Abstract
Breast cancer treatment often includes Doxorubicin as adjuvant as well as neoadjuvant chemotherapy. Despite its cytotoxicity, cells can develop drug resistance to Doxorubicin. Uncovering pathways and mechanisms involved in drug resistance is an urgent and critical aim for breast cancer research oriented to improve treatment efficacy. Here we show that Doxorubicin and other chemotherapeutic drugs induce the expression of ETV7, a transcriptional repressor member of ETS family of transcription factors. The ETV7 expression led to DNAJC15 down-regulation, a co-chaperone protein whose low expression was previously associated with drug resistance in breast and ovarian cancer. There was a corresponding reduction in Doxorubicin sensitivity of MCF7 and MDA-MB-231 breast cancer cells. We identified the binding site for ETV7 within DNAJC15 promoter and we also found that DNA methylation may be a factor in ETV7-mediated DNAJC15 transcriptional repression. These findings of an inverse correlation between ETV7 and DNAJC15 expression in MCF7 cells in terms of Doxorubicin resistance, correlated well with treatment responses of breast cancer patients with recurrent disease, based on our analyses of reported genome-wide expression arrays. Moreover, we demonstrated that ETV7-mediated Doxorubicin-resistance involves increased Doxorubicin efflux via nuclear pumps, which could be rescued in part by DNAJC15 up-regulation. With this study, we propose a novel role for ETV7 in breast cancer, and we identify DNAJC15 as a new target gene responsible for ETV7-mediated Doxorubicin-resistance. A better understanding of the opposing impacts of Doxorubicin could improve the design of combinatorial adjuvant regimens with the aim of avoiding resistance and relapse.
Abbreviations: ETV7, Ets Variant Gene 7; ETS, E26 Transformation-Specific; DNAJC15, DNAJ Heat Shock Protein family (Hsp40) member C15; ABCB1, ATP Binding Cassette subfamily B member 1; DNMT3A, DNA Methyl-Transferase 3A; 5-Aza, 5-Aza-2′-deoxycytidine; TNBC, Triple Negative Breast Cancer; ADR, Adriamycin
Introduction
Chemotherapy is commonly adopted for the pre- and post-surgical treatment of many solid tumors, including breast cancer, and it is still the only therapeutic option for most cases of metastatic spread. Among the drugs used in different regimens, Doxorubicin is often employed and is one of the most effective [1]. This drug is an anthracyclin intercalator that poisons topoisomerase II, thereby causing DNA damage and subsequent cytotoxicity [2]. As for most chemotherapeutic agents, several unwanted side-effects have been reported and, unfortunately, Doxorubicin is also known for late-onset cardiotoxicity determined by a complex cascade of events [3]. Despite the efficacy of Doxorubicin and other chemotherapeutic drugs, cancer cells may develop chemoresistance, resulting in treatment failure and recurrence. Indeed, there are many survival strategies available to cancer cells and some of them can even be activated by chemotherapy itself. Some activation mechanisms can lay the groundwork for unwanted future resistance to treatment, which can be driven or stimulated by the activation of pro-survival or pro-tumorigenic transcription factors, such as NF-κB [4], [5] and FOXM1 [6], [7]. Moreover, Doxorubicin can lead to an increased expression of drug efflux pumps such as MRP1 in breast cancer [8] and to the activation of the anti-apoptotic cascade HER3-PI3K-AKT in ovarian cancer [9].
ETV7 (Ets Variant Gene 7) is a transcription factor belonging to the ETS (E26 transformation-specific) family of transcriptional regulators. Various ETS factors, such as ETS1, ETS2, PU1, FLI1, and ERG, are distinguished for their pro-tumorigenic functions and are involved in chromosomal translocations often associated with Ewing's sarcoma and prostate cancer [10]. ETV7 is a poorly characterized protein that can act as a transcriptional repressor, which presents an 85-amino acid ETS domain responsible for binding a purine-rich GGAA core motif in the regulatory regions of target genes. The protein also has a conserved pointed (PNT) protein–protein interaction domain, required for the formation of homo−/hetero-dimers and oligomers and involved in transcriptional repression [11], [12]. Given the presence of the PNT domain, ETV7 can either self-associate or form heterodimers/oligomers with ETV6/TEL, a highly related ETS family member with tumor suppressor functions that also acts as transcriptional repressor [13]. In contrast, ETV7 is generally acknowledged to be an oncoprotein, and some of its pro-tumorigenic functions result from its ability to directly bind and inhibit ETV6-mediated gene repression [14].
Deregulated high ETV7 expression levels has been linked to hepatocellular carcinoma [15] and to leukemia [10], [16]. Over-expressed ETV7 can also cooperate with Eμ-MYC in promoting lymphomagenesis and blocking Myc-induced apoptosis [17]. Furthermore, ETV7 is able to enhance the Ras-driven transformation in fibroblasts and shows pro-proliferative and anti-differentiation roles observed in myeloid and lymphoid cells [10], [17]. In contrast, ETV7 can act as a tumor suppressor in nasopharyngeal carcinoma through binding SERPINE1 promoter and decreasing its expression [18]. Further, ETV7 down-regulation has been reported in drug-resistant gastric cancer cells [19].
We recently observed in human breast cancer cells that ETV7 can be transcriptionally activated upon Doxorubicin treatment and synergistically induced by the combined treatment with Doxorubicin and TNFα. Among the possible activators of its transcription, we identified tumor suppressor p53 and NFκB (p65) as transcription factors able to directly bind to ETV7 promoter [20].
Interestingly, ETV7 and DNAJC15 expression appear to inversely correlate upon Doxorubicin treatment and also upon interferon gamma expression. ETV7 is recognized as an interferon-stimulated gene, whereas down-regulation of DNAJC15 has been reported in interferon gamma treated macrophages [21]. DNAJC15 plays an intriguing role among the tumor suppressor genes whose repression is associated with tumor aggressiveness and chemoresistance. It belongs to the HSP40/DNAJ family of co-chaperones, mostly involved in helping ATP hydrolysis and thus the activation of the HSP70 chaperone with its roles in protein folding, trafficking, interaction, import and export [22], [23].
DNAJC15 is often hyper-methylated and repressed in malignant pediatric tumors [24], neuroblastoma [25], Wilm's tumor and melanoma [26]. Furthermore, its down-regulation associates with increased drug resistance in ovarian and breast cancer [27], [28]. Using MCF7 breast cancer cells, Hatle and colleagues observed that in Doxorubicin-resistant clones, the low expression of DNAJC15 in the Golgi network was responsible for the degradation of some proteins including the transcription factor c-JUN [29]. Therefore, inhibition of DNAJC15 resulted in increased levels of c-JUN protein, which was ultimately responsible for increased transcription of the multidrug transporter ABCB1/MDR1 [29]. Other studies have reported the localization of DNAJC15 inside the mitochondrial inner membrane where it can control the respiratory chain and thus the production of ROS [30]. Inside the mitochondria, it can also help mitochondrial import of proteins by favoring the ATP hydrolysis of a chaperone member of the TIMP23 translocase [26]. DNAJC15 exerts its tumor suppressor role also by promoting the release of pro-apoptotic molecules through the mitochondrial permeability transition pore complex [31].
In this study, we identify a novel circuitry for Doxorubicin resistance in breast cancer cells where ETV7 acts as a major player. Given the pro-tumorigenic roles of ETV7, its activation upon Doxorubicin treatment represents one of the unwanted side-effects that could possibly unleash a drug resistance mechanism. In particular, ETV7 appears to trigger the activation of a resistance circuitry by directly binding and, therefore, repressing the transcription of some tumor suppressor genes. Specifically, we demonstrate that ETV7 can repress DNAJC15 in a methylation-dependent manner. We propose a novel drug resistance mechanism directly driven by Doxorubicin whereby Doxorubicin itself induces the up-regulation of ETV7 that, in turn, down-regulates DNAJC15 expression giving rise to Doxorubicin resistance in breast cancer cells.
Materials and Methods
Cell Culture Conditions and Treatments
MCF7 cells were obtained from Interlab Cell Line Collection bank (Genoa, Italy). A549 and U2OS cell lines were from ATCC (Manassas, VA, USA), while A375M and MDA-MB-231 cells were a gift from, respectively, Dr. D. Bergamaschi (Centre for Cell Biology and Cutaneous Research, Blizard Institute, Barts and The London School of Medicine & Dentistry, UK) and Prof. A. Provenzani (Laboratory of Genomic Screening, CIBIO, University of Trento, Italy). MCF7 and U2OS cells were grown in DMEM medium supplemented with 10% FBS, 2 mM L-Glutamine and 2 mM of Penicillin/Streptomycin; MDA-MB-231 cells in the same medium with the addition of 1% Non-Essential Amino acids. A549 and A375M cells were cultured in RPMI medium +10% FBS, 2 mM L-Glutamine and 2 mM of Penicillin/Streptomycin. BJ1-hTERT cells (immortalized normal fibroblasts) were obtained from Dr. K. Lobachev (Georgia Institute of Technology, GA, USA) and were grown in MEM medium supplemented with 10% FBS, 2 mM L-Glutamine, 2 mM of Penicillin/Streptomycin and Puromycin. MCF10A cells (immortalized normal mammary epithelial cells) were received from Dr. S. Soddu (Unit of Cellular Networks and Molecular Therapeutic Targets, Regina Elena National Cancer Institute-IRCCS, Rome, Italy) and cultivated in DMEM/F12 1:1 medium supplemented with 5% Horse Serum, 17 ng/ml human epidermal growth factor (hEGF), 10% Mammary Epithelial Growth Supplement (MEGS: 0.4% bovine pituitary extract; 1 μg/ml recombinant human insulin-like growth factor 1; 0.5 μg/ml hydrocortisone, 3 ng/ml hEGF), 100 ng/ml cholera toxin (Sigma-Aldrich, Milan, Italy). When not explicitly stated, cell culture reagents were obtained from Gibco (Life Technologies, Thermo Fisher Scientific, Milan, Italy).
When needed cells were authenticated from the DNA Diagnostic Center (DDC, Fairfield, OH, USA) or Eurofins Genomics (Ebersberg, Germany). Mycoplasma test was done monthly as scheduled by our CIBIO Cell Technology Facility (PlasmoTest™ — Mycoplasma Detection Kit, InvivoGen, Toulouse, France). Experiments were performed within one month from the thawing procedure.
