes in macrophages is observed. This is consistent with previous findings in mouse LK cells and ESCs [14], [40], [41], [42], [43]. Recent studies have revealed that Tet2 can exert its function through both catalytic activity-dependent and -independent fashions [29], [44], [45]. Recently, we have demonstrated that the catalytic activity of Tet2 is essential for its myeloid tumor suppressive role in HSPCs [46]. However, it remains to be elucidated whether the Tet2 catalytic activity is critical for osteoclastogenesis. In addition, the distinction between the catalytic activity-dependent and -independent roles of Tet2 in gene transcription regulation needs to be investigated. Given that 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) are further oxidation products of 5hmC and excised by thymine DNA glycosylase (TDG)/base excision repair (BER) [47], [48], [49], it might be important to determine whether alterations in 5fC and 5caC levels in macrophages of Tet2−/− mice are correlated with gene expression changes. In addition, base resolution analysis of 5mC, 5hmC, 5fC, and 5caC in WT and Tet2−/− macrophages would provide more detailed information of Tet2-mediated genome-wide DNA demethylation in osteoclastogenesis and reveal the correlation between specific cytosine modification alterations and gene expression changes caused by Tet2 loss.
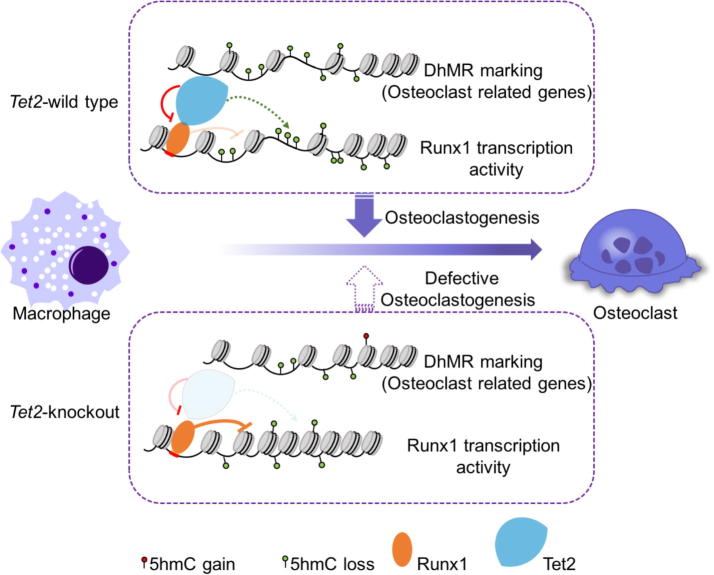
In summary, our results reveal a novel and critical role of Tet2 in osteoclastogenesis and provide significant insight into its underlying mechanisms. We propose a mechanism of Tet2 involvement in osteoclast differentiation through (1) interaction with Runx1 to modulate its transcriptional activity and regulate expression of genes critical for osteoclastogenesis; (2) maintenance of genomic 5hmC levels; and (3) regulation of important transcription factors for osteoclast differentiation such as Cebpa and Nfkbiz (Figure 7). Since Tet2 loss impairs osteoclast differentiation in vitro and Tet2-deficient mice exhibit increased bone mass, TET2 may be a potential target for therapeutic approaches in bone disorders associated with abnormal osteoclast function. Our findings have potential implications for these individuals with TET2 mutations, indicating a necessity to screen for alterations in bone mass.
Figure 7.
Model proposed for mechanisms underlying the control of osteoclast differentiation and function by Tet2 in macrophages
Materials and methods
Analyses of mice
Tet2:GFP and Tet2−/− mice were generated as we reported previously [11], [14], [22]. Animal maintenance and care was performed in accordance with the institutional guidelines and was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Miami Miller School of Medicine. WT control mice were obtained from the same colony as knockout mice.
Constructs and transduction
The codon-optimized cDNA sequence of mouse WT Tet2 (Tet2opt) was synthesized by SynBio Corp (Monmouth Junction, NJ) and ligated into pCDF1-IRES-GFP lentiviral vector. MEL cells were transduced with lentivirus encoding GFP and Tet2opt/GFP as described previously [27]. IL3-promoter luciferase construct and pCMV-Runx1 plasmid were gifts from Dr. Stephen Nimer of the University of Miami Miller School of Medicine. The DNA sequence of Tet2opt can be provided upon request.
