Significance
The outer membrane of Gram-negative bacteria presents a selectively permeable barrier to the environment and is the first line of defense against antibiotics and other antimicrobial agents. Maintenance of the outer membrane relies on lipoproteins delivered by the LolABCDE system, making the Lol proteins attractive targets for the development of new antimicrobial compounds. During trafficking, lipoproteins are extracted from the cytoplasmic membrane by the LolCDE complex, transported across the periplasm by LolA, and integrated into the outer membrane by LolB. Here, we describe structural features underpinning the interaction between LolA and LolCDE. The structure of LolA bound to the periplasmic domain of LolC provides an arresting molecular snapshot of a key intermediate in the bacterial lipoprotein trafficking pathway.
Keywords: lipoprotein trafficking, protein interactions, membrane biogenesis, X-ray crystallography, ABC transporter
Abstract
In Gram-negative bacteria, outer-membrane lipoproteins are essential for maintaining cellular integrity, transporting nutrients, establishing infections, and promoting the formation of biofilms. The LolCDE ABC transporter, LolA chaperone, and LolB outer-membrane receptor form an essential system for transporting newly matured lipoproteins from the outer leaflet of the cytoplasmic membrane to the innermost leaflet of the outer membrane. Here, we present a crystal structure of LolA in complex with the periplasmic domain of LolC. The structure reveals how a solvent-exposed β-hairpin loop (termed the “Hook”) and trio of surface residues (the “Pad”) of LolC are essential for recruiting LolA from the periplasm and priming it to receive lipoproteins. Experiments with purified LolCDE complex demonstrate that association with LolA is independent of nucleotide binding and hydrolysis, and homology models based on the MacB ABC transporter predict that LolA recruitment takes place at a periplasmic site located at least 50 Å from the inner membrane. Implications for the mechanism of lipoprotein extraction and transfer are discussed. The LolA–LolC structure provides atomic details on a key protein interaction within the Lol pathway and constitutes a vital step toward the complete molecular understanding of this important system.
In Gram-negative bacteria, the outer membrane provides an important physical barrier to the extracellular space, protecting against osmotic shock, noxious compounds, and antibiotics (1, 2). Lipoproteins, anchored by N-terminally linked acyl groups, are a crucial structural component of the outer membrane maintaining attachment to the peptidoglycan layer (3, 4). Other lipoproteins underpin assembly of integral β-barrel proteins at the outer membrane (1, 5, 6), insertion of lipopolysaccharide (7, 8), maintenance of outer-membrane lipid asymmetry (9, 10), and regulation of peptidoglycan synthesis (11). Lipoproteins are therefore central to the physiology of the cell envelope. Mislocalization of outer-membrane lipoproteins on the inner membrane results in cell death (12, 13), and proteins responsible for lipoylation and trafficking of outer-membrane lipoproteins are essential for bacterial viability (14–18). The relative accessibility of proteins involved in lipoprotein maturation and trafficking, combined with their essential functions, have made these systems attractive targets for developing new antimicrobial agents (19–21).
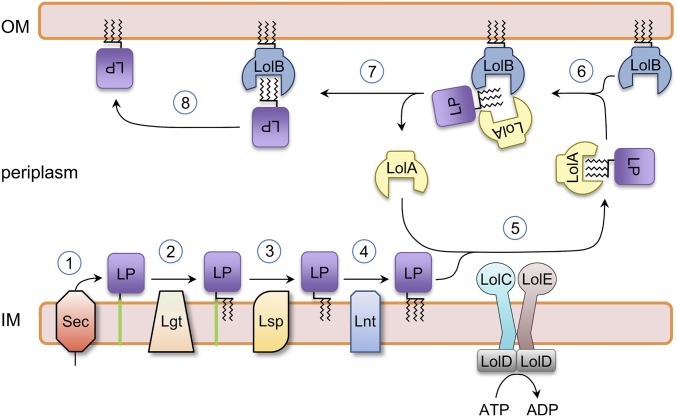
Maturation of bacterial lipoproteins is a multistep process (Fig. 1). Lipoproteins are first produced in “prepro” form in the cytoplasm and require transport across the inner membrane by the Sec pathway (22). Once integrated into the membrane, preprolipoproteins are subject to a series of modifications by enzymes recognizing a cluster of four sequential amino acids, termed the lipobox (22). Addition of the fatty acyl groups is accomplished by the sequential action of three enzymes: Lgt, Lsp, and Lnt. First, Lgt catalyzes addition of diacylglycerol to the lipobox cysteine residue before Lsp removes the N-terminal transmembrane anchor. Finally, Lnt acetylates the N-terminal amino group of the cysteine resulting in the mature, triacylated form (22). Lipoproteins bearing an aspartate at position 2 of the lipobox are retained in the inner membrane (23), and mature lipoproteins destined for the outer membrane are transported by the Lol system, which, in Escherichia coli, is composed of five proteins, LolABCDE (14, 15, 24).
Fig. 1.
Lipoprotein maturation and trafficking in E. coli. Steps 1–8 show a generic lipoprotein (LP) undergoing maturation and transport to the bacterial outer membrane (OM). Immature lipoprotein is secreted by the Sec system and integrated in the inner membrane (IM) (1). Lgt adds diacylglycerol to the lipobox cysteine residue (2). Lsp removes the transmembrane signal peptide (3). Lnt acylates the lipoprotein N terminus amino group (4). LolCDE transfers the mature (triacylated) lipoprotein to the LolA chaperone (5). Lipoprotein is passed from LolA to LolB by a mouth-to-mouth mechanism (6). LolA is recycled, leaving lipoprotein bound to LolB (7). LolB releases lipoprotein to the outer membrane (8).
