Abstract
Cell migration involves complex physical and chemical interactions with the substrate. To probe the mechanical interactions under different regions of migrating 3T3 fibroblasts, we have disrupted cell-substrate adhesions by local application of the GRGDTP peptide, while imaging stress distribution on the substrate with traction force microscopy. Both spontaneous and GRGDTP-induced detachment of the trailing edge caused extensive cell shortening, without changing the overall level of traction forces or the direction of migration. In contrast, disruption of frontal adhesions caused dramatic, global loss of traction forces before any significant shortening of the cell. Although traction forces and cell migration recovered within 10–20 min of transient frontal treatment, persistent treatment with GRGDTP caused the cell to develop traction forces elsewhere and reorient toward a new direction. We conclude that contractile forces of a fibroblast are transmitted to the substrate through two distinct types of adhesions. Leading edge adhesions are unique in their ability to transmit active propulsive forces. Their functions cannot be transferred directly to existing adhesions upon detachment. Trailing end adhesions create passive resistance during cell migration and readily redistribute their loads upon detachment. Our results indicate the distinct nature of mechanical interactions at the leading versus trailing edges, which together generate the mechanical interactions for fibroblast migration.
INTRODUCTION
The migration of cultured fibroblasts has been studied in detail for many decades (Abercrombie et al., 1970; Harris et al., 1980). The process consists of cycles of frontal extension and tail retraction, coupled to complex mechanical interactions between the cellular contractile apparatus and the substrate through integrin-mediated adhesion sites (Yamada and Miyamoto, 1995; Lauffenburger and Horwitz, 1996; De Beus and Jacobson, 1998; Sheetz et al., 1998; Horwitz and Parsons, 1999). It is generally agreed that frontal protrusion serves as a means to extend forward and establish new adhesion structures, whereas the transmission of contractile forces to the substrate at adhesion sites causes the cell body to move (Lauffenburger and Horwitz, 1996; Sheetz et al., 1998; Burton et al., 1999; Elson et al., 1999; Svitkina and Borisy, 1999). However, the exact mechanism of fibroblast migration remains controversial.
To begin to understand the physical principle of cell migration, it is important to characterize the spatial and temporal pattern of mechanical interactions between the cell and the substrate. We have recently developed a method, traction force microscopy, for imaging the distribution of mechanical stresses exerted by a cultured cell (Dembo and Wang, 1999). Traction images of migrating 3T3 cells indicated a general centripetal pattern of forces, with strong stresses located at the leading edge, lateral protrusions, and sometimes at the trailing edge (Dembo and Wang, 1999; Munevar et al., 2001). There are also weak, diffuse forces distributed under most of the cell body with a forward bias. These observations indicate that the cell body is indeed under tension and that constant mechanical cross-talk takes place among widely separated regions of the cell.
From the centripetal pattern of traction forces, one can conclude that propulsive actions for cell migration are concentrated at the leading edge (Dembo and Wang, 1999; Munevar et al., 2001). However, unregulated centripetal contractions would simply cause the cell to collapse. Sustained cell migration can take place only through a tight coordination of the protrusive activities, the assembly and disassembly of adhesion structures, and the distribution of traction forces (Galbraith and Sheetz, 1997; Svitkina et al., 1997; Oliver et al., 1999). Different theories, not necessarily exclusive of one another, emphasize different mechanical aspects and differ with respect to the scenario of events. In a simple tug-of-war mechanism, peripheral regions of the cell try to walk away from the cell center, and the region that generates the strongest forces determines the overall polarity. Thus, forces in different regions are qualitatively identical but differ in their magnitudes. From the retraction of tails coupled to the surge in protrusion (Chen, 1981), it has also been suggested that the elasticity of the cell body may play a role in cell migration, by storing potential energy like a rubber band. The direction of cell migration is determined primarily by the preferential rupture of adhesive bonds at the tail, followed by the conversion of stored potential energy in a stretched cell body into kinetic energy for forward migration. In the “frontal towing” mechanism, the leading edge represents a unique region that provides active forces for dragging a passive cell body across the substrate. This model contends that mechanical interactions at the leading edge differ both quantitatively and qualitatively from those under the rest of the cell body.
