Abstract
Yeast libraries revolutionized the systematic study of cell biology. To extensively increase the number of such libraries and the type of information that can be gleaned from them, we previously devised the SWAp-Tag (SWAT) approach that enables rapid, easy and efficient creation of yeast strain collections. Here we present the construction and investigation of a full genome library of ~5500 strains carrying the SWAT NOP1promoter-GFP module at the N terminus of proteins, as well as its use in creating six additional libraries that either restore the native regulation, create an overexpression library with a Cherry tag or enable protein complementation assays from two fragments of an enzyme or fluorophore. We show methods to utilize these SWAT collections to systematically characterize the yeast proteome on multiple levels spanning protein abundance, localization, topology and interactions. Our findings demonstrate how diverse full-genome SWAT libraries facilitate obtaining insights into numerous aspects of the proteome.
Introduction
Among the most important tools that the model organism S. cerevisiae (from here yeast) offers are systematic collections of strains, or libraries, in which each gene is modified in a similar manner to enable genome-wide studies 1–4. To overcome the major hurdles of library construction and to open the possibility for many more library variants to be made simply, rapidly and in a cost-effective way, we have recently developed a new methodology termed SWAp-Tag (SWAT) 5,6. SWAT acceptor libraries enable conversion to other libraries by replacement of the acceptor module with a new tag or genomic sequence of choice, introduced via crossing with a donor strain. The various arising libraries can then be utilized for systematic assays or as a strain reservoir for individual protein studies 5.
Here, we create the first whole-genome SWAT library with an N’ tag (For the complementary C’ SWAT library see 7). Our whole-genome library includes ~90% of yeast genes 8. Imaging of this library allowed us to reveal the localization of 796 yeast proteins that could not be visualized before with a C’ fluorescent tag.
By swapping the original library to six additional versions we could explore many aspects of yeast cell biology: the role of promoters in regulation of protein expression, the mitochondrial protein roster, protein interactions on a whole-organelle level and systematic assessment of protein topology. By constructing seven diverse libraries, as well as the tools to easily generate new ones, we provide possibilities for systematic exploration of eukaryotic cell biology.
Results
Generation of a SWAT full-genome collection
We recently established the SWAp Tag (SWAT) approach that enables rapid creation of yeast libraries 5. To enable proteome-wide investigations, we have now expanded the original N’ SWAT collection to include the majority of yeast genes. We compiled the sequences of all yeast genes using the S. cerevisiae genome database (SGD) 8 and attempted to tag 3916 proteins to complete the N’, genome-wide, SWAT library with the NOP1pr-GFP tag (which encodes for Superfolder GFP 9) (Supplementary table 1). Since the SWAT cassette is added to the N’ of proteins, we had previously used a tailored cassette encoding a strong Signal Peptide (SP) for all endomembrane system proteins that harbor such a targeting signal 5. In the current extended library, we similarly used a cassette encoding a strong mitochondrial targeting signal (MTS) for the several hundred proteins that have such a targeting signal at their N’ (NOP1pr-MTS-GFP) (Figure 1 and Supplementary table 2). Overall, the final SWAT full-genome library contains 5457 strains that underwent several quality control steps (Figure 1, Supplementary tables 1 and 3, Online Methods).
Figure 1. Library generation of genome wide N’ yeast collections by SWAT technology.
Composition of the SWAT full-genome library. Proteins were tagged at the N terminus with an acceptor SWAT module containing the NOP1 constitutive promotor and GFP. Proteins predicted to harbor a mitochondrial targeting signal (MTS) were tagged with a specific acceptor SWAT module containing an MTS upstream of GFP (MTSSu9). Proteins predicted to harbor a signal peptide (SP) were previously created 5 with a similar acceptor SWAT module containing a SP upstream of GFP (SPKar2). Numbers next each tag type name denote how many strains were created/attempted. The SWAT parental library underwent native promoter/regulation swapping: NOP1pr-GFP turned into NATIVEpr-GFP, NOP1pr-MTSSu9-GFP turned into NATIVEpr-MTSnative-GFP and NOP1pr-SPKar2-GFP turned into NATIVEpr-SPnative-GFP. The SWAT parental library was also swapped to create five additional libraries – TEF2pr-mCherry, TEF2pr-VC, CET1pr-VN, NATIVEpr-DHFR F[1,2] and NATIVEpr-DHFR F[3].
Using the SWAT approach to analyze the role of promoters in regulating protein abundance
We first used our library to investigate the relative contribution of promoters to protein expression levels. For this, we created two additional full genome libraries using the SWAT approach (Figure 1). First, a native promoter GFP library termed NATIVEpr-GFP, which restores the natural promoter and endogenous 5’ untranslated regions (UTR) (as well as the native MTS or SP for the relevant proteins) (Figure 1 and Supplementary figure 1). Second, a TEF2pr-mCherry library that introduces one of the strongest promoters in yeast 10,11. In addition, the TEF2pr-mCherry library provides a collection that is tagged with a different fluorophore, thus enabling co-localization studies 5.
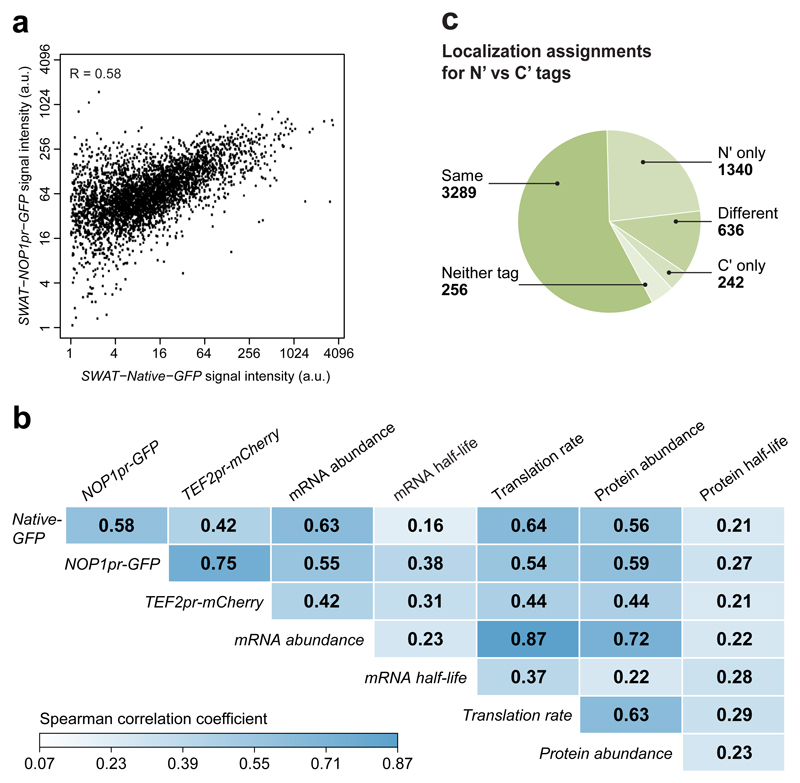
We imaged all three libraries with an automated microscopy system, and each strain was analyzed for fluorescence intensity. Surprisingly, in both the TEF2pr-mCherry and NOP1pr-GFP libraries, proteins had extremely diverse expression levels, spanning over two orders of magnitude (Figure 2a), despite harboring an identical promoter. This highlighted that the promoter only contributed a fraction of the expression regulation.
Figure 2. Genome wide N’ tagged collections enables investigation of abundance regulation and localization assignment.