Cell culture and tartrate-resistant acid phosphatase staining
HEK293T cells were grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. MEL cells were maintained in RPMI1640 supplemented with l-glutamine, 10% FBS, and 1% penicillin–streptomycin. Murine macrophages and osteoclasts were obtained from in vitro culture of bone marrow mononuclear cells (BMMNCs) as described previously [21]. To generate macrophages, BMMNCs were cultured in alpha minimum essential medium (α-MEM) supplemented with 10% FBS and macrophage colony-stimulating factor (M-CSF; 20 ng/ml, PeproTech, Rocky Hill, NJ) for 3 days. These cells were used as bone marrow derived macrophages. To generate osteoclasts, BMMNCs were cultured in α-MEM supplemented with 10% FBS, 20 ng/ml RANKL, and 20 ng/ml M-CSF for 2 days; the cytokines were changed to 30 ng/ml M-CSF and 60 ng/ml RANKL on day 3 and cells were cultured for additional 3 days. For tartrate-resistant acid phosphatase (TRACP) staining, adherent cells or femur sections were fixed and stained using the acid phosphatase kit (Sigma-Aldrich, Saint Louis, MO) according to the manufacturer’s instructions. TRACP-positive cells were captured with a microscope (Nikon TE2000-S, Nikon, Melville, NY).
Bone resorption assays
BMMNCs were seeded on dentine slices (ALPCO Diagnostic, Windham, NH) for osteoclast culture in the presence of M-CSF and RANKL at 37 °C with, 5% CO2 for 7 days. Cells were removed from the dentine slices using a sonicator. The area of resorptive “pits” was quantified using NIH ImageJ software. Pit area was normalized to the whole area of the field of dentine slices.
Micro-computed tomography
Bone volume and microarchitecture in the distal femoral metaphysis were evaluated as described previously [21]. Briefly, formalin-fixed femora were scanned with a micro-computed tomographer VivaCT40 (Scanco Medical AG, Bassersdorf, Switzerland). Images were obtained at 55 kV and 145 mA with a voxel size of 10.5 μm. The region for quantification was defined as 100 slices 250 μm below the growth plate and toward proximally.
Quantitative RT-PCR analysis
Total RNA was extracted with treatment of RNase-free DNase to remove contaminating genomic DNA. Quantitative RT-PCR was performed and analyzed as described previously [50]. The primers used for the amplification of each gene are shown in Table S10.
RNA-seq
Transcriptome differences were assessed using RNA-seq, performed at the Sequencing Core of John P. Hussman Institute of Human Genomics at the University of Miami as described previously [50]. The TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA) was used for sample preparation. Bioinformatic and statistical analysis was performed using a pipeline that was reported previously, including differential analysis using edgeR, DESeq2, and CuffDiff [51], [52], [53]. Only transcripts with an adjusted P below 0.05 as tested by all three algorithms were taken to be differential for further analysis [50].
Genome-wide 5hmC profiling and high-throughput sequencing
5hmC selective chemical labeling (hMe-Seal) was performed as described previously [24]. Enriched DNA from hMe-Seal was subjected to library construction using the NEBNext® ChIP-Seq Library Prep Reagent Set from Illumina according to the manufacturer’s protocol. 50-cycle single-read sequencing was performed using an Illumina HiSeq 2000. Image processing and sequence extraction were done using the standard Illumina pipeline.
Bioinformatic analysis of hMe-Seal was performed with strict quality control using our previously-published pipeline [50], which includes alignment using BWA, peak calling using MACS2 [54], and statistical analysis using edgeR. Peaks with a minimum fold change of 2 and an adjusted P below 0.05 were considered differential. De novo motif screening was performed using our recently published genome-wide data for Tet2 binding sites identified in MEL cells overexpressing FLAG-tagged Tet2 [27] . The significant motifs (E value ≤0.05) found by the programs MEME, DREME, and CentriMo were clustered based on similarity and ordered by their E values [55]. Genome-wide datasets generated for this study are deposited at GEO under the accession number GSE104828.
