Abstract
In this study, the composition of the microbial community on endive lettuce (Cichorium endivia) was evaluated during different postharvest processing steps. Microbial community structure was characterized by culture-dependent and culture-independent methods. Endive lettuce was sampled exemplarily at four different stages of processing (raw material, cut endive lettuce, washed endive lettuce, and spin-dried (ready to pack) endive lettuce) and analysed by plate count analysis using non-selective and selective agar plates with subsequent identification of bacteria colonies by matrix-assisted laser desorption/ionization time-of light mass spectrometry (MALDI-TOF MS). Additionally, terminal-restriction fragment length polymorphism (TRFLP) analysis and 16S rRNA gene nucleotide sequence analysis were conducted.
The results revealed structural differences in the lettuce microbiomes during the different processing steps. The most predominant bacteria on endive lettuce were detected by almost all methods. Bacterial species belonging to the families Pseudomonadaceae, Enterobacteriaceae, Xanthomonadaceae, and Moraxellaceae were detected in most of the examined samples including some unexpected potentially human pathogenic bacteria, especially those with the potential to build resistance to antibiotics (e.g., Stenotrophomonas maltophilia (0.9 % in cut sample, 0.4 % in spin-dried sample), Acinetobacter sp. (0.6 % in raw material, 0.9 % in cut sample, 0.9 % in washed sample, 0.4 % in spin-dried sample), Morganella morganii (0.2 % in cut sample, 3 % in washed sample)) revealing the potential health risk for consumers.
However, more seldom occurring bacterial species were detected in varying range by the different methods. In conclusion, the applied methods allow the determination of the microbiome's structure and its dynamic changes during postharvest processing in detail. Such a combined approach enables the implementation of tailored control strategies including hygienic design, innovative decontamination techniques, and appropriate storage conditions for improved product safety.
Keywords: Food safety, Food technology, Microbiology
1. Introduction
Fresh produce is minimally processed (i.e. often only washed, cut, and packaged) and commonly consumed raw. In recent years, the consumption of fresh fruits and vegetables increased by 4.5 % each year worldwide [1], with packaged lettuces as the mainly consumed fresh-cut products. Currently, packaged lettuce possesses a fresh-cut market volume of 50 % [2]. Concurrently, an increasing number of outbreaks of human diseases could be associated with the consumption of contaminated fresh products such as fruits and vegetables [3]. In this context, the combination of leafy greens eaten raw and Salmonella spp. was the top food/pathogen combination for foodborne diseases in Europe between 2007 and 2011 [4]. Therefore, microbial safety of fresh-cut produce while maintaining high product quality is mandatory and poses a high challenge for the fresh-cut industry.
The adhesion of pathogenic bacteria on fresh produce surfaces, as example, on lettuce leaves, the penetration of these bacteria into the tissue, as well as the presence of multi-resistant bacteria hamper the reduction of microorganisms during washing processes and disinfection treatments. The most relevant pathogenic microorganisms in fresh produce are verotoxigenic Escherichia coli strains (occurring e.g. in sprouts and leafy greens), Listeria monocytogenes (occurring e.g. in melons and fresh cut salad), Salmonella spp. (occurring e.g. in tomato, seeds sprouts, spices), Shigella (occurring e.g. in green onion), and Norovirus (occurring e.g. in berries) [5]. Hence, routinely applied microbiological sampling along the food processing chain is mainly focused on selected indicator microorganisms even so the composition of microbial diversity on fresh produce is not known in detail [6]. In consequence, unexpected potentially human pathogenic bacteria can remain undetected and could result in foodborne outbreaks.
The evaluation of microbial diversity on fresh produce is mainly focused on the processed products [6, 7, 8, 9]. Only few studies are dealing with the impact of the processing steps on the microbial communities of fresh-cut products [10, 11]. It is known that contamination of vegetables can occur at different steps of processing (e.g., during primary production, processing, distribution, and preparation) [12]. Detailed knowledge of the community structure and development of the microbial load along the processing chain will support the implementation of decontamination strategies and this way increase the product safety.
Qualitative testing of the absence/presence of pathogenic bacteria is sufficient regarding the microbial product safety and product releases, whereas for a complete risk assessment quantitative testing such as enumeration assays is necessary [5]. The structure of the microbial community on food can be evaluated by culture-independent and culture-dependent methods. Culture-independent methods used for the evaluation of microbial communities in food samples are based on analysing the microbial genomic DNA. Such methods can be divided into genetic fingerprinting analysis (e.g., terminal-restriction fragment length polymorphism, TRFLP), in situ hybridisation, amplification techniques [13] and high-throughput sequencing approaches based on DNA [14]. Advantages of these nucleic acid based methods are the high sensitivity and specificity, reliable results, the automation, and the short time required [15]. However, DNA-based approaches are hardly able to distinguish between viable and non-viable microorganisms [16]. Especially in terms of foodborne pathogens the knowledge of bacterial viability is indispensable to estimate the potential risk of foodborne pathogens.
Culture-dependent methods include the conventional plating technique, which is more time-consuming than most culture-independent methods because it relies on the ability of bacteria to proliferate and form visible colonies on specific agar plates at specific temperatures and particular atmospheres which can strongly vary between different groups of bacteria. Additionally, for the specific characterization of bacterial colonies, a biochemical screening and serological confirmation is required [15]. In this context, matrix-assisted laser desorption/ionization time-of light mass spectrometry (MALDI-TOF MS) enables a rapid identification of bacteria up to the strain level and is a promising tool for the rapid and reliable identification of foodborne bacteria [17]. The spectra of bacterial cells are characteristic for a given taxon because they are dominated by the peaks of ribosomal proteins and changes in cultivation conditions only marginally influence the spectra [18].
The application of MALDI-TOF MS in food microbiology is mainly focused on the identification of known foodborne bacteria [17, 19, 20, 21, 22], however, the application of MALDI-TOF MS for the evaluation of microbial communities in food or environmental samples is of growing interest [23, 24, 25, 26, 27, 28, 29]. Basically, the applicability of MALDI-TOF MS in bacterial diversity studies was shown by Spitaels et al. [30].
The microbial community of fresh produce depends on the specific product properties as well as on the preharvest and postharvest processing conditions and varies from case to case. Hence, to obtain knowledge about the composition of the microbial community of endive lettuce, the aim of this study was to evaluate exemplarily the microbial community of endive lettuce, a frequently used lettuce in ready-to-eat salads, and its changes along a commercial processing chain using a combination of culture-dependent and culture-independent methods. To evaluate the viable microbial diversity as accurate as possible, microbial plate count analysis with selective and non-selective media was conducted with subsequent identification of grown bacterial colonies by MALDI-TOF MS. In addition, to obtain information on the non-cultivable part of the microbial community and its dynamics along the processing chain of lettuce, 16S rRNA gene based TRFLP fingerprint analyses supported by a cloning/sequencing approach were conducted.
