Abstract
Gli-similar 3 (Glis3) is Krüppel-like transcription factor associated with the transcriptional regulation of insulin. Mutations within the Glis3 locus have been implicated in a number of pathologies including diabetes mellitus and hypothyroidism. Despite its clinical significance, little is known about the proteins and posttranslational modifications that regulate Glis3 transcriptional activity. In this report, we demonstrate that the SUMO-pathway associated proteins, PIASy and Ubc9 are capable of regulating Glis3 transactivation function through a SUMO-dependent mechanism. We present evidence that SUMOylation of Glis3 by PIAS-family proteins occurs at two conserved lysine residues within the Glis3 N-terminus and modification of Glis3 by SUMO dramatically inhibited insulin transcription. Finally, we provide evidence that Glis3 SUMOylation increases under conditions of chronically elevated glucose and correlates with decreased insulin transcription. Collectively, these results indicate that SUMOylation may serve as a mechanism to regulate Glis3 activity in β cells.
Keywords: Biochemistry, Cell biology, Molecular biology
1. Introduction
Insulin, a hormone produced and secreted by pancreatic β cells, is critical for the proper regulation of blood glucose homeostasis. Transcriptional regulation of the insulin gene is under complex controls and is mediated by a host of transcription factors that form a transcriptional regulatory complex through binding to specific enhancer elements within the insulin proximal promoter (Andrali et al., 2008; Melloul et al., 2002; Ohneda et al., 2000). Among these are pancreatic and duodenal homeobox protein 1 (Pdx1), v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MafA), and Gli-similar 3 (Glis3), which bind to A-boxes, C-boxes, or GlisBS within the insulin promoter, respectively. These factors have been shown to act synergistically to activate insulin transcription both through direct interactions and through their ability to recruit the co-activator CREB-binding protein (CBP/p300) (Qiu et al., 1998; ZeRuth et al., 2013; Zhao et al., 2005). These transcription factors additionally have roles in the specification, maturation, and maintenance of insulin-producing β cells (Ackermann and Gannon, 2007; Cerf, 2006; Lichti-Kaiser et al., 2012). In humans, GLIS3 deficiency has been linked to the development of a rare syndrome characterized by neonatal diabetes and congenital hypothyroidism that can additionally include polycystic kidney disease and hepatic fibrosis (Dimitri et al., 2011; Senee et al., 2006). Genome-wide association studies have further identified GLIS3 as a risk locus for both type 1 and 2 diabetes as well as Alzheimer's disease (Boesgaard et al., 2010; Cho et al., 2012; Cruchaga et al., 2013; Hu et al., 2010; Liu et al., 2011; Rees et al., 2011; Santin and Eizirik, 2013).
A yeast-two-hybrid analysis using the Glis3 N-terminus as bait and a pancreatic β cell prey library indicated that small ubiquitin-like modifier (SUMO) pathway associated proteins putatively associate with the transcription factor (ZeRuth et al., 2015). Posttranslational modifications (PTMs), including SUMOylation, are critical in the regulation of the activity and function of many proteins. SUMOylation has been shown to modify numerous proteins that play a role in diverse physiological processes (Flotho and Melchior, 2013). The consequences of SUMOylation are diverse and can include changes in sub-cellular localization, changes in protein stability, interplay with other PTMs, or regulation of protein-protein interactions (Geiss-Friedlander and Melchior, 2007). Additionally, SUMOylation has been implicated in both the positive and negative regulation of numerous transcription factors (Chymkowitch et al., 2015; Cox et al., 2010; Gong et al., 2014; Ihara et al., 2005; Kanai et al., 2010; Kishi et al., 2003; Shao and Cobb, 2009). SUMOylation requires a series of events that lead to the conjugation of SUMO to target substrates. These include activation of pro-SUMO by an E1 activating protease, conjugation by an E2 enzyme, Ubc9 (UBE2I) (Johnson and Blobel, 1997; Lee et al., 1998), and covalent attachment of SUMO to the ε-amino group of a target lysine by an E3 SUMO ligase (Gareau and Lima, 2010). Among the SUMO E3 ligases are a family of RING-domain-containing proteins that includes the protein-inhibitor-of-activated-STAT (PIAS) proteins, PIAS1, PIAS3, PIASXα, PIASXβ, and PIASy (PIAS4) (Rytinki et al., 2009).
In this report, it is shown that PIAS-family proteins and Ubc9 are capable of modifying Glis3 with SUMO. SUMOylation of Glis3 occurs at two conserved lysine residues located within the N-terminus of the protein, which can be SUMOylated by SUMO1 as well as by SUMO2/3. SUMOylation of Glis3 resulted in a dramatic downregulation of insulin transcription due to inhibition of its transcriptional activity with no observed changes in subcellular localization or loss of protein stability. Finally, we show that Glis3 is modified by SUMO2/3 in BRIN BD11 and INS1 832/13 cells following sustained levels of elevated glucose.
2. Materials and methods
2.1. Cells and growth conditions
Rat insulinoma INS-1832/13 cells, a generous gift from Dr. H. Hohmeier (Duke University), were maintained in RPMI 1640 supplemented with 10% fetal calf serum, 10 mM HEPES, 2 mM glutamine, 1 mM sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol. HEK293T cells were purchased from ATCC and cultured in DMEM containing 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. BRIN BD11 cells were purchased from the European Collection of Authenticated Cell Cultures (ECACC) and were maintained in RPMI 1640 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. BRIN BD11 and INS1 832/13 cells were used between passages 30–47.
2.2. Generation of plasmids and constructs
The generation of p3xFLAG-CMV10-Glis3 was described previously (Kang et al., 2009; ZeRuth et al., 2011, 2015) and expresses full-length murine Glis3 transcript variant 1 (ACCESSION: NM_175459). The luciferase reporter constructs p-mIP-696-Luc, p-(UAS)5-Luc, and pM-Glis3(1–653) were also described previously (Kang et al., 2009; ZeRuth et al., 2011, 2013, 2015). pCMV-Myc-PIAS1, PIASy, and Ubc9 were generated by PCR amplifying the full length cDNA and directionally cloning into pCMV-Myc (Clontech) using EcoRI and XhoI restriction enzymes. pM and VP16 PIAS1 and PIASy constructs were made by PCR amplifying the indicated regions and cloning into pM or VP16 vector (Clontech) using EcoRI and BamHI restriction enzymes. p3xFLAG-CMV10-MafA was described previously (ZeRuth et al., 2013). pcDNA3 HA-Sumo1 WT was a gift from Guy Salvesen (Addgene plasmid # 48966) and Flag-SENP1 was a gift from Edward Yeh (Addgene plasmid # 17357) and were described previously (Bekes et al., 2011; Cheng et al., 2007). 3xFLAG-Glis3 K224R and K430R mutants were generated by site-directed in vitro mutagenesis using p3xFLAG-CMV10-Glis3 as template. All mutants were verified by sequencing. FLAG-Glis3:SUMO fusion constructs were generated by overlap-extension-synthesis PCR (OES-PCR) using primer sets shown in Table 1. Briefly, the region encoding Glis3 amino acids 1–223 or 1–429 were amplified by PCR with a 5′ EcoRI overhang and 3′ overhangs overlapping the 5′ portion of SUMO1 using primers: Glis3 EcoRI F, SUMO224R, and 430-SUMO-R.
Table 1.
List of primers used for OES cloning.
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| Glis3 EcoRI F | ATGCGAATTCAAATGGAAGGTCATGT |
| 224-SUMO-R | TGCCTCCTGGTCAGACACACTCAAGGCTGA |
| 430-SUMO-R | TGCCTCCTGGTCAGAGAGCATGTTGGCTGT |
| SUMO1-224 F | TCAGCCTTGAGTGTGTCTGACCAGGAGGCA |
| SUMO1-224R | CTGAGACCACTCTTGTTGTTCCTGATAAAC |
| SUMO1-430F | ACAGCCAACATGCTCTCTGACCAGGAGGCA |
| SUMO1-430R | CTCCAGGCGCTCTGTTTGTTCCTGATAAAC |
| 224-SUMO F | GTTTATCAGGAACAACAAGAGTGGTCTCAG |
| 430-SUMO F | GTTTATCAGGAACAAACAGAGCGCCTGGAG |
| Glis3-BamHI R | ATGCGGATCCTCAGCCTTCGGTATA |
SUMO1 was PCR amplified with a 5′ and a 3′ overhang overlapping Glis3 at the indicated regions using primers: SUMO1-224 F and SUMO1-224R or SUMO1-430F and SUMO1-430R.
And the 3′ portion of Glis3 encoding amino acids 225–935 or 431–935 was amplified with a 5′ overhang overlapping the 3′ portion of SUMO1 and a 3′ BamHI overhang using primers: 224-SUMO F or 430-SUMO F and Glis3-BamHI R.
PCR products were cleaned up using a GenElute PCR Cleanup Kit (Sigma Aldrich) and added in equimolar concentrations along with dNTPs and Pfu TURBO (Agilent Technologies) and run in a thermal cycler programmed 95 °C 2 min, followed by 20 cycles of 95 °C 30 sec; 60 °C 30 sec; 72 °C 3 min. 5 ul of the resulting product was removed and used as template for a subsequent PCR reaction this time including 0.4 uM FLAG Glis3 EcoRI F primer and Glis3-BamHI R primer using conditions 95 °C 2 min followed by 40 cycles of 95 °C 30 sec; 55 °C 30 sec; 72 °C 3 min. The correct sized bands were gel purified in a 1% agarose gel and subsequently digested with EcoRI and BamHI. Digested inserts were directionally cloned into p3xFLAG-CMV10 plasmid (Sigma Aldrich) cut with identical enzymes. Positive clones were analyzed by restriction analysis and verified by sequencing.