(a) Histogram of the expression levels of fluorophore-protein fusions of the TEF2pr-mCherry, NATIVEpr-GFP and NOP1pr-GFP libraries. a.u., arbitrary units. (b) Scatter plots showing the correlation between protein abundance of NATIVEpr-GFP tagged strains versus C’ GFP tagged strains 12 (left) or protein abundance under generic regulation (NOP1pr-GFP) (right). R represents two-sided Spearman correlation test score. a.u., arbitrary units. (c) Spearman correlation scores of the expression levels of fluorophore-protein fusions of the TEF2pr-mCherry, NATIVEpr-GFP and NOP1pr-GFP libraries to mRNA abundance as measured by RNA-seq 14, protein translation rates as measured by ribosome profiling 15, mRNA half-lives 16, protein half-lives 17 and protein abundance as measured by mass spectrometry 13. (d) Comparison of N’ NOP1pr-GFP library localization assignments to assignments made from the C′-tagged library 3,12. A complete list of localization assignments is presented in Supplementary table 1. Quantitation of abundance was preformed once. Strains with a final abundance score lower than 1, were excluded from the data analysis and Spearman correlation tests.
Indeed, when comparing expression level of proteins in the NATIVEpr-GFP library they had no higher a correlation to strains that preserve the native promoter such as the C’ GFP library12 , correlation of 0.43 (Figure 2b left); or native abundance as measured by mass spectrometry 13, correlation of 0.56 (Figure 2c) relative to the abundance of proteins under the NOP1 correlation of 0.58 (Figure 2b right) or the TEF2 promoters correlation of 0.42 (Figure 2c).
To identify what other elements are affecting protein abundance, we compared our observed protein abundance measurements to published systematic data sets such as mRNA abundance as measured by RNA-seq 14, protein translation rates as measured by ribosome profiling 15, mRNA half-lives 16, and protein half-lives 17 (Figure 2c). Little correlation was found to RNA or protein half-lives. Poor correlation may be a result of different experimental set-ups but can also suggest that RNA and protein degradation may be a means to regulate the abundance of specific proteins rather than serving as a global regulatory mechanism. The highest correlation was found with mRNA abundance and translation rates, suggesting that chromatin state 18 together with translation both act as global effectors of abundance regulation across the proteome.
The SWAT-GFP library identifies a cellular localization for hundreds of proteins
We then annotated protein localization in all strains of the NATIVEpr-GFP, NOP1pr-GFP and TEF2-mCherry libraries. Assignments were given only to organelles or cellular locations that could be unequivocally determined without the need for co-localization (Supplementary table 1). Since punctate localization can represent a variety of compartments that can only be distinguished by co-localization studies 3,19, we dubbed such proteins as “punctate”.
We then compared the current tally of localizations in the N’ NOP1pr-GFP library to previously assigned localizations with the C’ GFP library 3,12 (Figure 2d). We found that 3289 proteins showed the same localization, strongly supporting the previously assigned location as the correct one for these proteins. Other proteins (242) could only be localized in the C’ library or could not be tagged or visualized in either the N’ or C’ libraries (256). Out of these, 63 are essential 2. Many proteins (636) displayed a localization that was different between N’ and C’ tagging. Additional work will be required to distinguish whether one tag is superior, both locations are correct or neither. Importantly, we could now assign a localization for an additional 796 proteins that were not visualized before in libraries. Taking into consideration also the 544 new localizations from our previous study 5 – altogether 1340 protein localizations were assigned based on the NOP1pr-GFP library.
Using the SWAT libraries to define a more complete mitochondrial proteome
Mitochondria in yeast have already been assigned over 900 high-confidence resident proteins 20. Our N’ libraries provided an opportunity to complete the mitochondrial proteome roster through visualization of mitochondrial proteins that were not recognized before due to either tag interference or conditional expression.
To ensure that we would capture the maximal repertoire of mitochondrial proteins, we built a comprehensive list of proteins with either a high-probability predicted MTS (according to Mitofates 21 and TargetP version 1.1 22) or an experimentally verified one 23,24 (Supplementary table 2), and planned the full genome N’ SWAT library to cater to the 420 proteins that were designated by our analysis as having an MTS (359/420 proteins are represented in our library, for a complete list see Supplementary table 1). MTS-containing proteins were not tagged with the normal NOP1pr-GFP cassette but rather using a specific N’ tagging cassette that has a generic, Su9-MTS before the GFP tag inserted downstream of the MTS cleavage site (similarly to the SP-containing proteins in the endomembrane SWAT library 5). By inserting the cassette 15 nucleotides (5 amino acids) downstream of the original MTS cleavage site the synthetic MTS could direct the protein into mitochondria and after being cleaved, would leave the GFP moiety fused to the mature protein.
We first verified that the Su9-MTS was sufficient to establish a mitochondrial localization (Supplementary figure 2a) and that the NOP1pr-MTS-GFP cassette supports the SWAT approach to return to a native MTS and promoter (Supplementary figure 2b).
We previously showed that N’ tagging of predicted mitochondrial proteins without an MTS can uncover mitochondrial localization for even very low-abundance proteins 20. An additional 15 new mitochondrial proteins that did not have an MTS were found in the whole-genome library either when tagged with the NOP1pr-GFP cassette (not including Su9-MTS) or in the native promoter version (Figure 3a). We verified one such new protein, Ysa1 (Supplementary figure 3).
Figure 3. Characterization of the mitochondrial proteome.
(a) New mitochondrial proteins not containing an N’ MTS that could only be visualized when N’ tagged with NOP1pr-GFP (Left) or were already visible under the NATIVEpr-GFP (Right). (b) The NATIVEpr-(NATIVE)MTS-GFP library (Not including Su9-MTS) uncovered new mitochondrial proteins containing an N’ MTS. (c) Several proteins can still be seen on mitochondria even after swapping their NOP1pr-Su9MTS-GFP cassette to a TEF2pr-mCherry without an MTS, suggesting alternate targeting information is found in these proteins. (d) MTS and TMD prediction analysis for proteins showing mitochondrial localization with the NOP1pr-GFP, NATIVEpr-GFP, and/or C’ GFP tags. Cx(9)C - cysteine-rich domain 26, β-barrel domain 27. All scale bars are 5µm. Imaging of strains was performed a single time. Images represent entire field.
We could also verify mitochondrial localization for several of the MTS containing proteins. Since the Su9-MTS is dominant and could mis-target non-mitochondrial proteins into mitochondria, we only assigned mitochondrial localization to proteins if we could verify their mitochondrial targeting in the NATIVEpr-GFP library when targeted by their native MTS. While in this library some proteins could not be visualized due to low expression levels, we found ten new mitochondrial proteins, most of which are encoded by genes without an annotated function or name (Figure 3b).
Interestingly, deleting the MTS from all MTS-containing proteins (by using a TEF2pr-mCherry cassette without an MTS) uncovered five proteins that robustly localized to mitochondria even in the absence of their predicted MTS (Figure 3c). We investigated whether the MTS truncated versions of two of them, Tam41 and Coq2, were still translocated into mitochondria using in vitro translocation assays (Supplementary figure 4). Indeed, Tam41 was efficiently imported into mitochondria even when its MTS was truncated. A TargetP analysis of Tam41 for internal MTS-like signals (iMTS-Ls) 25 suggests a C’ iMTS that could be involved in this process (Supplementary figure 4a). On the other hand, Coq2 lacking its MTS, which was targeted to the mitochondrial membrane (Figure 3c), could no longer be imported in vitro into isolated organelles (Supplementary figure 4b), suggesting that for this protein targeting and translocation information are found in distinct regions.
More broadly, when looking at all mitochondrial proteins visualized to date using a GFP tag (Total of 635 proteins as annotated by the C’ GFP, N’ NATIVEpr-MTS-GFP or the NOP1pr-GFP libraries), it appears that 70 such proteins have neither a predicted MTS nor a trans membrane domain (TMD) that might help in their targeting to mitochondria (Figure 3d). While six proteins out of these rely on the MIA complex (through a Cx(9)C cysteine-rich domain) 26 and four rely on the SAM/TOB complex (through a β-barrel domain) 27 for their targeting and translocation to mitochondria, the rest might target to mitochondria using alternate, yet uncharacterized, signals.
