Abstract
The lung is the entry site for Bacillus anthracis in inhalation anthrax, the most deadly form of the disease. Spores must escape through the alveolar epithelial cell (AEC) barrier and migrate to regional lymph nodes, germinate and enter the circulatory system to cause disease. Several mechanisms to explain alveolar escape have been postulated, and all these tacitly involve the AEC barrier. In this study, we incorporate our primary human type I AEC model, microarray and gene enrichment analysis, qRT-PCR, multiplex ELISA, and neutrophil and monocyte chemotaxis assays to study the response of AEC to B. anthracis, (Sterne) spores at 4 and 24 hours post-exposure. Spore exposure altered gene expression in AEC after 4 and 24 hours and differentially expressed genes (±1.3 fold, p ≤ 0.05) included CCL4/MIP-1β (4 hours), CXCL8/IL-8 (4 and 24 hours) and CXCL5/ENA-78 (24 hours). Gene enrichment analysis revealed that pathways involving cytokine or chemokine activity, receptor binding, and innate immune responses to infection were prominent. Microarray results were confirmed by qRT-PCR and multiplex ELISA assays. Chemotaxis assays demonstrated that spores induced the release of biologically active neutrophil and monocyte chemokines, and that CXCL8/IL-8 was the major neutrophil chemokine. The small or sub-chemotactic doses of CXCL5/ENA-78, CXCL2/GROβ and CCL20/MIP-3α may contribute to chemotaxis by priming effects. These data provide the first whole transcriptomic description of the human type I AEC initial response to B. anthracis spore exposure. Taken together, our findings contribute to an increased understanding of the role of AEC in the pathogenesis of inhalational anthrax.
Keywords: Bacillus anthracis, anthrax, type I alveolar epithelial cells, transcriptome microarray analysis, chemotaxis
1. Introduction
Anthrax, a virulent and zoonotic disease recognized since early human history, is caused by Bacillus anthracis, a Gram-positive, aerobic, spore-forming, rod-shaped bacterium. Anthrax disease manifests in three primary forms resulting from three different mechanisms of exposure to spores: Ingestion (gastrointestinal), skin contact (cutaneous), or inhalation (inhalational or pneumonic) [1]. Although all forms of disease can lead to fatal systemic infection, inhalation anthrax is the most life-threatening form, with significant mortality even with treatment, and nearly 100% mortality in the absence of early treatment [1, 2]
The lung is the site of entry for B. anthracis spores in inhalation anthrax, however, the organism does not cause primary disease in the lung [3–5]. Following exposure to spores, there is a significant delay, as long as 43 days, before clinical disease. This implies that there is temporary containment of the pathogen, which is likely mediated by the innate immune system [2]. In order for disease to occur, spores must escape the alveolus and then pass through the lymphatic system to the mediastinal lymph nodes. Vegetative bacteria subsequently disseminate via lymphatic ducts with subsequent appearance in the bloodstream [1, 6, 7]. Thus, a critical step in disease pathogenesis is alveolar escape: overcoming local containment of the pathogen in the alveolus and the traversing the epithelial barrier.
Alveolar escape of the pathogen has been a focus of many studies, and has resulted in the formulation of several proposed mechanisms for how this occurs. Four mechanisms for alveolar escape have been postulated, and two of these pathways involve a lung carrier cell. It should be noted that AEC are stationary and not considered carrier cells, yet they are tacit players in all four pathways because they form the alveolar epithelial barrier through which the spores must pass.
The initial two proposed mechanisms of alveolar escape incorporate a migratory or “carrier” cell, often referred to as a “Trojan horse”, for spore uptake and subsequent dissemination. First, macrophages were proposed as the Trojan horse, because alveolar macrophages (AM) ingest spores and because studies suggested that B. anthracis virulence toxins damage or impair these cells [7, 8]. However, other studies have shown spore dissemination from the lung in macrophage-depleted mice [4], and we have shown that primary human AM are not susceptible to toxins [9], indicating that macrophages may not play a key role in alveolar escape, assuming that the Trojan horse is sensitive to toxins. This work and the work of others has suggested that dendritic cells (DC) may be the Trojan horse. DC and other cells containing spores were observed in the thoracic lymph nodes of mice [3, 10]. DC sample the airway luminal surface [11]. Human lung DC internalize B. anthracis spores [12].
A third possibility is that AEC alone are involved in alveolar escape and that a Trojan horse is not required. Isolated cell culture models suggest a transcellular route of spore passage through the lung epithelium [13]. Using the human lung epithelial cell line A549 and primary human small airway cells (SAEC), both of which internalize spores, Russell et al. [14] showed that spores survived and were translocated from the apical to basolateral side of the cell. Spore internalization in A549 cells involves interaction of the spore BclA protein with cell integrin α2β1 and complement C1q [15]. These results suggest that spores may cross the alveolar epithelial barrier and disseminate without the assistance of migratory cells [14]. Finally, in the “Jailbreak” model, it is proposed that the clustering and germination of spores within the alveolus followed by production of various B. anthracis virulence toxins causes epithelial damage which permits free spore passage not involving a Trojan horse [16].
All of these theories demonstrate that the interaction of spores with alveolar epithelium is important in disease pathogenesis. This interaction would occur between the pathogen and one of two alveolar epithelial cell types that line the alveolus. These cell types are Type I alveolar epithelial cells (AEC I) and type II AEC (AEC II). AEC II cells are classically described as the precursors of AEC I though there may be a common precursor to type I and type II AEC [17]. AEC II are cuboidal in shape, produce surfactant, and line approximately 5% of the surface area of the alveoli. AEC I have flattened 45 to 60 nm thick cytoplasm, are terminally differentiated, and cover the remaining 95% of the alveolar epithelial surface [18]. An AEC I-like cell can be derived from human lungs by specialized isolation and culture techniques facilitating investigation of the role of these cells in anthrax pathogenesis [19].
We used an unbiased "top-down" transcriptomic approach to evaluate the transcriptional response to the initial interaction of B. anthracis spores with AEC I-like cells. There were no prerequisites or biases towards particular functions or pathways. This differs from kits and platforms that assess individual processes (e.g., apoptosis or inflammation) based on previous knowledge, and therefore, introduce preexisting bias by exclusion of other genes. The method of microarray data analysis we employed used not only statistical comparisons, but also considered biological properties of the data [20] in order to find unbiased, but reliable, results. The approach us to identify stable differentially expressed genes, i.e. genes whose expression was consistently low among microarrays in one case and consistently high in another. This increased the specificity of analysis and allowed us to identify the most robust "beacons" driving response to the pathogen. The results provided us with information about the major processes affected. Examination of the whole dataset based on this knowledge identified additional highly dynamic co-players that were selected by strict statistical criteria.
Our earlier studies examined the interaction of anthrax spores with intact lung epithelia in a human lung slice model. We studied the initial events up to 48 hours after exposing intact lung tissue slices to B. anthracis spores. Spore exposure caused transcriptional activation of cytokines and chemokine genes and the data was confirmed at the level of translation. Immuno-staining for IL-6 and IL-8 in spore-exposed lung slices revealed that alveolar epithelial cells and macrophages and a few interstitial cells were the source of the cytokines and chemokines. [21].
In the current study, we infected human type I-like AEC with B. anthracis (Sterne) spores and performed transcriptome analysis to provide a comprehensive view of the innate immune response of the type I-like AEC to the pathogen. In spore-exposed AEC, cytokine and chemokine transcripts were more highly expressed compared to mock-infected cells. We also found that pathways involved in these processes were identified by functional and pathway enrichment analysis. We then confirmed this response at the transcriptional and translational level, and assessed the relative contribution of select chemokines on monocyte and neutrophil chemotaxis.
2. Materials and Methods
2.1. Isolation of primary human type I alveolar epithelial cells
The isolation and culture of human AEC, type II, have been described previously [19]. Briefly, three human lungs rejected for transplant were obtained from the International Institute for the Advancement of Medicine (IIAM; Jessup, PA, USA) under a protocol approved by the University of Oklahoma Health Sciences Institutional Review Board. Following perfusion of 1 lobe with PBS without Ca2+ or Mg2+ and lavage with physiological (150 mM) NaCl containing 100 Kunitz units (KU) per mg protein of DNase from bovine pancreas (Sigma-Aldrich Co., St. Louis, MO, USA), 57 g tissue was digested for 60 min at 37°C in a sealed bag containing 200 ml HBSS (without Ca2+ or Mg2+, Corning Life Sciences/Mediatech, Inc., Manassas, VA, USA) with 8 U/ml porcine elastase (Worthington Biochemical Corp., Lakewood, NJ, USA) and 150 KU/ml DNase. After inactivation of enzyme by addition of 120 ml fetal bovine serum (FBS) (HyClone, GE Healthcare Life Sciences, Piscataway, NJ, USA) and 0.42% (wt/vol) DNase, the softened tissue was minced into a slurry and passed sequentially through gauze and increasingly smaller pore size nylon mesh until a single cell dispersion was obtained.
AEC II were purified from the mixture by 3 enrichment protocols: 1) Total cells were concentrated into DMEM + 150 g/ml DNase, then panned for 60 min at 37°C in flasks coated with 19.4 µg/cm2 human IgG (Sigma-Aldrich Co., St. Louis, MO, USA); 2) Unattached cells were concentrated, then layered onto a ρ 1.040/1.089 Percoll gradient (GE Healthcare Life Sciences, Piscataway, NJ, USA); and, 3) Cells in the interface band were negatively selected against α-CD45 coated magnetic microbeads (Thermo Fisher /Invitrogen/Dynal, Waltham, MA, USA). The average yield was 80 × 106 live, single cells with 93 % viability. AEC II identity was confirmed by a modified Papanicolaou stain using Harris hematoxylin that colored lamellar bodies blue.
2.2. Culture of primary human type I alveolar epithelial cells
Isolated AEC II were added to 6-well BioCoat plates pre-coated with collagen I extracellular matrix and overlaid with 5 µg/cm2 human fibronectin (both Corning Life Sciences., Corning, NY, USA) in DMEM supplemented with 10% FBS, 1% solution of penicillin/streptomycin/amphotericin B (Corning/Mediatech, Corning, NY, USA), 5.0 µg/liter vancomycin, and 10.0 µg/liter ceftazidime (both from Sigma-Aldrich Co., St. Louis, MO, USA) at a concentration of 20,000 cells/cm2. Cell density was optimized to drive differentiation toward an AEC, type I-like, phenotype. Freshly cultured cells were used for all experiments.