Human lymphocytes were isolated and cultured from the blood of healthy donors as previously described, in accordance with an NIEHS IRB-approved protocol IRB#10-E-0063 and in compliance with the Helsinki Declaration. Prior to participation in the study, subjects were informed of the procedures and potential risks and each signed a statement of informed consent [32].
Doxorubicin (Sigma-Aldrich) was used at the concentration of 1.5 μM (for all cells except for U2OS and BJ1-hTERT cells, which were treated with 0.5 μM Doxorubicin) for 16 hours treatment in the case of qPCR analysis and western blotting and at different concentrations for 72 hours for MTT viability assays.
Twenty-four-hour treatments were performed with the following drugs and concentrations: Etoposide 50 μM (Enzo Life Science, Rome, Italy), Nutlin-3a 10 μM, 5-FluoroUracil (5FU) 375 μM, Camptothecin 0.5 μM, Everolimus 50 nM, Tamoxifen 1 μM, Imatinib 3 μM (Selleckchem, Aurogene, Rome, Italy). Compounds were purchased from Sigma-Aldrich when not specifically indicated.
5-Aza-2′-deoxycytidine (Sigma-Aldrich) treatment was performed for 48 hours at the concentration of 5 μM.
Quercetin and Genistein were purchased from Extrasynthese (Genay, Lyon, France), and treatments were performed for 16 hours at the concentration of 50μM for Quercetin and 30μM for Genistein.
Plasmids
The expression vectors pCMV6-Entry-Empty, pCMV6-Entry-ETV7 and pCMV6-Entry-DNAJC15 C-terminally tagged with DDK-Myc tags were purchased from Origene (Tema Ricerca, Bologna, Italy).
pGL4.26-DNAJC15 reporter was obtained by cloning the promoter region of DNAJC15 (−299 to +512 bp from TSS according to the Eukaryotic Promoter Database, http://epd.vital-it.ch/) amplified with Q5 High Fidelity DNA Polymerase (New England Biolabs, Euroclone, Milan, Italy) and the following primers (Eurofins Genomics): Fw: GCCTCGAGCAGCACAAACTCATTTGAGGG and Rv: GCAAGCTTAGGCGGCCCGGAGACTCAAG. Purified PCR product was inserted into pGL4.26 backbone using Xho I and Hind III restriction endonucleases. Cloning was checked by restriction analysis and direct sequencing (Eurofins Genomics). For site-directed mutagenesis of this vector please refer to the section below. The pRL-SV40 (Promega) vector constitutively expressing the Renilla reniformis luciferase cDNA was used as transfection efficiency control for gene reporter assays.
Generation of Stable pCVM6-Entry-ETV7 and Empty MCF7 and MDA-MB-231 Cells
In order to get MCF7, MDA-MB-231 and U2OS cells stably over-expressing ETV7 and the empty control, cells were seeded in 6-well plates and subsequently transfected for 48 hours with 1 μg of pCMV6-Entry-Empty or pCMV6-Entry-ETV7 (Origene) using Lipofectamine LTX and Plus Reagent (Life Technologies) or FuGene HD (Promega, Milan, Italy) respectively for MCF7 or MDA-MB-231 and U2OS cells. Afterwards, cells were split and Geneticin (Life Technologies) was added at a concentration of 600 and 800 μg/ml respectively for MCF7 or MDA-MB-231 and U2OS cells; each 3 days medium was replaced and after 4 cycles of selection, single cell cloning was performed according to the Corning protocol for cell cloning by Serial dilution in 96 well plates. During the single cell cloning procedure Geneticin concentration was gradually reduced to 300 (MCF7) and 400 μg/ml (MDA-MB-231 and U2OS).
RNA Isolation and RT-qPCR
Total RNA was extracted using the Illustra RNA spin Mini Kit (GE Healthcare, Milan, Italy), converted to cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) and RT-qPCR was performed with 25 ng of template cDNA in 384 wells-plate (BioRad, Milan, Italy) using the Kapa Sybr Fast qPCR Master Mix (Kapa Biosystems, Resnova, Ancona, Italy) and the CFX384 Detection System (BioRad). YWHAZ and B2M genes were used as housekeeping genes to obtain the relative fold change by the ΔΔCt method as previously described [33]. Primer sequences were designed using Primer-BLAST designing tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), checked for specificity and efficiency, and are listed in Supplementary Table 1 (Eurofins Genomics).
Western Blot
Total protein extracts were obtained by lysing the cells in RIPA buffer and proteins were quantified by the BCA method (Pierce, Thermo Fisher Scientific); 20 to 50 μg of protein extracts were loaded on 7.5% and 12% polyacrylamide gels, and western blotting was performed as previously described [34]. Transferred proteins were probed over-night at 4°C with specific antibodies diluted in 1% non-fat skim milk-PBS-T solution: GAPDH (6C5, sc-32,233), ETV7/TEL2 (F-8, sc-376,137X), ETV7/TEL2 (H-88, sc-292,509), Histone H3 (FL-136, sc-10,809), α-Actinin (H-2, sc-17,829), DNMT3A (GTX129125, GeneTex, Prodotti Gianni, Milan, Italy). Antibodies were obtained from Santa Cruz Biotechnologies (Milan, Italy) when not specifically indicated. Detection was performed with ECL Select reagent (GE Healthcare) using a ChemiDoc XRS+ (BioRad) or UVITec Alliance LD2 (UVITec Cambridge, UK) imaging systems.
In order to separate cytoplasmic and nuclear fractions of proteins, MCF7 cells were seeded in p150 dishes and treated with Doxorubicin for either 6 or 16 hours. Cytoplasmic proteins were extracted following the instructions of NE-PER kit (Thermo Fisher Scientific). Alternatively, in order to enrich the nuclear extracts for chromatin-associated proteins, pellets remaining from cytoplasmic extraction were directly resuspended in 1x Loading Buffer and boiled. Approximately, 150 μg of nuclear protein extracts and 50 μg of cytoplasmic protein extracts were loaded on a polyacrylamide gel, blotted and detected as described above. Histone H3 and GAPDH were used respectively as controls for nuclear and for cytoplasmic extracts.
Gene Reporter Assay
24 hours prior to transfection, 7 × 104 MCF7 cells were seeded in 24 well-plate. Cells were transfected with Lipofectamine LTX and Plus Reagent (Thermo Fisher Scientific) along with 250 ng pGL4.26-DNAJC15 reporter, 250 ng pCMV6-Entry-Empty or pCMV6-Entry-ETV7 vectors, and 50 ng pRL-SV40 for each well. After 48 hours, cells were washed once in PBS and lysed in 1X PLB buffer and luciferase activity measurements were performed using the Dual-Luciferase Reporter Assay System (Promega) as previously described [35], [36] and detected using the Infinite M200 plate reader (Tecan, Milan, Italy). Renilla luciferase activity was used as an indicator of transfection efficiency and used to obtain the Relative Light Unit (RLU) measure.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed using GENEART Site-Directed Mutagenesis kit (Life Technologies) according to manufacturer's instructions. In order to mutate ETV7 binding sites within pGL4.26-DNAJC15 (substituting the GGA conserved bases with ATC random sequence), the reporter plasmid was first methylated and then amplified with AccuPrime Pfx DNA Polymerase (InVitrogen, Life Technologies) in a mutagenesis reaction with the following primers (Eurofins Genomics):
BS1_Fw: GGGAAGAAAGGCTGCCCatcAGGGGGTCAGGAAAGC;
BS1_Rv: GCTTTCCTGACCCCCTgatGGGCAGCCTTTCTTCCC;
BS2_Fw: GGTGAGAAGGGTATCTgatGGGAACCTCGCCTTTAA;
BS2_Rv: TTAAAGGCGAGGTTCCCatcAGATACCCTTCTCACC.
Mutagenesis was then followed by an in vitro recombination reaction to enhance efficiency and colony yield. Mutated plasmids (pGL4.26-DNAJC15-BS1 and -BS2) were subsequently transformed into DH5α-T1R E. coli competent cells, which circularize the linear mutated DNA and exploits McrBC endonuclease activity to digest methylated DNA. Complete and correct mutagenesis was verified by direct sequencing (Eurofins Genomics).
Bisulfite-Conversion
Genomic DNA was extracted from MCF7 cells left untreated, treated with Doxorubicin or over-expressing pCMV6-Entry-Empty or -ETV7 vectors. DNA and RNA extractions were obtained from the same samples using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Milan Italy).
Purified DNA was then denaturated and subjected to bisulfite conversion with the EZ DNA Methylation-Lightning™ Kit (Zymo Research, Euroclone) according to manufacturer's recommendations. The resulting product was subsequently PCR amplified and sequenced using the following bisulfite-specific primers (Eurofins Genomics): Fw: TTGGTAGGATTTATTAGTTTTTGTTGG; Rv: CACCCAACTAATCTTTATATTTTTAATAAA.
Doxorubicin Efflux Analysis
1.5 x 104 MDA-MB-231 cells were seeded in a 96 well-plate; the subsequent day, 10 or 20 μM Doxorubicin was added for 3 hours and cells were analyzed with the Operetta High Content Imaging System (Perkin Elmer, Milan, Italy) at CIBIO High Throughput Screening (HTS) Facility exploiting the intrinsic fluorescence of Doxorubicin. By using the Harmony 4.1 PhenoLOGIC software (Perkin Elmer) nuclear and cytoplasmic regions were detected; successively, the relative ratio of nuclear respect to cytoplasmic fraction from Doxorubicin signal was calculated. To measure the Doxorubicin efflux area, the Doxorubicin spot area into the cytoplasm was measured (see Suppl. Figure S2A for details).
Viability Assay
Cells were seeded in a 96 well-plate and treated with different concentrations of Doxorubicin for 72 hours. Medium was removed and wells were washed with 1X PBS to avoid possible reduction effects of the added compound with MTT reagent (Sigma-Aldrich). Ten μl of MTT (5 mg/ml solution in 1X PBS) was added to 100 μl of fresh medium and left in incubation for 3 hours. Afterwards, medium was accurately removed and cells were lysed in 100 μl of DMSO (Sigma-Aldrich), and a colorimetric measure was performed at the Infinite M200 plate reader (Tecan). Viability was calculated as a % ratio of viable cells treated with the indicated drug respect to an untreated control.
