Abstract
Circulating cancer exosomes are microvesicles which originate from malignant cells and other organs influenced by the disease and can be found in blood. The exosomal proteomic cargo can often be traced to the cells from which they originated, reflecting the physiological status of these cells. The similarities between cancer exosomes and the tumor cells they originate from exhibit the potential of these vesicles as an invaluable target for liquid biopsies. Exosomes were isolated from the serum of eight osteosarcoma-bearing dogs, five healthy dogs, and five dogs with traumatic fractures. We also characterized exosomes which were collected longitudinally from patients with osteosarcoma prior and 2 weeks after amputation, and eventually upon detection of lung metastasis. Exosomal proteins fraction were analyzed by label-free mass spectrometry proteomics and were validated with immunoblots of selected proteins. Ten exosomal proteins were found that collectively discriminate serum of osteosarcoma patients from serum healthy or fractured dogs with an accuracy of 85%. Additionally, serum from different disease stages could be distinguished with an accuracy of 77% based on exosomal proteomic composition. The most discriminating protein changes for both sample group comparisons were related to complement regulation, suggesting an immune evasion mechanism in early stages of osteosarcoma as well as in advanced disease.
Introduction
Canine osteosarcoma (OSA) exhibits morphologic and genomic resemblance to pediatric OSA in humans, with dogs exhibiting 10 times the incidence rate. The treatment options for both species are limited, and the prognosis is poor [1], [2]. The standard for diagnosis has not changed over the last several decades and includes primarily radiographic imaging of the affected limb and lungs followed by cytological or histological sample for confirmation and grading.
In humans, osteosarcoma is the most common primary malignant tumor of the bone. The standard of care includes surgical resection and multiagent chemotherapy; however, due to early micrometastasis to the lungs, for a majority of patients, the prognosis is considered poor. Furthermore, the evidence of radiographic metastasis was associated with disease progression and often an even worse prognosis. Therefore, improved diagnostic tools are needed to better detect and stage the disease. One strategy is to use the canine model to develop these diagnostic tools. The course of the disease and etiology are similar between humans and canines which, when combined with the high incidence rate of osteosarcoma in dogs, make canine osteosarcoma a robust spontaneous model for human disease [1], [2], [3].
Exosomes are 30- to 160-nm–diameter microvesicles secreted by the great majority of cells [4], [5]. They contain proteins, lipids, fragments of DNA, RNA, and microRNA, and are essential for trafficking of biological material and cellular cross talk [4], [6]. The exosomal proteomic cargo is variable and inclined to the physiological stressors exerted on the cell of origin [7], [8]. Exosome-bound proteins and genomic materials are packaged in a selective manner and can exhibit a profile which is often vastly different from the cell type of origin [8], [9].
Once secreted, exosomes can be carried through the circulatory system to their target cells, where they can transfer proteins, lipids, and nucleic acids that play influential roles in tumorigenesis, cancer progression, metastasis, response to treatment, and immune suppression [10], [11], [12]. The lipid bilayer of exosomes makes a stable structure which is resistant to enzymatic degradation in blood and other body fluids and an attractive source for the investigation of biomarkers.
The roles of exosomes in the disease course of osteosarcoma or response to therapeutic treatments have not been fully characterized [13], [14]. Exosomal proteins derived from human osteosarcoma cell line supernatants are enriched with proteins associated with angiogenesis, cell adhesion, immune evasion, and cell migration in comparison to proteins in exosomes from nonmalignant cells and non–exosome-derived blood proteins [13], [14], [15]. Recent investigations in human pediatric OSA have demonstrated significant differences in serum protein content; proteins which differ can be used as biomarkers for early detection of cancer, metastasis, and response to therapy [16]. We hypothesized that canine serum-derived exosomes will contain a unique protein signature that can be ascribed to OSA patients, analogous to what has been demonstrated in pediatric OSA. Furthermore, we postulated that exosomes derived from sera of OSA patients will exhibit a unique protein signature throughout the disease stages, including at time of diagnosis, at 2 weeks postamputation, and at the onset of lung metastasis. These signatures will have the potential to serve as a noninvasive diagnostic tool for the detection, staging of disease, and prognostication.
Materials and Methods
Patient Sera Samples
Sera were collected and stored at −20°C. Approximately 600 μl of serum was obtained from a total of 18 adult (>1 year) medium- and large-breed dogs (>20 kg) in the following categories: appendicular osteosarcoma that was verified by histopathology (OSA; n = 8); otherwise healthy animals that have experienced traumatic bone fractures (FX; n = 5); and healthy, size-matched canine controls (N; n = 5). Dogs were deemed healthy based on complete blood count, chemistry panel, urinalysis, normal physical examination, and no current history of illness or treatment with prescription medication. Samples were obtained from canine OSA patients at initial diagnosis, prior to standard treatment of limb amputation and carboplatin chemotherapy (270-300 mg/m2 administered every 3 weeks for a total of 4 doses). Patients were excluded from the study if they had evidence of metastasis at time of diagnosis, had nonappendicular disease, or did not receive standard treatment.
Based on empirical quantitative analysis of LC/MS-based measurement of serum protein abundances, we modeled protein abundances as log normal-distributed and on log2 scale. For the purpose of a power analysis, we assumed an intrasample group measurement standard deviation of approximately 6.0. Thus, a study size of 18 samples would give a power of 0.80 for detecting a log-abundance ratio of at least 4.0 between the sample groups, with significance level of .05.
Longitudinal samples were obtained from five additional patients with appendicular osteosarcoma at diagnosis, at 2 weeks postamputation (before initiation of chemotherapy), and upon first detection of metastatic disease.
