Abstract
Wilms' tumor is a pediatric malignancy that is thought to originate from faulty kidney development during the embryonic stage. However, there is a large variation between tumors from different patients in both histology and gene expression that is not well characterized. Here we use a meta-analysis of published microarray datasets to show that Favorable Histology Wilms' Tumors (FHWT's) fill a triangle-shaped continuum in gene expression space of which the vertices represent three idealized “archetypes”. We show that these archetypes have predominantly renal blastemal, stromal, and epithelial characteristics and that they correlate well with the three major lineages of the developing embryonic kidney. Moreover, we show that advanced stage tumors shift towards the renal blastemal archetype. These results illustrate the potential of this methodology for characterizing the cellular composition of Wilms' tumors and for assessing disease progression.
Introduction
Wilms' tumor is a common pediatric malignancy of the kidney appearing in children under the age of five. It is thought to be caused by faulty kidney development since it typically contains disorganized and immature nephron structures that histologically resemble structures found in the nephrogenic zone—the region near the cortex of the developing embryonic kidney where new nephrons are formed. As such, Wilms' tumor is considered as a model system for studying the relationship between development and tumorigenesis [1].
To date, Wilms' tumor heterogeneity is not well understood. Wilms' tumors can vary widely from patient to patient in both histology and gene expression, and many tumors contain heterogeneous cell types. Therefore, although Wilms' tumors are generally responsive to treatment and have a relatively good prognosis, there remains a need for measures to better classify them into subtypes and assess the progression of the tumor in each patient in order to design a more personalized treatment. We therefore set out to characterize the heterogeneity of Wilms' tumors according to their geometry in gene expression space and their genomic similarity to cells within the developing kidney.
Kidney development occurs during the embryonic stage from week 5 to week 36 of gestation in humans and from day E10.5 to day 3 after birth in mice [1], [2], [3], [4], [5]. It starts as an interaction between two lineages that originate from the intermediate mesoderm: the ureteric/nephric duct—an epithelial tubular structure, and the metanephric mesenchyme—which is composed of loosely connected mesenchymal cells (Figure 1, A). Signaling interactions between these two components induce the formation of a structure that branches out of the nephric duct—the “ureteric bud”. The ureteric bud invades the metanephric mesenchyme, branches, and grows to form a tree-like structure that will later become the collecting duct system for draining urine from the nephrons into the ureter.
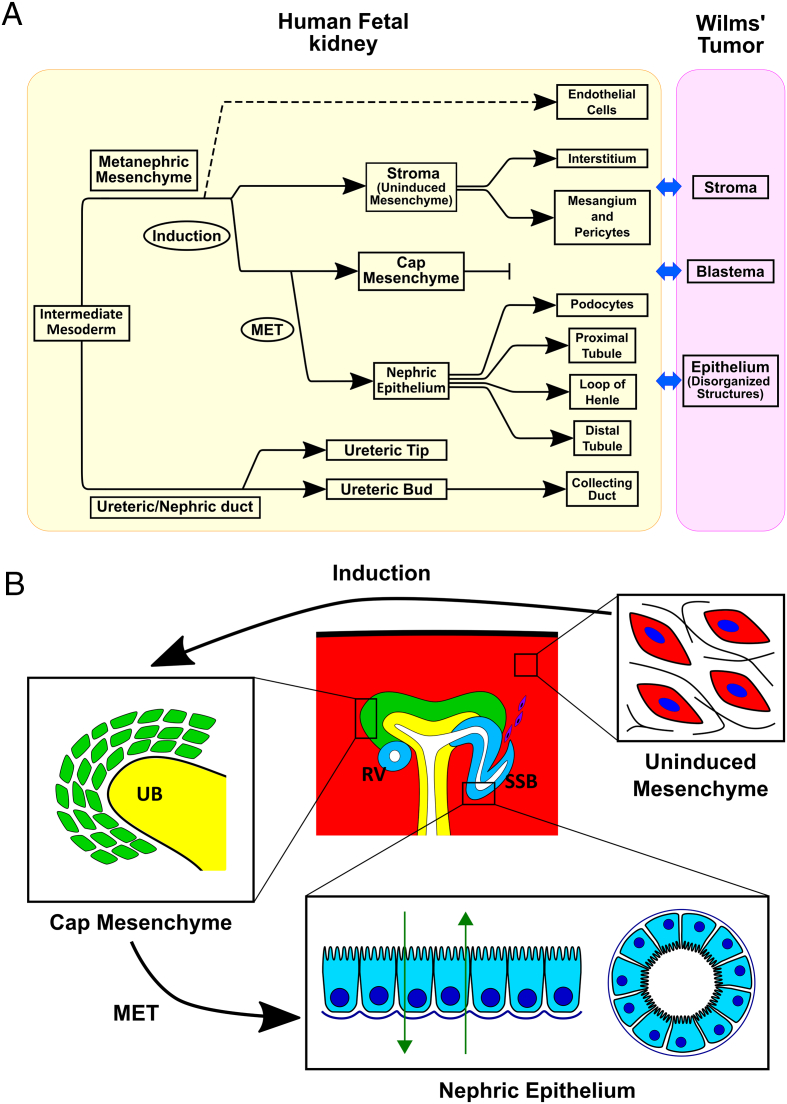
Figure 1.
Wilms' tumors typically consist of three cell types that correspond to the progenitor lineages that co-exist in the nephrogenic zone of the developing fetal kidney. (A) Shown is the developmental hierarchy of the various cell types involved in kidney development. (B) A sketch of nephron formation in the cortex of a fetal kidney with representative nephric structures (UB: Ureteric bud, RV: Renal Vesicle, SSB: S-Shaped body). Three cell types co-exist within the nephrogenic zone: (i) The un-induced metanephric mesenchyme: Mesenchymal cells that are loosely associated, have no polarity, are relatively motile, and are surrounded by an extracellular matrix. A subset of these cells are induced to condense around the ureteric tip and become the Cap mesenchyme. (ii) The Cap mesenchyme: Cells in a transient self-renewing progenitor state that in mammals normally exists only during embryonic stages and is depleted after birth. A subset of these cells undergo a Mesenchymal to Epithelial Transition (MET) and, through a series of differentiation events, differentiate to create the various tubular epithelial segments of the nephron. (iii) Early nephric epithelial structures: Epithelial cells that have a well-defined polarity, are connected by tight junctions, and create two-dimensional surfaces and tubules capable of transport (absorption and secretion). Wilms' tumor is a pediatric malignancy thought to arise from faulty kidney differentiation since it contains cell types that resemble the above three populations of the nephrogenic zone: Stromal cells that correspond to the un-induced metanephric mesenchyme, Blastemal cells that correspond to the Cap mesenchyme, and disorganized epithelial structures that correspond to the early nephric epithelium.
