Introductory paragraph
The angiosperm seed is composed of three genetically distinct tissues: the diploid embryo that originates from the fertilized egg cell, the triploid endosperm that is produced from the fertilized central cell, and the maternal sporophytic integuments that develop into the seed coat1. At the onset of embryo development in Arabidopsis thaliana, the zygote divides asymmetrically producing a small apical embryonic cell, and a larger basal cell that connects the embryo to the maternal tissue2. The coordinated and synchronous development of the embryo and the surrounding integuments, and the alignment of their growth axes suggest communication between maternal tissues and the embryo. In contrast to animals, however, where a network of maternal factors that direct embryo patterning have been identified3,4, only a few maternal mutations have been described to affect embryo development in plants5–7. Early embryo patterning in Arabidopsis requires accumulation of the phytohormone auxin in the apical cell by directed transport from the suspensor8–10. However, the origin of this auxin has remained obscure. Here we investigate the source of auxin for early embryogenesis and provide evidence that the mother plant coordinates seed development by supplying auxin to the early embryo from the integuments of the ovule. We show that auxin response increases in ovules upon fertilization, due to upregulated auxin biosynthesis in the integuments, and this maternally-produced auxin is required for correct embryo development.
Keywords: Arabidopsis thaliana, maize, auxin, auxin biosynthesis, embryogenesis, maternal sporophytic effect
The different developmental perspectives of the two daughter cells (Fig. 1a) of the Arabidopsis zygote are controlled by several factors. Among them, polarized auxin flow from the basal daughter cell via the PIN7 auxin transporter leads to auxin accumulation and activation of downstream responses in the apical daughter cell, which ultimately contributes to its specification as the founder of the proembryo9,10.
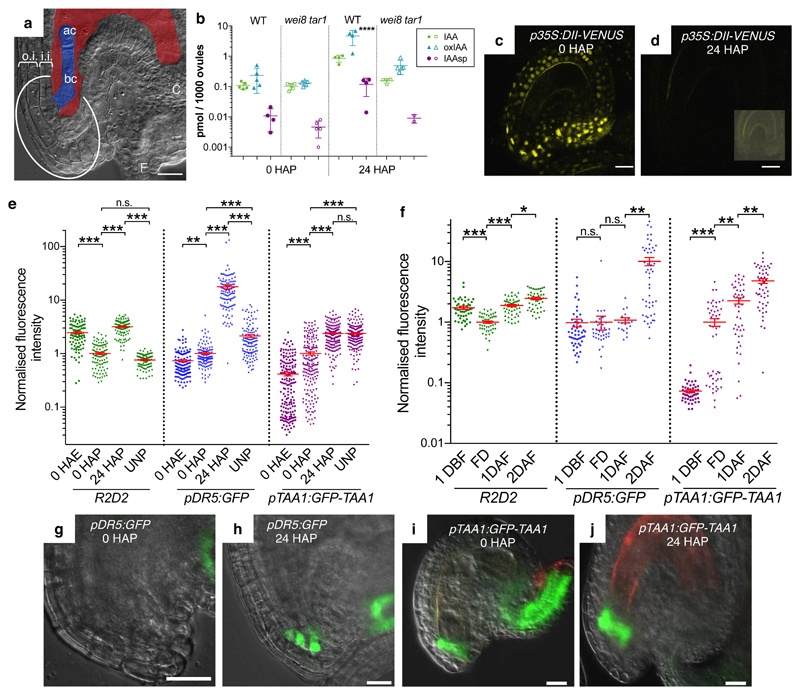
Fig. 1. Auxin accumulation in integuments.
a, Micropylar region of an Arabidopsis fertilized seed. Embryo and endosperm are colored in blue and red, respectively. ac, apical cell; bc, basal cell; C, chalaza; F, funiculus; o.i., outer integument; i.i., inner integument; the embryo attachment region is circled. b, Quantification of IAA, IAA-Aspartate (IAAsp), and oxidized IAA (oxIAA) in wild-type (WT) and wei8 tar1 ovules. c-d, p35S:DII-VENUS expression (yellow signal) in unfertilized (c) and 24 HAP (d) ovules. Inset in (d) shows the same ovule with enhanced brightness. e-f, Quantification in the embryo attachment region of GFP fluorescence (pDR5:GFP and pTAA1:GFP-TAA1) and mDII/DII ratio, normalized to 0 HAP (e) and FD (f). Data presented as individual points with a horizontal bar at the mean ± s.e.m at the vertical bar. g-j, pDR5:GFP and pTAA1:GFP-TAA1 expression in the wild-type embryo attachment region. Green GFP signal, merged with the brightfield image. Reddish signal, autofluorescence. Scale bars, 20 µm.
