Abstract
Enhanced gene transfer efficiencies and higher yields of transplantable transduced human hematopoietic stem cells are continuing goals for improving clinical protocols that use stemcell-based gene therapies. Here, we examined the effect of the HSC agonist UM171 on these endpoints in both in vitro and in vivo systems. Using a 22-hr transduction protocol, we found that UM171 significantly enhances both the lentivirus-mediated transduction and yield of CD34+ and CD34+CD45RA- hematopoietic cells from human cord blood to give a 6-fold overall higher recovery of transduced hematopoietic stem cells, including cells with long-term lympho-myeloid repopulating activity in immunodeficient mice. The ability of UM171 to enhance gene transfer to primitive cord blood hematopoietic cells extended to multiple lentiviral pseudotypes, gamma retroviruses, and non-integrating lentiviruses and to adult bone marrow cells. UM171, thus, provides an interesting reagent for improving the ex vivo production of gene-modified cells and for reducing requirements of virus for a broad range of applications.
Keywords: lentivirus, gene transfer, hematopoietic stem cells
Introduction
Transplantation of genetically modified autologous hematopoietic stem cells (HSCs) has become an approach with established therapeutic benefit for many inherited and acquired hematologic disorders of the blood and immune system. These include X-linked severe combined immunodeficiency (SCID-X1),1 Wiskott-Aldrich syndrome (WAS),2, 3 beta-thalassemia,4 X-linked adrenaleukodystophy5, 6 and metachromatic leukodystrophy.7, 8 Clinical success with this modality relies on the transplantation of sufficient modified cells and, for the diseases cited, cells with permanent normal blood cell output capacity. The introduction of lentivirus (LV)-based gene transfer has proven a significant advance for meeting this requirement, given the increased propensity of LV vectors to infect quiescent cells and the relatively shorter culture period required for infection compared to gamma-retrovirus (γRV)-based methods.9 Nevertheless, methods to transduce human HSCs with long-term in vivo repopulating activity (LT-HSCs) remain suboptimal and dependent on the use of high vector doses that are costly and accompanied by an increased risk of genotoxicity.10 Coupling enhanced gene transfer to improved culture conditions to increase transduced LT-HSC recovery could also have a major impact on the efficacy and safety of gene therapy-based approaches by accelerating the reconstitution of transplanted patients.
Various small molecules targeting specific steps of the retroviral life cycle have been tested to improve the permissiveness of HSCs to lentiviral vectors. Rapamycin increased LV-mediated, but not γRV-mediated, transduction of human and mouse HSCs while preserving their engraftment potential by enhancing postbinding endocytic events via mammalian target of rapamycin (mTOR) inhibition.11, 12 Cyclosporin A (CsA), at high concentrations, also increased LV-mediated transduction by a different mechanism, i.e., by relieving a viral capsid (CA)-dependent early block and by enhancing virus integration.12 Proteosome inhibition by MG-132 was also reported to increase LV-mediated transduction of human and mouse HSCs and hematopoietic stem and progenitor cells (HSPCs) independently of the cyclophilin A-CA interaction.13, 14 However, a drawback in the use of all of these strategies is their targeting of proteins that are broadly critical to cell survival.15
The recent discovery of small molecules stimulating the expansion of HSPCs in vitro, such as SR116 and UM171,17 or enhancing engraftment, such as PGE2,18 raises the possibility of their application also to the clinical gene transfer setting. Combined addition of dmPGE2 and SR1 to human cord blood (CB) CD34+ cell cultures shown to increase the recovery of cells with in vivo repopulating potential following zinc-finger nuclease-mediated gene editing is one such example.19 The demonstrated ability of the pyrimidoindole derivative, UM171, to stimulate a more-than-10-fold expansion of LT-HSCs in short-term cultures17 prompted us to examine its potential utility in the context of LV-mediated transduction of HSPCs.
Our findings provide evidence that short-term culture with UM171 significantly enhances HSPC transduction efficiency and yield. These newly defined properties of UM171 point to the potential advantageous application of this approach to future gene transfer protocols.
Results