2. Material and methods
2.1. Sampling of endive lettuce from a fresh-produce production facility
Endive lettuce (Cichorium endivia) from four different sections of a large-scale fresh-cut salad process chain in Germany were sampled exemplarily during processing: (A) raw material, (B) cut, (C) washed, and (D) spin-dried (ready to pack) lettuce. From the selected batch about 500 g product were taken from the before defined steps along the processing line. Washing of the lettuce was conducted without the addition of disinfectants. After sampling, the lettuce samples were stored at temperatures below 5 °C, and analyses were performed within 24 h. The samples were mashed by an immersion blender (Gastroback 40974 Stabmixer Advanced, 800 W, Gastroback GmbH, Germany) to allow the evaluation of microorganisms attached to the surface as well as internalized microorganisms. A total of 25 g of each sample was added to 225 ml peptone salt solution (EN ISO 6887-1 [31]) and homogenized in a shaker (Labotron, Infors AG, Switzerland) at 400 rpm for 20 min. Then, the mashed samples were filtered through a folded filter (pore size five – eight μm). The filtrated samples were used for plate count analyses and the DNA extraction. Before DNA extraction the samples were stored at – 20 °C.
2.2. Viable cell counts and MALDI-TOF MS analysis
Plate count analyses using selective and non-selective media were conducted according to the respective German and European reference standards (Table 1) to ensure the best possible detection of all cultivable microorganisms in the samples. Each sample was serially diluted and subsequently analysed in triplicates.
Table 1.
European and German standards applied for the evaluation of viable cell counts.
| Target | Norm resp. analytical method |
|---|---|
| Aerobic mesophilic total viable cell count | EN ISO 4833:2 [32] |
| Aerobic lactic acid bacteria | EN ISO 15214: 1998 [33] |
| Lactobacilli | BVL L 06.00-31:1992-06 [34] |
| Yeast and moulds | ISO 21527-1:2008 [35] |
| Enterobacteriaceae | EN ISO 21528-2:2009-12 [36] |
| E. coli | ISO 16649-2:2001:2001-04 [37] |
| Bacillus cereus | EN ISO 7932:2005-03 [38] |
| Pseudomonas spp. | EN ISO 13720:2010-12 [39] |
| Coagulase-positive staphylococci | IS0 6888-1:1999/Amd.l:2003(E) [40] |
| Enterococcus sp. | BVL L 06.00-32:1992-06 [41] |
| Clostridium perfringens | ISO 7937:2004-08 [42] |
| Mesophilic sulphite-reducing bacteria | BVL L 06.00-39:1994-05 [43] |
| Salmonella spp. | EN ISO 6579:2007-10 [44] |
| Listeria monocytogenes | EN ISO 11290-1:2005-01 [45] |
| Yersinia enterocolytica | EN ISO 10273:2003-06 [46] |
| Mesophilic spore-forming bacteria | EN ISO 6887-1: [31] |
To ensure that the variety of grown colonies was included in MALDI-TOF MS identification, colonies with different colour and morphology as well as randomly selected colonies from all inoculated selective and non-selective agar plates were sampled with the aim to obtain the best possible summary of the microbial diversity.
Prior to MALDI-TOF MS analysis, cell material of the colonies was transferred to a target and was overlaid with α-cyano-4-hydroxy cinnamic acid (CHCA) matrix (RIPAC-LABOR GmbH, Germany). After air drying, the samples were analysed by MALDI-TOF MS (Axima Confidence, Shimadzu Deutschland GmbH, Germany). Recording of the spectra was conducted in the linear mode with a laser frequency of 50 Hz. The mass range of the spectra was between 3,000 and 20,000 m/z. Calibration of the spectra was performed using E. coli ribosomal proteins. For identification of the bacterial colonies, the obtained mass spectra were compared with the reference mass spectra of the AnagnosTec SARAMIS™ database (Spectral ARchive And Microbial Identification System, bioMérieux Deutschland GmbH, Germany). All spectra were exported to BioNumerics (version 7.6; Applied Maths NV, Belgium) and cluster analysis (UPGMA clustering) was conducted using the peak based similarity coefficient ‘Dice’. The linear tolerance was set to 800 ppm, the constant tolerance was set to two m/z [47]. Clusters were reliable classified as identified if a spectrum matched with reference spectra of the AnagnosTec SARAMIS™ database with a confidence level ≥90 %. Clusters with spectra matched with reference spectra within a confidence level between 75 and 89.9 % were only identified as microorganisms belonging to the bacterial or fungi domain whereas clusters with spectra matched with reference spectra with a confidence level below 75 % were classified as unidentified.
2.3. DNA extraction
Prior to DNA extraction, the endive samples were concentrated. Therefore, four ml filtered endive sample was centrifuged at 14,000 × g for two min. After removing the supernatant, the pellet was resuspended in one ml 1x PBS (pH 7.4). Then, the samples were centrifuged again, and the supernatants were discarded. The pellet was resuspended in one ml 1 % KCl and centrifuged again. The residual pellet was resuspended in distilled H2O and was completely transferred to the Lysing Matrix E tube of the FastDNA™ SPIN Kit for Soil (MP Biomedicals LLC, USA). The subsequent extraction of microbial DNA was conducted according to the manufacturer's guidelines in duplicates for each process step. DNA was stored at 4 °C until further analyses.
2.4. TRFLP analysis
To determine the changes within the bacterial community along the endive process chain, a TRFLP fingerprint analysis was performed. Therefore, the two extracted DNA samples from each processing step were amplified twice applying a Bacteria-specific PCR using the following primers: forward primer 27f (5′- AGA GTT TGA TCM TGG CTC AG -3′) labelled with Cy5, and reverse primer 926r (5′- CCG TCA ATT CMT TTR AGT TT -3′) (Biomers.net GmbH, Germany). The thermal amplification protocol started with an initial step at 95 °C for three min, followed by 25 cycles consisting of a denaturation step at 94 °C for 30 sec, an annealing step at 51 °C for 30 sec and elongation step at 72 °C for 90 sec. After that, a final elongation at 72 °C for eight min was conducted. The reagent mixture consisted of 1x Taq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.4 μM each primer, 1 U recombinant Taq polymerase, and 1 μl template DNA in a total volume of 25 μl (all reagents purchased from Thermo Fisher Scientific Inc, Germany). Both PCR products of one DNA sample were pooled and cleaned up using the NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel GmbH & Co. KG, Germany). Approximately 300 ng PCR product, estimated by the NanoPhotometer® (Implen GmbH, Germany), were used for restriction enzyme digestion applying 10 U of MspI and Hin6I one after the other in 1x Tango buffer (all reagents: Thermo Fisher Scientific Inc, Germany) in a total volume of 20 μl at 37 °C for 4 h for each enzyme. After ethanol precipitation of the digestate, 0.5 μl of each restriction digest was analysed on GenomeLab™ GeXP Genetic Analysis System (Beckman Coulter GmbH, Germany) together with 0.2 μl 600 bp standard and 29.3 μl sample loading solution (Beckmann Coulter GmbH, Germany) applying following conditions: denaturation at 90 °C for two min, injection at 2 kV for 20 sec, and separation at 4.8 kV for a minimum of 70 min. For each DNA sample resp. pooled PCR product, two restriction digests resp. restriction fragment analyses were performed.
The results were exported to BioNumerics (version 7.6; Applied Maths NV, Belgium) and analysed. The search threshold parameters for the band search were set to 0.5 % OD range and two % curve range. Then, band matching was conducted using an optimization of 0.05 % and a position tolerance of 0.1 %.