Several N’ proteins that localized to mitochondria also showed localization to another organelle in the same cell or localized differently when C’ tagged. This suggests that some of these proteins are dually targeted 28. We imaged a subset of these proteins from the NATIVEpr-GFP library under several growth conditions (Supplementary figure 5) and found that they indeed could reside in a variety of organelles depending on the medium. Knowledge on the dynamics of such mitochondrial proteins may help to better understand the crosstalk of mitochondria with other cellular compartments 29,30.
Protein-fragment complementation SWAT libraries can be used for systematic measurement of protein-protein interactions
We next used our SWAT parental library to build four new libraries for assaying protein-protein interactions. We based our new libraries on two protein-fragment complementation assay (PCA) approaches: split Venus 31 and the split Dihydro Folate Reductase (DHFR) enzyme 4.
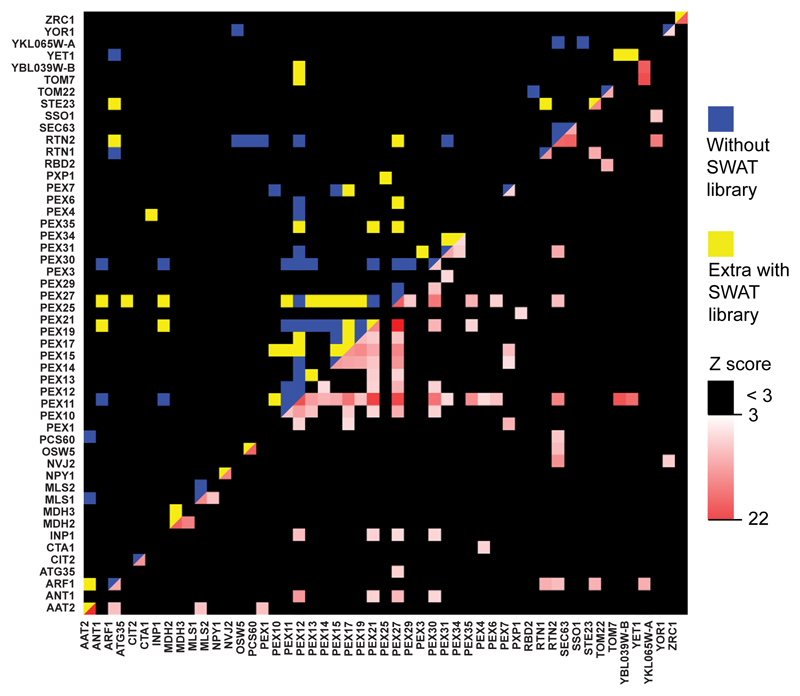
The DHFR PCA reporter confers resistance of cells to the cytostatic drug methotrexate allowing growth when the two proteins tagged with the two fragments of DHFR interact (the N’ fragment termed DHFR F[1,2] and C’ fragment termed DHFR F[3]) (Figure 4a). A previous large-scale protein interactome was determined in yeast with two C’-tagged DHFR fragment libraries 4. Since the C’ tag does not always enable the correct localization of proteins and since interaction between the two fragments requires specific topology of membrane proteins (the two fragments have to be facing the same side of the membrane to enable the enzymatic function to be reconstituted), we wished to investigate how complementary N’ DHFR PCA libraries could improve the coverage of protein-protein interactions. We therefore made two new libraries: NATIVEpr-DHFR-F[1,2] and NATIVEpr-DHFR-F[3]. As a test case of these libraries, we focused on peroxisome proteins to perform a whole organelle interactome. We utilized strains of peroxisomal proteins from all four DHFR libraries (the two previously published C’ ones and both new N’ ones). Then, strains from all four libraries were employed for a pairwise DHFR PCA screen to test for interactions among the 89 peroxisome-related proteins (each library was mated with the two others from the opposing mating type and assayed) (Figure 4a).
Figure 4. Protein-fragment complementation libraries enable systematic analysis of protein-protein interactions.
(a) Scheme of peroxisome DHFR PCA. Four yeast strain arrays were constructed, each representing 89 peroxisome-related genes tagged with either DHFR F[1,2] or DHFR F[3] fragments of methotrexate-resistant DHFR at either their N’ or C’. These strains were mated to test every protein pair in all permutations. An interaction between two proteins brings the DHFR fragments together resulting in their folding, reconstitution of activity and growth of strains in the presence of methotrexate. (b) The peroxisome interactome. Blue squares represent interactions discovered by the C’ tagged strains alone. Yellow squares represent interactions that required at least one N’ tagged strain for their discovery. The white-to-red spectrum squares correspond to the Z scores for the interaction. Only proteins with at least one interaction are depicted. A complete list of Z scores for the interaction is presented in Supplementary table 4.
The DHFR PCA revealed 230 positive results (Figure 4b and Supplementary table 4). We examined reproducibility by comparing the two strains with the same tagged proteins and the same position of the DHFR fragments (N’ or C’) but with the DHFR fragments swapped (e.g., X-DHFR-F[1,2] + DHFR-F[3]-Y compared to X-DHFR-F[3] + DHFR-F[1,2]-Y). From 165 that had such a paired setup, 120 were reproducible, which put the reproducibility at 73%. Due to the paired setup, the number of unique interactions was 109. From these 55.9% were previously reported in the literature including many known complexes on the peroxisome membrane 32 while only 4.7% of the non-interactions were previously reported supporting the validity of our scoring system.
Indeed, it seems like having both the C’ and N’ libraries is of importance for grasping the entire interactome as of the 109 unique interactions, 48 required the inclusion of N’ strains (Figure 4b) and 75 would not be found without the presence of the C’ strains. Unfortunately, the majority of proteins residing within the peroxisome matrix did not show any interaction in either library suggesting that DHFR substrate availability or DHFR reconstitution efficiency are too limited within the peroxisome.
In addition, we built two libraries based on the Venus PCA. In these libraries, two complementary N- and C-terminal fragments of the fluorescent protein Venus (YFP) (the C’ fragment termed VC and N’ fragment termed VN) were N’ fused to all proteins in opposite yeast mating types. Simple crossing of the two mating type strains resulted in diploids that were used to detect protein association by reconstitution of the full Venus fluorescence. Using these libraries, we repeated the whole-peroxisomal interactome (Supplementary table 5) and found that, in general, this approach is less specific than the DHFR PCA. However, we did find several high-confidence, newly predicted, interactions such as the one between Inp1 and Pex17 that we could verify using a yeast two-hybrid (Y2H) assay (Supplementary figure 6).
Split Venus libraries can be used to assay N’ topology
The strength of the Venus PCA library is that, due to the intrinsic affinity of the Venus fragments, in addition to protein-protein interactions it can be used to study membrane topology (Figure 5a). To do this we used the N’-tagged library including the C’ fragment of the split Venus cassette (VC) under the TEF2 constitutive promoter termed TEF2pr-VC (excluding SP- and MTS-bearing proteins) and mated it with a strain containing the N’ half of the split Venus cassette (VN) not conjugated to any other protein and therefore freely distributed in the cytosol (termed cyto-VN). This configuration should allow complementation of the VC and VN fragments, which results in a fluorescent signal, only if the N’ end of the VC-tagged protein faces the cytosol where the VN is prevalent (Figure 5a). After selection for diploids, the fluorescence of each strain was quantified and localization was assigned using fluorescence microscopy (Supplementary table 1). Cells that had fluorescence above a threshold level were termed as N’ “in” (facing the cytosol) (Because topology of “out” could also result from a technical error giving lack of signal, this assignment could not be made unequivocally) (Supplementary table 6). We verified this assignment for one protein, Scm4 (Supplementary Figure 7). As a more general quality control step, we compared the abundance of proteins in the TEF2pr-mCherry library with the signal intensities we measured in this complementation assay, since both libraries use the same promoter and should give a similar intensity profile. Indeed, the intensity of proteins that showed an “in” signal had a 0.82 two-sided Spearman correlation score with their TEF2pr-mCherry counterparts (Figure 5b). For a subset of proteins in which the orientation of the C’ had been experimentally verified 33 we could use our topology predictions to also resolve TMD number (Supplementary Figure 8). Our method, at present, can only clearly define proteins whose N’ faces the cytosol. By anchoring the complementary Venus fragment in the lumen of organelles, it would be possible to extend our method to define the topology of proteins whose N’ faces the interior of their respective organelles.