UM171 Enhances LV-Mediated Transduction of Primitive Human Hematopoietic Cells
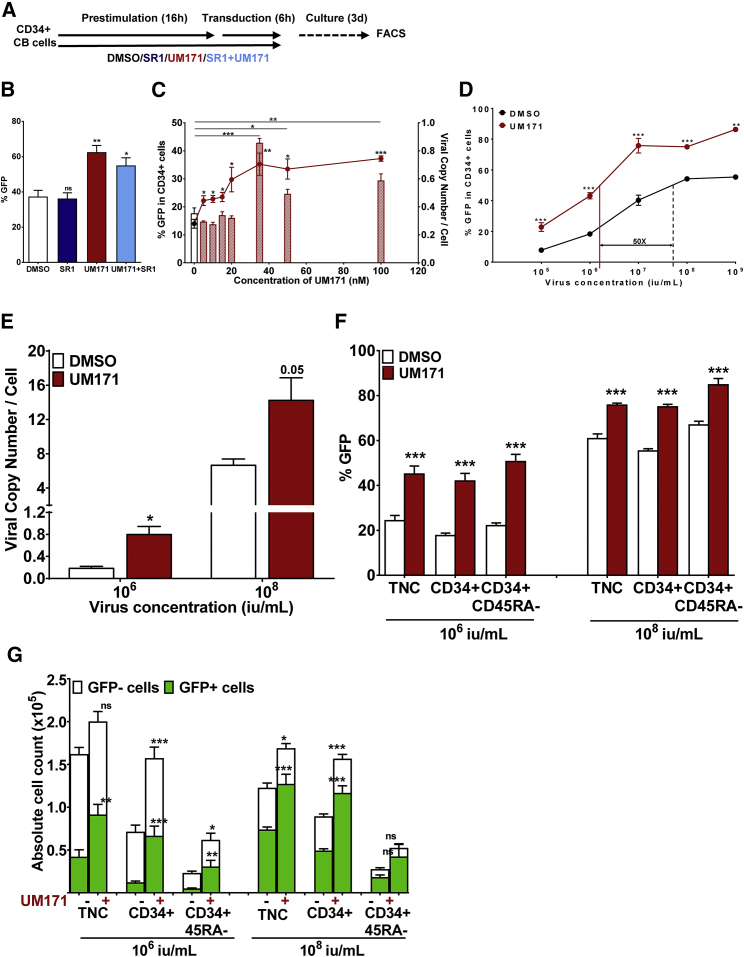
In a first series of experiments, we sought to determine how UM171 would affect LV-mediated gene transfer. To address this question, CD34+ CB cells were prestimulated for 16 hr with 100 ng/mL FLT3 ligand (FL), 100 ng/mL Steel Factor (SF), 20 ng/mL interleukin (IL)-3, IL-6, and granulocyte colony-stimulating factor (G-CSF) in a serum-free medium in the presence of UM171, the AhR antagonist SR1, or a combination of both (or neither) and then were transduced for 6 hr with green fluorescent protein (GFP)-containing lentiviral particles (MOI = 5) in the presence of the same compounds (Figure 1A). Transduction efficiency was determined by flow cytometry after an additional 3-day culture period in the same cytokine-supplemented medium but without either UM171 or SR1. UM171 enhanced transduction efficiency by ∼2-fold compared to control conditions (62 ± 4% versus 37 ± 4%, p = 0.001; Figure 1B). In contrast, the small molecule SR1, tested under the same conditions, did not have any effect on transduction efficiency, either alone or in combination with UM171 (Figure 1B). The ability of UM171 to stimulate gene transfer was dose dependent and reached plateau levels at 35 nM, as evidenced by a 2-fold increase in the proportion of GFP+ cells and as further supported by a 2-fold increase in the viral copy number (VCN) per cell assessed by qPCR (Figure 1C). UM171 also increased transduction efficiency over a broad range of virus concentrations (105 to 109 IU/mL, MOI = 0.5–5000), as shown by both measures of GFP+ cells (Figure 1D) and VCN (Figure 1D). Further highlighting UM171’s stimulatory effect is the observation that transduction efficiencies equivalent to those of control could be achieved with a 50-fold reduction in virus concentration (Figure 1D). Importantly, similar magnitudes of UM171-enhanced gene transfer to the primitive CD45RA− subset of CD34+ CB cells were also observed over a wide range of viral titers, as shown by the increased frequency of marked cells with this phenotype (Figure 1F).
Figure 1.
UM171 Enhances Lentiviral Transduction of Primitive Human Hematopoietic Cells
(A) Outline of experimental design. 20,000 CD34+ CB cells were prestimulated and transduced with a GFP LV (106 IU/mL, MOI = 5) in the presence of DMSO, SR1, UM171, or UM171+SR1. Cells were then washed and cultured further for 72 hr to assess transduction efficiency by FACS. (B) The proportion of GFP+ CD34+ cells at the end of 3-day culture. (C) CD34+ CB cells were prestimulated and transduced, as outlined in (A), in the presence of DMSO or various concentrations of UM171 (1, 5, 10, 20, 35, 50, and 100 nM). The percentage of GFP+ cells (curve) is indicated on the left y axis, and viral copy number per cell (bar graphs) is indicated on the right y axis. (D) Transduction efficiency in CD34+ cells exposed to various concentrations of virus ranging from 105 to 109 IU/mL (MOI = 0.5–5000). (E) LV copy number per cell in cells transduced in the presence of 106 or 108 IU/mL virus. (F) Transduction efficiency in various primitive hematopoietic subsets, transduced in the presence of 106 or 108 IU/mL. (G) Absolute yield of total and transduced cells across the range of hematopoietic subsets transduced in the presence of 106 or 108 IU/mL virus. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant. The data are shown as mean ± SD.
In addition to the enhanced gene transfer stimulated by UM171, significant increases were observed in the absolute yields of total and transduced CD34+ and CD34+CD45RA− cells (Figure 1G). In combination with the effect of UM171 on gene transfer efficiency, this resulted in a net 6-fold increase in transduced CD34+ and CD34+CD45RA− cells (p = 0.0002 and <0.0001 for CD34+ and CD34+CD45RA− cells at 106 IU/mL, respectively; Figure 1G).
In summary, these initial experiments demonstrated that prestimulation and transduction of HSPCs in the presence of UM171 allows for substantial and additive increases in the efficiency of lentiviral gene transfer and in the yield of transduced HPSCs subjected to a 22-hr transduction protocol.
UM171 Enhances LV-Mediated Transduction Efficiency of Functionally Defined Human HSCs
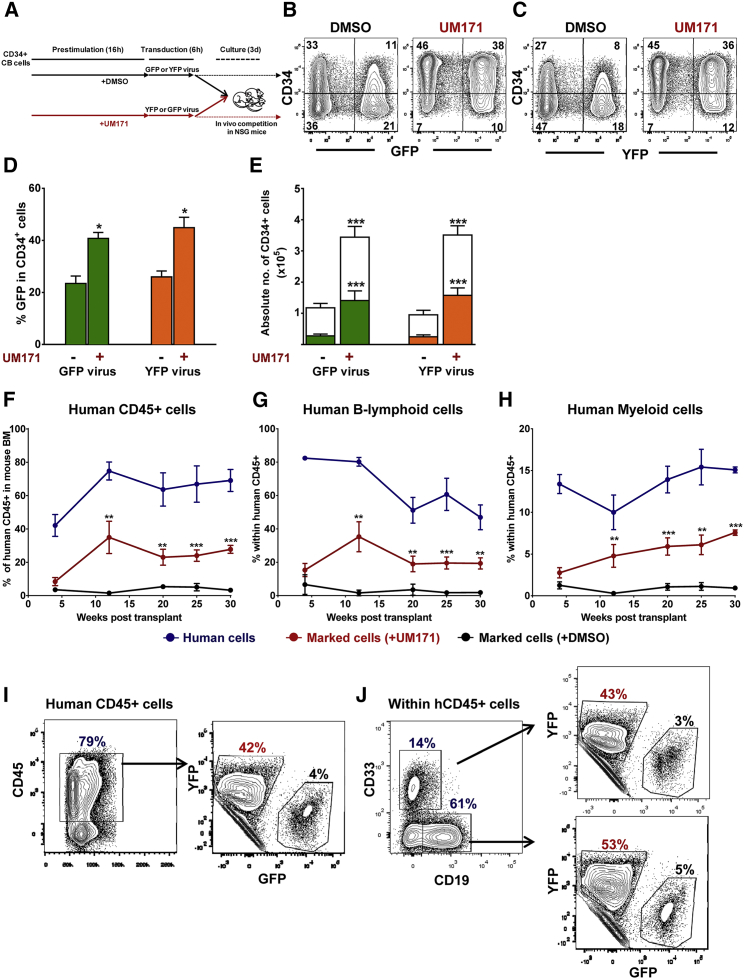
