Abstract
Circadian timekeeping systems drive oscillatory gene expression to regulate essential cellular and physiological processes. When the systems are perturbed, pathological consequences ensue and disease risks rise. A growing number of small-molecule modulators have been reported to target circadian systems. Such small molecules, identified via high-throughput screening or derivatized from known scaffolds, have shown promise as drug candidates to improve biological timing and physiological outputs in disease models. In this review, we first briefly describe the circadian system, including the core oscillator and the cellular networks. Research progress on clock-modulating small molecules is presented, focusing on development strategies and biological efficacies. We highlight the therapeutic potential of small molecules in clock-related pathologies, including jet lag and shiftwork; various chronic diseases, particularly metabolic disease; and aging. Emerging opportunities to identify and exploit clock modulators as novel therapeutic agents are discussed.
Keywords: circadian clock, high-throughput screen, chemical derivatization, chronotherapy, clock-related diseases, aging
INTRODUCTION
In biology, as in society, timing is crucial for optimal performance. In response to the daily environmental changes of our planet, a biological timer called the circadian clock has evolved in virtually all organisms (1, 2). These clocks are characterized by both stable periodicity and robust entrainment. The periodicity is primarily synchronized by the physical rhythm of Earth’s rotation. To align with the 24-h external cycle, the circadian system is endowed with exceedingly complex mechanisms to ensure period constancy. These include feedback gene regulation at all levels (ranging from transcriptional initiation to protein degradation), genetic redundancy (gene duplication), and coupling of intracellular and intercellular oscillators (3). By contrast, the clock can be readily entrained by cellular and external cues to adjust to environmental and (patho)physiological signals. Light is a dominant zeitgeber (time giver) for photosynthetic organisms and light-sensing organs in animals. Since the seminal study establishing peripheral oscillators in cell lines (4), a wide spectrum of external factors and endogenous metabolites capable of resetting the intrinsic clocks have been described (5–7). Analogous to a well-manufactured watch, the circadian system is both precise and adjustable.
In the natural environment, intact clock functions are required for optimal growth, predator avoidance, and protection against environmental challenges such as redox and irradiation (8–12). Studies conducted with laboratory animals and human subjects have shown acute and chronic adverse effects on health and fitness when circadian systems are misaligned or attenuated by genetic or environmental perturbation (13, 14). Although it is clearly important for species to survive and thrive, circadian timekeeping in the realm of drug development is mainly limited to chronotherapeutic applications, namely optimizing the schedule for established therapies to attain maximum therapeutic index (15). As our knowledge of the organization, regulation, and function of circadian systems grows, researchers are increasingly interested in developing small molecules to target the circadian system directly for therapeutic gains. In addition to jet lag, circadian dysregulation is implicated in various chronic diseases as well as age-related decline (16–19). The purpose of this review is to synthesize key research advances in small-molecule modulators of the mammalian clock and illustrate opportunities for further small-molecule identification and therapeutic developments.
MAMMALIAN CIRCADIAN SYSTEM
Hierarchical Organization and the Cell-Autonomous Oscillator
At the apex of the hierarchical mammalian circadian system (Figure 1) is the suprachiasmatic nucleus (SCN) master pacemaker, a pair of neuron clusters bilaterally located in the anterior of the hypothalamus (20). Blue light activates melanopsin-containing intrinsically photosensitive retinal ganglion cells (ipRGCs) in the retina, transmitting signals via the retinohypothalamic tract to the SCN (21). In postsynaptic SCN neurons, the N-methyl-D-aspartic acid (NMDA) glutamatergic and pituitary adenylate cyclase-activating polypeptide (PACAP) signaling pathways involving Ca2+ and cAMP (22) play predominant roles to induce immediate early genes, including the core clock gene Period1 (Per1), and subsequently reset SCN cellular oscillators. Compared with peripheral tissues, SCN oscillators are tightly coupled (23). Several neurotransmitters have been implicated in intercellular SCN coupling, with the vasoactive intestinal peptide (VIP) being the most prominent (24). The SCN functions to synchronize tissue and cellular clocks throughout the body via neural and hormonal signals (25). For example, the SCN directly or indirectly projects to several cell bodies in the hypothalamus, regulating the rhythmic release of glucocorticoid hormone (26).
Figure 1.
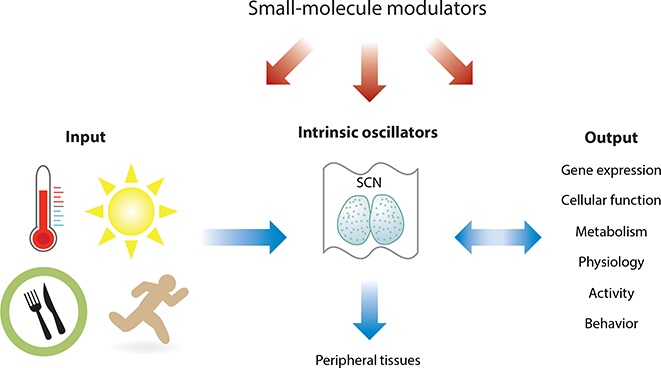
Mammalian circadian system. The circadian system consists of input pathways, the clock, and output functions. Light is a major input signal for the circadian system and activates the master pacemaker SCN. The SCN coordinates the cellular oscillator network throughout the body via neuronal and hormonal signals. The clock drives expression of clock-controlled genes in a tissue-specific manner, thereby regulating tissue physiologies and systemic functions. Many output functions such as activity, metabolism, and temperature also reciprocally regulate the clock. Small-molecule modulators can alter the input pathway, oscillator, and output functions with feedback activities. Abbreviation: SCN, suprachiasmatic nucleus.
The operational unit of the mammalian circadian system is the cell-autonomous molecular oscillator (Figure 2), found in not only SCN pacemaker neurons but also virtually all other peripheral cells (4, 27). The molecular oscillator is characterized by interconnected negative feedback loops (1). In the core loop, each of the paralogous basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) transcription factors circadian locomotor output cycles kaput (CLOCK) and neuronal pas protein 2 (NPAS2) heterodimerizes with brain and muscle ARNT-like 1 (BMAL1) through their bHLH and PAS domains to activate expression of Period (Per) and Cryptochrome (Cry) genes via E-box promoter elements. The PER and CRY proteins in turn heterodimerize and translocate into the nucleus to inhibit transcriptional activities of CLOCK/BMAL1 and hence their own transcription. In the stabilization loop, two families of nuclear receptors, REV-ERBα/β and RAR-related orphan receptor α/β/γ (RORα/β/γ), compete for binding to shared consensus elements (RORE and RevDR2) on the promoter of Bmal1 and other target genes throughout the genome (28). The negative feedback is mediated via robust circadian expression of the genes encoding REV-ERBs driven by CLOCK/BMAL1 (26). Other feedback loops, including a third transcriptional loop involving the proline and acidic amino acid-rich basic leucine zipper (PARzip) protein, D-box binding protein (DBP), and E4 promoter-binding protein 4 (E4BP4)/NFIL3 (29), as well as post-translational (NAD+-dependent SIRT1) mechanisms (30–32), also intersect with these two loops to confer further regulation (Figure 2).
Figure 2.
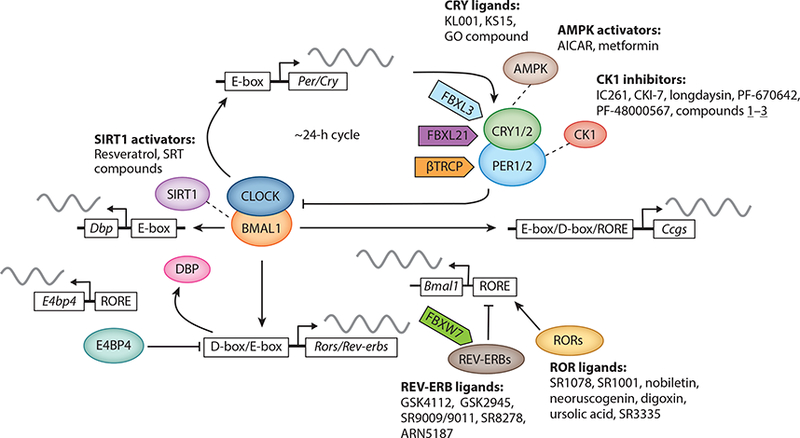
Core and regulatory components of the circadian oscillator and representative small-molecule modulators. The circadian clock oscillator is composed of a network of transcriptional-translational feedback loops, including the core loop (BMAL1/CLOCK/NPAS2 and PERs/CRYs), the stabilization loop (BMAL1/CLOCK, REV-ERBs, and RORs), and the auxiliary loop (DBP, E4BP4, REV-ERBs, and RORs). Various protein regulators and ligands have been determined to regulate core clock components and circadian functions. Modified from Reference 50 under Creative Commons Attribution license. Abbreviations: BMAL1, brain and muscle ARNT-like 1; CLOCK, circadian locomotor output cycles kaput; CRY, cryptochrome; DBP, D-box binding protein; E4BP4, E4 promoter-binding protein 4; NPAS2, neuronal PAS protein 2; PER, period; ROR, RAR-related orphan receptor.