Cell Culture Samples
For exosome isolation from HMPOS and POS (canine OSA cell lines) culture media, subconfluent cells were incubated in media containing 10% exosome-depleted FBS (Exo-FBS, System Biosciences, Inc., Palo Alto, CA) for 24 hours, and supernatants were removed prior to cells reaching 100% confluence. The collected media were subsequently centrifuged for 10 minutes at 2000×g, and the cell-free supernatants were collected. For exosome isolation from patient serum samples, 200 μl of serum was centrifuged for 10 minutes at 2000×g to remove residual cells or debris that remained in the sample as a preliminary step.
Once the cell culture supernatants and serum were clear of debris, exosomal extraction was carried out with Total Exosome Isolation reagent (Invitrogen, Thermo Fisher Scientific) following the manufacturer's instructions. Briefly, the reagent was vigorously mixed with the cell-free media or serum at a ratio of 1:2 and 1:5, respectively. After overnight incubation at 4°C, exosomes were then collected after centrifugation at 10,000×g for 60 minutes and resuspended in 1× phosphate-buffered saline (PBS). Concentration of protein content was determined using a Pierce BCA kit (Thermo Fisher Scientific) on exosome aliquots lysed in 1× RIPA (Pierce RIPA Buffer, Thermo Fisher Scientific).
NanoSight Analyses
Once purified, exosomes were suspended in 100 μl filtered PBS, and protein concentrations were determined with the Pierce BCA protein assay. Prior to size validation assay, the suspension was diluted again with filtered PBS at a 1:100 concentration and analyzed using the NanoSight LM10 system (NanoSight Ltd., Amesbury, United Kingdom, NTA 3.00060 software). The Nanoparticle Tracking Analysis software version 2.3 was used to analyze 30-second videos to determine vesicle size and concentration. The analysis of exosomes from cell culture supernatants was done on samples which were diluted to a concentration of 50-150 particles per frame. The chamber conditions and system settings were adjusted as previously described [14].
Proteomic Analysis
An aliquot of isolated exosomes from all 18 canine serum samples along with exosomes isolated from the serum of 5 distinct canine OSA patients at different time points in their disease (diagnosis, 2 weeks postamputation, and onset of lung metastases) was evaluated by mass spectrometry (LC-MS/MS) at the Oregon State University Mass Spectrometry Center. Additionally, exosome preparations that were isolated from culture media of carboplatin-resistant cell lines (as described in the exosome isolation methods section) and protein lysates from carboplatin-resistant cells were submitted for proteomic analysis. The proteins were digested by mass spectrometry–grade trypsin (Trypsin Gold, Promega Corporation, Madison, WI). The proteolytic samples were analyzed using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fischer Scientific) coupled to a Waters nanoAcquity UPLC system (Waters, Milford, MA). The mobile phase was composed of LC-MS–grade water containing 0.1% formic acid as solvent A and LC-MS–grade acetonitrile containing 0.1% formic acid as solvent B. The peptides were loaded onto a nanoAcquity UPLC 2G Trap Column (100 Å, 180 μm × 20 mm, 5 μm) for 5 minutes with 3% mobile phase B at a flow rate of 5 μl/min. For peptide separation, a nanoAcquity UPLC Peptide BEH C18 column (130 Å, 100 μm × 100 mm, 1.7 μm) was used. Peptide elution was achieved by applying a 120-minute gradient in which B was increased from 3% to 10% at 3 minutes, reached 30% at 105 minutes and then 90% at 108 minutes, held at 90% B until 112 minutes, then decreased to 3% at 113 minutes, and held until 120 minutes. A flow rate of 0.5 μl/min was used. Mass spectral data were acquired in the positive ion mode. The spray voltage was 2400 V, and the ion transfer tube temperature was 300°C. The Orbitrap analyzer acquired all MS spectra with 120 K resolution at m/z 200, and the ion trap analyzer acquired all MSMS spectra in collision-induced dissociation fragmentation mode using top speed method. Automatic gain control target was set to 4.0 × 105 for precursor ions and 104 for product ions. Thermo Scientific Proteome Discoverer 2.1 software was used for processing and analyzing the raw data files and for performing searching against the Uniprot Homo sapiens protein database using Sequest HT search engine. The mass tolerance of precursor and fragment ions was set at ±10 ppm and ± 0.6 Da, respectively. A maximum of two missed cleavage sites was allowed. Carbamidomethylation of cysteine was specified as static modification; oxidation of methionine was specified as dynamic modification. The overall false discovery rate was less than 1%.
Immunoblots
To confirm presence of the differentially expressed proteins indicated by mass spectrometry, Western blot analyses were performed. Patient serum exosomes (n = 9; 5 OSA, 2 FX, and 2 N) were lysed by adding 5× RIPA buffer containing phosphatase and proteinase inhibitors to a final 1× concentration. Homogenate aliquots (containing 10 μg protein) were mixed at a volume ratio of 1:1 with Laemmli buffer and 5% β-mercaptoethanol, boiled for 10 minutes at 100°C, and electrophoresed on a 10% SDS-PAGE gel at a constant current of 10 mA/plate. The separated proteins were transferred to a nitrocellulose membrane, blocked for 1 hour with Odyssey blocking buffer (LI-COR, Lincoln, NE) diluted 1:1 in TBS (20 mM Tris-HCl pH 7.4 and 150 mM NaCl), followed by overnight incubation at 4°C with rabbit anti-HSP90 antibody (Cell Signaling Technology, Danvers, MA) diluted 1:1000 in TBST (TBS with 0.1% Tween-20). Replicate blots were probed with rabbit anti-QSOX1 antibody (Proteintech Group, Inc., Rosemont, IL) and rabbit anti-ITIH1 antibody (Abcam, Cambridge, MA) diluted 1:1000 in TBST. All antibodies used had been previously validated for canine samples by Western blot. Proteins labeled with primary antibody were detected with goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Dallas, TX) diluted 1:40,000 in TBST. Membranes were developed with Supersignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and scanned on an ImageQuant LAS4000 (GE Healthcare Life Sciences, Marlborough, MA).