To assess the impact of UM171 on functionally defined HSCs, we used a competitive repopulation experimental design in which equal numbers of CD34+ CB cells were first transduced separately in the presence or absence of UM171 with matched GFP-containing and yellow fluorescent protein (YFP)-containing LVs and then mixed in equal ratios and co-transplanted into recipient non-obese diabetic/severe combined immunodeficiency (NOD/SCID)-IL2Rγ−/− (NSG) mice (shown schematically in Figure 2A). UM171, again, increased the overall transduction efficiency by approximately 2-fold for both types of virus (Figures 2B–2D) and enhanced the yield of transduced CD34+ cells recovered from the same 3-day post-transduction cultures (Figure 2E). Analysis of bone marrow (BM) aspirates obtained from the mice injected with cells harvested immediately after the 22-hr transduction protocol showed that cells transduced in the presence of UM171 consistently outcompeted those transduced under control conditions for up to 30 weeks of follow-up (Figures 2F–2H). At the time of sacrifice (at 30 weeks), the level of human CD45+ cells derived from human cells transduced in the presence of UM171 was an order of magnitude higher than that obtained from those transduced under the control condition (28 ± 2% versus 3 ± 0.7%, p < 0.001; Figures 2F and 2I), 8-fold for B-lymphoid cells (19 ± 3% versus 2 ± 0.4%, p < 0.01; Figures 2G and 2J), and 10-fold for myeloid cells (8 ± 0.3% versus 1 ± 0.2%, p < 0.001; Figures 2H and 2J). The reproducibility of these findings was documented in a second independent experiment that gave similar results (Figures S1A–S1C) and in a parallel experiment in which the vectors used for the experimental and control groups were exchanged (data not shown).
Figure 2.
UM171 Enhances LV Transduction of Transplantable Human HSPCs
(A) Outline of experimental design. CD34+ CB cells were prestimulated and transduced in the presence or absence of UM171 with a GFP- or a YFP-expressing vector (106 IU/mL, MOI = 1). Immediately after transduction, equal aliquots of YFP-marked and GFP-marked cells were mixed and coinjected into lethally irradiated NOD/SCID-IL2Rγ−/− (NSG) (n = 8 mice). A small aliquot of cells was cultured for 3 additional days to assess gene transfer into CD34+ cells. (B and C) Representative FACS blots showing transduction efficiency in CD34+ cells transduced with a GFP-expressing (B) or a YFP-expressing (C) virus. (D) Percentage of GFP or YFP in CD34+ cells (n = 3 experiments, each in triplicate). (E) Absolute yield of total and GFP- or YFP-transduced cells (n = 3 experiments, each in triplicate). The bar graphs in (D) and (E) represent mean values ± SD. (F) Percentage of total human engraftment in mouse BM over 30 weeks (blue). Proportion of cells transduced under UM171-stimulated condition is displayed in red, and proportion of cells transduced under control conditions is displayed in black. (G and H) Normal B-lymphoid (G) and myeloid (H) cell production over 30 weeks. Proportion of cells transduced under UM171 stimulated conditions in red and proportion of cells transduced under control conditions in black. The error bars in (F)–(H) are shown as mean values ± SEM. (I and J) FACS blots from a representative mouse BM at 30 weeks post-transplant. (I) CD45 cells. (J) CD33 and CD19 cells. *p < 0.05, **p < 0.01, ***p < 0.001.
In summary, a 22-hr exposure of CD34+ CB cells to UM171 enhances LV-mediated transduction efficiency of human HSCs with long-term-repopulating potential and does not alter their normally changing ratio over time of B-lymphoid to granulopoietic cell outputs.
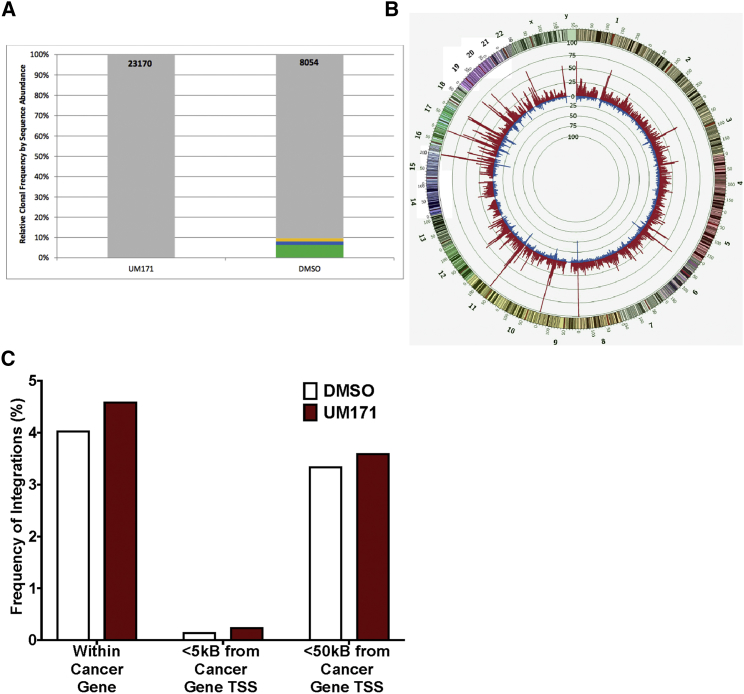
UM171 Induces Polyclonal LV-Mediated Transduction of Human HSPCs and Does Not Alter Their Chromosomal Integration Profile
We next examined whether the observed increased gene transfer to HSPCs was associated with an altered LV integration profile using modified genomic sequencing (MGS)-PCR20 to compare the retrovirus integration sites (RISs) present in CB CD34+ cells transduced in the presence or absence of UM171. Cells transduced in the presence or absence of UM171 displayed the same high degree of clonal diversity but with nearly 3-fold more unique RISs (clones) being present in cells transduced in the presence of UM171, compared to cells transduced under control conditions (23,170 and 8,054 for UM171 and DMSO, respectively, Figure 3A). Analysis of the genomic distribution of LV integration sites identified in both groups also revealed a mirror image of the histograms of site locations across the genome (Figure 3B), and the frequency of LV insertions within and proximal to cancer genes in particular was not altered by transduction of the cells in the presence of UM171 (Figure 3C).
