Abstract
Skeletal muscle is ideal for passive vaccine administration as it is easily accessible by intramuscular injection. Recombinant adeno-associated virus (rAAV) vectors are in consideration for passive vaccination clinical trials for HIV and influenza. However, greater human skeletal muscle transduction is needed for therapeutic efficacy than is possible with existing serotypes. To bioengineer capsids with therapeutic levels of transduction, we utilized a directed evolution approach to screen libraries of shuffled AAV capsids in pools of surgically resected human skeletal muscle cells from five patients. Six rounds of evolution were performed in various muscle cell types, and evolved variants were validated against existing muscle-tropic serotypes rAAV1, 6, and 8. We found that evolved variants NP22 and NP66 had significantly increased primary human and rhesus skeletal muscle fiber transduction from surgical explants ex vivo and in various primary and immortalized myogenic lines in vitro. Importantly, we demonstrated reduced seroreactivity compared to existing serotypes against normal human serum from 50 adult donors. These capsids represent powerful tools for human skeletal muscle expression and secretion of antibodies from passive vaccines.
Keywords: AAV, passive vaccine, directed evolution, skeletal muscle, human, screen, functional transduction
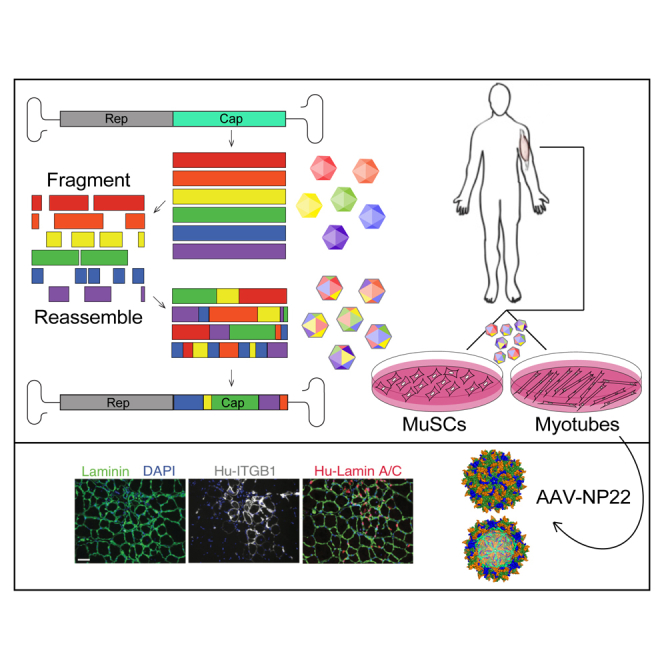
Graphical Abstract
Introduction
Skeletal muscle is the largest internal organ in the human body. It represents a desirable platform for expressing and secreting proteins1 of interest into the general circulation. Recombinant adeno-associated virus (rAAV) has received considerable attention for its proposed and current use in human clinical trials as a delivery vector for passive vaccines against HIV2, 3, 4 and influenza5, 6, 7, 8, 9 most notably. Passive vaccination is a therapeutic technology where antibodies against pathogenic agents are administered to patients, rather than the pathogenic agent itself. The cDNA encoding these antibodies must be effectively and safely delivered to humans via a vector, like rAAV, for continuous expression and secretion into the bloodstream at therapeutic levels.
Numerous characteristics define the desirability of rAAV as a passive vaccine vector; most importantly, it has an impressive safety record after demonstrated use in nearly 200 clinical trials.10 When vectorized, it transduces both dividing and non-dividing cells and shows stable expression in quiescent tissues like skeletal muscle through unintegrated episomes.11 Despite the ability of rAAV to transduce a variety of tissues, skeletal muscle has historically been one of the most challenging to transduce at levels sufficient to provide therapeutic expression of delivered products. Indeed, naturally occurring rAAV serotypes have seen limited success clinically for intramuscular delivery of transgene products in gene therapy trials for skeletal muscle disorders.12, 13, 14, 15, 16, 17, 18 This likely stemmed from the fact that preclinical modeling with rAAV to determine the best serotypes for transducing skeletal muscle was done in mice, which often don’t recapitulate the tissue tropism or transduction capabilities that can be expected in human patients at treatment.
Several preclinical studies and one ongoing clinical trial have begun piloting the use of existing rAAVs as vectors to deliver antibody expression cassettes in passive vaccine approaches for HIV/simian immunodeficiency virus (SIV),2, 3, 4 influenza,5, 6, 7, 8, 9 henipavirus,19 and human papilloma virus.20, 21, 22, 23 The promise of passive immunoprotection against pathogenic viruses has renewed the interest in obtaining rAAV capsids capable of highly efficient human intramuscular delivery. Given the modest human skeletal muscle transduction with existing rAAV serotypes,12, 13, 14, 15, 16, 17, 18 we sought to bioengineer a clinical candidate that could efficiently transduce human skeletal muscle at levels sufficient to express therapeutic levels of broad-spectrum antibodies for passive vaccine approaches. Using skeletal muscle as a platform for antibody expression, we would bypass the need to produce an endogenous adaptive immune response, as is needed with traditional vaccination. To accomplish this goal, we utilized directed evolution by DNA gene shuffling to screen for and evolve capsids with high-efficiency human skeletal muscle transduction in primary human skeletal muscle cells specifically.
Our evolved AAV vectors AAV-NP22 and AAV-NP66 show significantly increased human skeletal muscle transduction over existing muscle-tropic AAV vectors. AAV-NP22 and AAV-NP66 can be immediately implemented into passive vaccine workflows to enable significantly greater transduction for patients from a single intramuscular administration of a passive vaccine.
Results
Screening Diverse AAV Capsid Libraries in Primary Human Skeletal Muscle Cells
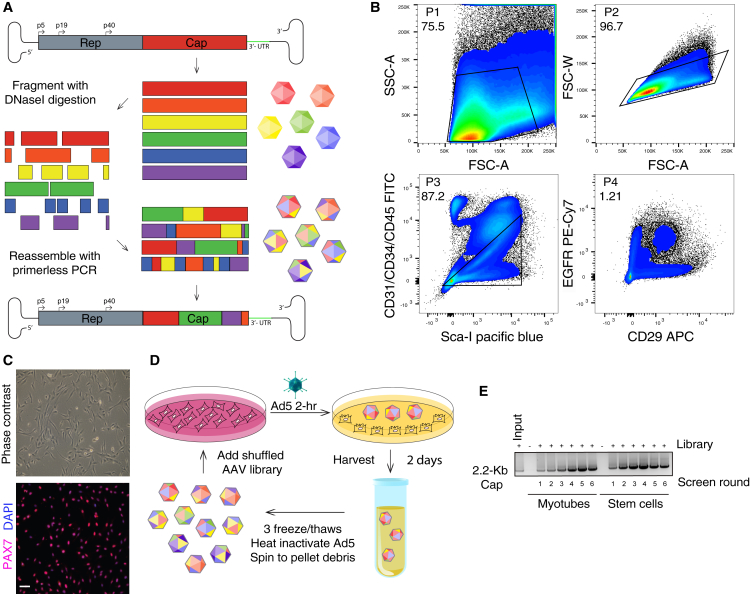
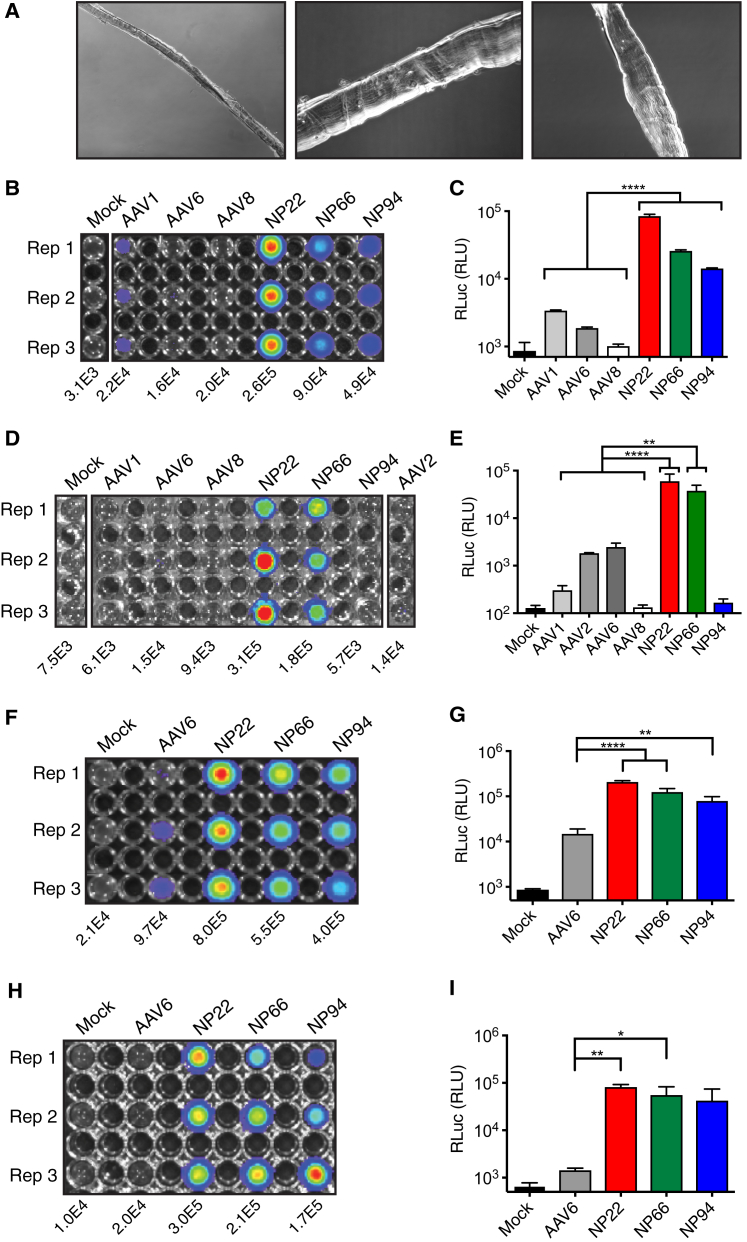