In specific tissues, the master pacemaker and local oscillators are normally coordinated to drive systemic and tissue-autonomous gene expression programs (33). The clock-controlled genes (CCGs) vary greatly from tissue to tissue, with the overlap of CCGs between tissues at approximately 10% (34). However, when 12 different tissues were examined, 43% of all genes were found to oscillate in at least 1 tissue in mice (35). Such rhythmic gene expression programs dictated by the circadian clock in turn govern the daily oscillation of metabolism, physiology, and behavior.
Physiological Inputs and Molecular Mechanisms to Reset the Clock
As mentioned above, the circadian clock is receptive to a diverse array of input signals (5), providing the initial hint that novel small-molecule modulators can be identified to manipulate the clock. The underlying regulatory network is emerging, including transcription cofactors, epigenetic regulators, and enzymatic activities involved in various steps of gene expression (33). Diverse regulators with histone modification (e.g., acetylation, methylation, and monoubiquitination and their respective reverse modifications) and nucleosome remodeling activities are involved in circadian regulation (36, 37), particularly on the E-box during the transcriptional cycle (1, 38). On the RORE elements, several cofactor complexes have also been identified, including HDAC3 and NCOR1 (36). Recent studies have also shown that lineage-specific enhancers and transcription factors cooperate with the core circadian factors to govern circadian transcription in individual tissues (28, 39). For example, in mouse liver, DNA binding-deficient REV-ERBα remained tethered to promoter regions enriched for the consensus sites for the liver transcription factor HTF6 (28), suggesting an indirect strategy to recruit core circadian factors to cell type-specific regulatory regions. Posttranscriptional and posttranslational mechanisms also play an important regulatory role (40–42). In particular, degradation of PERs and CRYs is a key mechanism governing circadian period length. PERs are phosphorylated by casein kinase 1 (CK1) prior to their proteasomal degradation by the F-box proteins ß-TRCP1/2 (41). Likewise, CRY proteins are substrates of AMPK (43), and their subsequent degradation is coordinately mediated by paralogous F-box proteins FBXL3 and FBXL21 in an antagonistic manner (44–48). As exemplified by the CK1 inhibitors detailed below, these key regulators of circadian clocks are excellent targets for small-molecule development.
Circadian robustness, or amplitude, serves an important physiological function, as dampened amplitude is strongly correlated with chronic diseases and aging (16, 49, 50). Genetic redundancy, intercellular coupling, and stoichiometric balance between positive and negative factors are important to sustain robust cycling. Paralogous circadian components play overlapping roles to safeguard rhythmicity. For example, CLOCK and NPAS2 are paralogous transcription factors with redundant functions in both SCN and peripheral cells (51–53). For both SCN and peripheral tissues (3, 23), coupling of oscillators [e.g., by VIP in the SCN (54)] renders greater resistance to genetic perturbation and noise compared with single-cell oscillators (27, 55, 56). Within the oscillator, stoichiometric ratios between positive and negative limbs are important for high-amplitude gene oscillation (57). In particular, REV-ERBs and RORs of the stabilization loop play important roles in amplitude regulation. REV-ERBs show strong oscillation in both mRNA expression and protein degradation and compete with RORs to regulate occupancy and activity at target gene promoters (28, 54). REV-ERBs and RORs also engage a facilitated recruitment mechanism in which a rate-limiting step mediated by ROR/BMAL1 and transcription cofactors SRC-2/PBAF permits REV-ERB binding to open chromatin (58). Recruitment of REV-ERB by the activators ROR/BMAL1 likely facilitates transcriptional repression at the end of the activation phase, ensuring robust gene oscillation.
SMALL-MOLECULE MODULATORS OF CIRCADIAN SYSTEMS
Identification of Small-Molecule Clock Modulators
The circadian system is highly amenable to resetting signals, including environmental changes, intracellular mechanisms, and endogenous metabolites [e.g., flavin adenine dinucleotide (FAD), heme, and cholesterols act as natural ligands for CRYs, REV-ERBs, and RORs] (59, 60). Novel small molecules can modulate the circadian clock directly on the oscillator, via input pathways or through feedback mechanisms from output functions (Figure 1). If the entry point is too far removed from the core oscillator, the small molecules would likely exhibit pleiotropic effects separate from its circadian function. Therefore, we focus our discussion on chemical compounds targeting core clock components (Table 1), key regulators, and the oscillator (Table 2).
Table 1.
Representative small-molecule modulators of core clock proteinsa
| Clock proteins |
Modulators | Circadian or target activities | Physiological effects | References |
|---|---|---|---|---|
| CRY1/2 | KL001 and CRY-stabilizing derivative |
Stabilize CRY Lengthen period Reduce amplitude |
Improve glucose tolerance in obese mice |
65, 72 |
| GO044 GO200 GO211 |
Variable effects on CRY Shorten period |
NA | 160 | |
| 2-ethoxypropanoic acid KS15 |
Inhibit CRY Activate E-box transcription Shorten period Reduce amplitude |
Inhibit breast cancer cell growth | 70, 161 | |
| REV-ERBs | GSK4112 | Enhance REV-ERB and NCOR peptide interaction |
Inhibit gluconeogenesis and inflammatory response in primary cells |
59,71,85 |
| SR9009 SR9011 |
Derived from GSK4112 Selective agonists for REV-ERB Alter circadian behavior and gene expression |
Improve glucose homeostasis in obese mice Promote wakefulness Reduce anxiety |
76,127 | |
| GSK2945 GSK0999 GSK5072 GSK2667 |
Derived from GSK4112 Selective agonists for REV-ERB Reduce Bmal1 expression |
Inhibit inflammatory response | 79 | |
| SR8278 | GSK4112-derived antagonist | Reduce glucagon secretion from mouse alpha cells Reduce anxiety and promote maniac-like behavior |
59, 128, 162 | |
| ARN5187 | REV-ERB β agonist | Cytotoxic against cancer cells | 163 | |
| RORs | SR1078 | ROR agonist | Induce apoptosis Inhibit hepatoma cell growth Improve autistic behavior in mice |
164, 165 |
| Nobiletin | Agonist Enhance amplitude and lengthen period |
Improve metabolic homeostasis in obese/diabetic mice Show broad efficacies against tumors, inflammation, and cardiovascular disease |
68,100–102 | |
| Neoruscogenin | Agonist promoting ROR interaction with NCOA2/TIF2 Activate Bmall expression |
Activate hepatic expression of ROR metabolic target genes such as Cyp7b1, G6Pase, Lpin2, and Angptl4 |
166 | |
| Compound 1a | RORγ agonist | Promote Th17 cell differentiation | 167 | |
| T0901317 | Inverse agonists of RORs and agonist of LXR |
Diverse biological activities regulated by LXR |
118 | |
| SR1001 | Derived from T0901317 with high inverse agonist activity and selectivity for RORα and RORγ | Inhibit Th17 cell differentiation and autoimmunity | 117 | |
| SR2211 SR1555 Digoxin Ursolic acid ML209 |
RORγ inverse agonist | Inhibit Th17 cell differentiation | 59 | |
| SR3335 | RORα inverse agonist | Reduce blood glucose in obese mice | 93 |
Abbreviations: BMAL1, brain and muscle ARNT-like 1; CRY, cryptochrome; LXR, liver X receptor; NA, not applicable; ROR, RAR-related orphan receptor.
For additional related compounds and references, please refer to a recent review (59).
Table 2.