Figure 3.
LV Genomic Integration Profile in Cells Transduced with UM171
(A) Clonal distribution of LV in CB CD34+ cells transduced in the presence of UM171 versus DMSO. The total number of unique RISs (clones) identified in each sample is listed at the top of each bar (23,170 in the presence of UM171 and 8,054 in the presence of DMSO). Each bar represents the frequency at which each clone in the sample was sequenced from greatest (bottom) to least (top). Clones sequenced at a frequency of ≥1% in the pool are designated by colored boxes. All other clones are grouped into a single gray box at the top of each bar. (B) Genome distribution of identified integration sites. Circos (www.circos.ca/) was used to plot the genomic distribution of integration sites identified in DMSO- and UM171-treated CD34+ CB cells as an inverted histogram plot. All integration sites identified are included for each experimental arm (red indicates the presence of UM171; blue indicates the presence of DMSO). The human genome (outer band, version hg19) from chromosome 1 through chromosome Y is shown with each chromosome color coded and corresponding G-banding patterns indicated. The genome was divided into 1-Mb bins. The number of unique integration sites identified in each 1-Mb bin is reflected in the height of the bin histogram. Each histogram gridline represents 25 unique integration sites. The mirror imaging of these two histograms across the genome indicates no significant difference in the global genomic pattern of integration between these two experimental arms. (C) Frequency of LV inserts within and proximal to cancer genes in cells transduced in the presence of DMSO (clear bars) and UM171 (red bars). Bar graph represents the percentage of insertions found within cancer genes or near cancer gene promoters, as determined by analysis using QuickMap. 95% CI for insertions: within cancer genes, 3–24% for DMSO and 3–10% for UM171; <5 kB from cancer genes, 0.1–21% for DMSO and 0.2–8% for UM171; and <50 kB from cancer genes, 3–24% for DMSO and 3–9.7% for UM171. *p < 0.05, **p < 0.01, ***p < 0.001.
Together, these results indicate that exposure of human CB HSPCs to UM171 during a 22-hr prestimulation and transduction protocol enhances polyclonal LV-mediated transduction without attendant alterations in the LV genomic integration profile. Thus, use of UM171 to enhance LV transduction appears unlikely to raise concerns of increased genotocixity.
UM171 Enhances Transduction of HSPCs Using a Spectrum of Viral Pseudotypes and Virus Types
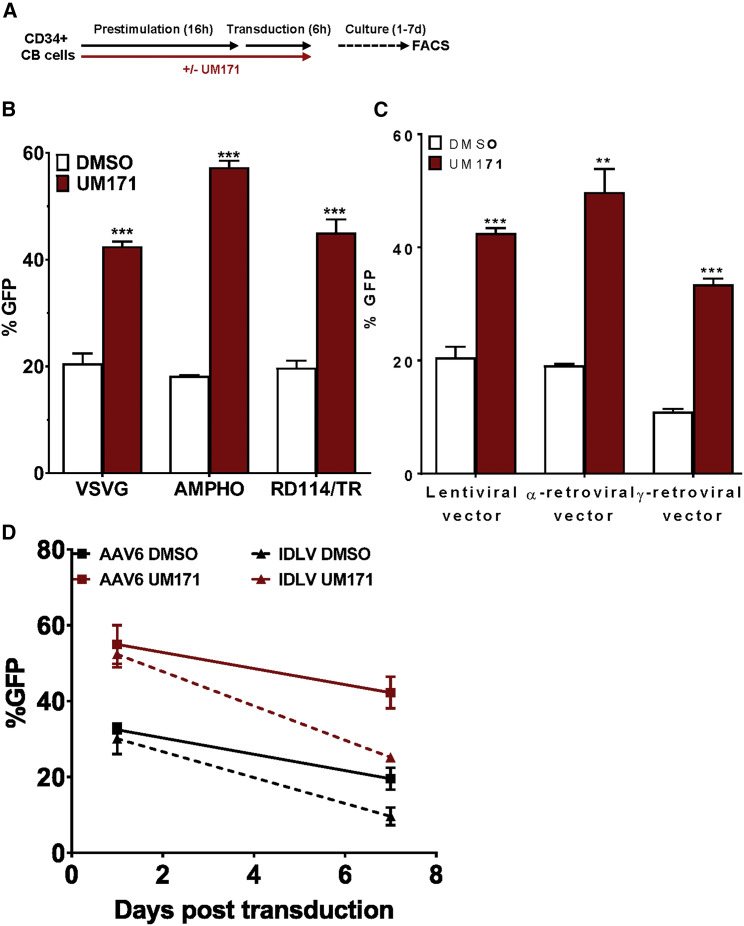
To investigate the mechanism by which UM171 increases LV-mediated transduction of human HSPCs, we first determined whether receptor-mediated viral uptake was affected. To assess this possibility, we transduced CD34+ CB cells with identical LV-GFP-encoding vectors that were pseudotyped either with the vesicular stomatitis virus glycoprotein (VSV-G) envelope,21 the amphotropic murine leukemia virus (MuLV) envelope,22 or the feline endogenous retroviral RD114-TR envelope,23 each of which targets a different known receptor. low-density lipoprotein (LDL) receptors (LDLRs) are the major entry port of VSV-G-pseudotyped lentiviral vectors21 and are dramatically upregulated on HSPCs upon cytokine treatment24 and well expressed on CB and mobilized peripheral blood (mPB) CD34+ cells (Figure S2A). The amphotropic MuLV receptor SCL20A222 and the SCL1A5 receptor for chimeric RD114-TR23 are also expressed on human HSPCs (Figures S1C and S2B). For all pseudotypes tested, UM171 enhanced transduction efficiencies to similar levels (Figures 4A and 4B), suggesting that the impact of UM171 on transduction does not occur at the level of receptor-mediated viral uptake by the cell.