Cell Death Analysis
6 × 103 MCF7 cells were seeded in a 96 well-plate; 24 hours after seeding, cells were treated with different concentrations of Doxorubicin; 72 hours after treatment cells were incubated with Hoechst 33,342 2 μg/ml (Life Technologies) for 30 min (to stain nuclei, therefore both viable and dead cells) and Topro-3 0.1 μM (Life Technologies) for 15 minutes (to visualize dead cells). Fluorescent images were obtained with the Operetta High Content Imaging System (Perkin Elmer) at CIBIO HTS Facility. The Topro-3 and Hoechst 33,342 positive objects were detected using the Harmony 4.1 PhenoLOGIC software (Perkin Elmer); subsequently, the relative ratio of Topro-3 positive objects on the total number of objects (Hoechst 33,342 positive) was calculated.
Chromatin Immunoprecipitation Assay
MDA-MB-231-ETV7 and MDA-MB-231-Empty cells were seeded in p150 dishes (four dishes each condition) and ChIP-PCR was performed following a revised version of Myers Lab protocol. Mouse monoclonal anti-ETV7/TEL2 antibody (F-8, sc-376,137X, Santa Cruz Biotechnologies) and normal mouse IgG (sc-2025, Santa Cruz Biotechnologies) were used for immunoprecipitation. Two μl of purified immmunoprecipitated DNA was then used for qPCR analysis and calculation was performed using the ΔCt method in respect to non-immunoprecipitated DNA (% of input) as previously detailed described [37]. A genomic region within GTF2H5 gene was used a negative control. A list of the primers sequences that were used for ChIP-PCR analysis is presented in Supplementary Table 1 (Integrated DNA Technologies, Coralville, IA, USA and Eurofins Genomics).
MCF7 cells were seeded in p150 dishes and transiently transfected with 10 μg of pCMV6-Entry-Empty or -ETV7 vectors using Lipofectamine LTX and Plus Reagent (Thermo Fisher Scientific) for 48 hours. ChIP was performed using the same protocol used for MDA-MB-231 using anti-ETV7/TEL2 (H-88, sc-292,509) and normal rabbit IgG (sc-2027, Santa Cruz Biotechnologies) for immunoprecipitation. qPCR on purified immunoprecipitated DNAs was performed as indicated above.
Co-Immunoprecipitation
MCF7 were seeded in p150 dishes and transiently transfected with pCMV6-Entry-ETV7 as above (Origene). 48 hours post-transfection cells were lysed in CHAPS buffer and then incubated over-night with an anti-ETV7 antibody (H-88, sc-292,509) or normal rabbit IgG (sc-2027) previously bound to Dynabeads protein A magnetic beads (Life Technologies). Beads were then washed and the immunoprecipitated lysate was eluted and loaded on a polyacrylamide gel for SDS-PAGE.
Analysis of Genome-Wide Data
Available expression arrays from our group (GSE24065, Agilent-014850 Whole Human Genome Microarray 4x44K G4112F) were analyzed for the specific genes of interest as previously described [20], [38].
Expression data of MCF7 cells resistant to Adriamycin -MCF7/ADR- (e.g. Doxorubicin) were obtained from Affymetrix Human Genome U133 Plus 2.0 Array platform and downloaded from GEO (GSE76540). Two transcripts for each gene of interest (ETV7, DNAJC15, and ABCB1) were available and expression averages were calculated.
Expression levels of ETV7 and DNAJC15 were obtained from microarray data of Triple Negative Breast Cancer patients who underwent neoadjuvant chemotherapy protocols (GSE43502, Affymetrix Human Genome U133 Plus 2.0 Array). The study included 25 patients (out of 47) showing recurrence.
Statistical Analysis
When appropriate, Student's t-test was applied for statistical significance. We selected throughout the manuscript the two-sample Student's t-test for unequal variance.
Results
ETV7 is Activated by Doxorubicin and other DNA Damaging Drugs in Cancer and Normal Cells
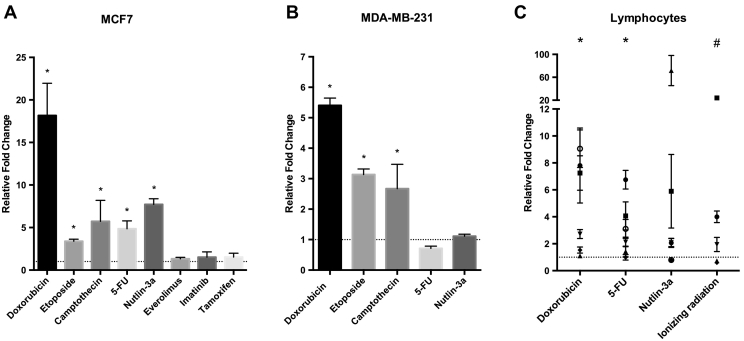
To investigate the differential expression of ETV7 in response to various stimuli in breast cancer cells we tested a panel of cytotoxic drugs in MCF7 cells. We observed a substantial induction of ETV7 expression with many of the treatments, especially DNA damaging drugs, among which Doxorubicin was the most effective inducer of ETV7 expression in comparison with 5FU, Camptothecin, and Etoposide (Figure 1A). The treatment with Nutlin-3a, a p53 specific activator [39] also triggered an increment in ETV7 mRNA levels while Everolimus (mTOR inhibitor), Imatinib (tyrosine kinase inhibitor) and Tamoxifen (estrogen modulator) had no effect (Figure 1A). Moreover, ETV7 transcriptional activation by Doxorubicin in MCF7 cells was reflected by an increase in protein levels in the nuclear compartment (Suppl. Figure S1A), highlighting its role as a transcriptional regulator.
Figure 1.
DNA damaging drugs promote ETV7 transcriptional activation. RT-qPCR analysis of ETV7 expression upon different chemotherapeutics treatment in breast cancer-derived MCF7 (A) and MDA-MB-231 cells (B), and in healthy donor-derived lymphocytes (C). Bars represent average Fold Changes relative to the untreated condition and standard deviations of at least three biological replicates. * = P-value <0.01.
We extended the analysis to the breast cancer cell line MDA-MB-231 and confirmed the induction of ETV7 upon treatments with DNA damaging agents, especially Doxorubicin (Figure 1B). Nevertheless, the levels of ETV7 induction in this cell line were not as high as in MCF7. The reduced level might be explained by the presence of a mutant nonfunctional form of p53 in MDA-MB-231 since p53 is an activator of ETV7 transcription [20]. Doxorubicin treatment also induced ETV7 in other cancer-derived cell lines: lung adenocarcinoma (A549) and osteosarcoma (U2OS) and melanoma (A375M) (Suppl. Figure S1B). Given the activation in the various cancer cell lines, we investigated the ETV7 expression in normal cells. Therefore, we treated lymphocytes obtained from healthy donors and two non-cancerous cell lines (immortalized normal fibroblasts, BJ1-hTERT, and immortalized normal mammary cells, MCF10A). These results along with those from cancer cell lines establish that ETV7 induction is a conserved response to DNA damaging treatments (Figure 1C and Suppl. Figure S1B).
ETV7 Can Promote Resistance to Doxorubicin in Breast Cancer Cells
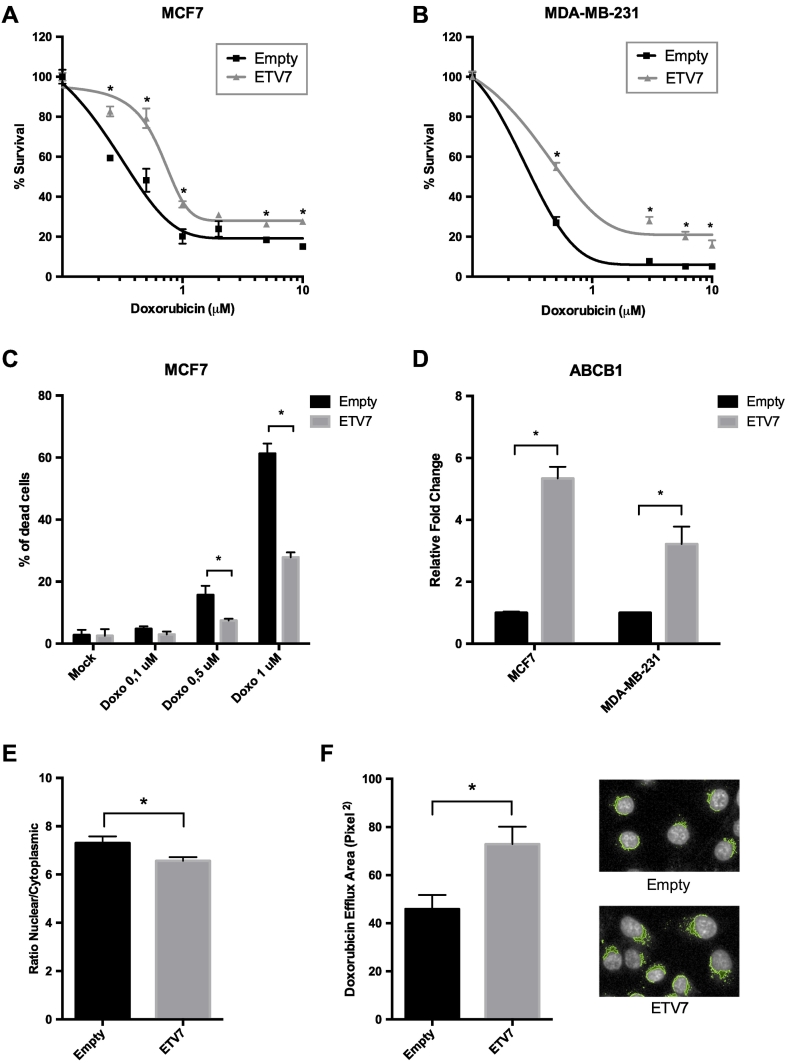
Given the observation that ETV7 is potently activated in response to Doxorubicin treatment, we hypothesized that it may be involved in drug resistance. To test this, we generated stable MCF7 and MDA-MB-231 cell lines over-expressing this transcription factor (Suppl. Figure S2A and B, respectively) and evaluated whether this could influence the survival upon Doxorubicin treatment. Importantly, ETV7 over-expression exerted a protective role against Doxorubicin-induced cell viability in both cell lines (Figure 2A-B). Interestingly, this effect was also visible in non-breast cancer cells as shown for the osteosarcoma U2OS cells stably over-expressing ETV7 (Suppl. Figure S2C-D). The effect of ETV7 over-expression on Doxorubicin-driven cell death was analyzed using the cell-impermeable dye Topro-3 (a representative image is presented in Supplementary Figure S2E). MCF7 cells over-expressing ETV7 resulted in a remarkably reduced sensitivity to Doxorubicin in comparison to cells stably transfected with an empty vector (Figure 2C). Since drug efflux is one of the most common mechanisms responsible for increased chemoresistance [40], [41], we hypothesized that it could be affected by ETV7 expression. We therefore checked for the expression of ABCB1/PgP, an ABC transporter frequently over-expressed in doxorubicin-resistant cells [42], [43], [44], and we observed a significant up-regulation of ABCB1 upon ETV7 over-expression in both MCF7 and MDA-MB-231 cells (Figure 2D). To further verify the aforementioned hypothesis, we monitored the nuclear efflux of Doxorubicin exploiting its light emission property in the ETV7 over-expressing MDA-MB-231 cells relative to their empty-vector counterpart. By measuring the ratio of nuclear to cytoplasmic Doxorubicin and the area of Doxorubicin efflux from the nuclei, we found a statistically significant decrease of nuclear Doxorubicin in the MDA-MB-231 cells over-expressing ETV7 that corresponded to an increased nuclear efflux of Doxorubicin (Figure 2E-F and Suppl. Figure S2F, showing details regarding the selection of nuclear and cytoplasmic regions).