The LolCDE complex is an ABC transporter that comprises a heterodimer of the transmembrane proteins LolC and LolE, and a homodimer of cytoplasmic LolD, which forms the nucleotide binding domain that hydrolyzes ATP. LolCDE is responsible for the energetically costly extraction of lipoproteins from the inner membrane and their transfer to LolA, a periplasmic chaperone. Lipoproteins bound to LolA are transported across the periplasm and accepted by the outer membrane receptor LolB, itself a lipoprotein, which mediates substrate integration into the outer membrane (14, 16). Although E. coli LolA and LolB have similar β-barrel folds (25), they perform distinct roles (26).
Transfer of lipoproteins between LolA and LolB is proposed to proceed by “mouth-to-mouth” exchange between the central barrels of these proteins (27). NMR and in vivo cross-linking experiments support the mouth-to-mouth model through identification of contacting residues in LolA and LolB that map to the rim of the barrel during complex formation (27, 28). In vivo cross-linking studies have also demonstrated that in E. coli, LolC and LolE have distinct roles. LolC interacts with the LolA chaperone while LolE binds lipoproteins, but the molecular details of these interactions are not clear (27, 29). In other organisms, including pathogens such as Francisella tularensis and Acinetobacter baumannii, such division of labor does not exist and LolF replaces both LolC and LolE in a symmetric, LolDF assembly (30).
The LolCDE complex belongs to the ABC3 superfamily of ABC transporters, which includes the tripartite efflux pump component MacB and the FtsEX cell division machinery (31, 32). Unlike canonical ABC transporters, ABC3 members (also known as type VII ABC transporters) (33) are not proposed to transport substrates across the membrane in which they are embedded. At present, MacB, a toxin and antibiotic transporter (33–35), is the only representative of the ABC3 superfamily to be structurally characterized (33, 36–38). Each monomer of the MacB homodimer has a distinctive four-transmembrane helix topology and an N-terminally fused nucleotide binding domain. A large periplasmic domain, composed of so-called Porter and Sabre subdomains, is elevated approximately 25 Å above the membrane by a helical “stalk” composed of extensions of the first and second transmembrane helices (TM1 and TM2). A shorter periplasmic loop, termed “the shoulder,” links TM3 and TM4. Comparison of ATP-bound (33) and nucleotide-free (37) structures indicates that MacB undergoes impressive conformational changes, termed “mechanotransmission,” during its ATP binding and hydrolysis cycle. Mechanotransmission couples cytoplasmic ATP hydrolysis with conformational changes used to perform work in the extracytoplasmic space (33). LolC and LolE have the same transmembrane topology as MacB (39), and the periplasmic domain has the same fold, with both Sabre and Porter domains evident. It is therefore likely that the mechanotransmission mechanism also underpins extraction and transfer of lipoproteins from the inner membrane to the periplasmic LolA chaperone (33).
In the present study, we define the interaction between LolCDE and LolA using a combination of structural, biochemical, and microbiological techniques. Atomic details of LolA–LolC interaction are captured by X-ray crystallography and the mode of binding is probed and validated using site-directed mutagenesis. We also analyze the nucleotide dependence of LolA binding to LolCDE and evaluate existing biochemical data in context of the complete LolCDE model based on the structure of MacB. Our data provide fundamental insights into bacterial lipoprotein trafficking and may assist the development or improvement of existing Lol-pathway inhibitors.
Results
Structure of LolA Bound to the Periplasmic Domain of LolC.
We determined the crystal structure of LolA in complex with the periplasmic domain of LolC at 2-Å resolution. Crystals of the LolA–LolC complex belong to space group P21212 and contain two complexes per asymmetric unit. Representative electron density for the LolA–LolC structure is shown in Movie S1 and X-ray data and refinement statistics are given in SI Appendix, Table S1. The buried surface area of the LolA–LolC complex is 1,950 Å2, which equates to 9% of the total LolA surface.