Figure 4.
UM171 Enhances Transduction of CD34+ CB Cells over a Spectrum of Viral Pseudotypes and Viral Types
(A) Outline of the experimental design. (B) Proportions of GFP+ cells 3 days after transduction with an LV pseudotyped with VSV-G, AMPHO, or RD114/TR envelopes. (C) Proportion of GFP+ cells 3 days after transduction with α- and γ-RVs pseudotyped with the VSV-G envelope. (D) Proportion of GFP+ cells 1 and 7 days after transduction with nonintegrating viral vectors such as AAV6 (black and red squares) and integrase-defective LV (IDLV; black and red triangles). **p < 0.01, ***p < 0.001. The error bars represent mean values ± SD.
We next tested whether UM171-mediated enhanced transduction efficiency might be specific to LVs that can transduce nondividing cells due to their ability to transport proviral DNA through the nuclear pore complex (NPC). The avian sarcoma-leukosis alpha-retrovirus (α-RV) is of particular interest, because it has a neutral chromosomal integration pattern,23 which is a significant safety feature for HSC-based gene therapy strategies. Accordingly, we first pseudotyped the α-RV and γ-RV vectors with a VSV-G envelope to change their target cell spectrum to that dictated by VSV-G, and we then used these pseudotyped vectors to transduce CB HSPCs in the same short 22-hr protocol used for the LV transduction experiments. The presence of UM171 in this protocol with VSV-G pseudotyped RVs gave a significant increase in the transduction efficiency obtained with both the α- and γ-retroviral vectors (Figure 4C).
We then tested whether UM171 could also enhance the efficiency of transduction obtained using nonintegrating viral vectors, such as integrase-defective LV (IDLV)25 and adeno-associated virus (AAV) serotype 6 (AAV6).26 In these experiments, UM171, again, produced a significant increase in transduction efficiencies of IDLV and AAV6 to CB HSPCs (Figure 4D).
UM171-Mediated Transduction Enhancement Extends to Adult Hematopoietic Cells
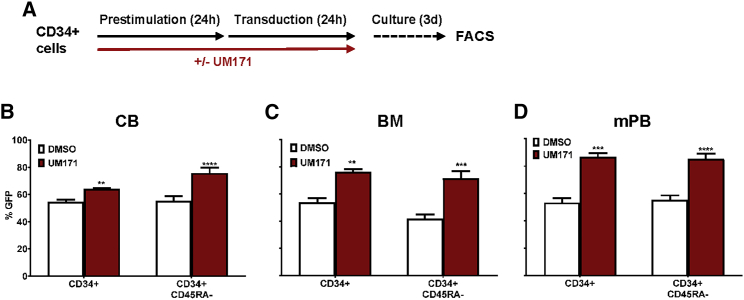
To determine whether the UM171 effect on gene transfer to CB HSPCs is dependent on their early developmental stage, we undertook additional experiments of the same design using CD34+ cells isolated from normal human adult BM and G-CSF-mobilized mPB cell samples. Preliminary data indicated that longer exposure to higher concentrations of virus is required for the transduction of HSPCs from human BM and mPB. Therefore, we prestimulated the adult cells for 24 hr with the same 5-growth factor-supplemented serum-free medium (SFM) as that used for the CB transduction experiments prior to transducing them for an additional 24 hr in the same growth factor-supplemented medium with an LV-GFP virus (107 IU/mL, MOI = 50), either with or without UM171 throughout the full 48 hr (Figure 5A). As observed for CD34+ CB cells, UM171 enhanced approximately 1.5-fold the transduction of both the CD34+ cells and the CD45RA− subset of CD34+ cells from normal adult human BM and mPB harvests (Figures 5B and 5C).
Figure 5.
UM171 Enhances Lentiviral Transduction to Adult HSPCs
(A) Outline of experimental design. CD34+ CB, adult BM, and mPB were prestimulated for 24 hr and transduced for 24 hr at a virus concentration of 107 IU/mL (MOI = 50) in the presence or absence of UM171 (n = 3 experiments, each in triplicate). (B–D) The proportion of GFP+ cells at the end of a 3-day culture shown for (B) CB, (C) adult BM, and (D) mPB HSPCs. **p < 0.01, ***p < 0.001, and ****p < 0.0001. The bar graphs represent mean values ± SD.
Taken together, these results suggest that a simple ex vivo manipulation protocol, as short as 48 hr, can achieve substantial increases in LV gene transfer levels in human HSPCs from adult as well as CB sources.
Discussion
Achieving efficient gene transfer to human HSPCs under culture conditions that preserve primitive HSCs is a prerequisite for clinical gene therapy applications. Previous studies identified the small molecule UM729 and its related derivative UM171 to be highly effective in promoting the in vitro expansion of primitive human CB HSCs, including those with long-term repopulation activity.17 Here, we show that very brief in vitro exposure to UM171 also enhances LV transduction of human LT-HSCs, as assessed both phenotypically and functionally in xenotransplant experiments. Another small molecule, PGE2, was shown to enhance transduction of human HSPCs by VSV-G-pseudotyped LV in 24- to 36-hr cultures; however, this compound was found to be effective only at saturating virus concentrations.27 Other pharmacological agents, such as rapamycin and CsA, have also been reported to enhance transduction of human HSPCs; however, since these target proteins involved in cell survival, HSPC toxicity could be an issue.11 UM171, on the other hand, preserves HSC activity and mediates its effects over a wide range of virus titers.
UM171 also enhances the repopulating potential of adult CD34+CD38- mPB cells27 and, as shown here, likewise enhances gene transfer to the CD34+ and CD34+CD45RA− cells from this source or adult BM. This could have an important impact on future gene therapy trials, since many of these use autologous HSPCs from adult BM or mPB and require greater yields of transduced HSCs to enable gene editing approaches to become feasible.