Figure 2.
ETV7 can trigger breast cancer resistance to Doxorubicin. A-B) MTT Assays for survival analyses upon Doxorubicin treatment in MCF7 (A) and in MDA-MB-231 (B) cells over-expressing ETV7 with respect to their empty control. C) Cell death analysis on Doxorubicin-treated (three different doses) MCF7 cells over-expressing ETV7 in comparison to the ones stably transfected with an empty vector. Percentage of dead cells was obtained through fluorescence studies (at Operetta Perkin Elmer) calculated as a ratio between the amount of Topro-3 positive cells (dead cells) and the total number of cells (Hoechst 33,342 positive cells). D) RT-qPCR analysis of ABCB1 expression in MCF7 and MDA-MB-231 cells over-expressing ETV7. E) Analysis of the ratio between the nuclear and cytoplasmic intensity of Doxorubicin in MDA-MB-231 cells over-expressing ETV7 compared with their empty control, performed through Operetta Perkin Elmer Software. F) Analysis of the cytoplasmic area of Doxorubicin efflux in MDA-MB-231 cells over-expressing ETV7 in comparison to their empty counterpart. Images are reporting one representative analyzed by Operetta PerkinElmer Software. Experiments are done in quadruplicate. * = P-value <0.01.
DNAJC15 is a Good Target for ETV7-Mediated Drug Resistance
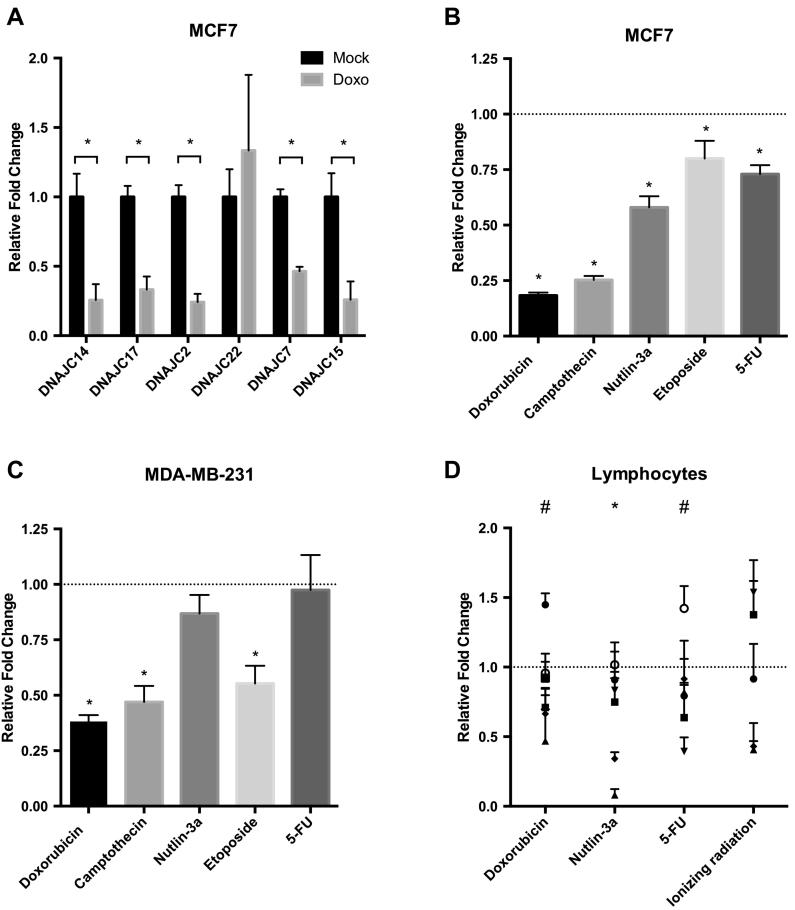
To further understand how ETV7, as a transcriptional repressor, could influence drug resistance, we searched for its putative targets by restricting the analysis to genes whose silencing is already known to be involved in Doxorubicin resistance in breast cancer cells. In particular, we considered a list of six genes (BRCA1, ESR1, DNAJC15, CDH1, RAB6C and SULF2) whose hyper-methylation correlates with Doxorubicin resistance in breast cancer (Table 1 from Boettcher et al., 2010 [45] and available at the Archive of Functional Genomics Data, accession number #E-MEXP-2698, using the ArrayExpress tool). To restrict the search to the most promising ETV7 targets, we analyzed the expression of this group of genes in microarray data that we previously described with Doxorubicin treated MCF7 cells available (GSE24065, Gene Expression Omnibus, GEO, NCBI [20], [38]). Given that Doxorubicin potently activated ETV7 expression, we expected to observe significant down-regulation of its targets upon the same treatment condition. Out of the six genes, three of them -DNAJC15, BRCA1, and ESR1- displayed a strong down-regulation pattern upon Doxorubicin treatment (Suppl. Figure S3A). No significant effects were observed for CDH1 and RAB6C, while SULF2 was induced after Doxorubicin treatment. Moreover, most DNAJC family members were repressed upon Doxorubicin in MCF7 cells, based on the previously mentioned microarray data (Suppl. Figure S3B). Therefore, we validated some of the highly down-regulated members of DNAJC family with RT-qPCR experiments in Doxorubicin-treated MCF7 cells and confirmed the repression of DNAJC2, C7, C14, C15, and C17 in response to Doxorubicin treatment (Figure 3A). Furthermore, DNAJC15 has already been reported to be involved in the negative regulation of ABCB1 transcription, thereby potentially explaining the ETV7-mediated ABCB1 up-regulation and, at least partially, the drug resistance mechanism associated with ETV7 [29]. We decided to focus our attention on DNAJC15 as a putative mediator of the ETV7-dependent Doxorubicin resistance. We extended the analysis of DNAJC15 repression to other DNA damaging agents in MCF7, MDA-MB-231 cells and in lymphocytes (Figure 3B, C, and D, respectively), and verified DNAJC15 down-regulation in response to most of these agents.
Figure 3.
DNAJC15 expression is repressed by DNA damaging drugs. A) RT-qPCR analysis in MCF7 cells of the expression of a selected group of DNAJC family members repressed upon Doxorubicin treatment according to microarray analysis (GSE24065). B-C) Expression analysis of DNAJC15 mRNA upon different chemotherapeutics treatment in breast cancer-derived MCF7 (B) and MDA-MB-231 cells (C), and in healthy donor-derived lymphocytes (D). Bars represent averages Fold Changes relative to the untreated condition of at least three biological replicates and standard deviations. * = P-value <0.01.
ETV7 Transcriptionally Regulates DNAJC15 Expression
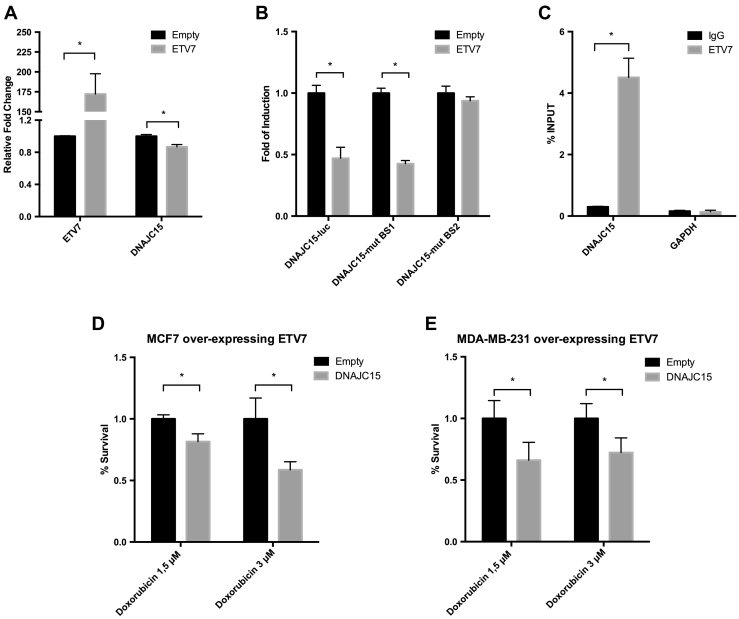
Since we observed that ETV7 and DNAJC15 expression were inversely correlated in response to several stimuli and given the presence of two putative ETV7 binding sites in the DNAJC15 promoter, we investigated the possibility of direct ETV7 influence on DNAJC15 expression. First, we demonstrated that the modulation of ETV7 expression inversely affected the mRNA levels of DNAJC15. Specifically, ETV7 over-expression led to a small but significant, repression of DNAJC15 both in MCF7 (Figure 4A) and MDA-MB-231 (Suppl. Figure S4A) cells.
Figure 4.
ETV7 can repress DNAJC15 expression at the transcriptional level and DNAJC15 over-expression can rescue Doxorubicin sensitivity. A) RT-qPCR analysis of ETV7 and DNAJC15 expression in MCF7 cells transfected with pCMV6-Entry-Empty or pCMV6-Entry-ETV7 plasmids. B) Gene reporter assay of MCF7 cells transiently over-expressing pCMV6-Entry-Empty or pCMV6-Entry-ETV7 along with pGL4.26-DNAJC15 reporter plasmid or the pGL4.26-DNAJC15-BS1 or -BS2 plasmids mutated in the putative ETV7 binding sites. Data are normalized using pRL-SV40 and are shown as fold of induction relative to the empty control. C) ChIP-PCR of DNAJC15 and GAPDH (control) promoter regions in MCF7 transfected with pCMV6-ETV7. Shown is the percentage of enrichment of ETV7 or control (IgG) bound to DNAJC15 promoter region in respect to INPUT DNA. For panels A-C, bars represent averages and standard deviations of at least three biological replicates. D-E) MTT Assay of ETV7-over-expressing MCF7 (D) and MDA-MB-231 (E) cells transiently transfected with pCMV6-Entry-Empty or pCMV6-Entry-DNAJC15 plasmids and treated with Doxorubicin 1.5 μM or 3 μM for 72 hours. Experiments are done in quadruplicate. * = P-value <0.01.
