Abstract
Background
The Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) is a commonly used risk tool for predicting the outcome on biopsy based on the established risk factors.
Objective
To determine whether incorporation of the novel urinary markers prostate cancer antigen 3 (PCA3) and TMPRSS2:ERG (T2:ERG) into the PCPTRC improves its discrimination, accuracy, and clinical net benefit.
Design, setting, and participants
Since PCA3 and T2:ERG were not measured as part of the PCPTRC, a Bayesian modeling approach was used to combine data where the markers were measured in a Michigan cohort with the PCPTRC as prior probabilities to form an updated PCPTRC. This update was compared to the existing PCPTRC on an independent Early Detection Research Network cohort in terms of discrimination, calibration, and decision curve analysis.
Results and limitations
Among the 1225 Michigan biopsies, 57.7%, 24.0%, and 18.3% were negative, with low- and high-grade (Gleason grade ≥ 7) prostate cancer, respectively. Evaluatedon the Early Detection Research Network validation set comprising 854 biopsies, areas under the curve (95% confidence interval) for predicting high-grade cancer in the 854 biopsies comprising the validation set were 70.0% (66.0–74.0%), 76.4% (72.8–80.0%), and 77.1% (73.6–80.6%) for the PCPTRC alone, with PCA3 added, and PCA3 and T2:ERG added, respectively. Net benefit was improved for the updated PCPTRC, while calibration was not. Limitations are that the updated PCPTRC is based on two different cohorts, the PCPT and Michigan, and that 20% of the validation set came from the Michigan center. More validation is required; hence, the updated risk tool is posted online.
Conclusions
Incorporation of PCA3 into the PCPTRC improved validation on an independent cohort, whereas T2:ERG offered negligible utility in addition to PCA3.
Patient summary
After passing external validation, prostate cancer antigen 3 has been added to the online Prostate Cancer Prevention Trial Risk Calculator for use by patients in deciding whether to proceed to biopsy. TMPRSS2:ERG did not improve prediction on the external validation set, but is included for further validation.
Keywords: Prostate cancer, Prostate-specific antigen, PCA3, T2:ERG
1. Introduction
Since its establishment in 2006, the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) has become a widely validated and accessed tool for clinical decision-making concerning prostate biopsy [1–4]. It was updated in 2014 in response to increasing recommendations for active surveillance for management of low-versus high-grade (Gleason score ≥ 7) disease [5,6]. An analysis of 6664 prostate biopsies from the PCPT forming this update determined that prostate-specific antigen (PSA) and age were associated with higher risks of both low- and high-grade cancer, abnormal digital rectal exam (DRE) and African-American race were significantly associated with risk of high-grade but not low-grade prostate cancer, and family history and history of a prior negative biopsy were only significantly associated with low-grade prostate cancer [5]. Against concerns of overdetection and overtreatment of low-risk cancers, the PCPTRC risk of high-grade disease has been recommended for decisions to proceed to biopsy.
As these changes occurred, novel serum markers, including percent free PSA, and genetic markers, including single nucleotide polymorphisms, were discovered and validated in multiple cohorts [7,8]. When they were proven to add independent predictive power to the established risk factors already in clinical use, they were incorporating into prostate cancer risk tools, including the PCPTRC [5,9]. Recently, attention has turned to the urinary biomarkers prostate cancer antigen 3 (PCA3) and TMPRSS2:ERG (T2: ERG), with multiple validation studies confirming their potential utility for clinical practice and additive value to the leading screen marker, PSA [10–12]. Given these developments, the purpose of this study was to investigate whether PCA3 and T2:ERG added independent predictive information to the risk factors included in the PCPTRC, and if so, incorporate them into the risk tool.
2. Patients and methods
The online PCPTRC includes the following established risk factors: PSA, DRE, age, African-American race, history of a prior negative biopsy, and first-degree family history of prostate cancer. To update the PCPTRC to include the urinary markers, a Bayesian methodology converting PCPTRC prior odds to posterior odds via likelihood ratios (LR) was used [13]. Prior odds of low-grade cancer versus no cancer were computed as the ratio of risks of low-grade disease to no cancer from the PCPTRC model, and prior odds of high-grade cancer versus no cancer were analogously computed. LRs for PCA3 and T2:ERG were calculated on data from a Michigan cohort, and were then multiplied by the PCPTRC prior odds to achieve updated PCPTRC risks incorporating all markers.