2.3. Reporter assays
Cells were plated in 12-well dishes at 1 × 105 cells/well and incubated for 24 h at 37 °C. Cells were subsequently transfected with the indicated reporter, pCMV-β-galactosidase, and the indicated expression vector in serum-free medium without antibiotic using Lipofectamine 3000 (Invitrogen) per the manufacturer's instructions. Each transfection was carried out in triplicate. Cells were harvested after 48 h by scraping them directly into 125 ul of reporter lysis buffer, and luciferase activity was measured using a luciferase assay kit (Promega). β-Galactosidase levels were measured using a luminometric β-galactosidase detection kit (Clontech) following the manufacturer's protocol. Each data point was assayed in triplicate, and each experiment was performed at least twice. Relative luciferase activity was calculated. All values underwent analysis of variance and Tukey-Kramer comparison tests using InStat software (GraphPad Software Inc.), and data from representative experiments are presented as mean ± S.D. Mammalian two-hybrid assays were performed with HEK293T cells plated in 12-well dishes at 1 × 105 cells/well and incubated for 24 h at 37 °C. Cells were subsequently transfected with pM or VP16 empty vector (Clontech) or the indicated chimera, pFR-Luc, and pCMV-β-gal diluted in serum-free media lacking antibiotic and incubated with Lipofectamine 3000 reagent according to the manufacturer's protocol (Invitrogen). Cells were harvested, and luciferase assays were conducted and analyzed as reported above.
2.4. Co-immunoprecipitation assays
Cells were transiently transfected with the specified plasmids using Lipofectamine 3000 reagent (Invitrogen) following the manufacturer's protocol. 48 h after transfection, cells were harvested by scraping in radioimmune precipitation assay buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 20 mM sodium molybdate, and 0.5% Nonidet P-40) containing protease inhibitor cocktails I and II (Sigma). Cell lysates were centrifuged at 16,000 x g for 10 min at 4 °C, and a fraction of the supernatant was stored at −80 °C for the input fractions. The remaining supernatant was incubated at RT for 15 min with DynaBeads Protein G (Invitrogen) conjugated to the indicated antibody. Beads were washed three times with 200 μl of ice-cold PBS containing protease inhibitor and proteins were released from the beads by boiling for 5 min in the presence of 1x Laemmli buffer supplemented with 2.5% 2-Mercaptoethanol. For IPs examining SUMOylation, 20 mM N-ethylmaleimide (NEM) was added to lysis buffer and all subsequent wash steps. Input and immunoprecipitated proteins were examined by Western blot analysis using mouse anti-FLAG M2 (Sigma Aldrich), mouse anti-Myc antibody (Invitrogen), rat anti HA antibody (Roche), ChIP grade rabbit anti-GFP (AbCam) rabbit anti-PIAS1 antibody (Invitrogen), rabbit anti-PIAS4 antibody (Cell Signaling Technologies), rabbit anti-SUMO1 (Cell Signaling Technologies), or mouse anti-SUMO2/3 antibody (Cytoskeleton) and the appropriate HRP-conjugated secondary antibodies. Blots were processed and analyzed using an Amersham Imager 600 digital imager (GE Life Sciences). All experiments were performed at least two times. Representative blots are shown.
2.5. Western blot analysis and protein quantification
Proteins were resolved by SDS-PAGE and then transferred to PVDF membrane (Invitrogen) by electrophoresis. Immunostaining was performed with the indicated antibody at either 4 °C for 18 h or 22 °C for 1 h in BLOTTO reagent (5% nonfat dry milk dissolved in 50 mM Tris, 0.2% Tween 20, and 150 mM NaCl). Blots were subjected to three 10-min washes in TTBS (50 mM Tris, 0.2% Tween 20, and 150 mM NaCl), and bands were detected by enhanced chemiluminescence following the manufacturer's protocol (GE Healthcare). Proteins were quantified using ImageQuantTL software (GE Life Sciences) The mean intensity and pixel count of experimental bands were multiplied and divided by the mean intensity multiplied by the pixel count of GAPDH bands used for normalization. All samples were run in duplicate and all experiments performed at least twice. Data shown are the average of duplicate samples ± S.D.
2.6. Quantitative reverse transcriptase real-time PCR analysis
INS1 832/13 or BRIN BD11 cells were transiently transfected with the indicated constructs using Lipofectamine 3000 reagent following the manufacturer's protocol (Invitrogen). RNA was isolated from the cells after 48 h using a GenElute Mammalian Total RNA miniprep kit (Sigma Aldrich) according to the manufacturer's specifications. Equal amounts of RNA were used to generate cDNA using a high capacity cDNA kit (Applied Biosystems), and cDNA was analyzed by quantitative real-time PCR using PowerUP SYBR green master mix (ThermoFisher Scientific). All qRT-PCR was performed in triplicate using an Applied Biosystems 7500 real time PCR system. The average Ct from triplicate samples was normalized against the average Ct of 18S rRNA. All experiments were performed two or more times and representative experiments are shown. Primers used are listed in Table 2.
Table 2.
List of primers used for RT-PCR.
| Gene target | Primer sequence (5′ to 3′) |
|---|---|
| rIns1 | F – CCTGCTCGTCCTCTGGGAGCCCAAG R – CTCCAGTGCCAAGGTCTGAAGATCC |
| rIns2 | F – CCTGCTCATCCTCTGGGAGCCCCGC R – CTCCAGTGCCAAGGTCTGAAGGTCA |
| rPIAS1 | F – CTACCAGCCTACGGGTTTCG R – GAACAGGTAAGTGCCCGACA |
| rPIASy | F – ATAGATGGGCTGCTGTCGAAGA R – ATTGGGCGCCATGAACCTT |
| rGlis3 | F – GTGAAGGCACATTCTTCCAAAGA R – GGAGATCTGGATGGAGCTCAGT |
| FLAG-Glis3 | F – TGGACTACAAAGACCATGACGG R – GGGCTCTGATGGGAGGGATA |
| rUcp2 | F – GCATTGGCCTCTACGACTCT R – CTGGAAGCGGACCTTTACC |
| 18s rRNA | F – GTAACCCGTTGAACCCCATT R – CCATCCAATCGGTAGTAGCG |
2.7. Immunocytochemistry and microscopy
BRIN BD11 cells were grown on 22 mm poly-L-lysine coated glass coverslips in 35 mm culture dishes for 48 h until cells were 70–80% confluent. The coverslips were washed in PBS and fixed for 15 min in 4% p-formaldehyde before being permeabilized in 0.2% Triton-X-100 for 7 min. Cells were blocked in Superblock (ThermoFisher Scientific) for 15 min and then stained with the indicated antibody overnight at 4 °C. Cells were stained with anti-mouse AlexaFluor 594 secondary antibody (ThermoFisher Scientific) for 30 min and mounted on glass microscope slides using Prolong Diamond anti-fade with DAPI (Life Technologies) and sealed with clear nail polish. Coverslips were observed on a Leica DMi8 fluorescence inverted microscope (Leica Microsystems) and images were captured using a DFC7000T cooled fluorescence camera and LAS X Expert software (Leica Microsystems).
2.8. Measurement of total insulin levels
BRIN BD11 or INS1 832/13 cells were transfected as described above. After 24 h, cells were washed twice with ice-cold PBS and lysed with ice-cold RIPA buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 20 mM sodium molybdate, and 0.5% Nonidet P-40) containing protease inhibitor cocktails I and II (Sigma). Lysates were centrifuged and supernatant was diluted to determine insulin content using a rat insulin ELISA kit (Thermo Scientific) following the manufacturer's protocol. Each sample was measured in triplicate and compared to a rat insulin standard.
3. Results
3.1. Glis3 interacts with PIAS-family proteins
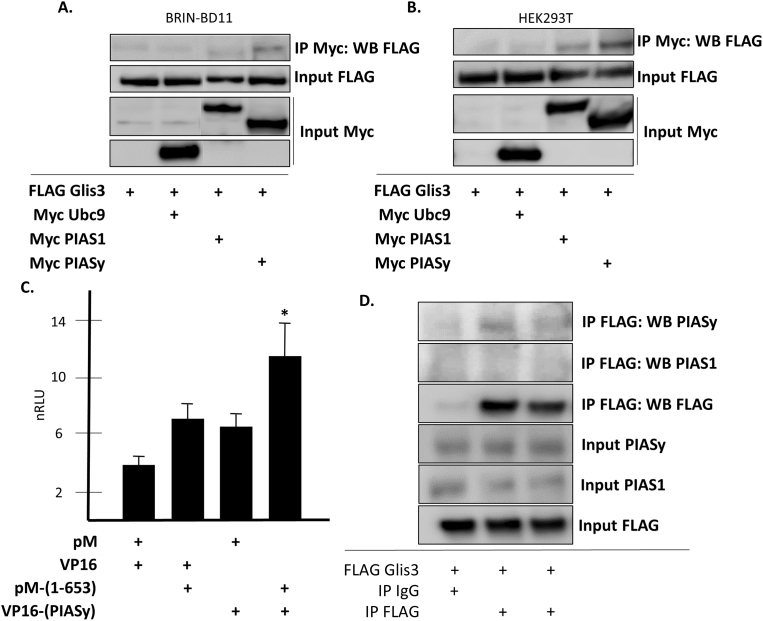
In order to identify proteins that interact with the Glis3 N-terminus, a yeast-two-hybrid analysis was conducted using the Glis3 N-terminus as bait to identify interacting proteins from a murine pancreatic β cell prey library (ZeRuth et al., 2015). This study identified several proteins implicated in the SUMO conjugation pathway as putative Glis3-interacting proteins including the SUMO-conjugating protein Ubc9 (UBE2I) and members of the protein inhibitor of activated STAT (PIAS)-family, PIAS1 and PIASy (PIAS4), which have been shown to act as E3 SUMO-ligases (Rytinki et al., 2009). Glis3 is normally expressed in both kidney and the β-cells of the pancreas (Kang et al., 2016; Lichti-Kaiser et al., 2012). The transcript (ENST00000324333.10) encoding a 775 amino acid protein has been identified in the human pancreas (The Genotype-Tissue Expression (GTEx) Portal; https://www.gtexportal.org/home/gene/GLIS3). The relatively low expression rate reported (median TPM = 3.52) is likely due to GLIS3 expression being limited to the islets and ducts, which comprise a small proportion of total pancreatic mass (Kang et al., 2016; Senee et al., 2006). The same transcript is also found in the human kidney cortex (median TPM = 4.989). Co-immunoprecipitation experiments performed in BRIN BD11 and in the human kidney derived HEK293T cell line co-expressing full length FLAG-Glis3 and Myc-tagged Ubc9, PIAS1, or PIASy, showed a weak interaction between Glis3 and the PIAS-family proteins (Fig. 1A and B). Similarly, a mammalian 2-hybrid assay performed in HEK293T cells using the N-terminus of Glis3 (aa 1–653) fused to the Gal4 DBD and full length PIASy fused to the Gal4 AD to drive luciferase expression under control of the Gal4-responsive UAS region showed a 1.5-fold increase in luciferase expression when the chimeric proteins were co-expressed (Fig. 1C). Endogenous PIAS1 or PIASy was not detected when FLAG Glis3 was immunoprecipitated in BRIN BD11 or HEK293T cells. It is possible however that the interaction between the SUMO ligase and its substrate is transient and therefore the amount of PIAS protein stably associated with Glis3 may be below the threshold of detection for the respective antibodies. In support of that supposition, immunoprecipitation of FLAG-Glis3 in BRIN BD11 cells following formaldehyde cross-linking produced a very weak band representing of the correct molecular weight using a PIASy antibody while no PIAS1 was evident (Fig. 1D). Collectively, these data indicate that Glis3 may form interactions with PIAS-family proteins.