By day 10 of culture, plated AEC, type II, had morphologically differentiated into confluent AEC, type I-like, monolayers. The confirmation of this change has been described previously, as has the lack of contaminating cells such as fibroblasts and endothelial cells in the cultures [19]. Briefly, test wells were fluorescently stained for the presence of purinergic receptor P2X7, an AEC I, phenotypic marker, and the presence of ZO-1 (zona occluens-1), the tight junction protein that forms between AEC I pneumocytes as the monolayer matures. The following antibodies were used: Rabbit α-human P2X7-FITC IgG (Sigma-Aldrich Co., St. Louis, MO, USA), mouse α-human ZO-1 IgG, and Alexa Fluor 555-conjugated donkey α-mouse IgG (both from Thermo Fisher /Invitrogen, Waltham, MA, USA).
2.3. Preparation of B. anthracis spores
B. anthracis, Sterne strain 7702 (pX01+, pX02−) spore stocks were prepared as previously described [21]. Briefly, bacteria were grown with continuous shaking in Luria–Bertani broth medium overnight at 37°C, then the suspension was spread on AK sporulation agar dishes and incubated for three weeks at 30°C. At time of harvest, each dish was washed by pipetting with chilled, sterile, deionized water to dislodge spores. The pooled collections were spun at 10,000 × g for 10 min., resuspended in 10 ml of chilled, sterile, deionized water, and the suspensions were heated at 65°C for 60 min to kill any vegetative bacteria. Next, the spores were washed a total of 5 times by centrifugation and resuspension, as above, to remove contaminating cell debris. The viable spore titer (cfu/ml) of the preparations was determined by spread plate counts. There was no detectable endotoxin in the final spore dilutions used in the experiments as determined by limulus amebocyte lysate assay (Lonza Group, Basel, Switzerland). The spores were diluted to 1 × 109 spores/ml and stored at 4°C, and the spore preparations were used in experiments before 30 days post-harvest.
2.4. Infection of AEC with B. anthracis spores and sample collection
After differentiation in 6-well plates, the type I AEC from the three donors were infected with 1 × 106 cfu/ml (MOI ≈ 2) B. anthracis (Sterne) endospores in fresh DMEM-10 and incubated for 4 and 24 h at 37°C in 5% CO2. DMEM-10 lacking phenol red pH indicator was used for chemotaxis assays. A volume of spore diluent (sterile di-water) equal to that of the spores was used as a negative control, and a mixture of phorbol 12-myristate 13-acetate (PMA) (100 ng/ml) and lipopolysaccharide (LPS) (1 µg/ml) served as a positive control. At the end of incubation, medium supernatants were collected and stored at −80°C for subsequent use in multiplex immunoassay of human cytokines and chemokines or in real-time chemotaxis assays. The cell monolayers to be used for microarray analysis of the RNA were washed twice with 2 ml PBS, lysed by addition of 700 µl/well QIAzol Lysis Reagent (Qiagen Inc., Germantown, MD, USA), and immediately stored at −80°C.
2.5. RNA extraction and purification
The QIAzol cell lysates were bead mill homogenized in a Qiagen TissueLyser II, then purified by the QIAcube automated workstation using miRNeasy Mini Purification Kits (all from Qiagen, Inc.). The resulting 50 µl of total RNA was quantitated with a NanoDrop 2000 spectrophotometer, averaging 7.0 µg/monolayer. Quality control for several randomly chosen RNA samples from each of the 3 lung preps was provided by the 2100 Electrophoresis Bioanalyzer (Agilent Technologies, Santa Clara, CA). Finally, to avoid batch effects, samples were randomized into 3 different amplification groups. Each group contained 2 additional samples, a well-characterized positive control made from the blood of 3 human volunteers by the PAXgene Blood miRNA Kit (Qiagen) on the QIAcube system, and a corresponding negative control having 20 pg/µl RNA.
2.6. cDNA preparation and labeling
The initial total used was 50 ng. Aliquots of purified RNA were adjusted to 20 ng/µl. Primer annealing, first-strand synthesis, and second-strand synthesis steps were accomplished with the NuGEN Ovation® Pico WTA V2 System (NuGEN, San Carlos, CA). The products were purified by the Agencourt RNAClean XP bead protocol (contained within the NuGen Ovation® kit), followed immediately by amplification using Ribo-SPIA® (Single Primer Isothermal Amplification) methodology from reagents also within the NuGEN Ovation kit. SPIA yield was determined by NanoDrop 2000 measurement. Up to this point, the amplification groups had been processed separately, however, fragmentation and biotinylation of all samples was performed in a single batch with the NuGen Encore® Biotin Module. Efficacy of fragmentation was tested with several random samples on the Agilent Bioanalyzer before labeling.
2.7. Microarray chip hybridization and scanning
The samples were randomized into hybridization groups, each containing members from all 3 previous amplification groups and each processed as a unit. The GeneChip Hybridization, Wash & Stain Kit (Affymetrix, Thermo Fisher Scientific Inc.) contained all the necessary reagents. The Hybridization Module of the kit supplied controls and master mix that was combined with the fragmented, labeled ssDNA, and added to pre-warmed Affymetrix GeneChip HuGene 2.0 ST Array, Format 100, chips for hybridization overnight. The Stain Module of the kit provided high-stringency wash solutions to be used in the Affymetrix GeneChip Fluidics Station 450. After washing, the array chips were scanned on the Affymetrix GeneChip Scanner 7G.
2.8. Microarray data analysis
The data were normalized using RMA [PMID: 12925520]. Probes were annotated on a transcript level using hugene20sttranscriptcluster v. 8.4.0 Bioconductor package. Quality control included hierarchical clustering, principal components analysis. Only protein-coding genes were analyzed. Differentially expressed genes were detected using limma R package v. 3.28.21 [PMID: 25605792] controlling for the “Amp group” variability. Functional enrichment analysis (Gene Ontology Molecular Function, Biological Process, Cellular Component pathways) was performed using GOStats R package v. 2.42.0 [PMID: 17098774]. Volcano plots and heatmap visualization was performed in R/Bioconductor environment v. 3.3.1. The microarray data presented in this article were submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and were assigned accession number GSE102106 for the series.
2.9. Real-time qPCR validation of cytokine/chemokine gene upregulation
Four and 24 hour SPIA generated cDNA samples from the AEC of three lung donors were diluted 10× in nuclease-free H2O and amplified on a CFX96 Real-Time System thermal cycler using CFX Manager software, Prime PCR run type, and iQ SYBR Green Supermix chemistry (all from Bio-Rad Life Science Research, Hercules, CA, USA). Bio-Rad primers were selected for those cytokine/chemokine genes revealed to be most differentially expressed by microarray: CXCL8/IL-8, CXCL5/ENA-78, CXCL2/GROβ, CCL20/MIP-3α, CCL2/MCP-1, and CCL4/MIP-1β. The reference gene was actin/ACTB. The cycling protocol used was vendor recommended for the Supermix. Results were expressed as 2ΔΔCT fold increase in gene expression of spore-treatment normalized with actin and the no-treatment samples.
2.10. Cytokine and Chemokine ELISA and multiplex immunoassay
Chemokine and cytokine protein levels in the AEC media supernatants were determined by multiplex immunoassay using a Bio-Plex 100 multiplex system with Bio-Plex Manager and Bio-Plex Data Pro software (Bio-Rad, Hercules, CA, USA). The chemokines and cytokines examined by multiplex assay include: IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, CXCL8/IL-8, IL-9, IL-10, IL-12 p70, IL-13, IL-15, IL-17, IL-23, Basic FGF, Eotaxin, G-CSF, GM-CSF, CXCL2/Gro-β, IFN-γ, IP-10, CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CCL20/MIP-3α, PDGF-BB, RANTES, TNF-α, and VEGF. Sandwich ELISA of CXCL5/ENA-78 was performed with a commercially available ELISA kit (R&D Systems, Minneapolis, MN, USA).
2.11. Monocyte and neutrophil isolation and fluorescent staining
Heparinized whole blood was obtained by venipuncture from healthy donors, and the peripheral blood monocytes and neutrophils were separated by density gradient centrifugation over Polymorphprep (Axis-Shield, Oslo, Norway), as we described previously [21]. To isolate monocytes, the upper, mononuclear cell layers were pooled, washed in in PBS + 0.1% FBS, and depleted of CD3, CD7, CD16 (CD16a and CD16b), CD19, CD56, CDw123 and CD235a (Glycophorin A)-positive cells by incubation with specific biotinylated antibodies and streptavidin-magnetic particle beads, and the monocytes were isolated from a series of washes using a magnetic particle concentrator according to the manufacturer’s recommendations (ThermoFisher/Invitrogen/Dynal, Waltham, MA, USA). The viability of the isolated monocytes was 98%, as determined by trypan blue exclusion, and the purity was greater than 90%, based on morphology as determined using preparations stained with Diff-Quick stain reagents.
To purify neutrophils, the lower, polymorphonuclear cell layers from the density gradient centrifugation were pooled, mixed with an equal volume of sterile, 0.45% NaCl solution, further diluted in 1.5 volumes with sterile PBS, centrifuged at 400 × g for 10 min., and then washed once in sterile, PBS + 0.1% FBS. To remove residual red blood cells the neutrophil cell pellet was suspended in 2 ml sterile, PBS + 0.1% FBS, followed by addition of 6 ml sterile, 4°C, de-ionized water. After incubation for 60 to 80 seconds at room temperature, 2 ml of sterile 5× PBS was mixed with the cells, this cell suspension was further diluted in 4 volumes sterile PBS + 0.1% FBS, and the cells were concentrated by centrifugation. The neutrophil viability was 95% based on trypan blue exclusion and the purity was greater than 95% based on morphology from Diff-Quick staining.
To label the cells with fluorescent dye, monocytes and neutrophils were resuspended at a concentration of 2 × 107 cells/ml in PBS + 0.1% FBS and incubated in 5 µM carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE, ThermoFisher, Waltham, MA, USA) for 10 min at room temperature. The cells were washed once with 25 ml PBS + 2% FBS, then washed twice in 25 ml PBS + 0.1% FBS to remove unbound dye label. The cells were then resuspended to a concentration of 1 × 107 cells/ml in either unamended Gey’s salt solution for monocytes, or Gey’s salt solution plus 2% FBS for neutrophils. After labeling, the cells were > 95% viable as determined by trypan blue exclusion. Purified, labeled monocytes and neutrophils were used in chemotaxis assays immediately after the labeling procedure.