Representative small-molecule modulators with other or unknown targetsa
| Small molecules | Target activity | Circadian phenotype | Physiological effects | References |
|---|---|---|---|---|
| Opsinamides | Melanopsin inhibitors |
Suppress light input to the SCN |
Generally suppress nonvisual photoresponse |
81 |
| SB432542 | ALK5 inhibitor | Attenuate phase delay | Inhibit TGF-β signaling | 82 |
| AICAR, metformin | AMPK activator | Lengthen period Reduce amplitude |
AICAR: exercise mimetic Metformin: antidiabetic |
43, 168 |
| CKI-7, IC261, D4476, PF-670462, longdaysin, LH846, Compound 1-3, and others |
CK1 inhibitor | Lengthen period | CK1 is broadly involved in various pathophysiologies, including familial sleep and mood disorders |
75, 78 |
| PF-4800567 | Selective CK1 ε inhibitor |
Blocks PER2 degradation but shows minimal effects on circadian period |
Increases sensitivity to psychostimulants and opioids |
73 |
| DRB, DMAT | CK2 inhibitor | Lengthen period | Antineoplasia | 78, 169 |
| Lithium | GSK3 β inhibitor | Lengthen period Enhance amplitude |
Mood disorders | 125, 170 |
| Chir99021, 1-azakenpaullone, indirubin |
GSK3 β inhibitor | Shorten period | Improve glucose metabolism, anti-inflammatory and antiangiogenesis |
62 |
| L-Methyl selenocysteine | Enhance Bmal1 transcription |
Induction of BMAL1 protein level and activity |
Improved survival of cyclophosphamide-sensitive Clock mutant mice |
69 |
| Resveratrol | Sirt1 activator | Modulate physiological rhythms and clock gene expression |
Broad physiological and antiaging efficacies |
140 |
| SRT2183, SRT1720, SRTCD1023, SRTCL1015 |
Sirt1 activator | Reduce circadian expression Lengthen period Reduce amplitude |
Improve circadian clock gene expression Reduce inflammation in chronic obstructive pulmonary disease |
143, 171 |
| OPC-21268, SSR 149415 | Antagonists for arginine vasopressin receptors V1a and V1b, respectively |
Accelerate entrainment to jet lag |
Accelerate reentrainment after jet lag and shift work |
109 |
| Compound 5, a benzodiazepine derivative |
GABA receptor agonist Acts on an unknown target |
Lengthens period in both SCN and peripheral clocks |
NA | 66 |
| Compound 10/CEM3 | Unknown | Enhance amplitude and shorten period in both SCN and peripheral clocks |
NA | 61,66 |
Abbreviations: AMPK, AMP-activated protein kinase; CEM, clock-enhancing small molecule; CK1, casein kinase 1; CK2, casein kinase 2; GABA, γ-aminobutyric acid; GSK3 β, glycogen synthase kinase 3 β; NA, not applicable; PER, period; SCN, suprachiasmatic nucleus; TGF-β, transforming growth factor-β.
For additional related compounds and references, please refer to a recent review (78).
Small-molecule clock modulators (Figure 3) have been identified via chemical screening of libraries of varying sizes using different primary screening assays (17, 61). In phenotypic functional assays, stable cell lines express either luciferase from an exogenous clock gene promoter (62–65) or PER2::luciferase fusion from the endogenous Per2 promoter (66–68), reporting circadian transcription and protein oscillation, respectively. Following cell synchronization, circadian bioluminescence rhythms are monitored continuously over several days (a so-called kinetic assay), and cardinal circadian parameters, including period, phase, and amplitude, are quantified by data analysis tools. Such screens using cell-based assays have several advantages. They are not restrictive with regard to compound targets and mechanisms (Figure 2), and circadian effects are well defined for the reporter assay. The most significant challenge, however, is the elucidation of the direct targets for novel small molecules. The second cell-based screening strategy entails endpoint assays measuring specific transcription activities. In these assays, reporter cell lines are constructed to express luciferase under the control of the circadian gene promoter or promoter element (69, 70), and compounds are screened for endpoint transcriptional readout (up- or downregu-lated). Secondary assays (such as the kinetic assays mentioned above) can be carried out to validate their circadian effects. Finally, in vitro biochemical assays can identify compounds that specifically target certain clock components. For example, GSK4112 (71), the first synthetic agonist of REV-ERB, was identified via a fluorescence assay measuring the biochemical interaction between REV-ERB and an interacting peptide of its cofactor NCOR. Compared with cell-based assays, biochemical assays directly identify compounds for the known protein targets.
Figure 3.
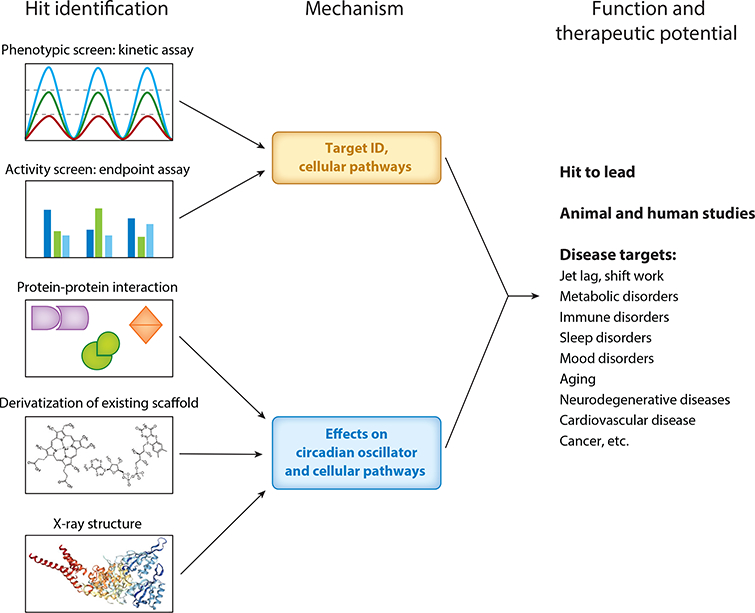
Development of clock-modulating small molecules. Two general strategies can be deployed to identify clock-modulating small molecules. The modulators can be identified via kinetic phenotypic screens measuring circadian parameters or endpoint activity screens measuring transactivation. Elucidation of the protein target is important for mechanistic and functional studies. Small molecules can also be developed via biochemical assays measuring protein-protein interaction, known ligands or scaffolds, or structure-based design. Circadian activities and mechanistic pathways can be characterized for prioritized hits. Depending on pharmacokinetic profile, target, and circadian function, small molecules can be applied to in vivo models of clock-associated diseases and aging.
Primary hits are prioritized through secondary and counterscreen assays. Validated modulators suggest privileged scaffolds that can be subjected to chemical modification, aided by available knowledge of ligand structure, ligand-protein interaction, or both (59, 72). Improved pharmacokinetics and pharmacodynamics are typically required before in vivo applications, particularly those of therapeutic potential. In some instances, the scaffolds are available without the initial screening. For example, whereas IC261 and CKI-7 are CK1 inhibitors known to lengthen the clock period with moderate potency and minimal selectivity (Table 2), a focused medicinal chemistry effort led to a selective inhibitor of CK1ε, PF-4800567, which confers >20-fold greater inhibition over that of CK1δ (73–75).
Circadian Efficacies and Targets of Small-Molecule Modulators
Phenotypic screens have identified a diverse array of compounds showing effects on circadian period, phase, and/or amplitude (63, 65, 76–79). A major group of clock-modulating small molecules are the ones that alter, and especially lengthen, circadian period length. Consistent with genetic evidence showing a key role of PERs and CRYs in period regulation, compounds that affect PER/CRY levels were found in several chemical screens to alter circadian periods. For example, screens of known drugs have revealed numerous kinase inhibitors that potently lengthened circadian period (62, 63, 67). Although many drugs are known to target other kinases, they showed robust inhibition of CK1 kinase activity (63). It is possible that CK1 inhibition plays a predominant role in their period-lengthening activity. A central role of CK1 in determining circadian period is further illustrated by two studies that used pull-down or activity assays to identify new period-lengthening compounds that serve as CK1 inhibitors (64, 66). Moreover, KL001, a car-bazole derivative, was identified in a cell-based screen to lengthen period and reduce amplitude, and pull-down experiments revealed CRY proteins as its target (65) (Table 1). Structural studies demonstrated that KL001 binding to the FAD-binding pocket of CRY interferes with its recognition by FBXL3 (80), thereby repressing CRY ubiquitination and proteasomal degradation. Compounds that target other cellular proteins or unknown targets with period-lengthening or -shortening effects have also been reported (61, 62).
Whereas period changes lead to long-term phase alteration, acute circadian phase resetting is primarily mediated via cAMP signaling, which leads to induction of the immediate early genes, including Per1 (26, 66). Compounds that acutely advance or delay circadian phase act at different points of the pathway, including upstream kinases [e.g., extracellular signal-regulated kinase (ERK) and Ca2+/calmodulin-dependent protein kinase II (CamKII)], adenylyl cyclase, and phosphodiesterase (61). Furthermore, researchers used high-throughput screening to discover a group of sulfonamide compounds, opsinamides, that antagonize retinal binding to melanopsin and inhibit its photoreceptor functions in vitro and in vivo (81). Per-independent phase resetting pathways have also been described (82, 83), and modulatory compounds (e.g., SB432542) may function to reset circadian phase (Table 2).
