Abstract
Delivery of plasmids for gene expression in vivo is an inefficient process that requires improvement and optimization to unlock the clinical potential of DNA vaccines. With ease of manufacture and biocompatibility in mind, we explored condensation of DNA in aqueous solution with a self-crosslinking, endosome-escaping lipopeptide (LP), stearoyl-Cys-His-His-Lys-Lys-Lys-amide (stearoyl-CH2K3), to produce cationic LP/DNA complexes. To test whether poly(ethylene glycol) (PEG)-ylation of these cationic complexes to neutralize the surface charge would improve the distribution, gene expression, and immune responses poly(ethylene glycol), these LP/DNA complexes were combined with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG2000). Fluorescence imaging illustrated that the cationic complexes exhibited the highest degree of localization and lowest degree of dispersion throughout the injected muscle, suggesting impaired mobility of cationic particles upon administration. Nanoluciferase reporter assays over a 90-day period demonstrated that gene expression levels in muscle were highest for PEGylated particles, with over a 200-fold higher level of expression than the cationic particles observed at 30 days. Humoral and cell-mediated immune responses were evaluated in vivo after injection of an ovalbumin expression plasmid. PEGylation improved both immune responses to the DNA complexes in mice. Overall, this suggests that PEGylation of cationic lipopeptide complexes can significantly improve both the transgene expression and immunogenicity of intramuscular DNA vaccines.
Keywords: DNA vaccine, PEGylation, lipopeptide, intramuscular, non-viral gene delivery, gene expression, humoral and cell-mediated immunity
Graphical Abstract

Introduction
The advancement of non-viral DNA vaccines has been hindered by poor immunological responses after intramuscular injection in vivo. Lack of efficacy may be related to low transfection efficiency in vivo, although the optimal mechanisms that lead to efficient antigen presentation by dendritic cells has not been established. Dendritic cells could take up DNA themselves, express protein, and subsequently process antigens for presentation to T cells. Alternatively, immune response may be dependent on dendritic cell sampling of protein expressed elsewhere (i.e., in muscle or other tissues). The relative importance of each mechanism is poorly understood and may be specific to each delivery system. The use of cationic materials to form complexes with plasmid DNA or RNA, to promote endocytosis into cells, is a common strategy for in vitro transfections.1, 2, 3, 4 However, a disparity exists between in vitro and in vivo findings because effective, commercialized in vitro transfection agents do not translate into effective in vivo transfection agents.
Because of its ease of administration, the intramuscular route of injection is commonly explored in the field of DNA vaccines. The extracellular matrix (ECM), however, is thought to serve as a major extracellular barrier to the delivery of intramuscular cationic DNA complexes because it consists of many negatively charged proteins or polysaccharides that may bind cationic complexes and restrict their mobility through the tissue.5, 6 Indeed, Ruponen et al.7 found that glycosaminoglycans, such as heparin sulfate and chondroitin sulfate, were able to completely block the transfection of various cationic DNA liposomes in cells. The restricted mobility of the DNA complexes in the tissue is thought to result in low transgene expression because it would restrict the number of cells the complex can interact with. Furthermore, the dissolved species in the extracellular environment may also create a hurdle in non-viral DNA delivery. Salt-induced aggregation of cationic complexes upon injection has been reported, occurring when the complexes are exposed to the isotonic environment in vivo, potentially further restricting their mobility.8, 9
A strategy to reduce unwanted electrostatic interactions has been to shield the surface charge of cationic complexes with poly(ethylene glycol) (PEG), increasing the stability and distribution of complexes in vivo. Typically, PEGylation has been used extensively to produce “stealth” particles to avoid recognition by the reticuloendothelial system (RES) and to reduce clearance times of drugs or DNA particles in vivo.8, 10, 11, 12, 13, 14, 15, 16 The use of PEGylated gene carriers, however, has also been shown to produce higher levels of transgene expression at the site of injection in comparison with cationic carriers.17, 18 Chang et al.17 attributed the high transfection efficiency of their non-ionic PEG-PLGA-PEG/DNA complexes in the skeletal muscle of rats to increased diffusivity. This was due to their reduced interactions with the ECM in comparison with cationic polyethyleneimine (PEI)/DNA complexes, which were found to be largely restricted to the site of injection. Both Palumbo et al.9 and van den Berg et al.19 have explored similar strategies for their intradermal DNA complexes, finding that PEGylation was able to increase the local tissue distribution and antigen expression of their complexes. Overall, this suggests that PEGylation may be increasing the transfection efficiency of DNA complexes in vivo by increasing the distribution of the complexes to a higher proportion of cells in the tissue. In the context of intramuscular DNA vaccination, however, little is known about the effectiveness of PEGylation in shielding cationic DNA complexes or whether this is a useful strategy to improve the efficacy of such non-viral DNA vaccines. In the present study, the use of two types of DNA complexes was compared in vivo: a cationic lipopeptide (LP)/DNA complex and a neutralized PEGylated LP/DNA complex.
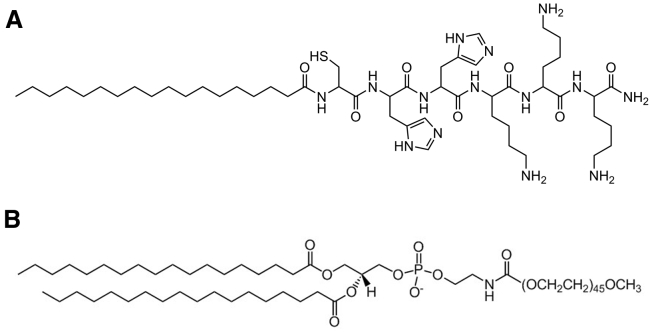
The cationic LP used in this study was the self-crosslinking stearoyl-Cys-His-His-Lys-Lys-Lys-amide (stearoyl-CH2K3), selected based on our previous knowledge and characterization of this class of LPs, carried out by Tarwadi et al.20 and Ho et al.21 (Figure 1A). These LPs offer the advantage of ease of manufacture because they can readily self-assemble with plasmid DNA in aqueous solution, circumventing the need for organic-based solvent preparations, limiting the chance of carry-over of the unwanted solvent. The lysine residues of the LP allow plasmid DNA condensation at physiological pH.22, 23, 24, 25 The inclusion of weakly alkaline (pKa = 6) histidine residues promotes endosomal escape of DNA in cells by taking advantage of the proton sponge effect.23, 26, 27, 28 The cysteine residue allows disulfide bonds to form between peptide molecules, stabilizing the DNA complexes by preventing rapid dissociation of the DNA and condensing agent in solution.29, 30, 31, 32 Last, conjugation of the stearoyl hydrophobic moiety assists with the stabilization of the DNA complexes in a particulate form in aqueous solutions.
Figure 1.
Structures of the LP and PEGylated Lipid Used to Form DNA Complexes
Stearoyl-CH2K3 lipopeptide as C-terminal amide (A) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG2000) (B).
The PEGylated LP/DNA complex was developed via addition of 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG2000) to the LP/DNA complex during self-assembly (Figure 1B). This contained an anionic moiety and a hydrophilic lipid to complex with the cationic LP to drive particle formation. To assist with understanding the effect of shielding the cationic surface charge of complexes in DNA vaccine delivery, the relationships between the distribution, transfection, and immunogenicity of the two DNA complexes were investigated. We hypothesized that PEGylation would increase the distribution, transgene expression, and immunogenicity of the cationic LP/DNA complexes in vivo.
Results
Formation, Characterization, and In Vitro Transfection of Self-Assembled LP/DNA Complexes
A dye exclusion assay of the LP/DNA complexes revealed a steep decline in fluorescence with increasing addition of the LP to DNA between (+/−) charge ratios of 1:1 to 2.5:1. Fluorescence reached a minimum at ratios greater than 2.5:1, indicating that no further condensation of the DNA occurred beyond this stage (Figure 2A). Substitution of the cysteine in stearoyl-CH2K3 with serine, a residue of similar polarity, resulted in a higher observed fluorescence intensity from a charge ratio of 2:1 onward (n = 3, p < 0.05) (Figure S1A). Similarly, substitution with the more non-polar alanine also resulted in higher fluorescence at these later charge ratios (n = 3, p < 0.05). The steep decline in fluorescence across low charge ratios, observed with stearoyl-CH2K3, was not observed with these two substituted LPs. Instead, a more gradual decrease in fluorescence was observed. This indicated that the cysteine residue played an important role in assisting with the condensation of plasmid DNA. Addition of the reducing agent DTT to the stearoyl-CH2K3 LP before DNA complexation significantly increased the degree of fluorescence observed for the LP/DNA particles at a charge ratio of 2.5:1 (n = 3, p < 0.05) (Figure S1B). This indicates that it is the formation of a disulfide bond between a pair of LPs that assisted with condensation of the DNA in the LP/DNA complexes.
Figure 2.