2.5. 16S rRNA gene nucleotide sequence analysis
To receive detailed information about the bacterial community along the endive postharvest processing chain, a bacterial 16S rRNA gene library was constructed for each sampled process step. Therefore, a Bacteria-specific PCR was performed using the unlabelled forward primer 27f and the reverse primer 1492r (5′- TAC GGY TAC CTT GTT ACG ACT T -3′) (Biomers.net GmbH, Germany) in order to amplify a nearly full length 16S rRNA gene sequence. The thermal amplification protocol started with an initial step at 95 °C for three min, followed by 25 cycles consisting of a denaturation step at 94 °C for 30 sec, an annealing step at 51 °C for 30 sec and elongation step at 72 °C for 90 sec. After that, a final elongation at 72 °C for eight min was conducted. The reagent mixture consisted of 1x Taq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.4 μM each primer, 1 U recombinant Taq polymerase and 1 μl template DNA in a total volume of 50 μl (all reagents purchased by Thermo Fisher Scientific Inc, Germany). DNA bands of expected size were cut out of the agarose gel and were cleaned up using the NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel GmbH & Co. KG, Germany). Ligation of PCR fragments into the pGEM®-T vector system and the transformation of vectors into JM109 competent cells was done according to the manufacturer's protocol (Promega Corporation, USA). White colonies were picked and grown over night at 37 °C in LB broth with ampicillin (0.05 mg ml−1). Plasmids were isolated applying the NucleoSpin® plasmid kit (Macherey-Nagel GmbH & Co. KG, Germany) and afterwards all plasmids were checked for inserts originated from chloroplasts by RFLP analysis. On average, 69 % of plasmids showed a chloroplast typical RFLP fingerprint pattern and were omitted from the subsequent sequencing. This led in an uneven number of analysed plasmids or sequences, respectively, concerning the four 16S rRNA gene sequence libraries, i.e. (A) raw material 50, (B) cut 87, (C) washed 13, and (D) spin-dried (ready to pack) 10.
Sequencing of the selected inserts with the correct size and RFLP pattern was performed by GATC Biotech AG (Germany). Obtained nucleotide sequences were analysed with BioNumerics (version 7.6; Applied Maths NV, Belgium). After the assembly of the forward and reverse sequence, the nearly full length sequences were multiple aligned applying the Needleman-Wunsch algorithm and a clustal w similarity calculation followed by a Kimura-2 correction [48]. The obtained alignment was the basis for a cluster analysis (multiple alignment, UPGMA clustering) resulting in 29 OTUs with a sequence similarity of ≥97%. Then, up to three sequences of each OTU were classified by the Ribosomal Database Project (RDP) (rdp.cme.msu.edu) (confidence threshold 80 %). All nucleotide sequences were deposited in the EMBL-EBI European Nucleotide Archive with the accession numbers: LT595724–LT595883. The assignment of sequences to operational taxonomic units (OTUs) and samples origin with corresponding EMBL accession numbers is given in the supplemental material (Table S1).
3. Results
3.1. Microbial community structure and dynamics along the endive postharvest processing chain as revealed by TRFLP analyses and 16S rRNA gene sequence analyses
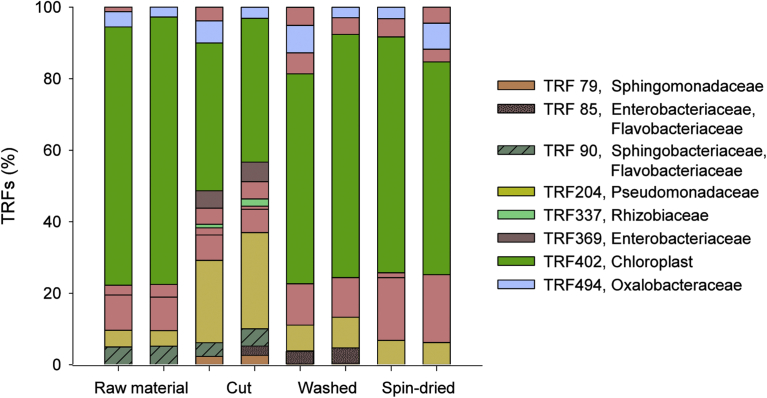
Along an endive postharvest processing chain, four different sections (raw material, cut, washed and spin-dried (ready to pack) lettuce) were sampled and analysed by TRFLP (Fig. 1) and 16S rRNA gene sequence analysis focusing on the bacterial load (Fig. 2A).
Fig. 1.
TRFLP analyses of the bacterial community structure on endive along a processing chain. Affiliations of terminal restriction fragments (TRFs) were obtained by in silico and in vivo analyses. Each TRF profile was averaged from two technical replicates.
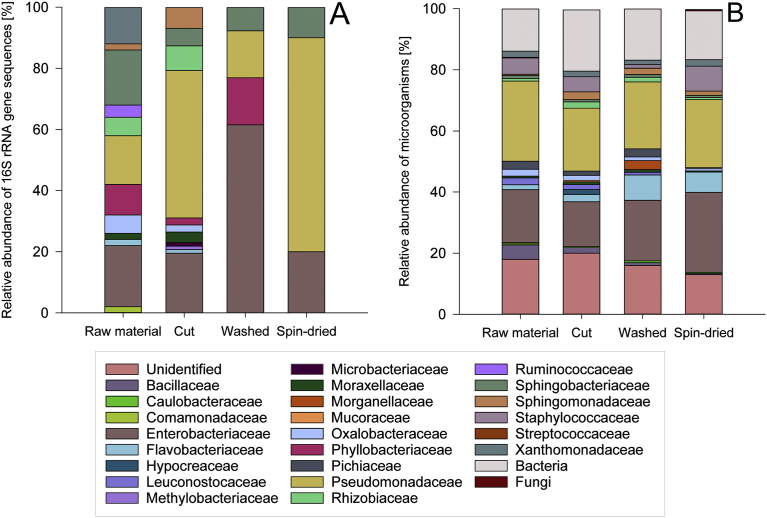
Fig. 2.
16S rRNA gene nucleotide sequence (A) and MALDI-TOF MS (B) analysis of the bacterial community structure on endive along the processing chain. The number of analysed sequences (raw material – 50 sequences, cut lettuce – 87 sequences, washed lettuce – 13 sequences, spin-dried (ready to pack) lettuce – 10 sequences) and colonies (raw material – 492 colonies, cut lettuce – 428 colonies, washed lettuce – 338 colonies, spin-dried (ready to eat) lettuce– 284 colonies) varied between samples.
During the processing of the endive lettuce, members of in total 14 bacterial families (29 OTUs) were identified by gene library construction as well as 15 TRFs by TRFLP analysis. The most prominent ones, Pseudomonadaceae (Pseudomonas), Enterobacteriaceae (e.g., Erwinia, Pantoea, Pectobacterium), and Sphingobacteriaceae (Pedobacter), were detected in both analyses along the whole process chain. However, some changes within the bacterial community could be identified upon processing from raw material to spin-dried (ready to pack) lettuce. Beside the predominant families, on the raw material bacteria belonging to the families Xanthomonadaceae, Phyllo- and Oxalobacteriaceae, Rhizobiaceae, and Sphingomonadaceae have also been identified.
After cutting, members of the family Pseudomonadaceae were more prevalent than before as indicated by TRFLP and 16S rRNA gene sequence analyses. Furthermore, both analyses indicated an increase of Sphingomonadaceae (Sphingomonas). In contrast, members of the Sphingobacteriaceae were reduced.
After washing, the relative abundance of Pseudomonadaceae was reduced, whereas the relative abundance of Enterobacteriaceae (e.g., Erwinia) and Phyllobacteriaceae (Phyllobacterium) increased. The bacterial load of the spin-dried (ready to pack) endive lettuce showed again a reduced relative abundance of Enterobacteriaceae. Herein, analysed 16S rRNA gene sequences could only be identified as unclassified Enterobacteriaceae as revealed by RDP ribosomal database.