Data for computation of LRs were provided by a Michigan community-based cohort [10]. Prospectively collected post-DRE urine samples were obtained prior to diagnostic biopsy, along with PSA and other risk factors included in the PCPTRC. Patient characteristics were compared across the no-, low-, and high-grade cancer outcome groups using chi-square tests. Areas underneath the receiver-operating characteristic curves (AUC) and 95% confidence intervals (CIs) were computed for PCA3 and T2:ERG for predicting high-grade cancer versus no cancer, low-grade cancer versus no cancer, and high-versus low-grade cancer.
To form LRs for PCA3, first a linear regression of log-base-2 PCA3 on PSA, which was also log-base-2 transformed, DRE, race, prior biopsy, and the indicator of biopsy outcome group (no prostate cancer vs low grade vs high grade) was performed with a subsequent Bayesian Information Criterion-assisted model selection step to derive the optimal model. The LRs were then computed as the ratio of the Normal regression density functions for high-grade cancer compared with no cancer, and similarly for low-grade cancer compared with no cancer. These LRs were multiplied by LRs for T2:ERG, the latter computed with the additional inclusion of log-base-2 PCA3 as a predictor in the regressions. To account for a high degree of nonexpression, a combined logistic-normal regression hurdle model was assumed for T2:ERG, whereby the probability of nonzero expression was modeled using logistic regression and conditional on nonzero expression, log-base-2 transformed T2:ERG values were assumed to follow a normal distribution.
The updated PCPTRC to include the urinary markers was validated on an independent National Cancer Institute Early Detection Research Network (EDRN) reference set, which included patients from Harvard (50%), Michigan (20%), and Cornell (30%) University hospitals; patients in the Michigan cohort were different from those in the Michigan cohort used to build the LRs [12]. Biopsies in the cohort were performed using systematic templates, with typically 12 cores, reviewed by site-specific genitourinary pathologists. Biopsy results, one per patient, were classified into three outcomes: no cancer, low-grade, and high-grade cancer.
Validation was quantified by receiver operating characteristic, calibration, and net benefit curves for the prediction of high-grade disease on the EDRN cohort. The R Shiny application was used to post the updated risk calculators on the PCPTRC website. Statistical significance for all tests was determined at the two-sided 0.05 level.
3. Results
Among the 1225 Michigan biopsies, 707 (57.7%) were negative for prostate cancer, 294(24.0%)were diagnosed with low-grade prostate cancer, and 224 (18.3%) with high-grade prostate cancer. Age, prior biopsy, and PSA were statistically significantly associated with cancer outcome, while DRE, family history, and African-American race were not (Table 1). For high-grade versus no cancer, AUCs (95% CIs) were 75.8 (72.4, 79.3) for PCA3 versus 69.3 (65.0, 73.6) for T2:ERG, for low-grade versus no cancer, 67.7 (64.1, 71.3) versus 64.5 (60.7, 68.3), respectively, and for high-versus low-grade cancer, 58.8 (53.9, 63.7) versus 56.8 (51.7, 61.9), respectively.
Table 1.
Characteristics of Michigan patients at time of biopsy (n, % across rows); p values for testing the null hypothesis of no difference across the three outcome groups of no cancer versus low-grade versus high-grade cancer.