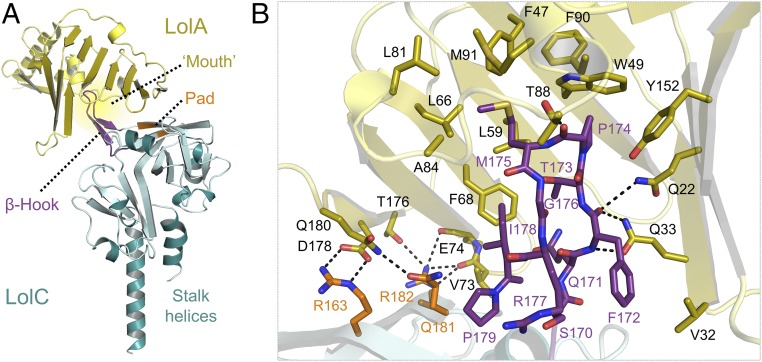
The structure of LolA in complex with the periplasmic domain of LolC is shown in Fig. 2A. The structures of isolated LolA (25) and the LolC periplasmic domain (33) have been described previously. LolA has a barrel-like fold comprised of an 11-stranded antiparallel β-sheet with a short helix located within its center (25), and the LolC periplasmic domain shares its fold with the MacB ABC transporter (33). In the complex, LolC binds to LolA by means of a distinctive β-hairpin structure formed by residues P167–P179 (full-length LolC numbering) and a trio of surface residues (R163, Q181, and R182). We define the hairpin loop of LolC as the “Hook” and the additional surface residues as the “Pad.” The tip of the Hook constitutes a classic type I reverse-turn with M175 and P174 at its apex (Fig. 2B). The tip residues make numerous hydrophobic interactions with LolA, including the side chains of F47, W49, L59, L66, L81, A84, F90, M91, and Y152. The backbone carbonyl of P174 forms a hydrogen bond with T88 of LolA. Hook residues F172, T173, and I178 also interact with residues in the LolA interior, but other residues in the Hook do not. The main chain of F172 is also involved in a hydrogen bonding network with Q22 and Q33 of LolA. The three residues of the Pad contribute to several intermolecular hydrogen bonds and R163 forms a salt bridge with D178 of LolA.
Fig. 2.
Crystal structure of LolA bound to LolC periplasmic domain. (A) Overall structure of the LolA–LolC complex. (B) Close-up view of the interaction interface. LolC and LolA are shown in cyan and gold, respectively. LolC residues belonging to the Hook and the Pad are shown in purple and orange. LolA residues interacting with LolC are shown in stick representation.
The conformation of LolC is not perturbed by interaction with LolA (rmsd of 0.77 Å over 224 residues). Conversely, as a consequence of the interaction with LolC, the LolA chaperone undergoes several conformational changes that are revealed by structural superposition of the LolA–LolC complex with known structures of LolA determined in isolation. SI Appendix, Fig. S1 highlights four regions exhibiting large structural differences including per residue rmsd plots (SI Appendix, Fig. S1A), their mapping to the LolA structure (SI Appendix, Fig. S1B), and close-up structural comparisons (SI Appendix, Fig. S1 C–F). A molecular morph of LolA transiting between LolC-bound and -free states is shown in Movie S2. The key differences in the structures are the widening of the mouth of LolA and displacement of the central helix. Most structural displacements in LolA can be attributed to interactions with the LolC Hook (SI Appendix, Fig. S1 C–E), but residues in the LolA C terminus shift due to their interaction with the LolC Pad (SI Appendix, Fig. S1F).
The Hook and Pad of LolC Are Required for Interaction with LolA.
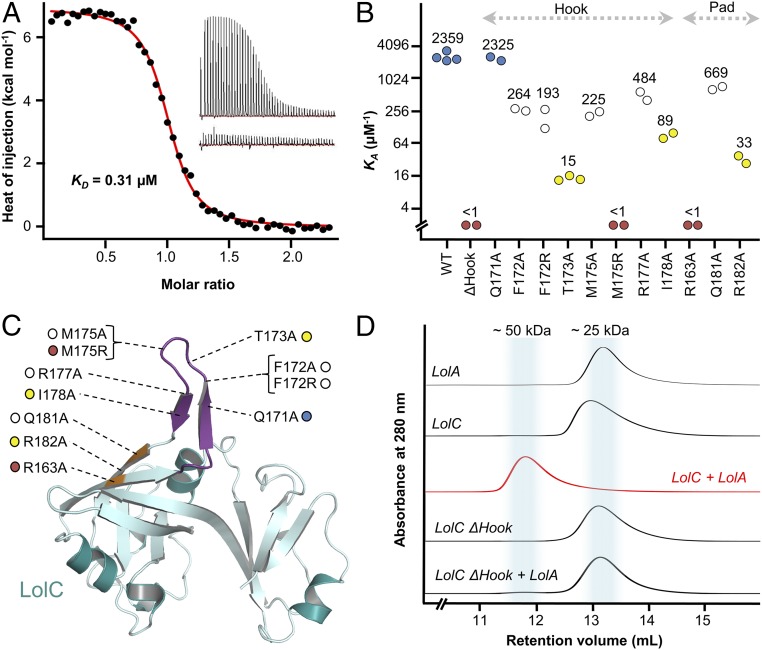
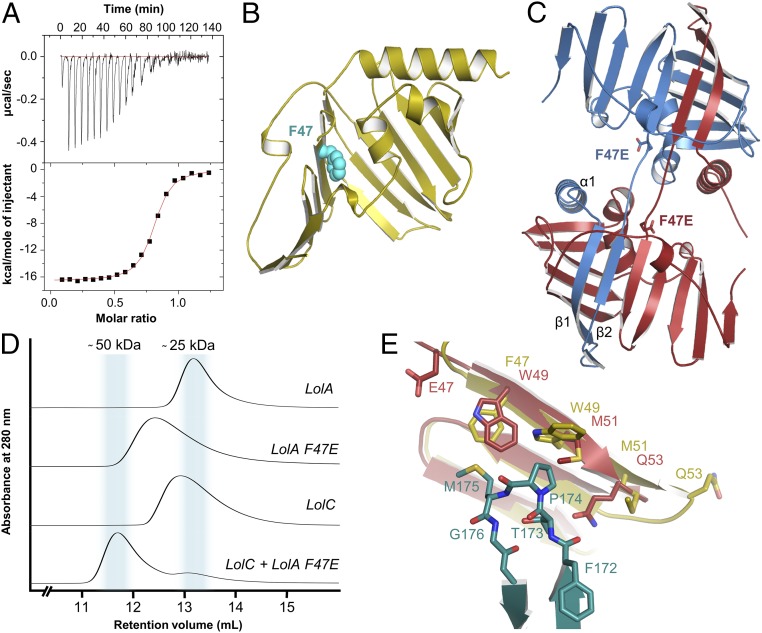
To assess the importance of the Hook and Pad in mediating complex formation between LolA and LolC, we made LolC periplasmic domain variants bearing point mutations in either the Hook or Pad and characterized their interaction with LolA using isothermal titration calorimetry (ITC) and size-exclusion chromatography (SEC). A representative ITC experiment for the interaction of LolA with wild-type LolC is presented in Fig. 3A, with ITC data for the variants summarized in Fig. 3 B and C. The thermodynamic properties extracted from each ITC experiment are given in SI Appendix, Table S2 and examples of raw ITC data and fitted curves for each LolC variant are in SI Appendix, Fig. S2. For wild-type LolC and LolA, we found that the complex is formed with high affinity (KD 405 nM) and has a one-to-one stoichiometry. ITC also shows that complex formation is entropy-driven (ΔH 7.3 and TΔS 16.0), confirming that hydrophobic interactions dominate the binding interface. SEC verifies complex formation between LolA and LolC, with an elution volume for LolA–LolC corroborating the equimolar stoichiometry of the crystal structure and ITC experiments (Fig. 3D).