3.2. Microbial community structure and dynamics along the endive postharvest processing chain as revealed by MALDI-TOF MS analyses
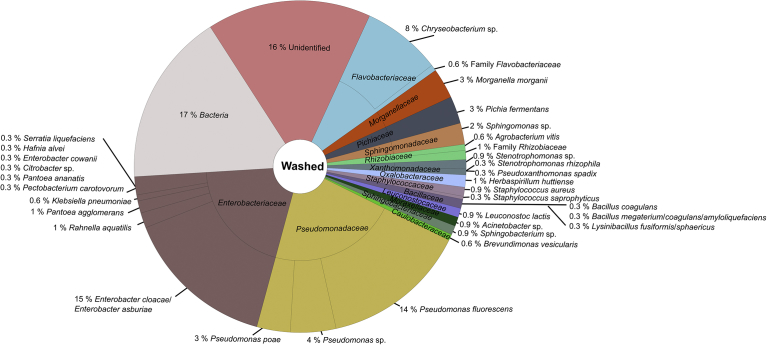
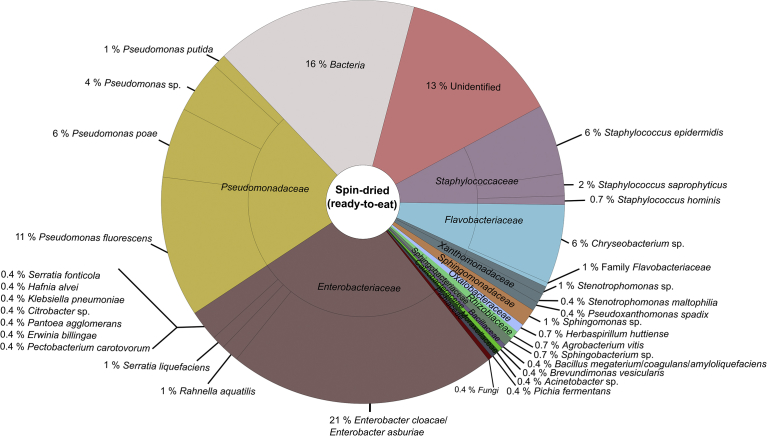
The aerobic mesophilic viable count of the raw material was 7.6 ± 0.3 log CFU/g and 6.1 ± 0.3 log CFU/g for the spin-dried (ready to pack) endive lettuce. The total number of analysed colonies grown on selective and non-selective media was uneven along the endive processing chain. 492 colonies resulting from the raw material sample, 428 colonies from cut endive, 338 colonies from washed endive, and 284 colonies from spin-dried (ready to pack) endive were analysed by MALDI-TOF MS. Along the processing chain of endive lettuce, 54 % of all analysed colonies could not be reliable identified by MALDI-TOF MS using the SARAMISTM database due to the lack of reference mass spectra. It cannot be fully excluded that these not identifiable bacterial species are potential human pathogenic bacteria. To receive further information about these species, analysis of the 16S RNA gene of unidentified colonies was conducted by direct colony PCR with subsequent sequence analysis and identification using the NCBI Megablast tool in combination with the NCBI Reference Sequence Database RefSeq. With these analyses, the number of unidentified bacteria could be reduced along the endive processing chain, up to now, i.e. 18 % of the colonies from the raw material sample, 20 % of the colonies from the cut endive, 16 % of the colonies from the washed endive, and 13 % of the colonies from the spin-dried (ready to pack) endive showed a match with reference spectra of the SARAMISTM database with a confidence level <75 % and were therefore classified as unidentified (Fig. 2B).
The classification of the identified spectra to the family level using the SARAMISTM database revealed that in all samples along the processing chain bacteria belonging to the families Pseudomonadaceae, Enterobacteriaceae, Flavobacteriaceae, Staphylococcaceae, Oxalobacteraceae, Rhizobiaceae, Xanthomonadaceae, Sphingomonadaceae, Sphingobacteriaceae, Moraxellaceae, Caulobacteraceae, Pichiaceae, and Bacillaceae occurred (Figs. 3, 4, 5, and 6). Thereby, bacteria belonging to the Pseudomonadaceae and Enterobacteriaceae were predominant along the processing chain.
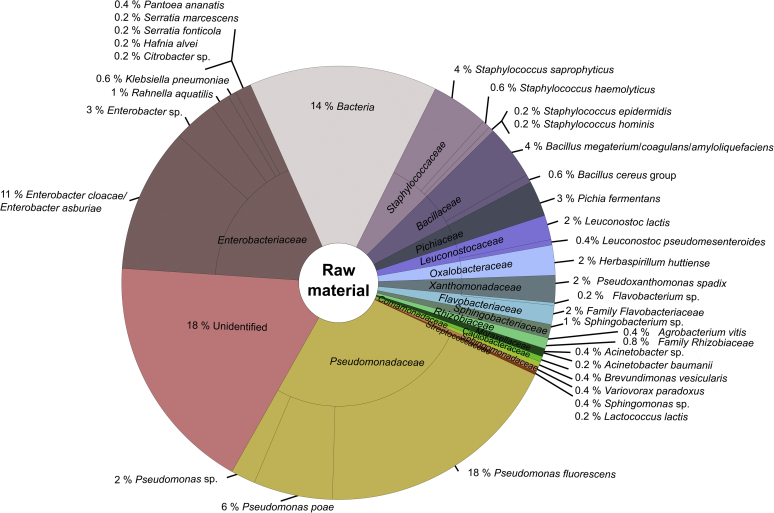
Fig. 3.
Microbial community structure of endive raw material obtained by MALDI-TOF MS analysis. In total, 492 colonies were analysed.
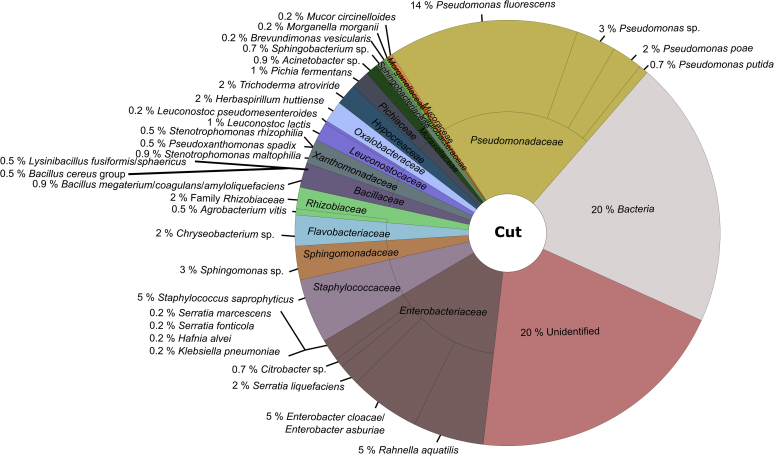
Fig. 4.
Microbial community structure of cut endive material obtained by MALDI-TOF MS analysis. In total, 428 colonies were analysed.
Fig. 5.
Microbial community structure of washed endive material obtained by MALDI-TOF MS analysis. In total, 338 colonies were analysed.
Fig. 6.
Microbial community structure of spin-dried (ready to pack) endive material obtained by MALDI-TOF MS analysis. In total, 284 colonies were analysed.