2.12. Monocyte and neutrophil real-time chemotaxis assays
Culture medium supernatants from three independent experiments with AEC exposed for 24 h to 1 × 106 B. anthracis spores or the mock-treated media from the same experiments were used to assess monocyte and neutrophil chemotaxis. A portion of the medium samples was preincubated overnight at 4°C with chemokine-specific, neutralizing antibodies (R&D Systems, Minneapolis, MN, USA). The amount of antibody needed to completely neutralize the chemokine present was based on the concentrations of chemokine proteins measured in AEC supernatants and antibody neutralization curves provided by the manufacturer. The following neutralizing antibodies were used to assess neutrophil chemotaxis: anti-CXCL8/IL-8, anti-CXCL5/ENA-78, and anti-CCL20/MIP-3α goat polyclonal antibodies, and anti-CXCL1/2/3/GRO-pan-specific mouse monoclonal antibody (Clone # 31716). The following neutralizing antibodies were used to assess monocyte chemotaxis: anti-CCL2/MCP-1, anti-CCL4/MIP-1β, and anti-CXCL10/IP-10 goat polyclonal antibodies. To assess the effect of antibody alone on cell migration, non-specific control antibodies, normal, non-immunized (naïve) goat IgG or mouse IgG1 kappa (Clone # 11711, both from R&D Systems), were prepared in fresh DMEM-10 at concentrations equivalent to the neutralizing antibodies. Preparations of recombinant chemokine proteins at concentrations equal to those measured in AEC supernatants with and without neutralizing antibody, and neutralizing antibody alone diluted in naïve DMEM-10 growth medium served as controls. Unamended DMEM-10 provided the assay background activity.
Chemotaxis was assayed using Corning Falcon 96-well HTS FluoroBlok™ multiwell plates with 3-µm pores (ThermoFisher, Waltham, MA, USA) by adding 5 × 105 purified, CFDA-SE-labeled cells (50 µl) to the upper insert chambers and 225 µl of the control or experimental media to the lower chambers. Each sample was assayed in duplicate. Immediately after the addition of cells and medium, the plates were placed in a Synergy HT fluorescent plate reader (Bio-Tek Instruments, Winooski, VT, USA) pre-heated to 37°C. The kinetics of fluorescence cell migration through the membrane was measured at 2 min intervals for 60 min using filter settings with an excitation wavelength of 440/40 nm and an emission wavelength of 508/20 nm. Slope analysis within the early, linear portion of the migration plot (fluorescent Intensity over time) was used to assess the rate of migration. Chemotactic activity was expressed as the adjusted fold change and was calculated as the fold increase in the migration rate of the experimental treatments (Exp) adjusted by subtraction of the unamended background rate (Bkg) versus the migration rate of the negative control (Neg), plus 1.
2.13. Statistics
Where applicable, the data are expressed as the means and standard deviation of the means (SD). All statistical analyses, except for those involving cDNA microarray data, were performed using Prism 6.0 software (GraphPad Software, La Jolla, CA, USA). For qRT-PCR, significance was determined by Student’s t-Test. For cytokine/chemokine protein multiplex ELISA and chemotaxis, statistical significance was determined by one-way ANOVA with the Tukey post hoc correction for multiple comparisons. In all cases, a P-value of ≤ 0.05 was considered significant [22, 23].
3. Results
3.1. B. anthracis spore exposure alters gene expression in primary human AEC I-like cells
Initial interactions of the B. anthracis spore with the alveolar epithelia are likely important in the pathogenesis of inhalational anthrax, the disease caused by this agent. We, therefore, sought to take an unbiased approach to examine the human AEC response to B. anthracis (Sterne) spores. Primary human AEC were purified from three normal human donor lungs, seeded on collagen and fibronectin-coated tissue plates at 25,000 cells/cm2, and grown for 10–14 days until confluence [19]. Culturing under these conditions resulted in an AEC I-like morphology. Primary AEC I cell cultures were exposed to 1 × 106 cfu/ml of B. anthracis spores for 4 and 24 hours, with cells exposed to an equal volume of spore diluent acting as a negative control. RNA was then extracted and converted to cDNA for analysis of the transcriptome response to spore exposure.
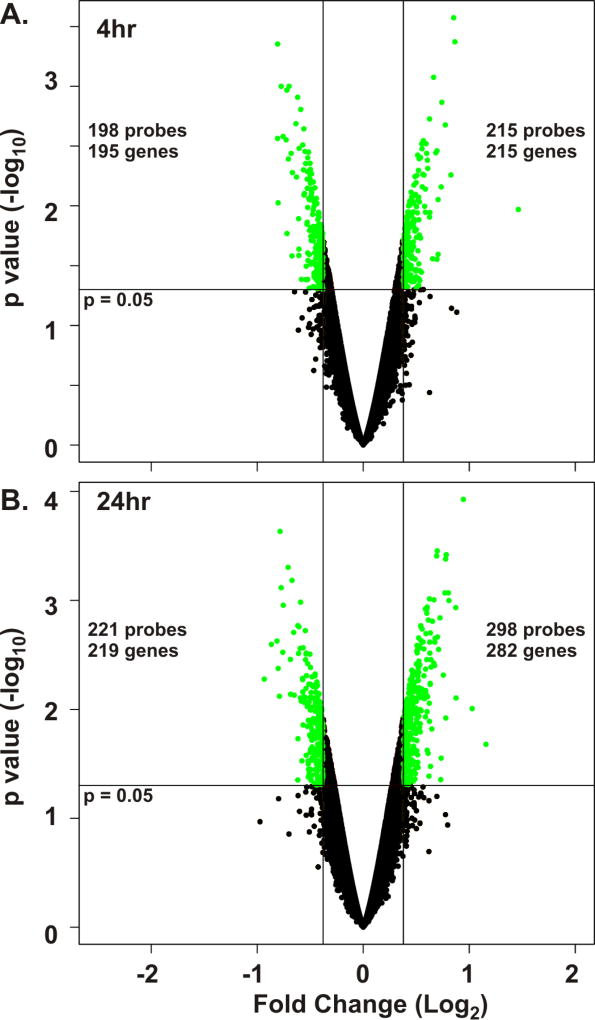
We hypothesized that only small changes in gene expression would reflect whole lung epithelial responses to spore contact. Therefore, we chose a 30% difference in gene expression (± 1.3 fold, base 10) as a cutoff to detect biological changes in the AEC response to spores. A volcano plot was created for both the 4 and 24 hour spore exposure experiments (Figure 1). The 4 hour plot demonstrated that there were 413 differentially expressed probesets that represented a total of 410 unique genes (215 upregulated by spore exposure and 195 downregulated by spore exposure, Figure 1A). Using the same analysis, the 24 hour plot demonstrated that there were 519 differentially expressed probesets at this time point representing a total of 501 unique genes (282 upregulated by spore exposure and 219 downregulated by spore exposure, Figure 1B).
Figure 1.
B. anthracis spore exposure alters gene expression in primary human type I-like AEC. cDNA prepared from type I-like AEC cells (n = 3 donors) exposed to 1 × 106 cfu/ml of B. anthracis spores or mock-exposed for 4 and 24 hours was used for whole-transcriptome microarray analysis using Affymetrix GeneChip HuGene 2.0 ST Array, Format 100 chips. Volcano plots of mock versus spore-exposed AEC cells for 4 hours (A) or 24 hours (B) using all probesets were generated. Differentially expressed genes were identified using the criteria of ≥ 1.3 or ≤ −1.3 (log 10) fold change from mock and an unadjusted p < 0.05 (green circles). Genes not meeting these criteria are shown as black circles. The 4 hour data (A) demonstrate a total of 413 spore-dysregulated probesets representing 410 unique genes, and the 24 hour data (B) demonstrate a total of 519 spore-dysregulated probesets representing 501 unique genes.
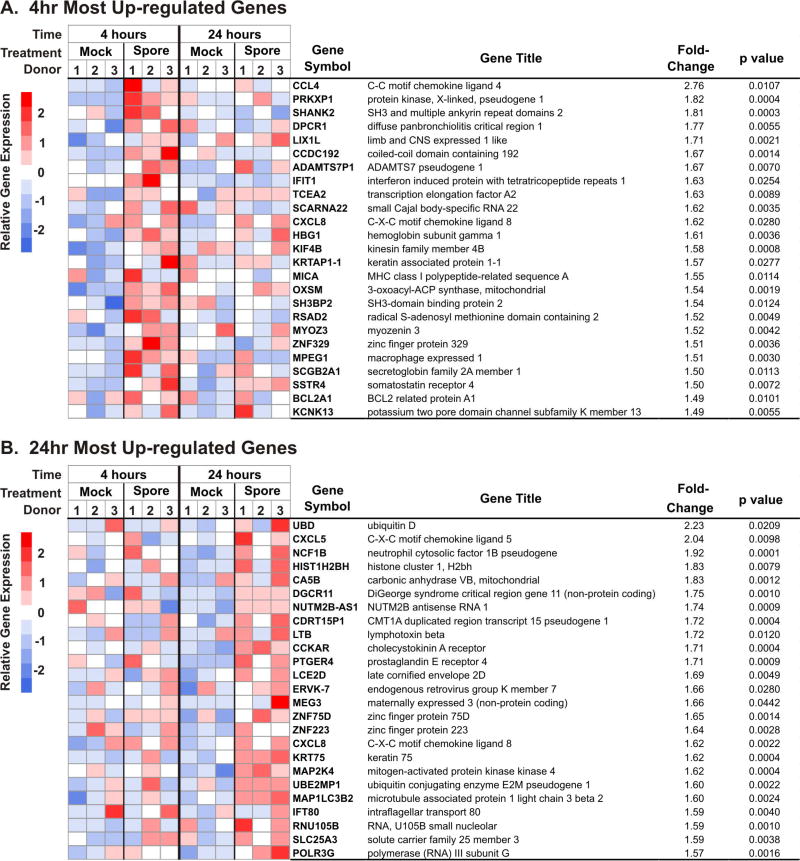
3.2. Characterization of the genes most upregulated by spores
To examine the potential implications of the effects of spore exposure on AEC I-like cells we reviewed the 25 genes most upregulated by spores at 4 and 24 hours. To facilitate analysis of the differentially expressed genes (DEGs), annotated heatmaps were generated for these genes (Figure 2). With the exception of CXCL8/IL-8, none of the most induced genes at 4 hours were represented in the 24 hour set. Also notable is that most of the genes upregulated by 4 hours of spore exposure (depicted in red hues) were either not upregulated (depicted in white) or down regulated (depicted in blue hues) after 24 hours of exposure (Figure 2A). This was also the case for the most upregulated genes at 24 hours, as the majority of these were either not upregulated or were down regulated at 4 hours of exposure (Figure 2B).
Figure 2.
Chemokine genes are among the 25 most upregulated genes in B. anthracis spore-exposed type I-like AEC at 4 and 24 hours. Depicted are heatmaps (left panel) and annotations (right panel) of the 25 genes most upregulated by spores after 4 hours (A) and 24 hours (B) of exposure. For comparison purposes, heat map expression information is shown at both 4 and 24 hours for each of the 25 genes selected at both times of exposure. The annotation list for each heat map row shows the gene symbol, gene title, average fold change (spore over mock, expressed in base 10, n = 3) and the unadjusted p value for these genes.