Purification of Tet2-associated proteins and mass spectrometry analysis
To survey the binding-partners of Tet2, FLAG-Tet2 expressing MEL cells (also empty vector transfected control cells) were expanded in 5 large square dishes (245 mm × 245 mm), washed with PBS, and collected for nuclear extract preparation. Briefly, cells were first re-suspended and lysed in Buffer I [10 mM HEPES (pH7.6), 1.5 mM MgCl2, 10 mM KCl], and then centrifuged to remove cytoplasmic proteins. Following that, nuclear fractions were re-suspended and lysed in Buffer II [20 mM HEPES (pH7.6), 25% glycerol (v/v), 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA]. After centrifugation, nuclear extracts were obtained. The salt concentration was subsequently decreased to 100 mM by dialyzing to Buffer III [20 mM HEPES (pH7.6), 20% glycerol, 1.5 mM MgCl2, 100 mM KCl, 0.2 mM EDTA] at 4 °C for 3 h, and precipitated proteins were removed by centrifugation. Buffer II and Buffer III were supplemented with benzonase to digest DNA and RNA. Freshly prepared nuclear extracts were then subjected to affinity purification. All buffers were supplemented with 0.2 mM PMSF and 1× protease inhibitor cocktail (Roche, Mannheim, Germany) before use. Nuclear extract immunoprecipitation, elution, and mass spectrometry analysis were performed as described previously [56]. LC–MS/MS was performed and the resulting data were analyzed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School).
Western blot and co-immunoprecipitation assays
Cells were lysed in lysis buffer containing 1% NP-40, 10% glycerol, 135 mM NaCl, and 20 mM Tris–HCl, pH 8.0, supplemented with 1× protease inhibitor cocktails (Roche). Whole cell lysates were pre-cleared with isotype IgG and Protein G Dynabeads™ (Life Technologies, Oslo, Norway). FLAG-TET2 was then immunoprecipitated from the pre-cleared whole cell lysate with anti-FLAG antibody-conjugated beads (Sigma-Aldrich, Saint Louis, MO) and washed with lysis buffer four times. For RUNX1 immunoprecipitation, the pre-cleared whole cell lysates were incubated with 4 µg anti-RUNX1 antibody (for control groups, 4 µg isotype IgG was added) overnight and then incubated with 40 µl Protein-G Dynabeads™ for 2 h and washed with lysis buffer four times. The beads were boiled directly with 1× SDS loading buffer. Precipitates were subjected to SDS–PAGE and Western blotting was performed using indicated antibodies. Antibodies used in this study were listed in Table S11.
Luciferase reporter assay
HEK293T cells cultured in 6-well plates were transiently transfected with Lipofectamine 2000 (Life Technologies, Canada) according to the standard protocol. Briefly, the cells were co-transfected with 1 μg IL3-promoter luciferase reporter plasmid, combined with 0.1 μg of pCMV-Runx1 plasmid and increasing concentrations of Tet2 expression constructs (pCDF-Tet2). Additionally, a set amount (50 ng) of Renilla luciferase plasmid (Promega, Madison, WI) was used as internal transfection control. Cells were collected 48 h after transfection and lysates were prepared and analyzed according to the manufacturers’ protocols using the Dual-Luciferase Reporter Assay System (Promega). Promoter-luciferase activity was normalized with Renilla luciferase values obtained in each sample. An equivalent quantity of DNA was transfected using the empty vectors for each construct as controls when necessary. Each assay was repeated at least three times.
Statistical analyses
GraphPad Prism 6.0 software was used for the statistical analysis. Data are presented as the mean ± SEM. Differences between two groups were determined using the Student’s t test. Differences were considered significant with P < 0.05.