| Characteristicsa | No cancer, n = 707 (57.7%) | Low-grade cancer, n = 294 (24.0%) | High-grade cancer, n = 224 (18.3%) | Total, n = 1225 | p value for association with cancer outcome within cohort |
|---|---|---|---|---|---|
| Age (yr) | <0.001 | ||||
| 33–49 | 43 (70.5) | 14 (23) | 4 (6.6) | 61 | |
| 50–59 | 205 (64.7) | 70 (22.1) | 42 (13.2) | 317 | |
| 60–69 | 303 (58.9) | 126 (24.5) | 85 (16.5) | 514 | |
| 70–79 | 146 (49.8) | 77 (26.3) | 70 (23.9) | 293 | |
| 80–92 | 10 (25.0) | 7 (17.5) | 23 (57.5) | 40 | |
| Race | 0.07 | ||||
| African American | 38 (46.3) | 27 (32.9) | 17 (20.7) | 82 | |
| Other | 668 (58.6) | 265 (23.2) | 207 (18.2) | 1140 | |
| Prior biopsy | 0.01 | ||||
| Yes | 163 (66.3) | 51 (20.7) | 32 (13.0) | 246 | |
| No | 542 (55.5) | 243 (24.9) | 192 (19.7) | 977 | |
| Digital rectal exam | 0.08 | ||||
| Normal | 554 (59.2) | 223 (23.8) | 159 (17.0) | 936 | |
| Abnormal | 152 (53.0) | 71 (24.7) | 64 (22.3) | 287 | |
| Family history of prostate cancer | 0.07 | ||||
| Yes | 118 (51.3) | 65 (28.3) | 47 (20.4) | 230 | |
| No | 560 (59.6) | 218 (23.2) | 162 (17.2) | 940 | |
| PSA (ng/ml) | <0.001 | ||||
| ≤4.0 | 298 (66.7) | 99 (22.1) | 50 (11.2) | 447 | |
| 4.0–10.0 | 352 (53.7) | 171 (26.1) | 132 (20.2) | 655 | |
| 10.0–20.0 | 47 (48.0) | 22 (22.4) | 29 (29.6) | 98 | |
| ≥20.0 | 10 (40.0) | 2 (8.0) | 13 (52.0) | 25 |
PSA = prostate-specific antigen.
Three participants had unknown race, one with no cancer, and two with low-grade cancer; two participants with no cancer had unknown prior biopsy; one participant with no cancer and one with high-grade cancer had unknown digital rectal exam. Family history was unknown for 29 participants with no cancer, 11 with low-grade cancer, and 15 with high-grade cancer.
Older age and African-American race were statistically significantly associated with higher PCA3 values (p values <0.001 and <0.001, respectively). Higher values of PCA3 was associated with a higher chance of T2:ERG expression (p values <0.001), and among men with expressed T2:ERG, increasing PCA3 scores were associated with increasing T2: ERG values (p value <0.0001). These regressions were combined with PCPTRC prior risks to form the updated PCPTRC incorporating PCA3 and T2:ERG (Fig. 1).
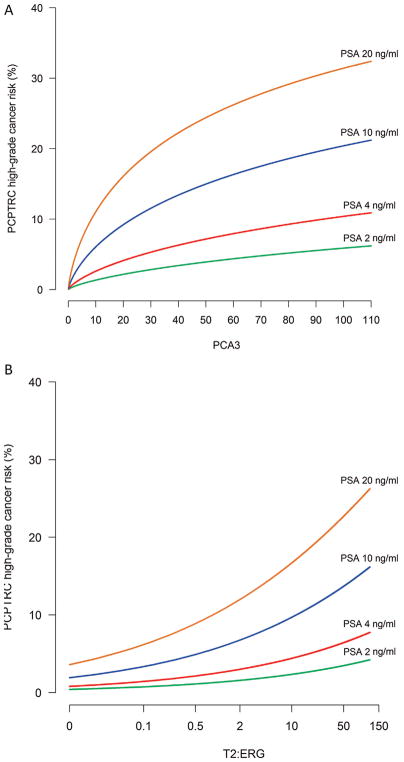
Fig. 1.
Risk of risk of high-grade prostate cancer detection within the context of the Prostate Cancer Prevention Trial Risk Calculator (PCPTR) for various prostate-specific antigen (PSA) levels for a 55-yr-old Caucasian man with no previous biopsy, normal rectal examination, and no family history: (A) according to prostate cancer antigen 3 (PCA3) and (B) according to TMPRSS2:ERG (T2:ERG) for a PCA3 value of 24. The updated risk tool is available at myprostatecancerrisk.com.