In search for the source of the auxin mediating early embryo development9, we measured free auxin and auxin derivatives in Arabidopsis ovules. We found that the levels of active auxin (IAA) and of the catabolic intermediates IAA-Aspartate (IAAsp) and oxidized IAA (oxIAA) strongly increase 24 h after pollination (24 HAP; Fig. 1b and Supplementary Table 1). At this stage, fertilization has usually occurred, as judged by the appearance of endosperm nuclei (not shown). To localize auxin accumulation in the ovule, we used the p35S-driven DII-VENUS auxin signaling sensor that rapidly degrades in response to auxin, and therefore negatively monitors relative cellular auxin levels11. 24 HAP, DII-VENUS signals become strongly reduced throughout the ovule compared to 0 HAP, indicating an increased amount of auxin (Fig. 1c-d). Control experiments confirm that sensor degradation was measured (Supplementary Fig. 1 and Supplementary Table 2). We quantified the DII signal using the R2D2 reporter that combines an pRPS5A-driven DII-VENUS with an pRPS5A-driven auxin-insensitive mDII-tdTomato as internal reference12. The mDII/DII signal ratio provides a quantitative measure of the cellular auxin signal. mDII/DII initially decreases before pollination (compare 0 HAE, h after emasculation, and 0 HAP), and then increases again after pollination (24 HAP) (Fig. 1e and Supplementary Fig. 2a).
Auxin mediates the degradation of Aux/IAA transcriptional repressors, resulting in transcriptional auxin output13, which we monitored with the pDR5:GFP reporter9. We observed three localized pDR5:GFP maxima at 24 HAP: (1) in the integument region where the embryo attaches, (2) at the opposite chalazal side of the embryo sac, and (3) in the funiculus that connects the ovule to the placenta (Fig. 1h and Supplementary Fig. 2b). By contrast, pDR5:GFP signal is weak or undetectable before pollination or in unpollinated controls (Fig. 1g and Supplementary Fig. 2b). Quantification in the embryo attachment region (outlined in Fig. 1a and Supplementary Fig. 2b) revealed that pDR5:GFP signal increases about 7 fold after pollination (Fig. 1e). We confirmed the observed changes of DII and pDR5:GFP expression in self-pollinated flowers, excluding that the upregulation was caused by emasculation or hand-pollination (Fig. 1f). Collectively, these results indicate that pollination leads to increased auxin levels in the maternal tissues surrounding the embryo and a localized upregulation of auxin response in the embryo attachment region.
To address whether upregulation of auxin response in the ovule by pollination is evolutionary conserved, we studied pDR5:GFP expression dynamics in the monocotyledonous maize. Unlike in Arabidopsis, the embryo sac in maize is entirely surrounded by several layers of nucellar tissue and thus is not in direct proximity to the integuments in the micropylar region. We find that pDR5 expression is strongly upregulated by pollination in the tips of the integuments and in the nucellus cells at the micropylar region (Supplementary Fig. 3). Thus, despite their different ovule organization, we observed a similar increase of auxin response in maternal tissues in Arabidopsis and maize.
In Arabidopsis, auxin production via the indole-3-pyruvic acid (IPyA) pathway14 is the main biosynthetic pathway and is catalyzed by the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA1) and related (TAR1 and TAR2) aminotransferases and by YUCCA (YUC) flavin monooxygenases15–17. Before pollination (0 HAP), we detected expression of a functional pTAA1:GFP-TAA1 reporter in the embryo attachment region, in the chalaza, and funiculus (Fig. 1i, Supplementary Fig. 2c). Notably, TAA1 expression is increased 24 HAP compared to 0 HAP in the embryo attachment region, whereas it is reduced in chalaza and funiculus (Fig. 1e,j and Supplementary Fig. 2c). TAA1 expression in the embryo attachment region is also increased in self-pollinated flowers at 1 DAF (Days After Flowering) compared to FD (Flowering Day) (Fig. 1f), again confirming that the observed upregulation is not caused by hand-pollination. Unlike auxin response, however, TAA1 expression increases with and without pollination to similar levels (compare UNP, unpollinated, and 24 HAP in Fig. 1e and Supplementary Fig. 2c). In addition, several YUC reporter genes are also expressed in the integuments (Supplementary Fig. 4 and Supplementary Table 2), of which YUC8 and YUC9 reporters appear upregulated at 16 HAP. In summary, the expression of the auxin biosynthetic machinery and the auxin signal output are differentially regulated during ovule maturation and pollination.