Figure 5. Protein-fragment complementation libraries enable systematic analysis of membrane protein topology.
(a) Scheme of split Venus analysis to determine topology for the N’ of proteins. The N’ TEF2pr-VC library was mated with a strain containing a cytosolic VN, and only if the N’ of a membrane protein faces the cytosol will complementation occur and a fluorescent signal appear suggesting the topology of the protein’s N’. A complete list of topology assignments is presented in Supplementary table 6. (b) Correlation graph between the TEF2pr-mCherry library and the TEF2pr-VC library with a cytosolic-VN (only “in” assignments). Correlation score is two-sided Spearman correlation. Quantitation of abundance was preformed once. Strains with a final abundance score lower than 1, were excluded from the data analysis and Spearman correlation tests.
Perspectives
Our new N’ tag genome-wide libraries have enabled us to explore the proteome on several levels – abundance, localization, topology and protein-protein interaction. We hope that the new information introduced here for uncharacterized proteins together with the presence of these genes in our new libraries will promote the investigation of their functions.
Currently our library is intended for use in an arrayed format. However, for future uses pooled experiments may be of value. For such cases, in principle, our pooled approach for sequencing the strains, which relies on the sequence of the L2 linker followed by the gene sequence, could serve as a pseudo-barcode, but would have to be developed into a quantitative assay 34. An alternative and easy strategy for using SWAT-derived libraries in a pooled fashion is mating them with a barcoder library 35.
The parental N’ SWAT library with its easy-to-use swapping ability enables endless new possibilities for array-wide protein investigation. In a short time period and at a fraction of the cost incurred to date using other approaches, any yeast laboratory can make its very own library harboring a diversity of selection markers, promoters, untranslated regions, targeting signals, fluorophores, affinity tags or any other genetic element of choice. Using this platform, the systematic exploration of any protein is no longer restricted and can be done either with N’ or C’ tagging 7. Together, these approaches should contribute greatly to our knowledge on how the living cell works.
Online Methods
Plasmid construction
We constructed plasmids using restriction-free cloning methods 36. For a complete list of plasmids, see Supplementary table 7. The I-SceI restriction site sequence was agttacgctagggataacagggtaatatag. The protein linker sequences (which also served as the generic recombination sites) were as follows: L1, 5′-cgtacgctgcaggtcgacggtggcggttctggcggtggcggatcc-3′; L2, 5′-ggcggttcctctggtggtggtggtgcgacagagaattcatcgatg-3′. Underlined sequences are the primer sequences used for amplification of the tagging module, corresponding to the pYM series sequences (S1and S4 respectively) 6. The use of these sequences ensured compatibility with existing oligo collections for these popular module sets.
The tagging modules included the constitutive promoter of the SpNOP1 gene 6 to drive the fusion tag–protein expression. This promoter confers medium-level expression compared to stronger promoters such as ScTEF1pr and ScGPDpr.
The GFP used in the cassettes of both the NOP1pr-GFP and NATIVEpr-GFP libraries is Superfolder GFP 9.
The Kar2 SP sequence used in the SWAT-SP-GFP module was: atgtttttcaacagactaagcgctggcaagctgctggtaccactctccgtggtcctgtacgcccttttcgtggtaatattacctttacagaattctttccactcctccaatgttttagttagaggtgccgat.
The codon-modified (to avoid altered recombination) Kar2 SP sequence used in the donor NAT::TEF2pr-SPKar2-mCherry plasmid was:
atgttcttcaatagattgtcagctgggaagcttcttgtgccactgtctgtagttctttacgcactgttcgtagtgatactacccctgcaaaactcctttcactcttctaatgtcctggtcagaggcgcagac.
The MTS of Neurospora crassa OR74A ATP synthase protein 9 sequence used in the SWAT-MTS-GFP module was:
atggcctccactcgtgtcctcgcctctcgcctggcctcccggatggctgcttccgccaaggttgcccgccctgctgtccgcgttgctcaggtcagcaagcgcaccatccagactggctcccccctccagaccctcaagcgcacccagatgacctccatcgtcaacgccaccacccgccaggctttccagaagcgcgcctac.
The codon-modified (to avoid altered recombination) MTS of N.crassa OR74A ATP synthase protein 9 sequence used in the donor NAT::TEF2pr-MTS-mCherry plasmid was:
atggcttctaccagagttttggcttctagattggcttctagaatggcagctagtgctaaggttgctagaccagctgttagagttgcacaagtttctaagagaacaatacaaaccggttctccattgcaaaccttgaagagaacccaaatgacttctatcgttaacgctactaccagacaagcatttcaaaagagagcttac.
The ORFs of Tam41 and Coq2 versions lacking the N-terminal 28 or 35 amino acids were amplified and cloned into the SacI and SalI or SacI and HindIII sites (respectively) of pGEM4 (Promega, Mannheim, Germany).
Primer choice and design
Total number of genes annotated in SGD currently stands at 6075 excluding dubious ones. Due to the structure of our tagging cassette we did not attempt to tag the 62 yeast proteins that include an N’ intron or the 250 genes which have identical homologues in the genome. From the remaining 5763 genes that can accurately be tagged with our N’ SWAT cassette, 1847 were already attempted in our previous work 5. For all rest we designed primers and attempted to create them.
Primers for amplification of transformation cassettes and gene-specific targeting were designed with the Primers-4-Yeast web tool 37 (http://wws.weizmann.ac.il/Primers-4-Yeast) using the pYM plasmid type 36. All tagging primers include a 40-bp homology sequence followed by 20 or 18 bp of cassette amplification sequence. The homology sequences were upstream and downstream of the protein start codon for normal N′ tagging, as described in the Primers-4-Yeast web tool. For N′ tagging of SP or MTS containing proteins, homology sequences were designed to insert the cassette five amino acids downstream from the predicted cleavage point. Primers for validation of tagging transformations were also designed with the Primers-4-Yeast web tool, using the appropriate “Check primers” option. Primers were manufactured by Sigma-Aldrich in 96-well plates. A full list of primers used in this study is presented in Supplementary table 8.
High-throughput yeast transformations
The BY4741 laboratory strain 38, which is the basis for most systematic yeast libraries, was used as the master strain for the collection. The SWAT-GFP, SWAT-MTS-GFP and SWAT-SP-GFP acceptor modules (Supplementary table 7) were PCR amplified (KAPA Hi-Fi or KOD Hot Start DNA polymerase) in 96-well plates (Thermo Fisher Scientific) and transformed into BY4741. Transformations were carried out via a modified PEG-LiAc protocol 39 in a high-throughput manner. Each reaction was composed of 2.1 OD600 of cells (3 ml of cells at 0.7–0.8 OD600), 120 μl of 50% PEG 3500 (wt/vol), 18 μl of 1 M LiAc, 25 μl of boiled SS-carrier DNA, 7 μl of double-distilled water and 20 μl of PCR-amplified transformation cassette DNA. Heat shock was applied in a PCR machine for 15 min at 30°C followed by 30 min at 42°C. Transformed cultures were plated on synthetic defined (SD)-URA media in 48-well divided agar plates (Bioassay X6029) and were incubated for 2–3 d at 30°C. All procedures were carried out using an automated liquid handler (Janus, PerkinElmer).