Important insights into the mechanism of fibroblast migration can be gained by determining not only the distribution but also the nature of traction forces in relation to the cell morphology and migration. Forces exerted on the substrate could reflect either active pulling or passive resistance to forces exerted elsewhere, and it is the active forces that drive the migration of the cell. In the present study, we combined traction force microscopy with focal release of the GRGDTP peptide, a competitive inhibitor of integrin-extracellular matrix (ECM) binding, to disrupt cell adhesions at defined regions. The results indicate the distinct nature of mechanical interactions at the leading versus trailing edges and suggest that the frontal towing mechanism plays a major role in fibroblast migration.
MATERIALS AND METHODS
Preparation and Characterization of Polyacrylamide Substrates
Thin sheets of polyacrylamide gel were prepared from acrylamide (40% wt/vol; Bio-Rad, Hercules, CA) and N,N-methylene-bis-acrylamide (2% wt/vol; Bio-Rad) and adhered to activated coverslips as described previously (Wang and Pelham, 1998). All the substrates used in this study contained 5% acrylamide, 0.1% N,N-methylene-bis-acrylamide, and 1:100 dilution of fluorescent latex beads (0.2-μm FluoSpheres; Molecular Probes, Eugene, OR). Fifteen microliters of the acrylamide solution was spread onto the surface of an activated large rectangular coverslip (45 × 50 mm) and induced to polymerize under a 22-mm-diameter circular coverslip. Type I collagen was covalently attached to the surface of the polyacrylamide gel with the use of photoactivatable heterobifunctional reagent sulfosuccinimidyl 6 (4-azido-2-nitrophenyl-amino) hexanoate, as described previously (Wang and Pelham, 1998).
Steady-state thickness of the polyacrylamide sheets at 37°C was estimated to be ∼75 μm, by focusing a microscope with a calibrated focusing knob from the glass surface to the surface of the polyacrylamide gel. Young's modulus of the polyacrylamide sheets was determined as described previously (Lo et al., 2000), with the use of a method based on the Hertz theory (Radmacher et al., 1992). This method yielded a Young's modulus of 2.8 × 104 N/m2.
Calculation and Rendering of Traction Stress
Traction force microscopy was performed as described recently (Munevar et al., 2001). Briefly, deformation of the substrate caused by cellular traction forces was determined relative to the relaxed substrate with the use of a pattern recognition algorithm. Coordinates defining the deformation field and cell boundary were analyzed with a Bayesian method to determine maximum likelihood traction vectors at preassigned nodes throughout the cell (Dembo and Wang, 1999). Pseudocolor images were obtained by first determining the magnitude of the traction stress at each pixel within the cell boundary and then converting the magnitude into different red-green-blue color combinations.
Cell Culture and Microscopy
Polyacrylamide substrates were equilibrated with culture medium for ∼30 min at 37°C. NIH 3T3 cells were cultured in DMEM (Sigma, St. Louis, MO), supplemented with 10% donor calf serum (JHR Biosciences, Lenexa, KS), 2 mM l-glutamine, 50 μg/ml streptomycin, and 50 U/ml penicillin (Invitrogen, Carlsbad, CA). Phase images of cells and fluorescent images of substrate-embedded beads were collected with a 40×/numerical aperture 0.75 Plan-Neofluar phase objective on a Zeiss Axiovert S100TV microscope equipped with a custom stage incubator. Bead images of relaxed substrates were collected at the end of time-lapse recording by removing the cell with a microneedle. All images were collected with a cooled charge-coupled device camera (ST133 controller with an EEV Type57 back-illuminated frame-transfer charge-coupled device chip; Roper Scientific, Trenton, NJ) and processed for background subtraction with the use of custom programs.
Local Application of GRGDTP Peptide
A synthetic peptide Gly-Arg-Gly-Asp-Thr-Pro (GRGDTP) (G-5646; Sigma) was used to disrupt cell substrate attachment (Dedhar et al., 1987; Gehlsen et al., 1988). An inactive synthetic peptide, Gly-Arg-Gly-Glu-Ser-Pro (GRGESP) (H-3136; Bachem, Torrance, CA), was used as a control (Pytela et al., 1986). Both peptides were dissolved to a concentration of 5 mg/ml in phosphate-buffered saline containing 5 mg/ml rhodamine dextran (Molecular Probes), which served as an inert fluorescent marker for visualizing the distribution of the released solution. Application of the rhodamine dextran marker alone had no discernable effect on migrating cells.