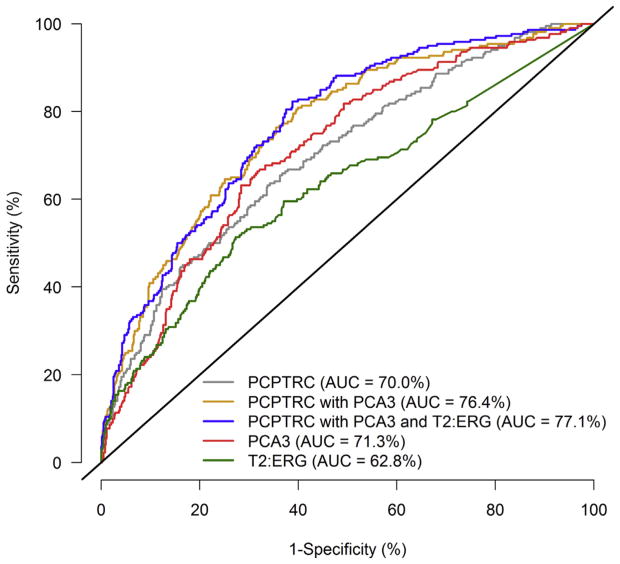
When evaluated in the external EDRN validation cohort of 854 patients, AUCs (95% CIs) for predicting high-grade cancer versus no or low-grade cancer were 70.0% (66.0–74.0%), 76.4% (72.8–80.0%), and 77.1% (73.6–80.6%) for the original PCPTRC risk calculator, the updated PCPTRC with PCA3 included, and the updated PCPTRC with both PCA3 and T2:ERG included, respectively. The PCPTRC incorporating PCA3 obtained higher sensitivities for all specificities 95% or less compared with the original PCPTRC, but there was negligible improvement from additionally incorporating T2:ERG to PCA3 in the PCPTRC (Fig. 2). The marker PCA3 alone with an AUC of 71.3% outperformed the PCPTRC on the validation set, but T2:ERG alone lagged behind with an AUC of 62.8%.
Fig. 2.
Receiver operating characteristic curves for the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) for high-grade disease risk, PCPTRC with prostate cancer antigen 3 (PCA3), and PCPTRC with PCA3 and TMPRSS2:ERG (T2:ERG) evaluated on the external validation set.
AUC = area under the curve.
The task of distinguishing high- from low-grade cancer is more difficult than the task of differentiating high- from low-grade or no cancer considered previously. For this, addition of the new markers diminished the performance of the PCPTRC, with AUCS (95% CIs) evaluated on the validation set of 67.1% (61.7%, 72.4%), 66.2% (60.8%, 71.7%), and 65.1% (59.7%, 70.6%) for the PCPTRC, PCPTRC incorporating PCA3, and PCPTRC incorporating PCA3 and T2:ERG, respectively.
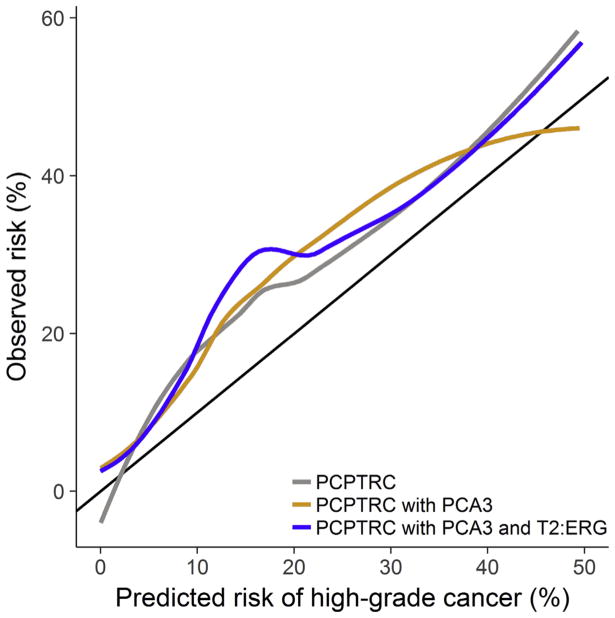
Calibration plots showed no improvement in accuracy for inclusion of PCA3 into the PCPTRC, or with both PCA3 and T2:ERG included compared with the original PCPTRC (Fig. 3). All three versions of the PCPTRC underpredicted the risk in the external EDRN validation cohort, ostensibly due to cohort differences. The EDRN validation cohort comprised of patients from tertiary referral clinics, compared with the Michigan development cohort, which included healthy men from community clinics, and the PCPT cohort, which was a highly screened prevention trial placebo arm of exclusively healthy men. Differences in the cohorts were reflected in their high-grade disease rates, 25.8%, 18.3%, and 3.8%, respectively.