In order to assess whether transcriptional repression was associated with ETV7 binding to DNAJC15, we cloned a region of the DNAJC15 promoter containing two putative binding sites for ETV7 into a pGL4.26 luciferase reporter vector. We found that ETV7 over-expression in MCF7 cells was able to decrease the expression of the luciferase reporter gene under the control of the DNAJC15 promoter (Figure 4B). We then performed site-directed mutagenesis to mutate the most conserved bases within the two putative ETV7-binding sites into the reporter vector in order to demonstrate the contribution of these two binding sites in DNAJC15 repression. The mutation of the binding site 1 (BS1 – chr.13:43′597’329–43′597’335) did not affect the ETV7-mediated down-regulation of luciferase activity. However, disruption of binding site 2 (BS2 – chr.13:43′597’624–43′597’632) prevented repression of the luciferase reporter induced by ETV7, demonstrating the importance of ETV7 binding to this site in the repression of DNAJC15 (Figure 4B).
Furthermore, we were able to demonstrate with chromatin immunoprecipitation the direct binding of ETV7 to the DNAJC15 promoter region (BS2) in both MCF7 and MDA-MB-231 cells (Figure 4C and Suppl. Figure S4B, respectively). In particular, in MDA-MB-231 cells over-expressing ETV7, the binding to the DNAJC15 promoter was markedly stimulated by Doxorubicin treatment. In order to better clarify this effect, we analyzed the distribution of ETV7 protein within the nucleus and found that upon Doxorubicin treatment the ETV7 protein was strongly enriched in the nuclear fraction especially in chromatin-associated structures (Suppl. Figure S4C).
DNAJC15 Over-Expression Partially Rescues ETV7-Mediated Drug Resistance
To address the idea that ETV7-mediated drug resistance is, at least partially, dependent on the repression of DNAJC15, we over-expressed DNAJC15 in MCF7 and MDA-MB-231 cells stably over-expressing ETV7 and analyzed cellular viability upon Doxorubicin treatment. Cells over-expressing DNAJC15 became more sensitive to Doxorubicin-mediated cell death, thus confirming this pathway as a mechanism exploited by ETV7 for drug resistance (Figure 4D-E). Furthermore, DNAJC15 over-expression was also able to down-regulate ABCB1 expression in MCF7 as well as MDA-MB-231 cells over-expressing ETV7, in accord with its reported negative role on ABCB1 levels (Suppl. Figure S5A-B).
ETV7 Represses DNAJC15 Expression by DNA Methylation
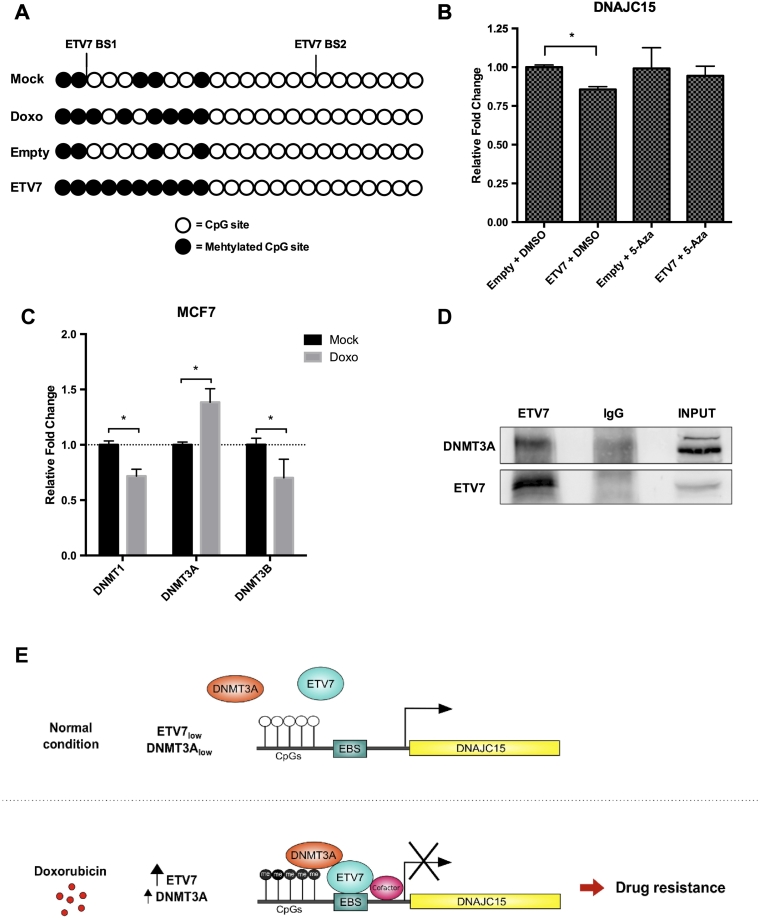
DNAJC15 is recognized to be a methylation-controlled gene, and its methylation-induced down-regulation has been associated with chemoresistance [29]. We investigated whether ETV7-mediated repression was dependent on DNA methylation. The methylation of CpGs in the promoter of DNAJC15 in MCF7 cells following ETV7 over-expression or Doxorubicin treatment was determined by bisulfite-conversion of genomic DNA followed by PCR and direct sequencing. In response to Doxorubicin, the promoter of DNAJC15 showed increased methylation of CpGs, which was even more evident upon ETV7 over-expression, as shown in Figure 5A. Moreover, ETV7-mediated effects on DNAJC15 transcript levels were abolished by treatment with the DNA methyltransferase (DNMT) inhibitor 5-Aza-2′-deoxycytidine (5-Aza) (Figure 5B), demonstrating that ETV7 repression of DNAJC15 expression is indeed methylation-dependent.
Figure 5.
ETV7 can regulate DNAJC15 expression in a methylation-dependent manner. A) Methylation status of CpGs within DNAJC15 promoter analyzed by bisulfite conversion followed by PCR and direct sequencing in MCF7 untreated, treated with Doxorubicin for 16 hours or transfected with pCMV6-Entry-Empty or pCMV6-Entry-ETV7 plasmids. Methylated CpGs are shown as black dots, whereas unmethylated CpGs as white dots. B) RT-qPCR analysis of DNAJC15 expression in MCF7 transfected with pCMV6-Entry-Empty or pCMV6-Entry-ETV7 and treated with DMSO or 5-Aza-2′-deoxycytidine for 48 hours. C) RT-qPCR analysis of DNMT1, DNMT3A and DNMT3B expression in MCF7 treated with Doxorubicin for 16 hours. D) Western blot of DNMT3A and ETV7 on the immunoprecipitation with an antibody against ETV7 or normal IgG as control and on INPUT lysates in MCF7 transfected with pCMV6-Entry-ETV7 plasmid. * = P-value <0.01. E) A graphical model for ETV7-dependent Doxorubicin resistance in breast cancer cells. In normal conditions, ETV7 and DNMT3A are maintained at basal levels (particularly low in case of ETV7) and DNAJC15 can be regularly expressed. In response to Doxorubicin treatment, ETV7 levels get elevated and DNMT3A slightly increases as well. Induced ETV7 can then accumulate into the nucleus and specifically to chromatin-enriched regions. In the nucleus, ETV7 recruits DNMT3A (through direct interactions with putative additional cofactors) on target DNA (DNAJC15 promoter in this case) that in turn it is responsible for the methylation of CpGs. This will result in DNAJC15 repression and ultimately will lead to chemoresistance, partly through the exclusion of the drug from the nucleus. EBS: ETV7 Binding Site. Methylated CpGs are shown as filled circles, whereas unmethylated CpGs as empty circles.
Given that DNMTs play key roles in Doxorubicin resistance as demonstrated for Adriamycin-resistant MCF7 cells [46], we hypothesized a possible direct interaction between ETV7 and DNMTs mediating the methylation and subsequent repression of the DNAJC15 promoter. Analysis of the expression of the DNMT1, DNMT3A and DNMT3B genes in our microarray data from Doxorubicin-treated MCF7 cells (GSE24065 [20], [38]) (Suppl. Figure S6A) and validation by RT-qPCR (Figure 5C), revealed the up-regulation of only DNMT3A among these DNMTs. Conversely, both DNMT1 and DNMT3B were down-regulated in response to the treatment. Moreover, a similar trend could be observed for DNMTs expression in response to ETV7 over-expression in MCF7 cells, even if the only statistically significant alteration in expression was for DNMT1 (Suppl. Figure S6B). To test the putative interaction of ETV7 with DNMT3A as a candidate mediator of DNAJC15 repression, we performed immunoprecipitation of ETV7 and found the direct interaction of ETV7 with DNMT3A in MCF7 cells transiently over-expressing ETV7 (Figure 5D).