Yeast strain validation and collection assembly
To select pristine strains to be included in the final library, we picked four clones for each gene, and performed several quality control steps. Transformations that failed to yield four clones were repeated (462), and those that still failed were redone using re-synthesized primer pairs (199). Following these efforts, we obtained coverage of 95% of the anticipated yeast genes. Out of the 251 proteins that could not be tagged, 33 have an over-expression growth inhibition 40, 77 are essential proteins 2, and 47 proteins showed a GO term enrichment for “cytoplasmic translation” (p-Value = 0.016448) 8(Supplementary table 9).
The quality control that each strain underwent was: (i) Validation of integration locus by PCR (Supplementary table 3): This was performed using a common forward primer from the 3′ end of the SWAT modules (S4 reverse complement) and a gene-specific reverse primer from the gene coding sequence (Supplementary table 8). Strains of proteins that have a signal peptide did not undergo a PCR check. Strains that did not have a positive PCR for any of the 4 clones were not included in the final clone library (251 proteins - This could mean that there were not clones obtained or that no clone gave a positive PCR). (ii) Detection of the fusion protein by fluorescence microscopy: Two clones were imaged by fluorescence microscopy and we reviewed images manually to assign up to three localizations to each clone. Strains with a non-distinct pattern were given the assignment of “ambiguous”. Strains with fluorescent signal that was quantified to be below background signal, were given the assignment of “below threshold”. Assignment categories were: Bud, Bud Neck, Cell Periphery, Cytosol, ER, Mitochondria, Nuclear Periphery, Nucleolus, Nucleus, Punctate, Vacuole and Vacuole Membrane. (iii) Determination of the SWAT swapping capability (See “Analysis of swapping-procedure efficiency” section); and finally (iv) Sequencing (Anchor-seq) to ensure correct reading frame: We employed a targeted-sequencing strategy detailed in 7 to verify the junction encompassing the 3’ end of the cassette and the 5’ end of each gene. Briefly, we pooled all strains from the SWAT library together, extracted their genomic DNA, sheared it into fragments of 300-800bp that were gel-purified, ligated to Anchor-seq adaptors and submitted to two rounds of PCR. The sequences of adapters and of oligonucleotides are detailed in Supplementary table 8. This protocol enriched specifically the junctions of interest, which we then sequenced by next-generation sequencing. The reads were subsequently analyzed to classify each ORF into one of three categories corresponding to either validated sequences, sequences containing a frameshift, or sequences containing a point mutation (Supplementary table 3). Sequencing was performed in two steps, first sequencing was performed on all 4 initial clones. In this round (Anchor-seq round 1, Supplementary table 3) the reads were 150 bp from the L2 linker into the coding sequence. Analysis was performed to find if strains were: "Positive" meaning that the cassette was inserted correctly and no mismatches or indels were observed / "Not detected" meaning that the sequence could not be found following sequencing / "Mismatch" meaning that either a mismatch or an indel was found/ "Half" meaning that there was an imbalance in read count between the first and second halves of the read. To complete the analysis also for the previously published strains 5, those were also sequenced. The reads were again 150 bp from the L2 linker into the coding sequence. The analysis was done as above and we added information of “Bad linker” which means there is only a mismatch in the L2 linker.
Finally, after the final clones were selected for the full genome SWAT library a second round of sequencing was carried out (Anchor-seq round 2, Supplementary table 3). This time the reads were only 92 bp from the L2 linker into the coding sequence. Analysis was done as above but differentiated between "Mismatch" which means that some bases did not match the expected sequence and "Indel" which means that an insertion or a deletion occurred within the sequence. We also annotated "Low read count" which means that the sequence was correct but observed less times than would be expected and we added information on the amount of bp altered in the mismatch or indel strains.
A strain was removed from the library if it had an indel in round 2 or if it could not be detected in round 2 than if it had a mismatch in round 1. Proteins bearing a signal peptide that did not undergo a PCR check were removed if they were not annotated as positive in the two Anchor-seq rounds. All other alterations from expected sequence were highlighted as remarks in Supplementary table 3.
Strains with a validated sequence, consistent localization assignment, swapping capacity and that had been validated by PCR were chosen to compose the final SWAT full-genome library that contains 5457 strains (Supplementary tables 1 and 3). The rigorous quality control should maximize the utility of this parental library, which will become a basis for multiple future N’ yeast full-genome libraries.
Donor strain construction
Donor strains were constructed on the background of an SGA 41 compatible query strain and contained a galactose-induced I-SceI endonuclease and a donor plasmid. To spare a selection marker in the donor strain, we introduced a K. lactis URA3 selection marker into the can1Δ locus, upstream of the STE2pr-SpHIS5 fragment (used for selection of MATa). A Gal1pr-I-SceI fragment was then introduced to replace the URA3 selection, resulting in can1Δ::GAL1pr-SceI::STE2pr-SpHIS5 (strain yMS2085).
Analysis of swapping-procedure efficiency
First, to see if the native promoter/regulation GFP swap can restore the regulation of the native promoter, we swapped three conditionally induced genes. NOP1pr-GFP-GAL2 and NATIVEpr-GFP-GAL2 were grown for 4 hours in either liquid SD (2% glucose) or SG (2% galactose) medium. NOP1pr-GFP-SUC2 and NATIVEpr-GFP-SUC2 were grown for 4 hours in either liquid SD glucose or synthetic medium with no glucose. NOP1pr-GFP-PHO5 and NATIVEpr-GFP-PHO5 were grown for 4 hours in either liquid SD complete or SD - phosphate medium (Supplementary figure 1). Next, we measured the swapping efficiency of NOP1pr-GFP strains to TEF2pr-mCherry for all 4 clones. The clones were imaged via a high-content screening platform in brightfield, GFP and Cherry channels. We reviewed images of all clones manually and assigned up to three localizations to each clone. Assignment categories were as above. Preference for inclusion in the SWAT-GFP library was given to clones that showed a similar localization to the NOP1pr-GFP tag.
Automated manipulation of yeast libraries
We conducted automated strain maintenance and manipulation using a RoToR benchtop colony arrayer 42 (Singer Instruments). We carried out SGA procedures 41 for mating of the parental SWAT-GFP collections with donor strains bearing the native promoter/regulation GFP donor (Supplementary Table 7; pSD-N9), the NAT:TEF2pr-mCherry donor (Supplementary Table 7; pSD-N15/16/21) the HYGRO:TEF2pr-VC donor (Supplementary Table 7; pSD-23), and the KAN:CET1pr-VN donor (Supplementary Table 7; pSD-N24). After double mutant selection all libraries were selected for MATα haploids except for the CET1pr-VN library that was selected for MATa. Then tag swapping was prompted by growth on yeast extract peptone (YEP)-galactose (2%) media for 1–2 d to induce I-SceI expression. Tag swapping was then selected by two cycles of growth over night on SD + 5-FOA (1 g/L) media for NATIVEpr-GFP library, yeast extract peptone dextrose (YEPD) + nourseothricin (NAT; 200 μg/mL) for the TEF2pr-mCherry library, SD + 5-FOA (1 g/L) + hygromycin B (200 μg/mL) for the TEF2pr-VC library, or SD + 5-FOA (1 g/L) + g418 (200 μg/mL) for the CET1pr-VN library.