Bacteria belonging to the families Streptococcaceae and Comamonadaceae were only found in the raw material, bacteria of the families Mucoraceae and Hypocreaceae were only detected in the cut endive sample, and bacteria belonging to the family Leuconostocaceae were only found in the spin-dried (ready to pack) endive.
A further classification to the species level using the SARAMISTM database revealed four different bacteria species within the family Xanthomonadaceae (Pseudoxanthomonas spadix, Stenotrophomonas sp., Stenotrophomonas rhiziphila, Stenotrophomonas maltophilia) along the endive processing chain, of these Pseudoxanthomonas spadix was found in all samples (Figs. 3, 4, 5, and 6). Regarding the complete endive processing chain, the highest diversity was found for the families Enterobacteriaceae followed by Bacillaceae and Pseudomonadaceae. Within the Enterobacteriaceae Rahnella aquatilis, Klebsiella pneumoniae, Enterobacter cloacae/Enterobacter asburiae, Citrobacter sp., Hafnia alvei and Serratia spp. (Serratia fonticola, Serratia marcescens, Serratia liquefaciens) were found along the endive processing chain, whereas Pantoea spp. (Pantoea ananatis, Pantoea agglomerans) were only found in the raw material and the washed and spin dried (ready-to-pack) endive sample. The predominant bacteria species of the Pseudomonadaceae was Pseudomonas fluorescens. Additionally, Pseudomonas poae, Pseudomonas putida, and Pseudomonas sp. were identified along the endive processing chain.
Acinetobacter baumannii (Moraxellaceae) was only identified in the raw material whereas Acinetobacter sp. was found in all samples along the processing chain (Figs. 3, 4, 5, and 6). Bacteria belonging to the Bacillus cereus group were only identified up to the cut endive sample.
The enrichment procedures of the bacteria from endive samples showed that bacteria belonging to the families Xanthomonadaceae (Stenotrophomonas maltophilia), Pseudomonadaceae (Pseudomonas putida), Enterobacteriaceae (Serratia fonticola, Serratia liquefaciens, Serratia marcescens, Klebsiella pneumoniae, Citrobacter sp., Hafnia alvei, Morganella morganii), Bacillaceae (Lysinibacillus fusiformis/sphaericus), and Staphylococcacae (Staphylococcus aureus, Staphylococcus haemolyticus, Staphylococcus saprophyticus) were present in the endive samples along the processing chain.
4. Discussion
The aerobic mesophilic viable count of the endive lettuce was in accordance with the findings of other surveys, showing that whole endive lettuce had an aerobic mesophilic viable count of 6.7–7.2 log CFU/g and ready-to-eat endive lettuce an aerobic mesophilic viable count of 4.3–7.2 log CFU/g [49]. However, the total viable count gives nearly no information on the microbial community of endive lettuce. Since leafy green vegetables can act as vehicles for the transmission of human pathogens [50], and it is known that they retain a majority of their indigenous microflora after processing, it poses a potential food safety problem [51]. Hence, the knowledge about the microbial community is of high interest for the food industry [52].
Using culture-dependent methods, information about the presence and viable abundance of bacterial populations are obtained. Additionally, culture-independent methods provide information about the entire bacterial community [53]. TRFLP analysis allows the monitoring of community changes (e.g., after each process step along the processing chain) and the detection of taxa that may be missed by culture-dependent methods [7]. However, 69 % of the plasmids obtained by 16S rRNA gene library construction showed chloroplast fingerprint pattern which led to an uneven number of analysed plasmids. This seems to be due to the amplification of the chloroplast and mitochondria specific 16S rRNA sequences by the primer pairs commonly applied for microbial community analysis which were also applied in this study. Even though the amount of non-bacterial DNA was reduced by filtration in preliminary experiments, the filtration step in these experiments was not sufficient resulting in such a high amount of plasmids with chloroplast sequence. Rudi et al. [8] also found a high frequency of chloroplasts performing a 16S rDNA array approach using the 16S rRNA gene targeting primer pairs 10-34f and 1485-1507r. They suggested that the microbial load of the sample was very low and therefore more chloroplast DNA than bacterial DNA was amplified by the used primers. This is also the most likely reason for the results obtained in our study. Randazzo et al. [54] used the 16S rRNA gene targeting primer pair 7f and 1510r for PCR-DGGE analyses of raw lettuce. In that study, a high frequency of chloroplasts was not reported. Further, Jackson et al. [55] used the primer pair Bac799f and Uni1492r for 454 pyrosequencing of leafy salad samples because no amplification of residual chloroplast DNA was expected and less than 0.05 % of the obtained sequences were originated from chloroplasts. However, the use of primer pairs resulting in 16S rRNA gene fragments or amplicons of shorter length leads to a minor resolution of bacterial taxa. Thus, the choice of primer pairs should be adapted for each investigation to obtain the best possible results.
For the identification of unidentified isolates by MALDI-TOF MS, a very good structured database with reference mass spectra of the target microorganisms is essential [56]. The higher the amount of habitat specific reference mass spectra within the database, the higher is the possibility to identify pathogenic bacteria. Due to the fact that MALDI-TOF MS is mainly used to identify pathogens in the medical field, databases with reference mass spectra from relevant food associated microorganisms are still missing. The analysis of the 16S RNA gene of unidentified colonies with subsequent sequence analysis and identification using the NCBI Megablast tool in combination with the NCBI Reference Sequence Database RefSeq enabled improve the a database with reference mass spectra from plant associated bacteria but a continuously expanding of the database is needed to enable a rapid identification of food-related bacteria in future investigations.
Both, TRFLP and 16S rRNA gene sequence analysis as well as the MALDI-TOF MS analysis, showed that the relative abundances of bacterial families changed along the processing chain. During processing contaminated wash water, equipment or improper handling can lead to a contamination of fresh produce [57]. In addition, also the persistence of pathogenic species during cleaning or seasonal shutdown is possible [58]. Cross-contamination during processing may be the reason for the changes of the microbial community along the processing chain. It has to be taken into account that the number of analysed sequences vary along the processing chain which may result into an underestimation or overestimation of bacterial abundances. However, differences in the microbial community of lettuce directly from farms and from supermarkets were also found by Jackson et al. [53]. It was also shown that different packaging and storage time also influences the bacterial community of lettuce [7]. 16S rRNA gene sequence analysis as well as the culture-dependent analyses with subsequent identification by MALDI-TOF MS showed that Pseudomonadaceae were the predominant bacteria on endive lettuce along the processing chain. This is in accordance with the literature where Pseudomonadaceae were found as predominant bacteria on lettuce [8, 9, 55, 59, 60, 61] which are not removed during washing [54].
Bacteria belonging to the family Xanthomonadaceae (e.g. Xanthomonas) can be plant pathogens [59] and some genera can include potential human pathogens (e.g. Stenotrophomonas spp.) [55]. Stenotrophomonas maltophilia was found in the cut endive sample and in the spin-dried (ready to pack) endive sample. Stenotrophomonas maltophilia is referred to as environmental global emerging multidrug resistant organism (MDRO) that is associated with wet surface and aqueous solution and is able to form biofilms [62]. Qureshi et al. [63] found Stenotrophomonas maltophilia in 78 % of tested lettuce samples, and all isolates were resistant or susceptible to antibiotics. It is emerging as nosocomial pathogen, especially for immunocompromised persons, but its importance in ready-to eat salads is still unknown.