At 24 hours of exposure, we noted that 4 out of the top 25 most induced genes were related to chemokines or chemokine induction (Figure 2B). CXCL5/ENA-78, a classic neutrophil chemokine, is the second most upregulated gene at 24 hours. As at 4 hours, CXCL8/IL-8 was also included in the group of most upregulated genes. Lymphotoxin β (LTB), a membrane protein of the TNF family, is also highly expressed at 24 hours, as is mitogen-activated protein kinase kinase 4 (MAP2K4), a serine/threonine kinase signaling pathway protein important in chemokine induction.
3.3 Pathway activation by spore exposure
To elucidate the processes activated by spore exposure, gene ontology (GO) functional and pathway enrichment analysis was performed on the significant DEG’s identified in the volcano plots (Tables I and II). GO molecular function analysis, which describes molecular level activities, of the 4-hour DEGs revealed 15 significant processes (p<0.05), and of these, 4 of the 7 most significant pathways involved cytokine or chemokine activity or receptor binding (Table I). Significant, upregulated molecular function pathways also included G-protein coupled receptor binding, signaling adaptor activity, and Ras guanyl-nucleotide exchange factor activity which are functional effectors of chemokine receptors. [24–26].
Table I.
Predominantly Enriched Gene Ontology Molecular Functions and Biological Processes from Upregulated Differentially Expressed Genes in Human Type I-like Alveolar Epithelial Cells after 4 hour Exposure to Bacillus anthracis spores.
| GO Domain* |
GO Term | Category Description | Gene Count# |
p value |
|---|---|---|---|---|
| MF | 0008009 | Chemokine activity | 5 | 0.0001 |
| MF | 0042379 | Chemokine receptor binding | 5 | 0.0004 |
| MF | 0005344 | Oxygen transporter activity | 2 | 0.0096 |
| MF | 0005126 | Cytokine receptor binding | 7 | 0.0119 |
| MF | 0001664 | G-protein coupled receptor binding | 7 | 0.0121 |
| MF | 0004222 | Metalloendopeptidase activity | 4 | 0.0276 |
| MF | 0005125 | Cytokine activity | 6 | 0.0279 |
| MF | 0008066 | Glutamate receptor activity | 2 | 0.0339 |
| MF | 0035591 | Signaling adaptor activity | 3 | 0.0355 |
| MF | 0035254 | Glutamate receptor binding | 2 | 0.0386 |
| MF | 0019894 | Kinesin binding | 2 | 0.0386 |
| MF | 0005267 | Potassium channel activity | 4 | 0.0386 |
| MF | 0008528 | G-protein coupled peptide receptor activity | 4 | 0.0417 |
| MF | 0001653 | Peptide receptor activity | 4 | 0.0439 |
| MF | 0005088 | Ras guanyl-nucleotide exchange factor activity | 4 | 0.0461 |
|
| ||||
| BP | 0051703 | Intraspecies interaction between organisms | 4 | 0.0008 |
| BP | 0035176 | Social behavior | 4 | 0.0008 |
| BP | 0009615 | Response to virus | 9 | 0.0016 |
| BP | 0071107 | Response to parathyroid hormone | 2 | 0.0031 |
| BP | 0051607 | Defense response to virus | 6 | 0.0052 |
| BP | 0071625 | Vocalization behavior | 2 | 0.0083 |
| BP | 0051705 | Multi-organism behavior | 4 | 0.0095 |
| BP | 0034123 | Positive regulation of toll-like receptor signaling pathway | 2 | 0.0096 |
| BP | 0072350 | Tricarboxylic acid metabolic process | 2 | 0.0096 |
| BP | 0051707 | Response to other organism | 11 | 0.0116 |
| BP | 0015671 | Oxygen transport | 2 | 0.0125 |
| BP | 0071345 | Cellular response to cytokine stimulus | 11 | 0.0133 |
| BP | 0033189 | Response to vitamin A | 2 | 0.0141 |
| BP | 0071356 | Cellular response to tumor necrosis factor | 4 | 0.0156 |
| BP | 0002011 | Morphogenesis of an epithelial sheet | 2 | 0.0157 |
| BP | 0015669 | Gas transport | 2 | 0.0192 |
| BP | 0034765 | Regulation of ion transmembrane transport | 8 | 0.0206 |
| BP | 0071773 | Cellular response to BMP stimulus | 2 | 0.0231 |
| BP | 0071772 | Response to BMP | 2 | 0.0231 |
| BP | 0045069 | Regulation of viral genome replication | 3 | 0.0234 |
| BP | 0006813 | Potassium ion transport | 5 | 0.0238 |
| BP | 0042330 | Taxis | 7 | 0.0240 |
| BP | 0006935 | Chemotaxis | 7 | 0.0240 |
| BP | 0034762 | Regulation of transmembrane transport | 8 | 0.0255 |
The top 25 terms in each GO domain were selected according to p < 0.05.
GO, gene ontology.
MF, molecular function.
BP, biological process.
Gene Count is the number of genes enriched in this term.
Table II.
Predominantly Enriched Gene Ontology Molecular Functions and Biological Processes from Upregulated Differentially Expressed Genes in Human Type I-like Alveolar Epithelial Cells after 24 hour Exposure to Bacillus anthracis spores.
| GO Domain* |
GO Term | Category Description | Gene Count# |
p value |
|---|---|---|---|---|
| MF | 0005125 | Cytokine activity | 9 | 0.0035 |
| MF | 0008009 | Chemokine activity | 4 | 0.0044 |
| MF | 0005126 | Cytokine receptor binding | 9 | 0.0055 |
| MF | 0015280 | Ligand-gated sodium channel activity | 2 | 0.0084 |
| MF | 0042379 | Chemokine receptor binding | 4 | 0.0087 |
| MF | 0032395 | MHC class II receptor activity | 2 | 0.0102 |
| MF | 0005355 | Glucose transmembrane transporter activity | 2 | 0.0121 |
| MF | 0015149 | Hexose transmembrane transporter activity | 2 | 0.0141 |
| MF | 0005272 | Sodium channel activity | 3 | 0.0154 |
| MF | 0001664 | G-protein coupled receptor binding | 8 | 0.0169 |
| MF | 0015145 | Monosaccharide transmembrane transporter activity | 2 | 0.0187 |
| MF | 0001848 | Complement binding | 2 | 0.0238 |
| MF | 0005248 | Voltage-gated sodium channel activity | 2 | 0.0353 |
| MF | 0051119 | Sugar transmembrane transporter activity | 2 | 0.0385 |
|
| ||||
| BP | 0045089 | Positive regulation of innate immune response | 11 | 0.0001 |
| BP | 0031349 | Positive regulation of defense response | 13 | 0.0001 |
| BP | 0002221 | Pattern recognition receptor signaling pathway | 9 | 0.0002 |
| BP | 0002758 | Innate immune response-activating signal transduction | 9 | 0.0002 |
| BP | 0045088 | Regulation of innate immune response | 12 | 0.0003 |
| BP | 0002218 | Activation of innate immune response | 9 | 0.0003 |
| BP | 0006954 | Inflammatory response | 14 | 0.0010 |
| BP | 0007254 | JNK cascade | 5 | 0.0011 |
| BP | 0032648 | Regulation of interferon-beta production | 4 | 0.0015 |
| BP | 0034142 | Toll-like receptor 4 signaling pathway | 6 | 0.0026 |
| BP | 0032655 | Regulation of interleukin-12 production | 4 | 0.0038 |
| BP | 0002757 | Immune response-activating signal transduction | 14 | 0.0043 |
| BP | 0032479 | Regulation of type I interferon production | 6 | 0.0044 |
| BP | 0032728 | Positive regulation of interferon-beta production | 3 | 0.0052 |
| BP | 0010533 | Regulation of activation of Janus kinase activity | 2 | 0.0053 |
| BP | 0000910 | Cytokinesis | 5 | 0.0053 |
| BP | 0002676 | Regulation of chronic inflammatory response | 2 | 0.0068 |
| BP | 0035871 | Protein K11-linked deubiquitination | 2 | 0.0068 |
| BP | 0045075 | Regulation of interleukin-12 biosynthetic process | 2 | 0.0068 |
| BP | 0051451 | Myoblast migration | 2 | 0.0068 |
| BP | 0002274 | Myeloid leukocyte activation | 5 | 0.0076 |
| BP | 0030261 | Chromosome condensation | 3 | 0.0079 |
| BP | 0002224 | Toll-like receptor signaling pathway | 6 | 0.0080 |
| BP | 0002237 | Response to molecule of bacterial origin | 9 | 0.0081 |
| BP | 0032735 | Positive regulation of interleukin-12 production | 3 | 0.0087 |
The top 25 terms in each GO domain were selected according to p < 0.05.
GO, gene ontology.
MF, molecular function.
BP, biological process.
Gene Count is the number of genes enriched in this term.
We then examined the GO biological process analysis results at 4 hours. This describes a series of events performed by one or many organized assemblies of molecular functions. For these 4 hour DEGs, the 25 most significant pathways showed a wide variety of biological processes, with 7 being involved in the immune response to infection or chemotaxis (Table I). Most notably, 2 processes were involved in the cellular response to cytokines or to tumor necrosis factor, and 2 different processes related to defense responses to infection. In addition, positive regulation of the toll-like receptor signaling pathway was identified, as were innate immune response pathways that involve these receptor signaling pathways [25].
Analysis of the 24 hour data revealed that DEG’s were involved in a number of GO molecular functions of which 14 were identified as being significant (p<0.05, Table II). Among these, 4 of the top 5 most significant pathways were for cytokine and chemokine activity and cytokine and chemokine receptor binding. Additionally, the G-protein coupled receptor binding pathway was identified, and several chemokine receptors use this pathway [24]. Of the top 25 identified GO biological process pathways at 24 hours (Table II), 20 relate to innate immune responses, or chemokine or chemokine induction pathways. As was the case at 4 hours, two of these pathways involved toll-like receptor signaling.
In short, gene ontology analysis of AEC I response to spore exposure revealed overrepresentation of pathways involving innate immune responses, particularly cytokine and chemokine signaling, responses, and production.