High-throughput microscopy
We carried out high-content screening of strain collections using an automated microscopy setup (ScanR system, Olympus) as previously described 12. We acquired images using a 60× air lens for GFP (excitation, 490/20 nm; emission, 535/50 nm), mCherry (excitation, 572/35 nm; emission, 632/60 nm), BFP (excitation, 402/15 nm; emission, 455/50 nm) and brightfield channels. Images were analyzed using the ScanR Analysis software 2.7.0 (r3429) x64 (Olympus), and single cells were recognized on the basis of the brightfield channel. Measures of cell size, shape and fluorescence signals were extracted. For localization assignments, we reviewed images manually using ImageJ (1.51p Java1.8.0_144 (64-bit)). As we did not use any co-localization markers, we assigned only those localizations that could be easily discriminated by eye: ER, nuclear periphery, cytosol, cell periphery, vacuole lumen, vacuole membrane, mitochondria, nucleus, bud or bud neck, and punctate (which includes structures such as the Golgi apparatus, peroxisomes, endosomes, p-bodies, inclusions, lipid droplets, other vesicular structures and subdomain compartments) (Supplementary table 1). All images of the N’ libraries strains can be found and downloaded at our Loqate database (http://www.weizmann.ac.il/molgen/loqate).
Computational quantification of single-cell fluorophore intensity
We measured the median GFP/mCherry intensity for each strain using single-cell recognition software (scanR Analysis software 2.7.0 (r3429) x64, Olympus) as previously described 12. Strains with fewer than 30 recognized cells were excluded. We obtained the baseline autofluorescence level of each plate from strains not expressing GFP. We then calculated each strain’s final score by subtraction of this value out of each strain median GFP/mCherry intensity. Strains with a final score lower than 1, were excluded from the data analysis and two-sided Spearman correlation tests that were performed with R studio version 0.99.486.
Data processing
We compared the subcellular-localization annotations of the NOP1pr-GFP, NOP1pr-MTS-GFP, NOP1pr-SP-GFP libraries with those of the C′-tag library (comprising data from two previously published data sets 3,12). The pairwise comparisons between N′-tagging and C′-tagging annotations were classified in the following manner: ‘Same’ was assigned when at least one N′ annotation corresponded to a C′ one. ‘N′ only’ was assigned if the C′ localization was classified as below threshold or ambiguous, or if no assignment existed. ‘C′ only’ was assigned if the N′ localization was classified as below threshold or ambiguous, or if no assignment existed. ‘Neither tag’ was assigned if both the N′ localization and C′ localization were classified as below threshold or ambiguous, or if no assignment existed. All other cases were classified as ‘different’ (Fig. 3a). All of the calculations were performed with Python 2.7 software.
Mitochondrial targeting signal predictions of yeast proteins
We compiled MTS determinations for all yeast proteins based on both experimental evidence (EE) from previous studies (EE1 23, EE2 24 and MTS prediction algorithms (Mitofates 21 and TargetP version 1.1 22)) (Supplementary table 2). As each of the four sources support different MTS designations we employed a scoring method to the analysis results where a prediction is accounted for if answers the following rules:
An MTS must be >6 amino acids in length based on experimental evidence or prediction.
Any protein identified in EE1 five times or more.
Any protein that Mitofates predicted as MTS containing with a score of >0.5 (reported precision of 0.83).
A few cases were manually assigned for known MTS containing proteins from literature review.
Bona fide non-mitochondrial proteins were removed after manual review.
Following these criteria we designated 420 proteins as having MTS. Then, the MTS cleavage site (distance in amino acids from N’) was selected by the experimental evidence and predictions. Selection of the cleavage site was done by the following hierarchy:
Site was consistent (i.e. within 5 amino acids apart) between EE1 and EE2.
Site was consistent between EE1 and Mitofates prediction.
Site was consistent between EE1 and TargetP prediction.
Site was consistent between EE2 and Mitofates prediction.
Site was consistent between EE2 and TargetP prediction.
When there was no consistency between experimental evidence the site was first determined by EE1 and only if not available than by EE2.
For MTSs classified only by predictions a site was given priority if it was consistent between Mitofates prediction and TargetP prediction. If no consistency was found than the site was chosen as determined by Mitofates.
For a few cases the cleavage site was picked manually based on previous evidence.
Subcellular fractionation and western blot analysis (For: Supplementary figure 3)
Yeast cultures were grown to an A600 of 1.5. mitochondria were isolated as described previously 43. Spheroplasts were prepared in the presence of zymolyase 20T (MP Biomedicals, Irvine, CA). Equivalent portions from fractions of the total (T), cytosol (C) and mitochondria (M) were analyzed by western blotting using αα to follow the tagged protein (either Ysa1 or Kgd2), αHsp60 as a mitochondrial marker and αHxk1 as a cytosolic marker. Full scans of all blots are shown in supplementary figures 9a-c.
Import of radiolabeled proteins into isolated mitochondria (For: Supplementary figure 4)
Isolation of yeast mitochondria and import reactions were essentially performed as described previously 44 in the following import buffer: 500 mM sorbitol, 50 mM Hepes, pH 7.4, 80 mM KCl, 10 mM magnesium acetate, and 2 mM KH2PO4. Mitochondria were energized by addition of 2 mM ATP and 2 mM NADH before radiolabeled precursor proteins were added. To dissipate the membrane potential, a mixture of 1 µg/ml valinomycin, 8.8 µg/ml antimycin, and 17 µg/ml oligomycin was added to the mitochondria. Precursor proteins were incubated with mitochondria for different times at 25°C before non-imported protein was degraded by addition of 100 µg/ml proteinase K. Full scans of all blots are shown in supplementary figures 9d and 9e.
Mitochondrial proteins dual localization analysis (For: Supplementary figure 5)
Strains showing mitochondrial dual localization in the N’ NATIVEpr-GFP library (Supplementary table 1) were arrayed into liquid 96-well polystyrene growth plates. Liquid cultures were grown overnight in SD medium at 30°C. Cells were back-diluted to ∼0.25 OD600 into 4 plates each containing a different medium: glucose 2%, glycerol 2%, galactose 2% or glucose 0.2%. Plates were then grown for 4 h at 30°C to reach logarithmic growth phase. Strains from all 4 plates in addition to the original overnight plate were transferred into glass-bottom 384-well microscope plates (Matrical Bioscience) coated with concanavalin A (Sigma-Aldrich) to allow cell adhesion. Wells were washed twice in appropriate medium to remove floating cells and reach cell monolayer. Manual microscopy was performed using VisiScope Confocal Cell Explorer system, composed of a Zeiss Yokogawa spinning disk scanning unit (CSU-W1) coupled with an inverted Olympus IX83 microscope. Images were acquired using a 60× oil lens and captured by a connected PCO-Edge sCMOS camera, controlled by VisView software, with wavelength of 488 nm (GFP). Images were transferred to ImageJ (1.51p Java1.8.0_144 (64-bit)), for slight, linear adjustments to contrast and brightness.
SWAT DHFR PCA library construction
Antibiotic resistance genes nat1 and hph were PCR amplified respectively from plasmids pAG25 and pAG32 45 with primers DHFR-F1 and DHFR-F2 (Supplementary table 8). Strain yMS2085 5 was transformed with the PCR products to create strains yMS2085-NAT1 and yMS2085-HPH where each resistance gene is integrated on chromosome V between genes CAJ1 and TPA1 46. Next, SWAT DHFR PCA donor plasmids were created. First, DHFR F[1,2] and DHFR F[3] were PCR amplified respectively from pAG25-linker-DHFR F[1,2] and pAG32-linker-DHFR F[3] 4 with primer pairs DHFR-F2 and DHFR-R2, and DHFR-F3 and DHFR-R3 (Supplementary table 8). The PCR products were cloned into pSD-N2 (Supplementary table 7) with restriction enzymes BamHI and SpeI to form pSD-N25 and pSD-N26 (Supplementary table 7). Strain yMS2085-NAT1 was transformed with pSD-N25 to create ySWAT-DHFR-F[1,2] and yMS2085-HPH was transformed with pSD-N26 to make ySWAT-DHFR-[F3].