Nephrons, the functional units of the kidney, are formed near the tip of each branch structure (the “ureteric tip”, Figure 1, B). Each ureteric tip induces the metanephric mesenchyme in its vicinity to condense around it and form a structure called the “Cap mesenchyme”, which is the transient self-renewing progenitor population from which new nephrons will be formed. Part of the Cap mesenchyme then ceases to self-renew and undergoes a mesenchymal to epithelial transition (MET) to form early nephric epithelial structures. The earliest epithelial structure is the “pretubular aggregate”, which transforms into the “renal vesicle”, which is the first structure to exhibit a polarized gene expression patterning in what will later become the proximal and distal segments of the nephron. At the end of this stage, the renal vesicle fuses with the ureteric tip to form a single continuous tube from the nephron to the ureter. The renal vesicle elongates and segments to form a “comma-shaped body” which further develops into an “S-shaped body”. The cells at the cleft of the S-shaped body, that will later become podocytes, attract endothelial and mesangial cell progenitors (from the un-induced metanephric mesenchyme, see below) to migrate into the cleft and create the glomerular vasculature. Eventually, through a series of differentiation events, the S-shaped body elongates and creates the various epithelial tubular segments of the nephron (podocytes, proximal tubule, loop of Henle, and distal tubule).
The un-induced metanephric mesenchymal cells—those that are further from the ureteric tip—eventually differentiate into other kidney supporting cell types. These include: interstitial fibroblasts, pericytes—cells that support non-glomerular kidney vasculature, and mesangial cells—specialized pericytes that support glomerular vasculature.
Thus, the nephrogenic zone contains three interacting cell lineages with distinct structural and functional characteristics (Figure 1, B, Table 1): The un-induced mesenchyme, the Cap mesenchyme, and the early nephric epithelium. The un-induced mesenchyme is made of cells that are loosely associated, are surrounded by an extracellular matrix, and have the capability to migrate. Major genes that are over-expressed include VIM, ZEB1, TWIST1, SNAI2, CDH11, as well as genes associated with the extracellular matrix (FN1, Collagens 1, 3, and 5). The Cap mesenchyme is a more condensed structure consisting of cells in a transient self-renewing nephron-progenitor state. Major gene markers are SIX2, EYA1, CITED1, OSR1, SALL2, and also the mesenchymal marker CDH11 to some extent [6]. The early nephric epithelium is composed of epithelial cells that are tightly interconnected by junctions and are highly polarized with distinct apical and basolateral membranes, thus creating a polarized 2-dimensional surface with an in/out distinction. These structures later develop into tubular structures capable of transport (reabsorption/secretion) through the tube walls. Major gene markers include [6]: CDH1, CDH6, KRT18, PAX2, PAX8, the gene NOTCH2 (associated with nephron tubular segmentation), as well as early podocyte progenitors markers that appear in the cleft of late S-shaped bodies (PODXL, NPHS1, NPHS2, PTPRO). Signaling interactions between these lineages are responsible for maintaining the continual process of nephron generation at the embryonic stage [7].
Table 1.
Selected Genes That are Known to be Predominantly Over-Expressed in specific Cell Types of the Developing Kidney
| HUGO gene symbol | Cell type in which this gene is predominantly expressed |
|---|---|
| SIX2 | Cap mesenchyme |
| EYA1 | Cap mesenchyme |
| OSR1 | Cap mesenchyme |
| SALL2 | Cap mesenchyme |
| CITED1 | Cap mesenchyme |
| PAX2 | Kidney epithelium |
| PAX8 | Kidney epithelium |
| SLC22A6 (OAT1) | Kidney epithelium |
| CDH6 | Kidney epithelium |
| CDH1 | Kidney epithelium |
| LRP2 | Kidney epithelium |
| AQP1 | Kidney epithelium |
| SLC5A1 | Kidney epithelium |
| ANPEP | Kidney epithelium |
| NOTCH2 | Kidney epithelium |
| KRT18 | Kidney epithelium |
| PODXL | Kidney epithelium (podocytes) |
| NPHS1 | Kidney epithelium (podocytes) |
| NPHS2 | Kidney epithelium (podocytes) |
| PTPRO | Kidney epithelium (podocytes) |
| SYNPO | Kidney epithelium (podocytes) |
| COL1A1 | Uninduced mesenchyme |
| COL1A2 | Uninduced mesenchyme |
| COL3A1 | Uninduced mesenchyme |
| COL5A2 | Uninduced mesenchyme |
| FN1 | Uninduced mesenchyme |
| ZEB1 | Uninduced mesenchyme |
| TWIST1 | Uninduced mesenchyme |
| SNAI2 | Uninduced mesenchyme |
| VIM | Uninduced mesenchyme |
| WT1 | Cap mesenchyme Kidney epithelium (podocytes) |
| CDH11 | Uninduced mesenchyme Cap mesenchyme |
Similar to the fetal developing kidney, Wilms' tumors can contain three cellular components (this is referred to as “triphasic histology”). These components are of similar properties to those of the nephrogenic zone (Figure 1, A): A blastema—composed of cells resembling the Cap mesenchyme, a stroma—composed of cells resembling the un-induced mesenchyme (and sometimes other mesenchyme-derived tissues), and an epithelium composed of disorganized and dysfunctional nephric tubular components. However, whereas the process of normal kidney development is quite similar in different individuals, Wilms' tumors can differ significantly between different patients both in their histology and behavior. Understanding which types of tumors exist, as well as their unique characteristics and clinical parameters, may pave the way for understanding their cellular composition, for assessing the progression of the tumor, and for tailoring a more personalized treatment that is optimized for each type. Therefore, we set out to characterize Wilms' tumor heterogeneity by meta-analysis of large gene expression datasets.
We chose a set of microarray measurements of Wilms' tumors from hundreds of different patients that were reported in a series of studies by the Children's Oncology Group [8], [9], [10], [11] (Figure 2, A). Treating each microarray measurement from a single tumor as a data point in the “space” of gene expression, we find that the tumors create a triangle-shaped continuum rather than discrete groups. The vertices of this triangle represent three idealized cell types, or “archetypes” [12], [13]. We show that these archetypes correspond to the blastemal, stromal, and epithelial cell types that constitute Wilms' tumors, and that their gene expression profiles correlate well with that of the three major lineages of the nephrogenic zone in the developing embryonic kidney.
