Abstract
Pancreatic ductal adenocarcinoma (PDAC) is among the deadliest cancers and overall survival rates have barely improved over the past five decades. The antimetabolite gemcitabine remains part of the standard-of-care, but shows very limited anti-tumor efficacy. Ataxia telangiectasia and Rad3-related protein (ATR), the apical kinase of the intra-S-phase DNA damage response (DDR), plays a central role in safeguarding cells from replication stress (RS) and can therefore limit the efficacy of antimetabolite drug therapies. We investigated the ability of the ATR inhibitor, AZD6738, to prevent the gemcitabine-induced intra-S-phase checkpoint activation and evaluated the anti-tumor potential of this combination in vitro and in vivo. In PDAC cell lines, AZD6738 inhibited gemcitabine-induced Chk1 activation, prevented cell cycle arrest and restrained RRM2 accumulation, leading to the strong induction of replication stress markers only with the combination. Moreover, synergistic growth inhibition was identified in a panel of 5 mouse and 7 human PDAC cell lines using both Bliss Independence and Loewe models. In clonogenic assays, the combination abrogated survival at concentrations for which single agents had minor effects. In vivo, AZD6738 in combination with gemcitabine was well tolerated and induced tumor regression in a subcutaneous allograft model of a KrasG12D; Trp53R172H; Pdx-Cre (KPC) mouse cancer cell line, significantly extending survival. Remarkably, the combination also induced regression of a subgroup of KPC autochthonous tumors, which generally do not respond well to conventional chemotherapy. Altogether, our data suggest that AZD6738 in combination with gemcitabine merits evaluation in a clinical trial in patients with PDAC.
Keywords: PDAC, Replication Stress, Combination Therapy
Introduction
With a mortality rate that nearly matches its incidence rate, pancreatic ductal adenocarcinoma (PDAC) is currently the 4th most common cause of cancer-death and is projected to become the 2nd leading cause by 2030(1). Late diagnosis, metastatic behavior and therapeutic resistance contribute to this poor prognosis and a 5-year survival rate of <8%. The antimetabolite gemcitabine, a nucleoside analogue that induces replication-associated DNA damage, was the first drug approved for this disease. In recent years, two combinations have been approved after showing significant benefit in clinical trials, FOLFIRINOX (folinic acid, fluorouracil, irinotecan, and oxaliplatin) and Nab-paclitaxel in combination with gemcitabine, but median survival remains less than 12 months for patients with PDAC(2,3). Also, both are cytotoxic-based therapies that many patients cannot tolerate. Intrinsic and acquired resistance limit the efficacy of these non-specific therapies and warrant the search for better, more targeted options.
Targeting the replication stress (RS) response is of particular interest because it has the potential to specifically kill cancer cells as well as enhance sensitivity to chemotherapies(4). Indeed, RS, which can be broadly defined as the perturbation of replication fork progression and stalling during S-phase, has emerged in recent years as one of the hallmarks of cancer(5). Although RS is inherent to DNA synthesis and can therefore affect normal replicative cells, it is particularly elevated in cancer cells for two reasons. First, the imposed and unrestrained proliferation underlying carcinogenesis greatly challenges the highly ordered nature of DNA replication, increasing reliance on RS pathways. Second, increased mutagenic exposure to extrinsic factors or by altered DNA repair mechanisms, which are common routes to transformation, result in DNA lesions impeding DNA synthesis. Reciprocally, persistent RS is thought to fuel genomic instability(6).
The need to tolerate high levels of RS is of particular relevance to PDAC biology. Indeed, canonical genetic alterations observed in human PDAC converge to promote cell cycle entry through KRAS activating mutations (90%), MYC amplification (14%), cyclins D and E amplification (40%), while G1/S cell cycle checkpoints are abrogated by deletion or inactivating mutation of tumor suppressor genes such as TP53 (75%) and CDKN2A (46%)(7–9). Besides alterations in canonical pathways, large scale sequencing efforts have recently uncovered the high frequency (>35% of cases) of mutations affecting DDR genes and their association with poor outcome in PDAC(9). This is in line with germline mutations in some DDR genes promoting familial pancreatic cancers and known interactions between altered DDR pathways and mutagenic exposure increasing the risk of developing PDAC(10). In addition, the high degree of genome instability frequently observed in PDAC is arguably a signature of an underlying defect in repair genes(11). Importantly, while loss of certain DDR components participates in generating high levels of RS, it also confers vulnerabilities in the form of synthetic lethality.
ATR is the apical kinase of the replication checkpoint and plays a critical role in safeguarding genome integrity from RS(4,5,12). ATR achieves this by inhibiting origin firing, preventing replication fork collapse and favoring the repair and restart of damaged forks. The global effect of ATR activation on cell cycle progression is mediated by its downstream target Chk1, leading to inhibition of Cdk2 in S-phase and of Cdk1 in G2. Meanwhile local effects are mediated by regulation of fork-repair/fork-remodeling proteins. In addition, ATR activation also helps sustain DNA synthesis by increasing dNTP pools through ribonucleotide reductase M2 (RRM2) upregulation(13,14). Activation of several oncogenes plus the inactivation of many tumor suppressors has been found to confer sensitivity to ATR inhibitors(15). Furthermore, because the RS response is an intrinsic resistance mechanism to chemotherapy, targeting the pathway is a promising strategy to sensitize cancer cells to cytotoxic drugs. Indeed, ATR inhibition has been shown to increase sensitivity to gemcitabine and radiation in vitro(16–18). Moreover, targeting Chk1, the downstream ATR effector has been shown to enhance gemcitabine anti-tumor activity in vitro and in vivo(19,20).
We describe the evaluation of the sensitivity of a panel of pancreatic cancer cell lines to the ATR inhibitor, AZD6738 (U.S. patent No. US 8,552,004 B2, Example 2.02) (21,22), alone or in combination with gemcitabine. We also investigated the ability of low concentrations of gemcitabine to sensitize cells to AZD6738, including the most resistant cell lines, and characterized the features and mechanisms of the apparent synergy. Finally, our in vivo studies demonstrate the anti-tumor effect of AZD6738 in combination with gemcitabine and provide a rational for a clinical trial to treat patients with PDAC.
Materials and Methods
Cell culture and chemicals
Human pancreatic cancer cells (MIA PaCa-2, Panc-1, SW1990, Capan-1, AsPC-1, HPAF-II, Capan-2), were obtained from either the European Collection of Cell Cultures or the ATCC. They were authenticated using either Promega GenePrint10 System or Promega PowerPlex 16HS kit and were grown in DMEM with 10% FBS (GIBCO). Murine pancreatic cancer cells K8484, DT8082, TB31456, TB32048 were previously established from KRasG12D; p53R172H; Pdx1-Cre mice and were grown in DMEM with 10% FBS(23). KPCFT79653 was from a KrasG12D; Trp53R270H; Brca2Tr/Δ11; Pdx-Cre (KPCB) mouse(24). All cell lines were grown up to a maximum of 20 passages and for fewer than 6 months following resuscitation. They were routinely verified to be mycoplasma-free using the Mycoprobe Mycoplasma Detection Kit (R&D Systems). Gemcitabine hydrochloride (Tocris), AZD6738 (AstraZeneca), were dissolved in DMSO, kept at -20°C, and used within 3 months. Final DMSO concentrations (0.2%) were kept constant in all experiments. Yoyo-3 (Life Technologies) was used at a final concentration of 0.1 mM.
SRB assay
Cells were grown in 96-well plates in their respective conditions for 72 or 120 hours. Medium was then aspirated and the cells washed once with phosphate-buffered saline (PBS), before the addition of 100 µL of 3% trichloroacetic acid (TCA) to each well. Following incubation at 4°C for 1 hour, the TCA was removed and each well was washed once with 100 µL of cold water. 50 µL of 0.057% Sulforhodamine B solution (Sigma-Aldrich, #230162-5G) in 1% acetic acid was added to each well to allow staining at room temperature for 30 minutes. Plates were rinsed four times with 1% acetic acid using an automated plate washer (Biotek Elx405 Select CW). After drying, 200 µL of 10 mM Tris base solution (pH 10.5) was added to each well and the solubilisation of protein-bound dye was achieved by placing plates on a gyratory shaker for 5 minutes. The SRB signal was measured at excitation and emission wavelengths of 488 nm and 585 nm respectively, using a PHERAstar FS plate reader (BMG Labtech). The fluorometric signal values were used as surrogates for cell number. Pharmacological growth rate inhibition was assessed using the GR metrics R package developed by Clark et al., 2017 (www.grcalculator.org). The metrics were based on a calculation that compares the cell number at the start and at the end of the assay in the absence of drug, to that in the presence of drug. This calculated a ratio between growth rates under treated and untreated conditions, normalized to a single doubling(25,26).