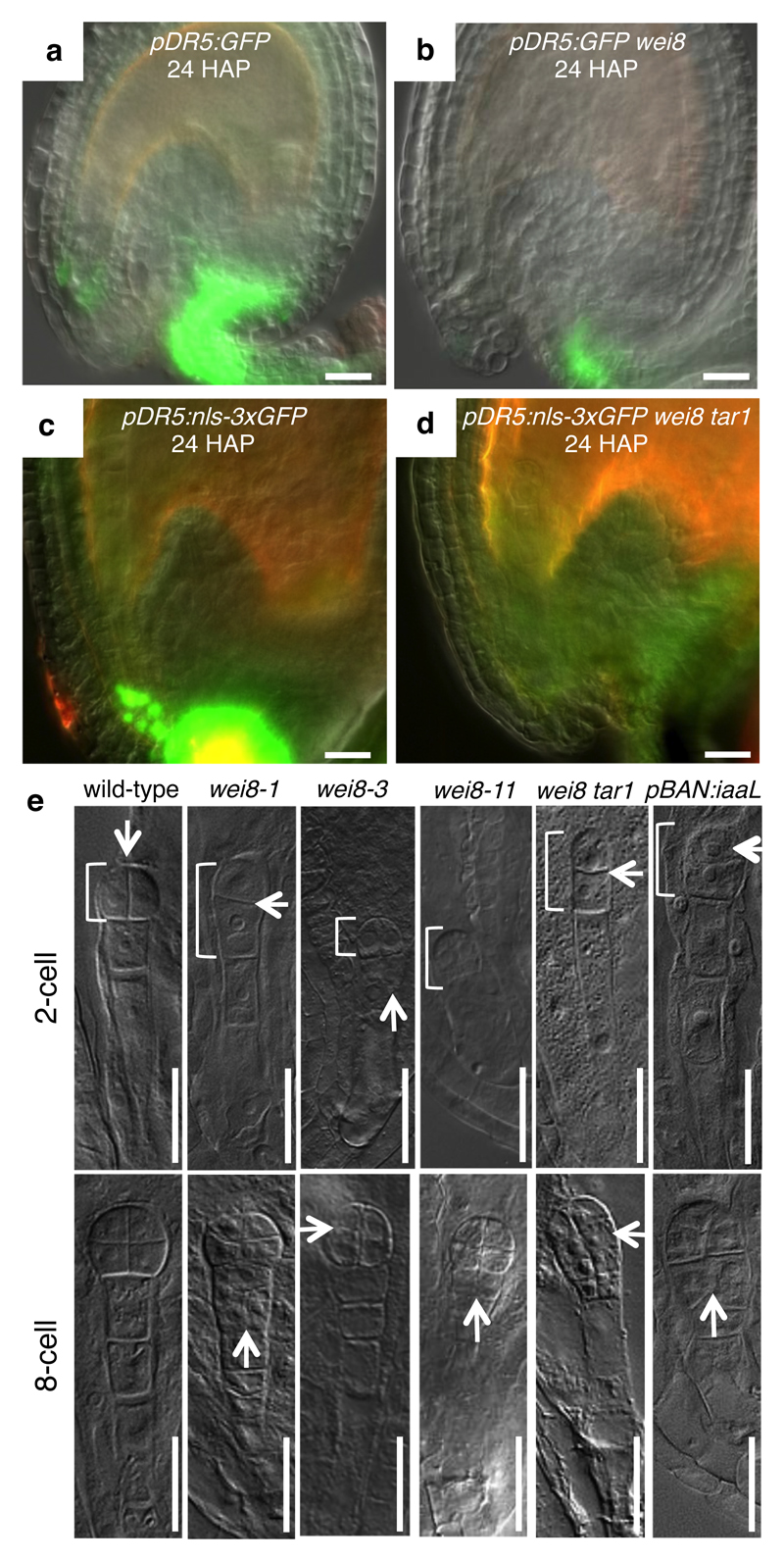
To address, whether upregulation of auxin biosynthesis is responsible for the elevated auxin response in the integuments, we studied auxin biosynthesis mutants. Profiling auxin metabolites revealed a strong reduction of IAA levels and its degradation products at 24 HAP in wei8 (weak ethylene insensitive8, a recessive allele of TAA1) tar1 ovules compared to wild type (Fig. 1b and Supplementary Table 1). Furthermore, pDR5 reporter expression remains undetectable in the embryo attachment region at 24 HAP, and is reduced at chalaza and funiculus of wei8 and more strongly in wei8 tar1 auxin biosynthetic mutants, compared to wild type (Fig. 2a-d and Supplementary Table 2). Collectively, these data indicate that an increased activity of the IPyA pathway is a major contributor of auxin accumulation in the integuments after pollination.
Fig. 2. Auxin biosynthesis mutants display early embryonic defects.
a-d, pDR5:GFP expression from indicated genotypes. Green GFP signal, merged with brightfield images. Autofluorescence appears reddish. e, Two- and eight-cell embryo phenotypes from indicated genotypes. Arrows, abnormal division planes. Brackets mark proembryos. Scale bars, 20µm.