Fig. 3.
ITC and SEC experiments probing the LolA–LolC interface. (A) Representative ITC experiment demonstrating interaction between LolA and LolC. The main figure shows background-corrected heats of injection and a fitted binding curve (red). (Inset) The two thermograms underpinning this curve are shown; injection of LolC into a cell containing LolA (Upper) and injection of LolC into buffer (Lower). (B) Association constants (KA) for wild-type and variant LolC periplasmic domains with LolA determined using ITC. Median values (μM−1) are indicated above each cluster of repeat experiments. Coloring is used to categorize binding strength of variants: wild-type–like binding, blue; modestly impaired, white; strongly impaired, yellow; and nonbinders, red. (C) Locations of amino acid substitutions in context of the LolC periplasmic domain (Hook, purple; Pad, orange). (D) SEC profiles for indicated proteins.
In contrast to the wild-type, a designed LolC protein construct lacking the Hook (LolC ΔHook) does not form a stable complex with LolA that is detectable by either ITC or SEC (Fig. 3 B and D). We solved the structure of this variant to demonstrate that the inability of LolC ΔHook to bind LolA is not due to loss of structural integrity (SI Appendix, Fig. S3A). Corresponding X-ray data and refinement statistics for the LolC ΔHook protein construct are given in SI Appendix, Table S1, and a close-up of the electron density defining residues in the shortened loop is given in SI Appendix, Fig. S3B. LolC wild-type and ΔHook can be superposed with an rmsd of 0.57 Å for 207 matched Cα positions and inspection of the atomic coordinates reveals no obvious structural differences beyond the absence of the Hook itself (SI Appendix, Fig. S3C).
Having established the importance of the Hook for LolA binding, we next tested the relative importance of its constituent residues. Alanine substitutions of F172, M175, and R177 in the LolC Hook each give nearly 10-fold reductions in affinity for LolA, as measured by ITC (Fig. 3 B and C and SI Appendix, Table S2). T173A and I178A LolC variants are more substantially impaired (160-fold and 25-fold reductions) but the Q171A variant retains wild-type binding characteristics. The pattern of reduced affinity among alanine-substituted Hook variants correlates strongly with the reduction of favorable interactions between LolC and LolA expected from inspection of the LolA–LolC crystal structure. Residues F172, T173, M175, and I178 all make important contributions to the LolA-binding interface that would be diminished by alanine substitution, while Q171 does not make meaningful contact with LolA. Reasons for impaired binding by the R177A variant are not clear as R177 does not contact LolA; however, interactions between R177 and other LolC Hook residues (F172 and S170) suggest a probable role in maintaining Hook structure.
No individual alanine substitution in the Hook was sufficient to prevent binding of LolA to LolC; however, an M175R variant lacks the capacity to bind LolA (Fig. 3 B and C and SI Appendix, Table S2). The location of M175 at the tip of the LolC Hook makes it a critical residue in the LolA–LolC interface, and substitution with arginine disrupts both the hydrophobic character and size of the Hook. The LolC M175R variant is stable and purified in similar yield to wild-type, suggesting that loss of LolA binding is due to steric hindrance and unfavorable electrostatics of the M175R substitution rather than protein misfolding.
The LolC Pad is significantly smaller than the Hook, but mutation of any of its three constituents (R163, Q181, and R182) reduces the affinity for LolA (Fig. 3 B and C and SI Appendix, Table S2). LolC Q181A and R182A variants exhibit 3- and 70-fold reductions in affinity, respectively. R163 is the most important Pad residue, as the alanine variant is unable to bind LolA. Indeed, in the LolA–LolC crystal structure, R163 forms a salt bridge with D178 of LolA, while Q181 and R163 support interfacial hydrogen bonds (Fig. 2B). Overall, the ITC and SEC results demonstrate the importance of the LolC Hook and Pad in mediating interaction with LolA and highlight the roles of M175, T173, I178, R163, and R182 of LolC in the LolA–LolC heterodimer interface.