Authors’ contributions
YC, ZZ, DWS, GZ, JCT, LL, JW, ZC, SC, XL, PZ, and XX performed the experiments and analyzed the data; YC, ZZ, SMG, JPW, GW, SN, and WY provided assistance in designing the study and revised the manuscript; FCY, ZZ, and MX designed and supervised the study, performed the experiments, analyzed data, wrote and finalized the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported by grants from the National Institutes of Health (Grant No. CA172408 to MX and FCY, Grant No. HL112294 to MX), the Leukemia & Lymphoma Society (LLS) (SCOR program to SN, FCY, and MX; translational grant to SN), and University of Miami Sylvester Comprehensive Cancer Center (SCCC to MX and FCY), the United States. The work was also supported by the Ministry of Science and Technology of China (Grant Nos. 2017YFA0103402to WY), National Natural Science Foundation of China (Grant Nos. 81629001 to MX, 81670102 to ZZ, 81600136 to YC, and 81421002 to WY), CAMS Innovation Fund for Medical Sciences (Grant Nos. 2017-I2M-3-015 to WY and 2016-I2M-1-017 to YC), Tianjin Application Foundation and Advanced Technology Research Program (Grant Nos. 16JCYBJC25200 to ZZ and 17JCQNJC09800 to YC), SKLEH-Pilot Research Grand (Grant No. ZK16-3 to ZZ), Peking Union Medical College Youth Fund (Grant No. 3332016092 to YC), China. We would like to thank the Satellite Histological Core of SCCC Core Facility for the histological processing and analysis services.
Handled by Yun-Gui Yang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.gpb.2018.04.005.
Contributor Information
Zhigang Zhao, Email: zzhao01@tmu.edu.cn.
Feng-Chun Yang, Email: fxy37@med.miami.edu.
Mingjiang Xu, Email: mxx51@miami.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Full scan images of reciprocal co-IPs and Western blots confirming the interaction between Tet2 and Runx1 in HEK293T cells overexpressing FLAG-tagged Tet2 A. Western blotting was performed using indicated antibodies for nuclear lysates from HEK293T cells overexpressing FLAG-vector (Ctrl) or FLAG-Tet2 (Tet2) with anti-Flag IP (related to Figure 5B). B. Western blotting was performed using indicated antibodies for nuclear lysates from HEK293T cells overexpressing FLAG-vector (Ctrl) or FLAG-Tet2 (Tet2) with anti-Runx1 IP (related to Figure 5C). The isotype IgG was used as negative control in panel B.
Enrichment of Ep300 and Pu.1 target genes with DhMRs, related to Figure 6 A. GSEA analysis showing decreased gene expression of the Ep300 signature in Tet2−/− macrophages compared with WT macrophages (NES = 1.483, FDR < 1 × 10−4). B. GSEA analysis showing decreased gene expression of the Pu.1 signature in Tet2−/− macrophages compared with WT macrophages (NES = 1.408, FDR < 1 × 10−4). C. and D. Distributions of average 5hmC enrichment at Ep300 (C) and Pu.1 (D) related genes in WT and Tet2−/− macrophages. GSEA, gene set enrichment analysis; NES, nominal enrichment score; FDR, false discovery rate.