Diversifying AAV capsid proteins begins with shuffling families of capsid genes from an array of AAV pseudo-species that are enzymatically shuffled and recombined to create a library of chimeras that can be cloned into an AAV shuttle vector to produce live replicating viral libraries (Figure 1A). We constructed a diverse library from 10 different parental serotypes (1, 2, 3b, 4, 5, 6, 8, 9_hu14, avian, and bovine). To maximize the likelihood that our eventual shuffled capsids could functionally transduce human skeletal muscle—as compared to those from model organisms typically used for pre-clinical evaluation—we performed two simultaneous screens in both primary human skeletal muscle stem cells (hMuSCs) and human muscle myotubes. Surgical skeletal muscle specimens from five patients were digested, and fluorescence-activated cell sorting (FACS) was used to isolate a defined hMuSC population that was CD31−CD34−CD45−EGFR+ITGB1+24 (Figure 1B). This hMuSC population can be maintained in an undifferentiated and proliferative state (Figure 1C), or it can be differentiated in short-term cultures to produce human myotubes.24 These cell types were separately pooled at equal ratios from five patient explants to maximize cellular/patient variation for screening. Replicating screens (Figure 1D) were carried out for six rounds of selection (Figure 1E), with diversity monitoring via Sanger sequencing until the end of the screens when selection pressure plateaued (Figures S1A and S1B).
Figure 1.
Directed Evolution of AAV Capsids by DNA Shuffling and Selection Screening on Primary Human Skeletal Muscle Stem Cells and Myotubes
(A) AAV capsid genes from ten parental serotypes (1, 2, 3b, 4, 5, 6, 8, 9_hu14, avian, and bovine) were PCR-amplified, fragmented with DNaseI digestion, and then randomly reassembled through self-priming PCR. Resultant shuffled Cap genes were cloned back into a replication-competent AAV production plasmid via flanking SwaI/NsiI sites downstream of AAV2 Rep. The resultant library production plasmid contained AAV2 ITRs and a modified AAV2 3′ UTR. The AAV library was packaged using standard production protocols as done previously,41 dot blot titered, and used for selection. (B) Sequential gating scheme Panels [P]1–P4) to isolate CD31−CD34−CD45−ITGB1+EGFR+ primary human skeletal muscle stem cells from surgical specimens. Numbers indicate percentage of total events falling within each gate. (C) Phase contrast and immunofluorescence PAX7 (pink) and DAPI (blue) nuclear staining of purified primary human muscle stem cell cultures. Scale bar, 100 μm. (D) Diagram illustrating the two selection screens with replicating AAV capsid libraries from (A) that was performed on both pooled primary human skeletal muscle stem cells and human myotubes. (E) Semiquantitative AAV Cap PCR (2.2-Kb product) was performed at each round of each selection screen to demonstrate active replication of AAV library genomes throughout each round of the screen.
Structural and Comparative Computational Modeling Identifies Key Functional Motifs from Parental Serotypes
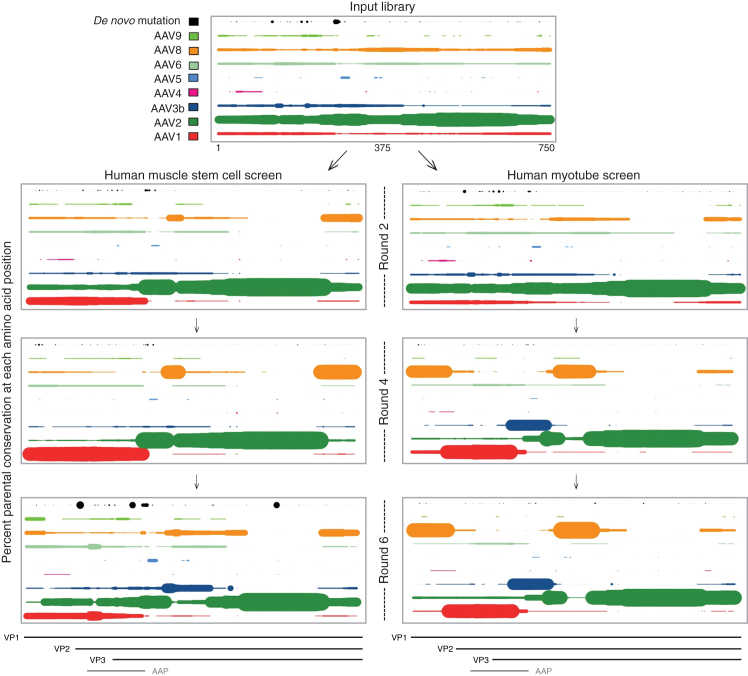
At the completion of both screens, the input library and every other selection round from each screen were analyzed using PacBio single-molecule high-throughput DNA sequencing for full-length capsid sequences. Round-to-round positional analyses for each screen identified the selection of key residues as the screens progressed (Figure 2), and they were far more informative than traditional phylogenetic representations that root the tree on the nearest full-length parental sequence, thereby masking functionally important residues within full-length capsid relatedness (Figure S2). For example, although AAV2 was the most highly represented parental sequence in the input library, evolved chimeras rapidly converged on non-AAV2 amino acids within just 2 rounds of screening. In the myotube screen, rapid selection occurred for the unique region of VP1 converging on AAV8, as well as the unique region of VP2 and nearly all of AAP from AAV1. VP3 selection heavily favored N-terminal AAV1 contributions, followed by AAV3b, AAV8, and lastly AAV2 at the C-terminal end. The muscle stem cell screen displayed a very different pattern wherein the unique region of VP1 was nearly all AAV1, even as early as round 2. Much of the VP2 and VP3 parental contributions were similar to the myotube variants. An exception occurred in the C-terminal end of VP3, which showed enrichment for AAV8 rather than AAV2 sequences.
Figure 2.
Percent Parental Conservation at Each Amino Acid Position during Screen Progression
Using PacBio single-molecule sequencing and bioinformatic analyses, positional assessments were performed to calculate percent conservation among amino acids from parental serotypes (AAVs 1, 2, 3b, 4, 5, 6, 8, and 9_hu14) or de novo mutations for each amino acid position among all capsids from key rounds of the hMuSC and myotube screens. Bovine and avian were removed from the plot as no variants showed contribution from those serotypes. Maximum dot size indicates 100% of variants share that amino acid from that parent at that position. All other dot diameters are proportional to the percent of variants from 0% to 100% having that amino acid at that position from that parent. Each parent is colored as shown in the legend (same color scheme in Figures 3A and 3B), and de novo mutations that evolved during the screens are black. VP1, VP2, VP3, and AAP ORFs are shown below for reference.