ETV7-DNAJC15 Clinical Relevance and Possible Therapeutic Strategy
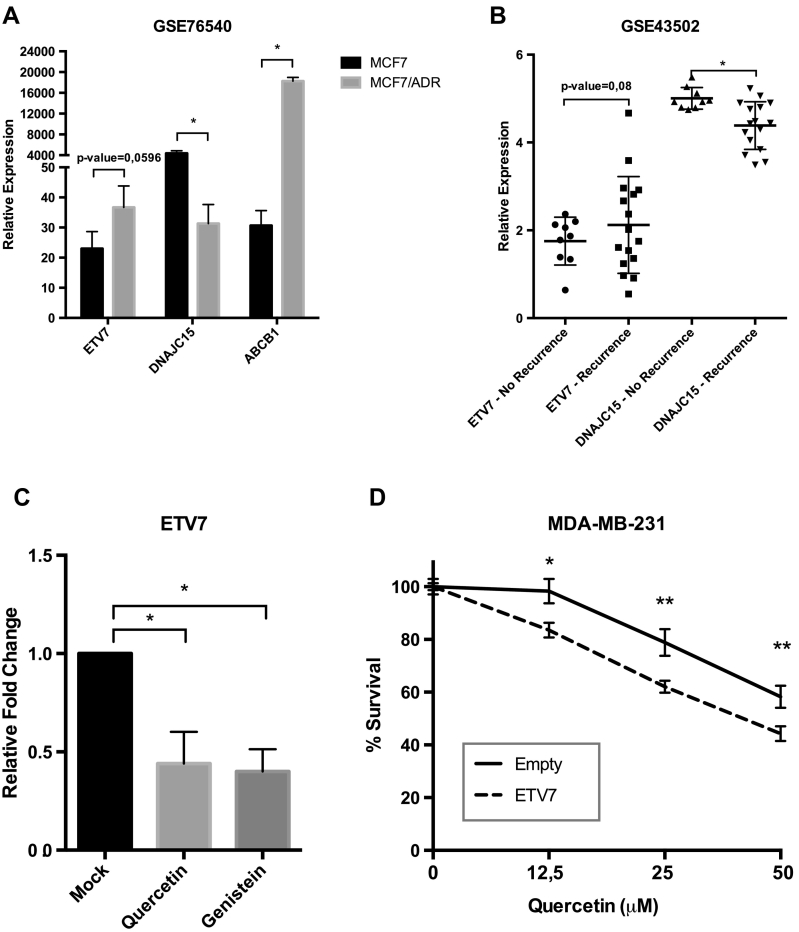
We confirmed a dramatic decrease in DNAJC15 and a corresponding increase in ETV7 expression in reported microarray analysis performed with Adriamycin (i.e. Doxorubicin) resistant MCF7 cells (MCF7/ADR, GSE76540, [47]) as shown in Figure 6A. Moreover, this effect was associated with a large increase in ABCB1 expression in MCF7/ADR cells, an observation consistent with what is observed in MCF7 and MDA-MB-231 cells over-expressing ETV7 (Suppl. Figure S2F-G).
Figure 6.
ETV7 and DNAJC15 levels inversely correlate with clinical status of breast cancer patients and ETV7 targeting could be exploited pharmacologically. A) ETV7, DNAJC15 and ABCB1 expression levels from microarray data (GSE76540) of MCF7 cells resistant to Adriamycin -MCF7/ADR- (e.g. Doxorubicin). Presented are the averages and standard deviations of at least three biological replicates. B) ETV7 and DNAJC15 expression levels from microarray data of Triple Negative Breast Cancer patients treated with neoadjuvant chemotherapy who were showing recurrence or not for the disease (GSE43502). C) ETV7 expression levels measured by RT-qPCR from MDA-MB-231 cells untreated (Mock) or treated with Quercetin 50μM or Genistein 30μM for 16 hours. Bars represent averages and standard deviations of at least three biological replicates. D) MTT assay in MDA-MB-231 cells over-expressing ETV7 or its empty vector and treated with increasing concentration of Quercetin. Experiments are done in quadruplicate. * = P-value <0.01.
To address possible clinical relationships between ETV7, DNAJC15 and Doxorubicin treatment, we evaluated data obtained from 25 chemoresistant samples among 47 neoadjuvant chemotherapy-treated triple negative breast cancer (TNBC) patients (GSE43502, [48]). We found an inverse correlation between the expression levels of ETV7 and DNAJC15 in TNBC patients associated with recurrence. Specifically, despite not stringently significant, it is visible an increase in ETV7 expression in patients with a recurrent disease that correlated with a remarkable decrease in DNAJC15 expression. These data indicate that ETV7-mediated repression of DNAJC15 could be linked to a worse prognosis in breast cancer patients (Figure 6B).
Given that the over-expression of a particular gene is still a challenging approach for therapeutic purposes, we tried to overcome ETV7-mediated drug resistance using Quercetin, a flavonoid recently shown to both increase therapeutic efficacy of Doxorubicin [49], [50], [51] and to reduce its cardiotoxicity [52], [53], and the isoflavone Genistein, which can inhibit topoisomerase II [54] and DNMTs [55] or act as a phytoestrogen [35]. The sensitizing action of flavonoids has not been fully elucidated yet, but it seems to involve the MDR transporter action. Surprisingly, we noticed that both flavonoids Quercetin and Genistein were able to reduce ETV7 expression in MDA-MB-231 cells, thereby suggesting a novel mechanism of sensitization for cancer cells (Figure 6C). Notably, MDA-MB-231 cells that over-express ETV7 were more sensitive to Quercetin relative to the empty counterpart, thereby unveiling a mechanism that could represent a promising target for ETV7-mediated resistance in cancer cells (Figure 6D).
Discussion
ETV7 has been recognized in the literature as an oncoprotein for blood cancers but its role in solid cancers is still poorly studied [10], [17]. In this work, we showed that ETV7 is activated in response to different DNA damaging agents in breast cancer cells, but its expression is not affected by other types of anti-cancer treatments such as estrogen antagonists, tyrosine kinase or mTOR inhibitors (Figure 1). We observed that this transcriptional activation is conserved in different cancer cell types and normal cells including lymphocytes obtained from healthy donors, thus highlighting its biological relevance. Moreover, we have demonstrated that ETV7 can directly promote resistance of breast cancer cells to standard-of-care chemotherapy, i.e. Doxorubicin (Figure 2). The ETV7-dependent mechanism of chemoresistance exploited by breast cancer cells involves the direct efflux of Doxorubicin from the nucleus of cells over-expressing ETV7 (Figure 2). This observation led us to hypothesize that the effect can be driven by membrane-associated transporters and, interestingly, we found that cells over-expressing ETV7 showed higher expression levels of ABCB1, a member of the family of ABC transporters (Figure 2). Despite being mainly expressed on the plasma membranes, ABCB1 protein has often been detected on nuclear membranes and Golgi compartments [56], possibly mediating the phenomenon of resistance to Doxorubicin observed in breast cancer cells in this study.
As a transcription factor, ETV7 can influence the expression of a complex range of targets that may result in the observed increased survival. Among the various possible ETV7 targets, we proposed DNAJC15, a co-chaperone member of the HSP40 family, reported to affect ABCB1 expression and anti-cancer drug efflux [29]. DNAJC15 has already been reported to be frequently hyper-methylated and repressed in breast cancer cells resistant to therapy [27,28]. However, which direct players were causing its transcriptional repression in breast cancer was not known. We confirmed the DNAJC15 repression triggered by Doxorubicin involves the direct binding of ETV7 on the DNAJC15 promoter. We were also able to identify the precise promoter region that ETV7 uses to reduce the expression of DNAJC15 located at +377 bp from the TSS (Figure 3, Figure 4). Given reports of DNAJC15 hyper-methylation and decreased expression in cancer [28], we investigated whether ETV7 could modulate DNAJC15 expression through this mechanism. By mapping the CpG islands that are methylated in response to Doxorubicin and ETV7 over-expression in breast cancer cells, we demonstrated that ETV7-dependent DNAJC15 transcriptional repression is methylation-mediated (Figure 5). We speculate that this may be achieved through the direct recruitment of the DNA methyltransferase DNMT3A on chromatin mediated by ETV7 given our observation of physical interaction between the two proteins (Figure 5).
In Figure 5E, we propose a model for the novel mechanism of Doxorubicin resistance in breast cancer cells that includes a pivotal role for ETV7, which is directly activated by this chemotherapeutic drug. The induced ETV7 acts as a direct negative regulator of DNAJC15 expression through the DNA methylation of the promoter region via DNMT3A. DNAJC15 repression leads to the efflux of the drug from the nucleus, a process possibly driven by the loss of the DNAJC15-dependent repression of ABCB1.
A better knowledge of the transcriptional repressors that impact DNAJC15 expression could help inform clinical treatment strategies in order to avoid or minimize the activation of one of its direct repressors such as ETV7. A combinatorial treatment could disrupt this resistance circuitry driven by ETV7. Based on our findings, we suggest considering the combined treatment of Doxorubicin with Quercetin as a therapeutic strategy, given its protective role against Doxorubicin cardiotoxicity and its negative action on ETV7 expression (Figure 6).
Taken collectively, our results uncovered a novel molecular mechanism that underlies the resistance to a standard-of-care treatment for breast cancer (Doxorubicin), providing insights on the players that take part in this process: ETV7, DNMT3A, and DNAJC15 all of which have the potential for pharmacological targeting. Moreover, it is worth noting that our findings provide the first evidence for a role of ETV7 expression and function in the resistance to Doxorubicin in breast cancer cells. We propose that further analyses on additional ETV7 targets could help investigations for novel breast cancer prognostic markers.
In general, given the complex universe beyond chemoresistance in cancer cells, it is of paramount importance to search for downstream master regulators like transcription factors. Despite the difficulties beyond their targeting, understanding how to restrict their activation and activity could provide a more promising therapeutic strategy than simply targeting a specific resistance effector.
Conclusions
With this study, we have uncovered a novel mechanism of resistance to Doxorubicin where ETV7 plays a major role. We propose a novel role for ETV7 in breast cancer, and we identify DNAJC15 as a new target gene responsible for ETV7-mediated Doxorubicin-resistance. The described molecular mechanism involves the ETV7-dependent repression of DNAJC15 through promoter methylation, a process that results in the increased expression of ABCB1. Overall, these findings can help to better understand how resistance to conventional chemotherapy can be hindered and possibly tackled pharmacologically.
Acknowledgements
We thank Lia Pinto, Michael Pancher, Dr. Valentina Adami and CIBIO High Throughput Screening Facility for technical assistance. We are also thankful to Dr. Bergamaschi, Prof. Provenzani, Dr. Lobachev, and Dr. Soddu for sharing cell lines. We appreciated Prof. Alberto Inga's past and present group members for sharing reagents and helpful discussions. We thank the Clinical Research Unit of NIEHS for providing human lymphocytes from healthy donors. This work was supported by CIBIO Start-up funds (to YC) and by Intramural NIH Research Program NIEHS Z01-ES065079 (to MAR).
Footnotes
Disclosure of Potential Conflicts of Interest: the authors declare no potential conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.06.008.
Contributor Information
Federica Alessandrini, Email: f.alessandrini@unitn.it.
Yari Ciribilli, Email: yari.ciribilli@unitn.it.
Appendix A. Supplementary data
Suppl. Figure S1: A). Western Blot analysis of nuclear and cytoplasmic extracts of Doxorubicin-treated MCF7 cells. Alpha-Actinin serves as loading control while Histone 3 is used as a control for nuclear fractionation. B) RT-qPCR analysis of ETV7 expression upon Doxorubicin treatment in non-breast cancer-derived cell lines, A375M, A549 and U2OS, and in non-cancerous cell lines MCF10A and BJ1-hTERT. Bars represent average Fold Changes relative to the untreated condition and standard deviations of at least three biological replicates. * = P-value < 0.01.