Fig. 1.
Glis3 interacts with PIASy and PIAS1. A. BRIN BD11 or B. HEK293T cells were transfected with FLAG-Glis3 and either Myc-empty vector, Myc-Ubc9, Myc-PIAS1, or Myc-PIASy as indicated. Co-immunoprecipitation was performed using a mouse anti-Myc antibody and immunoprecipitated proteins were examined by Western blot analysis using anti-FLAG M2 or anti-Myc primary and goat anti-mouse-HRP secondary antibodies. Note: the image in A was digitally cut between lanes 2 and 3 and flipped horizontally to match the loading order shown in B. C. HEK293T cells were co-transfected with the pFR-Luc reporter and the indicated pM and VP16 plasmid DNA. 48 h later cells were assayed for luciferase and β-galactosidase activities, and the relative Luc activity was calculated and plotted. A representative experiment is shown. Each bar represents mean ± S.D. *, statistically different from pM or VP16 empty vector controls p < 0.05. nRLU, normalized relative luc units. D. BRIN BD11 cells were transfected with FLAG-Glis3. After 48 h, cells were fixed using 4% paraformaldehyde in PBS for a total of 10 min. The reaction was quenched with 1.25 M glycine prior to cell lysis in RIPA buffer. Co-immunoprecipitation was performed using mouse IgG or a mouse anti-FLAG M2 antibody and input or immunoprecipitated proteins were examined by Western blot analysis using mouse anti-FLAG M2, anti-PIAS1, or anti-PIASy primary antibodies and the appropriate HRP-conjugated secondary antibodies. Unaltered blots are shown in Supplemental Fig S1.
3.2. PIAS family proteins mediate Glis3 SUMOylation
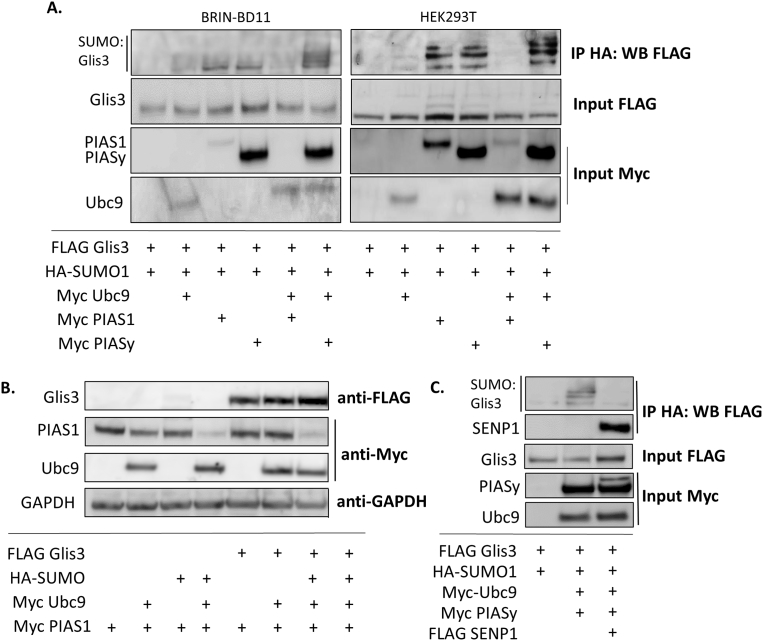
Given the role of PIAS proteins in mediating SUMOylation of target proteins, co-immunoprecipitation assays were performed in BRIN BD11 and HEK293T cells expressing FLAG-Glis3 and HA-SUMO1 in the presence or absence of Ubc9, PIAS1 or PIASy. To date, Ubc9 is the only SUMO-conjugating enzyme that has been identified in mammals and appears to be the sole E2 enzyme used in the SUMO-conjugation pathway (Gareau and Lima, 2010). Protein complexes were pulled down using an anti-HA antibody and SUMOylation of Glis3 was analyzed by Western blotting using anti-FLAG M2 antibody. As seen in Fig. 2A, the two members of the PIAS family promoted SUMOylation of exogenous Glis3 with relatively similar efficacy in both cell lines. Co-expression of Ubc9 with HA-SUMO and FLAG-Glis3 only modestly increased Glis3 SUMOylation compared to HA-SUMO and FLAG Glis3 alone (Fig. 2A). These results were not surprising as PIAS-family proteins are more likely to be limiting factors in regulating Glis3-SUMOylation than Ubc9, which is ubiquitously expressed (Kovalenko et al., 1996). Intriguingly, while simultaneous co-expression of Ubc9 and PIASy significantly increased the observed level of Glis3 SUMOylation compared to either protein alone, coincident expression of Ubc9 and PIAS1 virtually eliminated all detectable Glis3 SUMOylation. Nearly identical results were obtained whether the experiment was performed in BRIN-BD11 or HEK293T cells, suggesting the observation is not species-, cell- or tissue-specific. The lack of Glis3 SUMOylation under these conditions appeared to be the result of a significant decrease in PIAS1 expression in the presence of both exogenous Ubc9 and HA-SUMO (Fig. 2A and B). PIAS1 degradation did not appear to be affected by Glis3 as the protein was degraded in its presence or absence (Fig. 2B) and therefore the underlying mechanism of PIAS1 degradation was not investigated further. However, for further experiments requiring co-expression of Ubc9, SUMO, and PIAS proteins, we used PIASy to simplify data analysis. To determine whether Glis3 SUMOylation was reversible, the SUMO-specific protease SENP1 was co-expressed with FLAG-Glis3, HA-SUMO1, Ubc9, and PIASy and was found to be capable of effectively deconjugating SUMO from Glis3 as demonstrated in Fig. 2C. Together, these data suggest that the E2 SUMO conjugase, Ubc9 along with the E3 SUMO ligases, PIAS1 or PIASy are capable of reversibly modifying Glis3 with SUMO.
Fig. 2.
Glis3 is SUMOylated by PIAS-family proteins. A. BRIN BD11 or HEK293T cells were transfected with FLAG empty vector or FLAG Glis3 along with HA-SUMO1, Myc Ubc9, Myc PIAS1, or Myc PIASy as indicated. After 48 h, co-immunoprecipitation was performed using a rat anti-HA antibody and immunoprecipitated proteins were examined by Western blot analysis using mouse anti-FLAG M2 or anti-Myc primary and goat anti-mouse-HRP secondary antibodies. B. HEK293T cells were transfected with the indicated plasmids. Cells were treated with 10 ug/ml cycloheximide for 5 h prior to harvest. After 48 h, proteins were separated by SDS-PAGE and analyzed by Western blotting using the specified antibodies. GAPDH is shown as an internal control. C. HEK293T cells were transfected with the indicated plasmids and after 48 h, IP was performed and analyzed as described in A. Unaltered blots are shown in Supplemental Fig S2.