Clonogenic assay
Cells were plated at 300 cells/well in 6-well plates 5 hours prior to treatment. After 24 hours of treatment, the medium was replaced and the cells left to grow for 8 more days after the washout. Cells were then fixed and stained with SRB (as above). Colonies were imaged and quantified using the GelCount (Oxford Optronix). Plating efficiency was calculated from the ratio of the number of colonies to the number of cells seeded. The number of colonies that arose after treatment was expressed as surviving fraction. This was derived from the ratio of the number of colonies formed after treatment to the number of cells seeded multiplied by plating efficiency of the control.
IncuCyte time lapse imaging
Images were acquired with the IncuCyte Live Cell Imaging microscopy (Essen Bioscience) at every 3 hours under cell culture conditions with 10 objective. Averaged cell confluence was calculated from 3 random fields of view per well using the IncuCyte in-built algorithm. Cell death was measured by adding YOYO™-3 Iodide cell-impermeant dye to appropriate medium.
AZD6738 PK modeling
In vivo AZD6738 pharmaco-kinetics were simulated with the following standard two-compartmental model, reproducing gut, central, and peripheral clearance(27).
Model parameters are the ones reported(27) and drug concentration in the plasma is given by Simulations of AZD6738 dynamics were performed using the commercial software Matlab.
Immunostaining and immunoblotting
For immunostaining, cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, stained with antibodies, and counterstained with 4’,6-diamidino-2-phenylindole (DAPI). For immunoblotting, whole cell extracts were obtained by lysis in 50mM Tris-HCl PH7.5, 2% SDS, protease inhibitor, phosphatase inhibitors. Proteins were resolved using the SDS-PAGE gel system (Life Technologies). Blots were imaged and analyzed using the Odyssey Infrared Imaging System (LI-COR).
Antibodies
Primary antibodies used were from Cell Signalling unless otherwise mentioned: β-actin (Abcam #ab6276), cleaved PARP (#5625), CDK1 Y15 (#9111), CHK1 (#2360), CHK1 S345 (#2348), H2AX (#7631), H2AX S139 (Millipore #05-636), H3 (#9715), H3 S10 (#3377), RRM2 (ABNOVA #H00006241-M01), ATR (Santa Cruz #1887), ATR T1989 (Gene Tex #GTX128145), CHK2 (#2662), CHK2 T68 (Abcam #3501), KAP1 (Abcam #10484), KAP1 S824 (Abcam #133440), DNA-PKcs (Abcam #70250), DNA-PKcs S2056 (Abcam #18192), ATM (Abcam #78), ATM S1981 (Abcam #81292), RPA32 (Abcam #2175), RPA32 S4/8 (Bethyl Laboratories #A300-245A), RPA32 S33 (Bethyl Laboratories #A300-246A).
For secondary antibodies, Alexa 488 (#4408, #4412) and Alexa 647 (#4410, #4414) from Cell Signalling were used in immunostaining. IRDye800-conjugated (#925-32210, #926-33210) and IR680-conjugated (#926-68070, #926-68021) antibodies from LICOR were used in immunoblotting.
In vivo studies
All mouse experiments were carried out in the CRUK Cambridge Institute BRU, in accordance with the UK Animals (Scientific Procedures) Act 1986, with approval from the CRUK Cambridge Institute Animal Ethical Review and Welfare Body. Subcutaneous allografts of K8484 cells were conceived by implanting 106 cells in PBS, in the right flank of 7-12-week old female PC mice. For efficacy study, mice with established tumor (Average 374 mm3) were randomized and treated accordingly. Gemcitabine (LKT Laboratories, from Cambridge Bioscience) was dissolved in a saline solution (Vetivex) at 20 mg/mL and given to mice at 100 mg/kg intraperitoneally. AZD6738 (AstraZeneca) was dissolved at 2.5 mg/mL in 10% DMSO, 40% Propylene Glycol, 50% de-ionized sterile water and given to mice at 25 mg/kg by oral gavage.
PDAC was detected via ultrasound in KPC mice using a Vevo2100 imaging system (Visual Sonics, Toronto, Canada). Mice with 3 to 6 mm tumors were recruited for the efficacy study. Tumor diameter at each time point in the KPC study was calculated in a blinded manner using Vevo 2100 v1.5.0 by viewing ultrasound scans of the tumour in 3D mode, identifying in the z plane the widest part of the tumour then measuring the longest diameter at that point.
Quantitative fluorescence-based microscopy
Fixed and immuno-fluorescently labelled cells were imaged using the iCys laser scanning cytometer (CompuCyte) (40X objective). Analytical data were generated using the iCys Cytometric Analysis Software based on the acquired images.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were immunostained after heat-induced epitope retrieval by sodium citrate at 100°C for 10-20 minutes, using Bond Polymer Refine Detection kit on the automated Bond system according to manufacturer’s instructions (Leica). Slides were mounted using Leica CV5030 Coverslipper Workstation and scanned using a ScanScopeXT (Aperio Technologies). Quantification was performed using the ImageScope (Aperio Technologies).
Results
Genetic alterations driving RS in PDAC and sensitivity to AZD6738
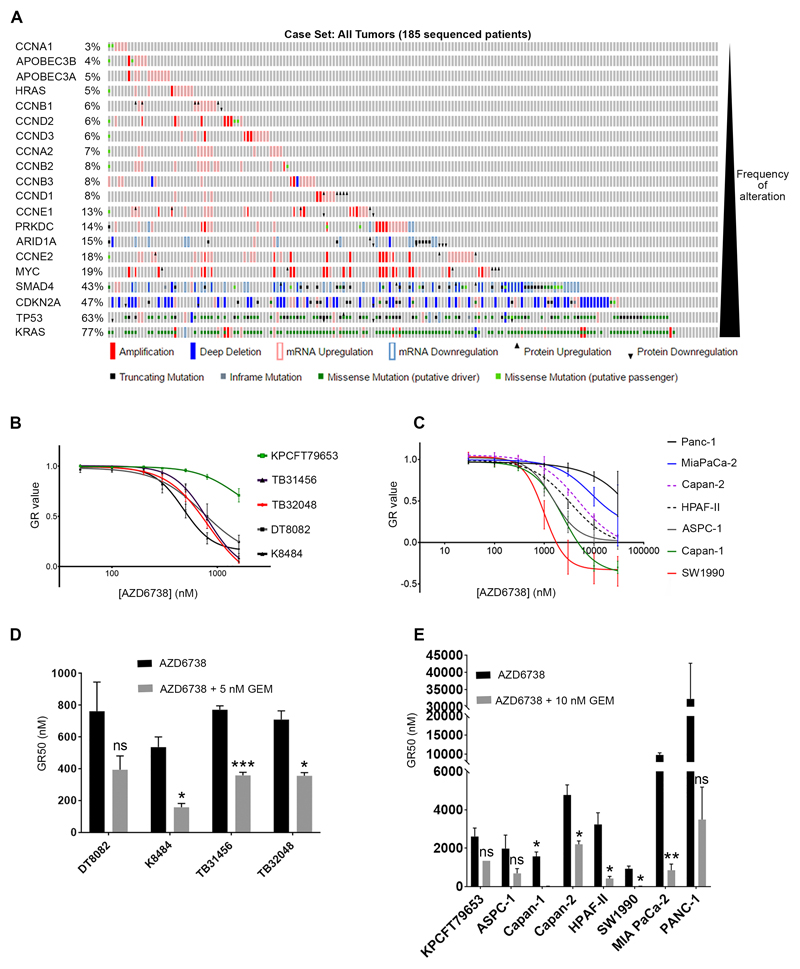
A number of studies, in various experimental settings in vitro and in vivo, have identified genetic alterations that increase the sensitivity to either downregulation or inhibition of ATR(15,28–33). We sought to interrogate this expanding list of genes, with respect to the frequency of their alteration in the complex genomic landscape of PDAC. For this purpose we explored the TCGA cohort using cBioPortal (http://www.cbioportal.org), also interrogating the database for alterations in genes likely to increase RS. Our analyses revealed that ~93% of PDAC samples exhibit at least one genetic alteration likely to sensitize to ATR inhibition (4% only one, 21% two and 68% three or more), validating this pathway as a potential therapeutic target (Fig. 1A).