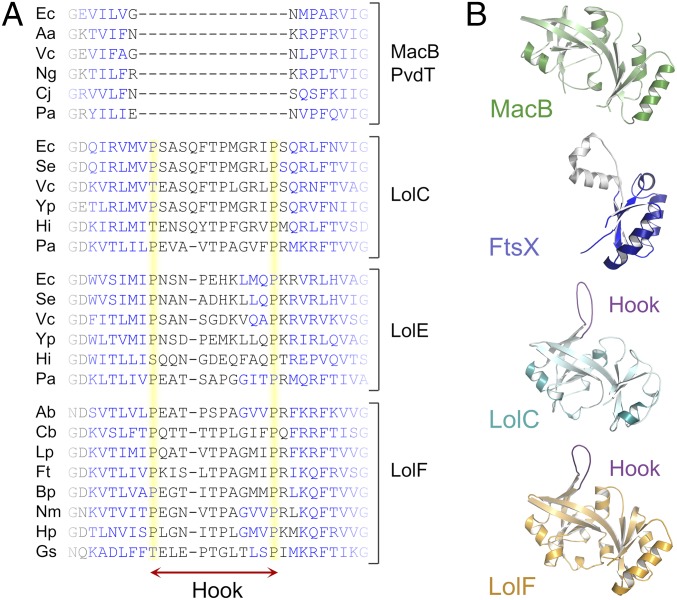
The Hook Is Conserved Among LolC, LolE, and LolF Proteins, but Is Absent from Other ABC Transporters Belonging to the MacB ABC Superfamily.
To establish the generality of the Hook for interaction between LolC and LolA, we examined the amino acid sequences of LolC homologs. We found that a stretch of residues equivalent to the Hook is present in all LolC, LolE, and LolF proteins analyzed, but is absent from the MacB family of efflux pumps (including PvdT) (40) and the FtsEX cell-division machinery (41, 42) (Fig. 4A). Inspection of periplasmic domain structures for LolC, LolF, FtsX, and MacB confirm that this result holds for all available structural data (Fig. 4B). In conclusion, analysis of available homologous sequences and protein structures shows that the Hook is a conserved feature of lipoprotein trafficking machinery that is absent from other members of the type VII ABC transporter superfamily.
Fig. 4.
Structural and bioinformatic evidence that the Hook is conserved among LolC, LolE, and LolF but absent from the wider type VII ABC transporter superfamily. (A) Multiple sequence alignment comparing Lol-family proteins (LolC, LolE, and LolF) with MacB and PvdT in the region of the Hook. Proline residues flanking the Hook are highlighted in yellow, and predicted β-sheets in blue. The full multiple sequence alignment is provided in SI Appendix, Fig. S4. Abbreviations are as follows: Aa, Aggregatibacter actinomycetemcomitans; Ab, Acinetobacter baumannii; Bp, Burkholderia pseudomallei; Cb, Coxiella burnetii; Cj, Campylobacter jejuni; Ec, Escherichia coli; Ft, Francisella tularensis; Gs, Geobacter sulfurreducens; Hi, Haemophilus influenzae; Hp, Helicobacter pylori; Lp, Legionella pneumophila; Ng, Neisseria gonorrhoeae; Nm, Neisseria meningitidis; Pa, Pseudomonas aeruginosa; Se, Salmonella enterica serovar Typhimurium; Vc, Vibrio cholerae; Yp, Yersinia pestis. (B) Comparison of the periplasmic domains of A. actinomycetemcomitans MacB (5LIL), Mycobacterium tuberculosis FtsX (4N8N), E. coli LolC (5NAA), and A. baumannii LolF (5UDF, annotated as LolE in the PDB). LolC and LolF Hooks are shown in purple. FtsX lacks a Sabre domain, the remaining Porter is shown in blue, and pair of helices at the location of the missing Sabre in gray.
The Hook in LolE Does Not Support LolA Binding.
The conservation of a loop of residues in LolE at an equivalent position to the LolC Hook compelled us to test whether LolE is also able to bind LolA. We performed SEC and ITC experiments using a LolE periplasmic domain construct to assess potential LolA binding under the same conditions we observed binding to LolC. We found no evidence that LolA is able to bind the LolE periplasmic domain (SI Appendix, Fig. S5). This result is consistent with previous work showing that E. coli LolC and LolE have different functions (27, 29), and suggests that the specific amino acid sequence of the LolC Hook is crucial for its interaction with LolA. Inspection of the LolE sequence reveals substantial sequence divergence in the Hook and absence of a residue equivalent to R163 of the LolC Pad, despite clear retention of both Porter and Sabre subdomains. We conclude that the interaction between LolA and the LolCDE complex occurs exclusively through LolC and not with LolE, even though LolE is likely to possess the same overall fold as LolC.
LolC Recognizes Features of LolA That Are Absent from LolB.