3.3. Glis3 is SUMOylated on Lys224 and Lys430
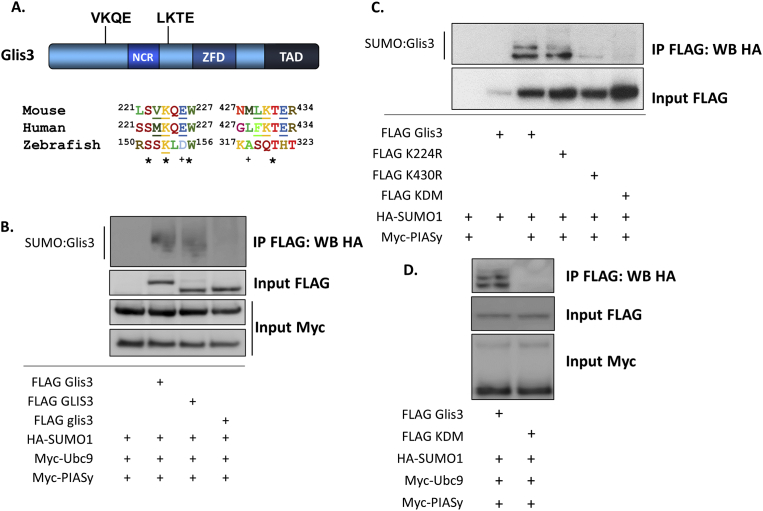
SUMO modification typically occurs at a lysine residue within the target substrate that conforms to the consensus sequence ψKxE where ψ is a hydrophobic residue and x is any amino acid (Duprez et al., 1999; Rodriguez et al., 2001; Sampson et al., 2001; Sternsdorf et al., 1999). Analysis of Glis3 using SUMOplot software (www.abgent.com) revealed two high probability consensus SUMO motifs located at Lys224 and Lys430 located within the N-terminus of the protein that are conserved between murine and human Glis3 but are absent in zebrafish glis3 (Fig. 3A). To determine whether Glis3 could be modified by SUMO, co-IP was performed using murine, human, or zebrafish Glis3 in HEK293T cells. Both human and mouse Glis3 were SUMOylated by PIASy and Ubc9, while no modification of zebrafish glis3 was observed (Fig. 3B). These results demonstrate that modification of Glis3 by SUMO appears to be conserved among mammals and may have evolved following the split that would give rise to tetrapods and teleost fishes. In order to determine whether SUMO was attached to the conserved residues within the Glis3 N-terminus, each lysine was mutated to arginine either alone or in combination. While co-expression of HA-SUMO1 and PIASy resulted in two distinct bands representing SUMOylated Glis3, mutation of Lys224 to Arg resulted in a decrease of the higher molecular weight band (Fig. 3C). Mutation of Lys430 to Arg similarly resulted in the elimination of the higher molecular weight band and additionally dramatically reduced the lower weight band representing SUMOylated Glis3. Simultaneous mutation of both lysine residues abolished all detectable Glis3 SUMOylation. Glis3 SUMOylation was similarly ablated in BRIN BD11 cells when Lys224 and Lys430 were mutated (Fig. 3D). These results indicate that PIAS family proteins can mediate the attachment of SUMO1 to Glis3 at either Lys224, Lys430, or both simultaneously.
Fig. 3.
Glis3 is SUMOylated on Lys224 and Lys430. A. Schematic representation of Glis3. Relative position and sequence of putative SUMOylation motifs are shown. NCR, N-terminal conserved region; ZFD, zinc finger domain; TAD, transactivation domain. Sequence alignment of mouse (NP_780668.3), human (NP_001035878.1), and zebrafish (AAI29174.1) Glis3 is shown in lower portion. Residues that match the consensus SUMOylation motif (ψKxE) are underlined. Amino acid numbers are indicated. B–C. HEK293T cells were transfected with FLAG-tagged murine Glis3, human GLIS3, or zebrafish glis3 and the indicated plasmids. After 48 h, co-immunoprecipitation was performed using a mouse anti-FLAG M2 antibody and immunoprecipitated proteins were examined by Western blot analysis using rat anti-HA or mouse anti-Myc primary and goat anti-mouse- or anti-rat-HRP secondary antibodies. D. BRIN BD11 cells were transfected with the indicated plasmids. After 48 h, co-immunoprecipitation was performed using a mouse anti-FLAG M2 antibody and input or immunoprecipitated proteins were examined by Western blot analysis using mouse anti-FLAG M2, rat anti-HA, or mouse anti-Myc primary antibodies and the appropriate HRP-conjugated secondary antibodies. Unaltered blots are shown in Supplemental Fig S3.
3.4. Glis3 is SUMOylated by SUMO1-3
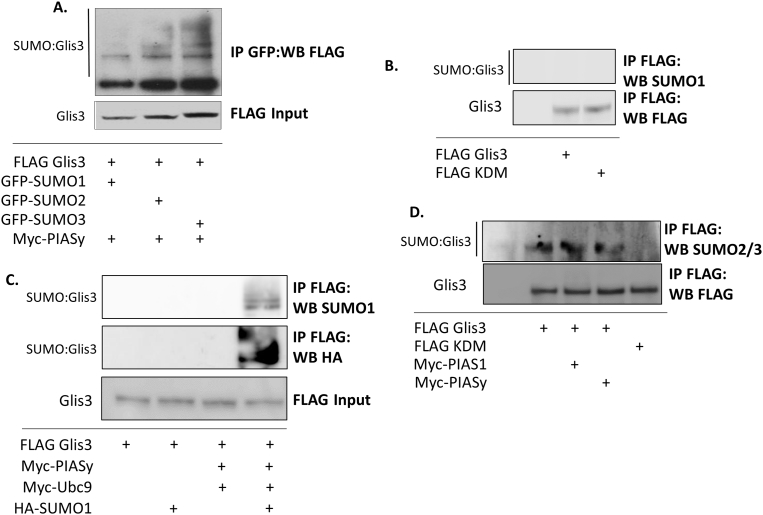
To investigate whether PIAS proteins preferentially modify Glis3 with SUMO 1, 2, or 3, co-IP was performed in HEK293T cells expressing FLAG-Glis3 and Myc-PIASy along with GFP-tagged SUMO1, SUMO2, or SUMO3. While Glis3 appeared to be mono- or di-SUMOylated by SUMO1, polySUMOylation was evident when Glis3 was co-expressed with PIASy and SUMO2 or SUMO3 (Fig. 4A). In order to determine whether Glis3 could be SUMOylated by endogenous SUMO proteins, FLAG-Glis3 or the Lys224/Lys430 double mutant was expressed in BRIN BD11 cells and pulled down using an anti-FLAG M2 antibody. Immunoprecipitated fractions were then analyzed by Western blotting using an antibody specific to SUMO1. Glis3 was not found to be modified by endogenous SUMO1 in the cells (Fig. 4B). SUMO1 modified Glis3 was also not detected in HEK293T cells in the presence or absence of exogenous PIASy and Ubc9 (Fig. 4C, lanes 2 and 3). SUMOylated Glis3 was evident, however when exogenous HA-SUMO1 was co-expressed along with PIASy and Ubc9 and could be detected in the immunoprecipitated fractions by antibodies targeting the HA tag or SUMO1 (Fig. 4C, lane 4). These data suggest that only a small proportion, if any of exogenous Glis3 is modified by endogenous SUMO1 under normal conditions in HEK293T or BRIN BD11 cells. In contrast, SUMO2/3 modified Glis3 could be observed in BRIN BD11 cells expressing FLAG-Glis3 in the presence or absence of exogenous PIAS1 or PIASy (Fig. 4D). No SUMO2/3 modified Glis3 could be detected however in BRIN BD11 cells overexpressing the Lys224/Lys430 double mutant. These results indicate that the PIAS-family proteins are capable of modifying Glis3 with SUMO1-3 and that exogenous Glis3 is modified by endogenous SUMO 2/3.
Fig. 4.
PIASy and Ubc9 can modify Glis3 with SUMO1-3. A. HEK293T cells were transfected with FLAG Glis3 and Myc PIASy along with the indicated GFP-tagged SUMO construct. Co-immunoprecipitation was performed using a mouse anti-GFP antibody and proteins were analyzed by Western blotting using mouse anti-FLAG M2 antibody. B. BRIN BD11 or C. HEK293T cells were transfected with the indicated plasmids. After 48 h, co-immunoprecipitation was performed using a mouse anti-FLAG M2 antibody and input or immunoprecipitated proteins were examined by Western blot analysis using mouse anti-FLAG M2, rabbit anti-SUMO1, or rat-anti-HA primary antibodies and the appropriate HRP-conjugated secondary antibodies. D. BRIN BD11 cells were transfected with the indicated plasmids and treated with 10 uM MG132 5 h prior to harvest. Co-immunoprecipitation was conducted using an anti-FLAG M2 antibody. Proteins were analyzed by Western blotting using a mouse anti-SUMO2/3 or anti-FLAG M2 antibody and the appropriate HRP-conjugated secondary antibodies. KDM, Lys224/Lys430 double mutant. Unaltered blots are shown in Supplemental Fig S4.