3.4. Upregulation of chemokine genes was confirmed by qRT-PCR and multiplex ELISA
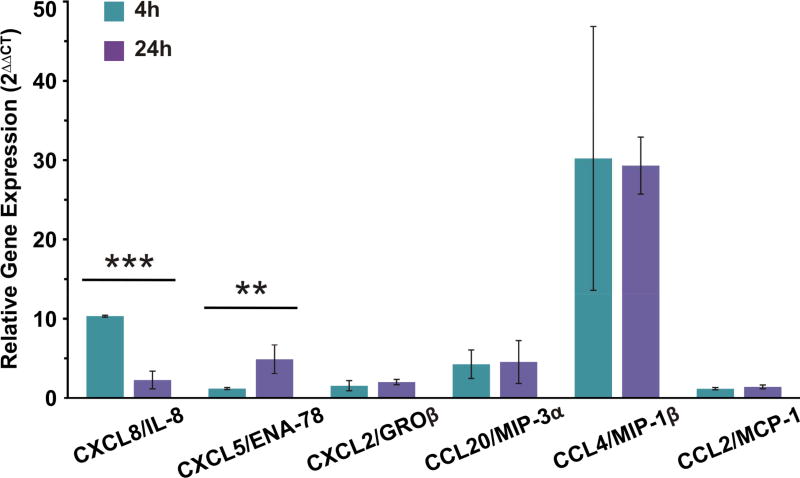
Analysis of microarrays implicated activation of cytokine and chemokine genes and gene pathways in the AEC I response to B. anthracis spores. We performed qRT-PCR of specific neutrophil and monocyte chemokines to determine if the microarray data accurately reflected the AEC response at the level of transcription. We chose for analysis the major neutrophil chemokines CXCL8/IL-8 and CXCL5/ENA-78, and the monocyte chemokine CCL4/MIP-1β which were also among the top 25 most upregulated genes by microarray. We also chose the additional neutrophil chemokines CXCL2/GROβ and CCL20/MIP-3α, and the monocyte chemokine CCL2/MCP-1 for analysis. The results demonstrated induction of chemokine mRNA by spores that varied by chemokine and by duration of exposure to spores (Figure 3). As in the microarray analysis, qRT-PCR demonstrated that spore treatment induced an average increase in mRNA expression (2ΔΔCT) for the all the tested genes. CXCL8/IL-8 and CXCL5/ENA-78 displayed significantly different levels of mRNA expression between 4 and 24 hours. At 4 hours, the average CXCL8/IL-8 expression was elevated 10 fold and decreased to 2 fold by 24 hours. Conversely, the average CXCL5/ENA-78 expression was initially low (1 fold) and became elevated to nearly 5 fold by 24 hours. CCL4/MIP-1β produced the largest fold difference at both 4 hour (30 fold) and 24 hours (29 fold) compared to mock treated AEC. CCL20/MIP-3α, which did not reach significance on the microarray, was induced at 4 and 24 hours at 4 and 5 fold by qRT-PCR. Both CXCL2/GROβ and CCL2/MCP-1 were only minimally induced to less than 2 fold at both time periods by qRT-PCR.
Figure 3.
qRT-PCR confirmed the upregulation of specific neutrophil and monocyte chemokine genes by B. anthracis. Four and 24 hour cDNA samples from mock and spore exposed AEC were analyzed by quantitative real time PCR. The samples were probed for CXCL8/IL-8, CXCL5/ENA-78, CXCL2/GROβ, CCL20/MIP-3α, CCL4/MIP-1β, and CCL2/MCP-1. Actin/ACTB served as a reference gene. Data are expressed as the relative gene expression (2ΔΔCT) of actin and mock normalized spore-exposed samples for each chemokine. Averages from the 4 and 24 hours within each probe set were compared by Student’s t-test. ** p ≤ 0.01, *** p ≤ 0.001.
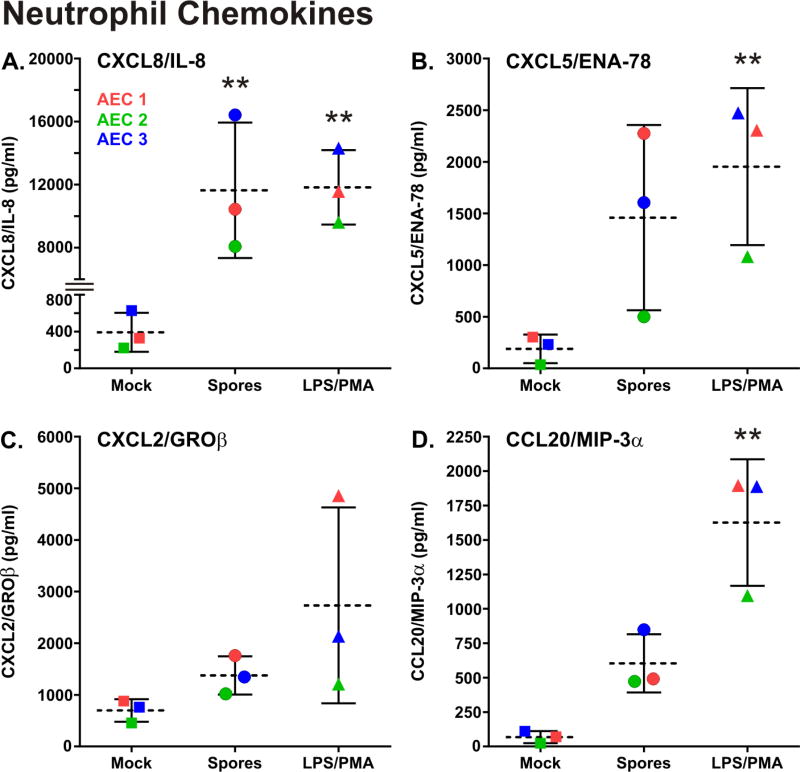
In order to confirm whether chemokine protein induction by spore exposure occurred and to potentially identify additional cytokines stimulated by spores, we performed multiplex ELISA, supplemented with additional individual assays on supernatants from AEC exposed to spores for 4 and 24 hours. Using these methods we assessed induction of 31 cytokines or chemokines. Exposure of cells to LPS/PMA (1µg/ml. and 0.1µg/ml., respectively) was used as a positive control, and exposure to an equal volume of spore diluent (sterile di-water) was used as a negative control for stimulation. The combination of LPS and PMA was used in combination rather than separately as a positive control because there were a limited number of cells available and this mixture induces a wide range of cytokines and chemokines [21]. There was either no induction, or minimal induction above background for all cytokines/chemokines tested at 4 hours of exposure. At 24 hours of spore exposure, there was significant induction of the neutrophil chemokines CXCL8/IL-8 (35 fold, P<0.0001) and CXCL5/ENA-78 (9.5 fold, P<0.01) (Figure 4, A, B). There was also induction of the neutrophil chemokines CXCL2/GROβ (2 Fold) and CCL20/MIP-3α (12 fold) though induction did not reach statistical significance (Figure 4, C, D). Exposure of AEC to PMA/LPS caused similar or greater induction of these chemokines than did spores (Fig. 4).
Figure 4.
B. anthracis spore exposure induces neutrophil chemokine production in primary human type I-like AEC. AEC were exposed for 24 hours to 1 × 106 cfu/ml of B. anthracis spores (circles), mock treated (squares) or exposed to LPS/PMA (1µg/ml and 0.1µg/ml, respectively) which served as a positive control (triangles). The media supernatants were harvested and the amount of neutrophil chemokine proteins in the media supernatants was measured by fluorescent multiplex or individual colorimetric sandwich ELISAs. The results from each of the three AEC donors is shown (donor 1 = red, donor 2 = blue, and donor 3 = green) and the data is expressed as the means (dashed line) ± SD. Means were compared to data for the mock (negative control) group, and statistical significance was determined by one-way ANOVA using Tukey multiple comparisons test. ** p ≤ 0.01.
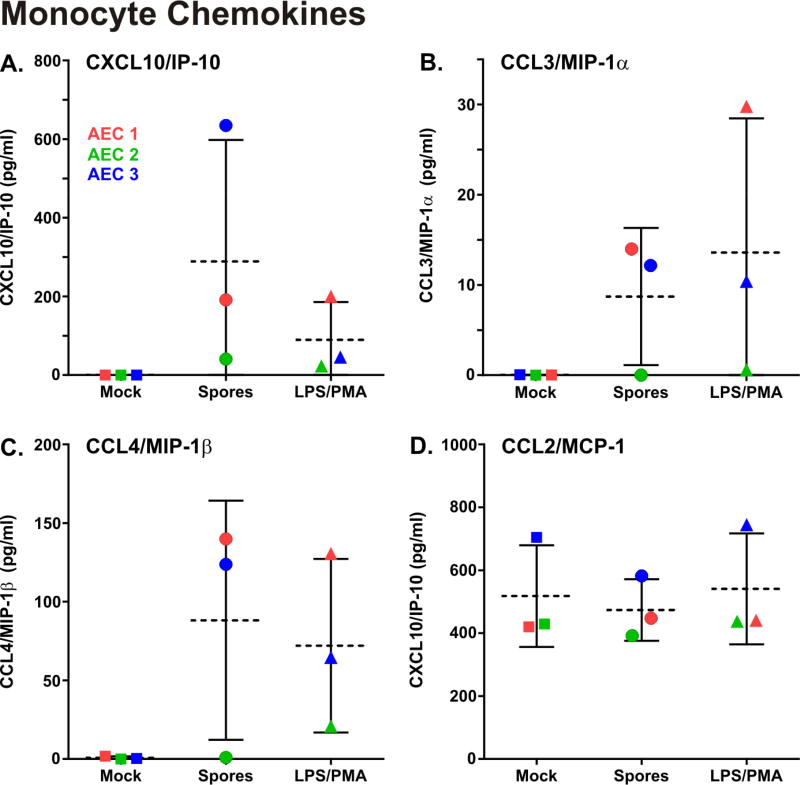
The monocyte chemokines CXCL10/IP-10, CCL3/MIP-1α, and CCL4/MIP-1β were only produced in stimulated cells and induction of these chemokines by spores did not reach statistical significance (Figure 5, A–C). CCL2/MCP-1 was constitutively produced in both unstimulated and stimulated cells, and not induced by spores (Figure 5, D). Of interest, spores induced a greater amount of IP-10 than PMA/LPS, although this did not reach statistical significance, while induction was similar for the other monocyte chemokines tested (Figure 5)
Figure 5.
B. anthracis spore exposure induces monocyte chemokine production in primary human type I-like AEC. AEC were exposed for 24 hours to 1 × 106 cfu/ml of B. anthracis spores (circles), mock treated (squares) or exposed to LPS/PMA (1µg/ml and 0.1µg/ml, respectively) which served as a positive control (triangles). The media supernatants were harvested and the monocyte chemokine proteins in the media supernatants were measured by fluorescent multiplex ELISA. The results from each of the three AEC donors is shown (donor 1 = red, donor 2 = blue, and donor 3 = green) and the data is expressed as the means (dashed line) ± SD. Means were compared to data for the mock (negative control) group, and statistical significance was determined by one-way ANOVA using Tukey multiple comparisons test.