Figure 1. PDAC sensitivity to AZD6738 monotherapy.
A, Frequency of alterations in genes reported to confer sensitivity to ATR inhibition, from the TCGA cohort of PDAC samples (http://www.cbioportal.org). B, C, human (B) and mouse (C) PDAC cell lines were grown in increasing concentrations of AZD6738 for 3 days to generate dose response curves using the SRB assay. The GR metrics R package (developed by Clarke et al., 2017) was used to generate normalized growth rate inhibition (GR) values, thus controlling for the different doubling times of the cell lines. Drug concentrations that bring the GR value to zero are cytostatic, while negative GR values represent cytotoxicity. Each point represents the mean of three independent experiments, +/- SEM. D, E, Comparison of GR50s for AZD6738 alone or in combination with low dose of gemcitabine (5 nM for the most sensitive mouse PDAC lines (D) or 10 nM for KPCFT79653 and human lines (E)). Data are represented as mean +/- SEM. Multiple t-tests were performed and statistical significance determined using the Holm-Sidak method **p≤0.01, ***p≤0.001, ns p>0.05.
We next assessed the growth inhibitory effect of AZD6738, a novel ATR inhibitor, using the SRB assay(34). In order to capture some of the genetic heterogeneity observed in PDAC, we tested AZD6738 in a panel of seven human cell lines (Supplementary Table 1), four lines from the KrasG12D; Trp53R172H; Pdx-Cre (KPC) mouse(35) and one line from a KrasG12D; Trp53R270H; Brca2Tr/Δ11; Pdx-Cre (KPCB) mouse(24). The growth rate (GR) metrics R package developed by Clarke et al. (www.grcalculator.org), was used to generate normalized GR inhibition values, thus controlling for the different doubling times of the cell lines(25,26). We observed that the 4 lines from the KPC model were very sensitive to AZD6738 with a submicromolar GR50, in line with our data mining (Fig. 1B). This degree of sensitivity has been shown to confer anti-tumor efficacy to AZD6738 monotherapy in human LoVo colorectal adenocarcinoma xenografts(21,36). Three of the human lines, SW1990 (GR50=0.94 μM), Capan-1 (GR50=1.57 μM) and AsPC-1 (GR50=1.98 μM), showed moderate sensitivity (Fig. 1C). Four human cell lines, HPAF-II (GR50=3.2 μM), Capan-2 (GR50=4.8 μM), MiaPaCa-2 (GR50=9.8 μM), Panc-1 (GR50=32.3 μM) and the mouse line KPCFT79653 (GR50=2.6 μM) from the KPCB model, exhibited various degrees of resistance.
As even for the most sensitive lines, GR90 values ranged from 1.4 μM (TB32048) to 1.97 μM (SW1990), we reasoned that achieving anti-tumor efficacy in vivo using AZD6738 monotherapy, would be unlikely for the majority of PDAC. Therefore, we sought to utilize gemcitabine to increase RS to sensitize PDAC cell lines to AZD6738 and also to overcome intrinsic gemcitabine resistance. Low concentrations of gemcitabine greatly reduced the GR50 of all cell lines, particularly the more resistant lines, demonstrating sensitization to AZD6738 by gemcitabine (Fig 1D, E).
AZD6738 prevents the checkpoint activation elicited by gemcitabine
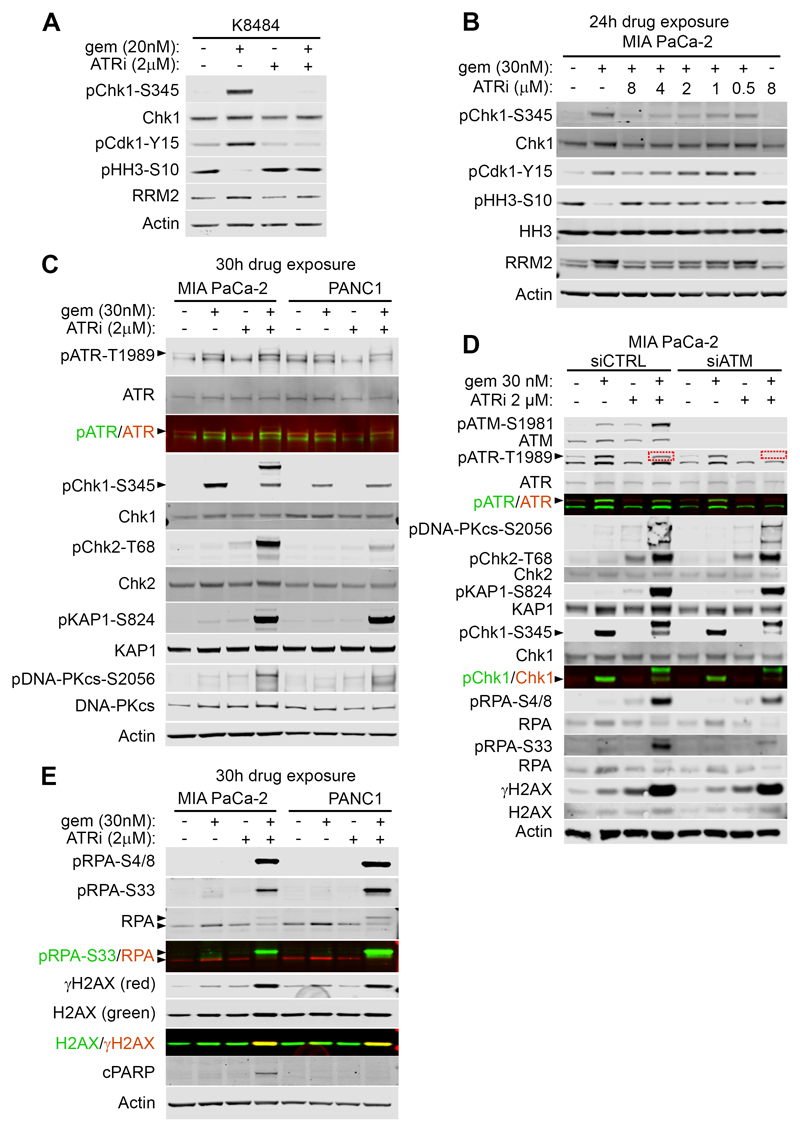
We next investigated whether AZD6738 could counteract the checkpoint activation elicited by gemcitabine. Importantly, in K8484 cells, AZD6738 at 2 µM completely prevented gemcitabine-induced Chk1 phosphorylation on Serine 345, the downstream ATR target (Fig. 2A). AZD6738 abrogated gemcitabine-induced inhibition of Cdk1, assessed by the inhibitory tyrosine 15 phosphorylation, and restored the mitotic marker Histone H3 phospho-Serine10 to normal levels (Fig. 2A). Finally, AZD6738 could prevent accumulation of RRM2 induced by gemcitabine (Fig. 2A). In the five mouse cell lines, AZD6738 at 0.5 μM also prevented gemcitabine-induced Chk1 phosphorylation on Ser345, leading to a strong induction of DNA damage, as visualized by γH2AX (Supplementary Fig.S1A). We also observed a potent abrogation of gemcitabine-induced Chk1 activation in the sensitive human lines SW1990 and AsPC-1(Supplementary Fig.S1B, C). We next explored the pathway abrogation by AZD6738 in the more AZD6738-resistant lines, MiaPaCa-2 and Panc-1. Although partial inhibition of the gemcitabine-induced checkpoint activation was detectable using AZD6738 at 0.5 μM, we found that a higher concentration was necessary to fully block the pathway in both cell lines (Fig. 2B and Supplementary Fig.S1D). To explore if the need for higher concentrations of AZD6738 was due to compensatory pathway activation in these cells, as opposed to lack of ATR target engagement, we probed for ATM and DNA-PK activation. We found that the combination of AZD6738 + gemcitabine strongly activated ATM downstream effectors, pChk2-Threonine 68, and pKap1-Serine 824, as well as auto-phosphorylation site Serine 1981 (Fig. 2C, D). DNA-PKcs phosphorylation on Serine 2056 was also strongly induced, only by the combination (Fig. 2C). Phosphorylation of RPA on S4/8, a consensus site for DNA-PK was also activated, plus S33 which is a consensus site for ATR, ATM, DNA-PK(37) (Fig. 2D)
Figure 2. AZD6738 abrogates the gemcitabine-induced checkpoint activation.