3.5. PIASy and Ubc9 negatively regulate Glis3-mediated activation of the insulin promoter
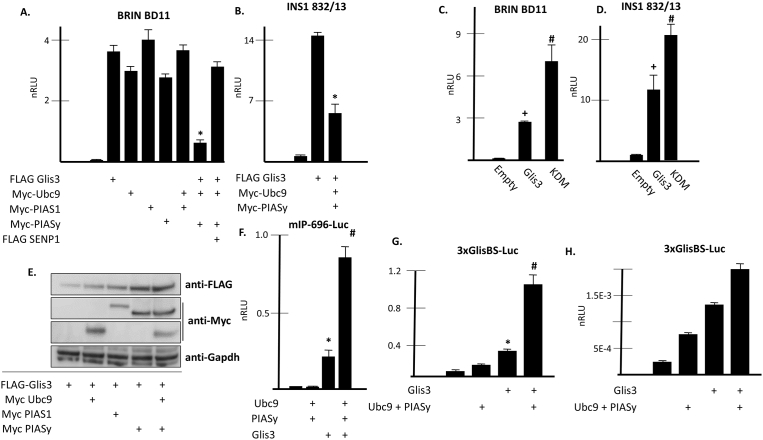
To ascertain whether SUMOylation affects Glis3 function, luciferase reporter assays were conducted in BRIN BD11 cells expressing a luciferase reporter under the control of the mIns2 promoter. As expected, FLAG-Glis3 expression resulted in a significant increase in luciferase activity (Fig. 5A). Co-expression of Ubc9, PIAS1, or PIASy alone did not have a significant impact on Glis3-directed activation of the insulin promoter. However, when PIASy and Ubc9 were both co-expressed with Glis3, a >70% decrease in luciferase activity was observed compared to expression of Glis3 alone. This effect was almost completely negated when SENP1 was co-expressed; suggesting the decrease in activity was SUMO dependent. Ubc9 and PIASy had a similar inhibitory effect in INS1 832/13 cells (Fig. 5B). Moreover, reporter activity increased roughly two-fold compared to the wild type Glis3 when the Lys224/Lys430 double mutant was expressed in BRIN BD11 cells (Fig. 5C) or INS1 832/13 cells (Fig. 5D) suggesting that SUMOylation of Glis3 negatively affected its transactivation function. The decreased Glis3 transactivation function was not associated with destabilization of Glis3 protein levels since Glis3 protein levels increased when PIASy was co-expressed in BRIN BD11 cells treated with cycloheximide for 6 h prior to lysis and immunoblot analysis (Fig. 5E). When reporter assays were performed in HEK 293T cells using either the mIns2 promoter or an artificial promoter driven by three tandem copies of the GlisBS (p3xGlisBS-Luc), co-expression of PIASy and Ubc9 resulted in an increase in Glis3-mediated reporter activation (Fig. 5F and G). Activation of p3xGlisBS-Luc was also enhanced by co-expression of PIASy and Ubc9 in BRIN BD11 cells (Fig. 5H). These data are consistent with increased Glis3 protein expression in the presence of PIASy and Ubc9 and suggest that the observed PIASy- and Ubc9-mediated inhibition of Glis3 transactivation function is relevant to Ins2 expression in β cells but does not occur using an artificial promoter or within the HEK293T environment.
Fig. 5.
PIASy and Ubc9 repress Glis3-mediated insulin transcription in BRIN BD11 cells. A–D. BRIN BD11 or INS1 832/13 cells were transfected with p-mIP-696-Luc, pCMV-β-Gal, along with the indicated plasmids. After 48 h, cells were assayed for luciferase and β-galactosidase activity and the normalized relative luciferase activity (nRLU) was calculated and plotted. A representative experiment is shown. Each bar represents the mean ± S.D. * indicates statistically different than corresponding Myc empty vector control. p < 0.01. + indicates statistically different from FLAG empty vector control p < 0.01. # indicates statistically different from FLAG empty vector control and FLAG Glis3 p < 0.05. KDM, Lys224/Lys430 double mutant. E. BRIN BD11 cells were transfected with the indicated plasmids and treated with 10 ug/ml cycloheximide for 5 h prior to harvest. Proteins were separated by SDS-PAGE and analyzed by Western blotting using the specified antibodies. GAPDH is shown as an internal control. F. HEK293T cells were transfected with p-mIP-696-Luc, pCMV-β-Gal, FLAG empty vector, FLAG-Glis3, Myc-Ubc9, or Myc-PIASy as indicated. After 48 h, cells were assayed for luciferase and β-galactosidase activity and the normalized relative luciferase activity (nRLU) was calculated and plotted. Each bar represents the mean ± SEM. * indicates significantly different from FLAG and Myc empty vector control. # indicates significantly different from FLAG and Myc empty vector control and cells expressing FLAG-Glis3 and Myc empty vector. P < 0.01. G. HEK293T cells or H. BRIN BD11 cells were transfected with p3xGlisBS-Luc and pCMV-β-Gal along with FLAG empty vector, FLAG-Glis3, Myc-Ubc9, or Myc-PIASy as indicated. After 48, cells were harvested and luciferase assay was performed as described in A. * indicates significantly different from FLAG and Myc empty vector control. # indicates significantly different from FLAG and Myc empty vector control and cells expressing FLAG-Glis3 and Myc empty vector. P < 0.05. Unaltered blots are shown in Supplemental Fig S5.
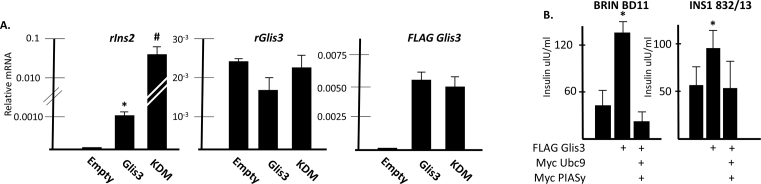
3.6. PIASy and Ubc9 decrease insulin production
To determine whether SUMOylation of Glis3 affected endogenous Ins2 transcription, BRIN BD11 cells expressing FLAG empty vector, FLAG Glis3, or the Lys224/Lys430 double mutant were analyzed by qRT-PCR for rIns2 expression. Consistent with luciferase reporter assays, endogenous insulin message was significantly increased in cells expressing the Glis3 mutant versus wild type Glis3 while the expression of both endogenous and exogenous Glis3 was not significantly different (Fig. 6A). The abundance of Glis3 protein was also similar between the wild type and Lys double mutant (Fig. 4B and D). Finally, total insulin levels increased in both BRIN BD11 and INS1 832/13 cells when Glis3 was overexpressed but failed to do so when Glis3 was co-expressed with exogenous Ubc9 and PIASy (Fig. 6B). These data establish that SUMOylation may serve as a mechanism to negatively regulate insulin transcription by Glis3 in pancreatic β cells.
Fig. 6.
SUMOylation of Glis3 decreases its transcriptional activity. A. BRIN BD11 cells were transfected with the indicated plasmids. After 48 h, total RNA was collected and the specified mRNA was measured by qRT-PCR analysis. A representative experiment is shown. Each bar represents relative mRNA levels normalized to 18s rRNA ± S.D. * indicates statistically different value compared to empty vector control p < 0.01. # indicates statistically different value from both empty vector control and wild type Glis3, p < 0.01. KDM, Lys224/Lys430 double mutant. B. BRIN BD11 and INS1 832/13 cells were transfected with the indicated plasmids. After 48 h, the cells were lysed and total cellular insulin content was measured by ELISA. Cells were transfected in triplicate and each bar represents the average insulin content of triplicate samples ± S.D. * indicates statistically different value compared to empty vector control p < 0.05.
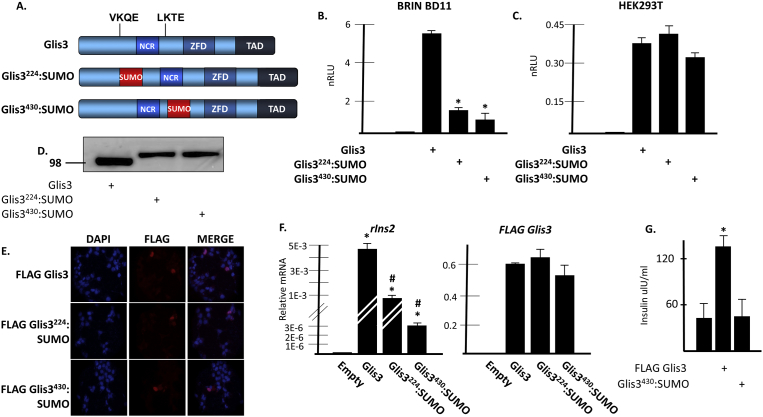
3.7. SUMOylation of Glis3 decreases its transcriptional activity
The fact that the Lys224/Lys430 double mutant exhibited roughly two-fold greater activity in BRIN BD11 cells than wild type Glis3 (Figs. 5B and 6A) suggested that PIASy and Ubc9 were negatively affecting Glis3-mediated insulin activity through a mechanism that relied upon their ability to directly SUMOylate the Glis3 N-terminus. Since varying proportions of Glis3 could be SUMOylated at any time within the cells, OES cloning was performed to generate chimeric proteins comprised of SUMO fused in-frame with Glis3 in place of lysine 224 or 430 to serve as an approximation for constitutively SUMOylated Glis3 (Fig. 7A). To prevent the SUMO fusion constructs from being able to interact with other proteins targeted for SUMO modification, the SUMO insert was truncated immediately prior to the C-terminal di-glycine motif. When expressed in BRIN BD11 cells along with a mIns2 reporter, both SUMO fusion chimeras were significantly less capable of activating the insulin promoter compared to wild type Glis3 (Fig. 7B). Consistent with what was observed following PIASy and Ubc9 overexpression, mIns2 promoter activation was not different using either of the SUMO fusion constructs relative to unmodified Glis3 in HEK293T cells (Fig. 7C). Western blot analysis using an anti-FLAG M2 antibody showed that both SUMO fusion constructs produced a single band migrating slightly slower than unmodified Glis3 and were expressed at generally similar levels (Fig. 7D). The reduced activity could not be attributed to defective nuclear localization as both chimeric proteins were localized exclusively to the nucleus in BRIN BD11 cells (Fig. 7E). To examine whether the in-frame addition of SUMO to Glis3 also had an effect on the regulation of endogenous insulin transcription, BRIN BD11 cells were transfected with empty vector, Glis3, Glis3224:SUMO, or Glis3430:SUMO, and rIns2 was measured by qRT-PCR. In-frame insertion of SUMO in place of Lys224 resulted in an 80% reduction in Ins2 transcription relative to cells expressing unmodified Glis3, while fusion of SUMO at Lys430 resulted in a 99% reduction in activity (Fig. 7F). Using primers specific to FLAG-Glis3, exogenous Glis3 transcripts were relatively quantified and found to be expressed at roughly equal levels between samples. Unlike unmodified Glis3, Glis3430:SUMO expression in BRIN BD11 cells did not promote an increase in total cellular insulin levels (Fig. 7G). Together, these results support the conclusion that SUMOylation of Glis3 at Lys224 and Lys430 negatively impact the ability of the protein to activate insulin transcription.
Fig. 7.