Cell-based phenotypic screens have identified several clock amplitude-enhancing small molecules (CEMs) (61). The first group of synthetic CEMs potentiated cellular and tissue reporter rhythms (66), and the benzimidazole compound CEM3 in particular displays a unique capability to potentiate SCN reporter rhythms. A more recent screen identified the natural flavonoid Nobiletin (NOB) and its analog Tangeretin as CEMs (68). NOB increased mRNA and protein oscillatory amplitude in both fibroblast cells and liver, and filter-binding and functional assays established ROR receptors as the direct targets of NOB (68), reaffirming an important function of RORs in circadian amplitude regulation. Despite a strong NOB-ROR interaction, NOB-mediated activation of ROR target genes, including both core clock (e.g., Bmal1) and clock-controlled output genes, was generally moderate, likely because of the autoregulatory feedback loop within the core circadian clock (68).
Chemical modification of privileged scaffolds has led to various ligands targeting REV-ERBs and RORs with improved specificity and potency (Table 1) (59). For example, a tertiary amine series was reported to function as agonists of REV-ERBα (71, 76, 84, 85). However, for many of these modified REV-ERB and ROR ligands, their effects on the circadian clock are not well characterized. An inverse agonist of REV-ERBs, SR9011, was found to diminish circadian amplitude (76) despite its similar function with NOB in tipping the balance between REV-ERBs and RORs toward the latter. Together with the moderate induction of NOB in association with amplitude enhancement, this finding is in agreement with the limit-cycle nature of the clock, in which positive and negative limbs must balance and synergize to achieve greater amplitude over a sustained period.
THERAPEUTIC POTENTIAL OF SMALL-MOLECULE CLOCK MODULATORS
Circadian Clock as Drug Target
A majority of top-selling drugs in the United States target proteins that are encoded by oscillatory genes, and almost half of these drugs have short half-lives (<6 h) (35). Molecular studies have also provided strong evidence that xenobiotic metabolism is subjected to circadian regulation (86). These observations are part of our growing knowledge on circadian pharmacokinetics and pharmacodynamics, indicating an important role of circadian dosing time for drug efficacy (15, 77). However, clock-modulating small molecules can directly manipulate the circadian system to improve clock-regulated output processes, alleviating disease symptoms and physiological decline (16–18, 61). Below, we begin our discussion of therapeutic potential with metabolic disease to highlight key lessons.
Distinct Clock-Modulating Small Molecules Against Metabolic Disorders
Circadian clocks and metabolism are interdependent (87, 88). In human subjects and laboratory mice, dysregulation of circadian timing by phase misalignment or genetic manipulation compromised energy homeostasis and increased the risk of metabolic disorders. Circadian clocks, both central and local, drive cyclic expression of metabolic genes in important metabolic tissues (32, 89). Mechanistic studies have unveiled molecular interaction between the clock and metabolic pathways. For example, it was recently found that the clock-NAD+ loop regulates the NAD+-dependent SIRT3 to modulate the acetylation state of key proteins involved in mitochondrial respiration (90). Together with previous studies on clock regulation by SIRT1 (30, 31), these observations underscore an important role of sirtuins in circadian metabolic control.
Several studies have reported distinct clock-modulating small molecules as potential therapeutics for metabolic disease. The first strategy targets circadian components with an established role in metabolic regulation, including REV-ERBs and RORs (36, 76, 91). Two synthetic agonists of REV-ERBs, SR9011 and SR9009, markedly improved energy homeostasis and acutely altered circadian behavior in diet-induced obese (DIO) mice (76). Consistent with the effect of these REV-ERB agonists, an inverse agonist of RORs, SR1555, was found to reduce weight and increase activity in DIO mice (92). Several other RORα/γ ligands can also modulate hepatic metabolism (93, 94). Another clock modulator, the CRY stabilizer KL001, lengthened circadian period and repressed amplitude (65), and a bioavailable derivative was recently shown to improve glucose tolerance in DIO mice (72). In addition to functioning as the major repressor in the circadian core loop, CRYs are also known to modulate gluconeogenesis in the liver by directly interacting with the Gsα subunit of G protein-coupled receptors to modulate cAMP signaling (95).
A different strategy focused on enhancing the clock amplitude. Several lines of evidence suggest a relationship between dampened amplitude and metabolic disorder, including human studies showing that attenuated oscillation in insulin secretion rhythm correlates with exaggerated diabetes risk (96). ClockΔ19/Δ19 mutant mice suffering a broad array of metabolic dysfunctions experienced a profound attenuation of circadian gene expression and feeding rhythm (97). Whereas ad libitum high-fat diet feeding repressed circadian gene oscillation in rodents, time-restricted feeding (TRF) during nighttime enhanced circadian amplitude and improved metabolic health (6, 98, 99). Consistent with previous studies (100), the clock-enhancing compound NOB showed considerable efficacy in mitigating weight gain and improving energy homeostasis in both DIO mice and db/db diabetic mice, corresponding to enhanced circadian functions (68, 101, 102). Together, these findings support the notion that clock enhancement by NOB contributes to metabolic improvement (103).
Several lessons have emerged from these studies. First, distinct strategies targeting either specific clock components or the overall circadian systemic feature (e.g., amplitude) can lead to metabolic improvements. The fact that these metabolic-promoting compounds showed distinct effects on circadian phenotypes (e.g., amplitude) suggests a circadian metabolic network that is both extensive and flexible. Second, ligand activity and physiological output may not follow a simple correlation, and each compound should be evaluated experimentally. Several ROR inverse agonists and REV-ERB agonists (59, 78) improved energy metabolism in metabolic disease models (92, 104), whereas NOB showed similar efficacies as an ROR agonist. Underscoring the need for detailed understanding of compound-specific mechanisms, researchers recently found that three antagonists of RORγt employ varying mechanisms to modulate RORγt promoter binding and target gene expression (105). Finally, genetic manipulation may not be extrapolated to pharmacological effects, and vice versa. The three RORγt antagonists mentioned above displayed different mimicry of genetic ablation (105). In a classical example (106), both peroxisome proliferator-activated receptor γ (ΡΡΑRγ) haplodeficiency and the ΡΡΑRγ agonists thiazolidine-diones (TZDs) led to insulin sensitizing, which was later found to involve distinct regulatory mechanisms of hepatic and adipose tissue functions, respectively. ROR activation by NOB seemingly presents a paradox because ROR deficiency appears to retard body weight gain (107). Of note, global ROR deficiency, although shown to improve certain metabolic parameters, is deleterious to overall fitness, including immunity, motor function, development, and cardiovascular health (91).
Jet Lag and Shift Work
Transmeridian long-distance flights lead to jet lag, characterized by misalignment of internal rhythms, which remain locked to the origin, and the destination solar and social cycles. When the circadian misalignment becomes chronic, as in shift work, prevalent health problems can arise, including serious sleep and gastrointestinal disturbances, increased risk of several cancers (e.g., breast cancer), metabolic syndrome (being prone to weight gain), cardiovascular disease, and mood disorders (14, 88). The pineal hormone melatonin can facilitate sleep adjustment after jet lag, likely involving its phase-resetting activity in the SCN, where its receptors are expressed (108). However, the effects of melatonin are typically minor, and several side effects, such as headache and daytime drowsiness, are common. Because the primary circadian phenotype in jet lag and shift work is phase misalignment, small molecules with appropriate phase-resetting activities are candidates for drug development. In an in vivo screen to identify signaling pathways for jet lag, arginine vasopressin and its receptors (V1a and V1b receptors) were found to play an important role in the intercellular cross talk in mouse SCN (109), and the receptor double knockout mice displayed immediate phase shift under experimental jet lag conditions. Treatment of mice with an antagonist mix (OPC-21268 and SSR 149415 for V1a and V1b, respectively) shortened the phase shift time by half, suggesting a therapeutic strategy for jet lag and shift work.
Sleep Disorders
Sleep is an essential function governed by both circadian and homeostatic mechanisms (110). Human and mouse genetic studies have shed light on clock-associated sleep disorders, particularly familial advanced sleep phase syndrome (FASPS) and delayed sleep phase syndrome (111). FASPS is characterized by circadian period shortening, and mutations in both PER2 (PER2 S662G) and CSNK1D (CK1δ T44A) have been identified as an underlying genetic cause (111). The CK1δ-specific inhibitor PF-670462 induced behavioral rhythms in arrhythmic mice either subjected to constant light exposure or harboring the Vipr2−/−mutation (75), suggesting a pharmacological agent could target CK1δ to prolong the period in FASPS patients or animal models. In a recent forward genetic screen (112), two mouse mutants expressing mutant forms of the SIK3 kinase and the sodium channel NALCN displayed long sleep and shortened REM sleep phenotypes. Interestingly, both genes exhibit circadian oscillatory expression under normal conditions (113), consistent with an integrative control of sleep by both the circadian clock and the homeostatic pathway. Circadian modulators that influence their expression can be evaluated for sleep improvement.