Pathogens with the potential to generate antibiotic resistance or multi resistance such as Enterobacter spp., Klebsiella pneumoniae, Klebsiella oxytoca, and Serratia marcescens [64] were also found in small quantities in the analysed endive samples. The presence of antibiotic resistant bacteria and their resistance rates on different vegetables were tested by Schwaiger et al. [65]. They found that resistance rates of bacteria from vegetable samples were lower than the resistance rates of bacteria of animal or human origin but since vegetables can be a source for the dissemination of antibiotic resistance. Schwaiger et al. [65] recommended thorough washing of raw vegetables before consumption. Antibiotic resistance of bacteria on salad was also shown for Pseudomonas fluorescence that was resistant to six antibiotics, and the occurrence of multi resistant bacteria is common in epiphytic bacteria [66]. Stenotrophomonas sp., Acinetobacter sp., Morganella morganii, Klebsiella sp., Enterobacter sp., and Serratia sp. with different antibiotic resistances were found in fruits and vegetables by Jones-Dias et al. [67]. They concluded that fresh produce is a relevant reservoir for Gram-negative bacteria with antibiotic resistance and a continuous monitoring is essentially required. However, the resistance rates of the bacteria were not tested in this study, and, due to its relevance, this topic need to be included in further studies.
In this study, bacteria belonging to the Bacillus cereus group were only found in samples of raw and cut endive. The German Society for Hygiene and Microbiology [68] recommends a warning value of 3 log CFU/g for presumptive Bacillus cereus in mixed salads which was not exceeded in this analyses. However, it has to be taken into account that the selective choice of colonies which were dominant and morphologically distinct instead of analysing all grown colonies may lead to an underestimation of bacterial presence.
The application of culture-dependent methods with subsequent identification by MALDI-TOF MS in combination with culture-independent methods such as TRFLP fingerprinting and 16S rRNA gene sequence analysis enables the evaluation of the microbial community of endive lettuce along the postharvest processing chain as accurate as possible. The predominant bacteria on endive lettuce were detected by almost all methods but there are also varying results indicating that the methods applied are complementary. The results of this study indicate that not only expected groups of microorganisms are detectable on lettuce but also unexpected potentially pathogenic bacteria (e.g., Stenotrophomonas maltophilia, Acinetobacter sp., Morganella morganii) can occur on fresh cut lettuce. The detection of unexpected pathogenic bacteria, especially those with antibiotic resistance is of great interest to avoid potential risks for consumers and to avoid the potential spread of antibiotic resistance in the food chain.
Naturally occurring phyllospheric and endophytic bacteria can act as commensals or symbionts [55]. In addition, they can act also as competitors for human pathogens limiting the presence of pathogenic bacteria [53]. Therefore, detailed knowledge of the microbial community and its dynamic changes during food processing is essential to allow the implementation of tailored control strategies including hygienic design, innovative decontamination techniques, and appropriate storage conditions.
Declarations
Author contribution statement
Antje Fröhling: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Antje Rademacher, Birgit Rumpold: Performed the experiments; Analyzed and interpreted the data.
Michael Klocke, Oliver Schlüter: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data;
Funding statement
This work was supported by the research project SAFEFRESH which was funded by the German Federal Ministry of Education and Research (BMBF) within the research program “Research for Civil Security” (grant no. 13N12427). The publication of this article was funded by the OpenAccess Fund of the Leibniz Association.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We gratefully acknowledge the excellent technical assistance of Beate-Kristin Kröck, Veronika Egert, Susanne Klocke, and Janett Schiffmann. The publication of this article was funded by the OpenAccess Fund of the Leibniz Association.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Olaimat A.N., Holley R.A. Factors influencing the microbial safety of fresh produce: a review. Food Microbiol. 2012;32:1–19. doi: 10.1016/j.fm.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Van Rijswick C. 2010. EU Fresh-cut Fruits and Vegetables Market Update. Rabobank Industry Note 246. [Google Scholar]
- 3.Lynch M.F., Tauxe R.V., Hedberg C.W. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 2009;137:307–315. doi: 10.1017/S0950268808001969. [DOI] [PubMed] [Google Scholar]
- 4.Da Silva Felício M.T., Hald T., Liebana E., Allende A., Hugas M., Nguyen-The C., Johannessen G.S., Niskanen T., Uyttendaele M., Mclauchlin J. Risk ranking of pathogens in ready-to-eat unprocessed foods of non-animal origin (FoNAO) in the EU: initial evaluation using outbreak data (2007–2011) Int. J. Food Microbiol. 2015;195:9–19. doi: 10.1016/j.ijfoodmicro.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Hoorfar J., Feng P., Duffy G., Malorny B., Binet R. 20-bottlenecks and limitations in testing for pathogens in fresh produce. In: Hoorfar J., editor. Global Safety of Fresh Produce. Woodhead Publishing; 2014. pp. 274–291. [Google Scholar]
- 6.Leff J.W., Fierer N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Carli M., De Rossi P., Paganin P., Del Fiore A., Lecce F., Capodicasa C., Bianco L., Perrotta G., Mengoni A., Bacci G., Daroda L., Dalmastri C., Donini M., Bevivino A. Bacterial community and proteome analysis of fresh-cut lettuce as affected by packaging. FEMS Microbiol. Lett. 2016;363 doi: 10.1093/femsle/fnv209. [DOI] [PubMed] [Google Scholar]
- 8.Rudi K., Flateland S.L., Hanssen J.F., Bengtsson G., Nissen H. Development and evaluation of a 16S ribosomal DNA array-based approach for describing complex microbial communities in ready-to-eat vegetable salads packed in a modified atmosphere. Appl. Environ. Microbiol. 2002;68:1146–1156. doi: 10.1128/AEM.68.3.1146-1156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldera L., Franzetti L. Effect of storage temperature on the microbial composition of ready-to-use vegetables. Curr. Microbiol. 2013;68:133–139. doi: 10.1007/s00284-013-0430-6. [DOI] [PubMed] [Google Scholar]
- 10.Zagory D. Effects of post-processing handling and packaging on microbial populations. Postharvest Biol. Technol. 1999;15:313–321. [Google Scholar]
- 11.Handschur M., Pinar G., Gallist B., Lubitz W., Haslberger A.G. Culture free DGGE and cloning based monitoring of changes in bacterial communities of salad due to processing. Food Chem. Toxicol. 2005;43:1595–1605. doi: 10.1016/j.fct.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira M., Usall J., Viñas I., Solsona C., Abadias M. Transfer of Listeria innocua from contaminated compost and irrigation water to lettuce leaves. Food Microbiol. 2011;28:590–596. doi: 10.1016/j.fm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Giraffa G., Neviani E. DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 2001;67:19–34. doi: 10.1016/s0168-1605(01)00445-7. [DOI] [PubMed] [Google Scholar]