A, Western blotting for K8484 cells treated as indicated for 7 hours. B, Western blotting for MiaPaCa-2 cells treated as indicated for 24 hours. C, Western blotting for MiaPaCa-2 and Panc-1 cells treated as indicated for 30 hours. D, MiaPaCa-2 cells were treated with scramble or ATM-specific siRNA. After 24h cells were treated as indicated for 24h and lysates were immunoblotted. E, Western blotting for MiaPaCa-2 and Panc-1 cells treated as indicated for 30 hours. Arrowheads indicate the bands of interest. Note that the upper band in the pChk1-S345 blots appearing in lysates of MiaPaCa-2 treated with the combination is most likely the result of a cross-reaction with p-Chk2.
This potent activation of ATM and DNA-PK by the combination strongly suggested an efficient ‘on target’ inhibition of ATR by AZD6738, and attempts by the cells to compensate. In order to confirm this hypothesis, we knocked-down ATM using siRNA in MiaPaCa-2 and repeated the drug exposures (Fig. 2D). Interestingly, gemcitabine-induced phosphorylation of ATR on Threonine 1989 was completely prevented by AZD6738 at 2 μM, only in the ATM knockdown condition (Fig. 2D, red dotted boxes). This suggested that the phosphorylation of Threonine 1989 on ATR in the presence of the combination was arising through ATM activity (ATM likely being activated as a consequence of fork collapse), and not through ATR auto-phosphorylation. This strongly indicated that AZD6738 at 2 µM was sufficient to completely prevent ATR activation.
We then assessed the RS generated by the combination. In MiaPaCa-2 and Panc-1, only the combination induced a strong phosphorylation of pRPA and γH2AX, indicating a strong induction of RS in these cells (Fig. 2E)(38). We could also detect apoptosis through cleaved PARP-1 in MiaPaCa-2, only with the combination, indicating induction of apoptosis even as early as 30h after drug exposure.
AZD6738 and gemcitabine synergize to inhibit cell growth
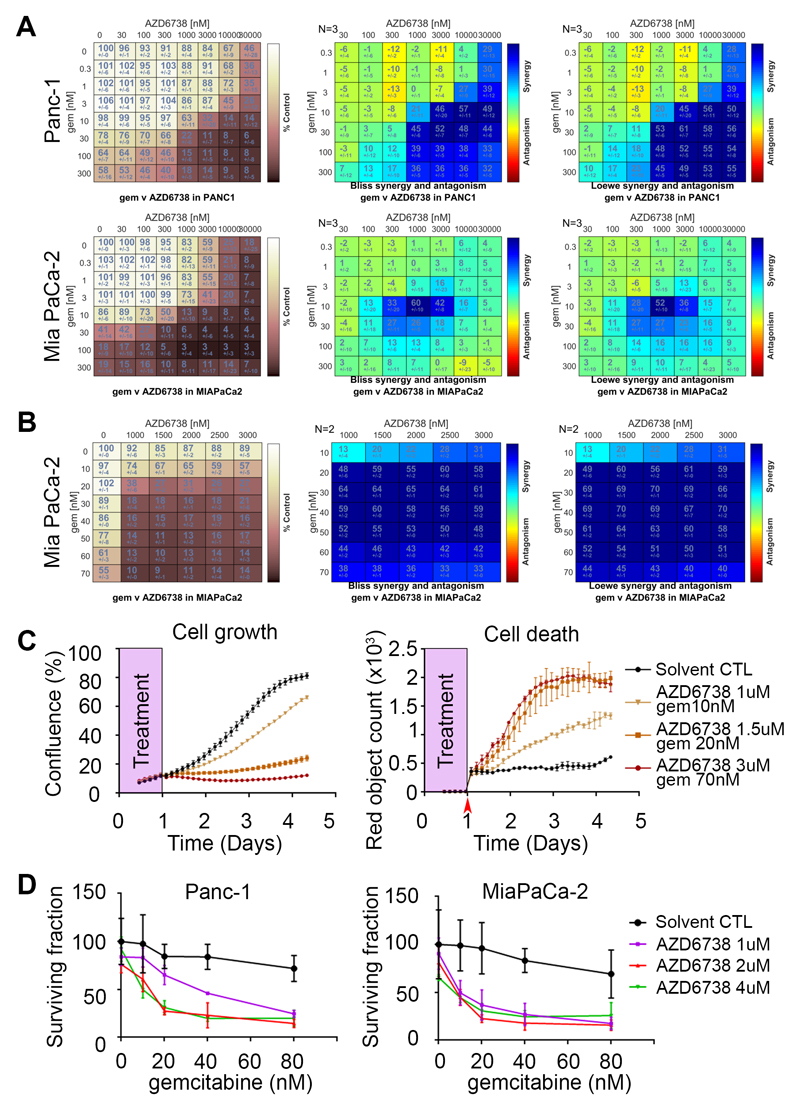
In order to analyze the degree of growth inhibition induced by the combination and identify areas of synergy across a wide range of concentrations, we used the SRB assay and calculated Loewe and Bliss drug synergy scores using Combenefit software(39). We identified areas of synergy in the 7 human and the 5 mouse PDAC cell lines (Fig. 3A and Supplementary Fig. S2). We used Metrics available within Combenefit to summarize and compare the features of synergy across the different cell lines (Table 1). As expected, the sum of synergy and antagonism was positive for all cell lines, attesting the overall synergy. Importantly, both the Loewe and the Bliss models similarly ranked the combination across the cell lines. Due to the differential sensitivity to single agents between mouse and human cells, we used 2 different concentration ranges, precluding direct comparison between the two species in the Metrics analyses. While few differences appeared in the mouse cells, it is noteworthy that Panc-1 cells, very resistant to both drugs alone, exhibited the highest degree and extent of synergy and were successfully sensitized by the combination. Similarly, the combination displayed high levels of synergy in MiaPaCa-2 cells. In contrast with the variability in the overall amount of synergy, the distribution of the observed synergy in the concentration space was relatively similar.
Figure 3. AZD6738 and gemcitabine synergistically inhibit cell growth in a panel of PDAC cell lines.
A, MiaPaCa-2 and Panc-1 cells were treated with AZD6738 and gemcitabine in an 8x8 concentration grid for 72 hours. Cell viability was determined by measuring the total protein content using the sulforhodamine B assay. The experimental data (left, values are percentage growth inhibition compared with control) were analyzed independently with the two synergy models (Bliss and Loewe) using the Combenefit software. Data, mean +/- SD, n = 3. B, C, MiaPaCa-2 cells were treated with AZD6738 and gemcitabine in an 6x8 concentration grid for 24 hours. The drugs were washed out and replaced with fresh medium containing the YOYO™-3 Iodide cell-impermeant dye to follow cell death. Three random fields per sample were imaged by time lapse microscopy every 3 hours for 4 days. Cell growth was analyzed using cell confluency data and expressed as percentage of growth inhibition compared with control (B, left panel) and Combenefit software for synergy analyses (B, 2 right panels). Growth curves (C, left panel) and cell death accumulation (C, right panel) from control cells or cells treated with indicated concentrations of AZD6738 and gemcitabine. Data, mean +/- SEM, n = 2. D, Clonogenic survival of Panc-1 and MiaPaCa-2 cells plated at very low density and exposed to the indicated drug combinations for 24 hours before washout. Cells were left to grow for 8 days after washout. Data, mean +/- SD, n = 3.