Immune Disorders
Both innate and adaptive immune functions are regulated by the circadian clock (114). For example, the clock regulates the timing and duration of the expression of various proinflammatory cytokines such as IL-6, IL-17, tumor necrosis factor, and CXCL1. REV-ERBα plays a regulatory role in macrophage transcription (59), and mice deficient in REV-ERBα expression lost the gating of IL-6 induction following lipopolysaccharide (LPS) challenge, as evidenced by a highly elevated trough level at CT0 (85). RORγt is a master regulator for the development of the T helper cell 17 (Th17), an important immune cell type for autoimmune disorders, and ligands of REV-ERBs and RORs have shown beneficial effects on immune functions. The REV-ERB ligand GSK4112 repressed LPS induction of IL-6, CXCL11, CXCL6, and CCL2 in primary human macrophages (85). Digoxin and ursolic acid, as RORγ inverse agonists, ameliorated autoimmune disorders, including arthritis and encephalomyelitis, via suppression of Th17 differentiation (115, 116). Several optimized RORα/γ ligands attenuated expression of downstream cytokines and strongly alleviated autoimmune disease symptoms (117, 118), suggesting that targeting these clock components constitutes a valid strategy against inflammation and autoimmune diseases.
Mood Disorders
Circadian occurrence of psychiatric episodes is well documented, often most pronounced in the morning or around sunset (119, 120). During winter months, depressed seasonal affective disorder patients (121) show dampened rhythms in feeding, sleep, body temperature, and hormone release. Consistent with disrupted circadian rhythms often found in human mania (122), similar behaviors (123) were observed in clock-disrupted ClockΔ19/Δ19 mice. Release of glucocorticoid hormones regulating stress response and mood balance is subject to circadian control. In a recent study, enhanced circadian amplitude of glucocorticoid rhythm, without increase of total glucocorticoid levels (124), showed anxiolytic effects, potentially due to the marked glucocorticoid reduction during the descending phase of its high-amplitude oscillation. In addition to environmental and behavioral therapies such as bright light therapy and sleep deprivation, small-molecule modulators of the clocks have also been used in mood disorders. Lithium, a mood stabilizer to treat bipolar disorder, showed complex effects on the circadian system, including enhancement of circadian reporter amplitude in both the SCN and the periphery (125). Another demonstrated molecular target of lithium is GSK-3β, a broad-acting kinase previously shown to phosphorylate and stabilize REV-ERBα (126). Agonists of REV-ERBs also displayed anxiolytic effects (127), consistent with genetic evidence showing enhanced anxiety in REV-ERBß knockout mice. Interestingly, acute administration of a REV-ERB antagonist, SR8278, to mouse ventral midbrain was also able to reduce anxiety and promote maniac-like behavior (128, 129). These findings are reminiscent of the aforementioned observations that both NOB, an agonist of ROR, and certain inverse agonists of RORs can improve energy metabolism, likely through distinct mechanisms. Given the strong correlation between circadian amplitude dampening and mood imbalance, the therapeutic potential of CEMs would be an interesting area for further investigation.
Aging
Aging, marked by progressive decline in metabolic, physiological, and behavioral functions, leads to widespread circadian changes such as sleep phase advance in humans. In both humans and rodents, the response to entraining cues is markedly weaker and slower with age (130), which correlates with age-related impediment in circadian synchronization and amplitude attenuation (19, 49). Clock-regulated physiological and behavioral processes, including SCN firing rate, hormone secretion (e.g., cortisol and melatonin), sleep architecture, and body temperature, display reduced amplitude with age (49, 131). At the molecular level, there is also broad dysregulation of clock gene expression (131), particularly in peripheral clocks (132, 133). In genetic studies, Bmal1 knockout mice and αMUPA transgenic mice provide contrasting examples of premature aging and longevity, with strong correlations between aging effects and respective circadian functions (134, 135).
Dietary interventions, including caloric restriction (CR) and TRF, have shown antiaging effects (19). CR universally promotes longevity; similar to TRF, timed CR is accompanied by highly consolidated food intake within a few hours and enhances the amplitude of circadian metabolic rhythms (136, 137) and core clock gene oscillation (138). CR involves several pivotal metabolic regulators, including SIRT1, AMPK, AKT, and mTORC1, all of which functionally interact with the clock (32, 137, 139). For example, SIRT1 was found to interact with PGC-1α to control Clock and Bmal1 gene expression in the SCN, regulating CLOCK/BMAL1 target genes (140). Various SIRT1-activating small molecules such as resveratrol show longevity effects (141, 142). Pharmacological agents shown to promote or mimic clock-enhancing manipulations, such as CR, TRF, and exercise, are a rich venue for discovery of additional clock-targeting agents (136, 137, 143).
FUTURE DIRECTIONS AND CONCLUDING REMARKS
Developing New Modulators
Opportunities to identify new clock-modulating small molecules include chemical screens to target regulatory steps (e.g., intracellular translocation via high-content screening), new circadian regulators or targets (e.g., epigenetic factors and cofactors, enzymes regulating mRNA and protein levels, and SIK3), and new cell types (e.g., disease-associated or neuronal cells). Focused ligand modification can be applied to existing ligands or factors such as REV-ERBs and RORs with natural ligand-binding domains. For example, multiple structural studies have revealed molecular details of ROR isoforms (α, β, and γ) binding to their natural ligands, including cholesterol and stearic acid. Both RORa and RORγ are widely expressed, and their ligand-binding domains are highly similar, with the former slightly smaller in volume (144). It is possible to develop isoform-specific ROR ligands and differentiate functional roles.
Structural studies have provided rich insight into the core oscillator and, importantly, novel regulatory surfaces involved in ligand binding or protein-protein interactions (PPIs) (Figure 3). Mammalian CRY proteins retain low-affinity FAD binding in the photolyase homology region (145), and KL001 competes with the FBXL3 C terminus for binding at the FAD pocket (80), exemplifying a structural motif for compound development. In the CLOCK-BMAL1 crystal structure, the PAS-B domains of these molecules engage an asymmetrical interaction between the BMAL1 β-sheet and CLOCK α-helix (146). This interaction creates a fairly large internal hydrophobic surface (approximately 700 Ǻ2). More recently, a detailed crystal structure study revealed ubiquitous internal pockets in the PAS-A and PAS-B domains of bHLH-PAS proteins, including CLOCK and BMAL1 (147). These pockets, varying in volume, can be targeted by functional ligands. For example, a buried pocket in the PAS-B domain of hypoxia-inducible factor 2α (HIF2α) is accessible by synthetic ligands to modulate HIF2α/ARNT interaction (148). PAS domains first evolved in bacteria and plant species for ligand binding. The structural studies described above revealed a remnant ligand-binding ability in mammalian PAS domains as a promising target for drug discovery (149).
Although the presence of cardinal or covert ligand-binding cavities or clefts on core clock proteins allows in silico screening or chemical optimization of known ligands, the discovery of GSK4112 exemplifies another drug discovery strategy targeting PPIs (71, 150). Compared with biologics, small-molecule modulators of PPIs pose greater challenges, especially in terms of tertiary sequences and allosteric sites. Finding the interaction hot spots that confer disproportionally high binding energy is important for small-molecule perturbation (151). In the PER-CRY crystal structure, several regulatory interaction surfaces can be distinguished from the extensive interaction throughout the CRY-binding domain of PER (152), including interaction near the CRY secondary pocket that binds to CLOCK/BMAL1 and a competitive binding with FBXL3 for access to the extreme C-terminal surface on CRY. As PPIs are important for both clock protein activities and core complex formation on chromatin, advanced structural understanding will offer a rich source of drug discovery targets.
Exploring Therapeutic Potential
The list of clock-associated diseases continues to grow. Cardiovascular functions, including blood pressure, heart rate, cardiac contractility, and metabolism, exhibit robust circadian oscillation (153), and several key regulators, including Hsd3b6 and KLF15, are subjected to direct clock control (154, 155). There is also growing evidence for a relationship between clock dysregulation and tumorigenesis (156, 157). For example, a chronic jet-lag paradigm induced several cancer types, including hepatocellular carcinoma, and metabolic dysregulation partly mediated by the constitutive androstane receptor promoted progression of tumorigenesis from the initial fatty liver phenotype (158). Neurodegenerative diseases (16, 159) have been shown to correlate with dampened circadian amplitude and may represent new venues for CEMs. Finally, for the existing chronotherapies, small-molecule modulators can be coadministered to manipulate circadian timing to better align with target expression or reduce drug metabolism.