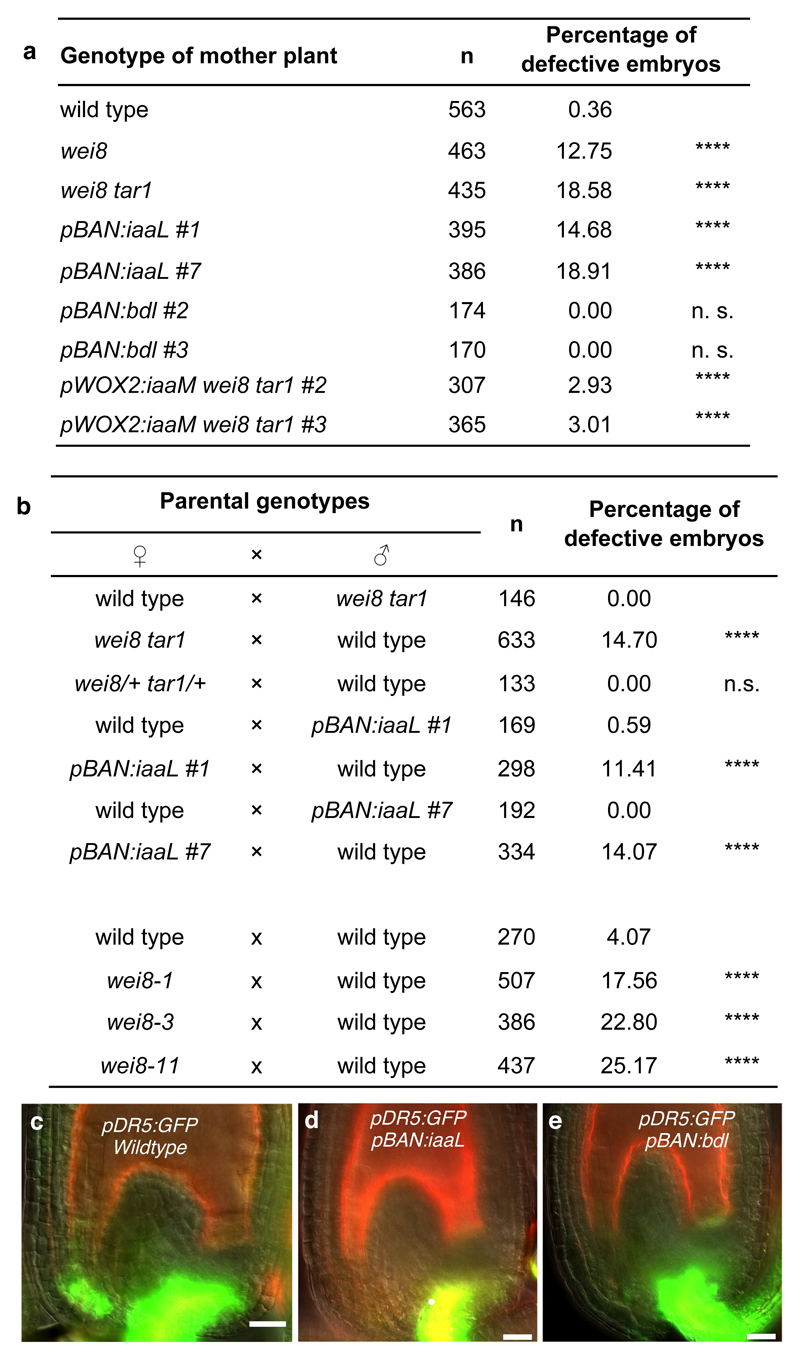
We then addressed the possible role of auxin production in the integuments for embryo development. Compared to wild type, embryos from selfed homozygous wei8-1 mother plants display abnormal cell division patterns and suspensor exhibit aberrant cell divisions and reduced length at 2-cell to 8-cell embryo stages (Figs 2e, 3a and Supplementary Fig. 5). The earliest observed phenotypic abnormality is a horizontal division of the embryo apical daughter cells instead of the regular vertical one (arrows in Fig. 2e, Supplementary Fig. 5), suggesting that initial steps in apical-basal embryo development are compromised in wei8 embryos. We observed similar defects in two independent alleles wei8-3 and wei8-11 (Figs 2e and 3b), confirming that they are caused by mutations in the TAA1 gene. Because the frequencies of defective embryos are increased in wei8 tar1 double mutants (Fig. 3a), we focused on wei8 tar1 embryos for further analysis.
Fig. 3. Sporophytic maternal early embryonic defects.
a-b, Frequencies of the embryo phenotypes in self-fertilized plants (a) and reciprocal crosses (b) of indicated genotypes. NA, not applicable; p-values by two-sided Fisher’s exact test compared to wild type, except for pWOX2:iaaM wei8 tar1 that was compared to wei8 tar1 in (a) and compared to the respective reciprocal crosses in (b). c-e, pDR5:GFP expression (green) at 24 HAP from indicated genotypes. Autofluorescence is reddish. Scale bars, 20µm.
Because TAA1 expression is undetectable in the embryo before the 16-cell stage10,18, we hypothesized that early embryo defects in selfed wei8 tar1 plants might be caused by compromised auxin biosynthesis in maternal tissues rather than in the embryo itself. To test this notion, we analysed early embryo development in reciprocal crosses between wild type and wei8 tar1 (Fig. 3b). When wei8 tar1 ovules were fertilized with wild-type pollen, but not in the reverse cross, we observed defective embryo phenotypes indistinguishable from the ones in selfed wei8 tar1 controls. We conclude that with regard to early embryogenesis, wei8 and tar1 are maternal effect mutations. Furthermore, pollinating double heterozygous wei8/+ tar1/+ plants with wild-type pollen resulted in normal developing embryos, suggesting that the maternal sporophytic tissue, such as the integuments, rather than the female gametophyte is responsible (Fig. 3b). We confirmed this finding by expressing the bacterial IAA-lysine synthetase (iaaL), which inactivates free auxin through conjugation19, specifically in the integuments using the BANYULS (BAN) promoter20. pBAN::iaaL expression causes a strong reduction of pDR5:GFP signal in this region after pollination with no detectable expression in the embryo attachment region (Fig. 3c-d and Supplementary Table 2) and results in defective embryo development similar to selfed wei8 tar1 controls (Fig. 3a). Reciprocal crosses confirm that pBAN:iaaL affects embryo development only when provided through the mother (Fig. 3b). Together, these observations provide evidence that maternal auxin production in the integuments contributes to early embryo development.
The incomplete penetrance of recognizable early embryo defects with a compromised maternal IPyA pathway suggests the existence of functionally redundant and compensatory mechanisms. For example, a recent study revealed that auxin synthesized in the developing endosperm is transported into the integuments, where it promotes development of the seed coat21,22. Because endosperm development initiates before the first division of the zygote, endosperm-produced auxin could also contribute to early embryo development, either directly or via the integuments. Our reciprocal crosses, however, do not suggest a prominent role of endosperm-derived auxin in early embryogenesis.