For visual assessment of uniform protein quantity loading between all samples, the 10% SDS-PAGE gel was stained with LabSafe GEL Blue (G-Biosciences, Geno Technology Inc., St. Louis, MO). Briefly, the gel was washed three times in deionized water for 5 minutes and then gently rocked in enough LabSafe GEL Blue to cover the gel; protein bands were visible at 5 minutes with maximum intensity at 60 minutes.
Proteomics Data Analysis
Statistical analysis on proteomic data was performed using Metaboanalyst 3.0 Software [17]. Peak intensity data were normalized and log transformed before multivariate analysis. Predictive accuracy for discriminating proteins was determined with R statistical software. Statistical significance was set at P = .05.
Classification Accuracy Analysis: Discrimination of OSA Versus Normal or Fracture
For each protein detected above background, its normalized, log-scale protein levels (see proteomics data analysis) in two sample groups (OSA-derived samples and samples from a non-OSA control group of healthy dogs and dogs with fractures) were scored for difference of means using a Welch's t test. Based on a minimum absolute t score of 2.0, 10 proteins were selected to include as features in a multivariate classifier (Table 1). The levels of these proteins were combined in an elastic-net-penalized binary logistic regression classifier (using the R package glmnet function cv.glmnet with α = 0.25 and with −[0.9 + log(λ)] taking all possible values in the range of 0.000 to 0.975 in increments of 0.025) to predict OSA versus non-OSA status. Using 100 replications of three-fold cross-validation, the classifier's accuracy was measured on samples held out from the training set. The average accuracy across the three folds and 100 replications was found to be 85%, with 6% standard error.
Table 1.
Proteins Used in the Binary Classifier for OSA Vs. Non-OSA Dogs Based on Serum Exosome Profiling
| Protein Number | Accession | Gene Name/Description | t |
|---|---|---|---|
| 1 | E9PGN7 | SERPING1 | 3.90 |
| 2 | A0A0K0K1H8 | HEL-S-71p | 3.75 |
| 3 | Q9BWU5 | HBB | 3.19 |
| 4 | P13645 | KRT10 | 2.81 |
| 5 | B6EDE2 | HEL180 | 2.63 |
| 6 | P19827 | ITIH1 | 2.22 |
| 7 | A8K5A4 | cDNA FLJ76826 (similar to human gene CP) | 2.08 |
| 8 | A0A075B6L1 | IGLC7 | 2.08 |
| 9 | B4E1Z4 | Uncharacterized protein | 2.06 |
| 10 | Q9H2L7 | DC33 | −3.80 |
Here, t is the test statistic for Welch's test for the mean protein level in samples from OSA dogs minus the mean protein level in samples from non-OSA dogs.
Given the small pilot study size, we used cross-validation in order to obtain a lower-variance estimate of classification accuracy on unseen cases and a confidence interval on the classifier estimate accuracy [18].
Detection of Disease Stage Using a Proteomic Signature
For each protein detected above background, its normalized, log-scale protein levels in three sample groups [at time of first OSA diagnosis (DX), at postamputation (PA), and at time when progressive disease (PD) was confirmed] were scored for using a one-way ANOVA F test. Based on the ANOVA F test (P < .05), two proteins (SERPIND 1 and a protein highly similar to human MHC class III complement component C6) were selected as the classifier. The levels of these proteins were combined in a LASSO-penalized multinomial logistic regression classifier (using the R package glmnet with −[0.9 + log(λ)] taking all possible values in the range of 0.000 to 0.975 in increments of 0.025) to predict the sample group (DX, PD, or PA). Using 1000 replications of three-fold cross-validation, the classifier's accuracy was measured on samples held out from the training set. The average accuracy across the three folds and 1000 replications was found to be 77% with 16% standard error.
Results
Evaluation of Cell Culture– and Serum-Derived Exosomes
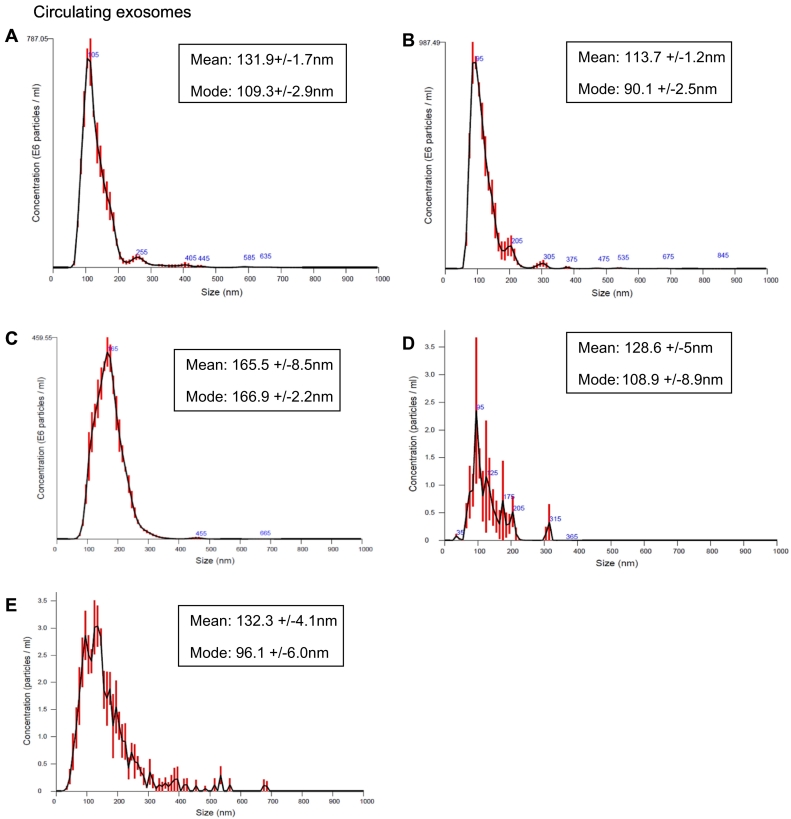
Exosomes were isolated from the serum of canine patients with histologically confirmed appendicular osteosarcoma (OSA); patients with traumatic bone fracture (FX); and normal, healthy size-matched controls (N). Samples were stored at −80°C for up to 14 months prior to being processed for exosomal extraction; no signs of degradation were evident on proteomic or Nanosight analysis. We also isolated exosomes from the supernatants of two canine osteosarcoma cell lines: POS and HMPOS. These cell lines, derived from canine osteosarcomas, were validated as osteosarcomas by proteomic analysis of alkaline phosphatase expression and calcification assays [19]. NanoSight light scatter analysis has shown a mean of 128.6 (+/−5) nm and mode of 108.9 (+/−8.9) nm for exosomes extracted from POS supernatants and mean of 132.3 (+/−4.1) nm and mode of 96.1 (+/−6.0) nm for the HMPOS exosomes (Figure 1).