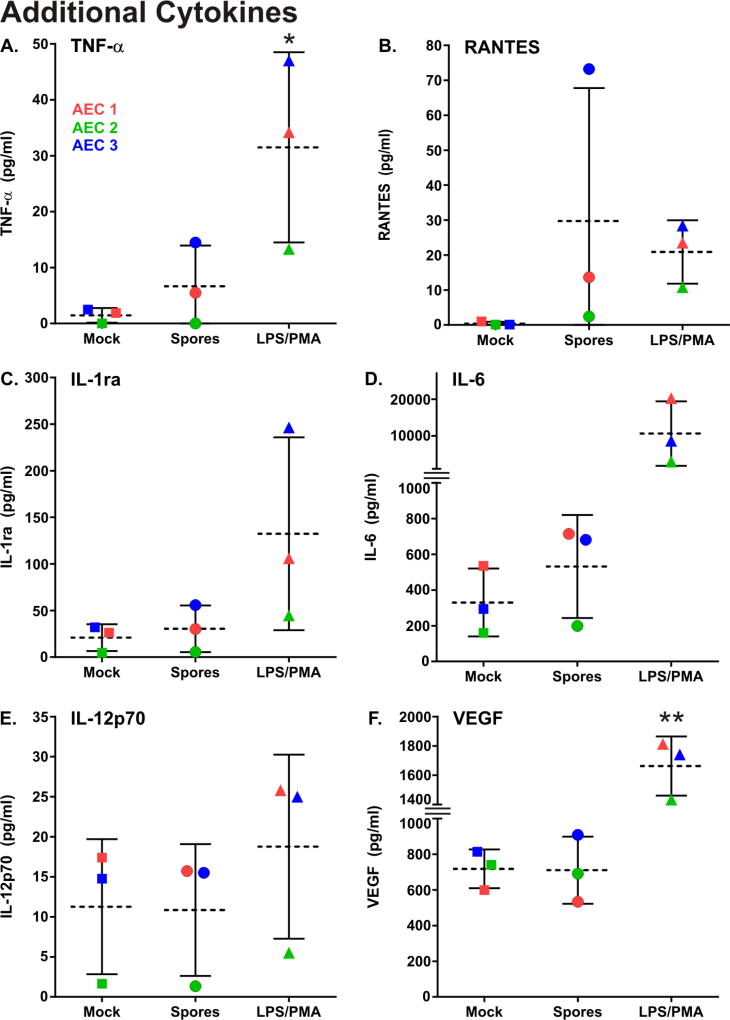
Although the primary purpose of the ELISA’s was to confirm induction of chemokine activity as evidenced by the transcriptional data, the assays revealed that spore exposure for 24 hours induced the cytokines TNFα (2.9 fold) and RANTES (30 fold). IL-1ra, IL-6, IL-12p70, and VEGF were constitutively produced in both unstimulated and stimulated cells, and not induced by spores. Exposure of AEC to PMA/LPS caused similar or greater induction of these chemokines than did spores (Fig. 6).
Figure 6.
B. anthracis spore exposure induces cytokine production in primary human type I-like AEC. AEC were exposed for 24 hours to 1 × 106 cfu/ml of B. anthracis spores (circles), mock treated (squares) or exposed to LPS/PMA (1µg/ml and 0.1µg/ml, respectively) which served as a positive control (triangles). The media supernatants were harvested and the cytokine proteins in the media supernatants was measured by fluorescent multiplex ELISA. The results from each of the three AEC donors is shown (donor 1 = red, donor 2 = blue, and donor 3 = green) and the data is expressed as the means (dashed line) ± SD. Means were compared to data for the mock (negative control) group, and statistical significance was determined by one-way ANOVA using Tukey multiple comparisons test. * p ≤ 0.05, ** p ≤ 0.01.
3.5. Spores induced neutrophil and monocyte chemotaxin release from AEC
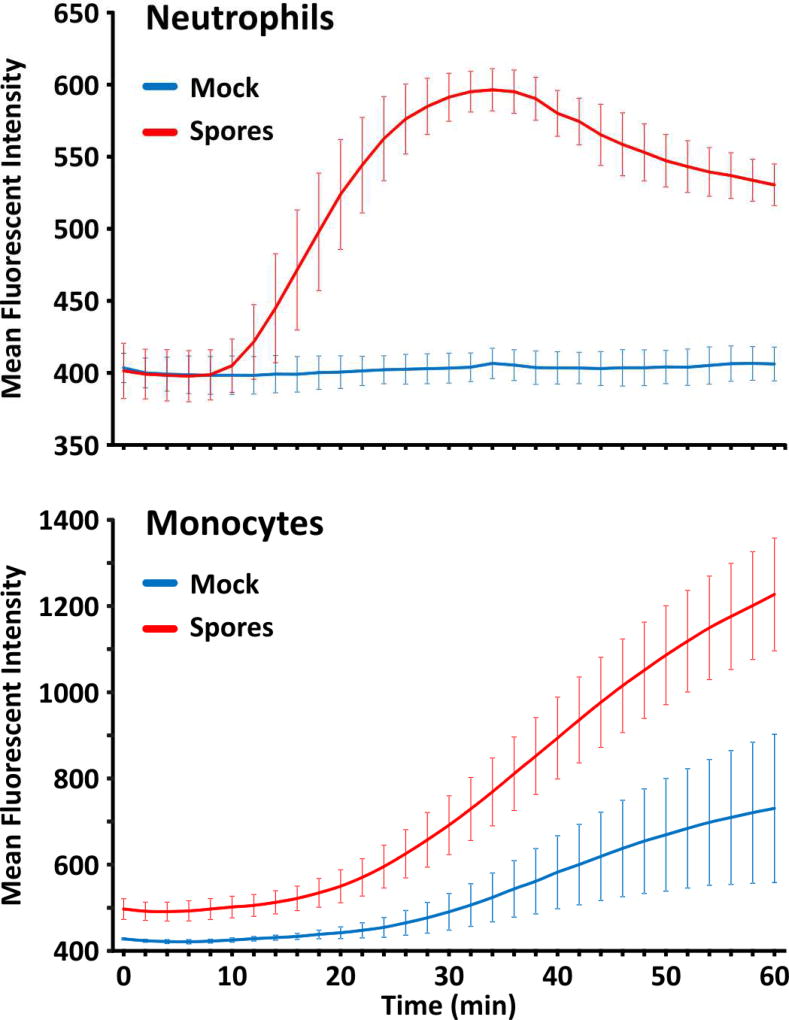
To determine whether or not the chemokines induced by spores were biologically active, chemotaxis of human neutrophils and monocytes towards supernatants of AEC exposed to spores for 24 hours was determined using fluorescently labeled cells. Chemotaxis towards supernatants of AEC exposed to an equal volume of spore diluent was used as a negative control. The results demonstrated the presence of chemokines active towards both neutrophils and monocytes as evidenced by a real-time fluorescent chemotactic assay system (Figure 7; see Material and Methods). The neutrophil chemotactic activity present in spore exposed cells resulted in an increase in the rate of neutrophil chemotaxis by greater than 10 fold over supernatant from untreated AEC, while the monocyte chemotactic activity increased the rate of monocyte chemotaxis by 1.7 fold over activity present in supernatant from untreated AEC.
Figure 7.
B. anthracis spores induced functional neutrophil and monocyte chemotaxin release from type I-like AEC. AEC were mock treated (blue line) or exposed for 24 hours to 1 × 106 cfu/ml of B. anthracis spores (red line), and the media supernatants were used to follow the kinetics of fluorescent cell migration using a real-time chemotaxis assay. AEC supernatants were assayed in duplicate, and the results for (A) Neutrophil chemotactic activity and (B) monocyte chemotactic activity are expressed as the means ± SD of three AEC donors.
We next sought to identify individual chemokines responsible for the chemotactic activity detected in spore-exposed AEC supernatants. We measured chemotactic activity in the presence or absence of neutralizing antibodies to the chemokines detected. Isotype control antibodies did not induce or inhibit significant chemotaxis. We also determined whether the amount of individual chemokines present in AEC supernatants was sufficient to induce chemotaxis, using recombinant chemokines added to naïve culture media.
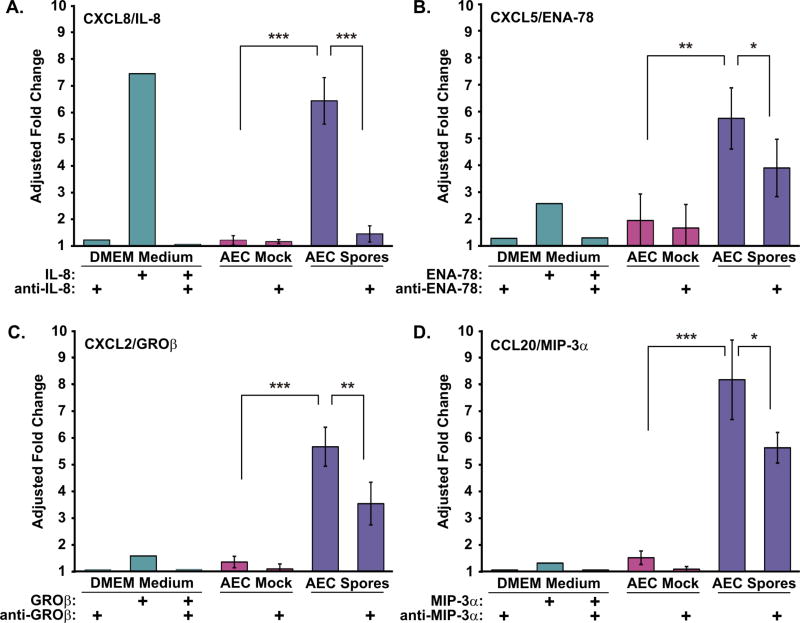
In terms of neutrophil chemotaxis, the major chemotaxin present in spore-exposed AEC supernatant was CXCL8/IL-8. Preincubation of this supernatant with neutralizing IL-8 antibody reduced chemotactic activity by 90% (P<0.001, Figure 8A). Activity was not inhibited by the addition of isotype control antibody (not shown), and naive media spiked with the same concentration of recombinant IL-8 as seen in spore-exposed AEC supernatants induced similar levels of chemotaxis as the spore-exposed media.
Figure 8.