SUMOylation of Glis3 inhibits its ability to activate insulin transcription. A. Schematic diagram of Glis3 and Glis3:SUMO fusion proteins generated by OES cloning. The conserved SUMOylation motifs within the Glis3 N-terminus are indicated. NCR, N-terminal conserved region; ZFD, zinc finger domain; TAD, transactivation domain. B. BRIN BD11 cells or C. HEK293T cells were transfected with p-mIP-696-Luc, pCMV-β-Gal, FLAG empty vector, FLAG-Glis3, or the indicated Glis3:SUMO fusion construct. After 48 h, cells were harvested and assayed as described in Fig. 5. * indicates statistically different from FLAG Glis3, p < 0.01. D. FLAG-stained Western blot of cell lysates from BRIN BD11 cells expressing FLAG Glis3 or the indicated Glis3:SUMO fusion protein. Molecular weight marker is indicated on the left. E. BRIN BD11 cells were grown on glass coverslips and transfected with FLAG Glis3 or the indicated FLAG-Glis3:SUMO fusion construct. After 48 h, cells were fixed and stained with an anti-FLAG M2 primary antibody and an anti-mouse Alexa-fluor 594 secondary antibody. Coverslips were mounted to slides using mounting media containing DAPI to visualize the nuclei and observed by fluorescence microscopy. F. BRIN BD11 cells were transfected with the indicated plasmids. After 48 h, total RNA was collected and the specified mRNA was measured by qRT-PCR analysis. Each bar represents relative mRNA levels normalized to 18s rRNA ± S.D. * indicates statistically different value compared to empty vector control p < 0.01. # indicates statistically different value from both empty vector control and wild type Glis3, p < 0.01. G. BRIN BD11 cells were transfected with the indicated plasmids. After 48 h, the cells were lysed and total cellular insulin content was measured by ELISA. Cells were transfected in triplicate and each bar represents the average insulin content of triplicate samples ± S.D. * indicates statistically different value compared to empty vector control p < 0.05. Unaltered blots are shown in Supplemental Fig S7.
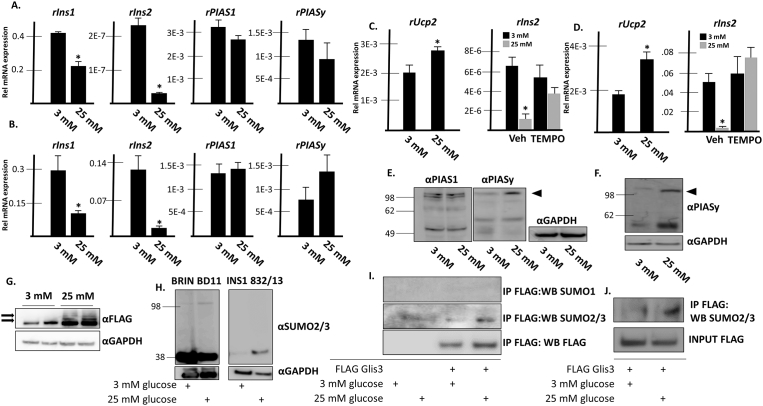
3.8. Glis3 is SUMOylated under conditions of chronically elevated glucose in pancreatic beta cells
Chronic exposure of β cells to high levels of glucose has previously been associated with decreased levels of insulin transcription mediated in part by decreased binding of MafA and Pdx1 to the insulin promoter (Harmon et al., 2005; Lee et al., 2012; Park et al., 2007). This so called glucose toxicity is also accompanied by decreased levels of MafA transcription (Olson et al., 1993; Poitout et al., 1996; Sharma et al., 1995). MafA SUMOylation was reported in β cells exposed to oxidative stress and SUMOylation of MafA negatively affected its transactivation function at the insulin promoter suggesting that modification of MafA by SUMO may serve as an additional mechanism to reduce insulin transcription under hyperglycemic conditions (Kanai et al., 2010; Shao and Cobb, 2009). To determine the effect of chronically elevated glucose concentrations on BRIN BD11 and INS1 832/13 cells, the cells were cultured in either 3 mM or 25 mM glucose for 48 hours and gene expression was analyzed by RT-PCR. As shown previously in other β cell lines, under conditions of prolonged exposure to high levels of glucose, insulin transcription was severely reduced in BRIN BD11 (Fig. 7A) and INS1 832/13 (Fig. 7B) cells. No statistically significant change in either PIAS1 or PIASy mRNA expression was observed in response to glucose levels in either cell line. Ucp2 has been shown to be upregulated in response to H2O2, free fatty acids, as well as in mice challenged with a high fat diet (Li et al., 2001; Medvedev et al., 2002; Surwit et al., 1998). Transcription of Ucp2 was elevated in both BRIN BD11 (Fig. 7C) and INS1 832/13 (Fig. 7D) cells subjected to chronically elevated glucose levels, suggesting that oxidative stress increased under these conditions. Further, decreased levels of Ins2 mRNA were at least partially rescued by the addition of the antioxidant, 4-hydroxy-TEMPO supporting the concept that oxidative stress underlies the effect (Fig. 7C and D). To determine whether PIAS-proteins were regulated posttranslationally in response to chronically elevated glucose levels, Western blots were performed from cells grown under high or low glucose conditions. While PIAS1 expression was similar under both conditions, a higher molecular weight band corresponding to a previously identified SUMOylated form of PIASy (Ihara et al., 2005) was significantly increased following prolonged exposure to 25 mM glucose in BRIN BD11 cells (arrowhead in Fig. 7E) and INS1 832/13 cells (Fig. 7F). Glis3 expression was examined at the protein level using BRIN BD11 cells stably expressing FLAG Glis3 and grown under low or high glucose concentrations. The results indicated that Glis3 protein expression was increased under chronically high glucose conditions (Fig. 8G). Importantly, under high glucose conditions, several higher molecular weight bands were observed that might represent SUMOylated Glis3 (Arrows in Fig. 8G). There was no discernible change in global SUMO2/3 levels in response to elevated glucose levels in BRIN BD11 cells however there were increased levels of a 40 kD band when INS1 832/13 cells were maintained in high glucose conditions (Fig. 7H). Finally, to determine whether Glis3 was SUMOylated under conditions of glucose toxicity, anti-FLAG M2 antibody was used to immunoprecipitate proteins in BRIN BD11 cells stably expressing FLAG Glis3 or empty vector grown under different glucose concentrations. The results indicated that SUMO2/3-modified Glis3 increased under high glucose conditions (Fig. 7I). Similar results were obtained in INS1 832/13 cells transiently overexpressing FLAG Glis3 under variable glucose concentrations (Fig. 7J).
Fig. 8.
Glis3 is SUMOylated in BRIN BD11 cells under conditions of chronically elevated glucose. A. BRIN BD11 cells were grown in media containing 3 mM or 25 mM D-glucose. Media was changed after 24 h and after 48 h, total RNA was collected and the specified mRNA was measured by qRT-PCR analysis. A representative experiment is shown. Each bar represents relative mRNA levels normalized to 18s rRNA ± S.D. * indicates statistically different value compared to cells grown in 3 mM glucose p < 0.05. B. INS1 832/13 cells were grown and assayed as described in A. C. BRIN BD11 or D. INS1 832/13 cells were grown as described in A. Vehicle or 50 uM 4-hydroxy-TEMPO was additionally added to media as indicated. After 48 h, RNA was collected and qRT-PCR was performed as described in A. E. BRIN BD11 or F. INS1 832/13 cells were grown as described in A. Cells were harvested, proteins were separated by SDS-PAGE, and analyzed by Western blotting using the specified antibodies. GAPDH is shown as an internal control. G. BRIN BD11 cells stably expressing FLAG-Glis3 were grown in the presence of either 3 mM or 25 mM glucose for 48 h. Cells were harvested and proteins were separated by SDS-PAGE and subsequently analyzed by Western blotting using the indicated antibodies. Arrows indicate higher molecular weight bands that may represent SUMOylated Glis3. GAPDH is shown as an internal control. H. BRIN BD11 or INS1 832/13 cells were grown as described in A. Cells were harvested, proteins were separated by SDS-PAGE, and analyzed by Western blotting using the indicated antibodies. GAPDH is shown as an internal control. I. BRIN BD11 cells stably expressing FLAG empty vector or FLAG Glis3 were grown in media containing 3 mM or 25 mM D-glucose. After 48 h, co-immunoprecipitation was performed using an anti-M2 FLAG antibody. Immunoprecipitated and input fractions were analyzed by Western blotting using a rabbit anti-SUMO1 antibody, mouse anti-SUMO2/3 antibody, or mouse anti-M2 FLAG antibody and the appropriate HRP-conjugated secondary antibodies. J. INS1 832/13 cells were transfected with FLAG-Glis3 and grown in media containing 3 mM or 25 mM D-glucose. After 48 h, co-immunoprecipitation was performed using an anti-M2 FLAG antibody. Immunoprecipitated and input fractions were analyzed by Western blotting using a mouse anti-SUMO2/3 antibody. Unaltered blots are shown in Supplemental Fig S8.
4. Discussion
In this report, we show that PIAS-family proteins are capable of modifying the transcription factor, Glis3 with the small ubiquitin-like modifier, SUMO1-3. Overexpression of PIASy in combination with Ubc9 had a significant negative influence on the transactivation function of Glis3 and greatly diminished its ability to activate Ins2 expression in BRIN-BD11 and INS1 832/13 cells. Data demonstrating that the mutation of both SUMO targeted lysines to arginine resulted in a significant increase in insulin transcription compared to wild type Glis3 is consistent with the idea that SUMOylation has an inhibitory effect on Glis3-mediated transactivation of insulin. The increased level of insulin transcription is not likely attributable to the creation of novel methylation sites that might subsequently affect transactivation function since similar observations were made when the Lys residues were mutated to Ala (data not shown).