Suppl. Figure S2: A-B). Western Blot analysis of ETV7 over-expressing clones, relative to MCF7 (A), MDA-MB-231 (B) and U2OS (C) cells stably transfected with the Empty vector. D) MTT Assay for survival analysis upon Doxorubicin treatment in U2OS cells over-expressing ETV7 with respect to their empty control. E) A representative image of cell death analysis on Doxorubicin-treated MCF7 cells over-expressing ETV7 or an empty vector obtained at Operetta Perkin Elmer (panels below and above, respectively). The total population of cells was obtained staining them with Hoechst 33342 (a cell-permeable nuclear dye). The amount of dead cells was attained via Topro-3 staining (a dye that is able to enter the nucleus only of damaged, and therefore dead, cells). To better visualize the effect of ETV7 over-expression on cell death, an example of a merge of both the staining is also presented. F) Doxorubicin nuclear efflux analysis using Operetta Imaging System, based on the detection of nuclear and cytoplasmic regions; the recognition of Doxorubicin efflux is done by calculating the fluorescence positive spots area (green spots in the panels on the left). This analysis was performed in MDA-MB-231 cells over-expressing ETV7 compared with their empty control cells. * = P-value < 0.01.
Suppl. Figure S3: A-B). Expression values from microarray data previously obtained by our group from MCF7 cells treated with Doxorubicin (GSE24065) of (A) the gene list the Boettcher group had obtained ( [45] as hypermethylated genes upon resistance to Doxorubicin) and of (B) the DNAJC family members. Results are presented as logarithm of Fold Change from Doxorubicin-treated samples calculated over Mock condition.
Suppl. Figure S4: A). RT-qPCR analysis of ETV7 and DNAJC15 expression in MDA-MB-231 over-expressing pCMV6-Entry-Empty or pCMV6-Entry-ETV7 plasmids. B) ChIP-PCR of DNAJC15 and GTF2H5 (control) promoter regions in MDA-MB-231 stably over-expressing ETV7 untreated or treated with Doxorubicin for 16 hours. C) Western Blot of chromatin and nuclear fractions of MDA-MB-231 over-expressing ETV7 upon treatment with Doxorubicin. Alpha-Actinin serves as loading control while Histone 3 is used as a control for chromatin-enriched nuclear fractions. * = P-value < 0.01.
Suppl. Figure S5: RT-qPCR analysis of DNAJC15 and ABCB1 expression in ETV7-over-expressing MCF7 (A) and MDA-MB-231 (B) cells transiently transfected with pCMV6-Entry-Empty or pCMV6-Entry-DNAJC15 plasmids. Bars represent averages and standard deviations of at least three biological replicates. * = P-value < 0.01.
Suppl. Figure S6: A). Expression of DNMT1, DNMT3A, and DNMT3B from microarray analysis, measured in MCF7 cells treated with Doxorubicin (GSE24065). B) RT-qPCR analysis of DNMT1, DNMT3A and DNMT3B expression in MCF7 transfected with pCMV6-Entry-Empty or pCMV6-Entry-ETV7 plasmids. * = P-value < 0.05.3.
Suppl. Table 1: Sequences of the primers used for qPCR measurements (mRNA expression and promoter occupancy after ChIP assays).
References
- 1.Cagel M, Grotz E, Bernabeu E, Moretton MA, Chiappetta DA. Doxorubicin: nanotechnological overviews from bench to bedside. Drug Discov Today. 2017;22(2):270–281. doi: 10.1016/j.drudis.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Bodley A, Liu LF, Israel M, Seshadri R, Koseki Y, Giuliani FC, Kirschenbaum S, Silber R, Potmesil M. DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res. 1989;49:5969–5978. [PubMed] [Google Scholar]
- 3.De Angelis A, Urbanek K, Cappetta D, Piegari E, Ciuffreda LP, Rivellino A, Russo R, Esposito G, Rossi F, Berrino L. Doxorubicin cardiotoxicity and target cells: a broader perspective. Cardio-Oncol. 2016;2(1):1–8. doi: 10.1186/s40959-016-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottero V, Busuttil V, Loubat A, Magne N, Fischel JL, Milano G, Peyron JF. Activation of nuclear factor kappaB through the IKK complex by the topoisomerase poisons SN38 and doxorubicin: a brake to apoptosis in HeLa human carcinoma cells. Cancer Res. 2001;61:7785–7791. [PubMed] [Google Scholar]
- 5.Wang CY, Mayo MW, Baldwin AS., Jr. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 6.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta. 2014;1839:1316–1322. doi: 10.1016/j.bbagrm.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim B, Stephen SL, Hanby AM, Horgan K, Perry SL, Richardson J, Roundhill EA, Valleley EM, Verghese ET, Williams BJ. Chemotherapy induces Notch1-dependent MRP1 up-regulation, inhibition of which sensitizes breast cancer cells to chemotherapy. BMC Cancer. 2015;15:635. doi: 10.1186/s12885-015-1625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezler M, Hengstler JG, Ullrich A. Inhibition of doxorubicin-induced HER3-PI3K-AKT signalling enhances apoptosis of ovarian cancer cells. Mol Oncol. 2012;6:516–529. doi: 10.1016/j.molonc.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carella C, Potter M, Bonten J, Rehg JE, Neale G, Grosveld GC. The ETS factor TEL2 is a hematopoietic oncoprotein. Blood. 2006;107:1124–1132. doi: 10.1182/blood-2005-03-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 12.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potter MD, Buijs A, Kreider B, van Rompaey L, Grosveld GC. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood. 2000;95:3341–3348. [PubMed] [Google Scholar]
- 14.Kawagoe H, Potter M, Ellis J, Grosveld GC. TEL2, an ETS factor expressed in human leukemia, regulates monocytic differentiation of U937 Cells and blocks the inhibitory effect of TEL1 on ras-induced cellular transformation. Cancer Res. 2004;64:6091–6100. doi: 10.1158/0008-5472.CAN-04-0839. [DOI] [PubMed] [Google Scholar]
- 15.Matos JM, Witzmann FA, Cummings OW, Schmidt CM. A pilot study of proteomic profiles of human hepatocellular carcinoma in the United States. J Surg Res. 2009;155:237–243. doi: 10.1016/j.jss.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quintana AM, Picchione F, Klein Geltink RI, Taylor MR, Grosveld GC. Zebrafish ETV7 regulates red blood cell development through the cholesterol synthesis pathway. Dis Model Mech. 2014;7:265–270. doi: 10.1242/dmm.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardone M, Kandilci A, Carella C, Nilsson JA, Brennan JA, Sirma S, Ozbek U, Boyd K, Cleveland JL, Grosveld GC. The novel ETS factor TEL2 cooperates with Myc in B lymphomagenesis. Mol Cell Biol. 2005;25:2395–2405. doi: 10.1128/MCB.25.6.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sang Y, Chen MY, Luo D, Zhang RH, Wang L, Li M, Luo R, Qian CN, Shao JY, Zeng YX. TEL2 suppresses metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma. Oncotarget. 2015;6:29240–29253. doi: 10.18632/oncotarget.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda O, Ando T, Ohmiya N, Ishiguro K, Watanabe O, Miyahara R, Hibi Y, Nagai T, Yamada K, Goto H. Alteration of gene expression and DNA methylation in drug-resistant gastric cancer. Oncol Rep. 2014;31:1883–1890. doi: 10.3892/or.2014.3014. [DOI] [PubMed] [Google Scholar]
- 20.Bisio A, Zamborszky J, Zaccara S, Lion M, Tebaldi T, Sharma V, Raimondi I, Alessandrini F, Ciribilli Y, Inga A. Cooperative interactions between p53 and NFkappaB enhance cell plasticity. Oncotarget. 2014;5(23):12111–12125. doi: 10.18632/oncotarget.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navasa N, Martin I, Iglesias-Pedraz JM, Beraza N, Atondo E, Izadi H, Ayaz F, Fernandez-Alvarez S, Hatle K, Som A. Regulation of oxidative stress by methylation-controlled J protein controls macrophage responses to inflammatory insults. J Infect Dis. 2015;211:135–145. doi: 10.1093/infdis/jiu389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang YQ, Charette N, Frazer J, Holland PJ, Attwood KM, Dellaire G, Dupre DJ. Dopamine receptor-interacting protein 78 acts as a molecular chaperone for CCR5 chemokine receptor signaling complex organization. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitra A, Shevde LA, Samant RS. Multi-faceted role of HSP40 in cancer. Clin Exp Metastasis. 2009;26:559–567. doi: 10.1007/s10585-009-9255-x. [DOI] [PubMed] [Google Scholar]
- 24.Lindsey JC, Lusher ME, Strathdee G, Brown R, Gilbertson RJ, Bailey S, Ellison DW, Clifford SC. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int J Cancer. 2006;118:346–352. doi: 10.1002/ijc.21353. [DOI] [PubMed] [Google Scholar]
- 25.Lau DT, Hesson LB, Norris MD, Marshall GM, Haber M, Ashton LJ. Prognostic significance of promoter DNA methylation in patients with childhood neuroblastoma. Clin Cancer Res. 2012;18:5690–5700. doi: 10.1158/1078-0432.CCR-12-0294. [DOI] [PubMed] [Google Scholar]
- 26.Schusdziarra C, Blamowska M, Azem A, Hell K. Methylation-controlled J-protein MCJ acts in the import of proteins into human mitochondria. Hum Mol Genet. 2013;22:1348–1357. doi: 10.1093/hmg/dds541. [DOI] [PubMed] [Google Scholar]