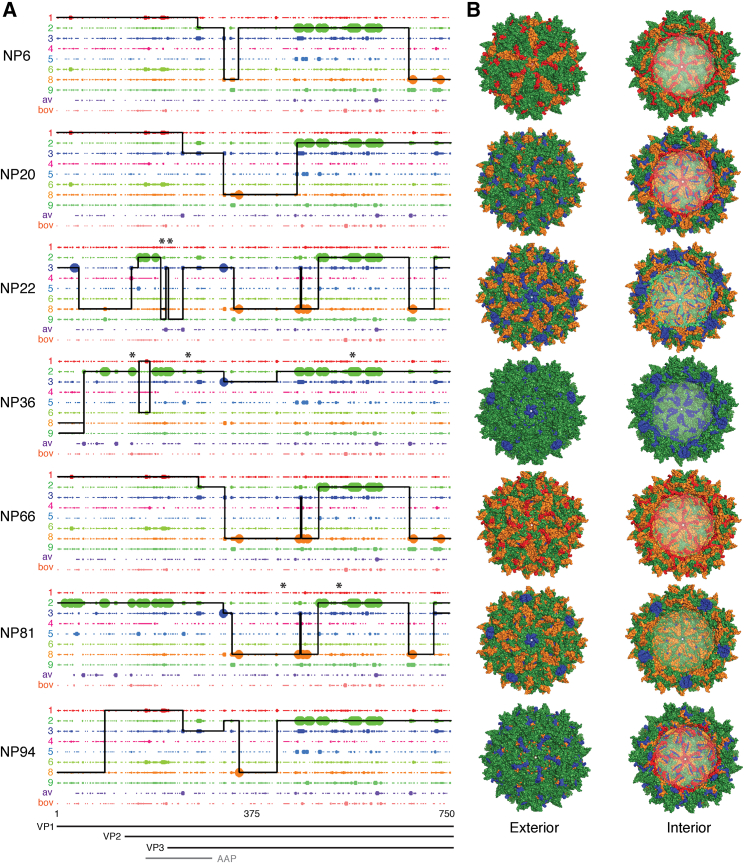
Several of the most highly selected variants from each screen were isolated and vectorized with Renilla and Firefly luciferase (RLuc/FLuc) expression constructs for subsequent validation experiments. To assess the genetic contribution of each parental AAV serotype to individual evolved capsids selected from each screen, we performed fragment crossover mapping (Figure 3A) and predictive fragment conservation analyses (Figure S1C) to calculate enrichment scores for the likelihood of parental contribution to shuffled fragments in the new capsids. These complementary methodologies demonstrated that diverse shuffling was achieved and maintained along the length of Cap, including VP1, VP2, VP3, and AAP. The parental serotypes that contributed most to the evolved variants included AAV1, 2, 3b, 6, 8, and 9_hu14. No variants had appreciable capsid fragment regions from AAV4, 5, bovine, or avian. Predictive three-dimensional structural capsid mapping of shuffled variants (Figure 3B) revealed not only diverse structural meta-heterogeneity in hypervariable regions but also micro-conservation within key structural domains, such as the cylinder, canyon, and various symmetry axes.
Figure 3.
Amino Acid Sequence and Structural Composition of Selected Shuffled AAV Capsid Variants
(A) Amino acid sequence mapping analysis of parental capsid fragment crossovers in vectorized shuffled capsids. Large dots are a 100% match for one parent and small dots represent that more than one parent matches at that amino acid position. The solid black line for each chimera represents the parental serotype match. Thin parallel lines indicate multiple parental matches at an equal probability. Vertical spikes indicate an amino acid mutation from within the parental sequence space, while an overhead asterisk indicates an evolved de novo mutation for which no parent has that amino acid at that position. VP1, VP2, VP3, and AAP ORFs are shown below. (B) Shuffled variants were 3D false color mapped onto the crystal structure of AAV8. Color coding indicates parental amino acid contribution using colors as in (A).
Enhanced Muscle Cell Transduction and Immunologic Properties of Bioengineered Variants
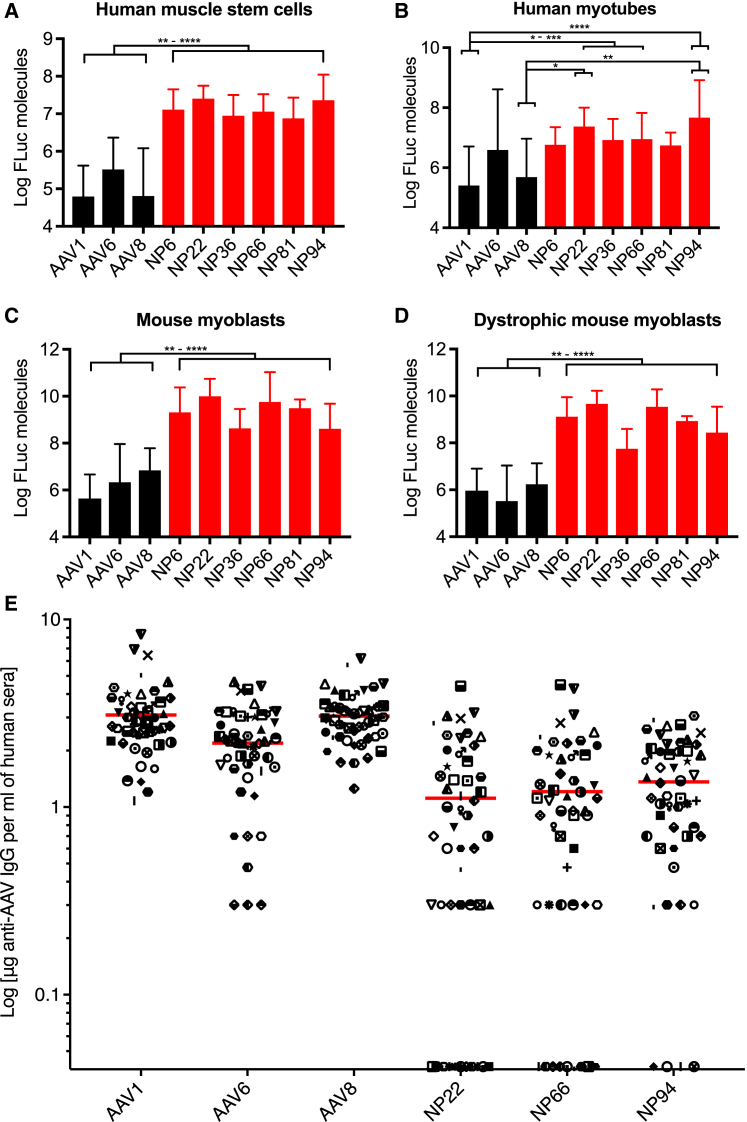
Large-scale productions of all vectorized variants were carried out, and those capable of producing high titers sufficient for eventual clinical use (stem screen variants NP6, NP22, NP36, and NP66 and myotube screen variants NP81 and NP94) were considered further for validation. We began with FLuc transduction efficiency assessments in vitro in primary hMuSCs and human myotubes, as well as mouse myoblasts from wild-type and dystrophic Mdx5cv mice, with comparisons to known muscle-tropic rAAV serotypes 1, 6, and 8. In pooled primary hMuSCs isolated from five patients (Figure 4A), all six shuffled variants showed significantly increased functional transduction over rAAV1 (p < 0.0001), rAAV6 (p < 0.01–0.0001), and rAAV8 (p < 0.0001). In pooled human myotube cultures differentiated from primary hMuSCs (Figure 4B), variants NP22, NP36, NP66, and NP94 showed significantly increased functional transduction over rAAV1 (p < 0.05–0.001), and NP22 and NP94 showed significantly increased functional transduction over rAAV8 (p < 0.05–0.01). No variants showed significant differences in transduction over rAAV6. However, the variability in differentiation state with human myotube cultures, particularly in pools from 5 patients, makes transduction assessments in these primary cells challenging, as can be seen in the large error bars within each treatment.
Figure 4.
In Vitro Validation of Superior Muscle Transduction and Seroreactivity
(A) Transduction assessments by FLuc expression of the best-performing vectorized variants (red) in primary human muscle stem cells compared to known muscle-tropic control rAAV serotypes 1, 6, and 8 (black). Each cell type was assayed in three separate experiments, each performed in technical duplicate or triplicate (n = 8–11 for each bar). Data represent mean log of FLuc molecules ± SD normalized to an rFLuc standard curve. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001. (B) Assessments as in (A) but in primary human myotube cultures. (C) Assessments as in (A) but in immortalized C2C12 mouse myoblasts. (D) Assessments as in (A) but in immortalized dystrophic Mdx5cv mouse myoblasts. (E) Seroreactivity ELISA for the presence of anti-AAV antibodies in normal human serum from 50 adults. Each patient was assayed in technical triplicates, with data points representing the mean minus background for each patient. Red line, mean. Symbols are consistent for each patient in each treatment column.