Figure 1.
Size and concentration of exosomes isolated from serum of three of the dogs representing the three cohorts in the study: (A) osteosarcoma,(B) traumatic fracture, (C) normal dogs, and exosomes isolated from the supernatants of two canine osteosarcoma cell lines: (D) POS and (E) HMPOS correspondently.
Circulating exosomes of five dogs were used for validation of exosome isolation and measured using NanoSight light scatter technology (Figure 1). The measured vesicles' size mean was in a range of 113.2-173.3 and a mode of 90.1-166.9, which is consistent with the size of exosomes measured in previous studies using similar technology [5], [14].
Proteomic Evaluation of Exosomes Derived from Sera of OSA, FX, and N Dogs
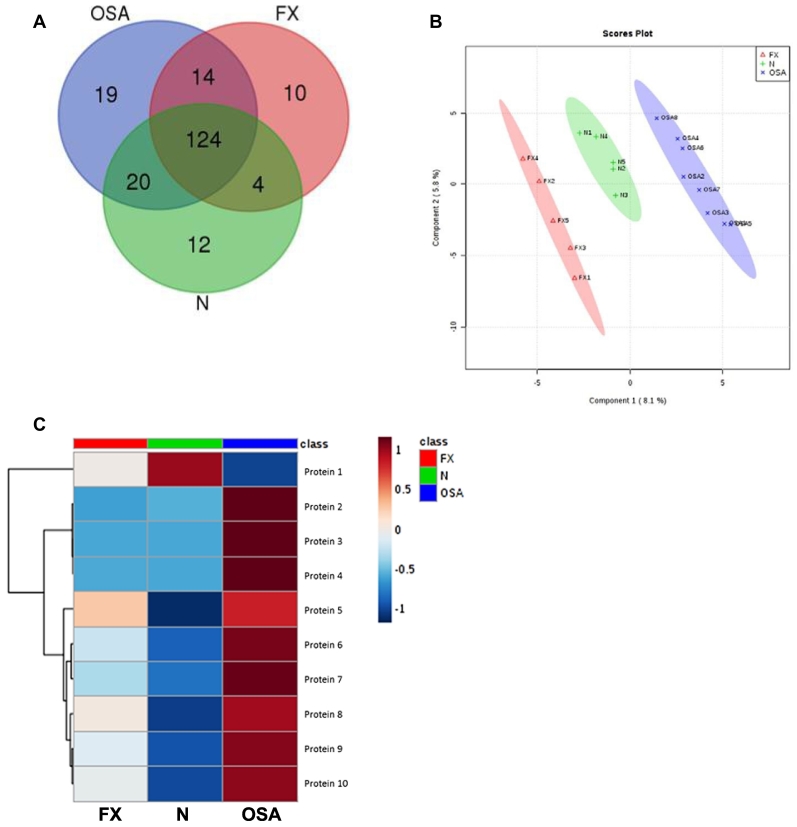
LC-MS/MS on purified serum-derived exosomes revealed 177 proteins contained within the exosomes of canine osteosarcoma patients (OSA), 152 proteins in the exosomes from patients with traumatic bone fracture (FX), and 160 proteins in the normal dogs (N) (Figure 2A). All exosomes shared 124 proteins. Serum-derived exosomes from OSA contained 19 unique proteins and shared 14 and 20 proteins with FX and N, respectively. FX and N had 10 and 12 unique proteins, respectively, and shared 4 proteins exclusively. A supervised partial least squares discriminate analysis was able to divide the patients into three distinct groups based on proteomic cargo isolated from circulating exosomes (Figure 2B). Based on logistic regression analysis, with penalized maximum likelihood and with feature selection based on a t test, we achieved 85% (±0.003811) accuracy for discriminating patients with OSA from normal (N) or traumatic fracture (FX) via evaluation of the top 10 discriminating circulating exosomal proteins. These top 10 proteins are diagrammed based on relative expression in the examined samples (Table 1, Figure 2C).
Figure 2.
Proteomic cargo of circulating exosomes was determined by LC-MS/MS. (A) Number of proteins shared by osteosarcoma (OSA), dogs with traumatic fracture (FX), and healthy controls (N) illustrated with a Venn diagram. (B) A supervised partial least squares discriminate analysis exhibits a distinct separation of the three groups based on their exosomal proteomic signature. (C) The 10 most significant proteins for discrimination between osteosarcoma (OSA) and fracture (FX) or normal (N) are listed along with relative protein expression (red = high, blue = low) (Table 1). OSA appears to have a distinct profile when compared with the other two groups, demonstrating primarily upregulated protein expressions.
Proteomic Evaluation of Longitudinally Collected Exosomes from Sera of Dogs with OSA
The overall disease-free interval for the longitudinally studied dogs was in the range of 58 to 257 days with an average of 134 days.