Type I-like AEC respond to B. anthracis spores by producing discrete neutrophil chemokines. Supernatants from AEC exposed to B. anthracis spores for 24 hours were tested for neutrophil chemotactic activity. Antibodies to specific chemotaxins were employed to confirm the dominant neutrophil chemokines present in the mock (violet bars) and spore-treated (blue bars) AEC supernatants. DMEM (AEC growth medium) amended with recombinant chemokines at the concentrations measured in AEC supernatants, chemokines plus specific neutralizing antibodies, or neutralizing antibodies alone as indicated were tested (green bars). Unamended DMEM provided the background activity (fold change = 1, not shown). DMEM and AEC supernatants were assayed in duplicate. Neutrophil chemotactic activity in the presence of (A) CXCL8/IL-8, (B) CXCL5/ENA-78, (C) CXCL2/GROβ, and (D) CCL20/MIP-3α neutralizing antibody. Chemotactic activity is expressed as the adjusted fold change over background and are the means ± SD of the three donors. Means of the AEC mock and spore-exposed supernatants were compared, and statistical significance was determined by one-way ANOVA with Tukey multiple comparisons test. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Using the same approach, we also assessed the contribution of CXCL5/ENA-78, CXCL2/GROβ, and CCL20/MIP-3α to the neutrophil chemotactic activity released by spore-exposed AEC. Preincubation of the AEC supernatants with neutralizing antibodies to these chemokines resulted in an inhibition of chemotaxis by 44%, 46%, and 36% respectively (Figure 8, B–D). Naïve media spiked with the same concentration of CXCL5/ENA-78 as seen in spore-AEC supernatants resulted in a 2.6 fold increase in neutrophil chemotaxis, and thus, this chemokine may contribute directly to the neutrophil chemotactic activity induced in AEC by spores. However, naive media spiked with the same concentration of CXCL2/GROβ and CCL20/MIP-3α recombinant chemokines as seen in spore-AEC supernatants resulted in only minimal induction of neutrophil chemotaxis (all ≤ 1.5 fold). Thus, the effects of these blocking antibodies on chemotaxis may have been due to inhibition of priming effects by sub-chemotactic doses of these chemokines [27].
We next sought to determine the contribution of specific chemokines to the monocyte chemotactic activity released from spore-exposed AEC. To determine whether the levels of chemotaxins detected could be responsible for the activity, we measured monocyte chemotaxis in response to levels of the chemokines CCL2/MCP-1, CXCL10/IP-10, CCL3/MIP-1α, and CCL4/MIP-1β present in spore-exposed AEC supernatants by adding recombinant chemokines to naïve media. For all of these chemokines, there was only minimal induction of monocyte chemotaxis (all < 1.5 fold) at levels seen in supernatants of spore-exposed AEC, and blocking isotype control antibodies caused approximately 1.2 fold non-specific induction of monocyte chemotaxis (not shown). Thus, we could not confirm the identity of the chemokine responsible for the differential monocyte chemotactic activity induced in spore exposed AEC.
4. Discussion
Escape of B. anthracis from the alveolar space of the lung is required for the pathogen to produce the fatal disease known as inhalation anthrax [1]. The time between spore exposure and symptoms can vary from days to weeks and the incubation time is inversely dependent on the dose of spores [1]. This latency period offers an opportunity to block progression of the disease. Therefore, understanding the initial interactions of the B. anthracis spore with the alveolar epithelium is important in discovering potential therapeutic interventions.
There have been several studies examining adherence, internalization, and transit of B. anthracis spores through epithelial cells. These studies primarily used epithelial cell lines or bronchial epithelial cell lines [13–15, 28, 29]. The only primary human cells that have been used are small airway epithelial cells [30, 31]. Although these studies have provided useful information regarding signaling events in internalization, it is of concern that none have used primary human alveolar epithelial cells. This is an important point because spore transport is thought to occur across the alveolar epithelial barrier [1].
We, therefore, sought to determine the initial response of primary human AEC to B. anthracis spores. We used microarrays to capture the transcriptional response of these cell to the pathogen. Although our sample size, and thus power, was limited due to the difficulty in obtaining purified AEC, we were able to perform statistical comparisons and evaluate biological properties of the data in order to find genome-wide unbiased results. A 30% difference in gene expression (± 1.3 fold, base 10) was chosen as a cutoff for biologically significant changes in gene expression. Many different fold-change values are used in these types of studies, and context, that is biological relevance, is frequently considered rather than an arbitrary value [32, 33]. Our cutoff of ± 1.3 fold appears biologically justifiable as this identified induction of similar cytokines at both 4 and 24 hours of exposure, whereas a higher cutoff (i.e. ± 1.5 fold) would have excluded chemokines from one or both of the time points. Specifically CXCL5/ENA-78 would have been excluded at 24hrs, and CXCL2/GROβ would have been excluded at both 4 and 24 hrs with a 1.5 fold threshold. In any case, we then confirmed the results at the level of transcription by qRT-PCR, at the protein level by multiplex ELISA, and at the level of biological activity by chemotaxis assays.
Transcriptome analysis demonstrated that there was an overrepresentation of differentially expressed genes involved in the immune response of AEC to spores, specifically cellular pathways involved in cytokine and chemokine activity. We also found induction of individual chemokine genes, including CCL4/MIP-1β, CXCL8/IL-8, and CXCL5/ENA-78. The most highly expressed gene at both 4 and 24 hours, CCL4/MIP-1β, is a chemokine that attracts NK cells, T-lymphocytes and monocytes, and is increased in respiratory secretions of patients with sarcoidosis, COPD and asthma [34]. CXCL8/IL-8, highly expressed at 4 hour, is a neutrophil chemokine that is increased in bronchoalveolar lavage fluid of patients with lung injury and infection [35, 36]. CXCL5/ENA-78, a classic neutrophil chemokine, is highly expressed at 24 hours, and is important in the immune response to lung infections [37–39].
Other highly expressed genes at 24 hours included lymphotoxin β (LTB) and mitogen-activated protein kinase kinase 4 (MAP2K4). Lymphotoxin β (LTB) a member of the TNF superfamily, is a subunit protein member that combines with other superfamily components, and signals through its cognate receptor LTBR. LTBR stimulation induces IL-8 production in human bronchial epithelial cells [40]. Mitogen-activated protein kinase kinase 4 (MAP2K4), is a signaling pathway protein that is a serine/threonine kinase family member upstream and activator of the JNK and P38 pathways. We have shown that activation of these pathways is important for chemokine induction by spores in a human lung model [21].
To confirm these results and to detect other innate immune response proteins induced by spores, we screened for induction of 31 different chemokines and cytokines. We found that only CXCL8/IL-8, and CXCL5/ENA-78 secretion into media were significantly induced by spores. Additional experiments demonstrated that IL-8 was the major neutrophil chemotaxin induced and that CXCL5/ENA-78 may also contributed a small amount to neutrophil chemotaxis, as both were active at the levels present in supernatants from spore-stimulated cells, while CXCL2/GROβ and CCL20/MIP-3α likely only act as priming agents for neutrophils. Monocyte chemotactic activity was induced by spores, but we were unable to identify the specific chemotaxin responsible.
Our data showing release of chemokines from AEC by spores is novel. The only data of which we are aware examining this issue showed modest (two-fold) induction of TNF-α by human SAEC, but only when these cells were pre-exposed with Bacillus cell wall preparations [31]. Our previous work has examined the induction of cytokines and chemokines by human precision cut lung slice cultures exposed to spores [21]. In those experiments, we demonstrated induction of many cytokines including IL-6, TNF-α, CXCL8/IL-8, CCL2/MCP-1, and MIP-1α/β. The assumption at that time was that epithelial cells and non-epithelial cells including antigen-presenting cells contributed to the innate immune response. With the exception of CXCL8/IL-8 (Figure 4A), our current work suggests that the source of these cytokines is primarily non-epithelial cells as the epithelial non-chemotactic cytokine response to spores is modest, and we have demonstrated vigorous induction by spores of all of these cytokines at the level of transcription or translation in human alveolar macrophages [41].
Induction of neutrophil chemotaxis by spores may explain some of the findings seen in examination of animal models of inhalational anthrax. In these models, neutrophils are present in the lungs shortly after exposure, some of which contain ingested spores [7]. In terminal phases of the illness, pulmonary edema with blood occurs and neutrophilic infiltration is minimal [42]. There is evidence that supports a more significant role for neutrophils in the pathogenesis of the disease, as depletion of neutrophils in mice lacking the anthrax toxin receptor CMG-2 increased mortality during spore infection [43]. In autopsies of human cases, pulmonary collections of neutrophils have been described in the vasculature, as well as minimal alveolar infiltration of neutrophils, and again, pulmonary edema and mild hemorrhage is frequent [5, 44].
One of the interesting findings of this work is what is not present. That is a generalized, vigorous response of AEC to spores. The cytokine responses are limited and modest compared to those seen with exposure of human alveolar macrophages to spores at a similar MOI [41]. The attenuated response of AEC to spores may explain how spores can persist in the lung after exposure in animal models by association with lung epithelia, and why there is a delay between exposure and the onset of symptoms [45].
Our data provide the first whole transcriptomic description of the human alveolar epithelial initial response to B. anthracis spore exposure. The primary response appears to be limited in intensity, and involves stimulation of neutrophil and monocyte chemotactic activity. This response may play a role in early events in the disease known as inhalation anthrax. Enhancing this response may facilitate elimination of the pathogen by the recruitment of immune cells. This benefit must be carefully weighed against the potential risk of attracting cells that enhance spore escape from the lung. Additional analysis of our data set may also reveal other unexplored targets for intervention in the early stages of inhalational anthrax.
Highlights.
Primary human type I alveolar epithelial cells (AEC) exposed to Bacillus anthracis, (Sterne) spores display altered gene expression at 4 and 24 hours.
In spore-exposed human type I AEC, the most highly upregulated differentially expressed genes include those for the chemokines CCL4/MIP-1β, CXCL8/IL-8, and CXCL5/ENA-78.
Gene enrichment analysis reveals that pathways involving cytokine or chemokine activity, receptor binding, and innate immune responses to infection are prominent.
Chemotaxis assays demonstrated that B. anthracis spores induced the release of neutrophil and monocyte chemokines, and that CXCL8/IL-8 is the major neutrophil chemokine.
Our results provide the first transcriptomic description of the initial response of human primary type I AEC to B. anthracis spore exposure, and contribute to an increased understanding of the role of AEC in the pathogenesis of inhalational anthrax.
Acknowledgments
The research described in this work was partially supported by the Federal Aviation Administration, Office of Aviation Medicine, by a Clinical Innovator Award from the Flight Attendant Medical Research Institute, by the Merit Review Program of the Department of Veterans Affairs, the National Institutes of Health [grant number 1 I01 BX001937], and by the National Institutes of Health, [grant numbers U19AI62629, to K.M.C] and GM103648 to J.P.M.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dixon TC, Meselson M, Guillemin J, Hanna PC. Anthrax. N. Engl. J. Med. 1999;341:815–26. doi: 10.1056/NEJM199909093411107. [DOI] [PubMed] [Google Scholar]
- 2.Inglesby TV, O'Toole T, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA. 2002;287:2236–52. doi: 10.1001/jama.287.17.2236. [DOI] [PubMed] [Google Scholar]
- 3.Cleret A, Quesnel-Hellmann A, Vallon-Eberhard A, Verrier B, Jung S, Vidal D, et al. Lung dendritic cells rapidly mediate anthrax spore entry through the pulmonary route. J. Immunol. 2007;178:7994–8001. doi: 10.4049/jimmunol.178.12.7994. [DOI] [PubMed] [Google Scholar]
- 4.Cote CK, Van Rooijen N, Welkos SL. Roles of macrophages and neutrophils in the early host response to Bacillus anthracis spores in a mouse model of infection. Infect. Immun. 2006;74:469–80. doi: 10.1128/IAI.74.1.469-480.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grinberg LM, Abramova FA, Yampolskaya OV, Walker DH, Smith JH. Quantitative pathology of inhalational anthrax I: quantitative microscopic findings. Mod. Pathol. 2001;14:482–95. doi: 10.1038/modpathol.3880337. [DOI] [PubMed] [Google Scholar]
- 6.Lincoln RE, Hodges DR, Klein F, Mahlandt BG, Jones WI, Jr, Haines BW, et al. Role of the lymphatics in the pathogenesis of anthrax. J. Infect. Dis. 1965;115:481–94. doi: 10.1093/infdis/115.5.481. [DOI] [PubMed] [Google Scholar]
- 7.Ross JM. The pathogenesis of anthrax following the administration of spores by the respiratory route. J. Path. Bact. 1957;73:485. [Google Scholar]
- 8.Guidi-Rontani C. The alveolar macrophage: the Trojan horse of Bacillus anthracis. Trends Microbiol. 2002;10:405. doi: 10.1016/s0966-842x(02)02422-8. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Mehta H, Chakrabarty K, Booth JL, Duggan ES, Patel KB, et al. Resistance of human alveolar macrophages to Bacillus anthracis lethal toxin. J. Immunol. 2009;183:5799–806. doi: 10.4049/jimmunol.0803406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brittingham KC, Ruthel G, Panchal RG, Fuller CL, Ribot WJ, Hoover TA, et al. Dendritic cells endocytose Bacillus anthracis spores: implications for anthrax pathogenesis. J. Immunol. 2005;174:5545–52. doi: 10.4049/jimmunol.174.9.5545. [DOI] [PubMed] [Google Scholar]
- 11.Jahnsen FL, Strickland DH, Thomas JA, Tobagus IT, Napoli S, Zosky GR, et al. Accelerated antigen sampling and transport by airway mucosal dendritic cells following inhalation of a bacterial stimulus. J. Immunol. 2006;177:5861–7. doi: 10.4049/jimmunol.177.9.5861. [DOI] [PubMed] [Google Scholar]
- 12.Patel VI, Booth JL, Duggan ES, Cate S, White VL, Hutchings D, et al. Transcriptional Classification and Functional Characterization of Human Airway Macrophage and Dendritic Cell Subsets. J. Immunol. 2017;198:1183–201. doi: 10.4049/jimmunol.1600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell BH, Vasan R, Keene DR, Xu Y. Bacillus anthracis internalization by human fibroblasts and epithelial cells. Cell. Microbiol. 2007;9:1262–74. doi: 10.1111/j.1462-5822.2006.00869.x. [DOI] [PubMed] [Google Scholar]
- 14.Russell BH, Vasan R, Keene DR, Koehler TM, Xu Y. Potential dissemination of Bacillus anthracis utilizing human lung epithelial cells. Cell. Microbiol. 2008;10:945–57. doi: 10.1111/j.1462-5822.2007.01098.x. [DOI] [PubMed] [Google Scholar]
- 15.Xue Q, Gu C, Rivera J, Hook M, Chen X, Pozzi A, et al. Entry of Bacillus anthracis spores into epithelial cells is mediated by the spore surface protein BclA, integrin alpha2beta1 and complement component C1q. Cell. Microbiol. 2011;13:620–34. doi: 10.1111/j.1462-5822.2010.01558.x. [DOI] [PubMed] [Google Scholar]
- 16.Weiner ZP, Glomski IJ. Updating perspectives on the initiation of Bacillus anthracis growth and dissemination through its host. Infect. Immun. 2012;80:1626–33. doi: 10.1128/IAI.06061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–5. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis. 1982;125:740–5. doi: 10.1164/arrd.1982.125.6.740. [DOI] [PubMed] [Google Scholar]
- 19.Langer M, Duggan ES, Booth JL, Patel VI, Zander RA, Silasi-Mansat R, et al. Bacillus anthracis lethal toxin reduces human alveolar epithelial barrier function. Infect. Immun. 2012;80:4374–87. doi: 10.1128/IAI.01011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dozmorov I, Centola M. An associative analysis of gene expression array data. Bioinformatics. 2003;19:204–11. doi: 10.1093/bioinformatics/19.2.204. [DOI] [PubMed] [Google Scholar]
- 21.Chakrabarty K, Wu W, Booth JL, Duggan ES, Nagle NN, Coggeshall KM, et al. Human lung innate immune response to Bacillus anthracis spore infection. Infect. Immun. 2007;75:3729–38. doi: 10.1128/IAI.00046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Zar JH. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice-Hall; 1996. [Google Scholar]
- 24.Marchese A. Endocytic trafficking of chemokine receptors. Curr. Opin. Cell. Biol. 2014;27:72–7. doi: 10.1016/j.ceb.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palsson-McDermott EM, O'Neill LA. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology. 2004;113:153–62. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Senoo H, Sesaki H, Iijima M. Rho GTPases orient directional sensing in chemotaxis. Proc. Natl. Acad. Sci. U S A. 2013;110:E4723–32. doi: 10.1073/pnas.1312540110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miralda I, Uriarte SM, McLeish KR. Multiple Phenotypic Changes Define Neutrophil Priming. Front. Cell. Infect. Microbiol. 2017;7:217. doi: 10.3389/fcimb.2017.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozue J, Moody KL, Cote CK, Stiles BG, Friedlander AM, Welkos SL, et al. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect. Immun. 2007;75:4498–505. doi: 10.1128/IAI.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell JD, Hutchison JR, Hess BM, Straub TM. Bacillus anthracis spores germinate extracellularly at air-liquid interface in an in vitro lung model under serum-free conditions. J. Appl. Microbiol. 2015;119:711–23. doi: 10.1111/jam.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popova T, Espina V, Bailey C, Liotta L, Petricoin E, Popov S. Anthrax infection inhibits the AKT signaling involved in the E-cadherin-mediated adhesion of lung epithelial cells. FEMS Immunol. Med. Microbiol. 2009;56:129–42. doi: 10.1111/j.1574-695X.2009.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radyuk SN, Mericko PA, Popova TG, Grene E, Alibek K. In vitro-generated respiratory mucosa: a new tool to study inhalational anthrax. Biochem. Biophys. Res. Commun. 2003;305:624–32. doi: 10.1016/s0006-291x(03)00830-1. [DOI] [PubMed] [Google Scholar]
- 32.Dalman MR, Deeter A, Nimishakavi G, Duan ZH. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13:S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters MS, De BP, Salit J, Buro-Auriemma LJ, Wilson T, Rogalski AM, et al. Smoking accelerates aging of the small airway epithelium. Respir. Res. 2014;15:94. doi: 10.1186/s12931-014-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barczyk A, Pierzchala E, Caramori G, Sozanska E. Increased expression of CCL4/MIP-1beta in CD8+ cells and CD4+ cells in sarcoidosis. Int. J. Immunopathol. Pharmacol. 2014;27:185–93. doi: 10.1177/039463201402700205. [DOI] [PubMed] [Google Scholar]
- 35.Mukaida N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;284:L566–77. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- 36.Zuniga J, Torres M, Romo J, Torres D, Jimenez L, Ramirez G, et al. Inflammatory profiles in severe pneumonia associated with the pandemic influenza A/H1N1 virus isolated in Mexico City. Autoimmunity. 2011;44:562–70. doi: 10.3109/08916934.2011.592885. [DOI] [PubMed] [Google Scholar]
- 37.Goodman RB, Strieter RM, Frevert CW, Cummings CJ, Tekamp-Olson P, Kunkel SL, et al. Quantitative comparison of C-X-C chemokines produced by endotoxin-stimulated human alveolar macrophages. Am. J. Physiol. 1998;275:L87–95. doi: 10.1152/ajplung.1998.275.1.L87. [DOI] [PubMed] [Google Scholar]
- 38.Cai S, Batra S, Lira SA, Kolls JK, Jeyaseelan S. CXCL1 regulates pulmonary host defense to Klebsiella Infection via CXCL2, CXCL5, NF-kappaB, and MAPKs. J. Immunol. 2010;185:6214–25. doi: 10.4049/jimmunol.0903843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nouailles G, Dorhoi A, Koch M, Zerrahn J, Weiner J, 3rd, Fae KC, et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J. Clin. Invest. 2014;124:1268–82. doi: 10.1172/JCI72030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikami Y, Matsuzaki H, Horie M, Noguchi S, Jo T, Narumoto O, et al. Lymphotoxin beta receptor signaling induces IL-8 production in human bronchial epithelial cells. PLoS One. 2014;9:e114791. doi: 10.1371/journal.pone.0114791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakrabarty K, Wu W, Booth JL, Duggan ES, Coggeshall KM, Metcalf JP. Bacillus anthracis spores stimulate cytokine and chemokine innate immune responses in human alveolar macrophages through multiple mitogen-activated protein kinase pathways. Infect. Immun. 2006;74:4430–8. doi: 10.1128/IAI.00446-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albrink WS, Goodlow RJ. Experimental inhalation anthrax in the chimpanzee. Am. J. Pathol. 1959;35:1055–65. [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, Miller-Randolph S, Crown D, Moayeri M, Sastalla I, Okugawa S, et al. Anthrax toxin targeting of myeloid cells through the CMG2 receptor is essential for establishment of Bacillus anthracis infections in mice. Cell Host Microbe. 2010;8:455–62. doi: 10.1016/j.chom.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albrink WS. Pathogenesis of inhalation anthrax. Bacteriol Rev. 1961;25:268–73. doi: 10.1128/br.25.3.268-273.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meselson M, Guillemin J, Hugh-Jones M, Langmuir A, Popova I, Shelokov A, et al. The Sverdlovsk anthrax outbreak of 1979. Science. 1994;266:1202–8. doi: 10.1126/science.7973702. [DOI] [PubMed] [Google Scholar]