Characterization of the LP/DNA Complexes
(A) Dye exclusion profile of LP/DNA complexes prepared with stearoyl-CH2K3. All data points are calculated as the percentage of fluorescence intensity of plasmid DNA in solution. *p < 0.05 and ****p < 0.0001 for significant differences in fluorescence intensity compared with DNA alone (one-way ANOVA, Dunnett’s post hoc test). (B and C) Zeta potential (B) and mean particle size and polydispersity index (C) of stearoyl-CH2K3/DNA complexes over a range of charge ratios. As the (+/−) charge ratio of LP to DNA increased, the zeta potential increased, whereas the Z-average remained low (generally between 10–100 nm), with the exception of complexes formed at charge ratios close to unity. All data are presented as mean ± SEM of n = 3 separate experiments. (D and E) Cryo-TEM images of the LP/DNA complexes prepared at a charge ratio of (D) 1.5:1 and (E) 2.5:1, illustrating aggregate complexes (denoted by the large white circle) and smaller, ∼30-nm particles (denoted by the small white circle), respectively. Scale bars, 100 nm (D) and 50 nm (E).
Dynamic light scattering (DLS) and zeta potential (ZP) measurements revealed that a generally polydisperse (polydispersity index [PDI] > 0.25) population of large, negatively charged and/or neutral population of particulates formed with apparent diameters of > 1 μm when the charge was close to neutral (Figures 2B and 2C). Cryogenic transmission electron microscopy (cryo-TEM) imaging of the complexes formed at a charge ratio of 1.5:1 showed formation of non-spherical, irregularly shaped particles in the solution, comprising predominantly large aggregates of smaller particles (Figures 2D and 2E). Interestingly, the aggregates appeared to comprise particles approximately 30 nm in diameter. At higher charge ratios (>2:1), a narrower dispersion of small, positively charged DNA complexes formed, with ZPs of ≥ +30 mV and particle diameters of approximately 100 nm (PDI ≤ 0.25). Cryo-TEM imaging of the LP/DNA complexes formed at a charge ratio of 2.5:1 revealed a more uniform population containing a large proportion of smaller particles approximately 30 nm in diameter, with occasional clusters that were approximately 60–90 nm in size. These aggregates also appeared to be comprised of a few primary ∼30-nm particles.
From the above characterizations, in vitro transfection of cationic LP/DNA complexes at selected charge ratios of 2:1 and 2.5:1 were tested against the standard Lipofectamine LTX transfection reagent along with non-particulate and non-luciferase coding plasmid particulate controls (Figure S2). Because of the differences of the effective in vitro mechanism of DNA uptake versus that in vivo, the complexes were transfected in two different types of medium: DMEM, to encourage particle aggregation in vitro, and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-glucose buffer (HGB), a formulation shown to be stable for our particulate formulations (Figure S2A). Interestingly, we found that, in DMEM, the lipofectamine (LFN)/DNA control showed the highest transgene expression levels in vitro (n = 3, p < 0.01). In HGB, the LP/DNA complexes formed at a charge ratio of 2.5:1 showed the most significant level of transgene expression (n = 3, p < 0.001) (Figure S2B).
Because of their relatively small size, high cationic surface charge, and relatively high in vitro transfection capabilities, the LP/DNA complexes formed at a charge ratio (+/−) of 2.5:1 were thus selected for evaluation in vivo. These complexes had a mean diameter of 112.5 ± 13.9 nm and a ZP of +34.2 ± 1.2 mV. Long-term evaluation of the in vitro stability of these LP/DNA formulations showed that these complexes retained their particle size and PDI over at least a 7-day period (Figure S3).
Formation and Characterization of PEGylated LP/DNA Complexes
DPSE-PEG with a PEG chain of 2,000 Da was used to test the PEGylated LP/DNA complexes in our study because we have shown that PEG chains of 1,000 Da or 5,000 Da do not form small uniform particle populations (Figure S4). Incremental addition of negatively charged DSPE-PEG2000 to the LP/DNA particles lowered their positive ZP so that the resulting particles reached a ZP close to ∼0 mV at ratios 0.75:2:1 and 1:2:1 (Figure 3A). Addition of the PEGylated coating to the LP/DNA complexes resulted in particles with a slightly larger hydrodynamic diameter (Figure 3B), although their diameter was not statistically different from that of the cationic LP/DNA particles, according to Student’s t test.
Figure 3.
Characterization of the DSPE-PEG2000/LP/DNA Complexes
(A and B) PEGylated LP/DNA complexes prepared at various charge ratios and the resultant (A) zeta potential and (B) particle size and polydispersity index. A reduction in zeta potential was observed with addition of negatively charged DSPE-PEG2000 to the LP/DNA complex. Data are presented as mean ± SEM, n = 3. (C and D) Cryo-TEM images of the DSPE-PEG2000/LP/DNA complexes at (C) pre-ultrafiltration and (D) post-ultrafiltration (5,000 × g for ∼5.5 hr at 4°C) with Amicon Ultra-15 (NMWL of 3 kDa). Filled-in white arrowheads denote example DSPE-PEG2000/LP/DNA particles, and the white arrow outline denotes free uncondensed plasmid. Scale bars, 100 nm.
It is important to note that increasing the addition of the DSPE-PEG2000 lipid resulted in the formation of biomidal populations of particles. A small ∼20-nm-sized peak was observed by DLS analysis in addition to the typical 80–100 nm peak (Figure S5). Hence, care was taken to ensure that only the lowest ratio of DSPE-PEG2000 to LP/DNA was used to generate the PEGylated LP/DNA complexes to limit variability within the particle population.
It is important at this stage to reveal the main reasons behind subjecting both DNA complexes to ultrafiltration before in vivo injection. The first reason is to concentrate the particles into a manageable small volume for in vivo administration. The second reason is to ensure that a more monodisperse population is achieved and to reduce the level of free naked plasmid present in the injection mixture, particularly with the PEGylated complexes, because cryo-TEM imaging of this particle mixture pre-ultrafiltration revealed the presence of free uncondensed plasmid (Figure 3C). Imaging post-ultrafiltration of the DNA complexes, however, revealed the absence of the free plasmid in solution (Figure 3D). Analysis of particle diameter and PDI pre- and post-ultrafiltration revealed no significant differences, although it should be noted that the PEGylated complexes did appear to be slightly larger post-ultrafiltration (presumably because of centrifugal forces overcoming the steric forces induced by the PEG chains) (Figure S6). However, no aggregation or expansive increase in the variability of the population was observed. Similar to the cationic LP/DNA complexes, a long-term in vitro stability assay showed no difference in particle size of the PEGylated LP/DNA complexes over time in the HGB formulation (Figure S3).
DSPE-PEG2000/LP/DNA complexes formed at a charge ratio of 0.75:2:1 were thus selected to test their stability against salt-induced aggregation and were subsequently used in in vivo studies. These complexes had a mean diameter of 198.6 ± 49.5 nm and a ZP of +0.76 ± 0.2 mV.
PEGylation Confers Stabilization against Salt-Induced Aggregation
Incubation of the LP/DNA complexes in an isotonic buffer to mimic the in vivo environment induced aggregation of the complexes. DLS analysis indicated that the apparent particle diameter increased significantly by 15-fold (p < 0.01) (Figure 4A). PEGylation of the LP/DNA complexes conferred salt stability to the DNA complexes. Particle size by DLS was not significantly different after addition of NaCl.
Figure 4.
Particle Stability on Addition of NaCl and Relative Gene Expression in Muscle after Intramuscular Injection
(A) The mean particle size and polydispersity index of LP/DNA prepared at a charge ratio of 2.5:1 and DSPE-PEG2000/LP/DNA prepared at a charge ratio of 0.75:2:1 with and without addition of 0.15M NaCl. *p < 0.05 for the increase in Z-average in comparison with the untreated control (paired Student’s t test). Data are presented as mean ± SEM, n = 3. (B) Relative gene expression determined as relative light units (RLUs) in the calf muscle 1, 30, and 90 days after injection of DNA. Data are presented as mean ± SEM of n = 3–6 mice. **p < 0.01 and ****p < 0.0001 for significant expression levels between treatments as indicated (two-way ANOVA, Tukey’s post hoc test). Background RLUs determined in saline-treated controls were less than 1.
PEGylation Increases Transgene Expression of LP/DNA Complexes In Vivo
An assay of nanoluciferase expression revealed that administration of LP/DNA and PEGylated LP/DNA complexes resulted in significant gene expression in the calf muscle at all assayed time points over the course of the 3-month study (Figure 4B). The PEGylated LP/DNA complexes exhibited higher gene expression levels in the muscle than the LP/DNA complexes at all time points, with maximal expression observed 1 month after injection (n = 3 to 6, p < 0.0001). At this time point, gene expression resulting from injection of the PEGylated complexes was over 200-fold higher than that of its non-PEGylated counterpart. To assist with standardizing the role of PEGylation in the improvement of gene expression levels of the cationic complexes, administration of both particulate formulations was also compared with that of injection of naked DNA alone. The results showed that naked DNA produced the overall highest level of gene expression in the calf muscle (n = 3–6, p < 0.0001 compared with the LP/DNA and DSPE-PEG2000/LP/DNA) (Figure S7). Although transgene expression was detected in the draining popliteal lymph node of mice injected with any of the three formulations, there were no significant differences in expression levels across all time points assayed. It should be noted that, for each of these time points, there were 1 or more mice that did not show any detectable nanoluciferase activity in this lymph node after administration of either the LP/DNA, DSPE-PEG2000/LP/DNA complexes or naked DNA (thus, no statistical significance could be observed with this variable dataset).
Quantitation of DNA Complexes Retained in Muscle Post-injection
To determine whether the differences in transgene expression levels of each of the formulations were due to clearance of the DNA or DNA complexes from the muscle after injection, a qPCR assay was carried out to determine the absolute plasmid levels present in the entire muscle at 30 min. The results of this assay revealed that similar concentrations of the plasmid were present, with no significant differences observed between the injected formulations (Figure 5A). The mass of plasmid detected in the entire muscle after injection of each of the three formulations ranged between ∼1% and ∼5% of the dose administered.
Figure 5.
Local Distribution of DNA Complexes In Vivo
(A) Total target plasmid detected by qPCR in the injected calf muscle 30 min after injection of naked DNA, LP/DNA, or DSPE-PEG2000/LP/DNA. Data are presented as mean ± SEM of n = 4 mice. (B) Total DNA pixel count in the muscle of naked DNA-AF594-, LP/DNA-AF594-, or DSPE-PEG2000/LP/DNA-AF594-injected mice at 30 min. Data are presented as mean ± SEM of n = 3 mice, with a total of 12 muscle sections (10 μm, every 800 μm) collected from each mouse. *p < 0.05 for significant differences in pixel count (one-way ANOVA, with Tukey’s post hoc test). (C–N) Example sections of skeletal muscle imaged after injection of saline (C–E), naked DNA-AF594 (F–H), LP/DNA-AF594 (I–K), or DSPE-PEG2000/LP/DNA-AF594 (L–N). Autofluorescence from tissue to illustrate muscle morphology is shown in green and AF594-labeled plasmid in red. The images in (C)–(E) were captured with a 4× objective lens.
Localization of DNA Complexes in Tissue
In our fluorescence accumulation assay, we found that injection of the cationic LP/DNA-AF594* complexes yielded the highest total pixel count and injection of the naked DNA-AF594 yielded the lowest (Figure 5B) (*p < 0.05 compared with naked DNA-AF594, n = 3, one-way ANOVA, Tukey’s post hoc test). Representative images of each muscle after injection of saline, naked DNA-AF594, LP/DNA-AF594, or DSPE-PEG2000/LP/DNA-AF594 can be seen in Figures 5C–5N. Additionally, corresponding H&E staining showed no signs of muscle damage and/or necrosis in the tissue after administration of either DNA complexes (Figure S8).
Humoral Immune Responses to Injection of pCMV-OVA
Immunization with 50 μg DNA in the form of LP/DNA complexes using the selected prime-boost regimen did not elicit significant ovalbumin (OVA)-specific immunoglobulin levels in mice. All mice exhibited activity below the positive index at all time points assayed after administration of these cationic LP/DNA complexes (Figure 6A). On the other hand, infection of the DSPE-PEG2000/LP/DNA complexes resulted in 50% positive mice, with two strong responders of four detected between 4 and 13 weeks (although this response was not sufficient enough to elicit significance according to the Kruskal-Wallis test). Similarly, mice injected with naked DNA resulted in 50% positive mice, although it should be noted that only one of these positive mice expressed similarly high levels as the DSPE-PEG2000/LP/DNA-injected mice, whereas the other positive mouse elicited lower antibody levels slightly above the positive index threshold. As expected, the positive control of the OVA-coated splenocytes co-administered with lipopolysaccharide (LPS) induced a robust response in all mice (3 of 3 mice).
Figure 6.
Immunogenicity of DNA Complexes
(A) Ovalbumin-specific antibody activity levels in sera of mice (relative to the positive index) after administration of DNA complexes or control formulations. Symbols represent the individual data points, whereas the line represents the mean. The dotted line denotes the positive index of 1. Values higher than 1 are positive for anti-OVA activity; values less than 1 are negative for anti-OVA activity. (B) Percentage of target epitope-specific cells eliminated in mice immunized with OVA-coated splenocytes (positive control), saline (negative control), LP/DNA, or DSPE-PEG2000/LP/DNA. Data are presented as the mean ± SEM. **p < 0.01 and ****p < 0.0001 for significant activity levels compared with saline-treated mice (one-way ANOVA with Dunnett’s post hoc test). (C–E) Representative histograms showing the distinct OVA257–264 pulsed (high CFSE concentration) and unpulsed (low CFSE concentration) population of splenocytes detected in mice immunized with saline (C), LP/DNA (D), or DSPE-PEG2000/LP/DNA (E) formulations.
Cell-Mediated Immune Responses to Injection of pCMV-OVA
Flow cytometry showed that injection of the pCMV-OVA plasmid in its complexed form induced highly significant epitope-specific T cell elimination of the target OVA-pulsed population of splenocytes in comparison with the saline control (Figures 6B–6E). Immunization with the LP/DNA complex induced the lowest response, with 14.3% ± 1.6% of cells eliminated from all three formulations. The response was significantly higher than the saline control (n = 8, p < 0.01, via Dunnett’s post hoc test). Addition of the PEG layer boosted this response, with mice immunized with the DSPE-PEG2000/LP/DNA complexes showing a significantly higher percentage of cells eliminated compared with the LP/DNA complexes (26.7% ± 2.2%, n = 8, p < 0.05, via Tukey’s post hoc test). Immunization with naked DNA alone elicited similar significant levels of cytotoxicity to the PEGylated LP/DNA complexes, 22.5% ± 2.3% (n = 8, p < 0.0001) compared with the saline controls. As expected, the positive control injection of OVA-coated splenocytes and LPS induced the most robust response, with 57.2% ± 5% of cells eliminated (n = 8, p < 0.0001).
Discussion
Commercial translation of non-viral DNA vaccines has been limited by the difficulty of achieving high levels of transfection in vivo, leading to disappointing immune responses. Inadequate biological responses can be attributed to the difficulty of overcoming the extracellular and intracellular barriers to DNA delivery that are present in vivo. To overcome these barriers, more focus needs to be directed to investigating how modifications of gene delivery systems affect vaccine response in vivo. Particle formulations may be less efficient than naked DNA in terms of gene expression in muscle tissue, but they should not necessarily be discounted as vaccine delivery systems. Rather, nanoparticle formulations are better regarded as a different gene delivery system than naked DNA because the mechanism of uptake and rate of degradation of naked DNA in vivo differ greatly from DNA presented in particulate form.33, 34 The present study is a good illustration; addition of a PEGylated lipid layer significantly reduced the degree of tissue entrapment, increased transgene expression, and boosted the immunogenicity of cationic LP/DNA particles so that the vaccine responses were at least as good as the responses to naked DNA. To our knowledge, this is the first illustration of the effects of PEGylation on physiochemical particle properties, degree of localization, tissue distribution, transgene expression, and immunogenicity of an intramuscular non-viral gene delivery system.
Ease of manufacture is an important factor when selecting a DNA delivery system. In this study, we deliberately designed agents for complexation that are soluble in water but could be assembled to form neutral or negatively charged particles. Thus, the use of alcoholic solutions was avoided. Assembly of particles was first investigated by forming complexes with LP and DNA and later including PE-PEG. ZP measurements of the LP/DNA complexes revealed that the particle charge was close to neutral when formed between (+/−) charge ratios of 1:1 and 1.5:1. DLS and cryo-TEM imaging of particles illustrated that large aggregates were formed at this stage because of the lack of charge repulsion between primary particles when the particles were assembled. In contrast, LP/DNA complexes prepared at a (+/−) charge ratio greater than 2.5:1, where the cationic LP was in excess, had ZPs above +30 mV. At these charge ratios, DLS showed that the particles were significantly smaller than under neutral charge conditions, supporting the well-known concept that high charge-to-charge repulsion limits aggregation during manufacture.35, 36, 37, 38 When DSPE-PEG2000 was included in appropriate proportions, we were able to produce neutral particles without extensive aggregation and chose to work with complexes of DSPE-PEG2000/LP/DNA produced at a charge ratio of 0.75:2:1. At this ratio, DSPE-PEG2000 was found to effectively neutralize the strong cationic surface charge of the LP/DNA complexes, whereas the particle diameters remained relatively low (∼200 nm). It is reasonable to assume that the DSPE-PEG2000 was able to provide steric stabilization of the LP/DNA complexes.
In support of the hypothesis that PEGylation may be an effective strategy for the intramuscular delivery of cationic complexes in vivo, our PEGylated complexes were able to induce higher transgene expression levels in the muscle than the cationic LP/DNA complexes. We observed that intramuscular administration of naked DNA was able to induce higher levels of transgene expression in muscle than particulate formulations, but we observed that higher gene expression did not translate into greater immune responses to the model antigen, OVA. The extent of enzymatic degradation and the mechanism of uptake of naked DNA in vivo is expected to differ significantly from DNA nanoparticles; hence, the discussion here is focused on comparison of the cationic DNA complexes and the neutral PEGylated DNA complexes. We hypothesized that the higher transfection efficiency of the PEGylated complexes, compared with their non-PEGylated counterparts, may have been due to widespread distribution of the PEGylated complexes throughout the muscle tissue, allowing them to come into contact with a larger number of cells. In contrast, the cationic complexes, which were likely to have been restricted by salt-induced aggregation and strong charge-charge interactions between particles and the ECM, were expected to remain in the vicinity of the site of injection, hence coming into contact with a limited number of cells. To test this hypothesis, imaging studies were carried out to investigate the degree of accumulation or localization of fluorescently labeled plasmid, examined 30 min after injection of each formulation into the muscle. Lateral sections throughout each muscle were collected for imaging, and 10-μm sections were examined every 800 μm throughout the tissue. A total pixel count of the fluorescently labeled plasmid was determined for each section.
Rather than focusing on the extent of dispersion of the formulation in muscle, the purpose of using fluorescently tagged DNA in our study was to look at the extent of localization of DNA close to the site of injection. Fluorescent imaging relies on detection of the given fluorophore above its effective limit of detection; i.e., above the auto-fluorescence observed in tissue sections of control animals that received saline injections. The practical problem faced when attempting to study dispersion of DNA is that a low level of fluorescence could reflect either absence of plasmid DNA-AF594/DNA-AF594 complexes in that given area of tissue or a high degree of dispersion of the plasmid DNA-AF594/DNA-AF594 complexes throughout the section so that the fluorescence is below the limit of detection or cannot be distinguished from tissue auto-fluorescence. This prevents the establishment of a mass balance and prevents comparisons of dispersion of particulate formulations and naked DNA. The fluorescence microscopy technique did allow comparison of the two particulate formulations to determine which formulation was observed to accumulate in a given region of tissue (i.e., the total pixel count in each section represented the accumulation of DNA in that particular region). Hence, a low total pixel count corresponded to a low level of DNA accumulated in each specific region of muscle tissue. In contrast to the fluorescence microscopy data, the qPCR data were able to confirm that similar total masses of DNA were present in the muscle at the time of sectioning, validating the use of fluorescence microscopy to investigate immobilization or accumulation of DNA in specific locations. It is important to note that this method refers to total pixel counts of muscle sections imaged only at low magnification (using a 4× objective) to gauge the accumulation throughout the entire muscle sections rather than at higher magnifications, when low signals can be detected. It would be impractical to analyze accumulation and/or localization throughout the entire muscle section at high magnification.
Overall, the localization studies illustrated that the LP/DNA complexes were associated with the highest total DNA pixel count, suggesting that the strong cationic surface charge and/or the salt-induced aggregation limited the mobility of these LP/DNA complexes in the ECM. It is also important to note that the LP/DNA complexes showed a higher variability in the level of retention of the fluorescent plasmid in the muscle, with the levels spanning 3 orders of magnitude, whereas the detected levels of PEGylated complexes spanned only 2 orders of magnitude, reflected by the coefficient variation of the two formulations. This high variability may be due to the uncontrollable size and nature of salt-induced aggregation of the LP/DNA complexes upon contact with biological fluid, generating variable degrees of mobility in vivo. DLS sizing confirmed the ability of the PEGylated particles to confer stability against this salt-induced aggregation, suggesting that PEGylation may reduce the overall variability of distribution of the complexes after administration in vivo.
Although PEGylated complexes have been tested in gene therapy studies,10, 18, 39, 40 the mechanism by which PEGylated complexes are taken up and processed by cells in vivo remains largely unknown. High-magnification imaging of our murine muscle tissue after injection of the DSPE-PEG2000/LP/DNA-AF594 complexes confirms that DNA-AF594 can be found in the nuclei of the myofibers (Figure S9). Given that our DSPE-PEG2000/LP/DNA complexes did show transfection activity in the injected muscle and DNA-AF594 nuclear uptake, it is likely that the complexes were taken up directly by cells, either by endocytosis or passive diffusion through transiently permeabilized plasma membranes. Injection of formulations into muscle could potentially cause mechanical stresses that cause temporary access across plasma membranes. Indeed, it has been known for decades that injection of naked DNA expression vectors in solution into muscle leads to gene expression in muscle, but the mechanisms of entry into the nucleus are not adequately understood. Another possible mechanism of uptake of the DSPE-PEG2000/LP/DNA complexes, often neglected in this field, is the possible shedding of the PEG lipid coating from the LP/DNA complex, leaving the cationic complex to be taken up into cells by adsorptive endocytosis. This mechanism is a possibility because the DSPE-PEG2000 used to assemble the DSPE-PEG2000/LP/DNA complexes was not covalently attached to either the DNA or cationic lipopeptide and may diffuse from the LP/DNA complex while circulating in vivo. Evidence for the spontaneous diffusion of various PEG-lipid conjugates from the membranes of liposomes has been illustrated in in vitro binding assays.41, 42 Some research groups have taken advantage of this, utilizing the PEG-lipids as “sheddable” coatings for the delivery of liposome-encapsulated plasmids or drugs to target tumors in vivo.43, 44 Indeed, Ambegia et al.44 found that stabilized DOPE/DODAC/DNA particles containing a sheddable PEG-diacylglycerol coating were able to extend circulation lifetimes and increase tumor-selective gene expression in vivo. Attempts in our study to investigate the degree of shedding of the DSPE-PEG2000 lipid tagged with the fluorescein isothiocyanate (FITC) fluorophore and used in conjunction with LP/DNA-AF594 proved difficult because we were unable to form uniform populations with DSPE-PEG2000-FITC/LP/DNA-AF594.
Previous studies have proposed three mechanisms by which cytotoxic T cells could be activated by DNA vaccines: transfection of the muscle may directly stimulate CD8+ T cells via presentation of major histocompatibility complex (MHC) class I molecules;45 Antigen-presenting cells (APCs) are directly transfected and rapidly migrate to draining lymph nodes for immune activation;46, 47 and cross-presentation of dendritic cells (DCs) via phagocytosis of transfected apoptotic or necrotic cell bodies.48, 49, 50 The in vivo cytotoxic T lymphocyte (CTL) assay utilized in our study measures epitope-specific elimination of target cells via activation of cytotoxic T lymphocytes. The assay suggested that the DSPE-PEG2000/LP/DNA particles induced a greater cellular response than the LP/DNA particles, reflecting the gene expression levels induced in the muscle. The antibody response assay indicated that the LP/DNA complex was not capable of eliciting significant levels of antibody activity in any of the assayed mice. Although not statistically significant in comparison with LP/DNA-immunized mice, it is evident that the DSPE-PEG2000/LP/DNA-immunized group contained a larger number of positive responses than the LP/DNA group. It is important to note that the C57BL/6J mouse strain used in our study is typically Th1-dominant; thus, a stronger bias toward cell-mediated immunity was expected.51, 52 Overall, our results suggest that PEGylation increases the transgene expression and immunogenicity of the cationic DNA complex and reduces the degree of tissue localization and variability of the intramuscular accumulation of the complexes.
van den Berg et al.19 tested the use of near-neutral PEGylated DOTAP lipoplexes and poly(amido amine) (PAA) polyplexes as vaccines in vivo and compared the effect with their non-PEGylated counterparts. Although they did not explore the distribution of these complexes, these authors found that the PEGylated nanoparticles induced significantly larger humoral and cellular responses than the cationic lipo- and poly-plexes. However, these and our results differ from the findings of Carstens et al.,53 who tested the effect of addition of DSPE-PEG2000 to cationic egg PC/DOPE/DOTAP DNA-encapsulated liposomes as vaccines in vivo. These authors found that PEGylation lowered the accumulation of liposomes at the site of injection and increased drainage to the lymph node and spleen but did not improve the humoral or cellular responses observed in mice. In our study, to assess the role of lymphatic drainage, the draining popliteal lymph node was also assayed for transgene expression. None of the formulations resulted in significant levels of expression compared with the saline control. From the tissue-calibrated qPCR assay carried out previously in our laboratories, intact plasmid was detected in the lymph node after administration of both complexes, at similarly low levels (<0.05% of the administered dose).21 A reason for these discordant results in our and published studies, aside from the different routes of administration used, may be the considerable difference in particle surface charge of the PEGylated liposomes used by Carstens et al.53 and our PEGylated complexes. The liposomes used in the former study were only partially shielded by the PEG layer (ZP of +28 mV), whereas our complexes were formulated to neutralize a positive charge. Taken together, the three studies suggest that formation of particles with a near-neutral ZP may be necessary to improve immune responses by PEGylation.
Additionally, the tissue-calibrated qPCR assay also showed no significant differences in the distribution of LP/DNA and DSPE-PEG2000/LP/DNA to the spleen or liver.21 Because a similar tissue distribution profile is observed post-administration of the two DNA complexes at both 30 min and 24 hr, it is possible that the significant differences in transgene expression and immunogenicity exhibited between the two particulates are due to the subtle differences in the degree of localization or mobility within the injected muscle. It should be noted that, 30 min after administration of all three formulations, it was evident that <5% of the administered dose of DNA or DNA complexes was retained as functional plasmid in the muscle (by qPCR). This indicates rapid clearance of all three formulations from the injected muscle. This is not surprising, considering that we observed that the injection of a 50-μL volume of fluid (which is the typical injection volume used in the calf muscles or the smaller tibialis anterior muscle in murine studies),54, 55, 56, 57, results in the immediate swelling of the tissue volume. The swelling reduces within 30 min of administration, presumably because of the excess fluid draining from the tissue. Interestingly, the level of DNA/DNA complexes detected in the injected muscle 30 min after administration in our study does appear to be lower than previous reports measuring naked DNA or other non-viral gene delivery complexes at their injected site.53, 58 An important consideration is that our tissue-calibrated qPCR assay measures absolute levels of intact plasmid in tissue. This is unlike other studies relying on radiolabeling or fluorescent labeling approaches to quantify their DNA/DNA complexes, when a signal from an intact labeled plasmid cannot be differentiated from a cleaved/degraded product. Hence, other gene delivery studies utilizing labeling approaches may have overestimated the mass of intact DNA present in the injected tissue.
In summary, we have shown that shielding the surface charge of cationic LP/DNA complexes via PEGylation can lead to reduced localization to the site of injection post-administration, reduced variability in intramuscular accumulation, increased transgene expression levels, and immunogenicity of the vaccine in vivo. Particle sizing and imaging experiments suggest that this was likely the result of decreasing non-specific electrostatic interactions of the cationic complex with extracellular components upon injection and the prevention of salt-induced aggregation, which, in turn, likely increased the complex’s distribution in vivo to a higher number of cells for uptake. This indicates that PEGylation is a promising approach to increase the efficacy of non-viral cationic DNA complexes for gene delivery, a novel finding in the context of intramuscular DNA vaccine delivery, which can assist with re-strategizing future non-viral intramuscular gene delivery formulations. DSPE-PEG2000/LP/DNA particles can be assembled in aqueous solution without the use of organic solvents, which, we suggest, offers a considerable manufacturing advantage over alternative approaches to formulation of DNA particles for vaccination.
Materials and Methods
Materials
The stearoyl-CH2K3 LP as C-terminal amide was custom-manufactured by G.L. Biochem (Shanghai, China), whereas DSPE-PEG2000 was obtained from Avanti Polar Lipids (Alabaster, AL). All materials were dissolved for use in HGB (15 mM HEPES and 5% w/v glucose [pH 7.4]). The plasmid DNA used in the fluorescence distribution studies was pcDNA3.1HygrolacZ (Thermo Fisher Scientific, Carlsbad, CA) containing the β-galactosidase expression cassette regulated by the cytomegalovirus promoter. For tissue-calibrated qPCR studies, we used pCMV-luc, constructed by ligating the firefly luciferase cDNA from pGL2 basic (Promega, Fitchburg, WI) into the multiple cloning site of pcDNA3 (Thermo Fisher Scientific, Carlsbad, CA). For in vivo gene expression studies, we used pNL1.1.CMV (Promega, Fitchburg, WI), which contains the nanoluciferase gene under control of the cytomegalovirus promoter. The plasmid used for immunogenicity studies was pCMV-OVA, which expresses a secreted form of OVA, provided by Dr. Andrew Lew from the Walter and Eliza Hall Institute (WEHI) in Melbourne, Australia.59 The OVA protein and OVA257–264 synthetic peptide (SIINFEKL) used in these studies were purchased from Sigma-Aldrich (St. Louis, MO). LPS, used as an immunostimulatory adjuvant60 co-injected with the OVA-positive control, was also purchased from Sigma-Aldrich (St. Louis, MO). Carboxyfluorescein succinimidyl ester (CFSE) dye was obtained from Thermo Fisher Scientific (Carlsbad, CA).
Mice
The animals used were C57BL/6J male mice aged between 8–12 weeks. After experimentation, mice were euthanized with gaseous CO2. All animal procedures were carried out under approval of ethics protocols MIPS.2011.20 and MIPS.2013.16, granted by the Monash Institute of Pharmaceutical Sciences Animal Ethics Committee. Injections were carried out intramuscularly into the left calf muscle, using a 50-μg dose of complexed plasmid DNA in a 50-μL volume, unless otherwise stated. All mice were anesthetized with 1%–4% titrated isoflurane gas prior to intramuscular injection.
Calculation of the Molar Charge Ratio
In the context of the LP/DNA complexes, the charge ratio refers to the number of positively charged amine groups (NH3+) provided by the cationic transfection agent per negatively charged phosphate group (PO4-−) of DNA (where 1 phosphate group = 1 nucleotide). For example, to obtain a theoretical charge ratio of 1:1 (LP:DNA) using stearoyl-CH2K3 (1,045.45 g/mol, 3 NH3+/molecule), 1 μg of plasmid DNA (3 nmol) is mixed with 1.06 μg of stearoyl-CH2K3 (1 nmol). An average mass of 330 g/mol per nucleotide of DNA was used for the calculations. In the context of the DSPE-PEG2000/LP/DNA complexes, the charge ratio refers to the number of negatively charged phosphate groups provided by the DSPE-PEG2000 per positively charged amine of the LP per phosphate group of the DNA, respectively. For example, to obtain a theoretical charge ratio of 1:1:1 (DSPE-PEG2000:LP:DNA) with DSPE-PEG2000 (2,805.54 g/mol, PO4−/molecule) and stearoyl-CH2K3, 1 μg of plasmid DNA (3 nmol) is mixed with 1.06 μg of stearoyl-CH2K3 (1 nmol) and 8.5 μg of DSPE-PEG2000 (3 nmol).
Preparation of DNA Complexes
LP/DNA complexes were prepared by mixing the LP and DNA over a range of (+/−) charge ratios from 0.5:1 to 5:1 (LP:DNA). DSPE-PEG2000/LP/DNA complexes were prepared by mixing DSPE-PEG2000, LP, and DNA over a range of charge ratios, from 0.5:2.5:1 to 5:2.5:1 (DSPE-PEG2000:LP:DNA). DNA complex mixtures were then incubated at room temperature for 30 min prior to ultrafiltration. To concentrate the DNA complexes for in vivo injections, ultrafiltration (5,000 g for ∼5.5 hr at 4°C) was carried out with an Amicon Ultra-15 centrifugal unit (Merck Millipore, Billerica, MA) with a nominal molecular weight limit (NMWL) of 3 kDa. Characterization and quantitation of the DNA complexes after ultrafiltration are described in Figure S5.
Particle Characterization
Dye Exclusion Assay
To determine the charge ratio at which the LP effectively condenses the DNA, a dye exclusion assay was carried out. In a 96-well black-bottomed plate, plasmid DNA (20 μg/mL) was prepared in 12 mM Tris-HCl (pH 7.4) buffer. To each well containing DNA, 0.05 μL of 10,000× concentrated stock of intercalating SYBR Gold (Thermo Fisher Scientific, Carlsbad, CA) was added, and the plate was incubated at room temperature for 15 min. The LP was then added to the wells at a series of charge ratios from 0.5:1 to 5:1. A well containing 20 μg/mL of naked DNA and SYBR Gold was used as a 100% fluorescence intensity standard (i.e., no LP added). A blank well containing only the SYBR Gold in buffer was used to subtract from the fluorescent readings of each sample. The plate was incubated at room temperature for 30 min for particle formation. The fluorescence (excitation, 492 nm; emission, 540 nm) of each sample was measured using a Wallac Envision 2102 Multilabel Reader (PerkinElmer, Waltham, MA).
Particle Sizing and ZP
The mean particle size (Z-average) and the ZP of each complex were measured via DLS using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). For particle sizing, measurements were carried out at 25°C, with each measurement consisting of 10 runs with a duration of 10 s each. The PDI of the size of the samples was recorded for each measurement. To ensure that the instrument was calibrated, a 60 nm ± 2.7 nm size standard (Malvern Instruments, Malvern, UK) was used prior to experimentation. For ZP measurements, to ensure that the instrument was calibrated, a −68 mV ± 6.8 mV ZP standard (Malvern Instruments, Malvern, UK) was analyzed prior to experimentation. All measurements were carried out at 25°C. All samples were prepared in triplicate.
Cryo-TEM
Cryo-TEM was carried out to image the DNA complexes. The complexes were prepared as stated above, and 5 μL of each sample was deposited onto a carbon-coated copper 300 mesh grid with holes. The grid was then rapidly frozen in a liquid ethane bath cooled with liquid nitrogen. The temperature of each sample was maintained in a cryo-holder until time of imaging. For imaging, a FEI Tecnai F30 transmission electron microscope (University of Melbourne, Melbourne, Australia) was used.
Salt Stability Assay
To test the stability of DNA complexes against salt-induced aggregation, particle sizing was carried out on the particles before and after addition of NaCl. Briefly, DNA complexes were prepared in HGB as outlined previously, and DLS was used to measure particle size. NaCl solution was then added to the particles to a final concentration of 0.15 M. The particles were incubated for 30 min before DLS measurements were carried out again.
Transgene Expression
The luciferase assay was carried out to assay the reporter nanoluciferase protein. 1, 30, and 90 days post-injection, the calf muscles of each mouse were dissected and rapidly frozen in liquid nitrogen. The muscle was then homogenized in buffer (50 mM potassium phosphate, 1 mM DTT, 1 mM EDTA, and 10% glycerol) using a rotor/stator-type tissue homogenizer (TissueTearor, Biospec, Bartlesville, OK). Quantification of the nanoluciferase reporter protein was carried out using the commercial Nano-Glo luciferase assay system with Glo Lysis Buffer (both from Promega, Fitchburg, WI), in accordance with the manufacturer’s instructions. Luminescence was measured using aLUMIStar Omega instrument (BMG Labtech, Ortenberg, Germany). To allow comparison between samples, a Bradford assay was employed to determine the total protein in the tissue lysates for the standardization of expression levels. The assay was carried out using the Quick Start Bradford assay (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions, using BSA as the protein standard. The resultant absorbance was measured using an EnVision Multilabel Reader (PerkinElmer, Waltham, MA) at a wavelength of 595 nm.
Tissue-Calibrated qPCR
Tissue-calibrated qPCR was carried out according to the procedure outlined by Ho et al.21 Briefly, mice received an intramuscular injection of the DNA complexes using pCMV-luc. Thirty minutes after injection, tissues were harvested and homogenized in DNAzol reagent (Thermo Fisher Scientific, Carlsbad, CA) using a handheld TissueTearor homogenizer (Biospec Products, Bartlesville, OK) or with a Pellet Pestles motor grinder (Sigma-Aldrich, St. Louis, MO) and incubated overnight with proteinase K (QIAGEN, Venlo, Netherlands) (100 μg/mL) at room temperature. Tissue extracts were then centrifuged for 10 min at 10,000 × g at 4°C, and total DNA was extracted from the supernatant using a Wizard DNA Prep Spin column (Promega, Fitchburg, WI) following the manufacturer’s instructions. Real-time qPCR was carried out with a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA). Forward (5′-CCTCATAAAGGCCAAGAAGG-3′) and reverse (5′-ACACCGGCCTTATTCCAAG-3′) primers were used to amplify a 114-bp fragment of pCMVluc. Each PCR reaction consisted of 5 μL of iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), 200 nmol/L of each primer, 1 ng of template DNA, and water to a total reaction volume of 10 μL. Reactions were carried out with an initial incubation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 47°C for 30 s, and extension at 72°C for 30 s. All reactions were carried out in triplicate, and “no template” controls, containing water instead of template DNA, were included in every PCR run. Absolute quantitation of the plasmid present in the tissue was carried out using standard curves constructed for each tissue assayed. Standard curves were prepared by addition of serially diluted plasmid standards to homogenates of each pre-weighed, excised tissue of interest.
Tracking Localization of DNA complexes in Tissue
To track the DNA complexes after administration, pcDNA3.1HygrolacZ was covalently labeled with fluorescent nucleotide ChromaTide Alexa Flour 594-5-dUTP (Thermo Fisher Scientific, Carlsbad, CA). Briefly, plasmid DNA was digested with the BglII restriction enzyme (New England Biolabs, Ipswich, MA) (2 U per 1 μg of DNA) and incubated overnight at 37°C to yield a linear plasmid with 5′ GATC overhangs. The plasmid was purified via phenol-chloroform extraction, followed by ethanol precipitation. The Klenow fragment (New England Biolabs, Ipswich, MA) was then added to the linearized plasmid in addition to 60 nM of each dNTP, substituting deoxyuridine triphosphate (dUTP) with the fluorescent Alexa Flour 594-5-dUTP, and the mixture was incubated at room temperature for 4 hr. The reaction was terminated by adding EDTA to a final concentration of 10 mM. Phenol-chloroform extraction and ethanol precipitation was carried out to purify the resultant labeled plasmid (DNA-AF594). To minimize the presence of any free unincorporated nucleotides in the solution, the DNA pellet was washed three times with 70% ethanol prior to DNA resuspension. Mice received an intramuscular injection of either of the DNA-AF594 complexes at a dose of 50 μg in 50 μL. The mice were then euthanized 30 min after injection. The muscle was dissected, fixed in 4% paraformaldehyde (PFA) and frozen with dry ice in Tissue-Tek optimum cutting temperature (OCT) Compound (Sakura Finetek USA, Torrance, CA). Cryosectioning was carried out at −20°C. 10-μm sections were collected for every 800 μm of tissue. Tissue sections were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and imaged using a Nikon A1R confocal microscope (Nikon, Minato, Japan). A total pixel count of DNA-AF549 was carried out for each image using ImageJ software (NIH), with a set threshold level of 60, as determined by imaging the vehicle-treated muscle, and the extent of localization of the fluorescent DNA in each of the images was classified as associated with either epimysium, perimysium, or endomysium. The classifications were as follows: +++ indicated a high degree of association, ++ indicated a medium degree of association, + indicated a low degree of association, and – indicated that no detectable levels of the labeled DNA were associated with this region. A value was then assigned to each category, with +++ equal to 3, ++ equal to 2, + equal to 1, and − equal to 0. The sum of these numbers (i.e., weighted pixel count) for each extracellular region was determined for each muscle, and the mean was plotted graphically.
In Vivo Cytotoxic T Lymphocyte Elimination Assay
To evaluate the overall cellular response elicited in mice injected with our DNA complexes, an in vivo CTL activity assay was carried out according to the procedure outlined in White et al.61
Mouse Injection and In Vivo Assay Schedule
On day 1, mice were primed with the DNA complexes or control formulations. LP/DNA and DSPE-PEG2000/LP/DNA complexes were prepared as described previously using the pCMV-OVA plasmid. The positive control formulation used in this experiment was an injection of 2 × 107 OVA-coated splenocytes with 1 μg of LPS. The OVA-coated splenocytes were prepared by incubating a single-cell suspension of isolated splenocytes (from naive C57BL/6J mice) with OVA. Splenocytes were suspended in red blood cell lysis buffer and incubated at 37°C for 2 min, washed, and passed through a 70-μm cell strainer to filter off any cell debris. Cells were then incubated with OVA (10 mg/mL) at 37°C for 10 min, washed, and then resuspended in PBS. On day 6 post-immunization, mice were injected intravenously with target splenocytes (preparation is in Preparation of Target Splenocytes). The following day, splenocytes were harvested from the mice and analyzed via flow cytometry (preparation is outlined in Flow Cytometry). The percentage of target OVA-pulsed splenocytes eliminated was then calculated for each mouse.
Preparation of Target Splenocytes
Target splenocytes were prepared by depleting a single-cell suspension of isolated splenocytes of red blood cells and dividing the population of cells equally into two tubes. To one tube, OVA257–264 peptide was added to a concentration of 1 μg/mL. Both tubes were then incubated at 37°C for 1 hr, washed, and resuspended in 107 cells/mL in PBS and 1% fetal bovine serum (FBS). To the OVA257–264 peptide-pulsed tube of splenocytes, 0.5 μL of 10 mM CFSE was added per 1 mL of cells (i.e., high CFSE concentration). To the unpulsed population of splenocytes, 0.5 μL of 1 mM CFSE was added per 1 mL of cells (i.e., low CFSE concentration). The two tubes were then incubated at 37°C for 10 min, washed, and then resuspended in PBS. The two populations of cells were then mixed at a 1:1 ratio (high CFSE:low CFSE populations), and each mouse received an intravenous injection of 2 × 107 mixed cells in 200 μL of PBS.
Flow Cytometry
Flow cytometry was carried out using the BD FACSCanto II (BD Biosciences, San Jose, CA) to identify the ratio of OVA-pulsed (high CFSE-labeled) population of splenocytes to unpulsed (low CFSE-labeled) splenocytes. For each sample, 2 million events were recorded. The percentage of cells eliminated from OVA-specific T cell activation in each mouse was then calculated using the following formula:
Antibody Response Assay
Mice were given an initial administration (prime) of either the LP/DNA or DSPE-PEG2000/LP/DNA complex (test formulations), OVA protein (positive control), or saline (negative control) on day 1 using OVA-expressing pCMV-OVA as the plasmid DNA. A booster injection was then administered 4 weeks later. Administration of the DNA complexes and saline were carried out intramuscularly into the calf muscles of mice using a dose of 50 μg in 50 μL. Administration of the OVA-positive control (50 μg OVA protein + 1 μg LPS) was carried out intramuscularly in a 50 μL injection volume. Blood (∼100 μL) was collected from mice via sub-mandibular bleeding of the mice using a 5-mm lancet on day 1 (prior to prime injection), at 4 weeks (prior to boost injection), and then 9, 11, and 13 weeks after the initial prime. Blood samples were then incubated at 37°C for 90 min and then stored at 4°C for ∼18 hr following the procedure outlined by White et al.61 Serum was then collected by centrifugation (10,000 × g for 15 min at 4°C) and stored at −20°C until further use. For quantitation of anti-OVA immunoglobulins (immunoglobulin G [IgG]-, IgM-, and IgA-class antibodies) in serum, the mouse anti-OVA Ig ELISA kit (Alpha Diagnostic International, San Antonio, TX) was used. The assay was carried out in accordance with the manufacturer’s instructions. The absorbance at 450 nm was measured with the Envison Wallac Plate Reader (PerkinElmer, Waltham, MA). Data were expressed in relation to the values of the positive index (i.e., mean + 2 SD of absorbance values of control pre-immunized mice). The quantitation of the anti-OVA activity level was calculated by the following equation, with values above 1.0 indicating positive antibody activity in mice, and values below 1.0 indicating negative antibody activity:
Statistical Analysis
For in vitro characterization of complexes, the data in graphs are represented as mean ± SEM, with each data point representing n = 3 samples, unless otherwise stated. For all in vivo experiments, the data in graphs are represented as mean ± SEM with each data point representing n = 6–8 samples, unless otherwise stated. A paired Student’s t test was used to determine the significant differences in the Z-average or PDI of the DNA complexes after addition of NaCl and too look at the differences in plasmid DNA retention levels in the muscle. Two-way ANOVA with Dunnett’s post hoc test was used to assess the statistical significance of the transgene expression levels of the DNA complexes in the nanoluciferase assay over time. Two-way ANOVA with Tukey’s post hoc test was used to assess the statistical significance of the differences in the weighted total pixel count of the fluorescent plasmid between the formulations and their extent of localization in the extracellular regions of the muscle. Kruskal-Wallis test, along with Dunn’s post hoc test, was used to compare significant differences in the number of anti-OVA-positive mice between each group. One-way ANOVA with Dunnett’s post hoc test was used to evaluate statistical significance in epitope-specific T lymphocyte activity in mice in comparison with saline. One-way ANOVA with Tukey’s post hoc test was used to evaluate the statistical significance of epitope-specific T lymphocyte activity in mice between groups and the differences in fluorescence intensities in the dye exclusion assay. For all tests, a p value of less than 0.05 was considered to be statistically significant.
Author Contributions
J.K.H. designed the experiments, carried out the experimental work, and wrote the paper. P.J.W. designed the experiments and supervised the work. C.W.P. conceived the work, designed the experiments, and wrote the paper.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We acknowledge the infrastructure provided by the Faculty of Pharmacy and Pharmaceutical Sciences (Monash University). J.K.H. acknowledges the receipt of a Ph.D. scholarship (Australian Postgraduate Award).
Footnotes
Supplemental Information includes nine figures can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.025.
Supplemental Information
References
- 1.Jeffs L.B., Palmer L.R., Ambegia E.G., Giesbrecht C., Ewanick S., MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- 2.Le Bihan O., Chèvre R., Mornet S., Garnier B., Pitard B., Lambert O. Probing the in vitro mechanism of action of cationic lipid/DNA lipoplexes at a nanometric scale. Nucleic Acids Res. 2011;39:1595–1609. doi: 10.1093/nar/gkq921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mochizuki S., Kanegae N., Nishina K., Kamikawa Y., Koiwai K., Masunaga H., Sakurai K. The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta. 2013;1828:412–418. doi: 10.1016/j.bbamem.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 5.Eggen K.H., Malmstrøm A., Kolset S.O. Decorin and a large dermatan sulfate proteoglycan in bovine striated muscle. Biochim. Biophys. Acta. 1994;1204:287–297. doi: 10.1016/0167-4838(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 6.Nakano T., Sunwoo H.H., Li X., Price M.A., Sim J.S. Study of Sulfated Glycosaminoglycans from Porcine Skeletal Muscle Epimysium Including Analysis of Iduronosyl and Glucuronosyl Residues in Galactosaminoglycan Fractions. J. Agric. Food Chem. 1996;44:1424–1434. [Google Scholar]
- 7.Ruponen M., Ylä-Herttuala S., Urtti A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. Biochim. Biophys. Acta. 1999;1415:331–341. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]
- 8.Finsinger D., Remy J.S., Erbacher P., Koch C., Plank C. Protective copolymers for nonviral gene vectors: synthesis, vector characterization and application in gene delivery. Gene Ther. 2000;7:1183–1192. doi: 10.1038/sj.gt.3301227. [DOI] [PubMed] [Google Scholar]
- 9.Palumbo R.N., Zhong X., Panus D., Han W., Ji W., Wang C. Transgene expression and local tissue distribution of naked and polymer-condensed plasmid DNA after intradermal administration in mice. J. Control. Release. 2012;159:232–239. doi: 10.1016/j.jconrel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi Y.H., Liu F., Kim J.-S., Choi Y.K., Park J.S., Kim S.W. Polyethylene glycol-grafted poly-L-lysine as polymeric gene carrier. J. Control. Release. 1998;54:39–48. doi: 10.1016/s0168-3659(97)00174-0. [DOI] [PubMed] [Google Scholar]
- 11.Hong K., Zheng W., Baker A., Papahadjopoulos D. Stabilization of cationic liposome-plasmid DNA complexes by polyamines and poly(ethylene glycol)-phospholipid conjugates for efficient in vivo gene delivery. FEBS Lett. 1997;400:233–237. doi: 10.1016/s0014-5793(96)01397-x. [DOI] [PubMed] [Google Scholar]
- 12.Lukyanov A.N., Hartner W.C., Torchilin V.P. Increased accumulation of PEG-PE micelles in the area of experimental myocardial infarction in rabbits. J. Control. Release. 2004;94:187–193. doi: 10.1016/j.jconrel.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Gref R., Lück M., Quellec P., Marchand M., Dellacherie E., Harnisch S., Blunk T., Müller R.H. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 14.Ogris M., Brunner S., Schüller S., Kircheis R., Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 15.Sonoke S., Ueda T., Fujiwara K., Sato Y., Takagaki K., Hirabayashi K., Ohgi T., Yano J. Tumor regression in mice by delivery of Bcl-2 small interfering RNA with pegylated cationic liposomes. Cancer Res. 2008;68:8843–8851. doi: 10.1158/0008-5472.CAN-08-0127. [DOI] [PubMed] [Google Scholar]
- 16.Peracchia M.T., Fattal E., Desmaële D., Besnard M., Noël J.P., Gomis J.M., Appel M., d’Angelo J., Couvreur P. Stealth PEGylated polycyanoacrylate nanoparticles for intravenous administration and splenic targeting. J. Control. Release. 1999;60:121–128. doi: 10.1016/s0168-3659(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 17.Chang C.-W., Choi D., Kim W.J., Yockman J.W., Christensen L.V., Kim Y.-H., Kim S.W. Non-ionic amphiphilic biodegradable PEG-PLGA-PEG copolymer enhances gene delivery efficiency in rat skeletal muscle. J. Control. Release. 2007;118:245–253. doi: 10.1016/j.jconrel.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Deng J., Gao N., Wang Y., Yi H., Fang S., Ma Y., Cai L. Self-assembled cationic micelles based on PEG-PLL-PLLeu hybrid polypeptides as highly effective gene vectors. Biomacromolecules. 2012;13:3795–3804. doi: 10.1021/bm3012538. [DOI] [PubMed] [Google Scholar]
- 19.van den Berg J.H., Oosterhuis K., Hennink W.E., Storm G., van der Aa L.J., Engbersen J.F.J., Haanen J.B., Beijnen J.H., Schumacher T.N., Nuijen B. Shielding the cationic charge of nanoparticle-formulated dermal DNA vaccines is essential for antigen expression and immunogenicity. J. Control. Release. 2010;141:234–240. doi: 10.1016/j.jconrel.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Tarwadi J., Jazayeri J.A., Prankerd R.J., Pouton C.W. Preparation and in vitro evaluation of novel lipopeptide transfection agents for efficient gene delivery. Bioconjug. Chem. 2008;19:940–950. doi: 10.1021/bc700463q. [DOI] [PubMed] [Google Scholar]
- 21.Ho J.K., White P.J., Pouton C.W. Tissue-specific Calibration of Real-time PCR Facilitates Absolute Quantification of Plasmid DNA in Biodistribution Studies. Mol. Ther. Nucleic Acids. 2016;5:e371. doi: 10.1038/mtna.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adami R.C., Collard W.T., Gupta S.A., Kwok K.Y., Bonadio J., Rice K.G. Stability of peptide-condensed plasmid DNA formulations. J. Pharm. Sci. 1998;87:678–683. doi: 10.1021/js9800477. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q.-R., Zhang L., Stass S.A., Mixson A.J. Branched co-polymers of histidine and lysine are efficient carriers of plasmids. Nucleic Acids Res. 2001;29:1334–1340. doi: 10.1093/nar/29.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenzie D.L., Collard W.T., Rice K.G. Comparative gene transfer efficiency of low molecular weight polylysine DNA-condensing peptides. J. Pept. Res. 1999;54:311–318. doi: 10.1034/j.1399-3011.1999.00104.x. [DOI] [PubMed] [Google Scholar]
- 25.Plank C., Tang M.X., Wolfe A.R., Szoka F.C., Jr. Branched cationic peptides for gene delivery: role of type and number of cationic residues in formation and in vitro activity of DNA polyplexes. Hum. Gene Ther. 1999;10:319–332. doi: 10.1089/10430349950019101. [DOI] [PubMed] [Google Scholar]
- 26.Leng Q., Scaria P., Ioffe O.B., Woodle M., Mixson A.J. A branched histidine/lysine peptide, H2K4b, in complex with plasmids encoding antitumor proteins inhibits tumor xenografts. J. Gene Med. 2006;8:1407–1415. doi: 10.1002/jgm.982. [DOI] [PubMed] [Google Scholar]
- 27.Midoux P., Kichler A., Boutin V., Maurizot J.-C., Monsigny M. Membrane permeabilization and efficient gene transfer by a peptide containing several histidines. Bioconjug. Chem. 1998;9:260–267. doi: 10.1021/bc9701611. [DOI] [PubMed] [Google Scholar]
- 28.Pichon C., Roufaï M.B., Monsigny M., Midoux P. Histidylated oligolysines increase the transmembrane passage and the biological activity of antisense oligonucleotides. Nucleic Acids Res. 2000;28:504–512. doi: 10.1093/nar/28.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok K.Y., Park Y., Yang Y., McKenzie D.L., Liu Y., Rice K.G. In vivo gene transfer using sulfhydryl cross-linked PEG-peptide/glycopeptide DNA co-condensates. J. Pharm. Sci. 2003;92:1174–1185. doi: 10.1002/jps.10384. [DOI] [PubMed] [Google Scholar]
- 30.McKenzie D.L., Kwok K.Y., Rice K.G. A potent new class of reductively activated peptide gene delivery agents. J. Biol. Chem. 2000;275:9970–9977. doi: 10.1074/jbc.275.14.9970. [DOI] [PubMed] [Google Scholar]
- 31.McKenzie D.L., Smiley E., Kwok K.Y., Rice K.G. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconjug. Chem. 2000;11:901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 32.van Hell A.J., Crommelin D.J., Hennink W.E., Mastrobattista E. Stabilization of peptide vesicles by introducing inter-peptide disulfide bonds. Pharm. Res. 2009;26:2186–2193. doi: 10.1007/s11095-009-9933-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budker V., Budker T., Zhang G., Subbotin V., Loomis A., Wolff J.A. Hypothesis: naked plasmid DNA is taken up by cells in vivo by a receptor-mediated process. J. Gene Med. 2000;2:76–88. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<76::AID-JGM97>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Satkauskas S., Bureau M.F., Mahfoudi A., Mir L.M. Slow accumulation of plasmid in muscle cells: supporting evidence for a mechanism of DNA uptake by receptor-mediated endocytosis. Mol. Ther. 2001;4:317–323. doi: 10.1006/mthe.2001.0465. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai F., Inoue R., Nishino Y., Okuda A., Matsumoto O., Taga T., Yamashita F., Takakura Y., Hashida M. Effect of DNA/liposome mixing ratio on the physicochemical characteristics, cellular uptake and intracellular trafficking of plasmid DNA/cationic liposome complexes and subsequent gene expression. J. Control. Release. 2000;66:255–269. doi: 10.1016/s0168-3659(99)00280-1. [DOI] [PubMed] [Google Scholar]
- 36.Saito Y., Kawakami S., Yabe Y., Yamashita F., Hashida M. Intracellular trafficking is the important process that determines the optimal charge ratio on transfection by galactosylated lipoplex in HEPG2 cells. Biol. Pharm. Bull. 2006;29:1986–1990. doi: 10.1248/bpb.29.1986. [DOI] [PubMed] [Google Scholar]
- 37.Guo X.D., Tandiono F., Wiradharma N., Khor D., Tan C.G., Khan M., Qian Y., Yang Y.Y. Cationic micelles self-assembled from cholesterol-conjugated oligopeptides as an efficient gene delivery vector. Biomaterials. 2008;29:4838–4846. doi: 10.1016/j.biomaterials.2008.07.053. [DOI] [PubMed] [Google Scholar]
- 38.Chen J.-X., Wang H.-Y., Quan C.-Y., Xu X.-D., Zhang X.-Z., Zhuo R.-X. Amphiphilic cationic lipopeptides with RGD sequences as gene vectors. Org. Biomol. Chem. 2010;8:3142–3148. doi: 10.1039/c003538f. [DOI] [PubMed] [Google Scholar]
- 39.Furuhata M., Izumisawa T., Kawakami H., Toma K., Hattori Y., Maitani Y. Decaarginine-PEG-liposome enhanced transfection efficiency and function of arginine length and PEG. Int. J. Pharm. 2009;371:40–46. doi: 10.1016/j.ijpharm.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Qi R., Gao Y., Tang Y., He R.-R., Liu T.-L., He Y., Sun S., Li B.Y., Li Y.B., Liu G. PEG-conjugated PAMAM dendrimers mediate efficient intramuscular gene expression. AAPS J. 2009;11:395–405. doi: 10.1208/s12248-009-9116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W.M., Xue L., Mayer L.D., Bally M.B. Intermembrane transfer of polyethylene glycol-modified phosphatidylethanolamine as a means to reveal surface-associated binding ligands on liposomes. Biochim. Biophys. Acta. 2001;1513:193–206. doi: 10.1016/s0005-2736(01)00351-0. [DOI] [PubMed] [Google Scholar]
- 42.Silvius J.R., Leventis R. Spontaneous interbilayer transfer of phospholipids: dependence on acyl chain composition. Biochemistry. 1993;32:13318–13326. doi: 10.1021/bi00211a045. [DOI] [PubMed] [Google Scholar]
- 43.Adlakha-Hutcheon G., Bally M.B., Shew C.R., Madden T.D. Controlled destabilization of a liposomal drug delivery system enhances mitoxantrone antitumor activity. Nat. Biotechnol. 1999;17:775–779. doi: 10.1038/11710. [DOI] [PubMed] [Google Scholar]
- 44.Ambegia E., Ansell S., Cullis P., Heyes J., Palmer L., MacLachlan I. Stabilized plasmid-lipid particles containing PEG-diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim. Biophys. Acta. 2005;1669:155–163. doi: 10.1016/j.bbamem.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Shirota H., Petrenko L., Hong C., Klinman D.M. Potential of transfected muscle cells to contribute to DNA vaccine immunogenicity. J. Immunol. 2007;179:329–336. doi: 10.4049/jimmunol.179.1.329. [DOI] [PubMed] [Google Scholar]
- 46.Casares S., Inaba K., Brumeanu T.-D., Steinman R.M., Bona C.A. Antigen presentation by dendritic cells after immunization with DNA encoding a major histocompatibility complex class II-restricted viral epitope. J. Exp. Med. 1997;186:1481–1486. doi: 10.1084/jem.186.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Condon C., Watkins S.C., Celluzzi C.M., Thompson K., Falo L.D., Jr. DNA-based immunization by in vivo transfection of dendritic cells. Nat. Med. 1996;2:1122–1128. doi: 10.1038/nm1096-1122. [DOI] [PubMed] [Google Scholar]
- 48.Corr M., von Damm A., Lee D.J., Tighe H. In vivo priming by DNA injection occurs predominantly by antigen transfer. J. Immunol. 1999;163:4721–4727. [PubMed] [Google Scholar]
- 49.Fu T.-M., Ulmer J.B., Caulfield M.J., Deck R.R., Friedman A., Wang S., Liu X., Donnelly J.J., Liu M.A. Priming of cytotoxic T lymphocytes by DNA vaccines: requirement for professional antigen presenting cells and evidence for antigen transfer from myocytes. Mol. Med. 1997;3:362–371. [PMC free article] [PubMed] [Google Scholar]
- 50.Ulmer J., Deck R.R., DeWitt C.M., Donnhly J.I., Liu M.A. Generation of MHC class I-restricted cytotoxic T lymphocytes by expression of a viral protein in muscle cells: antigen presentation by non-muscle cells. Immunology. 1996;89:59–67. doi: 10.1046/j.1365-2567.1996.d01-718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mills C.D., Kincaid K., Alt J.M., Heilman M.J., Hill A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe H., Numata K., Ito T., Takagi K., Matsukawa A. Innate immune response in Th1- and Th2-dominant mouse strains. Shock. 2004;22:460–466. doi: 10.1097/01.shk.0000142249.08135.e9. [DOI] [PubMed] [Google Scholar]
- 53.Carstens M.G., Camps M.G.M., Henriksen-Lacey M., Franken K., Ottenhoff T.H.M., Perrie Y., Bouwstra J.A., Ossendorp F., Jiskoot W. Effect of vesicle size on tissue localization and immunogenicity of liposomal DNA vaccines. Vaccine. 2011;29:4761–4770. doi: 10.1016/j.vaccine.2011.04.081. [DOI] [PubMed] [Google Scholar]
- 54.Bello-Roufaï M., Lambert O., Pitard B. Relationships between the physicochemical properties of an amphiphilic triblock copolymers/DNA complexes and their intramuscular transfection efficiency. Nucleic Acids Res. 2007;35:728–739. doi: 10.1093/nar/gkl860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehto T., Simonson O.E., Mäger I., Ezzat K., Sork H., Copolovici D.-M., Viola J.R., Zaghloul E.M., Lundin P., Moreno P.M. A peptide-based vector for efficient gene transfer in vitro and in vivo. Mol. Ther. 2011;19:1457–1467. doi: 10.1038/mt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng B., Zhao Y., Xu L., Xu Y. Electric pulses applied prior to intramuscular DNA vaccination greatly improve the vaccine immunogenicity. Vaccine. 2007;25:2064–2073. doi: 10.1016/j.vaccine.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z., Zhu J., Sriadibhatla S., Gebhart C., Alakhov V., Kabanov A. Promoter- and strain-selective enhancement of gene expression in a mouse skeletal muscle by a polymer excipient Pluronic P85. J. Control. Release. 2005;108:496–512. doi: 10.1016/j.jconrel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 58.Kawase A., Nomura T., Yasuda K., Kobayashi N., Hashida M., Takakura Y. Disposition and gene expression characteristics in solid tumors and skeletal muscle after direct injection of naked plasmid DNA in mice. J. Pharm. Sci. 2003;92:1295–1304. doi: 10.1002/jps.10392. [DOI] [PubMed] [Google Scholar]
- 59.Boyle J.S., Koniaras C., Lew A.M. Influence of cellular location of expressed antigen on the efficacy of DNA vaccination: cytotoxic T lymphocyte and antibody responses are suboptimal when antigen is cytoplasmic after intramuscular DNA immunization. Int. Immunol. 1997;9:1897–1906. doi: 10.1093/intimm/9.12.1897. [DOI] [PubMed] [Google Scholar]
- 60.Thompson B.S., Chilton P.M., Ward J.R., Evans J.T., Mitchell T.C. The low-toxicity versions of LPS, MPL adjuvant and RC529, are efficient adjuvants for CD4+ T cells. J. Leukoc. Biol. 2005;78:1273–1280. doi: 10.1189/jlb.0305172. [DOI] [PubMed] [Google Scholar]
- 61.White P.J., Anastasopoulos F., Church J.E., Kuo C.Y., Boyd B.J., Hickey P.L.C., Tu L.S., Burns P., Lew A.M., Heath W.R. Generic construction of single component particles that elicit humoural and cellular immune responses without the need for adjuvants. Vaccine. 2008;26:6824–6831. doi: 10.1016/j.vaccine.2008.09.087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.