Table 1. Summary of AZD6738 sensitivity and synergy with gemcitabine across the panel of PDAC cell lines.
GR50 for all cell lines were obtained using the GR metrics R package (developed by Clarke et al., 2017). Mouse and human lines are ranked by increasing GR50 for AZD6738. Summing of synergy and antagonism was employed to compare combinations using Metrics obtained from Combenefit. Both Bliss, Loewe models and the average (AVG) is shown. SUM_SYN_ANT is the sum of synergy and antagonism observed in concentration logarithmic space. For instance, an integrated synergy of 50 is equivalent to an extra synergistic effect of 50% which is spread over a square of 1 log x 1 log in the 2-d log-concentration space. Both antagonistic and synergistic effects are considered in this metric. The integrated weighted synergy SUM_SYN_ANT_WEIGHTED incorporates a weight based on the dose response which bias the total score towards synergy achieving highest effect. Hence, a synergy of 50% leading to a combined full effect (100%) will have more weight than if the corresponding effect was only 20 or 30%. Both antagonistic and synergistic effects are considered in this metric. SYN_MAX is the maximum level of synergy observed. SYN_SPREAD is a measure of synergy spread in the logarithmic concentration space.
| AZD6738 GR50 | SUM_SYN_ANT | SUM_SYN_ANT_WEIGHTED | SYN_MAX | SYN_SPREAD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| without gem | with gem | LOEWE | BLISS | AVG | LOEWE | BLISS | AVG | LOEWE | BLISS | AVG | LOEWE | BLISS | AVG | |||
| MOUSE LINES | K8484 | 536 | 158 | gem 5nM | 5 | 11 | 8 | 4 | 8 | 6 | 11 | 32 | 21 | 0.80 | 0.62 | 0.71 |
| TB32048 | 710 | 355 | 11 | 22 | 17 | 9 | 15 | 12 | 23 | 37 | 30 | 0.80 | 0.79 | 0.79 | ||
| DT8082 | 763 | 395 | 0 | 8 | 4 | 0 | 5 | 3 | 23 | 23 | 23 | 0.52 | 0.66 | 0.59 | ||
| TB31456 | 771 | 359 | 13 | 31 | 22 | 9 | 19 | 14 | 29 | 66 | 47 | 0.69 | 0.70 | 0.69 | ||
| KPCFT79653 | 2609 | 1346 | gem 10nM | 15 | 29 | 22 | 6 | 13 | 9 | 36 | 55 | 45 | 0.68 | 0.73 | 0.71 | |
| HUMAN LINES | SW1990 | 935 | 14 | 56 | 13 | 35 | 23 | 2 | 13 | 23 | 24 | 24 | 1.61 | 1.27 | 1.44 | |
| Capan-1 | 1572 | 21 | 65 | 31 | 48 | 36 | 16 | 26 | 35 | 35 | 35 | 1.52 | 1.28 | 1.40 | ||
| ASPC1 | 1983 | 691 | 51 | 31 | 41 | 24 | 13 | 18 | 29 | 26 | 27 | 1.43 | 1.32 | 1.38 | ||
| HPAFII | 3239 | 427 | 90 | 39 | 64 | 39 | 12 | 25 | 38 | 21 | 29 | 1.62 | 1.57 | 1.60 | ||
| Capan-2 | 4775 | 2208 | 62 | 57 | 60 | 12 | 10 | 11 | 33 | 33 | 33 | 1.52 | 1.48 | 1.50 | ||
| MiaPaCa-2 | 9844 | 854 | 91 | 85 | 88 | 57 | 49 | 53 | 52 | 60 | 56 | 1.37 | 1.23 | 1.30 | ||
| Panc-1 | >30000 | 3496 | 166 | 135 | 151 | 96 | 74 | 85 | 61 | 57 | 59 | 1.76 | 1.65 | 1.71 | ||
To gain more resolution in the synergy area for MiaPaCa-2, we narrowed the range of concentration around the most intense region of synergy. In addition, instead of using constant drug exposure, we washed out the drugs after 24h and followed cell proliferation by time-lapse microscopy. Profound synergy and durable growth inhibition was achieved across the full range of concentrations even with only 24 hour of drugs (Fig. 3B). Remarkably, total growth inhibition was obtained with the combination of 1.5uM AZD6738 and 20nM gemcitabine for 24h, while single agents had no effect at these concentrations. At these concentrations and higher, growth inhibition was total and maintained for at least 3 days after the washout and cell death, measured by YoYo-3 dye entry, increased in parallel (Fig. 3C).
In order to confirm the synergy between AZD6738 and gemcitabine in a different assay system, we used the clonogenic survival assay where the cells were exposed to the drugs for 24h before washout. We also found potent synergy and a profound reduction in the surviving fraction when combining both drugs in MiaPaCa-2 and Panc-1 (Fig. 3D). Intriguingly, we observed emergence of clones even at the highest concentrations that we used. This suggests that it might be relevant to repeat or increase frequency of treatment in a clinical setting, as well as identify potential mechanisms of resistance.
Scheduling of the AZD6738 + gemcitabine combination
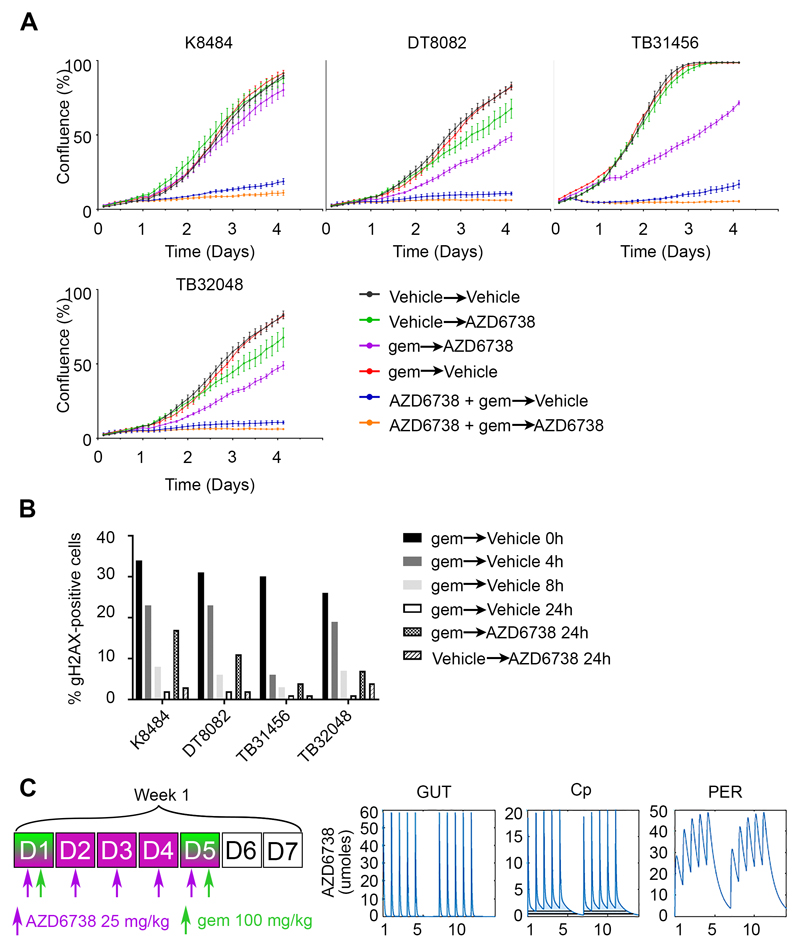
To gain insights into the best scheduling of the combination for subsequent in vivo efficacy, we tested sequential versus concurrent drug treatment in the four cell lines derived from the KPC model. We used drugs at the GI50 for each cell line and followed proliferation. Treating the cells for 16 hours with gemcitabine at GI50 concentrations had no impact on proliferation (Fig 4A). Treating the cells first with gemcitabine for 16 hours and replacing with AZD6738 had a modest effect. In contrast, the concurrent treatment of both drugs for 16h resulted in a profound and durable growth inhibition. Moreover, maintaining AZD6738 after the first concurrent treatment prevented the slow regrowth for at least 3 days. Mechanistically, we found that maintaining ATR inhibition after removal of gemcitabine increased the persistence of DNA damage (Fig. 4B). In order to translate these findings in vivo and optimize a tolerated dose/schedule for the combination, we employed in silico modelling of AZD6738 mouse pharmacokinetics (PK)(27). Guided by these simulations, we identified a dose/schedule predicted to sustain 136 hours of plasma [AZD6738]>1uM/week (Fig 4C).