In C2C12 mouse myoblasts (Figure 4C), all shuffled variants showed highly significant increased functional transduction over rAAV1 (p < 0.0001), rAAV6 (p < 0.001–0.0001), and rAAV8 (p < 0.01–0.0001). Similarly, in dystrophic mouse myoblasts from Mdx5cv mice (Figure 4D), all shuffled variants showed significantly increased functional transduction over rAAV1 (p < 0.001–0.0001), rAAV6 (p < 0.0001), and rAAV8 (p < 0.01–0.0001). Each of the four cell types used for in vitro transduction tests was assayed in three separate biological replicates, and each of those was performed in technical duplicate or triplicate. Importantly, results from these four different muscle lines highlighted the striking differences in transduction levels that can be seen in different species, different cell types, different disease states, and primary versus immortalized cell lines.
An advantage of the in vitro systems for characterization is the ability to perform binding seroreactivity assays to predict the likelihood of humoral neutralization by patients with pre-existing cross-reacting anti-AAV capsid antibodies. Individual human serum samples from 50 healthy adults of each sex (see Table S1 for details) were assessed for seroreactivity by indirect ELISA against the best-performing capsid variants from the in vitro transduction assays and control serotypes (Figures 4E, S3A, and S3B). Variants NP22, NP66, and NP94 all had highly significantly reduced mean seroreactivity compared to all three muscle-tropic control serotypes rAAV1, 6, and 8 (p < 0.0001). Based on the superior seroreactivity and in vitro transduction results from shuffled variants NP22, NP66, and NP94, we decided to move forward with these capsid serotypes for additional, more stringent, preclinical validation.
Enhanced Human Skeletal Muscle Explant Transduction by Evolved Variants Ex Vivo
To more accurately predict eventual skeletal muscle transduction in patients, we sought to transduce primary human skeletal muscle fiber explants from surgical resections ex vivo. Skeletal muscle fibers (Figure 5A) from five adult patients (three male and two female) were used for ex vivo explant transduction assessments. Initial transduction analyses began with NP22, NP66, and NP94 compared to muscle-tropic control serotypes rAAV1, 6, and 8. Primary skeletal muscle resections were digested, and muscle fibers were isolated within 1 hr of removal from each patient for 48-hr suspension culture and transduction comparisons with scAAV-CAG-RLuc (3E8 vector genomes [vg]/well) on 300 fibers per condition. Patient 1 was a 71-year-old male from whom fibers of latissimus dorsi muscle were excised and transduced in triplicate. Results showed that all three shuffled variants demonstrated significantly increased transduction over all control serotypes by live fiber imaging (Figure 5B). Further, to more accurately quantify the increased transduction, given the dense cellular structure of muscle fibers, luciferase assays were also performed on lysed muscle fibers (Figure 5C), and they were compared to all three control serotypes (p < 0.0001). NP22 demonstrated an 84-fold increase over rAAV8, a 45-fold increase over rAAV6, and a 25-fold increase over rAAV1. NP66 demonstrated a 26-fold increase over rAAV8, a 14-fold increase over rAAV6, and an 8-fold increase over rAAV1. Finally, NP94 demonstrated a 14-fold increase over rAAV8, an 8-fold increase over rAAV6, and a 4-fold increase over AAV1. Individual muscle fiber staining revealed rAAV uptake along the entire fiber length (Figure S4E).
Figure 5.
Validation of Human and Non-human Primate Primary Skeletal Muscle Transduction Ex Vivo
(A) Representative bright-field images of primary human skeletal muscle fibers used in (B)–(G). Magnifications were 10× and 20×. (B, D, and F) RLuc imaging at 48 hr of human skeletal muscle fibers transduced ex vivo with scAAV-CAG-RLuc. Mean radiance (p/s/cm2/sr) displayed below each treatment triplicate. (C, E, and G) RLuc assays on corresponding lysed human skeletal muscle fibers at 48 hr. Data represent mean ± SD. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001. (H) RLuc imaging at 48 hr of rhesus skeletal muscle fibers transduced ex vivo with scAAV-CAG-RLuc. Mean radiance (p/s/cm2/sr) displayed below each treatment triplicate. (I) RLuc assay on lysed rhesus skeletal muscle fibers at 48 hr. Data represent mean ± SD.
To ensure that the increased transduction was not simply the result of heparan sulfate proteoglycan (HSPG) binding—as rAAV1, 6, and 8 have variable (to no) HSPG-binding strength compared to AAV2, and our evolved variants inherited the AAV2 HSPG-binding domain (Table S2)—we performed the same sets of transduction experiments in another primary human skeletal muscle explant and compared to rAAV1, 6, and 8 as before, as well as rAAV2. Human patient 2 was a 72-year-old male from whom fibers of latissimus dorsi muscle were excised and transduced in triplicate. By live fiber luciferase imaging at 48 hr post-administration (Figure 5D), variants NP22 and NP66 again showed significantly increased transduction over all control serotypes, including rAAV2. Luciferase assays on lysed muscle fibers (Figure 5E) demonstrated that NP22 and NP66 had significantly increased transduction over all 4 control serotypes (p < 0.0001 and p < 0.004, respectively). Thus, HSPG binding was not the sole determinant for the increased human skeletal muscle explant transduction.
Having shown that rAAV6 was the best-performing control serotype in human skeletal muscle fibers ex vivo, we then transduced primary human skeletal muscle fibers from an additional 3 patients and compared to rAAV6. Human patient 3 was a 61-year-old female from whom fibers of pectoralis major muscle were excised and transduced in triplicate. By live fiber luciferase imaging at 48 hr post-administration (Figure 5F), variants NP22, NP66, and NP94 all showed significantly increased transduction over rAAV6. Luciferase assays were also performed on lysed muscle fibers (Figure 5G). Compared to rAAV6, variants NP22 (p < 0.0001), NP66 (p < 0.001), and NP94 (p < 0.005) showed significantly increased human skeletal muscle fiber transduction that ranged from a 5- to 14-fold average increase. Two additional patients of each sex and with varied muscle groups were used to confirm the results seen from the initial patients: patient 4 was a 58-year-old female and patient 5 was a 60-year-old male from whom fibers from the rectus abdominis and latissimus dorsi muscles, respectively, were excised and transduced (Figures S4A–S4D). Despite differences in their sex and muscle types, both patients also demonstrated 4- to 13-fold improvements in transduction with shuffled variants NP22, NP66, and NP94 over rAAV6 by live fiber imaging and luciferase assays on lysed muscle fibers.
Rhesus Skeletal Muscle Explant Transductions Support the Use of NP22 and NP66 for Preclinical Vaccine Testing
Given the superior transduction capabilities of shuffled variants NP22, NP66, and NP94 in human skeletal muscle settings in vitro and ex vivo, the next step in moving these variants to the clinic for use as passive vaccine vectors would be to determine efficacy in prophylactic experiments in non-human primates. To justify preclinical testing in non-human primates, we asked whether the same enhanced transduction observed in human skeletal muscle tissues could be reproduced in muscle from rhesus macaques. Mimicking our ex vivo transduction studies from human patients, we surgically resected biceps femoris skeletal muscle explants from a 20-year-old male rhesus macaque. The healthy skeletal muscle tissue (Figure S4F) was digested and individual muscle fibers were isolated for the administration of scAAV-CAG-RLuc within 1 hr of surgical excision, followed by 48-hr culture. Here again variants NP22 (p < 0.0002) and NP66 (p < 0.02) showed a highly significant 30- to 57-fold improvement in transduction over rAAV6 by live fiber luciferase imaging (Figure 5H) and luciferase assays on lysed fibers (Figure 5I).
Discussion
Muscle is increasingly being recognized as a key secretory organ,1 and it is among the most easily accessible tissues for localized vector administration. Therefore, muscle represents an ideal tissue platform for the expression of desired therapeutic factors like broadly neutralizing antibodies against pathogenic viruses. Indeed, an ideal passive vaccine would be administered intramuscularly in an EpiPen-style auto-injection format. This would bypass the need for intravenous administration by healthcare professionals in hospitals and instead allow for administration in the field. Patients, particularly in developing countries where the need for effective vaccines against pathogenic viruses is greatest, could administer their own passive vaccine at distribution centers. An added advantage to rAAV as a passive vaccine delivery modality is the remarkable heat tolerance that rAAV capsids display, with thermal stability up to 85°C for some serotypes,25 allowing for minimal refrigeration. Additionally, studies have demonstrated that rAAV can effectively be desiccated or lyophilized and later resuspended,26 offering additional ways to maintain stability and further reduce the cost of passive vaccine shipments to the developing world.