- 14.Garofalo C., Osimani A., Milanović V., Taccari M., Cardinali F., Aquilanti L., Riolo P., Ruschioni S., Isidoro N., Clementi F. The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol. 2017;62:15–22. doi: 10.1016/j.fm.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Mandal P.K., Biswas A.K., Choi K., Pal U.K. Methods for rapid detection of foodborne pathogens: an overview. Am. J. Food Technol. 2011;6:87–102. [Google Scholar]
- 16.Law J.W.-F., Ab Mutalib N.-S., Chan K.-G., Lee L.-H. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 2014;5:770. doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavlovic M., Huber I., Konrad R., Busch U. Application of Maldi-ToF MS for the identification of food borne bacteria. Open Microbiol. J. 2013;7:135. doi: 10.2174/1874285801307010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumann P., Maier T. Chapter 13-Maldi-ToF mass spectrometry applied to classification and identification of bacteria. In: Michael Goodfellow I.S., Jongsik C., editors. Methods in Microbiology. Academic Press; 2014. pp. 275–306. [Google Scholar]
- 19.Böhme K., Fernández-No I.C., Barros-Velázquez J., Gallardo J.M., Cañas B., Calo-Mata P. Species identification of food spoilage and pathogenic bacteria by Maldi-ToF mass fingerprinting. In: Kapiris Kostas., editor. Food Quality. InTech; 2012. https://www.intechopen.com/books/food-quality/species-identification-of-food-spoilage-and-pathogenic-bacteria-by-maldi-tof-mass-fingerprinting Available from: [Google Scholar]
- 20.Giebel R., Worden C., Rust S.M., Kleinheinz G.T., Robbins M., Sandrin T.R. Chapter 6-microbial fingerprinting using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Maldi-ToF MS): applications and challenges. In: Allen I.L., Sima S., Geoffrey M.G., editors. Advances in Applied Microbiology. Academic Press; 2010. pp. 149–184. [DOI] [PubMed] [Google Scholar]
- 21.Mazzeo M.F., Sorrentino A., Gaita M., Cacace G., Di Stasio M., Facchiano A., Comi G., Malorni A., Siciliano R.A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the discrimination of food-borne microorganisms. Appl. Environ. Microbiol. 2006;72:1180–1189. doi: 10.1128/AEM.72.2.1180-1189.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singhal N., Kumar M., Kanaujia P.K., Virdi J.S. Maldi-ToF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015;6:791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Böhme K., Fernández-No I.C., Gallardo J.M., Cañas B., Calo-Mata P. Safety assessment of fresh and processed seafood products by Maldi-ToF mass fingerprinting. Food Bioprocess Technol. 2010;4:907–918. [Google Scholar]
- 24.Böhme K., Fernández-No I.C., Pazos M., Gallardo J.M., Barros-Velázquez J., Cañas B., Calo-Mata P. Identification and classification of seafood-borne pathogenic and spoilage bacteria: 16S rRNA sequencing versus Maldi-ToF MS fingerprinting. Electrophoresis. 2013;34:877–887. doi: 10.1002/elps.201200532. [DOI] [PubMed] [Google Scholar]
- 25.Koubek J., Uhlik O., Jecna K., Junkova P., Vrkoslavova J., Lipov J., Kurzawova V., Macek T., Mackova M. Whole-cell Maldi-ToF: rapid screening method in environmental microbiology. Int. Biodeterior. Biodegrad. 2012;69:82–86. [Google Scholar]
- 26.Hausdorf L., Mundt K., Winzer M., Cordes C., Fröhling A., Schlüter O., Klocke M. Characterization of the cultivable microbial community in a spinach-processing plant using Maldi-ToF MS. Food Microbiol. 2013;34:406–411. doi: 10.1016/j.fm.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Agustini B.C., Silva L.P., Bloch C., Bonfim T.M.B., Da Silva G.A. Evaluation of Maldi-ToF mass spectrometry for identification of environmental yeasts and development of supplementary database. Appl. Microbiol. Biotechnol. 2014;98:5645–5654. doi: 10.1007/s00253-014-5686-7. [DOI] [PubMed] [Google Scholar]
- 28.Böhme K., Fernández-No I.C., Barros-Velázquez J., Gallardo J.M., Calo-Mata P., Cañas B. Species differentiation of seafood spoilage and pathogenic gram-negative bacteria by Maldi-ToF mass fingerprinting. J. Proteome Res. 2010;9:3169–3183. doi: 10.1021/pr100047q. [DOI] [PubMed] [Google Scholar]
- 29.Stets M.I., Pinto A.S., Jr., Huergo L.F., De Souza E.M., Guimarães V.F., Alves A.C., Steffens M.B.R., Monteiro R.A., Pedrosa F.D.O., Cruz L.M. Rapid identification of bacterial isolates from wheat roots by high resolution whole cell Maldi-ToF MS analysis. J. Biotechnol. 2013;165:167–174. doi: 10.1016/j.jbiotec.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Spitaels F., Wieme D.A., Vandamme P. Maldi-ToF MS as a novel tool for dereplication and characterization of microbiota in bacterial diversity studies. In: Demirev P., Sandrin R.T., editors. Applications of Mass Spectrometry in Microbiology: from Strain Characterization to Rapid Screening for Antibiotic Resistance. Springer International Publishing; Cham: 2016. pp. 235–256. [Google Scholar]
- 31.International Organization for Standardization . 1999. ISO 6887-1: Microbiology of Food and Animal Feeding Stuffs – Preparation of the Test Samples, of Initial Suspension and of Decimal Dilutions for Microbiological Examination – Part 1: General Rules for the Preparation of the Initial Suspension and Decimal Dilutions. [Google Scholar]
- 32.International Organization for Standardization . 2013. ISO 4833-2: Microbiology of the Food Chain — Horizontal Method for the Enumeration of Microorganisms — Part 2: Colony Count at 30 °C by the Surface Plating Technique. [Google Scholar]
- 33.International Organization for Standardization . 1998. ISO 15214:1998: Microbiology of Food and Animal Feeding Stuffs – Horizontal Method for the Enumeration of Mesophilic Lactic Acid Bacteria – colony-count Technique at 30 Degrees C. [Google Scholar]
- 34.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit . 1992. BVL L 06.00-31:1992-06: Untersuchung von Lebensmitteln; Bestimmung von Laktobazillen in Fleisch und Fleischerzeugnissen; Spatelverfahren (Referenzverfahren) [Google Scholar]
- 35.International Organization for Standardization . 2008. ISO 21527-1:2008: Microbiology of Food and Animal Feeding Stuffs – Horizontal Method for the Enumeration of Yeasts and Moulds – Part 1: Colony Count Technique in Products with Water Activity Greater than 0,95. [Google Scholar]
- 36.International Organization for Standardization . 2009. ISO 21528-2:2009-12: Microbiology of Food and Animal Feeding Stuffs - Horizontal Methods for the Detection and Enumeration of Enterobacteriaceae - Part 2: Colony-count Method (ISO 21528-2:2004) [Google Scholar]
- 37.International Organization for Standardization . 2001. ISO 16649-2:2001:2001–04: Microbiology of Food and Animal Feeding Stuffs — Horizontal Method for the Enumeration of -glucuronidase-positive Escherichia coli — Part 2: Colony-count Technique at 44 °C Using 5-bromo-4-chloro-3-indolyl -d-glucuronide. [Google Scholar]
- 38.International Organization for Standardization . 2004. EN ISO 7932:2005-03: Microbiology of Food and Animal Feeding Stuffs - Horizontal Method for the Enumeration of Presumptive Bacillus cereus – colony-count Technique at 30 °C (ISO 7932:2004); German Version EN ISO 7932:2004. [Google Scholar]
- 39.International Organization for Standardization . 2010. ISO 13720:2010-12: Meat and Meat Products – Enumeration of Presumptive Pseudomonas spp. (ISO 13720:2010); German version EN ISO 13720:2010. [Google Scholar]