Notably, similar embryo defects to those caused by the absence of maternal auxin production have been described for zygotic mutations affecting auxin transport from the suspensor to the embryo9,23,24 or auxin response within the embryo8. Therefore, we considered two hypotheses to explain the requirement of auxin biosynthesis in the integuments for embryo development. First, an auxin response in the integuments could cause the production of a downstream signal to the embryo, influencing its development. To address this hypothesis, we expressed an auxin-insensitive version of BODENLOS/AUX-IAA12 (BDL), a broad repressor of the transcriptional auxin response25, from the pBAN promoter. pBAN:bdl expression strongly reduces pDR5:GFP auxin response in the integuments (Fig. 3e and Supplementary Table 2). Nonetheless, this does not result in any observable embryo defects (Fig. 3a) suggesting that BDL-mediated auxin response and downstream processes in the integuments do not play a major role for embryo development.
An alternative hypothesis holds that the integuments provide auxin directly to the embryo. Consistent with this idea and a recent report26, we find that several auxin transport facilitators (PINs, AUX1, LAX1, ABCB1 and ABCB19) are expressed in the integuments (Supplementary Fig. 6). Of the facilitators tested, only AUX1 and LAX1 could be involved in the auxin flow between micropyle cells and early embryo based on their expression and intracellular localization. However, multiple mutants did not reveal any sporophytic maternal-dependent embryo phenotypes, possibly due to genetic redundancy, or confounding gametophytic defects as in aux1 mutants (not shown). We therefore analysed whether auxin production in the embryo might complement the maternal auxin defects to test our second hypothesis. To this end, we expressed the bacterial auxin biosynthetic enzyme iaaM27 from the WOX2 promoter throughout embryos of wei8 tar1 plants. We found that the embryo defects in wei8 tar1 mutants are significantly suppressed by the pWOX2:iaaM transgene (Fig. 3a), suggesting that ectopic auxin production in the embryo can overcome the reduction of maternal auxin supply.
This study provides insight into the mechanisms of how the mother plant contributes to the regulation of early embryogenesis. A straightforward interpretation of our results is that maternal auxin contributes to the reported auxin accumulation in the apical proembryo where it mediates early embryo development9. Supporting to this notion are predictions from recent mathematical modeling and experimental results showing that a local auxin source coupled to feedback regulation of PIN polarity28,29 in the embryo is sufficient to generate a robust auxin gradient that instructs apical-basal axis formation10,28. Although, we cannot exclude that auxin synthesized in the maternal integuments generates an additional, unknown downstream signal that affects embryogenesis, the observation that auxin-mediated transcriptional output in these tissues is not required for regulating embryogenesis did not provide evidence for this alternative model. Together with recent reports21,22,26, our data contribute to understanding auxin dynamics during development of the three compartments of the ovule; the integument, the endosperm, and the embryo: Before fertilization, stamen maturation restricts auxin export from the ovule26. Subsequently, fertilization causes further restriction of auxin export through the funiculus, resulting in the spread of auxin throughout the integuments26. The data presented here reveal that expression of the key biosynthetic enzyme TAA1 gradually increases in the integuments. Pollination then triggers a strong upregulation of local auxin accumulation in the integuments, which affect early steps in embryo development. Because fertilization has usually taken place at 24 hours after fertilization, future experiments will address which step during pollination and/or fertilization triggers these events. Finally, fertilization-induced auxin production in the endosperm signals back to the integuments, regulating seed coat development21,22. Thus, a circuitry of localized auxin accumulation, signaling, and activity coordinates embryo, endosperm and seed coat development.
Materials and Methods
Plant material, growth conditions and plant treatment
For all experiments, Arabidopsis plants were grown in long-day (16 h light/8 h dark) conditions at 18-20°C. The plant lines used in this study are listed in supplements. MG132 treatment was done on ovules at 24 HAP by incubation in liquid MS supplemented with 50 µM MG132 (Sigma-Aldrich, stock dissolved in DMSO) for 6 h. For backcrosses experiments, embryo development was analysed between 2 to 5 days after pollination. Embryos were counted as defective when the apical cell division diverged from the wild-type vertical division or the size of the apical cells or suspensor was affected. Genotypes of parental plants were confirmed by PCR. Hand pollination was done by emasculating the biggest unopened bud from the main shoot and by pollinating the stigma either immediately or after 24 h. For the experiment, R2D2 plants were selected by fluorescence in the main root tip.
Cloning
Coding sequences of iaaL and bdl were amplified (primer sequences in Supplementary Table 3) from pSDM7012 and pIC-myc-bdl and inserted into a blunted NcoI site of pBAN-pBS-SK. Using SalI site of pBAN-pBS-SK, pBAN:iaaL and pBAN:bdl are transferred after blunting into a SmaI site of a binary vector (pGIIK). Coding sequence of iaaM was amplified (primer sequences in Supplementary Table 3) from pSDM7010 and inserted into a blunted SalI site of a binary vector containing WOX2 promoter sequence (ZJ62). Constructs were transformed into Arabidopsis wild type by floral dip. pBAN:iaaL and pBAN:bdl lines are kanamycin resistant. pWOX2:iaaM line is MTX resistant.