Fig. 3.
Calibration plots for the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) for high-grade disease risk, PCPTRC with prostate cancer antigen 3 (PCA3), and PCPTRC with PCA3 and TMPRSS2:ERG (T2:ERG) evaluated on the external validation set.
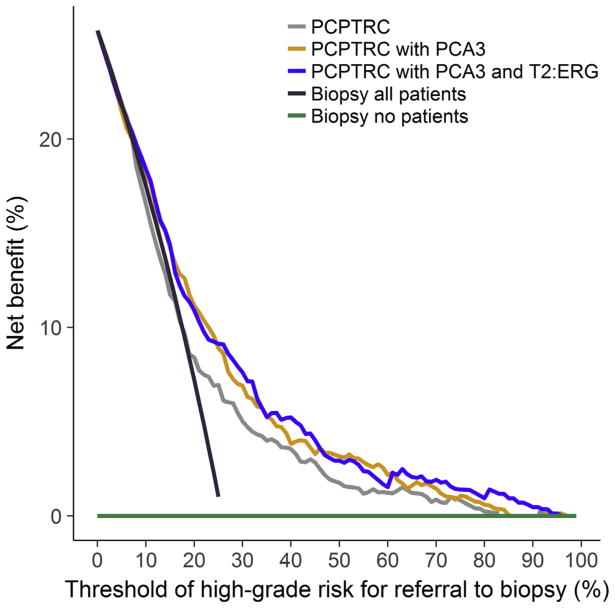
Net benefit curves quantify the weighted benefit to harm ratio of patient-selected thresholds for proceeding to biopsy, and all three versions of the PCPTRC demonstrated benefit compared with a regime that would send no patient to biopsy (Fig. 4). Use of the PCPTRC high-grade disease risk for referral to biopsy would show no benefit over a rule that sent all patients to biopsy for risk thresholds less than 25%, whereas the PCPTRC updated for the urinary markers showed incremental benefit beginning at risk thresholds of 10%. Across all thresholds from 10% to 50%, those for which a urologist would plausibly refer a patient to biopsy, incorporation of the urinary markers marginally increased the net benefit compared with use of the PCPTRC without the markers or compared with a strategy of sending all men to biopsy. But across these thresholds, there was no benefit to adding T2:ERG to PCA3, and this combined with the prior metrics led to the decision not to allow incorporation of T2: ERG without PCA3 into the PCPTRC. It should be noted that cost-to-benefit ratios for use of any tool are an individual decision and that the net benefit curves are not accompanied by 95% CIs, which could show that the differences are not statistically significant. While the independent value of T2:ERG was limited in this validation study, further validation is necessary to determine its potential, hence its inclusion in the online risk tool.
Fig. 4.
Decision curves for the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) for high-grade disease risk, PCPTRC with prostate cancer antigen 3 (PCA3), PCPTRC with PCA3 and TMPRSS2:ERG (T2:ERG) compared with a program that would refer all patients to biopsy to none (no) evaluated on the external validation set.
4. Discussion
Building on the established six risk factors for prostate cancer that are ubiquitously accessed in the clinic, the PCPTRC has become one of the most commonly used prostate cancer risk calculators for patients and their doctors. It currently is the only online prostate cancer risk tool that distinguishes the prediction of high- from low-grade disease. The PCPTRC has previously been updated to include other new markers that emerged in practice after the PCPT concluded, including serum percent free PSA, detailed family cancer history and prostate cancer associated genotypes [5,9,14]. After previous studies have detailed the independent predictive value of the urinary markers PCA3 and T2:ERG to the standard risk factors used for prostate biopsy, this study becomes the first to report on its differential value for predicting high-versus low-grade disease and to incorporate it into the online PCPTRC [10–12].