Statistical results and annotations for genes with altered expression between Tet2−/− and WT macrophages as determined by RNA-seq
Statistical results and gene annotations for DEGs between Tet2−/− and WT macrophages that are macrophage related genes (111 out of 206 genes)
Genome-wide 5hmC distributions in Tet2−/− and WT macrophages as determined by hMe-Seal
Correlation analysis of 5hmC and transcriptional changes in Tet2−/− mice
Tet2 LC-MS/MS data in MEL cells stably expressing mouse FLAG-Tet2
The consensus binding motifs enriched at FLAG ChIP-seq peaks via MEME-ChIP analysis from MEL cells expressing FLAG-Tet2 or FLAG-Ctrl
Statistical results and gene annotations for DEGs between Tet2−/− and WT macrophages that are also Runx1 target genes (70 out of 206 genes)
List of DEGs in Tet2−/− vs. WT LK cells that are Runx1 target genes (688 out of 3946)
Information for DhMRs in the genomic regions of Runx1 target genes
List of qPCR primers
Antibodies used in the current study
References
- 1.Delhommeau F., Dupont S., Della Valle V., James C., Trannoy S., Masse A. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 2.Langemeijer S.M., Kuiper R.P., Berends M., Knops R., Aslanyan M.G., Massop M. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 3.Tefferi A., Pardanani A., Lim K.H., Abdel-Wahab O., Lasho T.L., Patel J. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowska A.M., Szpurka H., Tiu R.V., Makishima H., Afable M., Huh J. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couronne L., Bastard C., Bernard O.A. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 6.Asmar F., Punj V., Christensen J., Pedersen M.T., Pedersen A., Nielsen A.B. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98:1912–1920. doi: 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meissner B., Kridel R., Lim R.S., Rogic S., Tse K., Scott D.W. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121:3161–3164. doi: 10.1182/blood-2013-01-478834. [DOI] [PubMed] [Google Scholar]
- 8.Busque L., Patel J.P., Figueroa M.E., Vasanthakumar A., Provost S., Hamilou Z. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie M., Lu C., Wang J., McLellan M.D., Johnson K.J., Wendl M.C. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z., Cai X., Cai C.L., Wang J., Zhang W., Petersen B.E. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quivoron C., Couronne L., Della Valle V., Lopez C.K., Plo I., Wagner-Ballon O. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Z., Chen L., Dawlaty M.M., Pan F., Weeks O., Zhou Y. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Rep. 2015;13:1692–1704. doi: 10.1016/j.celrep.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 16.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishikawa K., Iwamoto Y., Kobayashi Y., Katsuoka F., Kawaguchi S., Tsujita T. DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat Med. 2015;21:281–287. doi: 10.1038/nm.3774. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes S.D., Yang H., Dong R., Menon K., He Y., Li Z. Nf1 haploinsufficiency alters myeloid lineage commitment and function, leading to deranged skeletal homeostasis. J Bone Miner Res. 2015;30:1840–1851. doi: 10.1002/jbmr.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z., Chen C., Zhu X., Pan F., Ni H., Yang Y.C. The catalytic activity of TET2 is essential for its myeloid malignancy-suppressive function in hematopoietic stem/progenitor cells. Leukemia. 2016;30:1784–1788. doi: 10.1038/leu.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y., Chavez L., Chang X., Wang X., Pastor W.A., Kang J. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan F., Weeks O., Yang F.C., Xu M. The TET2 interactors and their links to hematological malignancies. IUBMB Life. 2015;67:438–445. doi: 10.1002/iub.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan F., Wingo T.S., Zhao Z., Gao R., Makishima H., Qu G. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat Commun. 2017;8:15102. doi: 10.1038/ncomms15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q., Chen Y., Bian C., Fujiki R., Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deplus R., Delatte B., Schwinn M.K., Defrance M., Mendez J., Murphy N. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Guallar D., Bi X., Pardavila J.A., Huang X., Saenz C., Shi X. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat Genet. 2018;50:443–451. doi: 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Growney J.D., Shigematsu H., Li Z., Lee B.H., Adelsperger J., Rowan R. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soung do Y., Kalinowski J., Baniwal S.K., Jacome-Galarza C.E., Frenkel B., Lorenzo J. Runx1-mediated regulation of osteoclast differentiation and function. Mol Endocrinol. 2014;28:546–553. doi: 10.1210/me.2013-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson N.K., Foster S.D., Wang X., Knezevic K., Schutte J., Kaimakis P. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yip K.Y., Cheng C., Bhardwaj N., Brown J.B., Leng J., Kundaje A. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruocco M.G., Maeda S., Park J.M., Lawrence T., Hsu L.C., Cao Y. I{kappa}B kinase (IKK){beta}, but not IKK{alpha}, is a critical mediator of osteoclast survival and is required for inflammation-induced bone loss. J Exp Med. 2005;201:1677–1687. doi: 10.1084/jem.20042081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W., Zhu G., Hao L., Wu M., Ci H., Li Y.P. C/EBPalpha regulates osteoclast lineage commitment. Proc Natl Acad Sci U S A. 2013;110:7294–7299. doi: 10.1073/pnas.1211383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki T., Shimizu Y., Furuhata E., Maeda S., Kishima M., Nishimura H. RUNX1 regulates site specificity of DNA demethylation by recruitment of DNA demethylation machineries in hematopoietic cells. Blood Adv. 2017;1:1699–1711. doi: 10.1182/bloodadvances.2017005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H., D'Alessio A.C., Ito S., Xia K., Wang Z., Cui K. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q., Zhao K., Shen Q., Han Y., Gu Y., Li X. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montagner S., Leoni C., Emming S., Della Chiara G., Balestrieri C., Barozzi I. TET2 regulates mast cell differentiation and proliferation through catalytic and non-catalytic activities. Cell Rep. 2016;15:1566–1579. doi: 10.1016/j.celrep.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Z., Chen S., Zhu X., Pan F., Li R., Zhou Y. The catalytic activity of TET2 is essential for its myeloid malignancy-suppressive function in hematopoietic stem/progenitor cells. Leukemia. 2016;30:1784–1788. doi: 10.1038/leu.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song C.X., Szulwach K.E., Dai Q., Fu Y., Mao S.Q., Lin L. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X., Han D., Zhao B.S., Song C.X., Zhang L.S., Dore L.C. Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 2015;25:386–389. doi: 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H., Zhang Y. Charting oxidized methylcytosines at base resolution. Nat Struct Mol Biol. 2015;22:656–661. doi: 10.1038/nsmb.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camarena V., Sant D.W., Huff T.C., Mustafi S., Muir R.K., Aron A.T. cAMP signaling regulates DNA hydroxymethylation by augmenting the intracellular labile ferrous iron pool. Elife. 2017;6:e29750. doi: 10.7554/eLife.29750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machanick P., Bailey T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full scan images of reciprocal co-IPs and Western blots confirming the interaction between Tet2 and Runx1 in HEK293T cells overexpressing FLAG-tagged Tet2 A. Western blotting was performed using indicated antibodies for nuclear lysates from HEK293T cells overexpressing FLAG-vector (Ctrl) or FLAG-Tet2 (Tet2) with anti-Flag IP (related to Figure 5B). B. Western blotting was performed using indicated antibodies for nuclear lysates from HEK293T cells overexpressing FLAG-vector (Ctrl) or FLAG-Tet2 (Tet2) with anti-Runx1 IP (related to Figure 5C). The isotype IgG was used as negative control in panel B.
Enrichment of Ep300 and Pu.1 target genes with DhMRs, related to Figure 6 A. GSEA analysis showing decreased gene expression of the Ep300 signature in Tet2−/− macrophages compared with WT macrophages (NES = 1.483, FDR < 1 × 10−4). B. GSEA analysis showing decreased gene expression of the Pu.1 signature in Tet2−/− macrophages compared with WT macrophages (NES = 1.408, FDR < 1 × 10−4). C. and D. Distributions of average 5hmC enrichment at Ep300 (C) and Pu.1 (D) related genes in WT and Tet2−/− macrophages. GSEA, gene set enrichment analysis; NES, nominal enrichment score; FDR, false discovery rate.
Statistical results and annotations for genes with altered expression between Tet2−/− and WT macrophages as determined by RNA-seq
Statistical results and gene annotations for DEGs between Tet2−/− and WT macrophages that are macrophage related genes (111 out of 206 genes)
Genome-wide 5hmC distributions in Tet2−/− and WT macrophages as determined by hMe-Seal
Correlation analysis of 5hmC and transcriptional changes in Tet2−/− mice
Tet2 LC-MS/MS data in MEL cells stably expressing mouse FLAG-Tet2
The consensus binding motifs enriched at FLAG ChIP-seq peaks via MEME-ChIP analysis from MEL cells expressing FLAG-Tet2 or FLAG-Ctrl
Statistical results and gene annotations for DEGs between Tet2−/− and WT macrophages that are also Runx1 target genes (70 out of 206 genes)
List of DEGs in Tet2−/− vs. WT LK cells that are Runx1 target genes (688 out of 3946)
Information for DhMRs in the genomic regions of Runx1 target genes
List of qPCR primers
Antibodies used in the current study