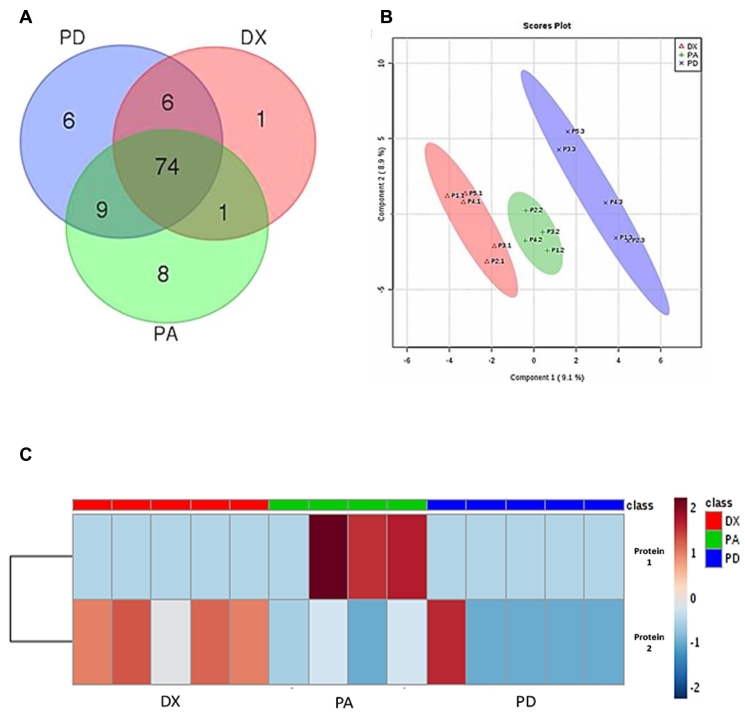
LC-MS/MS on purified serum-derived exosomes revealed 82 proteins in the exosomes of canine osteosarcoma patients at diagnosis (DX), 92 proteins in the exosomes from patients 2 weeks postamputation (PA), and 95 proteins from the same patients at time of progressive disease (PD). All exosomes shared about 74 proteins. Serum-derived DX exosomes contained one unique protein and shared one and six proteins with PA and PD, respectively. PA and PD had eight and six unique proteins, respectively, and shared nine proteins exclusively (Figure 3).
Figure 3.
Proteomic analysis of circulating exosomes extracted from osteosarcoma dogs at diagnosis and prior to amputation (DX), at 2 weeks after amputation (PA), and on detection of progressive disease (PD). (A) Venn diagram demonstrates the shared and unique proteins for each of the three cohorts. (B) A supervised partial least squares discriminate analysis discriminates the stages of disease into three distinct groups based on proteomic composition isolated from circulating exosomes; each mark (x, +, ∆) represents an individual time point for the same patient. (C) The two most significant proteins for discriminating between exosomes from the different disease stages of OSA are listed along with relative protein expression (red = high, blue = low). Protein 1 is SERPIND1 and protein 2 represents cDNA FLJ78071 which is highly similar to human MHC class III complement component C6.
A supervised partial least squares discriminate analysis revealed distinct protein clusters by which one could discriminate between the sera of dogs from the different disease stages (Figure 3B). Based on logistic regression analysis, we obtained 77% accuracy for distinguishing the different stages of OSA via evaluation of the top two discriminating circulating exosomal proteins. These two proteins, serpin D1 and major histocompatibility complex (MHC) class III-complement component C6, are diagrammed based on their relative expression (Figure 3C).
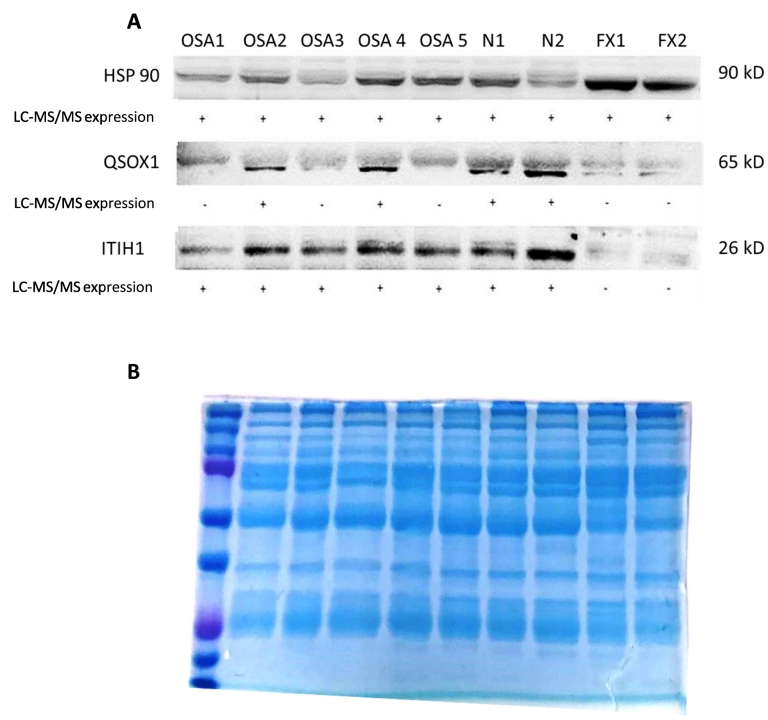
Validation of LC-MS/MS results was done by Western blot on selected proteins based on availability of antibodies validated for canines. The analysis of exosomal cargo confirmed the presence of select proteins, QSOX1 and ITIH1, along with a ubiquitous exosome marker protein, Hsp90 (Figure 4A). Visual evaluation of the blot demonstrates increased protein density in all patients where QSOX1 and ITIH1 were detected on mass spectrometry. This result provides independent support for the validity of the mass spectrometry results. Overall, QSOX1 and ITIH1 expressions were in agreement with the LC-MS/MS results. As expected, HSP90 which is commonly carried by exosomes did not dramatically differ between the groups. Total protein visualization on the 10% SDS-PAGE gel following electrophoresis demonstrated equivalent protein quantity loaded between all samples evaluated (Figure 4B).
Figure 4.