Auxin response in maize ovules
Maize pDR5:mRFP.er plants30 were grown under standard greenhouse conditions with 16 h light at 26°C and 8 h darkness at 22°C and a relative air humidity of 40 to 60%. Cobs were covered before ovule maturation and hand-pollinated when the silks reached a length of 2-3 cm outside the cobs. About 50% of silks were pollinated with fresh pollen (limited pollination). Before pollination as well as 2-5 days after pollination (DAP) ovules were longitudinally sectioned by hand along the embryo sac axis. Samples were mounted in 13% mannitol (w/v) solution on glass slides under a coverslip. Imaging was done using a Leica SP8 inverted confocal laser scanning microscope at 561 nm with a 570-666 nm band-pass emission filter.
Microscopy
For embryo phenotyping, embryos were cleared at mentioned stages in a chloral hydrate solution (chloral hydrate/water/glycerol, 8/3/1, w/v/v). Microscopy observations were done on a Zeiss Axioskop2 plus coupled to an AxioCam MRc camera, or an Olympus BX51 microscope, coupled to a Sony DS-5M-L1 camera, using differential interference contrast optics. For fluorescent visualization, ovules were mounted in 10% glycerol or 7% glucose with or without 0.1 mg/mL propidium iodide. Imaging was done on a Zeiss Axio Imager A1 coupled to an AxioCam MRm camera (wide-field fluorescence) or on a Zeiss LSM 700 or a ZEIS LSM 780 based on Zeiss Axio Observer Z1 microscopes, and a Zeiss LSM 700 based on a Zeiss Axio Imager Z2 microscope (confocal fluorescence). Acquisition with multiple channels was done by sequential scanning using 488 nm (GFP, YFP, propidium iodide) and 550 nm (TdTomato) with 505-530nm band pass (GFP, YFP) and 530-600 band pass (propidium iodide, TdTomato) emission filters.
Fluorescence signal quantification
We used wide-field fluorescence microscopy (see above) to quantify the expression pattern of fluorescent markers in the embryo attachment region in ovules of pTAA1:GFP-TAA1, pDR5:GFP and R2D2 plants. Flowers at stage 12c of pTAA1:GFP-TAA1, pDR5:GFP and R2D2 lines were emasculated 24 h before hand pollination. Respective pistils were pollinated with either pTAA1:GFP-TAA1, pDR5:GFP or R2D2 pollen. Pistils were collected at 0 h after emasculation (0 HAE), 0 h after pollination (0 HAP, equivalent to 24 HAE) and 24 HAP. Representative microscopic pictures are displayed in Supplementary Fig. 2. Unpollinated pistil (UNP) corresponding to a 48 HAE is used as control. From self pollinating flowers, ovules were collected one day before flowering (DBF), at flowering day (FD) and one, two, three or four days after flowering (DAF). In our conditions, FD corresponds to approximately 0 HAP of hand-pollinated ovules (four-celled female gametophyte), 1 DAF to approximately 12 HAP-24 HAP (1 to 8 nucleated endosperm and zygote), and 2 DAF to approximately 24 HAP-30 HAP (>8 nuclei in endosperm and zygote to one-cell embryo). Ovules were mounted in 10% glycerol for fluorescent visualization and quantification. For the quantification of GFP signal in pTAA:GFP-TAA1 and pDR5:GFP lines, we integrated the brightness of GFP multiplying the area with the pixels, and for R2D2 line we measured the n3xVENUS and ntdTomato pixels and calculated the mDII/DII ratio; the quantifications were done using AxioVisionLE 4.7.1.0 (Carl Zeiss Imaging Solutions GmbH). Presented data are means ± s.e.m, overlaid by the corresponding dot plots. Kruskal-Wallis Test (Nonparametric ANOVA) with Dunn's Multiple Comparisons Test was performed for all samples usimg GraphPad InStat 3.1 software, with the exception of Fig 3a-b using R version 3.4.5 (The R foundation for Statistical Computing). The presented data in Fig. 1 are normalized to the value for 0 HAP (hand pollination) or FD (self pollination). The graphs were prepared using GraphPad Prism software.
Auxin quantification
OxIAA levels were found to mirror auxin levels also in other tissues. It is an inactivate auxin molecule, the main production of auxin catabolism, and has no biological activity31. IAAsp is a conjugated product of the Asp amino acid of IAA. It is a reversible product of a GH3-dependent catabolic pathway used to balanced the levels of active IAA31. IAA and metabolites were quantified in four samples, including two genotypes (Col and wei8 tar1) and two time points (0 HAP [24 h after emasculation of unpollinated gynoecia] and 24 HAP [24 h after hand pollination of emasculated gynoecia]). Six-week old plants were grown in soil in controlled environment (PSI Fytoscope FS-WI, long-day regime with LED illumination, 21°C, 50% humidity). For each sample approximately 750 ovules were collected for each biological replicate, in five biological replicates. The exact number of siliques from which the ovules were collected, and the exact number of collected ovules per replicate are indicated in Supplementary Table 1. Ovules collection of one sample replicate took approx. 90 minutes. Collection was performed under a dissection microscope (SZ51 Olympus). A gynoecium was harvested from a plant and stuck on double-sided tape. Using a 30G hypodermic needle (Terumo) the gynoecium was cut along the septum and the carpel stuck to the tape. Using Dumont #5 tweezers ovules were harvested and collected into a 1.5 ml tube on dry ice. While collecting the exact number of ovules were counted using a manual cell counter.