LolA and LolB possess very similar protein folds (25) but it is not known how (or if) LolC is able to distinguish between these two proteins as binding partners. To address these questions, we evaluated the ability of soluble LolB to interact with the periplasmic domain of LolC by SEC (SI Appendix, Fig. S6A) and an immobilized metal affinity chromatography (IMAC)-based pull-down assay (SI Appendix, Fig. S6B). We did not observe binding between LolC and LolB in either case, even though LolC is able to bind LolA under the same conditions. Relative to LolB, LolA has an extended C terminus that is required for efficient LolA function (43). Our structure reveals that this C-terminal region contains the three residues—T176, D178, and Q180—that underpin interaction with the LolC Pad (Fig. 2B and SI Appendix, Fig. S6 C and D). Sequence alignments confirm that the presence of a C-terminal extension is conserved among LolA proteins but absent from LolB (SI Appendix, Fig. S6 C and D), suggesting that LolC does discriminate between LolA and LolB, and that interaction between the LolC Pad and the C terminus of LolA is essential for chaperone recruitment to the LolCDE complex.
Structural Determination of the F47E LolA Variant Reveals a Domain-Swapped Dimer.
Previous work has shown that an F47E LolA variant is defective in releasing lipoproteins from the bacterial inner membrane and tightly associates with proteoliposomes reconstituted with LolCDE (44). When expressed in vivo, F47E LolA impairs bacterial growth in a dominant-negative fashion. Intrigued by the unusual phenotypic effects of the F47E LolA variant and its effect on the interaction of LolA and LolCDE, we further scrutinized this protein using biophysical methods.
We first measured association of the LolA F47E variant with LolC using ITC and found a twofold higher affinity of LolC for the F47E variant (KD ∼200 nM) compared to that for wild-type LolA (KD ∼405 nM) (Fig. 5A). We then tried to rationalize this observation by inspecting the LolA–LolC crystal structure. The F47 side chain is located within the LolA interior (Fig. 5B), and in the LolA–LolC complex is ∼4 Å from M175 of LolC. A substitution of glutamate for phenylalanine at position 47 does not explain the higher affinity of the LolA variant for LolC because a polar residue would weaken otherwise favorable hydrophobic interactions with LolC. We therefore determined the crystal structure of the F47E variant (X-ray data and refinement statistics in SI Appendix, Table S1). To our surprise, we found that the LolA F47E variant is a domain-swapped dimer (Fig. 5C; electron density in Movie S3). The N-terminal α-helix and first two β-strands from one monomer replace the equivalent elements in the other monomer and the substituting glutamate is shifted away from the LolA cavity into solvent. The SEC elution profile of the F47E variant LolA confirms existence of the domain-swapped state in solution, although the peak is broader than that of the wild-type, and its apparent molecular mass (34 kDa) is smaller than expected from theory (46 kDa) (Fig. 5D). Hypothesizing that the domain-swapped state of the F47E variant may contribute to its unusual properties, we analyzed the F47E structure for features that explain its enhanced affinity for LolC. Inspection revealed that the β-strand on which F47E is located is shifted ∼6 Å relative to that of the wild-type (Fig. 5E). The displacement of this strand affects the position of residues F47, W49, M51, and Q53, all of which face the LolA barrel interior and two of which (F47 and W49) are involved in binding LolC in the wild-type protein. We therefore ascribe the “tight” binding properties of the F47E LolA variant to structural changes in the site that binds LolC resulting from a “strand slip” induced by domain-swap dimerization.
Fig. 5.
Structural and functional analysis of the tight-binding LolA F47E variant. (A) ITC experiment demonstrating binding of LolA F47E to the LolC periplasmic domain. (B) Location of residue F47 in wild-type LolA. (C) Crystal structure of LolA F47E revealing a domain-swapped dimer. (D) SEC experiment for wild-type and LolA F47E variant. (E) Close-up view of LolA F47E variant showing the strand-slip affecting the location of residues E/F47, W49, M51, and Q53. LolA wild-type and F47E are in yellow and red, respectively; LolC Hook is shown in teal.
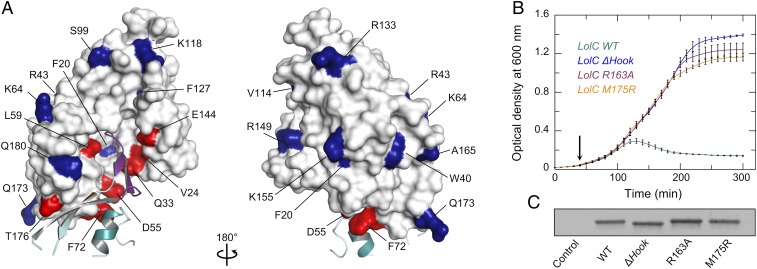
In Vivo Validation of the LolA–LolC Interaction by Mapping Cross-Link Data.
We mapped the locations of LolA residues previously tested for their capacity to form photo-inducible cross-links with LolCDE (27) to our crystal structure of the LolA–LolC complex (Fig. 6A). A full list of the Tokuda laboratory’s cross-linking results (27) alongside nearest-neighbor distances measured from our crystal structure can be found in SI Appendix, Table S3. There is excellent agreement between the in vivo cross-linking experiment and our crystal structure of the LolA–LolC complex. All seven LolA residues that crosslink to LolC are located within the binding interface (Fig. 6A, red). Conversely, residues identified as ineffective in forming cross-links are positioned in regions that do not contact LolC (Fig. 6A, blue). The mapping of previous cross-linking data to our crystal structure of the LolA–LolC complex validates both approaches and confirms that the interface derived here by X-ray crystallography is representative of the state found in vivo.