An exciting possibility with our new capsid variants that transduce human skeletal muscle at such high levels would be to decrease patient doses while still enabling therapeutic levels of antibody expression. This could bypass several hurdles to rAAV being an effective passive vaccine delivery tool: (1) reduced potential for the generation of anti-antibody responses to rAAV-delivered antibodies, like those shown for anti-HIV and anti-SIV broadly neutralizing antibodies;27 (2) decreased likelihood for neutralizing anti-capsid antibody binding28, 29 through intramuscular, rather than intravenous, administration; (3) reduced production and thus treatment costs, as current AAV therapies have ballooned to over $1 million dollars per patient30, 31 in some cases; and (4) reduced probability for capsid-specific T cell responses against transduced muscle fibers.28 In addition, given the favorable seroreactivity profiles of all three variants, NP22, NP66, and NP94, greater numbers of patients may be eligible to receive treatment than was possible with existing muscle-tropic serotypes. Variant NP66 is missing two highly antigenic residues (V70832 and N71733) present on rAAV1 and 6, known muscle-tropic parents under consideration for use in passive vaccines. For perspective, rAAV1 is second only to rAAV2 in terms of pre-existing neutralizing antibodies (35%–70% versus 60%–70%) in the human population, and rAAV6 is close behind at ∼50%.34, 35
Shuffled NP22, NP66, and NP94 capsid sequences (Figure S5) have many fragments from rAAV1, 6, and 8 parental sequences that have known muscle tropism, likely explaining why these variants were selected for so strongly in the muscle cell screens. Yet, each of these three shuffled capsids would be predicted to have unique comparative structural topologies given their divergent capsid sequence arrangements. NP22 is the most highly shuffled of the three, with the unique region of VP1 coming from rAAV3b and 8; the unique region of VP2 coming from rAAV8, 3b, and 6; and, finally, VP3 with contributions from rAAV9_hu14, 3b, 8, and 2 as well as de novo mutations coding for amino acids not present in the parental sequence space. NP66 is the least shuffled of the three, with the unique region of VP1 and VP2 coming solely from rAAV1 and a VP3 composed largely of AAV8 and 2 and several de novo mutations. NP94 has a rAAV8-based unique region of VP1, a unique region of VP2 entirely from rAAV1, and a mixed VP3 with contributions from rAAV1, 3b, 2, and 8. All three evolved variants would be predicted to bind HSPG, having inherited all five reported AAV2 HSPG-binding residues and several other residues key for transduction (Table S2).36, 37, 38, 39, 40 It is unclear whether variants NP22 and NP66 utilize sialic acid recognition for glycan binding given that they have half (A472 and N473 from AAVs 4 and 5 and W503 from AAVs 1 and 6) of the six known critical residues (Table S2). NP94 likely does not bind sialic acid with only one of the six sites (W503).
30 distinct amino acids differ between the two top variants, NP22 and NP66 (Figure S5), of which 19 exist within the non-ordered PLA2 domain in the unique region of VP1 (Table S3). However, of the remaining 11 amino acids that exist in regions with ordered crystal structures from parental capsid serotypes, several exist in surface-exposed regions (Table S3) and are likely key for human skeletal muscle transduction. Most notably, NP22 and NP66 utilize different acidic residues at position 327 at the top of the capsid cylinder (NP22 D327, NP66 E327) and different polar residues at the base of the 3-fold protrusion (NP22 N705, NP66 Y705; NP22 N709, NP66 S709). Interestingly, NP22 utilizes a hydrophobic V708 and polar T713 at the base of the 3-fold protrusion, while NP66 utilizes a polar T708 and hydrophobic A713 at the same positions. Lastly, within the canyon, NP22 has an acidic D715 while NP66 incorporated a polar N715. Additionally, the diverse shuffled capsid arrangements likely played a role in the reduced seroreactivity our shuffled variants had over control serotypes. Cumulatively, our results show the significantly increased human skeletal muscle transduction of shuffled AAV variants NP22 and NP66 (and in some cases NP94) over existing muscle-tropic serotypes, when assessed in various primary human muscle cell types in vitro and in human and non-human primate skeletal muscle resections ex vivo.
Materials and Methods
AAV Library Production, Vector Production, and Titration
The shuffled AAV capsid plasmid library was generated as described.41 AAV library productions were produced using a Ca3(PO4)2 transfection protocol (wild-type [WT] AAV library plasmid pool and pAd5 helper) in HEK293T cells (ATCC CRL-3216), followed by double cesium chloride density gradient purification and dialysis, as previously described,42 and resuspension in Dulbecco’s PBS (dPBS) with 5% sorbitol (w/v) and 0.001% Pluronic F-68 (v/v). AAV libraries were titered for Rep by TaqMan qPCR with the following primer/probe set: forward 5′-TTCGATCAACTACGCAGACAG-3′, reverse 5′-GTCCGTGAGTGAAGCAGATATT-3′, and probe 5′/FAM/TCTGATGCTGTTTCCCTGCAGACA/BHQ-1/-3′.
rAAV vector productions expressing FLuc or RLuc were similarly produced as above but as triple transfections with pAd5 helper, AAV transfer vector (either single-stranded AAV [ssAAV]-EF1α-GFP-P2A-FLuc cloned in-house or self-complementary AAV (scAAV)-CAG-RLuc from Addgene [83280]), and pseudotyping plasmids for each capsid of interest. AAV vectors were titered by TaqMan qPCR. The ssAAV-EF1α-GFP-P2A-FLuc vectors were titered on GFP with the following primer/probe set: forward 5′-GACGTAAACGGCCACAAGTT-3′, reverse 5′-GAACTTCAGGGTCAGCTTGC-3′, and probe 5′/FAM/CGAGGGCGATGCCACCTACG/BHQ-1/-3′.
The scAAV-CAG-RLuc vectors were titered on CAG with the following primer/probe set: forward 5′-GTTACTCCCACAGGTGAGC-3′, reverse 5′-AGAAACAAGCCGTCATTAAACC-3′, and probe 5′/FAM/CTTCTCCTCCGGGCTGTAATTAGC/BHQ-1/-3′.
Human Skeletal Muscle Isolation from Surgical Specimens
Human skeletal muscle specimens from both male and female patients were surgically isolated from latissimus dorsi, serratus anterior, pectoralis major, or rectus abdominis intraoperatively in accordance with the Stanford Institutional Review Board (IRB 15084). Muscle tissue was wrapped in sterile gauze and placed immediately on ice after isolation. Tissue processing for stem cell isolation or muscle fiber isolation always began within 1 hr of surgical excision.
Surgical Non-human Primate Skeletal Muscle Isolation
Rhesus macaque skeletal muscle specimens from the biceps femoris were surgically isolated by the Stanford University Veterinary Pathology team in accordance with the Stanford Institutional Animal Care and Use Committee. Tissue was placed immediately in Collagenase-II after isolation, and tissue processing for fiber isolation began within 1 hr of surgical excision.
hMuSC Isolation and Purification
Human skeletal muscle tissue was prepared as described24 for the isolation of pure populations of stem cells by FACS with the surface marker profile CD31−CD34−CD45−EGFR+ITGB1+.43 Sorted cells were assessed immediately post-sorting for adherence and viability as controls for sorting efficiency. In addition, a fraction of the sorted cells was routinely plated and stained for PAX7 to demonstrate purity of the sorted stem cell population, as previously described.24
hMuSC and Myotube Cultures
Plates were coated with extracellular matrix protein at 1:500 (v/v) in DMEM with 1% penicillin/streptomycin. The hMuSC medium was a 1:1 mixture of DMEM:MCDB media supplemented with 20% fetal bovine serum (FBS), 1% insulin-transferrin-selenium, 1% antibiotic/antimycotic, and 10 μM p38i (Cell Signaling Technology SB203580) to maintain the stem state, as described.24 Media for differentiating primary hMuSCs into myotubes lacked p38i and included a 2% horse serum starve instead of 20% FBS for 7 days. Immunofluorescence analysis of cultured hMuSCs was performed as described.24 All media was changed every 2 days.
Mouse Skeletal Muscle Myoblast Cultures
Wild-type C2C12 mouse myoblasts (ATCC CRL-1772) were maintained in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic. Dystrophic mouse myoblasts (Mdx5cv) were maintained in Nutrient mixture F10 supplemented with 20% FBS, 1% antibiotic/antimycotic, and 2.5 ng/mL recombinant human fibroblast growth factor (FGF) on the same extracellular matrix (ECM)-coated plates as the human cultures to improve adherence.