As discussed above, detailed mechanistic studies are important for the development and eventual application of therapeutically active small molecules (17, 18). For compounds isolated via phenotypic screening, it is invaluable to identify direct molecular targets. For compounds identified for specific targets, their circadian effects and cellular effectors should be characterized. Such mechanistic knowledge will have predictive values toward specific applications of the small molecules in certain disease areas. Chemical optimization of lead compounds is typically required to minimize polypharmacology and improve bioavailability (63, 79, 117) for in vivo animal and human studies. With the growing number of new clock modulators showing favorable pharmacokinetics and efficacies in mouse disease models, human trials will be the exciting goal in the near future to evaluate the therapeutic potential of circadian manipulation.
In conclusion, small-molecule modulators target clock components or clock-associated cellular pathways to modulate circadian characteristics and output functions. Target identification and mechanistic studies of new small-molecule modulators will unveil a key regulatory nexus in the circadian network and facilitate their therapeutic applications, alone or in combination, for clock-related diseases.
ACKNOWLEDGMENTS
This work was in part supported by the Robert A. Welch Foundation (AU-1731) and the National Institutes of Health/National Institute on Aging (NIH/NIA) (R01AG045828) to Z.C., NIH/National Institute of General Medical Sciences (R01GM114424) to S.-H.Y., and NIH/NIA (R01AG045795) to J.S.T. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
DISCLOSURE STATEMENT
J.S.T. is a cofounder and SAB member of Reset Therapeutics, Inc., a biotech company working on circadian rhythms and metabolism. The authors are not aware of any other affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Takahashi JS. 2017. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet 18:164–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, et al. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet 6:544–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogenesch JB, Herzog ED. 2011. Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett. 585:1427–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsalobre A, Damiola F, Schibler U. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929–37 [DOI] [PubMed] [Google Scholar]
- 5.Balsalobre A, Marcacci L, Schibler U. 2000. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol 10:1291–94 [DOI] [PubMed] [Google Scholar]
- 6.Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, et al. 2007. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6:414–21 [DOI] [PubMed] [Google Scholar]
- 7.Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GT. 2008. Phase resetting of the mammalian circadian clock by DNA damage. Curr. Biol 18:286–91 [DOI] [PubMed] [Google Scholar]
- 8.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. 2004. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol 14:1481–86 [DOI] [PubMed] [Google Scholar]
- 9.DeCoursey PJ. 2014. Survival value of suprachiasmatic nuclei (SCN) in four wild sciurid rodents. Behav. Neurosci 128:240–49 [DOI] [PubMed] [Google Scholar]
- 10.Gehring W, Rosbash M. 2003. The coevolution of blue-light photoreception and circadian rhythms. J. Mol. Evol 57(Suppl. 1):S286–89 [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, McKnight SL. 2007A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle 6:2906–12 [DOI] [PubMed] [Google Scholar]
- 12.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, et al. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bass J, Lazar MA. 2016. Circadian time signatures of fitness and disease. Science 354:994–99 [DOI] [PubMed] [Google Scholar]
- 14.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. 2009. Adverse metabolic and cardiovascular consequences of circadian misalignment. PNAS 106:4453–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi F, Schibler U. 2007. Circadian rhythms: mechanisms and therapeutic implications. Annu. Rev. Pharmacol. Toxicol 47:593–628 [DOI] [PubMed] [Google Scholar]
- 16.Schroeder AM, Colwell CS. 2013. How to fix a broken clock. Trends Pharmacol. Sci 34:605–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallach T, Kramer A. 2015. Chemical chronobiology: toward drugs manipulating time. FEBS Lett. 589:1530–38 [DOI] [PubMed] [Google Scholar]
- 18.Nohara K, Yoo SH, Chen ZJ. 2015. Manipulating the circadian and sleep cycles to protect against metabolic disease. Front. Endocrinol 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manoogian ENC, Panda S. 2016. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev 39:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. 2017. Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb. Perspect. Biol 9:a027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeGates TA, Fernandez DC, Hattar S. 2014. Light as a central modulator of circadian rhythms, sleep and affect. Nat. Rev. Neurosci 15:443–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colwell CS. 2011. Linking neural activity and molecular oscillations in the SCN. Nat. Rev. Neurosci 12:553–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. 2005. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci 8:476–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohawk JA, Green CB, Takahashi JS. 2012. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci 35:445–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu AC, Lewis WG, Kay SA. 2007. Mammalian circadian signaling networks and therapeutic targets. Nat. Chem. Biol 3:630–39 [DOI] [PubMed] [Google Scholar]
- 27.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, et al. 2004. PERIOD2::LUCIFERASE realtime reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. PNAS 101:5339–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, et al. 2015. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348:1488–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. 2001. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15:995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, et al. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317–28 [DOI] [PubMed] [Google Scholar]
- 31.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, et al. 2008. The NAD+-dependent deacety-lase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134:329–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass J 2012. Circadian topology of metabolism. Nature 491:348–56 [DOI] [PubMed] [Google Scholar]
- 33.Millius A, Ueda HR. 2017. Systems biology-derived discoveries of intrinsic clocks. Front. Neurol 8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, et al. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417:78–83 [DOI] [PubMed] [Google Scholar]
- 35.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. PNAS 111:16219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papazyan R, Zhang Y, Lazar MA. 2016. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat. Struct. Mol. Biol 23:1045–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckel-Mahan K, Sassone-Corsi P. 2013. Epigenetic regulation of the molecular clockwork. Prog. Mol. Biol. Transl. Sci 119:29–50 [DOI] [PubMed] [Google Scholar]
- 38.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, et al. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, et al. 2015. Pancreatic β cellenhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350:aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong K, He B, Nohara K, Park N, Shin Y, et al. 2015. Dual attenuation of proteasomal and autophagic BMAL1 degradation in ClockΔ19/+ mice contributes to improved glucose homeostasis. Sci. Rep 5:12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallego M, Virshup DM. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol 8:139–48 [DOI] [PubMed] [Google Scholar]
- 42.Kojima S, Shingle DL, Green CB. 2011. Post-transcriptional control of circadian rhythms. J. Cell Sci 124:311–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326:437–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, et al. 2007. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316:900–4 [DOI] [PubMed] [Google Scholar]
- 45.Godinho SIH, Maywood ES, Shaw L, Tucci V, Barnard AR, et al. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316:897–900 [DOI] [PubMed] [Google Scholar]
- 46.Siepka SM, Yoo SH, Park J, Song W, Kumar V, et al. 2007. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell 129:1011–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, et al. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152:1091–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M,et al. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152:1106–18 [DOI] [PubMed] [Google Scholar]
- 49.Brown SA, Pagani L, Cajochen C, Eckert A. 2011. Systemic and cellular reflections on ageing and the circadian oscillator: a mini-review. Gerontology 57:427–34 [DOI] [PubMed] [Google Scholar]
- 50.Gloston G, Yoo S, Chen Z.2017. Clock-enhancing small molecules and potential applications in chronic diseases and aging. Front. Neurol 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landgraf D, Wang LL, Diemer T, Welsh DK. 2016. NPAS2 compensates for loss of CLOCK in peripheral circadian oscillators. PLOS Genet. 12:e1005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertolucci C, Cavallari N, Colognesi I, Aguzzi J, Chen Z, et al. 2008. Evidence for an overlapping role of CLOCK and NPAS2 transcription factors in liver circadian oscillators. Mol. Cell. Biol 28:3070–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeBruyne JP, Weaver DR, Reppert SM. 2007. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci 10:543–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X, Hirota T, Han X, Cho H, Chong LW, et al. 2016. Circadian amplitude regulation via FBXW7-targeted REV-ERBα degradation. Cell 165:1644–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. 2004. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol 14:2289–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, et al. 2010. Emergence of noise-induced oscillations in the central circadian pacemaker. PLOS Biol. 8:e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y, Chen R, Lee HM, Lee C. 2011. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J. Biol. Chem 286:7033–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, et al. 2015. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol. Cell 60:769–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kojetin DJ, Burris TP. 2014. REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discov 13:197–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farrow SN, Solari R, Willson TM. 2012. The importance of chronobiology to drug discovery. Expert Opin. Drug Discov 7:535–41 [DOI] [PubMed] [Google Scholar]
- 61.Chen Z, Yoo SH, Takahashi JS. 2013. Small molecule modifiers of circadian clocks. Cell Mol. Life Sci 70:2985–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. 2008. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3 β. PNAS 105:20746–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, et al. 2009. CKIε/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. PNAS 106:15744–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, et al. 2010. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLOS Biol. 8:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirota T, LeeJ W, StJohn PC, Sawa M, Iwaisako K,et al. 2012. Identification of small molecule activators of cryptochrome. Science 337:1094–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, Yoo SH, Park YS, Kim KH, Wei S, et al. 2012. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. PNAS 109:101–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yagita K, Yamanaka I, Koinuma S, Shigeyoshi Y, Uchiyama Y. 2009. Mini screening of kinase inhibitors affecting period-length of mammalian cellular circadian clock. Acta Histochem. Cytochem 42:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He B, Nohara K, Park N, Park YS, Guillory B, et al. 2016. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 23:610–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Y, Spengler ML, Kuropatwinski KK, Comas-Soberats M,Jackson M, et al. 2011. Selenium is a modulator of circadian clock that protects mice from the toxicity of a chemotherapeutic drug via upregulation of the core clock protein, BMAL1. Oncotarget 2:1279–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chun SK, Jang J, Chung S, Yun H, Kim NJ, et al. 2014. Identification and validation of cryptochrome inhibitors that modulate the molecular circadian clock. ACS Chem. Biol 9:703–10 [DOI] [PubMed] [Google Scholar]
- 71.Grant D, Yin L, Collins JL, Parks DJ, Orband-Miller LA, et al. 2010. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbα. ACS Chem. Biol 5:925–32 [DOI] [PubMed] [Google Scholar]
- 72.Humphries PS, Bersot R, Kincaid J, Mabery E, McCluskie K,et al. 2016. Carbazole-containing sulfonamides and sulfamides: discovery of cryptochrome modulators as antidiabetic agents. Bioorg. Med. Chem. Lett 26:757–60 [DOI] [PubMed] [Google Scholar]
- 73.Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, et al. 2009. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J. Pharmacol. Exp. Ther 330:430–39 [DOI] [PubMed] [Google Scholar]
- 74.Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, et al. 2007. An inhibitor of casein kinase Iε induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther 322:730–38 [DOI] [PubMed] [Google Scholar]
- 75.Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, et al. 2010. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. PNAS 107:15240–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, et al. 2012. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 485:62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antoch MP, Kondratov RV. 2013. Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. Handb. Exp. Pharmacol. 217:289–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He B, Chen Z. 2016. Molecular targets for small-molecule modulators of circadian clocks. Curr. Drug Metab 17:503–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trump RP, Bresciani S, Cooper AW, Tellam JP, Wojno J, et al. 2013. Optimized chemical probes for REV-ERBα. J. Med. Chem 56:4729–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nangle S, Xing W, Zheng N. 2013. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 23:1417–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones KA, Hatori M, Mure LS, Bramley JR, Artymyshyn R, et al. 2013. Small-molecule antagonists of melanopsin-mediated phototransduction. Nat. Chem. Biol 9:630–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. 2008. Activation of TGF-ß/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat. Cell Biol 10:1463–69 [DOI] [PubMed] [Google Scholar]
- 83.Shim HS, Kim H, Lee J, Son GH, Cho S, et al. 2007. Rapid activation of CLOCK by Ca2+-dependent protein kinase C mediates resetting of the mammalian circadian clock. EMBO Rep. 8:366–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meng QJ, McMaster A, Beesley S, Lu WQ, Gibbs J, et al. 2008. Ligand modulation of REV-ERBα function resets the peripheral circadian clock in a phasic manner. J. Cell Sci 121:3629–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, et al. 2012. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. PNAS 109:582–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asher G, Schibler U. 2011. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 13:125–37 [DOI] [PubMed] [Google Scholar]
- 87.Rutter J, Reick M, McKnight SL. 2002. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem 71:307–31 [DOI] [PubMed] [Google Scholar]
- 88.Green CB, Takahashi JS, Bass J. 2008. The meter of metabolism. Cell 134:728–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harfmann BD, Schroder EA, Kachman MT, Hodge BA, Zhang X, Esser KA. 2016. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skeletal Muscle 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, et al. 2013. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342:1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jetten AM, Kang HS, Takeda Y. 2013. Retinoic acid-related orphan receptors α and γ: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front. Endocrinol 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang MR, He Y, Khan TM, Kuruvilla DS, Garcia-Ordonez R, et al. 2015. Antiobesity effect of a small molecule repressor of RORγ. Mol. Pharmacol 88:48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar N, Kojetin DJ, Solt LA, Kumar KG, Nuhant P,et al. 2011. Identification of SR3335 (ML-176): a synthetic RORα selective inverse agonist. ACS Chem. Biol 6:218–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y, Kumar N, Nuhant P, Cameron MD, Istrate MA, et al. 2010. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORα and RORγ ACS Chem. Biol 5:1029–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, et al. 2010. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med 16:1152–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boden G, Chen X, Polansky M. 1999. Disruption of circadian insulin secretion is associated with reduced glucose uptake in first-degree relatives of patients with type 2 diabetes. Diabetes 48:2182–88 [DOI] [PubMed] [Google Scholar]
- 97.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308:1043–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pendergast JS, Branecky KL, Yang W, Ellacott KL, Niswender KD, Yamazaki S. 2013. High-fat diet acutely affects circadian organisation and eating behavior. Eur. J. Neurosci 37:1350–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, et al. 2012. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15:848–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mulvihill EE, Burke AC, Huff MW. 2016. Citrus flavonoids asregulators oflipoproteinmetabolism and atherosclerosis. Annu. Rev. Nutr 36:275–99 [DOI] [PubMed] [Google Scholar]
- 101.Nohara K, Shin Y, Park N, Jeong K, He B, et al. 2015. Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid. Nutr. Metab 12:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shinozaki A, Misawa K, Ikeda Y, Haraguchi A, Kamagata M, et al. 2017. Potent effects of flavonoid nobiletin on amplitude, period, and phase of the circadian clock rhythm in PER2::LUCIFERASE mouse embryonic fibroblasts. PLOS ONE 12:e0170904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bass J 2016. Targeting time in metabolic therapeutics. Cell Metab. 23:575–77 [DOI] [PubMed] [Google Scholar]
- 104.Solt LA, Banerjee S, Campbell S, Kamenecka TM, Burris TP. 2015. ROR inverse agonist suppresses insulitis and prevents hyperglycemia in a mouse model of type 1 diabetes. Endocrinology 156:869–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, et al. 2014. Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity 40:477–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Knight ZA, Shokat KM. 2007. Chemical genetics: where genetics and pharmacology meet. Cell 128:425–30 [DOI] [PubMed] [Google Scholar]
- 107.Lau P, Fitzsimmons RL, Raichur S, Wang SCM, Lechtken A, Muscat GEO. 2008. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem 283:18411–21 [DOI] [PubMed] [Google Scholar]
- 108.Pevet P, Challet E. 2011. Melatonin: both master clock output and internal time-giver in the circadian clocks network. J. Physiol. Paris 105:170–82 [DOI] [PubMed] [Google Scholar]
- 109.Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K,et al. 2013. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342:85–90 [DOI] [PubMed] [Google Scholar]
- 110.Sehgal A, Mignot E. 2011. Genetics of sleep and sleep disorders. Cell 146:194–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones CR, Huang AL, Ptacek LJ, Fu YH. 2013. Genetic basis of human circadian rhythm disorders. Exp. Neurol 243:28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, et al. 2016. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539:378–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. 2013. Circa DB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res 41:D1009–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Man K, Loudon A, Chawla A. 2016. Immunity around the clock. Science 354:999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huh JR, Leung MWL, Huang P, Ryan DA, Krout MR, et al. 2011. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature 472:486–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu T, Wang X, Zhong B, Nurieva RI, Ding S, Dong C. 2011. Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of RORγt protein.J. Biol. Chem 286:22707–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, et al. 2011. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature 472:491–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, et al. 2010. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-α/γ inverse agonist. Mol. Pharmacol 77:228–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McClung CA. 2007. Circadian genes, rhythms and the biology of mood disorders. Pharmacol. Ther 114:222–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bedrosian TA, Nelson RJ. 2013. Sundowning syndrome in aging and dementia: research in mouse models. Exp. Neurol 243:67–73 [DOI] [PubMed] [Google Scholar]