Fig. 6.
In vivo validation of the LolA–LolC complex. (A) LolA positions determined to interact with LolC by in vivo cross-linking (27) mapped onto the LolA–LolC structure. LolC is represented in cyan with the Hook in purple. LolA is shown as a solid surface, residues reported to form cross-links to LolC are colored red, and those that do not are blue. (B) Growth curves for E. coli C43 (DE3) cells expressing the extracytoplasmic domain of wild-type LolC (or indicated variant) with a periplasmic targeting sequence. Protein expression was induced with 0.2% arabinose at the time point indicated by an arrow. Curves depict the mean ± SD for three independent cultures. (C) Immunoblot showing expression level of periplasmic extracts from E. coli C43 (DE3) cells bearing empty vector (control), or expressing the extracytoplasmic domain of wild-type LolC (WT) or indicated variant.
Mutations in the Hook and Pad of LolC Suppress Dominant-Negative lolD Alleles.
We reexamined data on previously reported LolC and LolE variants that suppress the dominant-negative effects of mutations in the LolCDE ATPase component, LolD (45). These mutants map primarily, although not exclusively, to the periplasmic region of LolC and to the cytoplasmic domains of LolE, suggesting they provide relief from growth arrest by different mechanisms. Our structure shows that two of the suppressor mutations, P174S and G176R, are located within the Hook of LolC and another two, R182C and R182H, are based within the Pad (Fig. 2B). Given the importance of the LolC Hook and Pad for LolA binding, our data predict that these four LolC periplasmic suppressors work by breaking the interaction between LolCDE and LolA to prevent accumulation of nonproductive LolA–LolCDE complexes that otherwise lead to growth arrest. Putting these LolC suppressor mutations into a structural context highlights the importance of the Hook and Pad in mediating LolA recruitment by LolCDE in vivo.
Disruption of the Lol System Using Knowledge of the LolA–LolC Interaction.
To further validate the interaction between LolA and LolC in vivo, we established an inducible plasmid-based system for expressing the LolC extracytoplasmic domain in the periplasm of E. coli with the intent of arresting growth through sequestration of LolA. Expression of the wild-type LolC extracytoplasmic domain in the periplasm produces growth arrest and cell lysis (Fig. 6B). Conversely, expression of variants lacking the Hook, or with single amino acid substitutions in the Hook (M175R) or Pad (R163A) that have been shown to abrogate the interaction between LolA and LolC in vitro, do not lead to growth defects (Fig. 6B), even though they are expressed at similar levels to the wild-type (Fig. 6C). These observations are consistent with growth arrest resulting from sequestration of periplasmic LolA by the overexpressed wild-type LolC periplasmic domain construct that can be relieved by mutations disrupting favorable interactions between LolA and the LolC Hook and Pad.
LolA Binding to LolCDE Is Mediated Purely by Access to the Hook and Pad and Is Independent of the ATP Binding and Hydrolysis Cycle.
To establish the behavior of LolA binding within the context of the LolCDE complex, we immobilized detergent-purified LolCDE variants on Ni-IMAC resin and tested their ability to bind LolA. We also assayed each variant’s ATPase activity using a spectrophotometric assay. Results are summarized for each variant in Table 1, with the supporting data presented in SI Appendix, Fig. S7. We found that LolA binds to the wild-type LolCDE complex irrespective of the presence of nucleotide (Table 1 and SI Appendix, Fig. S7 A and B) and that LolCDE exhibits equivalent ATPase activity in the presence and absence of LolA (SI Appendix, Fig. S7C). Binding to LolA was also unaffected by mutation of a catalytic glutamate in LolD, or by the presence of a nonhydrolyzable nucleotide analog (ATPɣS) (Table 1 and SI Appendix, Fig. S7A). These results suggest that LolA binding to LolCDE is not dependent on the transporter nucleotide status, or its ability to hydrolyze ATP. Purified LolCDE complexes in which the LolC Hook was removed, or in which the Hook or Pad were disrupted maintain their ability to hydrolyze ATP, but are unable to bind LolA (Table 1 and SI Appendix, Fig. S7 D and E). In contrast, deletion of the Hook in LolE does not impair LolA–LolCDE interaction (Table 1 and SI Appendix, Fig. S7D). We conclude that LolA binding to the LolCDE complex occurs exclusively through the Hook and Pad of LolC and is not regulated by nucleotide binding or hydrolysis.
Table 1.
LolCDE functional assays
| Protein | LolA binding | ATPase activity |
| LolCDE (WT) | + | + |
| LolC(ΔHook)DE | − | + |
| LolC(R163A)DE | − | + |
| LolC(M175R)DE | − | + |
| LolCDE(ΔHook) | + | + |
| LolCD(E171Q)E | + | − |
Raw data underpinning this table is provided in SI Appendix, Fig. S7.