Focal release of GRGDTP was carried out as described in O'Connell et al. (2001). The peptide was loaded into a release microneedle connected to a source of regulated compressed air. A suction micropipette was prepared by breaking and fire polishing the broken tip of a microneedle with the use of a microforge (Narishige, Tokyo, Japan) and was then connected to a regulated source of vacuum. The release and suction micropipettes were mounted on a custom micromanipulator that allowed both precise relative positioning of the two needles and their simultaneous movements. Generally, the suction pipette was positioned 20–40 μm behind the release needle at a slightly higher elevation. Because of the continuous liquid flow, the distribution of GRGDTP was determined by the flow pattern rather than its diffusion. By balancing the rates of release and suction, the distribution of GRGDTP was maintained in a region 10–15 μm in diameter as judged by the distribution of rhodamine dextran.
RESULTS
Imaging and Manipulating Cell-Substrata Mechanical Interactions
The basic approach for mapping traction stresses was described in Dembo and Wang (1999) and was recently modified by Munevar et al. (2001). The method is based on the use of flexible polyacrylamide substrates, coated with extracellular matrix proteins (type I collagen in the present study) for cell adhesion, and embedded with fluorescent beads for tracking the deformation caused by exerted forces. The magnitude of the traction stress was calculated at each pixel and displayed as pseudocolor images or movies.
Although the substrate was coated with type I collage, it is possible that additional components such as fibronectin were recruited to the surface. Cell–substrate interactions were disrupted by focal release of the GRGDTP peptide (see MATERIALS AND METHODS), which was reported to inhibit cell attachment to fibronectin, vitronectin, and type 1 collagen (Dedhar et al., 1987; Gehlsen et al., 1988), and caused the detachment of 3T3 cells in a highly controlled and localized manner. This manipulation served two purposes. The first was to probe the coordination of activities in different regions, by determining whether detachment of one region leads to changes in cell length or protrusive activities in other regions. The second purpose was to classify traction forces in specific areas. Because traction forces are globally balanced over the cell, local disruptions of adhesions must be compensated by changes in traction forces at distant sites. One possibility is that local disruption causes an immediate decrease in the magnitude of distal stresses pointing in the opposite direction. This occurs when the disrupted adhesions are directly connected to a contractile apparatus that