Extraction and purification were performed following the procedure as described32. 5 pmol of [13C6]IAA, [13C6]oxIAA and [13C6]IAAsp were added to each sample as an internal standards. IAA and IAA metabolites were quantified by LC-MS/MS method32. All data were processed using MassLynx™ software (ver. 4.1, Waters).
Statistics and Reproducibility
Fig 1a: Similar result was observed in around 1000 ovules in more than 50 plants by two experimenters.
Fig. 1b and Supplementary Table 1 (data source): The observations were independently repeated twice with similar output. Plotted data are the means of 5 replicates presented as pmol/1000 ovules, ± s.d. The exact number of ovules for each sample is indicated in Supplementary Table 1. Significant differences between 24 HAP wild-type ovules and either 0 HAP wild-type ovules, 0 HAP wei8 tar1 ovules or 24 HAP wei8 tar1 ovules were determined by two-way ANOVA followed by Tukey’s multiple comparison test (****p<0.0001) with a two-sided 95 % confidence interval of difference.
Fig. 1c-1d: Similar result was observed in around 200 ovules in more than 10 plants by two experimenters.
Fig. 1e-1f: The observations were independently repeated twice with similar results. Data represent individual points with means at horizontal bars ± s.e.m at vertical bars. Significance of differences was determined by Kruskal-Wallis Test (Nonparametric ANOVA) with Dunn's Multiple Comparisons Test: ***p < 0.001; **p < 0. 01; *p < 0. 05; n.s., not significant, with a two-sided 99.9 % confidence interval. In (e), n = 96 (R2D2), 120 (DR5), 180 (TAA1 – 0 HAE, 0 HAP, 24 HAP), 179 (TAA1 – UNP) biologically independent ovules. In (f), n = 49 (R2D2 – 1 DBF), 37 (R2D2 - FD), 20 (R2D2 – 1 DAF), 50 (R2D2 – 2 DAF), 48 (DR5), 50 (DR5 – 1 DBF), 51 (DR5 - FD), 48 (DR5 – 1 DAF), 51 (DR5 – 2 DAF) biologically independent ovules. Representative pictures are in Supplementary Fig. 2.
Fig. 2a-2d: Similar result was observed twice by two experimenters.
Fig. 3a-b: NA, test not applicable; p-values by two-sided Fisher’s exact test compared to wild type, except for pWOX2:iaaM wei8 tar1 that was compared to wei8 tar1 in (a) and compared to the respective reciprocal crosses in (b).
Fig. 2e, Fig. 3a-b, Supplementary Fig. 5: The observations were independently repeated more than 3 times with similar results in 3 different laboratories (Freiburg, Ghent, Brno), except for the data on wei8-3 and wei8-11 alleles repeated twice.
Fig. 3c-3e, Supplementary Figs 4, 6: The observations were repeated independently three times with similar results.
Supplementary Fig. 1: The observations were independently repeated twice with similar results
Supplementary Fig. 3: The similar results were independently observed on 10 independent biological samples.
Supplementary Material
Impact statement.
Early embryo development requires auxin production in the surrounding maternal integuments.
Acknowledgements
We thank Edwin Groot for assistance with statistical analyses and for critical reading of the manuscript. We thank J. Alonso for providing wei8-1, wei8-1 tar1-1, pDR5:GFP wei8-1, and pTAA1:GFP-TAA1 seeds, G. Jürgens for providing pDR5:nls-3xGFP and pDR5:nls-3xGFP wei8-1 tar1-1 seeds, T. Vernoux for p35S:DII-VENUS and p35S:mDII-VENUS seeds and plasmids, R. Offringa for pSDM7010 and pSDM7012, L. Lepiniec for pBAN-pBS-SK, Che-Yang Liao and Dolf Weijers for R2D2 seeds, and Thomas Friedrich for valuable suggestions. Seeds of wei8-3 and wei-11 seeds were obtained from the European Arabidopsis Stock Center (NASC). We acknowledge the CEITEC core facility CELLIM supported by the MEYS CR (LM2015062 Czech-BioImaging), and the CEITEC core facility Plant Sciences.
Funding
H.S.R. was supported by the SoMoProII program co-financed by the South-Moravian Region and the European Union, by the Ministry of Education, Youth and Sports of the Czech Republic within CEITEC 2020 (LQ1601) and by the Masaryk University.
C.P. was supported by the Kwanjeong Educational Foundation.
C.L.G. was supported by the Deutscher Akademischer Austauschdienst.
W.G. was a post-doctoral fellow of the Research Foundation Flanders.
B.W. was supported by the “NITKA” project under European Social Fund UDA-POKL.04.03.00-00-168/12, realized in the University of Silesia, Katowice, Poland.
A.P and O.N. were supported by the Czech Foundation Agency (GA17-21581Y) and the Ministry of Education, Youth and Sports of the Czech Republic via the National Program for Sustainability (LO1204).