Figure 4. Scheduling of the combination of AZD6738 with gemcitabine.
A, Four cell lines from the KPC mouse model were plated at low density and treated with the first condition for 16 hours, then the medium was replaced with the second condition and cell growth was monitored by time lapse microscopy every 3 hours for 4 days. Drugs were used at the GI50 (72h) concentrations. B, Cells were incubated with 10 nM gemcitabine for 16h, then fixed at the time point shown after washout into fresh medium or with 300 nM ATRi. Immunofluorescence followed by quantitative microscopy was realized to obtain the percentage of cells positive for γH2AX. C, Dose-schedule schematic (left) of the in silico modelling of AZD6738 mouse PK for the gut, circulating and peripheral compartments (right).
AZD6738/gemcitabine combination shows anti-tumor efficacy in vivo
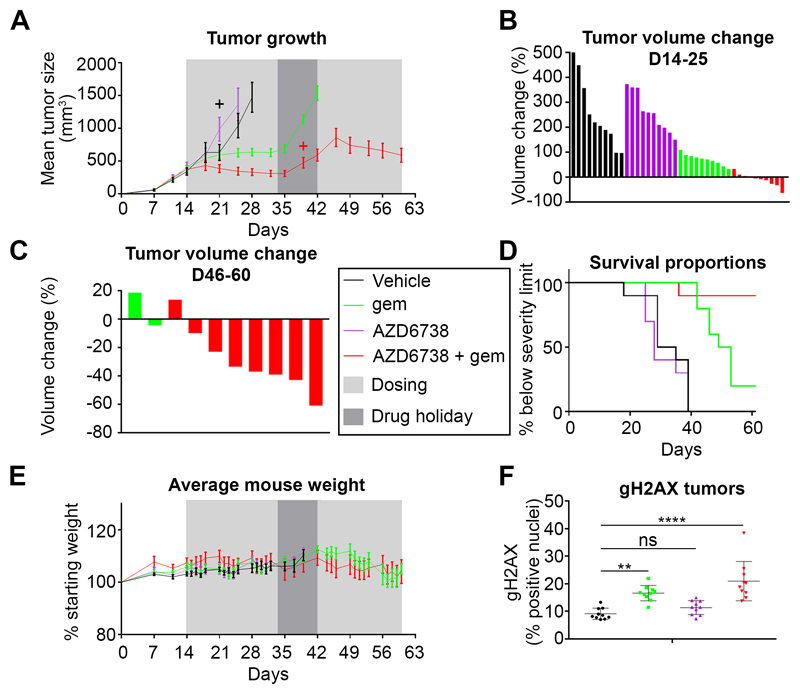
We next tested the efficacy and tolerability of this regimen in a four-arm study using the K8484 allograft model. All mice in the vehicle and AZD6738 (25 mg/kg x 5 days/week) single agent groups reached the maximum permitted tumor size before the end of the first three weeks of treatment. Gemcitabine (100 mg/kg x 2/week) was active as a single agent (as previously published(40)), and controlled tumor growth during the initial 3 weeks of treatment (Fig. 5A). However, none of the tumors shrank with gemcitabine alone (Fig.5B). In contrast, confirming the in vitro findings, combination of gemcitabine with AZD6738 was more efficacious, causing tumor shrinkage by the end of week 2 in 6/10 mice and 7/10 mice by the end of week 3. During a one week break in treatment, all tumors treated with gemcitabine alone regrew rapidly, and although treatment was resumed, most of the tumors reached the maximum permitted size, 2/10 remaining just below that threshold (Fig. 5C). One animal with a small tumor in the combination group was killed because of ill health during the ‘drug holiday’. In the combination group, tumors regrew during the break, but at a slower rate, allowing a further 3 weeks of treatment, leading to tumor shrinkage in 7/8 (one had completely resolved in the first 3 weeks) remaining mice (Fig. 5C), greatly extending survival (Fig 5D). Intriguingly, one tumor had resolved in the first three weeks of treatment and remained undetectable till the end. Importantly, the drug combination was tolerated without weight loss for the duration of the study, suggesting limited toxicity (Fig. 5E). In addition, γH2AX staining was elevated in the tumors of the mice treated with either single agent gemcitabine or the combination, but not in the tumors of the mice treated with single agent AZD6738, in line with the anti-tumor effects (Fig. 5F). Thus, the combination of gemcitabine + AZD6738 showed greater efficacy in this allograft model than gemcitabine alone.
Figure 5. The combination of AZD6738 + gemcitabine achieves anti-tumor effect and improves survival, in a subcutaneous allograft of a KPC cancer cell line.
A, Tumor volume of K8484 allografts. Ten mice per group were treated as indicated in the legend (middle of the figure) for 3 consecutive weekly cycles (see dose/schedule schematic in Fig.4 C). After a one week break, treatment was resumed for another 3 weeks. Data are represented as mean ± SEM. + Culled animals. B, Changes in individual tumor volume between D14 and D25 (when all mice were still alive). C, Changes in individual tumor volume between D46 and D60 (only two mice remaining in the gemcitabine alone group). D, Kaplan-Meier survival curves. E, Body weight change relative to pre-treatment body weight. Data are represented as mean ± SEM. F, Quantification of γH2AX-positive cells from tumors IHC. Data are represented as mean ± SD. A one-way ANOVA analysis with Tukey’s pairwise comparisons was performed, **p≤0.01, ****p≤0.0001, ns p>0.05.
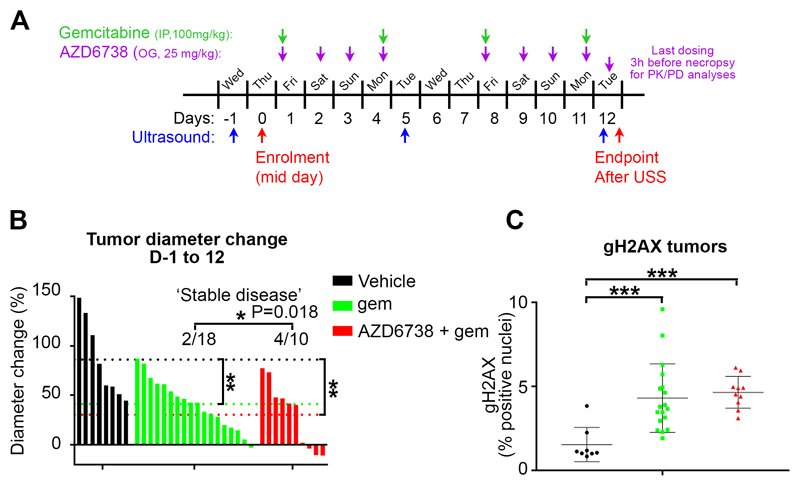
An efficacy study in the autochthonous KRAS G12D; p53 R172H; Pdx-cre (KPC) mouse model was then performed, according to the schedule in Figure 6A. Tumor growth was stabilized in 4/10 mice treated for 12 days with AZD6738 + gemcitabine (Fig. 6B). Remarkably, 3/10 tumors shrank, 2 by more than 10%. This effect was greater than with gemcitabine alone, which stabilized the growth of only 2/18 tumors (‘stable tumors’ defined as a tumor growth <10% during the 12-days treatment). KPC tumor regression has been very rarely seen historically. If the probability to observe a stable disease in the combination group is the same as in the gemcitabine single agent group (i.e. 2/18), we would expect to observe 4 or more stable diseases out of 10 animals only 1.84% of the time (P=0.0184). These data suggest that there may be a subset of KPC mice with tumors responsive to the AZD6738 + gemcitabine combination. Staining of the tumors for the pharmacodynamic biomarker γH2AX revealed a strong increase in the gemcitabine and combination groups in comparison to the vehicle group (Fig. 6C).