acts on the rest of the cell, and will be referred to as “active” tractions. Alternatively, local disruptions may not impair the average traction output but are compensated by an increase in the magnitude of proximal stresses that point at a similar direction. This happens when a group of adhesions shares the job of resisting contractile forces generated elsewhere, and will be referred to as “passive” or “reactive” tractions.
Responses of Traction Forces and Cell Migration to Spontaneous and Induced Tail Retraction
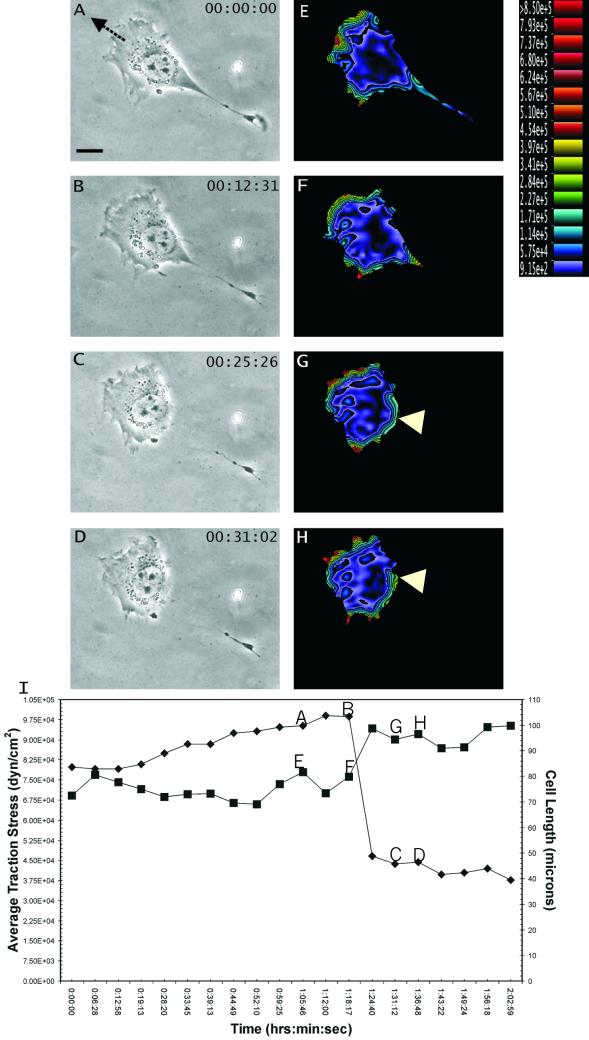
Most NIH 3T3 cells migrated on polyacrylamide substrates with stable, well-defined leading and trailing edges (Figure 1A). We first tested the involvement of the elasticity of cell body in cell migration. As predicted by some of the models such as the “elastic cell body” model (see INTRODUCTION), traction forces should increase in proportion to the cell length and decrease precipitously upon tail retraction. Contrary to this idea, however, we observed no change in the average magnitude of traction stress during cell elongation. Furthermore, spontaneous tail retraction was actually associated with a slight increase in frontal traction forces (Figure 1, E–H).
Figure 1.
Response of traction forces to spontaneous tail retraction. (A–D) Phase contrast images of a migrating NIH 3T3 fibroblast, recorded at time points indicated at the upper right corner. Arrow in A indicates the direction of cell migration. (E–H) Color rendering of the corresponding magnitude of traction stress, which ranges from 9.15 × 102 dynes/cm2 (violet) to >8.50 × 105 dynes/cm2 (red). Note the increase in traction stress at the leading edge after retraction of the trailing edge. In addition, traction at the tail shifted to a region not affected by the retraction (G and H, arrowhead). (I) Average magnitude of traction stress (▪) and cell length (♦) during spontaneous tail retraction. Letters A–H mark the corresponding panels. Note that there was no loss of average traction force as the cell shortened to <50% of the original length. Bar, 20 μm.
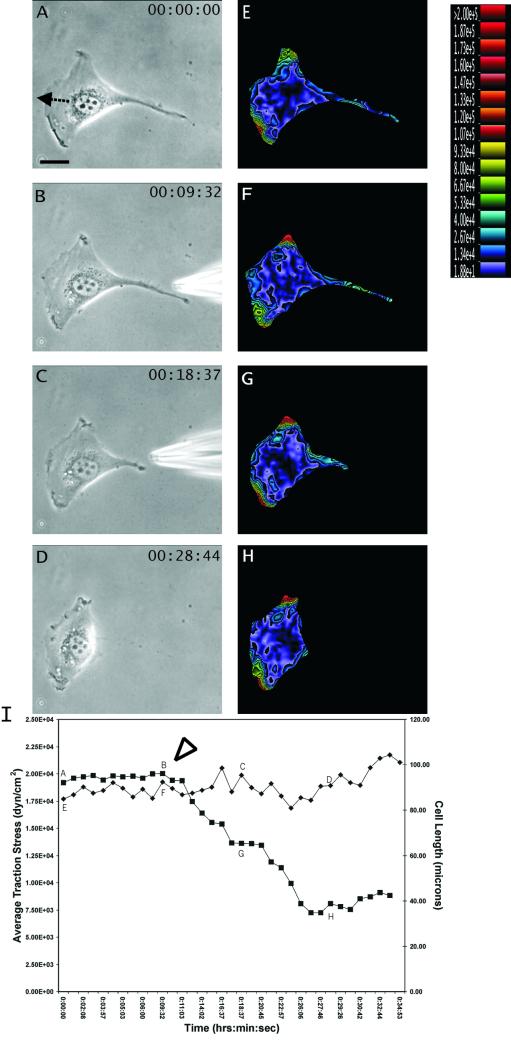
To substantiate the results with spontaneous tail retraction, we induced tail retraction with focal release of GRGDTP (n = 8 cells). This treatment again caused a dramatic decrease in cell length (>40%) accompanied by a slight increase in frontal traction stress (Figure 2, E–H), whereas the average magnitude of traction stress showed no significant change (Figure 2I). Both spontaneous and induced tail releases caused a transient increase in the rate and no change in the direction of cell migration. Application of GRGESP, a control peptide, caused no discernable detachment of the cell or change in substrate deformation.