J.C. is a post-doctoral fellow supported by a grant from the Deutsche Forschungsgemeinschaft (Dr334/10) to T.D.
This work was further supported by the European Research Council (FP7/2007-2013 / ERC-grant agreement n° 282300 to J.F.), and the Czech Science Foundation GACR (GA13-40637S) to J.F.; and by the Deutsche Forschungsgemeinschaft (La606/6, La606/13, La606/17 and ERA-CAPS program), and the EU 7th framework program (ITN SIREN) to T.L.
This material reflects only the author's views and the European Union is not liable for any use that may be made of the information contained therein.
Footnotes
Data availability
The data that supports the findings of this study are available from the corresponding authors upon request.
Accession numbers
ABCB1/PGP1 (At2g36910), ABCB19/PGP19 (At3g28860), AUX1 (At2g38120), BAN (At1g61720), BDL/IAA12 (At1g04550), LAX1 (At5g01240), PIN3 (At1g70940), R2D2 (NASC ID N2105637), TAA1 (At1g70560), TAR1 (At4g24670), YUC1 (At4g32540), YUC4 (At5g11320), YUC8 (At4g28720), YUC9 (At1g04180), WOX2 (At5g59340), p35S:DII-VENUS (NASC ID 799173), p35S:mDII-VENUS (NASC ID 799174).
Author contributions
H.S.R., C.P. and C.L.G. contributed equally to this work, performed experiments. H.S.R., B.W., A.P. and O.N. collected samples and performed the analysis for the auxin measurements. W.G. participated in the backcrosses experiments. J.C. and T.D. provided the maize data. H.S.R., C.P., C.L.G., J.F., and T.L. designed the experiments, analysed the data, and wrote the paper.
Competing interest
The authors declare no competing interest.
References
- 1.Figueiredo DD, Köhler CC. Auxin: a molecular trigger of seed development. Genes Dev. 2018;32:479–490. doi: 10.1101/gad.312546.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansfield SG, Briarty LG. Early embryogenesis in Arabidopsis thaliana 2. the developing embryo. Canadian Journal of Botany-Revue Canadienne De Botanique. 1991;69:461–476. [Google Scholar]
- 3.Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 4.Riechmann V, Ephrussi A. Axis formation during Drosophila oogenesis. Curr Opin Gen Dev. 2001;11:374–383. doi: 10.1016/s0959-437x(00)00207-0. [DOI] [PubMed] [Google Scholar]
- 5.Ray S, Golden T, Ray A. Maternal effects of the short integument mutation on embryo development in Arabidopsis. Dev Biol. 1995;180:365–369. doi: 10.1006/dbio.1996.0309. [DOI] [PubMed] [Google Scholar]
- 6.Prigge MJ, Wagner DR. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1280. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa LM, et al. Central cell-derived peptides regulate early embryo patterning in flowering plants. Science. 2014;344:168–172. doi: 10.1126/science.1243005. [DOI] [PubMed] [Google Scholar]
- 8.Möller BK, Weijers D. Auxin control of embryo patterning. Cold Spring Harbor Perspect Biol. 2009;1:a001545. doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friml J, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 10.Robert HS, et al. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol. 2013;23:2506–2512. doi: 10.1016/j.cub.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 11.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 12.Liao C-Y, et al. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods. 2015;12:207–210. doi: 10.1038/nmeth.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paciorek T, Friml J. Auxin signaling. Journal of Cell Science. 2006;119:1199–1202. doi: 10.1242/jcs.02910. [DOI] [PubMed] [Google Scholar]
- 14.Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- 15.Stepanova AN, et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashiguchi K, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Won C, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 19.Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998;116:455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debeaujon I, et al. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueiredo DD, Batista RA, Roszak PJ, Köhler CC. Auxin production couples endosperm development to fertilization. Nature Plants. 2015 doi: 10.1038/nplants.2015.184. 15184. [DOI] [PubMed] [Google Scholar]
- 22.Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler CC. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife. 2016;5:e20542. doi: 10.7554/eLife.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blilou I, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 24.Vieten A, et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132:4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- 25.Weijers D, et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10:265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Larsson E, Vivian-Smith A, Offringa R, Sundberg E. Auxin homeostasis in Arabidopsis ovules Is anther-dependent at maturation and changes dynamically upon fertilization. Front Plant Sci. 2017;8:315–14. doi: 10.3389/fpls.2017.01735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weijers D, et al. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell. 2005;17:2517–2526. doi: 10.1105/tpc.105.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wabnik K, Robert HS, Smith RS, Friml J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 2013;23:2513–2518. doi: 10.1016/j.cub.2013.10.038. [DOI] [PubMed] [Google Scholar]
- 29.Prat T, et al. WRKY23 is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet. 2018;14:e1007177. doi: 10.1371/journal.pgen.1007177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallavotti A, Yang Y, Schmidt RJ, Jackson DP. The relationship between auxin transport and maize branching. Plant Physiol. 2008;147:1913–1923. doi: 10.1104/pp.108.121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Peer WA. Auxin homeostasis: the DAO of catabolism. J Exp Bot. 2017:1–10. doi: 10.1093/jxb/erx221. [DOI] [PubMed] [Google Scholar]
- 32.Pencík A, et al. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J Exp Bot. 2018;69:2569–2579. doi: 10.1093/jxb/ery084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.