The resulting strains were used as SWAT donor strains to generate two libraries with DHFR-F[1,2] or DHFR-F[3] tagged N-terminally of 89 peroxisome-related genes, according to the procedure previously described 5.
SWAT DHFR PCA screen
Four libraries were employed for the DHFR PCA screen to test for interactions among the 89 peroxisome-related proteins. These libraries are the SWAT-based DHFR F[1,2] library (87 strains available out of 89), the SWAT-based DHFR F[3] library (88 strains), a library with C-terminal DHFR F[1,2] tags (65 strains) and a library with C-terminal DHFR F[3] tags (75 strains) 4. Each DHFR F[1,2] strain was mated with each of the DHFR F[3] strains by overnight incubation on YPD. The strains were organized in such a way that each row contained the same DHFR F[1,2] strain and each column the same DHFR F[3] strain, in a 1536-format on 24 plates in total (Figure 4a). After mating, diploid cells were selected for by incubation for 2 days on YPD medium with 100 µg/ml nourseothricin (Werner Bioagents, Jena, Germany) and 250 µg/ml hygromycin B (Wisent Bioproducts, Saint-Jean-Baptiste, Canada). This step was repeated once. Next, the strains were transferred to synthetic complete medium (4% (w/v) Noble agar) with 200 µg/ml methotrexate (Bioshop Canada, Montréal, Canada) and without adenine or ammonium sulfate. Pictures of the strains were taken at the start of the experiment and after 4 days incubation at 30°C. Every protein-protein interaction was tested in duplicate and all plate handling and imaging was done with a BioMatrix automated plate handler (S&P Robotics Inc., North York, Canada).
SWAT DHFR PCA data analysis
The integrated colony densities, which are an estimation of colony volume, were obtained with a custom-made ImageJ (1.51p Java1.8.0_144 (64-bit)) scripts that measures the integrated colony density by multiplication of the colony area with its mean intensity. To account for variation in the initial cell material deposited at the start of the experiment, the day 4 colony densities were corrected with their day 0 values through linear regression and normalization. Some crossed strains show higher overall background growth. To correct for this phenomenon, the mean of the median row (identical F[1,2] strains) and median column (identical F[3] strains) values where the strain is located were subtracted from the colony size. These two normalization steps improved the correlation between our results and those in the literature 47. The colony sizes on each plate showed a normal distribution (Shapiro test, p-values < 10-32) and the Z scores of the colony sizes were calculated on the basis of the distribution within each plate. A result was considered positive if the minimum Z score (of the two duplicates) was greater than 3. From the positive results, 48 % had been detected in previous protein-protein interaction studies (www.biogrid.org), not including results of the original C’ DHFR PCA screen 4. However, this agreement drops to less than 10 % (4/46) when considering results with relatively low Z scores (between 3 and 4.85) and at least one highly abundant protein. Therefore, those results with Z scores below 4.85 and at least one highly abundant protein were removed from the list of positive results. A highly abundant protein was defined as having a score above 30 according to GFP abundance data 3,5,12 with the GFP tagged in the same position as the DHFR PCA tag (N- or C-terminal). If this data was not available, GFP abundance values were taken from sources in which GFP was positioned on the other terminus. Calculations were preformed using R Studio version 0.99.486.
SWAT Venus PCA analysis (For: Supplementary table 5)
A yeast array of 92 strains each expressing a peroxisomal associated protein (and some control strains) was compiled from the NOP1pr-GFP library (Supplementary table 5). Strain manipulation was performed using a RoToR benchtop colony arrayer 42 (Singer Instruments). We carried out SGA procedures 41 with donor strains bearing either the KAN:CET1pr-VN donor (Supplementary Table 7; pSD-N24) or the HYGRO:TEF2pr-VC donor (Supplementary table 7; pSD-N23) with NAT:PEX3-mCherry as a peroxisomal marker. After double mutant selection the TEF2pr-VC array was selected for MATα haploids and the CET1pr-VN array was selected for MATa. Then tag swapping was prompted by growth on yeast extract peptone (YEP)-galactose (2%) media for 1–2 d to induce I-SceI expression. Tag swapping was then selected by two cycles of growth over night on SD + 5-FOA (1 g/L) + hygromycin B (200 μg/mL) + nourseothricin (NAT; 200 μg/mL) for the TEF2pr-VC array, or SD + 5-FOA (1 g/L) + g418 (200 μg/mL) for the CET1pr-VN array. All strains from the two arrays were then crossed, and selected for diploids. Strains were then imaged and analyzed for signal localization and intensity (See “High-throughput microscopy” and ”Computational quantification of single-cell fluorophore intensity” sections) (Supplementary table 5).
Yeast two-hybrid assay (For: Supplementary figure 6)
The yeast strain HF7c 48 and protocols were from Clontech Laboratories. The full-length PEX17 open reading frame was amplified from S. cerevisiae genomic DNA and inserted into the BamHI/SalI restriction sites of plasmids pGAD424 (AD) and pGBT9 (BD), respectively 49. pGAD424-INP1 and pGBT9-INP1 plasmids have been described 50. Plasmids were transformed into HF7c cells, and cells were cultured at 30°C in synthetic dropout medium to an OD600 of 0.5-0.8 before being harvested by centrifugation. The OD600 was adjusted to 1.0 for all cells, and 1 μL of a series of 1:10 dilutions (corresponding to an OD600 of 100, 10-1, 10-2, 10-3) were then spotted onto selective plates, which were incubated at 30°C for 3-5 days. -Leu -Trp medium selects for the presence of both pGAD424 and pGBT9 plasmids in cells, whereas -His -Leu -Trp medium selects for the presence of a protein-protein interaction.
TMD and N’ topology analysis
Transmembrane prediction was performed with the following programs using default parameters: TMHMM 51, HMMTOP 52, Phobius 53, Philius 54, TOPCONS 55. The TOPCONS results were taken for the TOPCONS algorithm itself, as well as all the component programs individually, Octopus, Polyphobius, Philius, Scampi and Spoctopus. As the results of Philius that were run separately and the results of Philius in the TOPCONS program were identical, only the results from the program run individually were used (Phobius and PolyPhobius gave different results). The annotation of TMD from the Uniprot database was taken from the website, uniprot.org for the whole yeast proteome (accession number: UP000002311) using the Subcellular location “Transmembrane”. Topology prediction was extracted from the same results of all the programs with the exception of Uniprot, where there is no topology prediction. Custom scripts (available upon request) and manual analysis were used to parse the results.
Topology analysis using Venus PCA
Using automated strain maintenance and manipulation with a RoToR benchtop colony arrayer 42 (Singer Instruments) we carried out mating of the entire TEF2pr-VC library with a BY4741 strain containing HO::KAN-CET1pr-VN, NAT:PEX3-mCherry and the URA:MTS-BFP plasmid (Supplementary table 7). After two rounds of diploid selection strains were imaged and analyzed for signal localization and intensity (See “High-throughput microscopy” and ”Computational quantification of single-cell fluorophore intensity” sections) (Supplementary tables 1 and 6). Strains showing a signal above the his3Δ1::GFPdC control strain were considered as having the tagged proteins N’ facing the cytosol (denoted “in”).
Proteinase K protection assay (For: Supplementary figure 7)
Mitochondria isolated from cells expressing tagged Scm4 were treated with the indicated amounts of proteinase K (PK) or Trypsin. After inhibiting the proteases, samples were precipitated with trichloroacetic acid and analyzed by SDS-PAGE followed by immunodecoration with antibodies against the HA-tag or the indicated mitochondrial proteins. Tom70, a MOM protein exposed to the cytosol; Aco1 and Hep1, matrix proteins. Full scans of all blots are shown in supplementary figures 9f and 9g.