Figure 6. The combination of AZD6738 + gemcitabine induces tumor regression in a subgroup of KPC tumors.
A, Schematic of the study and dosing schedule. B, Changes in individual tumor long diameter between Day -1 and Day 12. Dotted lines represent the mean of each group. A one-way ANOVA analysis was performed with Tukey’s pairwise comparisons, **p≤0.01, ns p>0.05. During the 12-day study, in the gemcitabine alone group, two mice had a ‘stable’ (<10% diameter change) disease, while 16 had a ‘progressive’ (>10% diameter change) disease. In the combination group, four mice had a stable disease, while six had a progressive disease. If the probability to have a stable disease in the combination group is the same as in the gemcitabine alone group (i.e. 2/18), we would expect to observe 4 or more stable disease out of 10 animals only 1.84% of the time (p=0.018). C, Quantification of γH2AX-positive cells from tumors IHC. Data are represented as mean ± SD. A one-way ANOVA analysis with Tukey’s pairwise comparisons was performed, ***p≤0.001.
Discussion
In recent years, signaling pathways activated by RS, one of the hallmarks of cancer, have attracted a lot of attention as therapeutic targets. Potent and selective small-molecule inhibitors of the apical kinase of the RS response, ATR, are now in early phase clinical trials. However, how to rationally combine ATR inhibition with genotoxic therapies and which genetic alterations truly confer sensitivity, are important questions that remain to be fully answered. Recent reports have demonstrated the causal role of certain oncogenic activation and/or tumor suppressor inactivation in increasing sensitivity to ATR inhibition. In addition, a number of studies have described synthetic lethality relationships with ATR, mostly involving alterations in DDR pathways(15,29,31,33,41–47). The most prominent genetic alterations found in PDAC (KRAS, TP53, SMAD4, ARID1A) have all been shown to increase sensitivity to ATR inhibition. In addition, among the long tail of less frequent mutations uncovered by recent studies, mutations in DDR genes are found at the highest frequency(9). Therefore, there is an urgent need to assess the utility of ATR inhibition in PDAC. In the present study, we explored whether the ATR inhibitor AZD6738, could be used to inhibit the growth of PDAC, alone or in combination with the antimetabolite gemcitabine, which remains a cornerstone for the treatment of patients with all stages of the disease. The results of this study demonstrate the improved anti-tumor effect of the combination compared to gemcitabine alone and warrant clinical investigation.
ATR is an essential protein and its absence is lethal both at the cellular and organismal level, although a profound downregulation is compatible with life(48–50). Early studies in yeast revealed that the lethality induced by Mec1 (ATR) ablation was due to fatal decrease in ribonucleotide reductase expression(13). The function of ATR in maintaining sufficient dNTP levels appears to be conserved in mammalian cells(14). In addition, lethality of ATR inhibition in cancer cells has also been shown to be the result of premature mitotic entry(51), or global exhaustion of the ssDNA binding protein RPA leading to replication catastrophe(52). In the present study we observed that 5 out of 12 pancreatic cancer cell lines were very sensitive to AZD6738 monotherapy. Interestingly, sub-GI50 levels of gemcitabine potently sensitized even the most resistant cell lines to AZD6738. Gemcitabine could sensitize to ATR inhibitors by elevating RS through at least two mechanisms. First, by poisoning the DNA polymerase and RRM2, further reducing nucleotide pools to compromise DNA synthesis. Second, by inducing DNA lesions, either due to dFdCTP incorporation or other misincorporations driven by nucleotide shortage. These lesions further increase the reliance on ATR during S-phase, driving RPA exhaustion upon ATR inhibition. Reciprocally, we observed that AZD6738 sensitized to gemcitabine by at least three mechanisms. Firstly, by preventing the cell cycle arrest that would otherwise antagonize gemcitabine incorporation and permit DNA repair. Secondly, by promoting fork collapse, through blocking restart or repair of damaged forks. And thirdly, by limiting endogenous dNTP pools competing with gemcitabine – this can occur through RRM2 downregulation as well as through the increase of unscheduled origin firing which further exhausts nucleotides. By providing evidence of replication checkpoint inhibition, generation of DSB and prevention of RRM2 upregulation, our results suggest that AZD6738 and gemcitabine synergize at several molecular levels.
In addition to overcoming inherent gemcitabine resistance mechanisms, ATR inhibition could potentially limit acquired resistance to gemcitabine. Indeed, a recent study revealed the reliance of gemcitabine-resistant cells on de novo pyrimidine synthesis(53). Importantly, in this study the authors did not find evidence of increased cytidine deaminase as a mechanism of acquired gemcitabine resistance and resistant cells were still capable of gemcitabine uptake. Therefore, combined action of RRM2 irreversible inhibition by gemcitabine metabolite dFdCDP and downregulation by AZD6738, could potentially help sustain favorable ratio of dFdCTP/dCTP needed to maintain efficacy. Accordingly, we found that AZD6738 restored gemcitabine sensitivity in intrinsically gemcitabine-resistant cells (e.g. AsPC-1 and PANC-1). This is also in line with our findings showing sustained efficacy of the combination over a period of 6-weeks treatment in vivo. However, other mechanisms of resistance to gemcitabine could still curtail the efficacy of the combination. The intriguing effect of the combination in our KPC study, showing ‘responders’ and ‘non responders’, could therefore reflect an intrinsic mechanism of gemcitabine resistance in the tumors that did not respond.
Low level of CDC25A has been found to confer resistance to ATR inhibition as monotherapy(51). However, the absence of toxicity in cells expressing low levels of CDC25A appeared to be due to the absence of premature entry into mitosis. Importantly the increased incorporation of EdU upon ATR inhibition was not affected by levels of CDC25A. Similarly, dFdCTP incorporation should not be affected, hence, combining gemcitabine with ATR inhibition could potentially overcome the resistance conferred by CDC25A deficiency.
We observed that the combination of AZD6738 + gemcitabine strongly induced ATM and DNA-PK activation. This is consistent with the role of ATR in preventing fork collapse and DSB-response. It is noteworthy that these kinases are members of the same family and display substantial promiscuity towards substrates (e.g. γH2AX, p-RPA). For example, DNA-PK can phosphorylate Chk1 independently of ATR, indicating that pChk1 is not the most appropriate biomarker for detection of target inhibition by AZD6738, especially as cell death and durable growth inhibition was achieved at doses lower than that required for maximal inhibition of pChk1-Ser345(14). ATM and HR repair factors are the first recruited components to collapsed forks, generating ssDNA that continue to exhaust RPA pools and push cells toward replication catastrophe(54). DNA-PK instead is recruited to the damaged forks generated by new origin firing. Therefore, the strong activation of DNA-PK by the combination of gemcitabine and AZD6738, indicates an increase in origin firing, strongly suggesting on-target ATR inhibition. Moreover, increased levels of DNA-PKcs has been shown to confer sensitivity to ATR inhibitor monotherapy(15). Therefore, although we cannot exclude compensatory roles for ATM and DNA-PKcs activation, our data suggest that activation of both kinases further propagates the RS induced by AZD6738 in combination with gemcitabine.
KPC tumor regression has been very rarely seen historically. Our data suggest that there may be a subset of KPC mice with tumors responsive to the combination of AZD6738 with gemcitabine. A number of genetic alterations have been reported in the literature to have synthetic lethal relationships with ATR inhibition, raising the possibility of an underlying genetic vulnerability in the responsive tumors. While it will be essential to gain insights into the mechanisms underlying responsiveness or resistance and possibly identify predictive biomarkers, this work suggests immediate clinical benefit for PDAC patients, and will serve as the basis to initiate a clinical trial.