Animals
Skeletal muscle from a healthy, 20-year-old, 14.5-kg, intact male rhesus macaque (Macaca mulatta) was harvested immediately following humane euthanasia during an end-of-study time point. The rhesus macaque study protocol, and all related animal care procedures, complied with NIH guidelines. Ethical standards included a psychological enrichment program, frequent contact with other animals (visual, auditory, olfactory, and, where appropriate, touch and grooming), regular veterinary supervision, and pharmacological amelioration of pain associated with surgeries. The Administrative Panel on Laboratory Animal Care of Stanford University approved the animal protocol for non-human primates.
Replication-Competent Shuffled AAV Capsid Library Selection
Pooled human muscle stem cells from 5 patients (2–3E6 cells) or differentiated myotubes were initially transduced with 1E11 vg (MOI 20,000 for round 1) AAV library in 20-mL media in 15-cm dishes. 5E8 plaque-forming units (PFUs) wild-type replication-competent human adenovirus-5 (hAd5) (in 500 μL volume, ATCC VR-5) was added to media 2 hr later. The media were changed 12 hr later after 3 dPBS washes. Cells were harvested 48 hr after hAd5 administration. Each sample was mixed with 1 mL PBS and underwent three freeze-thaw cycles to ensure complete cell lysis, followed by hAd5 heat inactivation (65°C for 30 min) and 5-min spin at 14,000 × g at 4°C. This supernatant (∼800 μL per round, MOI 1,000–5,000) was then used for subsequent in vitro selection steps and PCR analysis. For PCR analysis, 20 μL supernatant was used for AAV genomic DNA (gDNA) extraction using the MinElute Virus Spin Kit (QIAGEN 57704), followed by PCR amplification using forward 5′-TGGATGACTGCATCTTTGAA-3′ and reverse 5′-ATGGAAACTAGATAAGAAAGAA-3′. PCR to assess AAV amplification at each round was performed using Phusion high-fidelity polymerase and the following program: 98°C 2 min, 30 cycles of 98°C 20 s, 55°C 15 s, and 72°C 1 min. AAV capsid open reading frames (ORFs) from rounds 3–6 of the selection screens were cloned using a Zero Blunt TOPO Kit, and 100 clones were sent for full Sanger sequencing to assess remaining library diversity with primers: forward 1 5′-TGGATGACTGCATCTTTGAA-3′, forward 2 5′-ATTGGCATTGCGATTCC-3′, and reverse-1 5′-ATGGAAACTAGATAAGAAAGAA-3′.
Vectorization and Sequence Contribution Analysis of Evolved AAV Capsids
Contigs were assembled using Geneious R7 version (v.)7.1.9 software, and clones selected for vectorization were PCR-amplified using forward 5′-AAATCAGGTATGGCTGCCGATG-3′, reverse 5′-AACGCCCGGGCTGTAGTTAAT-3′, and 5′-GATTAAGCCGCCATGCTACTTATCTACGTAGCCATGGAAACTAGATAAGAAAG-3′. PCR amplicons were cloned in-frame, downstream of Rep, into predigested recipient pCap packaging plasmid containing AAV2 Rep without inverted terminal repeats (ITRs) using SwaI and XmaI restriction sites. AAV capsid genes were sequence verified, and the resultant contigs were analyzed using a custom Perl pipeline that assessed multiple sequence alignments using Clustal Omega (EMBL-EBI) to generate the overall serotype composition of the shuffled AAVs, by comparison of DNA and amino acid sequences with the parental AAV serotypes based on maximum likelihood. Xover 3.0 DNA/protein shuffling pattern analysis software was used to generate parental fragment crossover maps of shuffled variants.44 Each parental serotype was color coded as follows: AAV1, red; AAV2, forest green; AAV3b, marine blue; AAV4, magenta; AAV5, tv blue; AAV6, green cyan; AAV8, orange; AAV9, pale green; bovine, deep salmon; and avian, purple.
PacBio Library Preparation and Full-Length Single-Molecule Capsid Sequencing
Pacific Biosciences (PacBio) SMRTbell libraries were prepared following the “Procedure and Checklist-2 kb Template Preparation and Sequencing” protocol from PacBio using the SMRTbell Template Prep Kit v1.0 (100-259-100). PacBio binding and annealing calculator determined appropriate concentrations for annealing and binding of SMRTbell libraries. SMRTbell libraries were annealed and bound to the P6 DNA polymerase for sequencing using the DNA/Polymerase Binding Kit P6 v.2.0 (100-372-700). Bound SMRTbell libraries were loaded onto SMRT cells using standard MagBead protocols and the MagBead Buffer Kit v.2.0 (100-642-800). The standard MagBead sequencing protocol is followed with the DNA Sequencing Kit 4.0 v.2 (100-612-400, also known as P6/C4 chemistry). Sequencing data were collected for 6-hr movie times with Stage Start not enabled. Circular consensus sequence (CCS) reads were generated using the PacBio SMRT portal and the RS_ReadsOfInsert.1 protocol, with filtering set at Minimum Full Pass = 3 and Minimum Predicted Accuracy = 95.
Bioinformatic Assessment of PacBio Sequence Reads
CCS reads with sequence lengths from 2,300 to 2,350 bp were included in downstream bioinformatics analyses. Insertions or deletions (indels) in CCS reads were corrected using an in-house algorithm that first assesses parental fragment identity using Xover 3.0 DNA/protein shuffling pattern analysis software.44 Once the parental identity of each crossover fragment was determined, this information was used to determine indels for correction. SNPs that did not result in indels were maintained. The SNP error rate with the PacBio platform is 1.3%–1.7%.45, 46 SNP frequencies above this rate range were assumed to have arisen from de novo mutations. Corrected sequences in FASTA format were then aligned with MUSCLE.47 Phylogenetic analysis was conducted using the maximum-likelihood method in RAxML.48 Percent parental conservation was determined using an in-house algorithm that identifies the percentage of each parent on each aligned position in the shuffled library. The maximum dot size indicates that 100% of variants share that amino acid from that parent at that position. All other dot sizes are proportional to the percent of variants from 0% to 100% that have that amino acid at that position from that parent.
In Vitro Transduction Analysis in Human Muscle Stem Cells, Human Myotubes, and Mouse Myoblasts
18,000 cells of each type were plated in 48-well coated plates in 500 μL of the respective maintenance media. At 80% confluency, cells were transduced with ssAAV-EF1α-GFP-P2A-FLuc vectors at an MOI of 20,000 for 12 hr and then media were replaced. FLuc levels were measured 3 days post-AAV administration using a Luciferase 1000 Assay System kit (Promega E4550), per the manufacturer’s instructions, and read on a Veritas luminometer with the following settings: 100 μL luciferin injection, 2 s delay, 10 s measure. Experiments were performed in biological triplicate each with technical duplicates or triplicates. To compare signals across biological replicates, output RLU was normalized to molecules of FLuc using a dilution series of recombinant QuantiLum Firefly Luciferase (Promega E1701).
Ex Vivo Transduction Analysis in Human and Rhesus Skeletal Muscle Fiber Explants
Human or rhesus skeletal muscle specimens were incubated in collagenase-II (500 U/mL) in Ham’s F10 with 10% horse serum and 1% penicillin/streptomycin for 80 min in a 60-rpm shaking water bath. Post-digest, DMEM with 20% FBS and 1% penicillin/streptomycin was added in equal volume, and tissue was triturated with a glass pipet coated in horse serum to separate single muscle fibers. 300 fibers were counted under a dissecting microscope and placed in each well of a horse serum-coated 24-well plate in 1-mL media. Fibers were transduced with 3E8 vg/well scAAV-CAG-RLuc in triplicate. RLuc levels were measured 48 hr later using a Renilla Luciferase Assay System Kit (Promega E2820), per the manufacturer’s instructions, and read on a Veritas luminometer for quantitation as above and a Xenogen IVIS Spectrum imaging system for visualization. Immunofluorescence analysis of single transduced human fibers was performed as previously described24 using a rabbit anti-RLuc monoclonal immunoglobulin G (IgG) (Abcam 185926; 1:1,000 v/v) and goat anti-rabbit Alexa Fluor 488 secondary. Imaging was performed on a Leica TCS SP8-X WLL inverted confocal microscope with 40× oil immersion objective and imaged with Leica AF software v.3.3.0.10134. White light laser power was kept constant at 405 nm 48% and 488 nm 75% for all images. The z stacks were compressed using ImageJ v.2.0.0 and overlaid in Adobe Photoshop CS6 v.13.0. H&E staining on rhesus skeletal muscle sections was performed per standard protocols. To obtain uncontracted single human muscle fibers for bright-field imaging, skeletal muscle specimens were digested as described above, transferred to Ham’s F10 with 10% horse serum, and triturated with a glass pipet coated in horse serum to separate single fibers. Fibers were immediately fixed by transferring to 4% paraformaldehyde (PFA) (in PBS) and incubated for 10 min. Bright-field imaging was performed with a Keyence BZ-X700 microscope equipped with full BZ acquisition and analysis software.