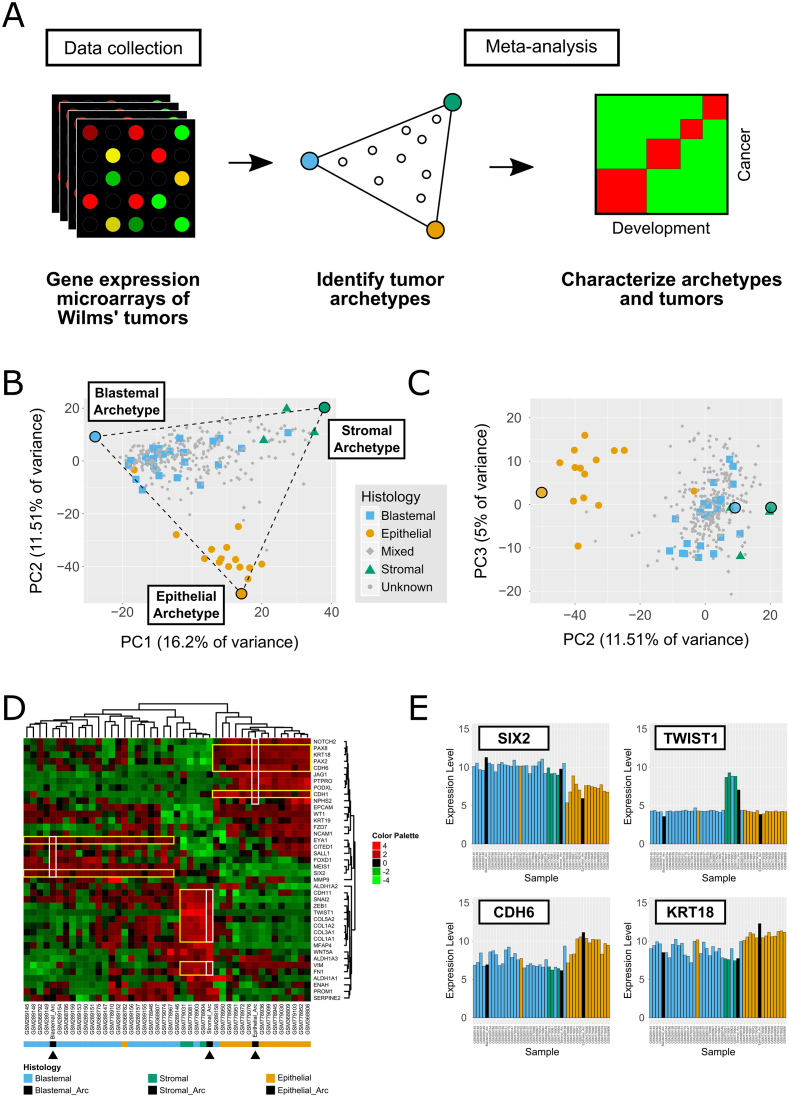
Figure 2.
Favorable Histology Wilms' Tumors (FHWT's) fill a triangle-shaped continuum in gene expression space in which the vertices represent three idealized “archetypes” with predominantly blastemal, stromal, and epithelial characteristics. (A) A sketch of the analysis workflow. (B) A PCA plot of microarray datasets from 4 studies [8], [9], [10], [11]. Each data point represents an individual Wilms' tumor. Tumors for which the histology was known are designated with a unique symbol (Epithelial: circle, Blastemal: square, Stromal: triangle, Mixed: diamond, Unknown: dot). PCA was performed on the 1000 most highly variable genes. It can be seen that tumors from different patients create a triangular-shaped continuum in PC1 and PC2. The best fitting triangle and the three archetypes were calculated using the ParTI Matlab package [12], [15]. (C) PC3 does not significantly distinguish between tumors of different known histologies. We therefore hypothesize that PC3 predominantly represents technical variability in sample collection and gene expression measurements rather than tumor phenotypic heterogeneity. (D,E) Expression analysis of selected genes confirms that the archetypes have blastemal, stromal, and epithelial phenotypes. The gene expression heatmap (D) shows 40 genes measured from Wilms' tumors with blastemal, stromal, and epithelial histologies as well as the three calculated archetypes (black arrowheads). We chose genes that are known to be associated with the three main cell lineages that are involved in kidney development and Wilms' tumors from the top 1000 most highly variable genes in all tumor samples. Each gene (=row) was standardized by subtracting the mean over all samples and dividing by the standard deviation. Hierarchical clustering was done using Pearson correlation distance and average linkage. It can be seen that the archetypes over-express gene sets associated with their respective phenotypes (Selected genes are highlighted in the heatmap with yellow and white rectangles). For example (E), Six2, a marker for the Cap-mesenchyme and blastemal Wilms' tumors [16], [23], [27], is overexpressed in the blastemal archetype. TWIST1, a marker for the uninduced mesenchyme and stromal tumors, is over-expressed in the stromal archetype. Likewise, the epithelial markers CDH6 and KRT18 are overexpressed in the epithelial archetype.
Methods
Microarray Data Preprocessing
Microarray data preprocessing was performed with the “simpleaffy” R package using the RMA procedure with default parameters. We created a gene expression table by choosing, for each gene, the probe-set with maximal mean value across all arrays using the “collapseRows” R function from the WGCNA package [14].
Archetype Analysis
Archetypes were calculated using the ParTI_lite matlab function (https://www.weizmann.ac.il/mcb/UriAlon/download/ParTI) with default parameters.
Data Analysis
PCA was performed in R using the “prcomp” function. Hierarchical clustering was performed in R using the “Heatmap” function with correlation dissimilarity and average linkage unless stated otherwise. Venn diagrams were prepared using the “VennDiagram” R package. Enrichment analysis was performed using the “clusterProfiler” R package using the functions “enrichGO”, “enrichKEGG”, and “enrichPathway”. The Kolmogorov–Smirnov test and the Wilcoxon-Mann–Whitney rank sum test were performed using the R functions “ks.test” and “wilcox.test” respectively.
A short program for data collection, preprocessing, and visualization is attached (Supplementary Information).
Results
Favorable Histology Wilms' Tumors (FHWT's) Create a Triangle-Shaped Continuum in Gene Expression Space, of Which the Vertices Represent Three Idealized “Archetypes”
To minimize experimental bias, we chose microarray datasets from a series of studies performed by the Children's Oncology Group [8], [9], [10], [11] (GEO Series accession numbers GSE31403, GSE14767, GSE11482, GSE10320). All Wilms' tumors in these studies were Favorable Histology Wilms Tumors (FHWT) and gene expression was measured using the same microarray platform (Platform identifier GPL96, HG-U133A Affymetrix Human Genome U133A Array). After removing duplicate samples we were left with 306 samples. We labeled the samples according to their reported histology metadata as “blastemal”, “stromal”, “epithelial”, “mixed”, and “unknown”.
In order to visualize the variability in gene expression space, we chose the 1000 most variable genes in our dataset and performed Principal Component Analysis (PCA). We find that PC1 and PC2 are able to distinguish between tumors of blastemal, stromal, and epithelial histologies (Figure 2, B). PC3, however, was much less capable of separating the different histologies (Figure 2, C). We therefore hypothesize that PC1 and PC2 capture most of the tumor heterogeneity while PC3 captures mostly the measurement noise, i.e., the differences between measurements due to different experimental conditions, operators, and facilities.
Next, we observed that the data points in PC1 vs. PC2 space fall within a triangle. Presumably, the three vertexes of this triangle represent idealized tumor cell types, or “archetypes” [12], [13]. We therefore performed archetype analysis using the ParTI Matlab package developed by Alon and colleagues [12], [15] to calculate the best fitting triangle that encloses as many data points as possible. This analysis was performed for the 1000 most highly variable genes and the archetypes too were presented along with the original data points in PCA space (Figure 2, B, C) (Note that the three additional archetypes did not change the PCA representation significantly). Since PC3 describes mostly the measurement noise, we ruled out higher dimensional (d > 2) polytopes such as tetrahedrons, so that the archetype analysis will not be confounded by the measurement noise. We identified three archetypes, corresponding to the three vertexes of the triangle. We labeled each archetype as the “Blastemal archetype”, “Stromal archetype”, or “Epithelial archetype”, according to the histologies of the majority of neighboring tumors. Gene expression profiles of the three archetypes can be found in Table S1.