Immunoblots of exosomal cargo selected proteins. (A) Immunoblots of exosomal lysates confirm universal expression of ubiquitous exosomal protein HSP90. Proteins QSOX1 and ITIH1 are expressed to a greater extent in exosomes from patients, concurrent with mass spectrometry. Patients with traumatic bone fracture did not contain QSOX1 and ITIH1 in their exosomal cargo. Positive (+) protein was detected by LC-MS/MS; negative (−) protein was not detected by LC-MS/MS. (B) Coomassie blue stain following protein electrophoresis demonstrates equivalent protein amounts loaded between all samples evaluated.
Comparison of Exosomal Proteomic Cargo from Serum and Osteosarcoma Cell Lines
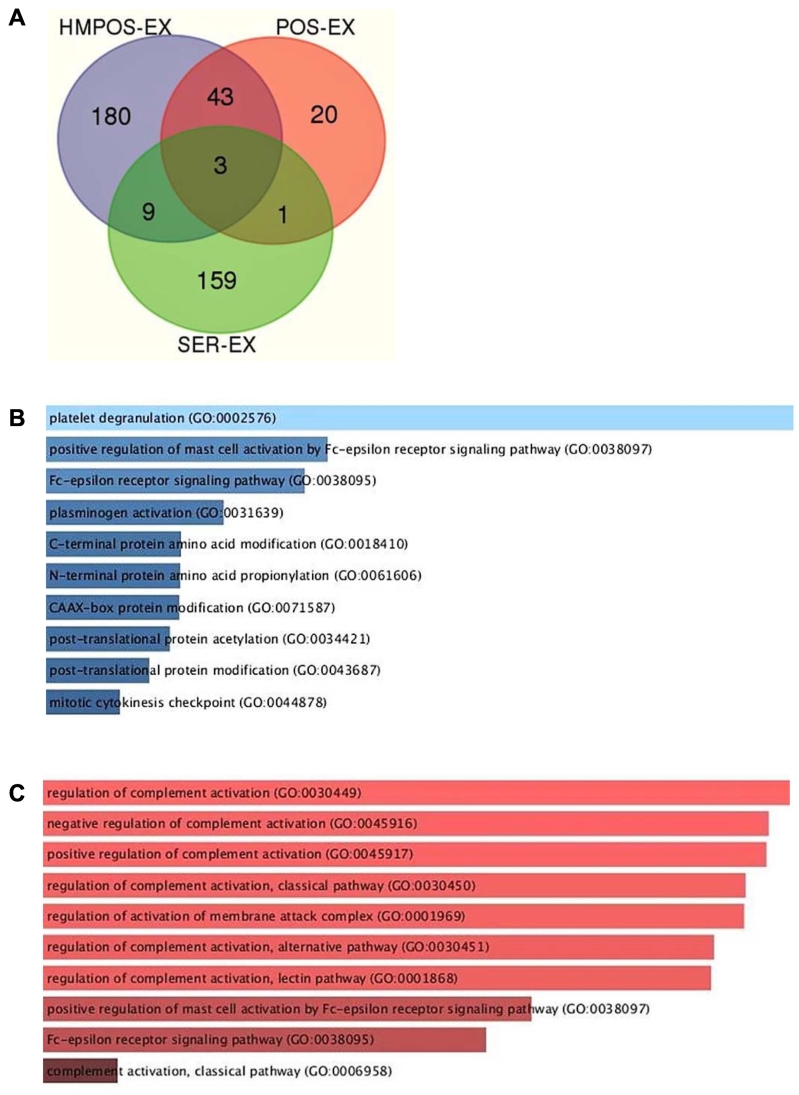
In order to determine whether the serum exosomal proteome from OSA dogs contains similar cargo to the exosomal proteome of OSA cells, mass spectrometry was conducted on exosomes from OSA cell lines HMPOS and POS along with exosomes from pooled OSA dog serum. LC-MS/MS analysis of exosomal proteomic cargo extracted from the supernatants of HMPOS and POS revealed a total of 235 and 67 proteins, respectively. HMPOS and POS shared 43 proteins between them and three more proteins with serum exosomes. HMPOS had 180 proteins exclusively and shared 9 more proteins with serum exosomes, while POS had 20 proteins and shared only 1 protein solely with serum exosomes (Figure 5A). GO biological processes analysis of top osteosarcoma discriminating 37 proteins was done to better characterize the exosomal proteome (Figure 5, B and C).
Figure 5.
(A) Venn diagram exhibiting the distribution of identified proteins between the OSA cell lines exosomes (HMPOS-EX, POS-EX) and the serum exosomes. (B) GO biological processes analysis of top 37 proteins exhibits an increase of platelets degranulation and mast cell activation by the Fc receptor in OSA circulating exosomes versus the two control cohorts (normal and traumatic fracture). (C) GO biological processes analysis of top 37 proteins exhibit a downregulation of multifacet deficiencies of complement regulation in OSA circulating exosomes versus the two control cohorts. The color of each raw reflects the relative amount of proteins in each category.
Discussion
In the current study, we have isolated exosomes from serum of healthy, fractured, and osteosarcoma-diagnosed dogs. Proteomic analysis of the exosomes revealed a characteristic protein signature of each of these groups and demonstrated the potential use of exosomes as a viable source for biomarkers discovery in cancer-bearing dogs that could be extended to human osteosarcoma. The abundant availability of exosomes in serum is a positive factor for the potential use of serum exosome profiling for diagnosis, as is the minimal invasiveness of sample collection. Thus, serum exosome profiling can be performed at different time points throughout the disease course, thereby improving the existing tools for early diagnosis and disease staging.