- 121.Magnusson A, Boivin D. 2003. Seasonal affective disorder: an overview. Chronobiol. Int 20:189–207 [DOI] [PubMed] [Google Scholar]
- 122.Souetre E, Salvati E, Belugou JL, Pringuey D, Candito M, et al. 1989. Circadian rhythms in depression and recovery: evidence for blunted amplitude as the main chronobiological abnormality. Psychiatry Res. 28:263–78 [DOI] [PubMed] [Google Scholar]
- 123.Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, et al. 2007. Mania-like behavior induced by disruption of CLOCK. PNAS 104:6406–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ikeda Y, Kumagai H, Skach A, Sato M, Yanagisawa M. 2013. Modulation of circadian glucocorticoid oscillation via adrenal opioid-CXCR7 signaling alters emotional behavior. Cell 155:1323–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li J, Lu WQ, Beesley S, Loudon AS, Meng QJ. 2012. Lithium impacts on the amplitude and period of the molecular circadian clockwork. PLOS ONE 7:e33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yin L, Wang J, Klein PS, Lazar MA. 2006. Nuclear receptor Rev-erbα is a critical lithium-sensitive component of the circadian clock. Science 311:1002–5 [DOI] [PubMed] [Google Scholar]
- 127.Banerjee S, Wang Y, Solt LA, Griffett K, Kazantzis M, et al. 2014. Pharmacological targeting of the mammalian clock regulates sleep architecture and emotional behaviour. Nat. Commun 5:5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chung S, Lee EJ, Yun S, Choe HK, Park SB,et al. 2014. Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell 157:858–68 [DOI] [PubMed] [Google Scholar]
- 129.Son GH, Chung S, Ramirez VD, Kim K. 2016. Pharmacological modulators of molecular clock and their therapeutic potentials in circadian rhythm-related diseases. Med. Chem 6:724–33 [Google Scholar]
- 130.Sellix MT, Evans JA, Leise TL, Castanon-Cervantes O, Hill DD, et al. 2012. Aging differentially affects the re-entrainment response of central and peripheral circadian oscillators. J. Neurosci 32:16193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Banks G, Nolan PM, Peirson SN. 2016. Reciprocal interactions between circadian clocks and aging. Mamm. Genome 27:332–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, et al. 2011. Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat. PLOS ONE 6:e27168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Luo W, Chen WF, Yue Z, Chen D, Sowcik M, et al. 2012. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell 11:428–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. 2006. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20:1868–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gutman R, Genzer Y, Chapnik N, Miskin R, Froy O. 2011. Long-lived mice exhibit 24 h locomotor circadian rhythms at young and old age. Exp. Gerontol 46:606–9 [DOI] [PubMed] [Google Scholar]
- 136.Longo VD, Panda S. 2016. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. 23:1048–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tevy MF, Giebultowicz J, Pincus Z, Mazzoccoli G, Vinciguerra M. 2013. Aging signaling pathways and circadian clock-dependent metabolic derangements. Trends Endocrinol. Metab 24:229–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, et al. 2016. Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab 23:143–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Finkel T 2015. The metabolic regulation of aging. Nat. Med. 21:1416–23 [DOI] [PubMed] [Google Scholar]
- 140.Chang HC, Guarente L. 2013. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153:1448–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hubbard BP, Sinclair DA. 2014. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci 35:146–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pifferi F, Dal-Pan A, Languille S, Aujard F. 2013. Effects of resveratrol on daily rhythms of locomotor activity and body temperature in young and aged grey mouse lemurs. Oxidative Med. Cell. Longev 2013:187301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bellet MM, Nakahata Y, Boudjelal M, Watts E, Mossakowska DE, et al. 2013. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. PNAS 110:3333–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, et al. 2001. X-ray structure of the orphan nuclear receptor RORß ligand-binding domain in the active conformation. EMBO J. 20:5822–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, et al. 2013. SCFfbxl3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496:64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, et al. 2012. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337:189–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu D, Su X, Potluri N, Kim Y, Rastinejad F. 2016. NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. eLife 5:e18790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. 2009. Principles of ligand binding within a completely buried cavity in HIF2αPAS-B. J. Am. Chem. Soc 131:17647–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McIntosh BE, Hogenesch JB, Bradfield CA. 2010. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol 72:625–45 [DOI] [PubMed] [Google Scholar]
- 150.Arkin MR, Tang Y, Wells JA. 2014. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem. Biol 21:1102–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Laraia L,McKenzie G, Spring DR, Venkitaraman AR, Huggins DJ. 2015. Overcoming chemical, biological, and computational challenges in the development of inhibitors targeting protein-protein interactions. Chem. Biol 22:689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, et al. 2014. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. eLife 3:e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsimakouridze EV, Alibhai FJ, Martino TA. 2015. Therapeutic applications of circadian rhythms for the cardiovascular system. Front. Pharmacol 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, et al. 2010. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat. Med 16:67–74 [DOI] [PubMed] [Google Scholar]
- 155.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, et al. 2012. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 483:96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, et al. 2016. Circadian rhythm disruption promotes lung tumorigenesis. CellMetab. 24:324–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fu L, Lee CC. 2003. The circadian clock: pacemaker and tumour suppressor. Nat. Rev. Cancer 3:350–61 [DOI] [PubMed] [Google Scholar]
- 158.Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, et al. 2016. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30:909–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kondratova AA, Kondratov RV. 2012. The circadian clock and pathology of the ageing brain. Nat. Rev. Neurosci 13:325–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Oshima T, Yamanaka I, Kumar A, Yamaguchi J, Nishiwaki-Ohkawa T, et al. 2015. C-H activation generates period-shortening molecules that target cryptochrome in the mammalian circadian clock. Angew. Chem. Int. Ed. Engl 54:7193–97 [DOI] [PubMed] [Google Scholar]
- 161.Chun SK, Chung S, Kim HD, Lee JH, Jang J, et al. 2015. A synthetic cryptochrome inhibitor induces anti-proliferative effects and increases chemosensitivity in human breast cancer cells. Biochem. Biophys. Res. Commun 467:441–46 [DOI] [PubMed] [Google Scholar]
- 162.Vieira E, Marroqui L, Figueroa ALC, Merino B, Fernandez-Ruiz R, et al. 2013. Involvement of the clock gene Rev-erb alpha in the regulation of glucagon secretion in pancreatic alpha-cells. PLOS ONE 8:e69939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.De Mei C, Ercolani L, Parodi C, Veronesi M, Lo Vecchio C, et al. 2015. Dual inhibition of REV-ERBß and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene 34:2597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Y, Billon C, Walker JK, Burris TP. 2016. Therapeutic effect of a synthetic RORa/y agonist in an animal model of autism. ACS Chem. Neurosci 7:143–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Byun JK, Choi YK, Kang YN, Jang BK, Kang KJ, et al. 2015. Retinoic acid-related orphan receptor alpha reprograms glucose metabolism in glutamine-deficient hepatoma cells. Hepatology 61:953–64 [DOI] [PubMed] [Google Scholar]
- 166.Helleboid S, Haug C, Lamottke K, Zhou Y, WeiJ, et al. 2014. The identification of naturally occurring neoruscogenin as a bioavailable, potent, and high-affinity agonist of the nuclear receptor RORα(NR1F1). J. Biomol. Screen 19:399–406 [DOI] [PubMed] [Google Scholar]
- 167.Zhang W, Zhang J, Fang L, Zhou L, Wang S,et al. 2012. Increasing human Th17 differentiation through activation of orphan nuclear receptor retinoid acid-related orphan receptor γ (RORγ) by a class of aryl amide compounds. Mol. Pharmacol 82:583–90 [DOI] [PubMed] [Google Scholar]
- 168.Barnea M, Haviv L, Gutman R, Chapnik N, Madar Z, Froy O. 2012. Metformin affects the circadian clock and metabolic rhythms in a tissue-specific manner. Biochim. Biophys. Acta 1822:1796–806 [DOI] [PubMed] [Google Scholar]
- 169.Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S,et al. 2009A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23:708–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Iwahana E, Akiyama M, Miyakawa K, Uchida A, Kasahara J, et al. 2004. Effect of lithium on the circadian rhythms of locomotor activity and glycogen synthase kinase-3 protein expression in the mouse suprachiasmatic nuclei. Eur. J. Neurosci 19:2281–87 [DOI] [PubMed] [Google Scholar]
- 171.Yao H, Sundar IK, Huang Y, Gerloff J, Sellix MT, et al. 2015. Disruption of sirtuin 1-mediated control of circadian molecular clock and inflammation in chronic obstructive pulmonary disease. Am. J. Respir. CellMol. Biol 53:782–92 [DOI] [PMC free article] [PubMed] [Google Scholar]