The Three Archetypes Have Predominantly Blastemal, Stromal, and Epithelial Characteristics
In order to characterize the three archetypes, we checked the expression levels of selected genes that are known to mark the main cell lineages involved in kidney development and tumorigenesis. We chose genes that we previously found to be useful for identifying the un-induced mesenchyme, the Cap mesenchyme, and the early nephric epithelium within human fetal kidney cells and Wilms' tumor xenografts [16]. To this list, we added genes that we manually selected from the GUDMAP database [17]. We intersected the resulting list (Table 1 and Table S2) with the 1000 most variable genes that were used for identifying the three archetypes. We checked the expression levels of these genes in those tumors whose histology was labeled as “blastemal”, “stromal”, and “epithelial”, as well as in the three corresponding archetypes. Hierarchical clustering (Figure 2, D) shows that the blastemal archetype over-expressed genes known to be over-expressed in the Cap-mesenchyme (SIX2, EYA1, CITED1, SALL1), the stromal archetype over-expresses genes whose expression is characteristic of the un-induced mesenchyme (CDH11, SNAI2, ZEB1, TWIST1, FN1, VIM, and collagens 1, 3, and 5), and the epithelial archetype over-expresses genes that are over-expressed in nephric epithelium (NOTCH2, PAX2, PAX8, KRT18, CDH6, CDH1) as well as genes that are over-expressed in podocyte progenitors that are known to appear in the clefts of late S-shaped epithelial bodies (PTPRO, PODXL, NPHS2). This is consistent with bar graphs showing expression level measurements for SIX2, TWIST1, CDH6, and KRT18 (Figure 2, E).
Next, we used a more global approach to characterize each archetype. For each archetype, we created a list of genes that are over-expressed 2-fold with respect to both other archetypes, and checked for over-represented gene ontologies (Figure 3). We find that the blastemal archetype is enriched for genes involved in early differentiation of the metanephric mesenchyme and mesenchymal to epithelial transition (MET)—properties that are also typical to the Cap mesenchyme. The epithelial archetype is enriched for genes involved in creating the nephric epithelium (e.g. kidney epithelial development, glomerulus development, cell–cell junction organization) —processes that occur in the early nephric epithelial structures. Finally, the stromal archetype is enriched for genes that are typical of the un-induced mesenchyme, for example, genes involved in the formation and organization of the extracellular matrix (of which collagen is a major component), genes involved in connective tissue development, and genes related to cell motility.
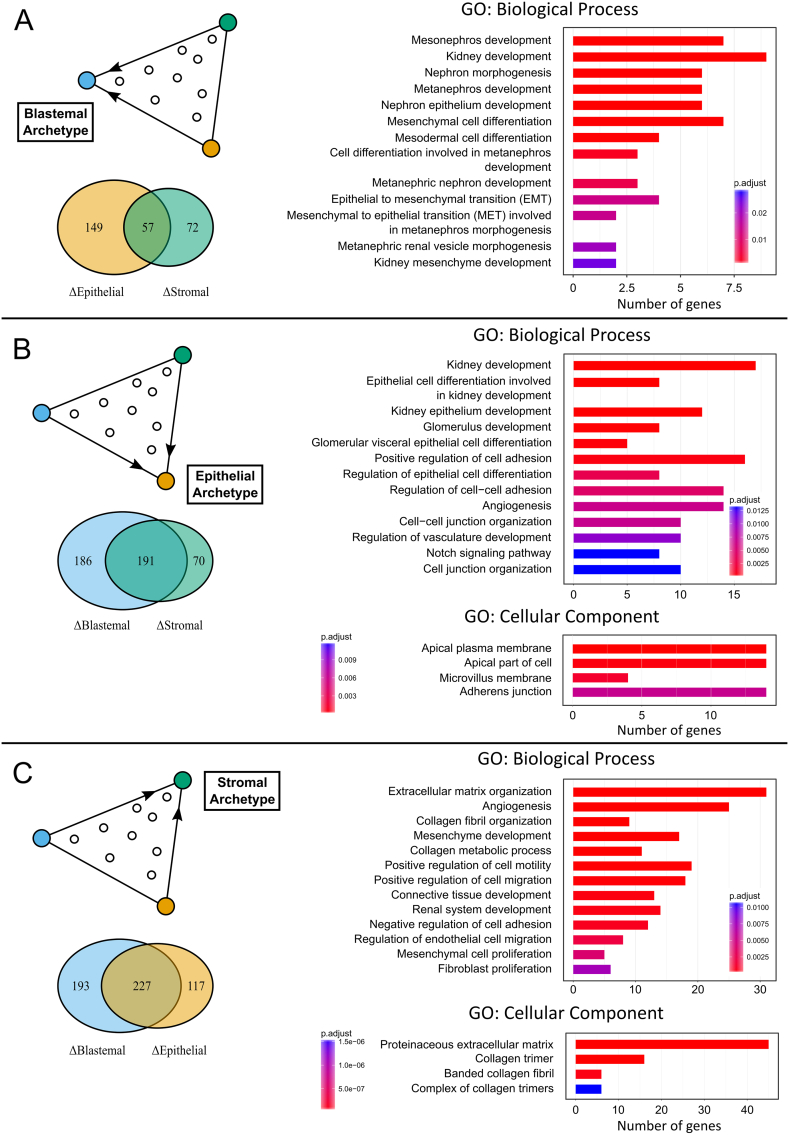
Figure 3.
The archetypes calculated from Favorable Histology Wilms' Tumors (FHWT's) expression datasets are enriched for genes that characterize the three major cell lineages that are involved in kidney development. (A) We found a set of >50 genes that are over-expressed two-fold in the blastemal archetype with respect to both the epithelial and stromal archetypes. This set is enriched for genes involved in early stages of kidney development, and in particular, the differentiation of the metanephric mesenchyme and Mesenchymal to Epithelial Transition (MET). These processes are characteristic to the Cap mesenchyme. (B) The set of >190 genes that are over-expressed two-fold in the epithelial archetype are enriched for genes involved in differentiation of the kidney epithelium (both tubular and glomerular visceral epithelia), as well as genes involved in creating junctions between cells and cell polarity (apical vs. basolateral membranes) that are characteristic of epithelial structures in the kidney. (C) The set of >220 genes that are over-expressed two-fold in the stromal archetype are enriched for genes involved in mesenchymal proliferation, maintaining an extracellular matrix, and cell migration, processes that are characteristic to the un-induced mesenchyme in the nephrogenic zone.
The Three Archetypes Correlate Well with the Three Major Lineages Found in the Nephrogenic Zone of the Developing Embryonic Kidney
We next checked how well each archetype correlates to cell types found in the nephrogenic zone of a developing kidney. Since a thorough gene expression profiling of the human embryonic kidney is lacking [16], we used selected mouse microarray datasets from the GUDMAP database [17] (www.gudmap.org). We chose samples representing major cellular components found in the nephrogenic zone: The cap mesenchyme (E15.5 Cap mesenchyme), the un-induced mesenchyme (E15.5 nephrogenic interstitium), early nephric epithelium (E12.5 Renal vesicle (RV), E15.5 S-shaped body, and E15.5 Early proximal tubule), podocytes (E15.5 renal corpuscle), and the ureteric bud (E11.5 ureteric bud). To avoid confounding effects of measurement noise, we chose datasets measured by the same microarray platform (Platform identifier GPL1261, Affymetrix Mouse Genome 430 2.0 Array).
Since the samples in the GUDMAP database were isolated using manual micro-dissection, Laser Capture Microdissection (LCM), or flow cytometry using fluorescent reporter genes in transgenic mice [17], we wanted to check to what degree they represent “pure” cell types rather than mixtures. We therefore chose a list of selected genes (Table S2) known to mark specific components during embryonic kidney development [16], [17] and performed hierarchical clustering (Figure 4, C). As expected, we find that the cap mesenchyme samples over-express the genes SIX2, SALL2, EYA1, OSR1, and CITED1. The un-induced mesenchyme over-expresses the genes VIM, FN1, COL1A1, COL1A2, COL3A1, COL5A2, TWIST1, SNAI2, and ZEB1. The epithelial components over-express the genes PROM1, EPCAM, CDH1, NOTCH2, and PAX8 in the early stages (renal vesicles and S-shaped bodies), and AQP1, SLC5A1, and SLC22A6—genes involved in reabsorption of water and organic anions—in more advanced stages (Proximal tubules). The podocytes over-express NPHS1, NPHS2, SYNPO, PTPRO, and PODXL (note that these samples consist of the whole renal corpuscle of which podocytes are a major component, but that also contain endothelial progenitors expressing the gene KDR (FLK1)). Likewise, the ureteric bud over-expresses the epithelial markers CLDN4 and CLDN8.
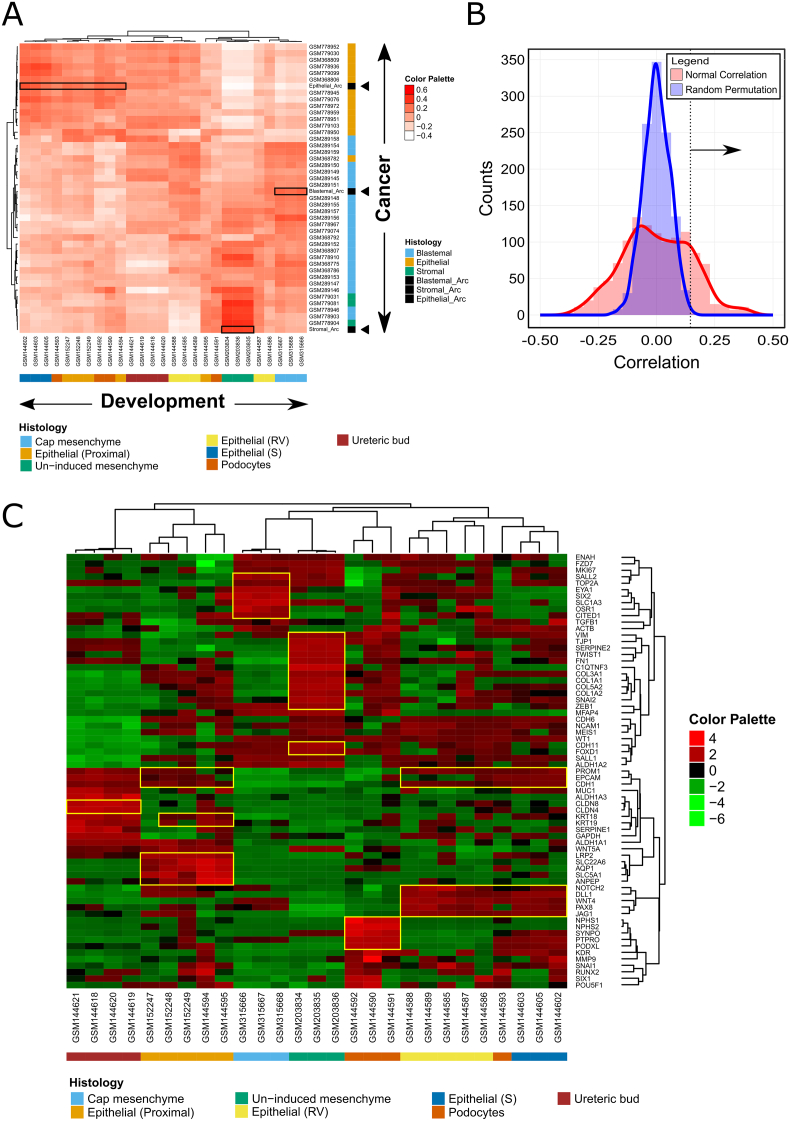
Figure 4.
The archetypes calculated from Favorable Histology Wilms' Tumors (FHWT's) expression datasets correspond to major lineages in the normal embryonic developing kidney. (A) A correlation heatmap between Wilms' tumors and selected developmental lineages from the mouse embryonic kidney. Expression profiles of Wilms' tumors with specific histologies and their respective archetypes (black arrowheads) were correlated to selected developmental lineages from the mouse embryonic kidney taken from the GUDMAP database [17], [28], [29] (http://www.gudmap.org/). For each human and mouse sample-pair we calculated the Spearman correlation using 235 genes that were obtained by intersecting the 1000 most variable genes in our Wilms' tumor samples and the 1000 most variable genes in the mouse fetal kidney samples, to which we added 70 genes known to be associated with the three main cell lineages that are involved in kidney development and tumorigenesis (Table S2). Before calculating the correlation, each gene from each human/mouse dataset was standardized by subtracting the mean and dividing by the standard deviation over all human/mouse datasets respectively. This was done in order to bring all genes into approximately the same range, since otherwise some genes are high in all samples (e.g. housekeeping genes) and others are always low (e.g. certain transcription factors). For visualization, the correlation matrix was hierarchically clustered in both dimensions using Pearson correlation distance and average linkage. We find that the blastemal archetype best correlates with the cap mesenchyme (r > 0.2), the stromal archetype best correlates with the un-induced mesenchyme (r > 0.35), and the epithelial archetype best correlates with the epithelial components (r > 0.14) such as S-shaped bodies, early proximal tubules, and developing podocytes. (B) A histogram of the correlation values from the correlation heatmap, as well as the corresponding histogram obtained from randomized datasets. Randomized datasets were obtained by randomly permuting the order of genes in each sample separately such that the distribution of expression levels are conserved. It can be seen that correlation values above 0.14 are significant, in the sense that they are unlikely to arise in randomized datasets (FDR< 0.0034). (C) An expression heatmap of 67 genes in selected mouse fetal kidney samples from selected lineages of the mouse embryonic kidney taken from the GUDMAP database. We chose genes that are known to be associated with specific cellular compartments during kidney development. Each gene (=row) was standardized by subtracting the mean over all samples and dividing by the standard deviation. Hierarchical clustering was done using Pearson correlation distance and average linkage. Selected genes are highlighted in the heatmap with yellow rectangles. Since bulk measurements sometimes contain a mixture of two or more cell types, we used this heatmap to confirm that each one of the selected mouse embryonic kidney datasets that we used represents a well-defined lineage, in the sense that it predominantly over-expresses genes that characterize a specific cell lineage.
The archetypes calculated from Favorable Histology Wilms' Tumors (FHWT's) expression datasets correspond to major lineages in the normal embryonic developing kidney. (A) A correlation heatmap between Wilms' tumors and selected developmental lineages from the mouse embryonic kidney. Expression profiles of Wilms' tumors with specific histologies and their respective archetypes (black arrowheads) were correlated to selected developmental lineages from the mouse embryonic kidney taken from the GUDMAP database [17], [28], [29] (http://www.gudmap.org/). For each human and mouse sample-pair we calculated the Spearman correlation using 235 genes that were obtained by intersecting the 1000 most variable genes in our Wilms' tumor samples and the 1000 most variable genes in the mouse fetal kidney samples, to which we added 70 genes known to be associated with the three main cell lineages that are involved in kidney development and tumorigenesis (Table S2). Before calculating the correlation, each gene from each human/mouse dataset was standardized by subtracting the mean and dividing by the standard deviation over all human/mouse datasets respectively. This was done in order to bring all genes into approximately the same range, since otherwise some genes are high in all samples (e.g. housekeeping genes) and others are always low (e.g. certain transcription factors). For visualization, the correlation matrix was hierarchically clustered in both dimensions using Pearson correlation distance and average linkage. We find that the blastemal archetype best correlates with the cap mesenchyme (r > 0.2), the stromal archetype best correlates with the un-induced mesenchyme (r > 0.35), and the epithelial archetype best correlates with the epithelial components (r > 0.14) such as S-shaped bodies, early proximal tubules, and developing podocytes. (B) A histogram of the correlation values from the correlation heatmap, as well as the corresponding histogram obtained from randomized datasets. Randomized datasets were obtained by randomly permuting the order of genes in each sample separately such that the distribution of expression levels are conserved. It can be seen that correlation values above 0.14 are significant, in the sense that they are unlikely to arise in randomized datasets (FDR< 0.0034). (C) An expression heatmap of 67 genes in selected mouse fetal kidney samples from selected lineages of the mouse embryonic kidney taken from the GUDMAP database. We chose genes that are known to be associated with specific cellular compartments during kidney development. Each gene (=row) was standardized by subtracting the mean over all samples and dividing by the standard deviation. Hierarchical clustering was done using Pearson correlation distance and average linkage. Selected genes are highlighted in the heatmap with yellow rectangles. Since bulk measurements sometimes contain a mixture of two or more cell types, we used this heatmap to confirm that each one of the selected mouse embryonic kidney datasets that we used represents a well-defined lineage, in the sense that it predominantly over-expresses genes that characterize a specific cell lineage.
Next, we measured the correlation between the datasets of the mouse fetal kidney samples and the Wilms' tumors. Again, we chose those tumors whose histology was labeled as “blastemal”, “stromal”, and “epithelial”, as well as the blastemal, stromal, and epithelial archetypes. Since the expression measurements were performed with different microarray platforms and are from different species (mouse vs. human), we chose a common list of genes by intersecting the 1000 most variable genes in our Wilms' tumor samples and the 1000 most variable genes in the mouse fetal kidney samples, to which we added a list of 70 genes that were known from previous studies to mark cell types involved in kidney development and tumorigenesis (Table S2). We calculated the correlation matrix (Figure 4, A, Figure S1) and found that the blastemal archetype best correlates with the cap mesenchyme (r > 0.2), the stromal archetype best correlates with the un-induced mesenchyme (r > 0.35), and the epithelial archetype best correlates with the epithelial components (r > 0.14), in particular, the S-shaped bodies, the proximal tubule, and podocytes (Note that podocyte markers are over-expressed in the clefts of S-shaped bodies).
In order to test for the significance of our results, we set out to reject the null hypothesis that the same correlation values can be obtained from corresponding randomized data. We therefore randomly permuted the order of genes in each human and mouse sample and recalculated the correlation matrix. We obtained values which were significantly lower in absolute value (Figure 4, B). In particular, correlation values above 0.14 were extremely rare in the randomized dataset (FDR<0.0034). We noticed that the correlation values are rather low. This is expected since although there is some resemblance, there are considerable differences between normal kidney development and Wilm's tumors, as well as differences between humans and mice [16].
Expression Profiles of Favorable Histology Wilms' Tumors with Higher Stage Tend to Localize Closer to the Blastemal Archetype in Gene Expression Space
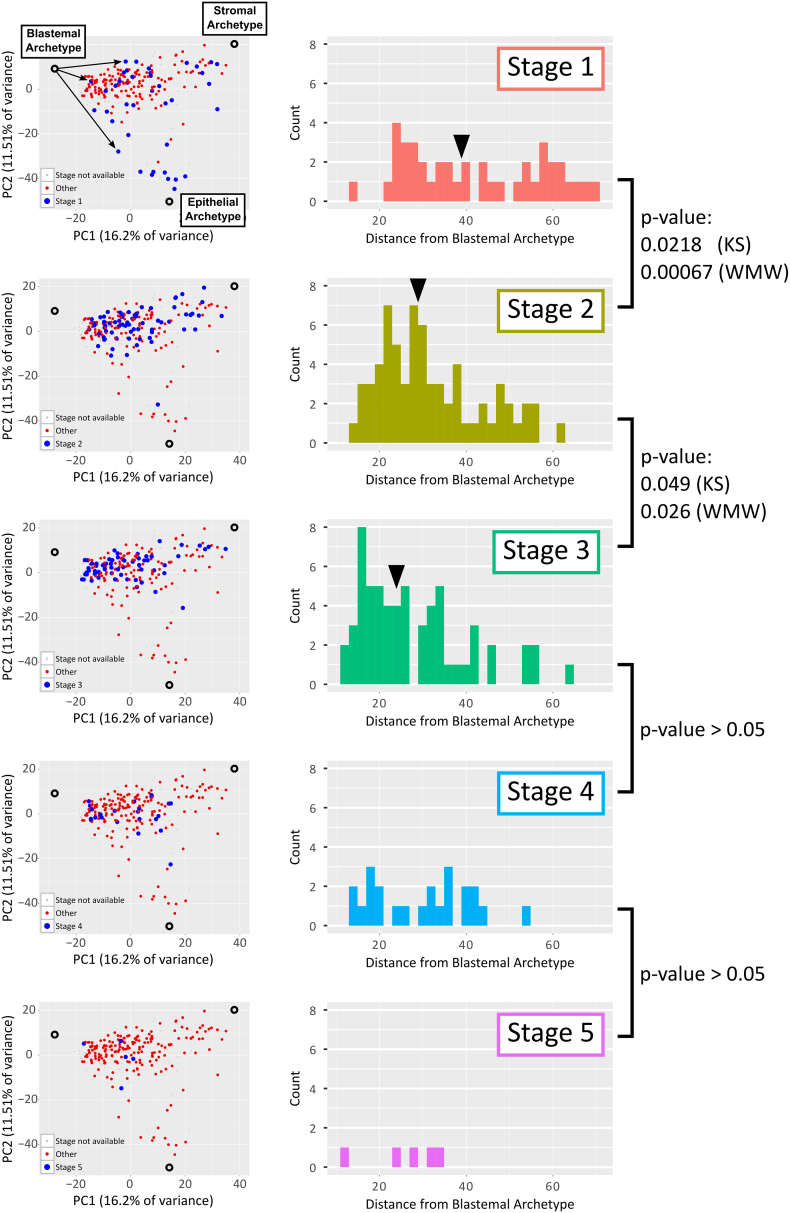
We checked if there is a relationship between clinical parameters and the localization of tumors in gene expression space. Most tumors in our datasets had three clinical parameters: Tumor stage (1,2,…,5), Patient age (months), and whether the tumor was relapsed or non-relapsed. We found that while tumors of stage 1 are spread uniformly within the triangle created by the three archetypes, tumors of higher stage tend to localize towards the blastemal archetype (Figure 5). We next measured the distance of each tumor to the blastemal archetype in principal component space and compared distributions of distances for tumors of each stage separately. We found that the distributions become more skewed towards the blastemal archetype with increasing stage, with a statistically significant difference between the distributions of stages 1, 2, and 3, for which we had a relatively large number of samples available. Likewise, the median distance from the blastemal archetype was found to decrease with increasing stage. A similar analysis of patient age and relapse/non-relapsed state visually showed a similar trend, but the differences were not statistically significant (Figures S2 and S3).
Figure 5.
Expression profiles of Favorable Histology Wilms' Tumors with higher stage tend to localize closer to the blastemal archetype in gene expression space. Shown are PCA plots highlighting tumors of selected stages (left panels, large blue circles), along with corresponding histograms of their distances to the blastemal archetype (right panels). It can be seen that, whereas tumors of stage 1 are widely distributed within the triangle created by the three archetypes, tumors from higher stages become increasingly over-represented in the vicinity of the blastemal archetype. The distance of each tumor from the blastemal archetype was calculated as the Euclidean distance in PC1 vs. PC2 space. Black arrowheads (right panels) represent the median distances from the blastemal archetype. The p-values testing for the differences between distributions were calculated using the Kolmogorov–Smirnov (KS) test and the Wilcoxon-Mann–Whitney (WMW) rank sum test. The large P values for the transitions to stages 4 and 5 can be attributed to the small number of samples available from these stages.
Discussion
Wilms' tumor is a model system for understanding the relationship between kidney development and tumorigenesis (e.g. [19], [20], [21], [22], [23]). In our paper, we show that this relationship can be used to better characterize the heterogeneity between tumors from different patients. Rather than forming distinct groups in gene expression space corresponding to discrete tumor subtypes, our analysis suggests that tumors from different patients fill a continuum whose extreme points—the “archetypes”—represent idealized cell types. We believe that this method provides a quantitative measure for characterizing the cellular composition of Wilms' tumors that is more objective and less prone to human error, and can therefore be used to complement manual pathological inspection for tumor classification. Moreover, it can be used to quantitatively characterize tumors for which the histology is unknown or not well defined, such as tumors of “mixed” histology that can contain varying proportions of different cell types.
In a recent study [16] we used single-cell qPCR to identify and characterize cell types within human fetal kidney cells cultured in mNPEM (Modified Nephron Progenitor Expansion Medium). We observed three distinct cells types: A cap-mesenchyme cell population, an un-induced mesenchyme cell population, and a nephric epithelial cell population. We also observed cells undergoing MET (Mesenchymal to Epithelial Transition) and transitioning from the cap-mesenchyme state to the early epithelial state. A similar analysis of a late blastemal Wilms' tumor xenograft revealed that it was composed mainly of two cell types: a cap-mesenchyme-like cell population and an un-induced mesenchyme-like cell population.
Our previous single-cell analysis of an individual Wilms' tumor and an individual human fetal kidney sample, together with our current microarray analysis of many (>300) tumors at the “bulk” level, suggest that the cell sub-population repertoire in Wilms' tumors is similar to that found in the developing fetal kidney. However, genetic and epigenetic distortions (which to date are not fully understood), result in imbalanced proportions of cell types [24]. As a result, each tumor contains a different mixture of the three basic cell types—represented by the three archetypes—with varying proportions: “blastemal” cells resembling the cap-mesenchyme, “stromal” cells resembling the un-induced mesenchyme, and “epithelial” cells that are similar to early nephric epithelial structures. In our present study, each “bulk” microarray measurement gives an averaged expression profile from all three cell types that co-exist within the tumor, resulting in a data point that lies somewhere in-between the three idealized archetypes. Since different tumors have different cellular mixtures, this results in a triangle-shaped continuum between the three archetypes in gene expression space. Thus, our meta-analysis of hundreds of tumors is consistent with (and complementary to) our previous single-cell analysis of hundreds of individual cells from a single tumor.
Moreover, our observation that Wilms' Tumors with higher stage tend to localize closer to the blastemal archetype conform to our previous findings in Wilms' tumor patient-derived xenografts [20], [25]. These xenografts, when propagated in immuno-deficient mice, were found to select for the subset of blastemal cells and to lose the more differentiated epithelial tubular structures with increasing passage. We hypothesize that in this sense, late passage xenografts mimic Wilms tumors of late stage.
A major challenge in stem cell biology is to identify and characterize cell types in regenerating tissues and tumors, and in particular, to find specific markers for tissue-specific progenitors and cancer stem cells. One approach is to break up the tissue or tumor into hundreds of individual cells and to measure the expression profile of each individual cell [16], [26]. Then, clustering algorithms are used to group together similar single cell profiles in order to identify and characterize all distinct cell types. This approach, however, typically requires fresh samples of tissues and tumors, which are often difficult to collect from human patients. Our study demonstrates that in certain cases we can use the geometry of gene expression space created by many “bulk” measurements from heterogeneous tumors to deconvolute their cellular heterogeneity and calculate “pure” expression profiles of the cell types from which they are composed. Moreover, once the archetypes have been well characterized, our analysis suggests that tumors might be better characterized by their distance to each archetype, which is related to their sub-population repertoire.
Conclusions
-
•
Microarray datasets of Wilms' tumors from different patients show that tumors span a continuum in gene expression space that is spread out between 3 extreme archetypes.
-
•
These archetypes correspond to 3 major cell types within the developing embryonic kidney.
-
•
Tumors with higher stage tend to localize closer to the blastemal archetype in gene expression space.
-
•
This may assist in better characterization and classification of Wilms' tumors and for assessing disease progression.
The following are the supplementary data related to this article.
Supplementary figures.
Gene expression profiles of the blastemal, stromal, and epithelial archetypes. The archetypes were calculated using the 1,000 most highly variable genes found in the combined expression datasets of Favorable Histology Wilms Tumors (FHWT) from >300 patients.
A list of selected genes that are known from the literature to be associated with the three main cell lineages that are involved in kidney development and tumorigenesis.
A compressed directory containing a short R program (and supporting files) for data collection, preprocessing, and visualization.
Acknowledgments
Acknowledgments
We wish to thank Peter Hohenstein, Uri Alon, Yael Korem, and all members of our labs for useful comments and suggestions.
Footnotes
Funding: T.K., A.T., and I.K. are supported by the Israel Science Foundation (ICORE no. 1902/12 and Grants no. 1634/13 and 2017/13), the Israel Cancer Association (Grant no. 20150911), the Israel Ministry of Health (Grant no. 3–10146), and the EU-FP7 (Marie Curie International Reintegration Grant no. 618592). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests: The authors have declared that no competing interests exist.
References
- 1.Hohenstein P, Pritchard-Jones K, Charlton J. The yin and yang of kidney development and Wilms' tumors. Genes Dev. 2015;29:467–482. doi: 10.1101/gad.256396.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert SF. Developmental Biology. Sinauer Associates; Sunderland (MA): 2000. Intermediate Mesoderm. (Available: http://www.ncbi.nlm.nih.gov/books/NBK10089/) [Google Scholar]
- 3.Little MH, McMahon AP. Mammalian kidney development: Principles, progress, and projections. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little M, Georgas K, Pennisi D, Wilkinson L. Kidney Development: Two Tales of Tubulogenesis. In: Thornhill BA, Chevalier RL, editors. Current Topics in Developmental Biology. 2010. pp. 193–229. [DOI] [PubMed] [Google Scholar]
- 5.Schell C, Wanner N, Huber TB. Vol. 36. Elsevier Ltd; 2014. Glomerular development—Shaping the multi-cellular filtration unit; pp. 39–49. (Semin Cell Dev Biol). [DOI] [PubMed] [Google Scholar]
- 6.Cho EA, Patterson LT, Brookhiser WT, Mah S, Kintner C, Dressler GR. Differential expression and function of cadherin-6 during renal epithelium development. Development. 1998;125:803–812. doi: 10.1242/dev.125.5.803. (Available: http://www.ncbi.nlm.nih.gov/pubmed/9449663) [DOI] [PubMed] [Google Scholar]
- 7.Brown AC, Deepthi S, Oxburgh L, Brown AC, Muthukrishnan SD, Oxburgh L. A Synthetic Niche for Nephron Progenitor Cells Technology A Synthetic Niche for Nephron Progenitor Cells. Dev Cell. 2015;34:229–241. doi: 10.1016/j.devcel.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadd S, Huff V, Huang C-C, Ruteshouser EC, Dome JS, Grundy PE, Breslow N, Jennings L, Green DM, Beckwith JB. Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: a Children’s Oncology Group Study. Neoplasia. 2012;14:742–756. doi: 10.1593/neo.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadd S, Sredni ST, Huang C-C, Perlman EJ. Rhabdoid tumor: gene expression clues to pathogenesis and potential therapeutic targets. Lab Invest. 2010;90:724–738. doi: 10.1038/labinvest.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang CC, Gadd S, Breslow N, Cutcliffe C, Sredni ST, Helenowski IB, Dome JS, Grundy PE, Green DM, Fritsch MK. Predicting relapse in favorable histology wilms tumor using gene expression analysis: A report from the renal tumor committee of the children’s oncology group. Clin Cancer Res. 2009;15:1770–1778. doi: 10.1158/1078-0432.CCR-08-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sredni ST, Gadd S, Huang CC, Breslow N, Grundy P, Green DM, Dome JS, Shamberger RC, Beckwith JB, Perlman EJ. Subsets of very low risk Wilms tumor show distinctive gene expression, histologic, and clinical features. Clin Cancer Res. 2009;15:6800–6809. doi: 10.1158/1078-0432.CCR-09-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart Y, Sheftel H, Hausser J, Szekely P, Ben-Moshe NB, Korem Y, Tendler A, Mayo AE, Alon U. Inferring biological tasks using Pareto analysis of high-dimensional data. Nat Methods. 2015:1–6. doi: 10.1038/nmeth.3254. [DOI] [PubMed] [Google Scholar]
- 13.Korem Y, Szekely P, Hart Y, Sheftel H, Hausser J, Mayo A, Rothenberg ME, Kalisky T, Alon U. Geometry of the Gene Expression Space of Individual Cells. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S. Strategies for aggregating gene expression data: The collapseRows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morup M, Hansen LK. Vol. 80. Elsevier; 2012. Archetypal analysis for machine learning and data mining; pp. 54–63. (Neurocomputing). [Google Scholar]
- 16.Pode-Shakked N, Gershon R, Tam G, Omer D, Gnatek Y, Kanter I, Oriel S, Katz G, Harari-Steinberg O, Kalisky T. Evidence of In Vitro Preservation of Human Nephrogenesis at the Single-Cell Level. Stem Cell Rep. 2017;9 doi: 10.1016/j.stemcr.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y. The GUDMAP database--an online resource for genitourinary research. Development. 2011;138:2845–2853. doi: 10.1242/dev.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pode-Shakked N, Shukrun R, Mark-Danieli M, Tsvetkov P, Bahar S, Pri-Chen S, Goldstein RS, Rom-Gross E, Mor Y, Fridman E. The isolation and characterization of renal cancer initiating cells from human Wilms’ tumour xenografts unveils new therapeutic targets. EMBO Mol Med. 2013;5:18–37. doi: 10.1002/emmm.201201516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekel B, Metsuyanim S, Schmidt-Ott KM, Fridman E, Jacob-Hirsch J, Simon A, Pinthus J, Mor Y, Barasch J, Amariglio N. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66:6040–6049. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- 21.Metsuyanim S, Harari-Steinberg O, Buzhor E, Omer D, Pode-Shakked N, Ben-Hur H, Halperin R, Schneider D, Dekel B. Expression of stem cell markers in the human fetal kidney. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metsuyanim S, Pode-Shakked N, Schmidt-Ott KM, Keshet G, Rechavi G, Blumental D, Dekel B. Accumulation of Malignant Renal Stem Cells Is Associated with Epigenetic Changes in Normal Renal Progenitor Genes. Stem Cells. 2008;26:1808–1817. doi: 10.1634/stemcells.2007-0322. [DOI] [PubMed] [Google Scholar]
- 23.Pode-Shakked N, Metsuyanim S, Rom-Gross E, Mor Y, Fridman E, Goldstein I, Amariglio N, Rechavi G, Keshet G, Dekel B. Developmental tumourigenesis: NCAM as a putative marker for the malignant renal stem/progenitor cell population. J Cell Mol Med. 2009;13:1792–1808. doi: 10.1111/j.1582-4934.2008.00607.x. [ http://www.ncbi.nlm.nih.gov/pubmed/20187302 (accessed December 14, 2014)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, a Leyrat A, Sim S, Okamoto J, Johnston DM, Qian D. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukrun R, Pode-Shakked N, Pleniceanu O, Omer D, Vax E, Peer E, Pri-Chen S, Jacob J, Hu Q, Harari-Steinberg O. Wilms’ tumor blastemal stem cells dedifferentiate to propagate the tumor bulk. Stem Cell Rep. 2014;3:24–33. doi: 10.1016/j.stemcr.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Brunskill EW, Potter SS, Dexheimer PJ, Salomonis N, Aronow BJ, Hong CI, Zhang T, Kopan R. Intrinsic Age-Dependent Changes and Cell-Cell Contacts Regulate Nephron Progenitor Lifespan. Dev Cell. 2015;35:49–62. doi: 10.1016/j.devcel.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 Defines and Regulates a Multipotent Self-Renewing Nephron Progenitor Population throughout Mammalian Kidney Development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 29.Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Grimmond S, McMahon AP. Atlas of Gene Expression in the Developing Kidney at Microanatomic Resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.
Gene expression profiles of the blastemal, stromal, and epithelial archetypes. The archetypes were calculated using the 1,000 most highly variable genes found in the combined expression datasets of Favorable Histology Wilms Tumors (FHWT) from >300 patients.
A list of selected genes that are known from the literature to be associated with the three main cell lineages that are involved in kidney development and tumorigenesis.
A compressed directory containing a short R program (and supporting files) for data collection, preprocessing, and visualization.