UM171-mediated enhanced LV transduction was observed for LVs pseudotyped with VSV-G, amphotropic MuLV, and feline endogenous retroviral RD114-TR envelopes. All of these envelope proteins mediate internalization via different entry pathways, suggesting that UM171 does not enhance gene transfer by altering the level of receptor-mediated viral uptake. UM171 also did not affect LDLR expression, even after 24 hr of UM171 treatment, when most of the viral entry has occurred (data not shown). UM171 has no mitogenic activity on human HSPCs,17 but it may, however, alter cellular metabolism and promote mitosis, thereby facilitating reverse transcription and integration even with α- and γ- retroviral vectors that lack the ability to transduce freshly isolated human HSPCs that are quiescent and variably enter S-phase over several more days. This contrasts with the more restricted ability of PGE2 to enhance gene transfer to adult HSPCs by viruses bearing endocytosis-dependent viral envelopes.28
In conclusion, our data support the idea that agents such as UM171’s dual properties of enhancing transduction and stimulating ex vivo expansion of human HSPCs, may provide potential benefits for gene therapy and gene editing applications.
Materials and Methods
Viral Vectors and Virus Production
The pCCl-c-MNDUSpgkGFP and pCCl-c-MNDUSpgkYFP lentiviral vector backbones used in these studies were described by Logan et al.29 High-titer lentiviral vectors were produced as previously described by standard CaPO4-mediated transfection of 293T cells using either VSV-G, RD114/TR, or Ampho envelopes. α- and γ-RV vector particles were produced as previously described.30, 31, 32 The phCMV-RD114/TR plasmid-encoding RD114/TR envelope was provided by Francois-Loic Cosset, and the plasmid phCMV-Ampho encoding the transgene of the amphotropic envelope was a gift from Miguel Sena-Esteves. Pseudotyped RV particles were concentrated by ultracentrifugation at 25,000 rpm and 4°C for 1.5 hr (VSV-G) or at 10,000 rpm and 4°C for 16 hr (when Ampho and RD114/TR envelopes were used). Aliquots of viral supernatants were titrated on HeLa or K562 cells and stored at −80°C. The concentrated supernatant of the integrase-defective lentiviral vector, CCLc-MND-GCP IDLV25 was kindly provided by Donald B. Kohn (Department of Microbiology, Immunology, and Molecular Genetics and Department of Pediatrics, University of California, Los Angeles). AAV6 was kindly provided by Aravind Asokan (Gene Therapy Center, University of North Carolina at Chapel Hill).26, 33 The α-RV and γ-RV were kindly provided by Dr. Axel Schambach (Hannover Medical School, Hannover, Germany).
Transduction of Human Hematopoietic Cells
Umbilical CB and G-CSF mPB cells were collected with informed consent according to procedures approved by the research ethics board of the University of British Columbia. CD34+ cell-enriched adult BM cells were purchased from STEMCELL Technologies. CD34+ CB and mPB cells were enriched to >90% purity using, first, the RosetteSep CD34 preenrichment cocktail (STEMCELL Technologies), followed by positive selection using magnetic beads (EasySep Kit, STEMCELL Technologies). In some cases, additional enrichment was achieved by sorting CD34+ cells using an Influx II sorter (BD Biosciences). CD34+ CB cells were pre-stimulated for 16 hr in SFM (Iscove’s medium supplemented with BSA, insulin, and transferrin [BIT]; STEMCELL Technologies), 10 μg/mL LDL (STEMCELL Technologies), 10−4 M 2-mercaptoethanol (Sigma-Aldrich), 10−4 M glutamax 500 (STEMCELL Technologies), penicillin and streptomycin plus 100 ng/mL FL, 100 ng/mL SF, 20 ng/mL IL-3, IL-6, and G-CSF (all from STEMCELL Technologies). Adult CD34+ BM and mPB cells were prestimulated for 24 hr in SFM supplemented with 100 ng/mL FL, 100 ng/mL SF, 100 ng/mL thrompopoietin (TPO), and 20 ng/mL IL-3. The cells were prestimulated in the presence or absence of in-house-prepared UM171 (35 nM), SR1 (0.75 μM; purchased from Alichem, catalog no. 41864), or DMSO (not exceeding 0.01%), as indicated. At the end of the prestimulation period, cells were resuspended in fresh growth factor-supplemented SFM with concentrated lentivirus (GFP or YFP) and 5 μg/mL protamine sulfate and incubated at 37°C for 6 hr for CB at virus concentrations ranging from 105 to 109 IU/mL, and for 24 hr for adult BM and mPB at a virus concentration of 107 IU/mL (MOI = 50), and placed in a 96-well plate coated with 5 μg/cm2 fibronectin (Sigma-Aldrich). In experiments using different viral vectors and pseudotypes, CD34+ CB cells were always transduced at virus concentrations of 106 IU/mL (MOI = 5).
Mice
NOD.Cg-Prkdcscid Il2rγtm1Wj1/SzJ (NSG) mice, originally obtained from Jackson Laboratory were bred in the animal resource center at the British Columbia Cancer Research Centre. All mouse experimental procedures were carried out in accordance with Canadian Council on Animal Care guidelines, with approval from the University of British Columbia.
Xenotransplantation and Tracking of Transplanted Transduced Human Cells
Eight- to 12-week-old NSG mice were sublethally irradiated (315 cGy of 137Cs γ-rays) 24 hr prior to transplantation and then injected intravenously with the progeny of 20,000 CD34+ CB cells transduced in the presence of UM171 and of 20,000 CD34+ CB cells transduced in the presence of DMSO. Human lympho-myeloid reconstitution in NSG mice was monitored over 30 weeks in BM aspirates obtained at 3, 12, 20, and 25 weeks post-transplant and in flushed femurs, tibias, and pelvic bones removed when animals were sacrificed at week 30.
Flow Cytometry
The efficiency of gene transfer to total CD34+ cells and the CD45RA− subset thereof was determined after staining the cells with the following anti-human-specific antibodies (all from eBioscience, unless noted otherwise): CD34-APC (clone 8G12, STEMCELL Technologies), and CD45RA-APC780 (clone HI100).