Figure 2.
Response of traction forces to GRGDTP-induced tail retraction. (A–D) Phase contrast images of a migrating NIH 3T3 fibroblast, recorded at time points indicated at the upper right corner. The needle releasing GRGDTP is seen in B and C. Arrow in A indicates the direction of cell migration. (E–H) Color rendering of the corresponding magnitude of traction stress, which ranges from 1.88 × 101 dynes/cm2 (violet) to >2.00 × 105 dynes/cm2 (red). Note the increase in traction stress at the leading edge after GRGDTP-induced retraction of the tail. (I) Average magnitude of traction stress (♦) and cell length (▪) during GRGDTP-induced tail retraction. Letters A–H mark the corresponding panels. The arrowhead indicates the time when the GRGDTP peptide was locally applied to the trailing edge. Bar, 20 μm.
On the loss of adhesion and traction forces at the tail, global balance was maintained by an immediate increase of tractions in other trailing regions (Figure 1, G and H, arrowheads). Together, these observations indicate that traction forces near the tail of a locomoting 3T3 cell are of the passive or reactive type.
Responses of Traction Forces and Cell Migration to Induced Frontal Retraction
The results with tail release indicated that a large portion of tail forces may be actively generated elsewhere, whereas the tail serves only as a site of passive anchorage. A similar approach with GRGDTP was thus applied to assess the effects of frontal release on traction forces and cell migration (n = 11 cells).
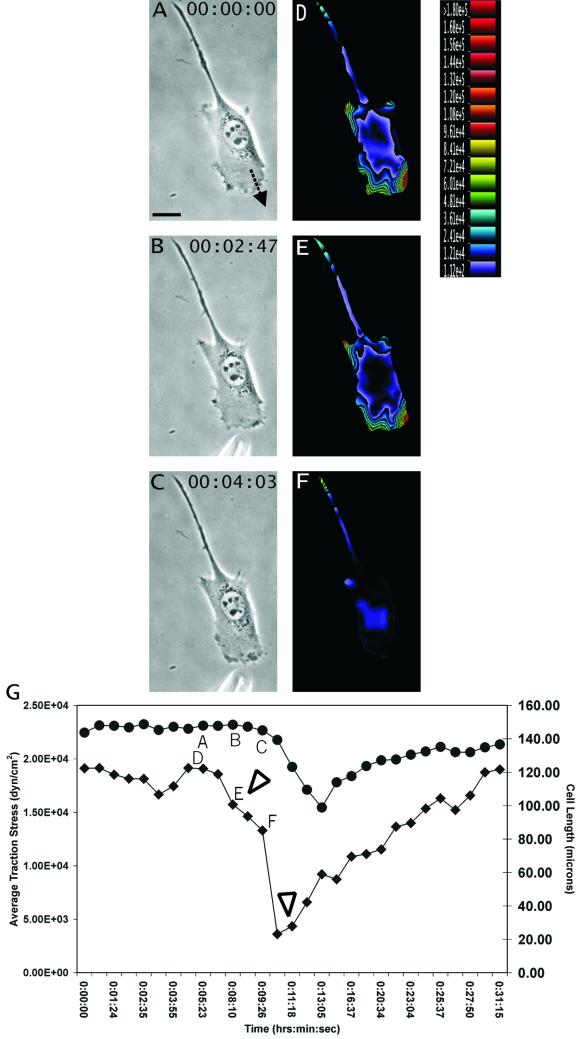
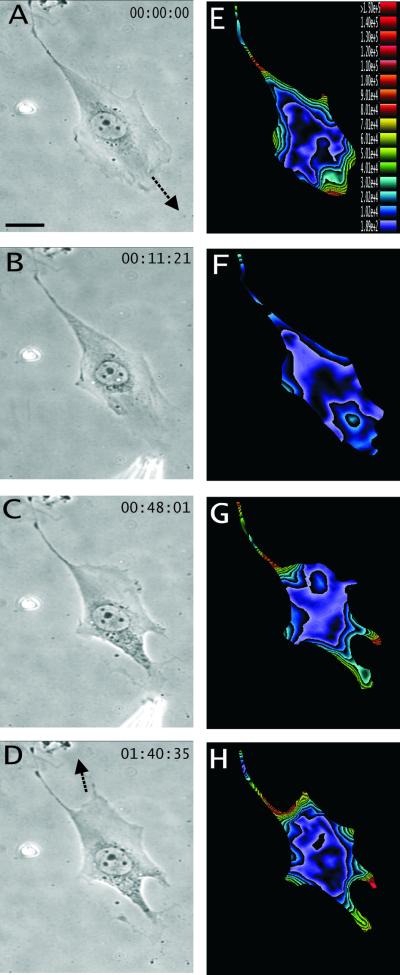
Limited detachment of the leading edge, induced by transient application of the GRGDTP peptide near the tip of the cell, resulted in an immediate and global decrease in traction stress by as much as 90%, before any significant decrease in cell length (Figure 3, D–F). Although the treatment caused the leading edge to collapse, upon removal of the peptide the lamellipodium and the traction forces returned within 10–20 min and cell migration resumed along the previous direction (Figure 3G). Sustained applications of GRGDTP caused the cell to switch its direction of migration by establishing a new leading lamellipodium in a distal area (Figure 4, A–D). Application of the control peptide GRGESP again had no effect. These results indicate that traction forces were actively applied to the protrusive regions of the cell independent of the length or elastic properties of the cell. Furthermore, unlike tail detachment, the loss of frontal adhesion cannot be compensated by simply shifting the traction forces to other preexisting adhesion sites. These “active” traction forces appeared to reestablish only by the formation of new adhesion sites.
Figure 3.
Response of traction forces to GRGDTP-induced frontal release. (A–C) Phase contrast images of a migrating NIH 3T3 fibroblast, recorded at time points indicated at the upper right corner. The needle releasing GRGDTP is located near the bottom of B and C. Arrow in A indicates the direction of cell migration. (D–F) Color rendering of the corresponding magnitude of traction stress, which ranges from 1.12 × 102 dynes/cm2 (violet) to >1.80 × 105 dynes/cm2 (red). (G) Average magnitude of traction stress (♦) and cell length (●) during GRGDTP-induced frontal release. Letters A–F mark the corresponding panels. The first arrowhead indicates the time when GRGDTP peptide was locally applied to the leading edge, causing a sharp drop in the average traction stress. The second arrowhead indicates the time when the needle releasing GRGDTP was removed. Note that upon local detachment of the leading edge, traction stress decreases dramatically across the entire cell, while the length of the cell decreased only slightly (G; time ∼11:00). The average traction stress recovers to the previous level over a period of 10–20 min. Bar, 20 μm.
Figure 4.
Redirection of cell migration after persistent treatment of the GRGDTP peptide at the front. (A–D) Phase contrast images of a migrating NIH 3T3 fibroblast, recorded at time points indicated at the upper right corner. Arrows in A and D indicate the initial and final direction of cell migration, respectively. (E–H) Color rendering of the corresponding magnitude of traction stress, which ranges from 1.89 × 102 dynes/cm2 (violet) to >1.50 × 105 dynes/cm2 (red). The cell changes its direction of migration after persistent frontal treatment with GRGDTP. A region near the original tail expands into a new lamellipodium with a concomitant increase in traction stress. Bar, 20 μm.
DISCUSSION
The first step toward a mechanistic understanding of cell migration is to determine the dynamic distribution of traction forces at the cell-substrate interface in relation to cell migration. We have extended the elastic substrate approach (Harris et al., 1980; Wang and Pelham, 1998; Pelham and Wang, 1999) by introducing computational methods capable of converting substrate displacements into high-resolution fields of traction vectors (Dembo et al., 1996; Dembo and Wang, 1999). As currently implemented, this methodology is capable of providing dynamic images of the traction stress under live cells, with a maximum temporal resolution on the order of few seconds and an estimated spatial resolution of 5–6 μm (Beningo et al., 2001; Munevar et al., 2001). Analysis of polarized migrating 3T3 fibroblasts indicated that strong inward tractions were localized at the leading edge, lateral protrusion, and sometimes at the trailing edge of the cell. However, although these images clearly indicated that the cytoskeleton is under tension and that propulsive actions are concentrated at the front, they shed limited light on where forces are actively exerted and how they are converted into directional cell migration.