- 27.Witham J, Vidot S, Agarwal R, Kaye SB, Richardson A. Transient ectopic expression as a method to detect genes conferring drug resistance. Int J Cancer. 2008;122:2641–2645. doi: 10.1002/ijc.23427. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Cabezudo MJ, Faour I, Jones K, Champagne DP, Jaloudi MA, Mohamed YA, Bashir G, Almarzooqi S, Albawardi A, Hashim MJ. Deficiency of mitochondrial modulator MCJ promotes chemoresistance in breast cancer. JCI Insight. 2016;1(7):e86873. doi: 10.1172/jci.insight.86873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatle KM, Neveu W, Dienz O, Rymarchyk S, Barrantes R, Hale S, Farley N, Lounsbury KM, Bond JP, Taatjes D. Methylation-controlled J protein promotes c-Jun degradation to prevent ABCB1 transporter expression. Mol Cell Biol. 2007;27:2952–2966. doi: 10.1128/MCB.01804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatle KM, Gummadidala P, Navasa N, Bernardo E, Dodge J, Silverstrim B, Fortner K, Burg E, Suratt BT, Hammer J. MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations. Mol Cell Biol. 2013;33:2302–2314. doi: 10.1128/MCB.00189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinha D, Srivastava S, Krishna L, D'Silva P. Unraveling the intricate organization of mammalian mitochondrial presequence translocases: existence of multiple translocases for maintenance of mitochondrial function. Mol Cell Biol. 2014;34:1757–1775. doi: 10.1128/MCB.01527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciribilli Y, Monti P, Bisio A, Nguyen HT, Ethayathulla AS, Ramos A, Foggetti G, Menichini P, Menendez D, Resnick MA. Transactivation specificity is conserved among p53 family proteins and depends on a response element sequence code. Nucleic Acids Res. 2013;41:8637–8653. doi: 10.1093/nar/gkt657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monti P, Ciribilli Y, Bisio A, Foggetti G, Raimondi I, Campomenosi P, Menichini P, Fronza G, Inga A. N-P63alpha and TA-P63alpha exhibit intrinsic differences in transactivation specificities that depend on distinct features of DNA target sites. Oncotarget. 2014;5:2116–2130. doi: 10.18632/oncotarget.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciribilli Y, Andreotti V, Menendez D, Langen JS, Schoenfelder G, Resnick MA, Inga A. The coordinated p53 and estrogen receptor cis-regulation at an FLT1 promoter SNP is specific to genotoxic stress and estrogenic compound. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monti P, Perfumo C, Bisio A, Ciribilli Y, Menichini P, Russo D, Umbach DM, Resnick MA, Inga A, Fronza G. Dominant-negative features of mutant TP53 in germline carriers have limited impact on cancer outcomes. Mol Cancer Res. 2011;9:271–279. doi: 10.1158/1541-7786.MCR-10-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciribilli Y, Singh P, Spanel R, Inga A, Borlak J. Decoding c-Myc networks of cell cycle and apoptosis regulated genes in a transgenic mouse model of papillary lung adenocarcinomas. Oncotarget. 2015;6:31569–31592. doi: 10.18632/oncotarget.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lion M, Bisio A, Tebaldi T, De Sanctis V, Menendez D, Resnick MA, Ciribilli Y, Inga A. Interaction between p53 and estradiol pathways in transcriptional responses to chemotherapeutics. Cell Cycle. 2013;12(8):1211–1224. doi: 10.4161/cc.24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 40.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 41.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 42.Tada Y, Wada M, Migita T, Nagayama J, Hinoshita E, Mochida Y, Maehara Y, Tsuneyoshi M, Kuwano M, Naito S. Increased expression of multidrug resistance-associated proteins in bladder cancer during clinical course and drug resistance to doxorubicin. Int J Cancer. 2002;98:630–635. doi: 10.1002/ijc.10246. [DOI] [PubMed] [Google Scholar]
- 43.Abolhoda A, Wilson AE, Ross H, Danenberg PV, Burt M, Scotto KW. Rapid activation of MDR1 gene expression in human metastatic sarcoma after in vivo exposure to doxorubicin. Clin Cancer Res. 1999;5:3352–3356. [PubMed] [Google Scholar]
- 44.Cox J, Weinman S. Mechanisms of doxorubicin resistance in hepatocellular carcinoma. Hepat Oncol. 2016;3:57–59. doi: 10.2217/hep.15.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boettcher M, Kischkel F, Hoheisel JD. High-definition DNA methylation profiles from breast and ovarian carcinoma cell lines with differing doxorubicin resistance. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segura-Pacheco B, Perez-Cardenas E, Taja-Chayeb L, Chavez-Blanco A, Revilla-Vazquez A, Benitez-Bribiesca L, Duenas-Gonzalez A. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J Transl Med. 2006;4(32) doi: 10.1186/1479-5876-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y, Wei L, Tao X, Gu D, Zhao F. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3beta/beta-catenin Signaling. Theranostics. 2016;6:1205–1219. doi: 10.7150/thno.15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu KD, Zhu R, Zhan M, Rodriguez AA, Yang W, Wong S, Makris A, Lehmann BD, Chen X, Mayer I. Identification of prognosis-relevant subgroups in patients with chemoresistant triple-negative breast cancer. Clin Cancer Res. 2013;19:2723–2733. doi: 10.1158/1078-0432.CCR-12-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Balasubramanian V, Bhat C, Vahermo M, Makila E, Kemell M, Fontana F, Janoniene A, Petrikaite V, Salonen J. Quercetin-Based Modified Porous Silicon Nanoparticles for Enhanced Inhibition of Doxorubicin-Resistant Cancer Cells. Adv Healthc Mater. 2017;6 doi: 10.1002/adhm.201601009. [DOI] [PubMed] [Google Scholar]
- 50.Lv L, Liu C, Chen C, Yu X, Chen G, Shi Y, Qin F, Ou J, Qiu K, Li G. Quercetin and doxorubicin co-encapsulated biotin receptor-targeting nanoparticles for minimizing drug resistance in breast cancer. Oncotarget. 2016;7:32184–32199. doi: 10.18632/oncotarget.8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minaei A, Sabzichi M, Ramezani F, Hamishehkar H, Samadi N. Co-delivery with nano-quercetin enhances doxorubicin-mediated cytotoxicity against MCF-7 cells. Mol Biol Rep. 2016;43:99–105. doi: 10.1007/s11033-016-3942-x. [DOI] [PubMed] [Google Scholar]
- 52.Dong Q, Chen L, Lu Q, Sharma S, Li L, Morimoto S, Wang G. Quercetin attenuates doxorubicin cardiotoxicity by modulating Bmi-1 expression. Br J Pharmacol. 2014;171:4440–4454. doi: 10.1111/bph.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cote B, Carlson LJ, Rao DA, Alani AW. Combinatorial resveratrol and quercetin polymeric micelles mitigate doxorubicin induced cardiotoxicity in vitro and in vivo. J Control Release. 2015;213:128–133. doi: 10.1016/j.jconrel.2015.06.040. [DOI] [PubMed] [Google Scholar]
- 54.Lopez-Lazaro M, Willmore E, Austin CA. Cells lacking DNA topoisomerase II beta are resistant to genistein. J Nat Prod. 2007;70:763–767. doi: 10.1021/np060609z. [DOI] [PubMed] [Google Scholar]
- 55.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 56.Molinari A, Calcabrini A, Meschini S, Stringaro A, Crateri P, Toccacieli L, Marra M, Colone M, Cianfriglia M, Arancia G. Subcellular detection and localization of the drug transporter P-glycoprotein in cultured tumor cells. Curr Protein Pept Sci. 2002;3:653–670. doi: 10.2174/1389203023380413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Figure S1: A). Western Blot analysis of nuclear and cytoplasmic extracts of Doxorubicin-treated MCF7 cells. Alpha-Actinin serves as loading control while Histone 3 is used as a control for nuclear fractionation. B) RT-qPCR analysis of ETV7 expression upon Doxorubicin treatment in non-breast cancer-derived cell lines, A375M, A549 and U2OS, and in non-cancerous cell lines MCF10A and BJ1-hTERT. Bars represent average Fold Changes relative to the untreated condition and standard deviations of at least three biological replicates. * = P-value < 0.01.
Suppl. Figure S2: A-B). Western Blot analysis of ETV7 over-expressing clones, relative to MCF7 (A), MDA-MB-231 (B) and U2OS (C) cells stably transfected with the Empty vector. D) MTT Assay for survival analysis upon Doxorubicin treatment in U2OS cells over-expressing ETV7 with respect to their empty control. E) A representative image of cell death analysis on Doxorubicin-treated MCF7 cells over-expressing ETV7 or an empty vector obtained at Operetta Perkin Elmer (panels below and above, respectively). The total population of cells was obtained staining them with Hoechst 33342 (a cell-permeable nuclear dye). The amount of dead cells was attained via Topro-3 staining (a dye that is able to enter the nucleus only of damaged, and therefore dead, cells). To better visualize the effect of ETV7 over-expression on cell death, an example of a merge of both the staining is also presented. F) Doxorubicin nuclear efflux analysis using Operetta Imaging System, based on the detection of nuclear and cytoplasmic regions; the recognition of Doxorubicin efflux is done by calculating the fluorescence positive spots area (green spots in the panels on the left). This analysis was performed in MDA-MB-231 cells over-expressing ETV7 compared with their empty control cells. * = P-value < 0.01.
Suppl. Figure S3: A-B). Expression values from microarray data previously obtained by our group from MCF7 cells treated with Doxorubicin (GSE24065) of (A) the gene list the Boettcher group had obtained ( [45] as hypermethylated genes upon resistance to Doxorubicin) and of (B) the DNAJC family members. Results are presented as logarithm of Fold Change from Doxorubicin-treated samples calculated over Mock condition.
Suppl. Figure S4: A). RT-qPCR analysis of ETV7 and DNAJC15 expression in MDA-MB-231 over-expressing pCMV6-Entry-Empty or pCMV6-Entry-ETV7 plasmids. B) ChIP-PCR of DNAJC15 and GTF2H5 (control) promoter regions in MDA-MB-231 stably over-expressing ETV7 untreated or treated with Doxorubicin for 16 hours. C) Western Blot of chromatin and nuclear fractions of MDA-MB-231 over-expressing ETV7 upon treatment with Doxorubicin. Alpha-Actinin serves as loading control while Histone 3 is used as a control for chromatin-enriched nuclear fractions. * = P-value < 0.01.
Suppl. Figure S5: RT-qPCR analysis of DNAJC15 and ABCB1 expression in ETV7-over-expressing MCF7 (A) and MDA-MB-231 (B) cells transiently transfected with pCMV6-Entry-Empty or pCMV6-Entry-DNAJC15 plasmids. Bars represent averages and standard deviations of at least three biological replicates. * = P-value < 0.01.
Suppl. Figure S6: A). Expression of DNMT1, DNMT3A, and DNMT3B from microarray analysis, measured in MCF7 cells treated with Doxorubicin (GSE24065). B) RT-qPCR analysis of DNMT1, DNMT3A and DNMT3B expression in MCF7 transfected with pCMV6-Entry-Empty or pCMV6-Entry-ETV7 plasmids. * = P-value < 0.05.3.
Suppl. Table 1: Sequences of the primers used for qPCR measurements (mRNA expression and promoter occupancy after ChIP assays).