Obtaining the libraries, plasmids, images and protocols
All strains, plasmids and libraries presented in this manuscript are freely available upon request. All protocols for using the SWAT strategy can be found on Protocol Exchange site (https://www.nature.com/protocolexchange/labgroups/1106525) and on our lab website (http://www.weizmann.ac.il/molgen/Maya/SWAT). All images of the N’ libraries strains can be found and downloaded at our Loqate database (http://www.weizmann.ac.il/molgen/loqate).
Supplementary Material
Acknowledgements
We would like to thank Yoav Peleg for plasmid construction, Genia Brodsky for graphics, Ron Rotkopf for support in statistical analysis, Kelly Tedrick for technical help with the Y2H experiments, and Chris Meisinger and Nora Vögtle for help with the MTS assignments. We thank Gat Krieger for helpful discussions and technical help. We would like to thank Christian Ungermann and Won-ki Huh for plasmids. The work in the Schuldiner lab was supported by an ERC CoG Peroxisystem (646604), an SFB 1190 from the DFG, a Mitzutani foundation grant and a VolksWagen foundation grant (93092). The collaborative work in this manuscript by the Schuldiner, Pines, Herrmann and Rapaport labs was supported by a DIP grant (P17516). Work at the Rachubinski lab was supported by Foundation Grant FDN-143289 from the Canadian Institutes of Health Research. Work in the Michnick lab was supported by a Canadian Institutes of Health Research grant MOP-GMX-152556. UW and DD are recipients of the Azrieli student-award grant. MS is an Incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics.
Footnotes
Data Availability
The data that support the findings of this study are available from the corresponding author upon request.
Code availability
All original code used in this study are publicly available at:
ORCID ID: Maya Schuldiner: 0000-0001-9947-115X
Author Contribution
Conceptualization: UW, IY, MS; Investigation: UW, IY, ES, BS, DD, JN, RB, ZA, OG, NF, SC, KK, BK, JL, FB, JK, SB; Writing: MS, UW; Review and editing: all authors; Supervision and funding acquisition: EZ, JMH, RAR, OP, DR, SWM, EDL, MS.
Competing Interests: The authors claim no competing interests.
References
- 1.Botstein D, Fink GR. Yeast: An Experimental Organism for 21st Century Biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 3.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 4.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 5.Yofe I, et al. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods. 2016;13:371–378. doi: 10.1038/nmeth.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khmelinskii A, Meurer M, Duishoev N, Delhomme N, Knop M. Seamless gene tagging by endonuclease-driven homologous recombination. PLoS One. 2011;6:e23794. doi: 10.1371/journal.pone.0023794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meurer M, et al. A genome-wide resource for high-throughput genomic tagging of yeast ORFs. bioRxiv. 2017 doi: 10.1101/226811. 226811. [DOI] [Google Scholar]
- 8.Engel SR, Cherry JM. The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database. 2013;2013 doi: 10.1093/database/bat012. bat012-bat012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 10.Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–22. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng. 2012;109:2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- 12.Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picotti P, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg DE, et al. Improved Ribosome-Footprint and mRNA Measurements Provide Insights into Dynamics and Regulation of Yeast Translation. Cell Rep. 2016;14:1787–1799. doi: 10.1016/j.celrep.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science (80-.) 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neymotin B, Athanasiadou R, Gresham D. Determination of in vivo RNA kinetics using RATE-seq. RNA. 2014;20:1645–1652. doi: 10.1261/rna.045104.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Zhang J. The Genomic Landscape of Position Effects on Protein Expression Level and Noise in Yeast. Cell Syst. 2016;2:347–354. doi: 10.1016/j.cels.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weill U, et al. Toolbox: Creating a systematic database of secretory pathway proteins uncovers new cargo for COPI. Traffic. 2018;19:370–379. doi: 10.1111/tra.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgenstern M, et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukasawa Y, et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14:1113–26. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 23.Vögtle F-N, et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–39. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 24.Venne AS, Vögtle F-N, Meisinger C, Sickmann A, Zahedi RP. Novel Highly Sensitive, Specific, and Straightforward Strategy for Comprehensive N-Terminal Proteomics Reveals Unknown Substrates of the Mitochondrial Peptidase Icp55. J Proteome Res. 2013;12:3823–3830. doi: 10.1021/pr400435d. [DOI] [PubMed] [Google Scholar]
- 25.Backes S, et al. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J Cell Biol. 2018 doi: 10.1083/jcb.201708044. jcb.201708044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chacinska A, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–46. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiedemann N, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Menachem R, Pines O. Methods in molecular biology (Clifton, N.J.) 2017;1567:179–195. doi: 10.1007/978-1-4939-6824-4_11. [DOI] [PubMed] [Google Scholar]
- 29.Eisenberg-Bord M, Schuldiner M. Mitochatting – If only we could be a fly on the cell wall. Biochim Biophys Acta - Mol Cell Res. 2017;1864:1469–1480. doi: 10.1016/j.bbamcr.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg-Bord M, Schuldiner M. Ground control to major TOM: mitochondria-nucleus communication. FEBS J. 2017;284:196–210. doi: 10.1111/febs.13778. [DOI] [PubMed] [Google Scholar]
- 31.Jin L, et al. Random insertion of split-cans of the fluorescent protein venus into Shaker channels yields voltage sensitive probes with improved membrane localization in mammalian cells. J Neurosci Methods. 2011;199:1–9. doi: 10.1016/j.jneumeth.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erdmann R. Assembly, maintenance and dynamics of peroxisomes. Biochim Biophys Acta. 2016;1863:787–9. doi: 10.1016/j.bbamcr.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Kim H, Melén K, Osterberg M, von Heijne G. A global topology map of the Saccharomyces cerevisiae membrane proteome. Proc Natl Acad Sci U S A. 2006;103:11142–11147. doi: 10.1073/pnas.0604075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2012;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 35.Douglas AC, et al. Functional Analysis With a Barcoder Yeast Gene Overexpression System. G3: Genes|Genomes|Genetics. 2012;2:1279–1289. doi: 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 37.Yofe I, Schuldiner M. Primers-4-Yeast: a comprehensive web tool for planning primers for Saccharomyces cerevisiae. Yeast. 2014;31:77–80. doi: 10.1002/yea.2998. [DOI] [PubMed] [Google Scholar]
- 38.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 40.Sopko R, et al. Mapping Pathways and Phenotypes by Systematic Gene Overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Hin Yan Tong A, Boone C. High-Throughput Strain Construction and Systematic Synthetic Lethal Screening in Saccharomyces cerevisiae. Methods Mol Biol. 2007;36:1–19. [Google Scholar]
- 42.Cohen Y, Schuldiner M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol. 2011;781:127–59. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- 43.Knox C, Sass E, Neupert W, Pines O. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J Biol Chem. 1998;273:25587–93. doi: 10.1074/jbc.273.40.25587. [DOI] [PubMed] [Google Scholar]
- 44.Weckbecker D, Longen S, Riemer J, Herrmann JM. Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 2012;31:4348–58. doi: 10.1038/emboj.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption inSaccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 46.Flagfeldt DB, Siewers V, Huang L, Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26:545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- 47.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 49.Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 50.Knoblach B, et al. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 2013;32:2439–2453. doi: 10.1038/emboj.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. Cohen F, editor. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 52.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–50. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 53.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reynolds SM, Käll L, Riffle ME, Bilmes JA, Noble WS. Transmembrane Topology and Signal Peptide Prediction Using Dynamic Bayesian Networks. PLoS Comput Biol. 2008;4:e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–8. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.