For phenotypic analysis of human cells from NSG mice, flow-cytometric analysis was performed on freshly collected BM cells. Cells were treated with red blood cell lysis buffer (STEMCELL Technologies), washed, and incubated with a blocking reagent (PBS with 2% fetal bovine serum (FBS), 5% human serum, and anti-CD16/CD32 antibody [2.4G2]). The cells were then stained with the following anti-human-specific antibodies: CD45-Alexa Fluor 700 (clone HI30, BioLegend), CD33-PECY7 (clone WM-53, eBioscience), CD19-PE (clone HIB19, BioLegend), CD20-PE (clone L27, StemCell Technologies). A minimum of 200,000 BM cells were analyzed per mouse. A negative control was set on non-transplanted mouse BM. In some cases, we utilized a fluorescence-minus-one (FMO) gating strategy to overcome the ambiguity created by artifacts that could be introduced by simultaneously compensating different fluorochromes.
LV Copy Number Determination
Human CB CD34+ cells (20,000 cells) were transduced in the presence or absence of UM171. After transduction, cells were washed and cultured for 10 days prior to extraction of genomic DNA using DNAzol Reagent (Invitrogen). Mean vector copy numbers were determined by multiplex TaqMan qPCR on an ABI 7900HT (Applied Biosystems), as previously described,34 with minor modifications. Integrated vectors were quantified by amplification of their GFP or YFP transgene using either primer pair GFP_FW (5′-GTA GCG GCT GAA GCA CTG-3′) and GFP_RV (5′-CTG CAC CAC CGG CAA-3′) in combination with a GFP probe (/56-FAM/CC ACC CTG A/Zen/C CTA CGG CGT G/3IABkFQ/) or primer pair GFP/YFP2_FW (5′-GAA CCG CAT CGA GCT GAA-3′) and GFP/YFP2_RV (5′-TGC TTG TCG GCC ATG ATA TAG-3′) in combination with a GFP/YFP2 probe (/56-FAM/ATCGACTTC/ZEN/AAGGAGGACGGCAAC/3IABkF). Values were normalized for input DNA by amplification of the PTBP2 reference gene using PTBP2_FW (5′-TCT CCA TTC CCT ATG TTC ATG C-3′) and PTBP2_RV (5′-GTT CCC GCA GAA TGG TGA GGT G-3′) primers and a PTBP2 probe (/5TET/AT GTT CCT C/Zen/G GAC CAA CTT G/3IABkFQ/). PCR efficiencies were determined by serial dilutions of a plasmid harboring GFP and PTBP2 target sequences and subsequent calculation of mean vector copy numbers according to Pfaffl.35
Lentivirus Integration Site Analysis by MGS-PCR
Genomic DNA (gDNA) was extracted from CB cells using the QIAGEN Blood DNA Mini Kit (QIAGEN). Lentivirus LTR-genome junctions were amplified by modified MGS-PCR, as described previously.20 The resulting sequence libraries were subjected to Ion Torrent semiconductor sequencing, and the resulting sequence reads were analyzed using the Vector Integration Site Analysis (VISA) server.36 Genomic sequences were mapped to the human genome (hg19) using a stand-alone version of BLAT available from the UCSC (University of California, Santa Cruz) Genome Browser. The total number of sequences corresponding to a mappable integration event was recorded. Sequences corresponding to the same integration locus were grouped together to determine the total number of unique integration sites (clones) identified in the sample. Relative contributions of each clone were determined by the number of insertion site (IS)-associated sequence reads corresponding to that clone. A quality control check was performed to reveal clones over-represented by PCR bias by comparing the number of IS-associated sequence reads with the number of different fragment lengths observed for each genomic locus. Circos was used to visualize global genomic integration patterns for aesthetic comparison.37 For statistical analysis of integrations within genomic elements, we used QuickMap.38
Statistical Analysis
Results are shown as mean ± SEM or SD. Differences between groups were assessed using the Student’s t test (paired or unpaired as appropriate) directly calculated on Prism GraphPad. Statistical significance is indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Author Contributions
M.N., T.M., S.I., and R.K.H. designed and carried out experiments relating to UM171’s effect; J.E.A. carried out integration analysis; C.E., J.C., I.F., G.S., and P.L. helped in the experimental designs and/or interpretation of results; M.N., S.I., C.E., and R.K.H. wrote and M.-E.B. and G.S. reviewed the manuscript.
Acknowledgments
All experiments were conducted in Vancouver, BC, Canada. This work was supported by a Terry Fox Foundation Program Project grant (TFF-122869) and by a Stem Cell Network Global Research grant 9/5264(CT10). We would like to thank Dr. Paul Miller for helpful discussions, Glen Edin for lentivirus production and generation of BM aspirates, and the staff of the Stem Cell Assay Laboratory, Flow Core, and the Animal Resource Center of the British Columbia Cancer Agency Research Centre for excellent technical support. The project management support of Amy Yu is also gratefully acknowledged.
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.06.009.