Supplementary Material
Acknowledgments
We thank Cancer Research UK Cambridge Institute Biological Resources Unit, Preclinical/Genome Editing, Biorepository, Histopathology/in situ Hybridization and Light Microscopy Core Facilities for technical provision; as well as members of the Pharmacology & Drug Development Group for discussions. Thanks to Prof Ashok Venkitaraman’s lab in the MRC Cancer Cell Unit, Cambridge for providing a KPCB cell line. YW, CRD, TIJ, SBK, SBQF, FMR and DIJ were funded by Cancer Research UK institute core grant C14303/A17197. CRD and SBK were also recipients of funding from the Pancreatic Cancer UK Future Research Leaders Fund. The Li Ka Shing Centre in which this research was performed was generously funded by CK Hutchison Holdings Limited, the University of Cambridge, Cancer Research UK, The Atlantic Philanthropies and a range of other donors
Financial Support:
YW, CRD, TIJ, SBK, SBQF, FMR and DIJ were funded by Cancer Research UK institute core grant C14303/A17197. CRD and SBK were also recipients of funding from the Pancreatic Cancer UK Future Research Leaders Fund. The Li Ka Shing Centre in which this research was performed was generously funded by CK Hutchison Holdings Limited, the University of Cambridge, Cancer Research UK, The Atlantic Philanthropies and a range of other donors
Footnotes
Conflicts of Interest
C.F., J.Y., A.L. are employees of AstraZeneca. D.I.J. has received research funding from AstraZeneca for projects unrelated to AZD6738
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. The New England journal of medicine. 2013;369(18):1691–703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. The New England journal of medicine. 2011;364(19):1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 4.Dobbelstein M, Sorensen CS. Exploiting replicative stress to treat cancer. Nature reviews Drug discovery. 2015;14(6):405–23. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- 5.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16(1):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magdalou I, Lopez BS, Pasero P, Lambert SA. The causes of replication stress and their consequences on genome stability and cell fate. Semin Cell Dev Biol. 2014;30:154–64. doi: 10.1016/j.semcdb.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 7.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016 doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 8.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nature communications. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Suzuki H, Liu B, Morris J, Liu J, Okazaki T, et al. DNA repair gene polymorphisms and risk of pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(2):740–6. doi: 10.1158/1078-0432.CCR-08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyer SB, Chang DK, Bailey P, Biankin AV. Pancreatic Cancer Genomes: Implications for Clinical Management and Therapeutic Development. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23(7):1638–46. doi: 10.1158/1078-0432.CCR-16-2411. [DOI] [PubMed] [Google Scholar]
- 12.Iyer DR, Rhind N. The Intra-S Checkpoint Responses to DNA Damage. Genes (Basel) 2017;8(2) doi: 10.3390/genes8020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Molecular cell. 1998;2(3):329–40. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 14.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Molecular cell. 2015;59(6):1011–24. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middleton FK, Patterson MJ, Elstob CJ, Fordham S, Herriott A, Wade MA, et al. Common cancer-associated imbalances in the DNA damage response confer sensitivity to single agent ATR inhibition. Oncotarget. 2015;6(32):32396–409. doi: 10.18632/oncotarget.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13(11):1072–81. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell death & disease. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Ge Y, Wang T, Edwards H, Ren Q, Jiang Y, et al. Inhibition of ATR potentiates the cytotoxic effect of gemcitabine on pancreatic cancer cells through enhancement of DNA damage and abrogation of ribonucleotide reductase induction by gemcitabine. Oncology reports. 2017;37(6):3377–86. doi: 10.3892/or.2017.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews DJ, Yakes FM, Chen J, Tadano M, Bornheim L, Clary DO, et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell cycle. 2007;6(1):104–10. doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- 20.Koh SB, Courtin A, Boyce RJ, Boyle RG, Richards FM, Jodrell DI. CHK1 Inhibition Synergizes with Gemcitabine Initially by Destabilizing the DNA Replication Apparatus. Cancer research. 2015;75(17):3583–95. doi: 10.1158/0008-5472.CAN-14-3347. [DOI] [PubMed] [Google Scholar]
- 21.Foote KM, Lau A, Nissink JW. Drugging ATR: progress in the development of specific inhibitors for the treatment of cancer. Future medicinal chemistry. 2015;7(7):873–91. doi: 10.4155/fmc.15.33. [DOI] [PubMed] [Google Scholar]
- 22.Yap TA, Krebs MG, Postel-Vinay S, Bang YJ, El-Khoueiry A, Abida W, et al. Phase I modular study of AZD6738, a novel oral, potent and selective ataxia telangiectasia Rad3-related (ATR) inhibitor in combination (combo) with carboplatin, olaparib or durvalumab in patients (pts) with advanced cancers. European Journal of Cancer. 2016;69(Supplement 1):S2. [Google Scholar]
- 23.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skoulidis F, Cassidy LD, Pisupati V, Jonasson JG, Bjarnason H, Eyfjord JE, et al. Germline Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer cell. 2010;18(5):499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Clark NA, Hafner M, Kouril M, Williams EH, Muhlich JL, Pilarczyk M, et al. GRcalculator: an online tool for calculating and mining dose-response data. BMC Cancer. 2017;17(1):698. doi: 10.1186/s12885-017-3689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 2016;13(6):521–7. doi: 10.1038/nmeth.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Checkley S, MacCallum L, Yates J, Jasper P, Luo H, Tolsma J, et al. Bridging the gap between in vitro and in vivo: Dose and schedule predictions for the ATR inhibitor AZD6738. Sci Rep. 2015;5:13545. doi: 10.1038/srep13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer research. 2014;74(10):2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikkila J, Kumar R, Campbell J, Brandsma I, Pemberton HN, Wallberg F, et al. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. British journal of cancer. 2017;117(1):113–23. doi: 10.1038/bjc.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nature genetics. 2009;41(10):1144–9. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoppy DW, Ragland RL, Gilad O, Shastri N, Peters AA, Murga M, et al. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. The Journal of clinical investigation. 2012;122(1):241–52. doi: 10.1172/JCI58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nature structural & molecular biology. 2011;18(6):721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nature communications. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nature protocols. 2006;1(3):1112–6. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 35.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell. 2005;7(5):469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 36.Dillon MT, Barker HE, Pedersen M, Hafsi H, Bhide SA, Newbold KL, et al. Radiosensitization by the ATR Inhibitor AZD6738 through Generation of Acentric Micronuclei. Molecular cancer therapeutics. 2017;16(1):25–34. doi: 10.1158/1535-7163.MCT-16-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci. 2009;122(Pt 22):4070–80. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuss JE, Patrick SM, Oakley GG, Alter GM, Robison JG, Dixon K, et al. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry. 2005;44(23):8428–37. doi: 10.1021/bi0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32(18):2866–8. doi: 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bapiro TE, Frese KK, Courtin A, Bramhall JL, Madhu B, Cook N, et al. Gemcitabine diphosphate choline is a major metabolite linked to the Kennedy pathway in pancreatic cancer models in vivo. British journal of cancer. 2014;111(2):318–25. doi: 10.1038/bjc.2014.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3(3):248–54. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304(5667):93–6. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 43.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nature structural & molecular biology. 2011;18(12):1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peasland A, Wang LZ, Rowling E, Kyle S, Chen T, Hopkins A, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. British journal of cancer. 2011;105(3):372–81. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nature chemical biology. 2011;7(7):428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 46.Regal JA, Festerling TA, Buis JM, Ferguson DO. Disease-associated MRE11 mutants impact ATM/ATR DNA damage signaling by distinct mechanisms. Hum Mol Genet. 2013;22(25):5146–59. doi: 10.1093/hmg/ddt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sultana R, Abdel-Fatah T, Perry C, Moseley P, Albarakti N, Mohan V, et al. Ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase inhibition is synthetically lethal in XRCC1 deficient ovarian cancer cells. PloS one. 2013;8(2):e57098. doi: 10.1371/journal.pone.0057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10(8):479–82. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 49.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes & development. 2003;17(5):615–28. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nature genetics. 2009;41(8):891–8. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz S, Mayor-Ruiz C, Lafarga V, Murga M, Vega-Sendino M, Ortega S, et al. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Molecular cell. 2016;62(2):307–13. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toledo L, Neelsen KJ, Lukas J. Replication Catastrophe: When a Checkpoint Fails because of Exhaustion. Molecular cell. 2017;66(6):735–49. doi: 10.1016/j.molcel.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer cell. 2017;32(3):392. doi: 10.1016/j.ccell.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, et al. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Molecular cell. 2015;59(6):998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.