The results of our study reveal a unique protein cargo expressed by circulating exosomes in canine osteosarcoma patients. This cargo contained >170 proteins, which were largely associated with proteolysis, regulation of the immune system, activation of stress response, and regulation of metabolic processes. The serum exosome proteomics-based classifier for detecting osteosarcoma that we developed in this study is largely based on proteins that are upregulated in the osteosarcoma cases vs. normal cases (9 out of 10). Although eight proteins were detected in at least one normal sample that were entirely undetected in osteosarcoma, these eight proteins all had sporadic expression in the normal samples and thus high variance that precluded their inclusion as features in the classifier. The top 10 discriminating proteins were utilized for logistic regression analysis and demonstrated that OSA-bearing dogs could be differentiated from control animals (healthy and traumatic fracture) with 85% accuracy.
We also demonstrated distinct protein clustering based on disease stage using circulating exosomes collected from five osteosarcoma patients at three time points over their disease course. Logistic regression analysis revealed that various disease stages in OSA could be differentiated based on two discriminating proteins, Serpin D1 and a protein highly similar to major histocompatibility complex (MHC) class III-complement component C6, with 77% accuracy.
Differences in circulating exosomal composition collected at different time points along the linear study were evident in our proteomic analysis. These differences may be explained by several factors associated with the complexity of tumorigenesis and disease progression. Secretions from a primary osteosarcoma may exhibit a vastly different proteomic composition from metastatic disease due to the changes associated with mutations and epigenetic variations of the tumor during its evolution. Other obvious changes of exosomal composition may be derived from stromal cells surrounding the tumor, whether it is bone, lung, or any other tissue. Additionally, the capacity of exosomes and other soluble mediators to impact distant tissues, which are not in direct contact with the tumor, may result in the secretion of diverse exosomes that will be typical to these organs and their sustained damage. The complement system plays a critical role in eradicating cancer cells by direct antitumor effects resulting in lysis of malignant cells and in the enhancement of antibody-dependent cell-mediated cytotoxicity [20]. Immunotherapy, which has become the mainstay for the treatment of several types of cancers in the recent years, has been shown to mediate complement activation; increased induction of complement activation may result in clinical benefit [21], [22], [23]. Evading immune destruction is one the hallmarks of cancer, and the expression of membrane-bound complement regulatory proteins such as CD35, CD46, CD55, and CD59, as well as the C1 inhibitor which prevents the activation of the classical pathway, has shown to be a potential evasion mechanism of tumor cells and can be found in serum and ascetic fluids of cancer patients [24], [25], [26], [27], [28]. In our study, the two of the top three discriminating proteins were plasma protease C1 inhibitor and C1qa, which are key conflicting factors of the classical pathway. Furthermore, these markers were expressed in opposing manner with an increased expression of the plasma protease C1 inhibitor and a decreased expression of the C1qa in the exosomes of osteosarcoma-bearing dogs.
When we examined the discriminating proteins associated with disease stage, we identified the expression of a protein highly similar to human MHC class III complement component C6 expressed by exosomes from serum collected prior to amputation. However, the expression of this protein was reduced at the progressive disease stage. This finding may be a reflection of immune suppression associated with disease progression.
Investigation of the overall impact associated with proteomic changes in complement factors of circulating exosomes was beyond the scope of this study. However, the decreased expression in factor C1qa and the increased expression of plasma protease C1 inhibitor, taken together with the decreased expression of complement component C6, may be a reflection of an altered complement system in OSA patients and patients with progressive disease. These findings reveal a novel set of biomarkers, and further investigation should examine the potential of these complement factors as viable immunotherapeutic targets for osteosarcoma.
The results of this current study confirmed our initial hypothesis that serum-derived exosomes contain a unique protein signature that can be ascribed to canine OSA patients. Additionally, this signature appears to evolve throughout the course of disease. Evaluation for these discriminating serum exosomal proteins could potentially be utilized as an easy, noninvasive liquid biopsy for canine patients with appendicular osteosarcoma, aiding in diagnosis and providing real-time information about disease progression without the need for invasive procedures, similar to what has been reported in human malignancies [29], [30], [31].
We also studied the proteomic exosomal cargo from two canine OSA cell lines, and as expected, the comparison of serum-derived exosomes from OSA patients with OSA cell line exosomes demonstrates very little overlap in protein content. The differences in protein cargo reflect the fact that serum exosomes originate from a multitude of cells in the body as opposed to purely the neoplastic population; as a result, serum exosomes provide information about the body's cumulative and distant response to presence of cancer. Another point is that serum samples represent responses to tumor in vivo, which likely exhibits significant variation from in vitro conditions.
The diversity represented in the proteomic composition of circulating exosomes can expand our understanding about the disease course and the impact that malignant cells exert on their stroma. The significance of the discriminating proteins we identified beyond their role as biomarkers should be further investigated.
In future studies, we will assess the sensitivity of these markers and investigate their expression prior to the radiographic evidence of metastatic disease. We also plan to initiate a study that will attempt to predict the response to treatment based on markers expressed in circulating exosomes prior to amputation.
Conclusions
The current study has demonstrated the capacity of the proteomic cargo of circulating exosomes to discriminate tumor-bearing from non–tumor-bearing dogs with high accuracy. Furthermore, we have shown how this proteomic cargo of exosomes changes with disease stage and can serve as a viable source for biostaging of patients during the disease course. The current study has highlighted several proteins of the complement system as key discriminators between osteosarcoma-bearing dogs and their controls which warrant a further investigation for their role in disease progression.
Footnotes
The Thermo Scientific Orbitrap Fusion Lumos mass spectrometer used for this work was partly supported by the National Institute of Health (grant number 1S10OD020111-01).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.07.004.