- 40.International Organization for Standardization . 2003. ISO 6888-1:1999/amd.L:2003(e): Microbiology of food and animal feeding stuffs – Horizontal method for the enumeration of coagulase-positive staphylococci (Staphylococcus aureus and other species) – Part 1: Technique using baird-parker agar medium, amendment 1: Inclusion of precision data. [Google Scholar]
- 41.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit . 1992. BVL L 06.00-32:1992-06: Untersuchung von Lebensmitteln – Bestimmung von Enterococcus faecalis und Enterococcus faecium in Fleisch und Fleischerzeugnissen; Spatelverfahren (Referenzverfahren) (Übernahme der gleichlautenden deutschen norm DIN 10106, Ausgabe September 1991) [Google Scholar]
- 42.International Organization for Standardization . 2004. ISO 7937:2004-08: Microbiology of Food and Animal Feeding Stuffs – Horizontal Method for the Enumeration of Clostridium perfringens – Colony-count Technique. [Google Scholar]
- 43.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit . 1994. BVL L 06.00-39:1994-05: Untersuchung von Lebensmitteln – Bestimmung von mesophilen sulfitreduzierenden Clostridien in Fleisch und Fleischerzeugnissen – Plattengussverfahren (Referenzverfahren) (Übernahme der gleichlautenden deutschen norm DIN 10103, Ausgabe August 1993) [Google Scholar]
- 44.International Organization for Standardization . 2007. ISO 6579:2007-10: Microbiology of Food and Animal Feeding Stuffs - Horizontal Method for the Detection of Salmonella spp. (ISO 6579:2002+amd 1:2007); German version EN ISO 6579:2002+a1:2007. [Google Scholar]
- 45.International Organization for Standardization . 2005. EN ISO 11290-1:2005-01: Microbiology of Food and Animal Feeding Stuffs – Horizontal Method for the Detection and Enumeration of Listeria monocytogenes – Part 1: Detection Method (iso 11290-1:1996 + amd 1:2004); German version EN ISO 11290-1:1996 + a1:2004. [Google Scholar]
- 46.International Organization for Standardization . 2003. ISO 10273:2003-06: Mikrobiologie von Lebensmitteln und Futtermitteln - Horizontales Verfahren zum Nachweis von präsumtiv pathogenen Yersinia enterocolitica. [Google Scholar]
- 47.Wieme A.D., Spitaels F., Aerts M., De Bruyne K., Van Landschoot A., Vandamme P. Effects of growth medium on matrix-assisted laser desorption-ionization time of flight mass spectra: a case study of acetic acid bacteria. Appl. Environ. Microbiol. 2014;80:1528–1538. doi: 10.1128/AEM.03708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klang J., Theuerl S., Szewzyk U., Huth M., Tölle R., Klocke M. Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage. Microb. Biotechnol. 2015;8:764–775. doi: 10.1111/1751-7915.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abadias M., Usall J., Anguera M., Solsona C., Vinas I. Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. Int. J. Food Microbiol. 2008;123:121–129. doi: 10.1016/j.ijfoodmicro.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 50.Berger C.N., Sodha S.V., Shaw R.K., Griffin P.M., Pink D., Hand P., Frankel G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 51.Mercanoglu Taban B., Halkman A.K. Do leafy green vegetables and their ready-to-eat [RTE] salads carry a risk of foodborne pathogens? Anaerobe. 2011;17:286–287. doi: 10.1016/j.anaerobe.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Nicholl P., McInerney S., Prendergast M. Growth dynamics of indigenous microbial populations on vegetables after decontamination and during refrigerated storage. J. Food Process. Preserv. 2004;28:442–459. [Google Scholar]
- 53.Jackson C., Stone B., Tyler H. Emerging perspectives on the natural microbiome of fresh produce vegetables. Agriculture. 2015;5:170. [Google Scholar]
- 54.Randazzo C.L., Scifò G.O., Tomaselli F., Caggia C. Polyphasic characterization of bacterial community in fresh cut salads. Int. J. Food Microbiol. 2009;128:484–490. doi: 10.1016/j.ijfoodmicro.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Jackson C.R., Randolph K.C., Osborn S.L., Tyler H.L. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkes J.G., Glover K.L., Holcomb M., Rafii F., Cao X., Sutherland J.B., Mccarthy S.A., Letarte S., Bertrand M.J. Defining and using microbial spectral databases. J. Am. Soc. Mass Spectrom. 2002;13:875–887. doi: 10.1016/S1044-0305(02)00390-2. [DOI] [PubMed] [Google Scholar]
- 57.Everis L. Review, Campden & Chorleywood Food Research Association; 2004. Risks of pathogens in ready-to-eat fruits, vegetables, and salads through the production process. No.44, vii + 94pp. [Google Scholar]
- 58.Hausdorf L., Neumann M., Bergmann I., Sobiella K., Mundt K., Fröhling A., Schlüter O., Klocke M. Occurrence and genetic diversity of Arcobacter spp. in a spinach-processing plant and evaluation of two Arcobacter-specific quantitative PCR assays. Syst. Appl. Microbiol. 2013;36:235–243. doi: 10.1016/j.syapm.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 59.Dees M.W., Lysøe E., Nordskog B., Brurberg M.B. Bacterial communities associated with surfaces of leafy greens: shift in composition and decrease in richness over time. Appl. Environ. Microbiol. 2015;81:1530–1539. doi: 10.1128/AEM.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez-Velasco G., Welbaum G.E., Boyer R.R., Mane S.P., Ponder M.A. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J. Appl. Microbiol. 2011;110:1203–1214. doi: 10.1111/j.1365-2672.2011.04969.x. [DOI] [PubMed] [Google Scholar]
- 61.Ramos B., Miller F.A., Brandão T.R.S., Teixeira P., Silva C.L.M. Fresh fruits and vegetables — an overview on applied methodologies to improve its quality and safety. Innov. Food Sci. Emerg. 2013;20:1–15. [Google Scholar]
- 62.Brooke J.S. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qureshi A., Mooney L., Denton M., Kerr K.G. Stenotrophomonas maltophilia in salad. Emerg. Infect. Dis. 2005;11:1157–1158. doi: 10.3201/eid1107.040130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robert-Koch-Institut (RKI) Surveillance nosokomialer Infektionen sowie die Erfassung von Krankheitserregern mit speziellen Resistenzen und Multiresistenzen. Bundesgesundheitsblatt - Gesundheitsforsch. - Gesundheitsschutz. 2013;56:580–583. doi: 10.1007/s00103-013-1705-6. [DOI] [PubMed] [Google Scholar]
- 65.Schwaiger K., Helmke K., Hölzel C.S., Bauer J. Antibiotic resistance in bacteria isolated from vegetables with regards to the marketing stage (farm vs. supermarket) Int. J. Food Microbiol. 2011;148:191–196. doi: 10.1016/j.ijfoodmicro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Hamilton-Miller J.M.T., Shah S. Identity and antibiotic susceptibility of enterobacterial flora of salad vegetables. Int. J. Antimicrob. Agents. 2001;18:81–83. doi: 10.1016/s0924-8579(01)00353-3. [DOI] [PubMed] [Google Scholar]
- 67.Jones-Dias D., Manageiro V., Ferreira E., Barreiro P., Vieira L., Moura I.B., Caniça M. Architecture of class 1, 2, and 3 integrons from gram negative bacteria recovered among fruits and vegetables. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.German Society for Hygiene and Microbiology (DGHM e.V.), Mikrobiologische Richt- und Warnwerte zur Beurteilung von Lebensmitteln (Stand: November 2015) - Entwürfe. http://www.dghm.org/wissenschaftlichethemenforschung/fachgruppen/lebensmittelmikrobiologie/m_748.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.