Indirect Seroreactivity ELISA for Anti-AAV Antibodies in Normal Human Serum
Off-clot serum collected from peripheral blood of 50 healthy adults (see Table S1) in the United States was used as the primary antibody. Human IgG (Baxter LE1500190, lot LE12P180AB) was used to prepare a standard curve (16 2-fold dilutions of 100 mg/mL stock intravenous immunoglobulin [IVIG] in blocking buffer). Chimeric and control AAV capsids served as the ELISA antigens (5E8 vg/well). Triplicates of human IgG standards and AAV samples were fixed to wells of 96-well immunoplates with 50 μL coating solution (13 mM Na2CO3 and 35 mM NaHCO3 in water [pH 9.6]), the plates were sealed, and then incubated overnight at 4°C. Plates were washed 2 times with PBS-T containing 0.05% Tween-20 and blocked with blocking buffer (PBS, 6% BSA, and 0.05% Tween-20) for 1 hr at 25°C. The plates were washed 2 times with PBS-T. Each human serum sample was diluted 1:400 in blocking buffer, and 50 μL was added to each of the experimental wells. Plates were incubated for 2 hr at 37°C and then washed 2 times in PBS-T. Polyclonal sheep anti-human IgG-horseradish peroxidase (HRP) secondary antibody (GE Bioscience NA933V) was diluted 1:500 in wash buffer, and 100 μL was added to the wells to detect bound antibodies in the human sera. Plates were incubated again for 2 hr at 37°C and washed 2 times in PBS-T. OPD substrate (o-phenylenediaminedihydrochloride, Sigma P4664) was added in 100 μL/well in a 0.1 M sodium citrate buffer and incubated at 25°C for exactly 10 min. The reaction was stopped with 50 μL/well 3M H2SO4 and the absorbance determined at 490 nm on a microplate reader (Bio-Rad, Hercules, CA, USA). A set of no AAV blank wells was used to subtract background for non-specific binding of human serum and antibodies to the microplate wells. Standards were plotted using Four Parameter Logistic (4PL) curve fitting to determine the concentration of samples that fall within the linear range.
False-Colored Structural Capsid Mapping
Chimeric capsids were false color mapped onto the AAV8 capsid structure 3RA849 using Pymol v.1.7.6.0. Mapped colors correspond to parental serotype colors used in the parental fragment crossover maps. Exterior capsid views have all chains represented, while cross-section views have chains surrounding a cylinder at the 5-fold symmetry axis removed exposing the capsid interior lumen.
Statistics
Statistical analyses were conducted with Prism v.7. Experimental differences in FLuc expression for Figures 4A–4D were assessed via one-way ANOVA using Tukey’s multiple comparisons test. Experimental differences in seroreactivity Figures 4E, S3A, and S3B were log+1 transformed before being assessed via two-way ANOVA using Tukey’s multiple comparisons test. Experimental differences in FLuc expression for Figure 5 were assessed via one-way ANOVA using Tukey’s multiple comparisons test. In all assessments, p values < 0.05 were considered statistically significant (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001).
Author Contributions
N.K.P., K.P., G.W.C., K.M.-N., T.A.R., and M.A.K. designed the experiments. N.K.P., G.W.C., K.M.-N., L.L., K.P., J.X., M.N.W., Y.Z., and B.Y. generated reagents, protocols, performed experiments, and/or analyzed data. G.K.L. and J.B.S. performed the surgeries for human skeletal muscle sample isolation. J.G.V.-M. performed the surgery for the rhesus skeletal muscle sample isolation. N.K.P. wrote the manuscript and generated the figures. All authors reviewed, edited, and commented on the manuscript.
Conflicts of Interest
N.K.P. and M.A.K. are inventors on patents for AAV serotypes used in this paper. M.A.K., L.L have commercial affiliations. N.K.P., L.L., and M.A.K. have consulted on technologies broadly related to this paper. All other authors declare no conflicts of interest.
Acknowledgments
The authors wish to acknowledge Megan Albertelli, David Judah, and Elias Godoy of the Stanford Department of Comparative Medicine for rhesus macaque help; as well as Derek Pouchnik and Mark Wildung of the Washington State University Laboratory for Biotechnology & Analysis for sequencing help. N.K.P. was supported by an independent career transition grant (K01-DK107607), as well as former postdoctoral fellowships from the NIH (F32-HL119059), the American Liver Foundation Hans Popper Memorial Fellowship, and the Stanford Dean’s Fellowship. G.W.C. was supported by a graduate fellowship (F30-AG035521) and the Stanford MSTP Program. M.N.W. was supported by a training grant (T32-AG000266) and a career transition grant (K99-AG053438). This work was supported by research grants to M.A.K. from the Bill & Melinda Gates Foundation (Project 112438) and the NIH (R01-AI116698); and to T.A.R. from the NIH (P01-AG036695 and R01-AR062185), the Department of Veterans Affairs, the Muscular Dystrophy Association, and the Dutch Parent Project. This project was also supported, in part, by an NIH Shared Instrumentation Grant (S10-OD010580) from the National Center for Research Resources (NCRR) with significant contribution from Stanford’s Beckman Center as well as the Stanford Small Animal Imaging Facility.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR, the NIH, or the respective universities. A patent has been filed for the newly evolved capsid variants. Packaging plasmids for academic use of any of the new capsids described herein must be obtained via a material transfer agreement (MTA) with Stanford University.
Footnotes
Supplemental Information includes five figures and three tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.06.001.