Appendix A. Supplementary data
Supplementary Figure
References
- 1.Morello E, Martano M, Buracco P. Biology, diagnosis and treatment of canine appendicular osteosarcoma: similarities and differences with human osteosarcoma. Vet J. 2011;189(3):268–277. doi: 10.1016/j.tvjl.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Carrle D, Bielack SS. Current strategies of chemotherapy in osteosarcoma. Int Orthop. 2006;30(6):445–451. doi: 10.1007/s00264-006-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan T, Khanna C. Comparative aspects of osteosarcoma pathogenesis in humans and dogs. Vet Sci. 2015;2(3):210. doi: 10.3390/vetsci2030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azmi A, Bao B, Sarkar F. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sokolova V, Sokolova A-K, Hornung S, Rotan O, Horn Peter A, Epple M, Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Fan G-C. Hypoxic exosomes promote angiogenesis. Blood. 2014;124(25):3669–3670. doi: 10.1182/blood-2014-10-607846. [DOI] [PubMed] [Google Scholar]
- 7.Wubbolts R, Leckie RS, Veenhuizen PTM, Schwarzmann G, Möbius W, Hoernschemeyer J, Slot J-W, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes - Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278(13):10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 8.Ramteke A, Ting H, Agarwal C, Mateen S, Somasagara R, Hussain A, Graner M, Frederick B, Agarwal R, Deep G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol Carcinog. 2015;54(7):554–565. doi: 10.1002/mc.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickman CTD, Lawson J, Jabalee J, MacLellan SA, LePard NE, Bennewith KL, Garnis C. Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget. 2017;8(9):15252–15266. doi: 10.18632/oncotarget.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Erviti L, Seow Y, Yin HF, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341. doi: 10.1038/nbt.1807. [U179] [DOI] [PubMed] [Google Scholar]
- 11.Plebanek MP, Angeloni NL, Vinokour E, Li J, Henkin A, Martinez-Marin D, Filleur S, Bhowmick R, Henkin J, Miller SD, Ifergan I. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat Commun. 2017;8(1):1319. doi: 10.1038/s41467-017-01433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao L, Wang L, Dai T, Jin K, Zhang Z, Wang S, Xie F, Fang P, Yang B, Huang H. Nat Immunol. 2018. Tumor-derived exosomes antagonize innate antiviral immunity. [DOI] [PubMed] [Google Scholar]
- 13.Garimella R, Washington L, Isaacson J, Vallejo J, Spence M, Tawfik O, Rowe P, Brotto M, Perez R. Extracellular membrane vesicles derived from 143B osteosarcoma cells contain pro-osteoclastogenic cargo: a novel communication mechanism in osteosarcoma bone microenvironment. Transl Oncol. 2014;7(3):331–340. doi: 10.1016/j.tranon.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Troyer RM, Ruby CE, Goodall CP, Yang L, Maier CS, Albarqi HA, Brady JV, Bathke K, Taratula O, Mourich D. Exosomes from osteosarcoma and normal osteoblast differ in proteomic cargo and immunomodulatory effects on T cells. Exp Cell Res. 2017;358(2):369–376. doi: 10.1016/j.yexcr.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Jerez S, Araya H, Thaler R, Charlesworth MC, López-Solís R, Kalergis AM, Céspedes PF, Dudakovic A, Stein GS, van Wijnen AJ. Proteomic analysis of exosomes and exosome-free conditioned media from human osteosarcoma cell lines reveals secretion of proteins related to tumor progression. J Cell Biochem. 2017;118(2):351–360. doi: 10.1002/jcb.25642. [DOI] [PubMed] [Google Scholar]
- 16.Savitskaya YA. Serum tumor markers in pediatric osteosarcoma: a summary review. Clin Sarcoma Res. 2012;2:9. doi: 10.1186/2045-3329-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia JG, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 18.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 1998;10(7):1895–1923. doi: 10.1162/089976698300017197. [DOI] [PubMed] [Google Scholar]
- 19.Barroga EF, Kadosawa T, Okumura M, Fujinaga T. Establishment and characterization of the growth and pulmonary metastasis of a highly lung metastasizing cell line from canine osteosarcoma in nude mice. J Vet Med Sci. 1999;61(4):361–367. doi: 10.1292/jvms.61.361. [DOI] [PubMed] [Google Scholar]
- 20.Meyer S, Leusen JHW, Boross P. Regulation of complement and modulation of its activity in monoclonal antibody therapy of cancer. MAbs. 2014;6(5):1133–1144. doi: 10.4161/mabs.29670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Meerten T. Complement-induced cell death by rituximab depends on CD20 expression level and acts complementary to antibody-dependent cellular cytotoxicity. Clin Cancer Res. 2006;12(13):4027–4035. doi: 10.1158/1078-0432.CCR-06-0066. [DOI] [PubMed] [Google Scholar]
- 22.Lundin J, Kimby E, Björkholm M, Broliden P-A, Celsing F, Hjalmar V, Möllgård L, Rebello P, Hale G, Waldmann H. Phase II trial of subcutaneous anti-CD52 monoclonal antibody alemtuzumab (Campath-1H) as first-line treatment for patients with B-cell chronic lymphocytic leukemia (B-CLL) Blood. 2002;100(3):768–773. doi: 10.1182/blood-2002-01-0159. [DOI] [PubMed] [Google Scholar]
- 23.Gelderman KA, Tomlinson S, Ross GD, Gorter A. Complement function in mAb-mediated cancer immunotherapy. Trends Immunol. 2004;25(3):158–164. doi: 10.1016/j.it.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20(1):34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 26.Gorter A, Meri S. Immune evasion of tumor cells using membrane-bound complement regulatory proteins. Immunol Today. 1999;20(12):576–582. doi: 10.1016/s0167-5699(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 27.Bjorge L. Ascitic complement system in ovarian cancer. Br J Cancer. 2005;92(5):895–905. doi: 10.1038/sj.bjc.6602334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okroj M, Hsu Y-F, Ajona D, Pio R, Bloma AM. Non–small cell lung cancer cells produce a functional set of complement factor I and its soluble cofactors. Mol Immunol. 2008;45(1):169–179. doi: 10.1016/j.molimm.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 29.Rupp A-K, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, Moldenhauer G, Marmé F, Sültmann H, Altevogt P. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122(2):437–446. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 30.Khan S, Jutzy JMS, Valenzuela MMA, Turay D, Aspe JR, Ashok A, Mirshahidi S, Mercola D, Lilly MB, Wall NR. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure