Significance
There has been an impassioned debate in recent years about whether biodiversity is negatively or positively correlated to wildlife and zoonotic disease transmission and risk, suggesting a “dilution” or “amplification” effect, respectively. Here, we demonstrate for an important zoonotic disease (hantavirus pulmonary syndrome) that species diversity can act differently on competing drivers of disease transmission (host density, contact rates, transmissibility) and may cause increases and decreases in transmission, concurrently in the same host–pathogen system. The net effect (dilution, amplification, or no effect) is determined by the strength of the competing mechanisms. Therefore, to move forward, researchers should focus on how biodiversity affects individual mechanisms separately and their net effects if we want to make generalizable predictions across systems.
Keywords: dilution effect, amplification effect, hantavirus, SIR modeling, zoonotic disease
Abstract
In this era of unprecedented biodiversity loss and increased zoonotic disease emergence, it is imperative to understand the effects of biodiversity on zoonotic pathogen dynamics in wildlife. Whether increasing biodiversity should lead to a decrease or increase in infection prevalence, termed the dilution and amplification effects, respectively, has been hotly debated in disease ecology. Sin Nombre hantavirus, which has an 35% mortality rate when it spills over into humans, occurs at a lower prevalence in the reservoir host, the North American deermouse, in areas with higher small mammal diversity—a dilution effect. However, the mechanism driving this relationship is not understood. Using a mechanistic mathematical model of infection dynamics and a unique long-term, high-resolution, multisite dataset, it appears that the observed dilution effect is a result of increasing small-mammal diversity leading to decreased deermouse population density and, subsequently, prevalence (a result of density-dependent transmission). However, once density is taken into account, there is an increase in the transmission rate at sites with higher diversity—a component amplification effect. Therefore, dilution and amplification are occurring at the same time in the same host–pathogen system; there is a component amplification effect (increase in transmission rate), but overall a net dilution because the effect of diversity on reservoir host population density is stronger. These results suggest we should focus on how biodiversity affects individual mechanisms that drive prevalence and their relative strengths if we want to make generalizable predictions across host–pathogen systems.
Biodiversity is being lost at an unprecedented rate; some say we have entered the sixth mass extinction (1). Meanwhile, zoonotic infectious disease outbreaks are increasing (2, 3). Therefore, it is imperative to understand how biodiversity affects disease dynamics in wildlife. Work on Lyme disease demonstrated how, under certain conditions, increased vertebrate species diversity can lead to decreased disease transmission and prevalence in the reservoir host and subsequently decreased spillover to humans—the so-called “dilution effect” (4, 5). The generality of the dilution effect has been the topic of much recent debate. If it is a general phenomenon, preserving biodiversity would be a win–win for animal conservation and control of zoonotic diseases. Mounting evidence suggests the dilution effect applies to several systems across taxa (6–8). However, the dilution effect does not appear to be universal but depends on the animal community composition, host and pathogen ecologies, and the scale at which the system is examined (9–12). In some circumstances, increased species diversity can lead to increased infection prevalence, the “amplification effect” (10–13). For most systems, exactly which circumstances and mechanisms lead to disease amplification or dilution have not been fully characterized, particularly for directly transmitted diseases (14).
Much of the research on this issue has focused on vector-borne (e.g., tick or mosquito-transmitted) or environmentally transmitted diseases, such as Lyme disease, West Nile virus, and Ribeiroia ondatrae, where the dilution effect is caused by more diverse communities having multiple species with a range of competencies with respect to pathogen replication and vector preferences (5, 15, 16). In these systems, when biodiversity is lost, the species that remain tend to be the most competent reservoirs. [The generality of this phenomenon is debated (13, 17).] Directly transmitted diseases have received little attention.
For directly transmitted pathogens with one main reservoir host, the SIR (susceptible–infected–recovered) framework is useful for conceptualizing how species diversity could affect infection dynamics. These models are called compartmental models, because individuals within a population are grouped into different compartments with respect to their disease status. Individuals are either uninfected and susceptible to infection (), infected and infectious (), or recovered and immune (). Over time, individuals flow through these compartments. The rate of new infections is determined by the density of susceptible individuals (), the density of infected individuals (), the transmission rate (), and whether contact rates increase with density (density-dependent transmission) or are independent of density (frequency-dependent transmission). Therefore, the rate of new infections is or , for density-dependent or frequency-dependent transmission, respectively.
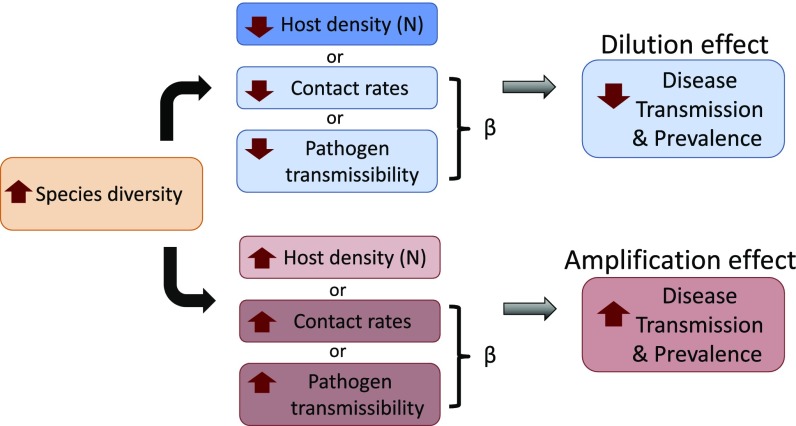
The transmission rate, , is a product of the contact rate and the probability of transmission given contact (or transmissibility). Therefore, three mechanisms determine how diversity could affect pathogen transmission and prevalence. Dilution will occur if increased species diversity leads to (i) decreased host population density (if transmission is density-dependent), (ii) decreased contact rates, and/or (iii) decreased transmissibility (Fig. 1, blue). Conversely, amplification could occur if increased diversity leads to increased host density, contact rates, and/or transmissibility (Fig. 1, red).
Fig. 1.
Conceptual diagram of how species diversity could affect transmission and prevalence of a directly transmitted disease.
Sin Nombre hantavirus (SNV) is a directly transmitted zoonotic pathogen whose reservoir host is the North American deermouse (Peromyscus maniculatus, hereafter deermouse). Humans are spillover hosts infected when they have direct contact with urine, feces, or saliva from an infected host rodent or inhale infectious aerosols of these substances. In humans, SNV causes hantavirus pulmonary syndrome (HPS), which has an overall 36% mortality rate. SNV prevalence in deermice is often lower in communities with higher small mammal diversity—the dilution effect (18–20).
Because SNV is a directly transmitted disease (no vector) that is highly host-specific and contacts of infected deermice with members of other species in the community are not likely to pass on the infection, all other hosts are essentially noncompetent. Spillover of SNV from deermice may occur (e.g., to other rodents and humans). However, they are not likely to be important in the reservoir dynamics except under unusual conditions (21). Thus, the mechanism for how small mammal diversity acts on SNV prevalence must be different from Lyme disease, West Nile virus, and R. ondatrae and has yet to be determined. The SIR framework with one reservoir host is appropriate for examining this question. Accordingly, the decrease in prevalence as small mammal diversity increases must be caused by at least one of the following mechanisms: (i) decreased host population density (), or (ii) decreased transmission rate () within the host population by either decreasing (a) intraspecific contact rates and/or (b) transmissibility of the pathogen.
For the first mechanism, host population density, to be the driving factor, there must be density-dependent transmission, which leads to decreased infection prevalence with decreased deermouse density. Some researchers concluded host density could not be the mechanism driving dilution because they found no simultaneous positive relationship between deermouse population density and SNV infection prevalence (19, 20). However, we recently clarified the density–prevalence relationship showing that in host–pathogen systems with highly variable dynamics (where the carrying capacity and host density vary widely over space and time), the system never reaches equilibrium. Therefore, host population density can have a strong effect on SNV prevalence but with time lags that themselves depend on density, thus making the relationship not apparent using statistical correlations (22). Due to these nonequilibrium dynamics, the relationship between density and prevalence can only be deciphered using a dynamical mathematical model of disease dynamics. We showed SNV does have density-dependent transmission (22); therefore, reduced host density could, indeed, be driving the dilution effect.
Alternatively, the dilution effect could be caused by increased small mammal diversity decreasing the transmission rate (), by decreasing the contact rate or the transmissibility of the virus between mice.
To determine the mechanism of the observed dilution effect for SNV, we examine the two components that determine prevalence (density and transmission rate) by fitting a mechanistic model to data from a multistate long-term study of hantavirus ecology. The US Centers for Disease Control and Prevention (CDC) and other sources funded longitudinal monitoring of small mammal communities at more than 40 sites in Arizona, Colorado, Montana, and New Mexico. Starting in 1994, rodents were trapped monthly at all sites (except every 6 wk in Colorado) for up to 15 y. To synthesize drivers of infection across sites with differing dynamics, we apply the same dynamical modeling framework to examine all sites with a time series of infection long enough to analyze. We apply a mechanistic dynamical SIR-type model that simulates infection dynamics given host density (22) to 18 sites in 4 US states, with varying species diversity, estimating a transmission rate for each site.
We hypothesize that the dilution effect is driven by host population density—sites with higher small mammal diversity have lower deermouse density, and SNV prevalence is lower as a result of density-dependent transmission. This leads to the prediction that when we fit our mechanistic model, driven by host density, to each site, the model will fit the observed host–pathogen dynamics, and the transmission rate () will not vary as a function of diversity. The alternative hypothesis is that the observed dilution effect is driven by contact rates and/or probability of transmission given contact, components of the transmission rate (). This predicts that when we fit our model to multiple sites, the estimated transmission rate () will be lower for sites with higher species diversity.
Materials and Methods
Study Sites.
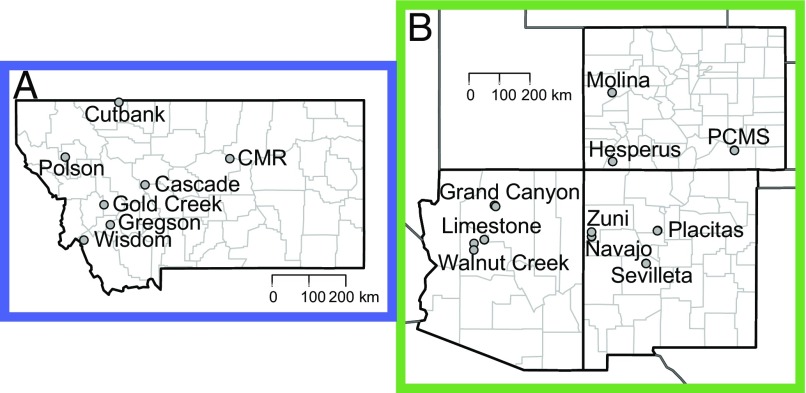
We analyze data from small mammal capture–mark–recapture SNV studies in Montana, New Mexico, Arizona, and Colorado (Fig. 2). Data span June 1994 through 2015, with sites trapped an average of 79 mo (range 3 to 179). SI Appendix, Table S1 summarizes data from all 40 sites. We applied our model to sites in which there was an average of 2 deermice/hectare, 4% SNV prevalence, and 12 mo of data (18 sites, Table 1); because our model is dynamical and prevalence depends on previous population density, we excluded sites with too few data points.
Fig. 2.
Map of study sites in (A) Montana and (B) the southwest (SW) United States, including Arizona, Colorado, and New Mexico.
Table 1.
Summary of longitudinal data from modeled sites
| State | Web or grid | Mo. | Dens. | Prev. | ||
| MT | Cascade.11 | 179 | 25.65 | 0.048 | 1.06 | 0.0040 |
| MT | CMR.18 | 69 | 8.70 | 0.044 | 1.51 | 0.0183 |
| MT | Cutbank.15 | 70 | 10.59 | 0.152 | 1.52 | 0.0202 |
| MT | GoldCreek.8 | 70 | 11.24 | 0.133 | 1.62 | 0.0091 |
| MT | GoldCreek.9 | 70 | 8.60 | 0.086 | 1.86 | 0.0152 |
| MT | Gregson.Upper | 134 | 10.49 | 0.137 | 1.01 | 0.0106 |
| MT | Gregson.Lower | 158 | 16.18 | 0.108 | 1.06 | 0.0073 |
| MT | Polson.5 | 96 | 53.64 | 0.213 | 1.12 | 0.0027 |
| MT | Polson.6 | 70 | 14.91 | 0.125 | 1.54 | 0.0084 |
| AZ | Grand Canyon.E | 43 | 20.15 | 0.277 | 1.72 | 0.0042 |
| AZ | Grand Canyon.M | 46 | 9.57 | 0.311 | 1.52 | 0.0120 |
| AZ | Grand Canyon.T | 46 | 8.73 | 0.152 | 1.80 | 0.0184 |
| CO | Hesperus.ha | 67 | 8.43 | 0.172 | 1.77 | 0.0138 |
| CO | Hesperus.hb | 68 | 9.28 | 0.183 | 1.34 | 0.0141 |
| CO | Molina.ma | 58 | 4.84 | 0.062 | 2.53 | 0.0277 |
| CO | Molina.mb | 47 | 4.49 | 0.099 | 2.21 | 0.0326 |
| NM | Navajo.1 | 92 | 3.27 | 0.065 | 2.47 | 0.0523 |
| NM | Zuni.2 | 92 | 2.06 | 0.089 | 3.76 | 0.0789 |
AZ, Arizona; β, estimated transmission rate, per hectare; CMR, Charles M. Russell Wildlife Refuge; CO, Colorado; D, Simpson’s D diversity index; dens., average deermouse density, per hectare; mo., number of months sampled; MT, Montana; NM, New Mexico; prev., average SNV antibody prevalence. See SI Appendix, Table S1 for more information and all sites.
In Montana, 10-by-10 trap grids contained 100 Sherman (8 × 9 × 23 cm; H.B. Sherman Trap Company) live-capture traps at 10-m intervals covering 1 ha. See ref. 23 for details. Grids were trapped monthly as weather allowed. In the SW (Arizona, Colorado, New Mexico), trapping webs were used rather than grids. Each web covered 3.14 ha and contained 12 100-m transects radiating from a central point. Each web contained 148 Sherman traps. Webs in Arizona and New Mexico were trapped monthly and in Colorado every 6 wk, as weather allowed. See SI Appendix, Table S1 for details about frequency and duration of trapping at sites, deermouse population densities, SNV prevalence, and small mammal diversity. See ref. 24 for more details of sites and protocols.
Trapping protocols were similar across sites. As a rule, researchers trapped for three consecutive nights. Traps were set in the evenings baited with peanut butter and oats, cracked corn, or mixed grain. Cotton or polyester fiberfill was added for nest material. Traps were checked in the mornings. Captured small mammals were anesthetized, ear-tagged, weighed, and a blood sample taken to test for SNV antibodies. Because SNV is a chronic infection in deermice, antibodies are an appropriate indicator of current infection. Serologic testing was conducted at CDC, Atlanta; the Montana Department of Public Health and Human Services; Montana State University; or Montana Tech using a standard enzyme-linked immunosorbent assay (ELISA) for IgG antibody to SNV (25).
We analyze each web/grid separately because often they were trapped for differing lengths and were sufficiently distant from one another that many of the grids’ community dynamics were not significantly correlated. Additionally, we group Montana and SW sites separately because of the different trapping structure and minor differences in data collection. For comparing data across Montana and the SW, we normalize density to 1 ha.
Model.
Deermouse population dynamics are highly variable over time and strongly tied to local environmental conditions (26, 27). We have developed a mechanistic mathematical model of SNV in deermice that allows for the environmental carrying capacity and population density to vary over time (essential in this highly variable, bottom–up system) (22). Here we use a version of our model, which is a type of SIR compartmental model; SNV is a chronic, probably life-long infection in deermice, and thus, there is no class. The model includes logistic growth, where the host population experiences density dependence partitioned across birth and death rates and determined by a time-varying carrying capacity, . The underlying model is
| [1] |
where is the density of susceptible individuals, is the density of infected individuals, and . [This is our published model (22), except without age structure, which did not significantly impact dynamics.] is the maximum birth rate (in absence of density dependence; i.e., when ), is the minimum death rate, and is the proportion of density dependence due to density dependence in birth rates. If , birth rates are density independent, and all of the density dependence is on the death rates; if , all of the density dependence is on the birth rates. is . is the disease-induced mortality rate. is the transmission rate, and is the immigration rate of infected individuals (see SI Appendix, Fig. S1 for life-cycle diagram).
This is the underlying model we use; however, for the purposes of our analyses—to understand the importance of deermouse population density in driving dynamics—we are interested in predicting assuming we know the population density, . Therefore, we only need to model , since .
Previously, we estimated parameters for study sites in Montana (22, 28). Here we assume that the disease-induced mortality rate (), maximum birth rate (), minimum death rate (), and how density dependence is partitioned () are the same across all sites [, , , and (22)]. However, the environmental carrying capacity () and therefore the realized birth and death rates vary across sites and over time at the same site. The quantities that vary across sites include the inputs for the model: , , and initial infected deermouse density (), all taken from the data. Parameters, transmission rate () and infected immigration rate (), are estimated using maximum likelihood for each site.
Parameter Estimation.
We previously found that setting to a smoothed spline of the deermouse population density 3 mo ahead approximated the time-varying carrying capacity (density lags behind carrying capacity by 3 mo) (22). Using this approximation, the model-predicted deermouse population dynamics matched observed deermouse population density. Needing future deermouse density to predict current infection levels makes this implementation of the model less useful for prediction. However, our goal is to understand what processes are leading to the observed dynamics, not prediction.
As an index of relative population density, we use minimum number known alive (MNA) (29). Similarly for relative density of infected individuals, we use minimum number known infected (MNI). See SI Appendix for justification of this index and comparisons with robust design population estimates that account for probability of detection using Program Mark.
We estimate a transmission rate () and infected immigration rate () for each site separately using trajectory matching (30). For this, we numerically minimize the negative log likelihood between the vector of model-predicted density of infected mice and the vector of observed MNI using the Nelder–Mead algorithm implemented in the “optim” function in R, assuming Poisson errors. The only inputs are initial (set to that observed in the data) and and (both smoothed splines from the data to accommodate the continuous-time framework).
We then compare the estimated transmission rate () for each site to the average small mammal diversity for that site, using Simpson’s diversity index, , where is the proportional abundance of species, , and is the small mammal species richness—that is, total number of small mammal species trapped at that site. We also compare results to the Shannon–Weiner index and species richness (see SI Appendix).
See SI Appendix for data and R code.
Results
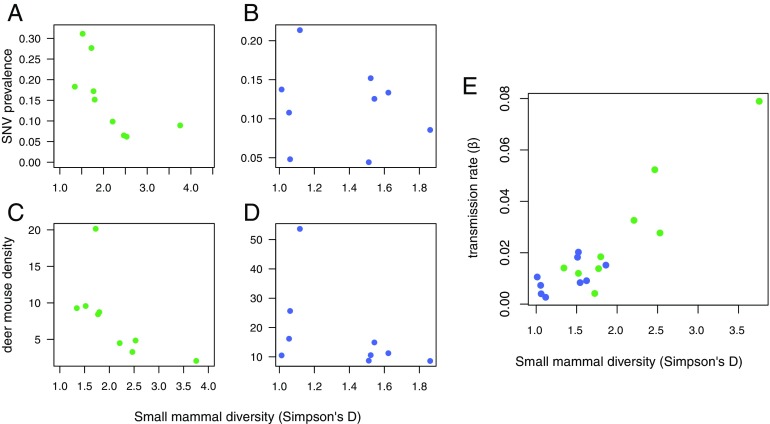
We observe a dilution effect—a decrease in average SNV prevalence in deermice with increased average small mammal diversity—in the SW (P = 0.002, = 0.64; Fig. 3A) but not in Montana (P = 0.60; Fig. 3B). However, our dynamical epidemiological (SI Appendix) model, which simulates infection based on deermouse density, fits both Montana and SW sites well (Fig. 4 and SI Appendix, Figs. S2 and S3). Once density is taken into account using the model, the estimated transmission rate () is significantly correlated to diversity. However, it is in the opposite direction to what is predicted if the dilution effect is driven by host contact rates or transmissibility. There is a positive relationship between diversity and transmission rate—a component amplification effect (Fig. 3E).
Fig. 3.
Relationship between average small mammal diversity by Simpson’s diversity index and (A) average SNV prevalence in deermice for SW sites and (B) Montana sites, (C) average deermouse population density for SW sites and (D) Montana sites, and (E) estimated transmission rate () among deermice. Throughout, SW sites are green, and Montana sites are blue.
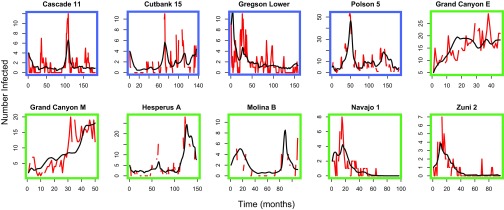
Fig. 4.
Fit of models to data from 10 of the 18 sites modeled. Observed number of infected individuals is in red. Model predicted number of infected individuals is in black. Montana sites (blue) are per hectare, and SW sites (green) are per 3.14 ha. See SI Appendix, Figs. S1 and S2 for all sites including total deermouse density per site.
An increase in small mammal diversity is associated with a decrease in deermouse density in the SW (Fig. 3C) and decrease in transmission and prevalence—the dilution effect (Fig. 3A; as described in Fig. 1, dark blue pathway). However, in Montana, increased small mammal diversity does not lead to decreased deermouse density (Fig. 3D), and there is not a dilution effect (Fig. 3B). However, overall small mammal diversity in Montana is lower than in the SW (see x axis for Fig. 3E). Although there is not a significant dilution effect in Montana (because the relationship between diversity and deermouse density is not the same as in the SW), the relationship between transmission rate () and diversity is similar across SW and Montana sites (Fig. 3E). Results are robust to different diversity measures (see SI Appendix).
Discussion
This study brings together dynamical ecological models with high-resolution, long-term data, spanning more than 20 y of data collection at 18 sites with different small mammal community composition and deermouse host population dynamics. Our model accurately predicts infection dynamics over time at these disparate sites (Fig. 4 and SI Appendix, Fig. S2 and S3), given only deermouse population density and the initial number of infected individuals, and estimating a transmission rate () and infected immigration rate () for each site. With these data and models, we can explore hypotheses about how small mammal diversity affects SNV transmission and disease risk. At our SW study sites, we observe a dilution effect—sites with higher small mammal diversity had decreased prevalence of infection (Fig. 3A). We do not observe a dilution effect at our Montana sites (Fig. 3B).
Given the SIR framework, the dilution effect in the SW must be driven by increased species diversity leading to (i) decreased host population density or (ii) a decrease in the transmission rate, which includes contact rates between deermice and transmissibility (Fig. 1, blue). If population density (mechanism 1) drove the dilution effect, our model, which predicts infection given host density, should predict the dynamics well, and the transmission rate () should not be negatively correlated to diversity. If transmission rate (mechanism 2) drove the dilution effect, our model should predict the dynamics, and each site’s estimated transmission rate should be negatively correlated to its diversity. We found support for mechanism 1—that deermouse population density drives the dilution effect in the SW. Our model fits the dynamics at disparate sites (Fig. 4 and SI Appendix, Figs. S1 and S2), and the transmission rate is not negatively correlated to diversity. In fact, it is positively correlated to small mammal species diversity ( = 0.83, P 0.001; Fig. 3E). This implies that density causes the observed dilution effect (dark blue, Fig. 1) but that there is an additional component amplification effect through changes in transmission rate (dark red, Fig. 1). Therefore, aspects of both dilution and amplification are occurring at the same time in the same system.
For models with density-dependent transmission, the transmission rate () gives the rate of transition from susceptible to infected for a given host density; as density increases, overall transmission increases. Therefore, our finding of increased transmission rates with increased diversity means that for a given host density, transmission occurs at a faster rate in more diverse communities. However, density is a stronger driver of infection dynamics in this system, and since increased diversity leads to lower host density in the SW, there is a net dilution effect in the SW (Fig. 3). In Montana, increased diversity does not lead to a decrease in host density, and there is not a significant dilution effect (Fig. 3).
The dilution effect has been demonstrated for hantaviruses, including SNV in Portland, OR (19) and Utah (20, 31); Puumala virus in bank voles in Finland (32); and hantaviruses (Choclo and Calabazo viruses) in their hosts in Panama (33). However, the mechanism driving the dilution effect has not been fully elucidated for any of these systems. The studies in Finland and Panama reported that increased diversity led to decreased host density and prevalence (32, 33), suggesting density-dependent transmission and host density as a potential mechanism for the observed dilution effect. Neither transmission rate nor its components, contact rates, or transmissibility were measured in these studies. However, the authors of the Portland and Utah studies discounted density as the mechanism for their observed dilution effect, because they did not see a positive (concurrent) relationship between deermouse density and SNV prevalence and concluded that, therefore, transmission was not density-dependent. However, both studies noted a significant negative relationship between small mammal diversity and deermouse density (19, 20, 31, 34). Clay et al. (34) found a negative association between intraspecific contact rate and species diversity; however, the relationship may have been largely driven by one outlier. A recent experimental study using enclosures and controlling for density showed no change in intraspecific deermouse contacts as small mammal diversity increased (35).
For deermouse population density to drive the dilution effect for SNV, there would need to be density-dependent transmission, which should lead to an observed positive relationship between deermouse density and SNV prevalence. Field studies have found mixed results, only rarely showing a concurrent positive relationship (36), and often showing either no relationship (21, 34, 36, 37) or a negative relationship (23, 38). Using our dynamical epidemiological model, we recently clarified how a concurrent negative relationship between deermouse population density and SNV prevalence can be observed with (positive) density-dependent transmission (22). This negative correlation observed at the Cascade, MT site is a result of the nonequilibrium dynamics (from a constantly changing carrying capacity) in deermouse population density leading to delayed density dependence, such that prevalence lags density by approximately 8 to 16 mo (22, 39–41). At that site, deermouse density peaked, and prevalence peaked later when population density had significantly declined again. Note that with delayed density dependence, a positive relationship or no relationship may also be observed between concurrent density and prevalence (e.g., refs. 21, 36, and 37), depending upon the pattern of population fluctuation. We also showed that the delays may not be fixed [e.g., at 1 y (39)] but depend on host density, potentially making the relationship not apparent using conventional statistical analyses or examining averages; it can only be deciphered using a mechanistic dynamical model of infection, such as ours (22). Therefore, we caution against inferring too much from point estimates or averages (e.g., Fig. 3); the true test is how the data match predictions from the dynamical model, which is driven by population density, because it can account for the transient dynamics and inherent variable time lags.
We see a component amplification effect through an increase in the transmission rate at sites with higher small mammal diversity (Fig. 3E). This could be due to increased diversity leading to an increase in contact rates or an increase in the probability of transmission given contact (transmissibility; Fig. 1, dark red). Increased species diversity could increase contact rates between deermice if, in more diverse areas, deermice cluster in refugia away from dominant competitors. Alternatively, increased species diversity could lead to increased transmissibility if host animals are stressed in the presence of competitors; increased stress can depress the immune system and lead to an increase in viral replication and increased susceptibility (42). Studies of how contact rates vary with species diversity give mixed results—one study suggested a decrease in contacts (34), but an experimental study (with enclosures controlling for density) showed no change in contacts with increased diversity (35). How transmissibility or immunity is affected by species diversity has yet to be examined and is a plausible mechanism for our observed increase in transmission rates.
Future studies should examine the importance of specific species or what aspects of “biodiversity” lead to both the dilution and component amplification. Host interactions with different species vary. In an enclosure experiment, deermice had more interspecific interactions with Merriam’s kangaroo rats than desert pocket mice or Chihuahuan grasshopper mice, although intraspecific contact rates among deermice remained unchanged (35). Clay et al. suggested that Ord’s Kangaroo rats and pinyon mice may be particularly important in determining SNV prevalence (31).
Although previous studies have shown that the dilution effect can be scale-dependent (11), we show that dilution and amplification can occur at the same time in the same system, at the same scale, and the resultant overall effect will be determined by which effect is stronger. A recent, impassioned debate has centered on whether loss of biodiversity should be more likely to lead to an increase (dilution), decrease (amplification), or idiosyncratic change in disease risk (6–8, 10, 11, 43, 44). Perhaps, the reason that responses to biodiversity seem “idiosyncratic” (10) is because there are differential effects of biodiversity on the different mechanisms driving transmission. Therefore, if we want to make generalizable predictions across systems, we should focus on how biodiversity affects individual mechanisms that drive transmission and prevalence separately [as called for by, e.g., Johnson et al. (45)] and the relative strength of these.
Supplementary Material
Acknowledgments
Long-term data collection was supported by the US Centers for Disease Control and Prevention, Atlanta and the NIH Institutional Development Award (IDeA) Networks of Biomedical Research Excellence (INBRE) program in Montana. A.D.L. was supported by the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science and Technology Directorate (US Department of Homeland Security) and the Fogarty International Center (NIH); the University of Montana, University Grant Program; and NSF Established Program to Stimulate Competitive Research (EPSCoR).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807106115/-/DCSupplemental.
References
- 1.Ceballos G, et al. Accelerated modern human–induced species losses: Entering the sixth mass extinction. Sci Adv. 2015;1:e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith KF, et al. Global rise in human infectious disease outbreaks. J R Soc Interface. 2014;11:20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostfeld RS, Keesing F. Biodiversity and disease risk: The case of Lyme disease. Conserv Biol. 2000;14:722–728. [Google Scholar]
- 5.LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci USA. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keesing F, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostfeld RS, Keesing F. Effects of host diversity on infectious disease. Annu Rev Ecol Evol Syst. 2012;43:157–182. [Google Scholar]
- 8.Civitello DJ, et al. Biodiversity inhibits parasites: Broad evidence for the dilution effect. Proc Natl Acad Sci USA. 2015;112:8667–8671. doi: 10.1073/pnas.1506279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood CL, McInturff A, Young HS, Kim D, Lafferty KD. Human infectious disease burdens decrease with urbanization but not with biodiversity. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160122. doi: 10.1098/rstb.2016.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salkeld DJ, Padgett KA, Jones JH. A meta-analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecol Lett. 2013;16:679–686. doi: 10.1111/ele.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood CL, Lafferty KD. Biodiversity and disease: A synthesis of ecological perspectives on Lyme disease transmission. Trends Ecol Evol. 2013;28:239–247. doi: 10.1016/j.tree.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol Lett. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- 13.Wood CL, et al. Does biodiversity protect humans against infectious disease? Ecology. 2014;95:817–832. doi: 10.1890/13-1041.1. [DOI] [PubMed] [Google Scholar]
- 14.Kilpatrick AM, Salkeld DJ, Titcomb G, Hahn MB. Conservation of biodiversity as a strategy for improving human health and well-being. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160131. doi: 10.1098/rstb.2016.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Host heterogeneity dominates West Nile virus transmission. Proc Biol Sci. 2006;273:2327–2333. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson PT, Preston DL, Hoverman JT, Richgels KL. Biodiversity decreases disease through predictable changes in host community competence. Nature. 2013;494:230–233. doi: 10.1038/nature11883. [DOI] [PubMed] [Google Scholar]
- 17.Ostfeld RS, Keesing F. Is biodiversity bad for your health? Ecosphere. 2017;8:e01676. [Google Scholar]
- 18.Mills JN. Regulation of rodent-borne viruses in the natural host: Implications for human disease. Arch Virol. 2005;19:45–57. doi: 10.1007/3-211-29981-5_5. [DOI] [PubMed] [Google Scholar]
- 19.Dizney L, Ruedas L. Increased host species diversity and decreased prevalence of Sin Nombre virus. Emerg Infect Dis. 2009;15:1012–1018. doi: 10.3201/eid1507.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dearing MD, Clay C, Lehmer E, Dizney L. The roles of community diversity and contact rates on pathogen prevalence. J Mammal. 2015;96:29–36. [Google Scholar]
- 21.Mills JN, et al. Patterns of association with host and habitat: Antibody reactive with Sin Nombre virus in small mammals in the major biotic communities of the southwestern United States. Am J Trop Med Hyg. 1997;56:273–284. doi: 10.4269/ajtmh.1997.56.273. [DOI] [PubMed] [Google Scholar]
- 22.Luis AD, Douglass RJ, Mills JN, Bjørnstad ON. Environmental fluctuations lead to predictability in Sin Nombre hantavirus outbreaks. Ecology. 2015;96:1691–1701. [Google Scholar]
- 23.Douglass RJ, et al. Longitudinal studies of Sin Nombre virus in deer mouse-dominated ecosystems of Montana. Am J Trop Med Hyg. 2001;65:33–41. doi: 10.4269/ajtmh.2001.65.33. [DOI] [PubMed] [Google Scholar]
- 24.Mills JN, Yates TL, Ksiazek TG, Peters CJ, Childs JE. Long-term studies of hantavirus reservoir populations in the southwestern United States: Rationale, potential, and methods. Emerg Infect Dis. 1999;5:95–101. doi: 10.3201/eid0501.990111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schountz T, et al. Rapid field immunoassay for detecting antibody to Sin Nombre virus in deer mice. Emerg Infect Dis. 2007;13:1604–1607. doi: 10.3201/eid1310.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luis AD, Douglass RJ, Mills JN, Bjornstad ON. The effect of seasonality, density and climate on the population dynamics of Montana deer mice, important reservoir hosts for Sin Nombre hantavirus. J Anim Ecol. 2010;79:462–470. doi: 10.1111/j.1365-2656.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 27.Glass GE, et al. Satellite imagery characterizes local animal reservoir populations of Sin Nombre virus in the southwestern United States. Proc Natl Acad Sci USA. 2002;99:16817–16822. doi: 10.1073/pnas.252617999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luis AD, Douglass RJ, Mills JN, Hudson PJ, Bjornstad ON. Sin Nombre hantavirus decreases survival of male deer mice. Oecologia. 2012;169:431–439. doi: 10.1007/s00442-011-2219-2. [DOI] [PubMed] [Google Scholar]
- 29.Krebs CJ. Demographic changes in fluctuating populations of Microtus californicus. Ecol Monogr. 1966;36:239–273. [Google Scholar]
- 30.Wood SN. Partially specified ecological models. Ecol Monogr. 2001;71:1–25. [Google Scholar]
- 31.Clay CA, Lehmer EM, Jeor SS, Dearing MD. Sin Nombre virus and rodent species diversity: A test of the dilution and amplification hypotheses. PLoS One. 2009;4:e6467. doi: 10.1371/journal.pone.0006467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voutilainen L, et al. Life-long shedding of Puumala hantavirus in wild bank voles (Myodes glareolus) J Gen Virol. 2015;96:1238–1247. doi: 10.1099/vir.0.000076. [DOI] [PubMed] [Google Scholar]
- 33.Suzan G, et al. Experimental evidence for reduced rodent diversity causing increased hantavirus prevalence. PLoS One. 2009;4:e5461. doi: 10.1371/journal.pone.0005461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clay C, Lehmer E, Jeor S, Dearing M. Testing mechanisms of the dilution effect: Deer mice encounter rates, Sin Nombre virus prevalence and species diversity. EcoHealth. 2009;6:250–259. doi: 10.1007/s10393-009-0240-2. [DOI] [PubMed] [Google Scholar]
- 35.Rubio AV, et al. Is species richness driving intra- and interspecific interactions and temporal activity overlap of a hantavirus host? An experimental test. PLoS One. 2017;12:e0188060. doi: 10.1371/journal.pone.0188060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boone JD, et al. Ecology and demographics of hantavirus infections in rodent populations in the Walker River Basin of Nevada and California. Am J Trop Med Hyg. 1998;59:445–451. doi: 10.4269/ajtmh.1998.59.445. [DOI] [PubMed] [Google Scholar]
- 37.Calisher CH, Sweeney W, Mills JN, Beaty BJ. Natural history of Sin Nombre virus in western Colorado. Emerg Infect Dis. 1999;5:126–134. doi: 10.3201/eid0501.990115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calisher CH, et al. Epizootiology of Sin Nombre and El Moro Canyon hantaviruses, southeastern Colorado, 1995–2000. J Wildl Dis. 2005;41:1–11. doi: 10.7589/0090-3558-41.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Yates T, Mills JN, Parmenter CA, Ksiazek TG. The ecology and evolutionary history of an emergent disease: Hantavirus pulmonary syndrome. Bioscience. 2002;52:989–998. [Google Scholar]
- 40.Madhav NK, Wagoner KD, Douglass RJ, Mills JN. Delayed density-dependent prevalence of Sin Nombre Virus in Montana deer mice (Peromyscus maniculatus) and implications for human disease risk. Vector Borne Zoonotic Dis. 2007;7:353–364. doi: 10.1089/vbz.2006.0605. [DOI] [PubMed] [Google Scholar]
- 41.Adler FR, Pearce-Duvet JMC, Dearing MD. How host population dynamics translate into time-lagged prevalence: An investigation of Sin Nombre virus in deer mice. Bull Math Biol. 2008;70:236–252. doi: 10.1007/s11538-007-9251-8. [DOI] [PubMed] [Google Scholar]
- 42.Brenner GJ, Moynihan JA. Stressor-induced alterations in immune response and viral clearance following infection with herpes simplex virus-type 1 in balb/c and c57bl/6 mice. Brain Behav Immun. 1997;11:9–23. doi: 10.1006/brbi.1997.0480. [DOI] [PubMed] [Google Scholar]
- 43.Randolph SE, Dobson A. Pangloss revisited: A critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology. 2012;139:847–863. doi: 10.1017/S0031182012000200. [DOI] [PubMed] [Google Scholar]
- 44.Lafferty KD, Wood CL. It’s a myth that protection against disease is a strong and general service of biodiversity conservation: Response to Ostfeld and Keesing. Trends Ecol Evol. 2013;28:503–504. doi: 10.1016/j.tree.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Johnson PT, Ostfeld RS, Keesing F. Frontiers in research on biodiversity and disease. Ecol Lett. 2015;18:1119–1133. doi: 10.1111/ele.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.