Addition of urinary biomarkers to the established risk factors will now allow physicians to entertain questions regarding how they help predict biopsy outcomes. For example, assume that a 58-yr-old White man with no family history, normal exam, no previous biopsies, and a PSA of 4.2 ng/ml enters the office. His current risk of low-grade cancer is 17% and high-grade cancer is 5%. A discussion of the risks and benefits of prostate biopsy ensues using the updated American Urological Association White Paper on prostate biopsy complications [15]. The patient is apprehensive and is wondering if any other testing could be performed; this could then prompt the prostate cancer biomarker discussion. With the incorporation of PCA3 and T2:ERG, the patient could now be provided additional information impacting his prostate cancer risk. Another potential way to use the updated calculator is in the context of considering the amount of risk the patient is willing to tolerate prior to changing their decision. For example, the physician could put examples of abnormal values for PCA3 (>35) in the calculator and view the change in prostate cancer risk. For example, a urologist could enter the value of a PCA3 score of 50 and the prostate cancer risk for our example patient would increase to 26% for low-grade and 8% for high-grade prostate cancer. This new information could lead to guidance regarding usefulness of additional biomarker testing in a patient or provide a means of how to interpret additional data in a personalized, patient-centric manner.
There are several limitations to the procedure used in this study to update the PCPTRC. Because PCA3 and T2:ERG were not measured in the PCPT, the approach for updating the PCPTRC to include them involved measurement on an external cohort comprising different patients, clinical follow-up, and biopsy techniques. This approach to binding information from different patient cohorts is clearly inferior to those building single models on single cohorts as was performed on the original PCPTRC. Single cohorts containing novel markers are typically limited in size, whereas the approach used in this study builds on the large PCPT cohort of 6664 biopsies combined with 1225 biopsies of the Michigan cohort. Biases due to differences in the two cohorts may arise when integrating them to form a single risk model. For example, to update the risk tool the Michigan cohort used primarily 12 core biopsies compared with only six used in the PCPT, and an increase in the number of cores has been associated with increased chance of detecting prostate cancer and high-grade disease [16]. For this reason, external validation on an independent EDRN validation set was performed, showing proof of principle. As has been the case for the PCPTRC since its inception, the update incorporating the novel urinary markers has been posted online to facilitate further external validation in addition to use by patients and their clinicians.
A limitation of the online PCPTRC is that while it now incorporates several new markers, including percent free PSA, 30 prostate cancer genetic markers, detailed family history, and now the urinary markers, there is only the option to incorporate one of these sets of markers at a time. This is due to instability concerns that would further mount as data from more than one cohort in addition to the PCPT would be integrated. Future well-sized biopsy cohorts measuring all novel validated markers in addition to the established prostate cancer risk factors are needed in order to access their independent predictive value so that the most parsimonious risk tools measuring only the minimal risk factors necessary are developed in a coherent manner.
Medical resonance imaging, either before or during the prostate biopsy, is increasingly being implemented in practice, which may generate more accurate predictors of biopsy outcome, as well as lead to more accurate classification of high-versus low-grade disease, both of which could increase the accuracy of risk tools in practice [17].
5. Conclusions
The PCPTRC offers the visual representation of the risk of no cancer, low-grade, and high-grade cancer. The current update provides the next step in the implementation of biomarkers in the evaluation of prostate cancer risk. PCA3 and T2:ERG are now being offered to men but may not be offered in the context of their current risk profile. The calculator now provides more options for men and their physicians to facilitate informed discussion of new prostate cancer biomarkers.
Acknowledgments
Funding/Support: Funded by U01 CA086402, P30 CA054174, U01 CA113913, U01 CA214170, and R01 CA179115. The work of Dr. Liss was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Prostate Cancer Research Program under Award Number W81XWH-15-1-0441.
Footnotes
role of the sponsor: Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.
Author contributions: Donna P. Ankerst had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Ankerst, Goros, Gelfond, Feng, Sanda, Thompson, Leach, Liss.
Acquisition of data: Ankerst, Goros, Tomlins, Patil, Wei, Sanda, Gelfond, Thompson, Leach, Liss.
Analysis and interpretation of data: Ankerst, Goros, Patil, Feng, Sanda, Gelfond, Leach, Liss.
Drafting of the manuscript: Ankerst, Goros, Sanda, Thompson, Leach, Liss.
Critical revision of the manuscript for important intellectual content: Ankerst, Goros, Tomlins, Patil, Feng, Wei, Sanda, Gelfond, Thompson, Leach, Liss.
Statistical analysis: Ankerst, Goros, Gelfond.
Obtaining funding: Ankerst, Tomlins, Thompson, Liss.
Administrative, technical, or material support: Ankerst, Thompson, Liss.