Supplemental Information
References
- 1.Pedersen B.K., Febbraio M.A. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 2.Johnson P.R., Schnepp B.C., Zhang J., Connell M.J., Greene S.M., Yuste E., Desrosiers R.C., Clark K.R. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balazs A.B., Chen J., Hong C.M., Rao D.S., Yang L., Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2011;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balazs A.B., Ouyang Y., Hong C.M., Chen J., Nguyen S.M., Rao D.S., An D.S., Baltimore D. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 2014;20:296–300. doi: 10.1038/nm.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balazs A.B., Bloom J.D., Hong C.M., Rao D.S., Baltimore D. Broad protection against influenza infection by vectored immunoprophylaxis in mice. Nat. Biotechnol. 2013;31:647–652. doi: 10.1038/nbt.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limberis M.P., Adam V.S., Wong G., Gren J., Kobasa D., Ross T.M., Kobinger G.P., Tretiakova A., Wilson J.M. Intranasal antibody gene transfer in mice and ferrets elicits broad protection against pandemic influenza. Sci. Transl. Med. 2013;5:187ra72. doi: 10.1126/scitranslmed.3006299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limberis M.P., Racine T., Kobasa D., Li Y., Gao G.F., Kobinger G., Wilson J.M. Vectored expression of the broadly neutralizing antibody FI6 in mouse airway provides partial protection against a new avian influenza A virus, H7N9. Clin. Vaccine Immunol. 2013;20:1836–1837. doi: 10.1128/CVI.00545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J., Calcedo R., Vandenberghe L.H., Bell P., Somanathan S., Wilson J.M. A new genetic vaccine platform based on an adeno-associated virus isolated from a rhesus macaque. J. Virol. 2009;83:12738–12750. doi: 10.1128/JVI.01441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sipo I., Knauf M., Fechner H., Poller W., Planz O., Kurth R., Norley S. Vaccine protection against lethal homologous and heterologous challenge using recombinant AAV vectors expressing codon-optimized genes from pandemic swine origin influenza virus (SOIV) Vaccine. 2011;29:1690–1699. doi: 10.1016/j.vaccine.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 10.Gray S.J., Blake B.L., Criswell H.E., Nicolson S.C., Samulski R.J., McCown T.J., Li W. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol. Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penaud-Budloo M., Le Guiner C., Nowrouzi A., Toromanoff A., Chérel Y., Chenuaud P., Schmidt M., von Kalle C., Rolling F., Moullier P., Snyder R.O. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 2008;82:7875–7885. doi: 10.1128/JVI.00649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowles D.E., McPhee S.W., Li C., Gray S.J., Samulski J.J., Camp A.S., Li J., Wang B., Monahan P.E., Rabinowitz J.E. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brantly M.L., Chulay J.D., Wang L., Mueller C., Humphries M., Spencer L.T., Rouhani F., Conlon T.J., Calcedo R., Betts M.R. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc. Natl. Acad. Sci. USA. 2009;106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flotte T.R., Brantly M.L., Spencer L.T., Byrne B.J., Spencer C.T., Baker D.J., Humphries M. Phase I trial of intramuscular injection of a recombinant adeno-associated virus alpha 1-antitrypsin (rAAV2-CB-hAAT) gene vector to AAT-deficient adults. Hum. Gene Ther. 2004;15:93–128. doi: 10.1089/10430340460732490. [DOI] [PubMed] [Google Scholar]
- 15.Flotte T.R., Trapnell B.C., Humphries M., Carey B., Calcedo R., Rouhani F., Campbell-Thompson M., Yachnis A.T., Sandhaus R.A., McElvaney N.G. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing α1-antitrypsin: interim results. Hum. Gene Ther. 2011;22:1239–1247. doi: 10.1089/hum.2011.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller C., Chulay J.D., Trapnell B.C., Humphries M., Carey B., Sandhaus R.A., McElvaney N.G., Messina L., Tang Q., Rouhani F.N. Human Treg responses allow sustained recombinant adeno-associated virus-mediated transgene expression. J. Clin. Invest. 2013;123:5310–5318. doi: 10.1172/JCI70314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendell J.R., Campbell K., Rodino-Klapac L., Sahenk Z., Shilling C., Lewis S., Bowles D., Gray S., Li C., Galloway G. Dystrophin immunity in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2010;363:1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendell J.R., Rodino-Klapac L.R., Rosales X.Q., Coley B.D., Galloway G., Lewis S., Malik V., Shilling C., Byrne B.J., Conlon T. Sustained alpha-sarcoglycan gene expression after gene transfer in limb-girdle muscular dystrophy, type 2D. Ann. Neurol. 2010;68:629–638. doi: 10.1002/ana.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ploquin A., Szécsi J., Mathieu C., Guillaume V., Barateau V., Ong K.C., Wong K.T., Cosset F.L., Horvat B., Salvetti A. Protection against henipavirus infection by use of recombinant adeno-associated virus-vector vaccines. J. Infect. Dis. 2013;207:469–478. doi: 10.1093/infdis/jis699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuck D., Lau T., Leuchs B., Kern A., Müller M., Gissmann L., Kleinschmidt J.A. Intranasal vaccination with recombinant adeno-associated virus type 5 against human papillomavirus type 16 L1. J. Virol. 2006;80:2621–2630. doi: 10.1128/JVI.80.6.2621-2630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieto K., Kern A., Leuchs B., Gissmann L., Müller M., Kleinschmidt J.A. Combined prophylactic and therapeutic intranasal vaccination against human papillomavirus type-16 using different adeno-associated virus serotype vectors. Antivir. Ther. (Lond.) 2009;14:1125–1137. doi: 10.3851/IMP1469. [DOI] [PubMed] [Google Scholar]
- 22.Nieto K., Stahl-Hennig C., Leuchs B., Müller M., Gissmann L., Kleinschmidt J.A. Intranasal vaccination with AAV5 and 9 vectors against human papillomavirus type 16 in rhesus macaques. Hum. Gene Ther. 2012;23:733–741. doi: 10.1089/hum.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L., Zhu T., Ye X., Yang L., Wang B., Liang X., Lu L., Tsao Y.P., Chen S.L., Li J., Xiao X. Long-term protection against human papillomavirus e7-positive tumor by a single vaccination of adeno-associated virus vectors encoding a fusion protein of inactivated e7 of human papillomavirus 16/18 and heat shock protein 70. Hum. Gene Ther. 2010;21:109–119. doi: 10.1089/hum.2009.139. [DOI] [PubMed] [Google Scholar]
- 24.Charville G.W., Cheung T.H., Yoo B., Santos P.J., Lee G.K., Shrager J.B., Rando T.A. Ex Vivo Expansion and In Vivo Self-Renewal of Human Muscle Stem Cells. Stem Cell Reports. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayaprolu V., Kruse S., Kant R., Venkatakrishnan B., Movahed N., Brooke D., Lins B., Bennett A., Potter T., McKenna R. Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 2013;87:13150–13160. doi: 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croyle M.A., Cheng X., Wilson J.M. Development of formulations that enhance physical stability of viral vectors for gene therapy. Gene Ther. 2001;8:1281–1290. doi: 10.1038/sj.gt.3301527. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Navio J.M., Fuchs S.P., Pedreño-López S., Rakasz E.G., Gao G., Desrosiers R.C. Host Anti-antibody Responses Following Adeno-associated Virus-mediated Delivery of Antibodies Against HIV and SIV in Rhesus Monkeys. Mol. Ther. 2016;24:76–86. doi: 10.1038/mt.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greig J.A., Calcedo R., Grant R.L., Peng H., Medina-Jaszek C.A., Ahonkhai O., Qin Q., Roy S., Tretiakova A.P., Wilson J.M. Intramuscular administration of AAV overcomes pre-existing neutralizing antibodies in rhesus macaques. Vaccine. 2016;34:6323–6329. doi: 10.1016/j.vaccine.2016.10.053. [DOI] [PubMed] [Google Scholar]
- 30.Han X., Ni W. Cost-Effectiveness Analysis of Glybera for The Treatment of Lipoprotein Lipase Deficiency. Value Health. 2015;18:A756. [Google Scholar]
- 31.Morrison C. $1-million price tag set for Glybera gene therapy. Nat. Biotechnol. 2015;33:217–218. doi: 10.1038/nbt0315-217. [DOI] [PubMed] [Google Scholar]
- 32.Lochrie M.A., Tatsuno G.P., Christie B., McDonnell J.W., Zhou S., Surosky R., Pierce G.F., Colosi P. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 2006;80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCraw D.M., O’Donnell J.K., Taylor K.A., Stagg S.M., Chapman M.S. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology. 2012;431:40–49. doi: 10.1016/j.virol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 36.Summerford C., Samulski R.J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opie S.R., Warrington K.H., Jr., Agbandje-McKenna M., Zolotukhin S., Muzyczka N. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 2003;77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Donnell J., Taylor K.A., Chapman M.S. Adeno-associated virus-2 and its primary cellular receptor--Cryo-EM structure of a heparin complex. Virology. 2009;385:434–443. doi: 10.1016/j.virol.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy H.C., Bowman V.D., Govindasamy L., McKenna R., Nash K., Warrington K., Chen W., Muzyczka N., Yan X., Baker T.S., Agbandje-McKenna M. Heparin binding induces conformational changes in Adeno-associated virus serotype 2. J. Struct. Biol. 2009;165:146–156. doi: 10.1016/j.jsb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kern A., Schmidt K., Leder C., Müller O.J., Wobus C.E., Bettinger K., Von der Lieth C.W., King J.A., Kleinschmidt J.A. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A. Bioengineered AAV Capsids with Combined High Human Liver Transduction In Vivo and Unique Humoral Seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimm D. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods. 2002;28:146–157. doi: 10.1016/s1046-2023(02)00219-0. [DOI] [PubMed] [Google Scholar]
- 43.Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W., Johnston W.A., Boden M., Gillam E.M. ReX: A suite of computational tools for the design, visualization, and analysis of chimeric protein libraries. Biotechniques. 2016;60:91–94. doi: 10.2144/000114381. [DOI] [PubMed] [Google Scholar]
- 45.Weirather J.L., de Cesare M., Wang Y., Piazza P., Sebastiano V., Wang X.J., Buck D., Au K.F. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res. 2017;6:100. doi: 10.12688/f1000research.10571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiao X., Zheng X., Ma L., Kutty G., Gogineni E., Sun Q., Sherman B.T., Hu X., Jones K., Raley C. A Benchmark Study on Error Assessment and Quality Control of CCS Reads Derived from the PacBio RS. J. Data Mining Genomics Proteomics. 2013;4:16008. doi: 10.4172/2153-0602.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatakis A., Ludwig T., Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 49.Nam H.J., Lane M.D., Padron E., Gurda B., McKenna R., Kohlbrenner E., Aslanidi G., Byrne B., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structure of adeno-associated virus serotype 8, a gene therapy vector. J. Virol. 2007;81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.