To probe into the functional role of traction forces in different regions, we have coupled traction force microscopy with focal release of GRGDTP. The responses of traction stress and cell migration to localized disruption of cell-extracellular matrix adhesions allowed us to gain unique insight into the mechanism of fibroblast migration. Our results indicated fundamental differences between traction forces at the front and rear of a 3T3 fibroblast. Release of frontal adhesions caused a drastic, global decrease in traction forces, indicating that integrins and the associated cytoskeleton in this region are rather specialized in their abilities to transmit active forces and to maintain the general tension throughout the cell. Moreover, the force-generating mechanism cannot be rapidly recycled to preexisting adhesion sites upon detachment, but appeared to function only on new adhesion sites in the protrusive region. On the contrary, release of adhesions at the tail caused no decrease in traction forces elsewhere, indicating that they are passive anchors resisting forces generated in other regions and transmitted through the cell body. Furthermore, although tensile stresses along the cell body were often focused disproportionally onto a small subset of adhesion sites at the tail, these resistive forces were spread dynamically among multiple adhesion sites and were able to redistribute immediately upon the formation and dissociation of substrate adhesions. This may ensure a high degree of mechanical leverage for tail retraction and could be an important factor in maintaining the front-back asymmetry that enables the cell to migrate forward.
Our results further indicate that frontal and rear adhesion sites play different roles in maintaining the cell length. Dissociation of frontal adhesions caused a drastic loss of traction forces before any cell shortening becomes apparent. Conversely, both spontaneous and induced tail retraction caused striking shortening without a concomitant decrease in the overall traction stress. These observations may be explained if the elongated cell body is maintained by anchoring sites located at the tail and along the cell body, and the leading edge itself constitutes a mechanically distinct entity that exerts strong forces to tow this anchored cell body forward (Munevar et al., 2001).
The present results also help address several potential mechanisms in cell migration (Bray, 2001). One common notion is that the cell consists of a network of contractile fiber attached to the substrate at various places. Directional migration arises as a result of differential distribution of traction forces. This “tug-of-war” mechanism predicts that disruption of frontal attachments should not cause a dramatic, global decrease in tension, but simply cause the cell to contract toward the center until the tension is taken up by remaining adhesions, much like the detachment of the tail. This is clearly at odds with our findings. To account for tail retraction, some models place the emphasis on the elastic characteristics of the cell body, and use the conversion between potential and kinetic energies as an important part of the migration mechanism. However, the present results showed no evidence for the correlation between cell length and traction forces, or the storage of a significant amount of elastic energy for propelling cell movement, although such changes in cell length and cytoskeletal tension may serve an indirect function in regulating the rate or direction of cell movements (Chen, 1981). Our observations are in general accord with the notion that the tension in the cell body is the result of continuous contraction, maintained at a more or less constant level regardless of cell length.
The present results are consistent with our recent finding that nascent focal adhesions near the leading edge are involved in generating transient propelling forces (Beningo et al., 2001), possibly coupled to the assembly of myosin II ribbons and minifilaments observed previously in this area (McKenna et al., 1989; Verkhovsky and Borisy, 1993; Svitkina et al., 1997). Important questions remain concerning the molecular interactions responsible for the generation and transmission of mechanical forces, the nature of chemical signals regulating the “active” and “passive” traction forces in different regions, and the possible differences between the migration of fibroblasts and other types of cells. This interface between physical forces and chemical signals lies at the very heart of the mechanism that drives the directed cellular migration in a physiologically responsive manner.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Boston University Center for Scientific Computing for the use of supercomputer facilities. This project is supported by a National Institutes of Health research grants GM-32476 (to Y.-L.W.) and GM-61806 (to M.D.). S.M. is supported by National Institutes of Health National Research Service Award predoctoral fellowship GM-20749.
Footnotes
Online version of this article contains video material for certain figures. Online version available at www.molbiolcell.org.
REFERENCES
- Abercrombie M, Heaysman JEM, Pegrum SM. The locomotion of fibroblasts in culture. Exp Cell Biol. 1970;59:393–398. doi: 10.1016/0014-4827(70)90646-4. [DOI] [PubMed] [Google Scholar]
- Beningo KA, Dembo M, Kaverina I, Small JV, Wang Y-L. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating Fibroblasts. J Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D. Cell movements from molecules to motility. New York: Garland Publishing; 2001. pp. 119–120. [Google Scholar]
- Burton K, Park JH, Taylor LD. Keratocytes generate traction forces in two phases. Mol Biol Cell. 1999;10:3745–3769. doi: 10.1091/mbc.10.11.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-T. Surface changes during retraction induced spreading of fibroblasts. J Cell Sci. 1981;49:1–13. doi: 10.1242/jcs.49.1.1a. [DOI] [PubMed] [Google Scholar]
- De Beus E, Jacobson K. Integrin involvement in keratocyte locomotion. Cell Motil Cytoskeleton. 1998;41:126–137. doi: 10.1002/(SICI)1097-0169(1998)41:2<126::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Dedhar S, Ruoslahti E, Pierschbacher MD. A cell surface receptor complex for collagen type 1 recognizes the arg-gly-asp sequence. J Cell Biol. 1987;104:585–593. doi: 10.1083/jcb.104.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70:2008–2022. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M, Wang Y-L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson EL, Felder SF, Jay PY, Kolodney MS, Pasternak C. Forces in cell locomotion. Biochem Soc Symp. 1999;65:299–314. [PubMed] [Google Scholar]
- Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA. 1997;94:9114–9118. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlsen RK, Argraves SW, Pierschbacher MD, Ruoslahti E. Inhibition of in vitro tumor cell invasion by arg-gly-asp containing synthetic peptides. J Cell Biol. 1988;106:925–930. doi: 10.1083/jcb.106.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KA, Wild P, Stopack D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- Horwitz AR, Parsons JT. Cell migration—Movin'on. Science. 1999;286:1102–1103. doi: 10.1126/science.286.5442.1102. [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lo C-M, Wang H-B, Dembo M, Wang Y-L. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NM, Wang Y-L, Konkel ME. Formation and movement of myosin-containing structures in living fibroblasts. J Cell Biol. 1989;109:1163–1172. doi: 10.1083/jcb.109.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munevar S, Wang Y-L, Dembo M. Traction force microscopy of migrating normal and h-ras transformed 3T3 fibroblasts. Biophys J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Warner AK, Wang Y-L. Distinct roles of equatorial and polar cortices in the cleavage of adherent cells. Curr Biol. 2001;11:702–707. doi: 10.1016/s0960-9822(01)00181-6. [DOI] [PubMed] [Google Scholar]
- Oliver T, Dembo M, Jacobson K. Separation of propulsive and adhesive traction stresses in locomoting keratocytes. J Cell Biol. 1999;145:589–604. doi: 10.1083/jcb.145.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Wang Y-L. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol Biol Cell. 1999;10:935–945. doi: 10.1091/mbc.10.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytela R, Pierschbacher MD, Ginsberg MH, Plow EF, Ruoslahti E. Platelet membrane glycoprotein Iib/IIIa: member of a family of Arg-Gly-Asp-specific adhesion receptors. Science. 1986;231:1559–1562. doi: 10.1126/science.2420006. [DOI] [PubMed] [Google Scholar]
- Radmacher M, Tillmann RW, Fritz M, Gaub HE. From molecules to cells: imaging soft samples with the atomic force microscope. Science. 1992;257:1900–1905. doi: 10.1126/science.1411505. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular matrix-integrin complexes. Trends Cell Biol. 1998;8:51–54. doi: 10.1016/s0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Progress in protrusion: the tell-tale scar. Trends Biochem Sci. 1999;24:432–436. doi: 10.1016/s0968-0004(99)01461-9. [DOI] [PubMed] [Google Scholar]
- Svitkina T, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky BA, Borisy GG. Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J Cell Biol. 1993;123:637–652. doi: 10.1083/jcb.123.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L, Pelham R. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods Enzymol. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- Yamada MK, Miyamoto S. Integrin transmembrane signaling and cytoskeletal control. Curr Opin Cell Biol. 1995;7:681–689. doi: 10.1016/0955-0674(95)80110-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.