Supervision: Ankerst, Thompson, Liss.
Other: None.
Financial disclosures: Donna P. Ankerst and Michael A. Liss certify that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: The University of Michigan has been issued a patent on the detection of ETS gene fusions in prostate cancer, on which Tomlins is listed as a coinventor. The University of Michigan licensed the diagnostic field of use to Hologic/Gen-Probe, Inc., which sublicensed some rights to Ventana Medical Systems. Tomlins has received honoraria from, and served as a consultant to Ventana Medical Systems.
References
- 1.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–34. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 2.Ankerst DP, Boeck A, Freedland SJ, et al. Evaluating the PCPT risk calculator in ten international biopsy cohorts: results from the Prostate Biopsy Collaborative Group. World J Urol. 2012;30:181–7. doi: 10.1007/s00345-011-0818-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auffenberg GB, Merdan S, Miller DC, et al. Evaluation of prostate cancer risk calculators for shared decision making across diverse urology practices in Michigan. Urology. 2017;104:137–42. doi: 10.1016/j.urology.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Poyet C, Nieboer D, Bhindi B, et al. Prostate cancer risk prediction using the novel versions of the European Randomised Study for Screening of Prostate Cancer (ERSPC) and Prostate Cancer Prevention Trial (PCPT) risk calculators: independent validation and comparison in a contemporary European cohort. BJU Int. 2016;117:401–8. doi: 10.1111/bju.13314. [DOI] [PubMed] [Google Scholar]
- 5.Ankerst DP, Hoefler J, Bock S, et al. Prostate Cancer Prevention Trial risk calculator 2.0 for the prediction of low- vs high-grade prostate cancer. Urology. 2014;83:1362–7. doi: 10.1016/j.urology.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson IM, Jr, Leach RJ, Ankerst DP. Focusing PSA testing on detection of high-risk prostate cancers by incorporating patient preferences into decision making. JAMA. 2014;312:995–6. doi: 10.1001/jama.2014.9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roddam AW, Duffy MJ, Hamdy FC, et al. Use of prostate-specific antigen (PSA) isoforms for the detection of prostate cancer in men with a PSA level of 2–10 ng/ml: systematic review and meta-analysis. Eur Urol. 2005;48:386–99. doi: 10.1016/j.eururo.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Kim ST, Cheng Y, Hsu FC, et al. Prostate cancer risk-associated variants reported from genomewide association studies: meta-analysis and their contribution to genetic variation. Prostate. 2010;70:1729–38. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grill S, Fallah M, Leach RJ, et al. A simple-to-use method incorporating genomic markers into prostate cancer risk prediction tools facilitated future validation. J Clin Epidemiol. 2015;68:563–73. doi: 10.1016/j.jclinepi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016;70:45–53. doi: 10.1016/j.eururo.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanda MG, Feng Z, Howard DH, et al. Association between combined TMPRSS2:ERG and PCA3 RNA urinary testing and detection of aggressive prostate cancer. JAMA Oncol. 2017;3:1085–93. doi: 10.1001/jamaoncol.2017.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei JT, Feng Z, Partin AW, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol. 2014;32:4066–72. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ankerst DP, Koniarski T, Liang Y, et al. Updating risk prediction tools: a case study in prostate cancer. Biom J. 2012;54:127–42. doi: 10.1002/bimj.201100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grill S, Fallah M, Leach RJ, et al. Incorporation of detailed family history from the Swedish Family Cancer Database into the PCPT risk calculator. J Urol. 2015;193:460–5. doi: 10.1016/j.juro.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liss MA, Ehdaie B, Loeb S, et al. An update of the American Urological Association white paper on the prevention and treatment of the more common complications related to prostate biopsy. J Urol. doi: 10.1016/j.juro.2017.01.103. In press. [DOI] [PubMed]
- 16.Ankerst DP, Till C, Boeck A, et al. The impact of prostate volume, number of biopsy cores, and American Urological Association symptom score on the sensitivity of cancer detection using the Prostate Cancer Prevention Trial risk calculator. J Urol. 2013;190:70–6. doi: 10.1016/j.juro.2012.12.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA. 2015;313:390–7. doi: 10.1001/jama.2014.17942. [DOI] [PMC free article] [PubMed] [Google Scholar]