In order to better understand the effect of SUMOylation on Glis3 function, Glis3:SUMO fusion chimeras, which should approximate SUMOylated Glis3, were created and analyzed. These experiments demonstrated a profound negative regulatory consequence of Glis3 SUMOylation that affected Glis3-mediated activation of the preproinsulin (Ins2) promoter in a pancreatic β-cell environment. The SUMO fusion with Glis3 was unlikely to have affected Glis3 function by causing improper folding or inhibiting binding to GlisBS since the constructs activated a mIns2 reporter in HEK293T cells similarly to full-length, wild type Glis3. The Glis3:SUMO chimeras additionally did not exhibit any defect in their ability to localize to the nucleus nor were protein expression levels dramatically different. These data are in agreement with experiments that demonstrated that PIASy and Ubc9 negatively regulated activation of a mIns2 reporter in BRIN BD11 cells but failed to do so in HEK293T cells. Furthermore, PIASy and Ubc9 did not have a repressive effect on the activation of an artificial promoter in BRIN BD11 cells driven by three tandem copies of the GlisBS. The mechanism by which SUMOylation regulates Glis3 function is not clear at this time. It is possible that the addition of SUMO moieties to the Glis3 N-terminus competes with other posttranslational modifications such as phosphorylation or acetylation that may be associated with positive transcriptional regulation of insulin within β cells. Alternatively, SUMOylation of Glis3 may interfere with Glis3 recruitment of pancreatic β-cell-specific co-activators. Indeed, previous reports have shown that the ability of Glis3 to activate insulin transcription was severely compromised when binding by Pdx1, NeuroD1, and MafA to the Ins2 promoter was abrogated (ZeRuth et al., 2013).
Previous studies have shown that the transcription factor, Pdx1, was SUMOylated in pancreatic β cells; however, unlike Glis3, the modification resulted in increased nuclear localization and positive regulation of insulin transcription (Kishi et al., 2003). In contrast, SUMOylation by SUMO1 or SUMO2 negatively regulated the transcriptional activity of the β cell transcription factor, MafA through an unknown mechanism that did not involve changes in subcellular localization or decreased protein stability (Kanai et al., 2010; Shao and Cobb, 2009). It has previously been shown that Glis3 acts synergistically at the insulin promoter with several other key insulin regulatory factors (Yang et al., 2009; ZeRuth et al., 2013). Whether or not SUMOylation of Glis3 interferes with these events and prevents synergistic activation of target genes such as insulin will be the subject of future studies.
Chronic exposure of β cells to high levels of glucose has previously been associated with decreased levels of insulin transcription mediated in part by decreased binding of MafA and Pdx1 to the insulin promoter (Harmon et al., 2005; Lee et al., 2012; Park et al., 2007). Much of the decrease in insulin transcription was likely due to the observed transcriptional downregulation of Pdx1 and MafA. However, MafA SUMOylation was reported in β cells exposed to oxidative stress suggesting that modification of MafA by SUMO may additionally serve as a mechanism to reduce insulin transcription under hyperglycemic conditions (Shao and Cobb, 2009). Under conditions of chronically elevated glucose in BRIN BD11 and INS1 832/13 cells, we observed increased expression of a high molecular weight species of PIASy that correlated with increased SUMOylation of exogenous Glis3 by SUMO2/3. The absence of a reliable Glis3 antibody restricts the ability to determine whether endogenous Glis3 is SUMOylated under conditions of prolonged glucose exposure, but further studies will be conducted to examine whether oxidative stress promotes SUMOylation of Glis3 and the subsequent downregulation of insulin transcription.
Despite the fact that PIASy and Ubc9 negatively affected Glis3-mediated Ins2 activation, the proteins appeared to increase Glis3 expression at the protein level. Previously, the tumor suppressor, SUFU, had been shown to have a similar effect on Glis3 in that it repressed insulin activation by Glis3 in INS1 cells while simultaneously stabilizing Glis3 protein levels (ZeRuth et al., 2011). Taken together, these studies suggest that Glis3 protein turnover may be linked to target gene activation. This hypothesis is consistent with the observation that the ZFD of Glis3 was required for proteolytic degradation by Itch (ZeRuth et al., 2015).
5. Conclusions
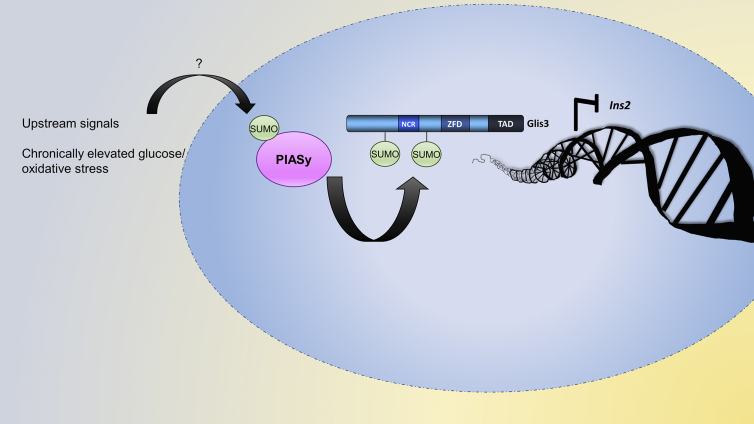
Collectively, these studies implicate SUMOylation as a mechanism that may be used to negatively regulate insulin transcription through inhibition of Glis3 transactivation function. A schematic representation of these conclusions can be seen in Fig. 9. Reversible posttranslational modifications of transcription factors could serve as an effective means to rapidly respond to environmental cues such as dynamic glucose levels. These modifications might also be a source of dysfunction in instances such as oxidative stress where their negative influence on insulin transcription can contribute to disease states such as diabetes. Investigation of Glis3 expression and posttranslational modifications under conditions of chronic hyperglycemia and oxidative stress will be a continued focus of research into the future.
Fig. 9.
Summary of research findings. Upstream signals including conditions of chronically elevated glucose levels, which promotes oxidative stress in BRIN BD11 and INS1 832/13 cells, PIASy is likely SUMOylated and can subsequently promote SUMOylation of Glis3 at Lys224 and Lys430. SUMOylation of Glis3 reduces the ability of the transcription factor to activate Ins2 transcription in the cell lines.
Declarations
Author contribution statement
Tyler M. Hoard: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Xiao-Ping Yang: Performed the experiments.
Anton M. Jetten: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Gary T. ZeRuth: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by NSF-KY EPSCoR grant [RSF-042-14] and the Intramural Research Program of the National Institute of Environmental Health Sciences, the National Institutes of Health [Z01-ES-100485] with additional support from a competitive research grant sponsored by the Committeee for Institutional Studies and Research at Murray State University.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to thank Erin Clayton for assistance with molecular cloning and Fumihiko Nakamura for technical assistance with Western blots and co-IP. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 12/13/17.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ackermann A.M., Gannon M. Molecular regulation of pancreatic beta-cell mass development, maintenance, and expansion. J. Mol. Endocrinol. 2007;38:193–206. doi: 10.1677/JME-06-0053. 38/2/193 [pii] [DOI] [PubMed] [Google Scholar]
- Andrali S.S., Sampley M.L., Vanderford N.L., Ozcan S. Glucose regulation of insulin gene expression in pancreatic beta-cells. Biochem. J. 2008;415:1–10. doi: 10.1042/BJ20081029. BJ20081029 [pii] [DOI] [PubMed] [Google Scholar]
- Bekes M., Prudden J., Srikumar T., Raught B., Boddy M.N., Salvesen G.S. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J. Biol. Chem. 2011;286:10238–10247. doi: 10.1074/jbc.M110.205153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesgaard T.W., Grarup N., Jorgensen T., Borch-Johnsen K., Hansen T., Pedersen O. Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia. 2010;53:1647–1655. doi: 10.1007/s00125-010-1753-5. [DOI] [PubMed] [Google Scholar]
- Cerf M. Transcription factors regulating beta-cell function. Eur. J. Endocrinol. 2006;155:671–679. doi: 10.1530/eje.1.02277. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kang X., Zhang S., Yeh E.T. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.S., Chen C.H., Hu C., Long J., Ong R.T., Sim X., Takeuchi F., Wu Y., Go M.J., Yamauchi T., Chang Y.C., Kwak S.H., Ma R.C., Yamamoto K., Adair L.S., Aung T., Cai Q., Chang L.C., Chen Y.T., Gao Y., Hu F.B., Kim H.L., Kim S., Kim Y.J., Lee J.J., Lee N.R., Li Y., Liu J.J., Lu W., Nakamura J., Nakashima E., Ng D.P., Tay W.T., Tsai F.J., Wong T.Y., Yokota M., Zheng W., Zhang R., Wang C., So W.Y., Ohnaka K., Ikegami H., Hara K., Cho Y.M., Cho N.H., Chang T.J., Bao Y., Hedman A.K., Morris A.P., McCarthy M.I., Takayanagi R., Park K.S., Jia W., Chuang L.M., Chan J.C., Maeda S., Kadowaki T., Lee J.Y., Wu J.Y., Teo Y.Y., Tai E.S., Shu X.O., Mohlke K.L., Kato N., Han B.G., Seielstad M. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat. Genet. 2012;44:67–72. doi: 10.1038/ng.1019. ng.1019 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chymkowitch P., Nguea P.A., Enserink J.M. SUMO-regulated transcription: challenging the dogma. BioEssays: News Rev. Mol. Cell. Dev. Biol. 2015;37:1095–1105. doi: 10.1002/bies.201500065. [DOI] [PubMed] [Google Scholar]