Supplemental Information
References
- 1.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacein-Bey Abina S., Gaspar H.B., Blondeau J., Caccavelli L., Charrier S., Buckland K., Picard C., Six E., Himoudi N., Gilmour K. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA. 2015;313:1550–1563. doi: 10.1001/jama.2015.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Veres G., Schmidt M., Kutschera I., Vidaud M., Abel U., Dal-Cortivo L., Caccavelli L. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 6.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., Bougnères P., Schmidt M., Kalle C.V., Fischer A., Cavazzana-Calvo M., Aubourg P. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol. 2012;507:187–198. doi: 10.1016/B978-0-12-386509-0.00010-7. [DOI] [PubMed] [Google Scholar]
- 7.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 8.Sessa M., Lorioli L., Fumagalli F., Acquati S., Redaelli D., Baldoli C., Canale S., Lopez I.D., Morena F., Calabria A. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet. 2016;388:476–487. doi: 10.1016/S0140-6736(16)30374-9. [DOI] [PubMed] [Google Scholar]
- 9.Naldini L., Trono D., Verma I.M. Lentiviral vectors, two decades later. Science. 2016;353:1101–1102. doi: 10.1126/science.aah6192. [DOI] [PubMed] [Google Scholar]
- 10.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 11.Wang C.X., Sather B.D., Wang X., Adair J., Khan I., Singh S., Lang S., Adams A., Curinga G., Kiem H.P. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124:913–923. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrillo C., Cesana D., Piras F., Bartolaccini S., Naldini L., Montini E., Kajaste-Rudnitski A. Cyclosporin a and rapamycin relieve distinct lentiviral restriction blocks in hematopoietic stem and progenitor cells. Mol. Ther. 2015;23:352–362. doi: 10.1038/mt.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santoni de Sio F.R., Cascio P., Zingale A., Gasparini M., Naldini L. Proteasome activity restricts lentiviral gene transfer into hematopoietic stem cells and is down-regulated by cytokines that enhance transduction. Blood. 2006;107:4257–4265. doi: 10.1182/blood-2005-10-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoni de Sio F.R., Gritti A., Cascio P., Neri M., Sampaolesi M., Galli C., Luban J., Naldini L. Lentiviral vector gene transfer is limited by the proteasome at postentry steps in various types of stem cells. Stem Cells. 2008;26:2142–2152. doi: 10.1634/stemcells.2007-0705. [DOI] [PubMed] [Google Scholar]
- 15.Maiese K. Stem cell guidance through the mechanistic target of rapamycin. World J. Stem Cells. 2015;7:999–1009. doi: 10.4252/wjsc.v7.i7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boitano A.E., Wang J., Romeo R., Bouchez L.C., Parker A.E., Sutton S.E., Walker J.R., Flaveny C.A., Perdew G.H., Denison M.S. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fares I., Chagraoui J., Gareau Y., Gingras S., Ruel R., Mayotte N., Csaszar E., Knapp D.J., Miller P., Ngom M. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345:1509–1512. doi: 10.1126/science.1256337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goessling W., Allen R.S., Guan X., Jin P., Uchida N., Dovey M., Harris J.M., Metzger M.E., Bonifacino A.C., Stroncek D. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8:445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beard B.C., Adair J.E., Trobridge G.D., Kiem H.P. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol. Biol. 2014;1185:321–344. doi: 10.1007/978-1-4939-1133-2_22. [DOI] [PubMed] [Google Scholar]
- 21.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlic D., Girard L.J., Jordan C.T., Anderson S.M., Cline A.P., Bodine D.M. The level of mRNA encoding the amphotropic retrovirus receptor in mouse and human hematopoietic stem cells is low and correlates with the efficiency of retrovirus transduction. Proc. Natl. Acad. Sci. USA. 1996;93:11097–11102. doi: 10.1073/pnas.93.20.11097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labenski V., Suerth J.D., Barczak E., Heckl D., Levy C., Bernadin O., Charpentier E., Williams D.A., Fehse B., Verhoeyen E., Schambach A. Alpharetroviral self-inactivating vectors produced by a superinfection-resistant stable packaging cell line allow genetic modification of primary human T lymphocytes. Biomaterials. 2016;97:97–109. doi: 10.1016/j.biomaterials.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 24.Amirache F., Lévy C., Costa C., Mangeot P.E., Torbett B.E., Wang C.X., Nègre D., Cosset F.L., Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 25.Joglekar A.V., Hollis R.P., Kuftinec G., Senadheera S., Chan R., Kohn D.B. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol. Ther. 2013;21:1705–1717. doi: 10.1038/mt.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madigan V.J., Asokan A. Engineering AAV receptor footprints for gene therapy. Curr. Opin. Virol. 2016;18:89–96. doi: 10.1016/j.coviro.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zonari E., Desantis G., Petrillo C., Boccalatte F.E., Lidonnici M.R., Kajaste-Rudnitski A., Aiuti A., Ferrari G., Naldini L., Gentner B. Efficient ex vivo engineering and expansion of highly purified human hematopoietic stem and progenitor cell populations for gene therapy. Stem Cell Reports. 2017;8:977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heffner G.C., Bonner M., Christiansen L., Pierciey F.J., Campbell D., Smurnyy Y., Zhang W., Hamel A., Shaw S., Lewis G. Prostaglandin E2 increases lentiviral vector transduction efficiency of adult human hematopoietic stem and progenitor cells. Mol. Ther. 2018;26:320–328. doi: 10.1016/j.ymthe.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logan A.C., Nightingale S.J., Haas D.L., Cho G.J., Pepper K.A., Kohn D.B. Factors influencing the titer and infectivity of lentiviral vectors. Hum. Gene Ther. 2004;15:976–988. doi: 10.1089/hum.2004.15.976. [DOI] [PubMed] [Google Scholar]
- 30.Schambach A., Mueller D., Galla M., Verstegen M.M., Wagemaker G., Loew R., Baum C., Bohne J. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- 31.Suerth J.D., Maetzig T., Galla M., Baum C., Schambach A. Self-inactivating alpharetroviral vectors with a split-packaging design. J. Virol. 2010;84:6626–6635. doi: 10.1128/JVI.00182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suerth J.D., Maetzig T., Brugman M.H., Heinz N., Appelt J.U., Kaufmann K.B., Schmidt M., Grez M., Modlich U., Baum C., Schambach A. Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol. Ther. 2012;20:1022–1032. doi: 10.1038/mt.2011.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song L., Kauss M.A., Kopin E., Chandra M., Ul-Hasan T., Miller E., Jayandharan G.R., Rivers A.E., Aslanidi G.V., Ling C. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy. 2013;15:986–998. doi: 10.1016/j.jcyt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maetzig T., Kuehle J., Schwarzer A., Turan S., Rothe M., Chaturvedi A., Morgan M., Ha T.C., Heuser M., Hammerschmidt W. All-in-One inducible lentiviral vector systems based on drug controlled FLP recombinase. Biomaterials. 2014;35:4345–4356. doi: 10.1016/j.biomaterials.2014.01.057. [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hocum J.D., Battrell L.R., Maynard R., Adair J.E., Beard B.C., Rawlings D.J., Kiem H.P., Miller D.G., Trobridge G.D. VISA--Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinformatics. 2015;16:212. doi: 10.1186/s12859-015-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Appelt J.U., Giordano F.A., Ecker M., Roeder I., Grund N., Hotz-Wagenblatt A., Opelz G., Zeller W.J., Allgayer H., Fruehauf S., Laufs S. QuickMap: a public tool for large-scale gene therapy vector insertion site mapping and analysis. Gene Ther. 2009;16:885–893. doi: 10.1038/gt.2009.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.