- Cox B., Briscoe J., Ulloa F. SUMOylation by Pias1 regulates the activity of the Hedgehog dependent Gli transcription factors. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C., Kauwe J.S., Harari O., Jin S.C., Cai Y., Karch C.M., Benitez B.A., Jeng A.T., Skorupa T., Carrell D., Bertelsen S., Bailey M., McKean D., Shulman J.M., De Jager P.L., Chibnik L., Bennett D.A., Arnold S.E., Harold D., Sims R., Gerrish A., Williams J., Van Deerlin V.M., Lee V.M., Shaw L.M., Trojanowski J.Q., Haines J.L., Mayeux R., Pericak-Vance M.A., Farrer L.A., Schellenberg G.D., Peskind E.R., Galasko D., Fagan A.M., Holtzman D.M., Morris J.C., Goate A.M. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer's disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P., Warner J., Minton J., Patch A., Ellard S., Hattersley A., Hawkes D., Wales J., Gregory J. Novel GLIS3 mutations demonstrate an extended multisystem phenotype. Eur. J. Endocrinol. 2011;164:437–443. doi: 10.1530/EJE-10-0893. [DOI] [PubMed] [Google Scholar]
- Duprez E., Saurin A.J., Desterro J.M., Lallemand-Breitenbach V., Howe K., Boddy M.N., Solomon E., de The H., Hay R.T., Freemont P.S. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 1999;112(Pt 3):381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- Flotho A., Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Gareau J.R., Lima C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gong L., Ji W.K., Hu X.H., Hu W.F., Tang X.C., Huang Z.X., Li L., Liu M., Xiang S.H., Wu E., Woodward Z., Liu Y.Z., Nguyen Q.D., Li D.W. Sumoylation differentially regulates Sp1 to control cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:5574–5579. doi: 10.1073/pnas.1315034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon J.S., Stein R., Robertson R.P. Oxidative stress-mediated, post-translational loss of MafA protein as a contributing mechanism to loss of insulin gene expression in glucotoxic beta cells. J. Biol. Chem. 2005;280:11107–11113. doi: 10.1074/jbc.M410345200. M410345200 [pii] [DOI] [PubMed] [Google Scholar]
- Hu C., Zhang R., Wang C., Wang J., Ma X., Hou X., Lu J., Yu W., Jiang F., Bao Y., Xiang K., Jia W. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M., Yamamoto H., Kikuchi A. SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol. Cell. Biol. 2005;25:3506–3518. doi: 10.1128/MCB.25.9.3506-3518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Kanai K., Reza H.M., Kamitani A., Hamazaki Y., Han S.I., Yasuda K., Kataoka K. SUMOylation negatively regulates transcriptional and oncogenic activities of MafA. Genes Cells: Devoted Mol. Cell. Mech. 2010;15:971–982. doi: 10.1111/j.1365-2443.2010.01431.x. [DOI] [PubMed] [Google Scholar]
- Kang H.S., Kim Y.S., ZeRuth G., Beak J.Y., Kilic G., Jensen J., Sosa-Pineda B., Jetten A.M. Transcription factor Glis3: a novel critical player in the regulation of pancreatic β-cell development. Mol. Cell. Biol. 2009;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., Takeda Y., Jeon K., Jetten A.M. The spatiotemporal pattern of Glis3 expression indicates a regulatory function in bipotent and endocrine progenitors during early pancreatic development and in beta, PP and ductal cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi A., Nakamura T., Nishio Y., Maegawa H., Kashiwagi A. Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation. Am. J. Physiol. Endocrinol. Metab. 2003;284:E830–E840. doi: 10.1152/ajpendo.00390.2002. [DOI] [PubMed] [Google Scholar]
- Kovalenko O.V., Plug A.W., Haaf T., Gonda D.K., Ashley T., Ward D.C., Radding C.M., Golub E.I. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.W., Melchior F., Matunis M.J., Mahajan R., Tian Q., Anderson P. Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- Lee K.M., Seo Y.J., Kim M.K., Seo H.A., Jeong J.Y., Choi H.S., Lee I.K., Park K.G. Mediation of glucolipotoxicity in INS-1 rat insulinoma cells by small heterodimer partner interacting leucine zipper protein (SMILE) Biochem. Biophys. Res. Commun. 2012;419:768–773. doi: 10.1016/j.bbrc.2012.02.098. [DOI] [PubMed] [Google Scholar]
- Li L.X., Skorpen F., Egeberg K., Jorgensen I.H., Grill V. Uncoupling protein-2 participates in cellular defense against oxidative stress in clonal beta-cells. Biochem. Biophys. Res. Commun. 2001;282:273–277. doi: 10.1006/bbrc.2001.4577. [DOI] [PubMed] [Google Scholar]
- Lichti-Kaiser K., ZeRuth G., Kang H.S., Vasanth S., Jetten A.M. Gli-similar proteins: their mechanisms of action, physiological functions, and roles in disease. Vitam. Horm. 2012;88:141–171. doi: 10.1016/B978-0-12-394622-5.00007-9. B978-0-12-394622-5.00007-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li H., Qi L., Loos R.J., Qi Q., Lu L., Gan W., Lin X. Variants in GLIS3 and CRY2 are associated with type 2 diabetes and impaired fasting glucose in Chinese Hans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021464. PONE-D-10-03608 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev A.V., Robidoux J., Bai X., Cao W., Floering L.M., Daniel K.W., Collins S. Regulation of the uncoupling protein-2 gene in INS-1 beta-cells by oleic acid. J. Biol. Chem. 2002;277:42639–42644. doi: 10.1074/jbc.M208645200. [DOI] [PubMed] [Google Scholar]
- Melloul D., Marshak S., Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- Ohneda K., Hooi E., German M. Regulation of insulin gene transcription. Semin. Cell Dev. Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- Olson L.K., Redmon J.B., Towle H.C., Robertson R.P. Chronic exposure of HIT cells to high glucose concentrations paradoxically decreases insulin gene transcription and alters binding of insulin gene regulatory protein. J. Clin. Investig. 1993;92:514–519. doi: 10.1172/JCI116596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.G., Lee K.M., Seo H.Y., Suh J.H., Kim H.S., Wang L., Won K.C., Lee H.W., Park J.Y., Lee K.U., Kim J.G., Kim B.W., Choi H.S., Lee I.K. Glucotoxicity in the INS-1 rat insulinoma cell line is mediated by the orphan nuclear receptor small heterodimer partner. Diabetes. 2007;56:431–437. doi: 10.2337/db06-0753. 56/2/431 [pii] [DOI] [PubMed] [Google Scholar]
- Poitout V., Olson L.K., Robertson R.P. Chronic exposure of betaTC-6 cells to supraphysiologic concentrations of glucose decreases binding of the RIPE3b1 insulin gene transcription activator. J. Clin. Investig. 1996;97:1041–1046. doi: 10.1172/JCI118496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Sharma A., Stein R. p300 Mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees S.D., Hydrie M.Z., O'Hare J.P., Kumar S., Shera A.S., Basit A., Barnett A.H., Kelly M.A. Effects of 16 genetic variants on fasting glucose and type 2 diabetes in South Asians: ADCY5 and GLIS3 variants may predispose to type 2 diabetes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024710. PONE-D-11-07898 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., Hay R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Rytinki M.M., Kaikkonen S., Pehkonen P., Jaaskelainen T., Palvimo J.J. PIAS proteins: pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci.: CMLS. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson D.A., Wang M., Matunis M.J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Santin I., Eizirik D.L. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and beta-cell apoptosis. Diabetes Obes. Metab. 2013;15(Suppl. 3):71–81. doi: 10.1111/dom.12162. [DOI] [PubMed] [Google Scholar]
- Senee V., Chelala C., Duchatelet S., Feng D., Blanc H., Cossec J.C., Charon C., Nicolino M., Boileau P., Cavener D.R., Bougneres P., Taha D., Julier C. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- Shao C., Cobb M.H. Sumoylation regulates the transcriptional activity of MafA in pancreatic beta cells. J. Biol. Chem. 2009;284:3117–3124. doi: 10.1074/jbc.M806286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Olson L.K., Robertson R.P., Stein R. The reduction of insulin gene transcription in HIT-T15 beta cells chronically exposed to high glucose concentration is associated with the loss of RIPE3b1 and STF-1 transcription factor expression. Mol. Endocrinol. (Baltimore, Md) 1995;9:1127–1134. doi: 10.1210/mend.9.9.7491105. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Reich B., Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- Surwit R.S., Wang S., Petro A.E., Sanchis D., Raimbault S., Ricquier D., Collins S. Diet-induced changes in uncoupling proteins in obesity-prone and obesity-resistant strains of mice. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4061–4065. doi: 10.1073/pnas.95.7.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Chang B.H., Samson S.L., Li M.V., Chan L. The Kruppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–2538. doi: 10.1093/nar/gkp122. gkp122 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZeRuth G.T., Takeda Y., Jetten A.M. The Kruppel-like protein Gli-similar 3 (Glis3) functions as a key regulator of insulin transcription. Mol. Endocrinol. (Baltimore, Md.) 2013;27:1692–1705. doi: 10.1210/me.2013-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZeRuth G.T., Williams J.G., Cole Y.C., Jetten A.M. HECT E3 ubiquitin ligase Itch functions as a novel negative regulator of Gli-similar 3 (Glis3) transcriptional activity. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZeRuth G.T., Yang X.P., Jetten A.M. Modulation of the transactivation function and stability of Kruppel-like zinc finger protein Gli-similar 3 (Glis3) by Suppressor of Fused. J. Biol. Chem. 2011;286:22077–22089. doi: 10.1074/jbc.M111.224964. M111.224964 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Guo M., Matsuoka T.A., Hagman D.K., Parazzoli S.D., Poitout V., Stein R. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J. Biol. Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. M409475200 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.