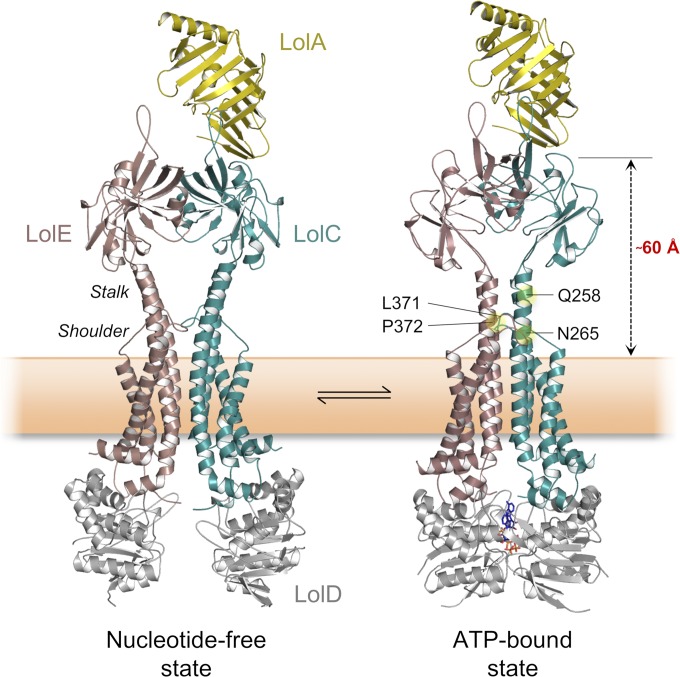
Modeling of the LolA–LolCDE Complex in ATP-Bound and Nucleotide-Free States.
Because of established homology (33), the structure of LolCDE (and its complex with LolA) can be modeled on the basis of available crystal structures of MacB, the LolC periplasmic domain, and the LolA–LolC complex. Such models are useful for contextualizing the LolA–LolC interaction in 3D space, giving clues as to the likely disposition of LolA relative to the membrane and other components of the LolCDE complex. We produced two distinct homology models of LolCDE corresponding to each of the different nucleotide states observed for the structural archetype of the family, MacB (33) (Fig. 7). The models show that binding of LolA to the LolCDE complex is feasible in both ATP-bound and nucleotide-free states, just as we found in our in vitro binding experiments (SI Appendix, Fig. S7). The models also predict LolA to be located ∼60 Å from the cytoplasmic membrane, with the “mouth” of the LolA barrel facing toward the LolE periplasmic domain. This result suggests that lipoproteins need not only be extracted from the inner membrane, but also passed a considerable distance to the waiting LolA chaperone on the top of LolCDE. While molecular details of lipoprotein transfer remain to be determined, the position and orientation of LolA are consistent with lipoprotein delivery via the central cavity between the periplasmic domains of LolC and LolE, perhaps aided by periplasmic conformational changes generated by mechanotransmission.
Fig. 7.
Homology-based models of the LolA–LolCDE complex. Models of full-length LolCDE generated from the nucleotide-free and ATP-bound structures of MacB (5NIL and 5LIL, respectively). LolA has been docked according to LolA–LolC crystallographic data (6F3Z). Positions at which mutations confer resistance to both compound 2 (21) and G0507 (19) inhibitors are shown mapped to the ATP-bound state.
Inhibition of LolCDE by Compound 2 Proceeds by Mechanotransmission Uncoupling.
Homology models of LolCDE and LolA–LolCDE facilitate physical mapping of LolCDE mutations reported to provide resistance to two antimicrobial compounds: pyrrolopyrimidinedione G0507 (19) and pyridineimidazole compound 2 (21) (hereafter, C2). G0507 and C2 are both purported inhibitors of LolCDE with potent antibacterial activity against E. coli strains lacking the tripartite efflux pump component, TolC. The majority of rescuing mutations for both G0507 and C2 cluster within the stalk and shoulder regions of the LolCDE complex, which are spatially close to one another despite separation in primary sequence (Fig. 7, Right). In MacB, the stalk structure is intimately connected with mechanotransmission, suggesting that G0507 and C2 exert their effects by interfering with analogous movements necessary for coupling LolCDE’s cytoplasmic ATPase activity with the lipoprotein transfer reaction. Consistent with this “mechanotransmission uncoupling” as a hypothesis for the action of these inhibitors, G0507 is known to stimulate ATPase activity of LolCDE while inhibiting the release of lipoproteins from the inner membrane (19). Because C2 also inhibits lipoprotein release, we tested its effect on LolCDE ATPase activity and found that, like G0507, C2 causes an increase in the rate of hydrolysis (SI Appendix, Fig. S8A). We also found that C2 does not have any detectable effect on LolA binding to the LolC periplasmic domain or LolCDE, as judged by IMAC-based pull-down experiments, ruling out competition between the inhibitor and the chaperone as an alternative hypothesis (SI Appendix, Fig. S8 B and C).
Discussion
We solved the crystal structure of the periplasmic lipoprotein chaperone, LolA, in complex with the extracytoplasmic domain of LolC (Fig. 2). LolC recruits LolA by means of a finger-like protrusion that we term the Hook and a patch of surface residues termed the Pad. ITC and SEC, coupled with structure-led amino acid substitutions in LolC, demonstrate the importance of these features (Fig. 3), and sequence-based analyses show that the Hook is conserved among LolC proteins but absent from homologous ABC transporters (such as MacB, PvdT, and FtsEX) that do not have a lipoprotein trafficking function (Fig. 4). We uncovered the structural basis for enhanced affinity of the LolA F47E variant (Fig. 5) and validated the native LolA–LolC interface in vivo using cross-linking data from the Tokuda laboratory (27) and a growth inhibition assay (Fig. 6). The interaction between LolC and LolA was confirmed for the detergent-purified LolCDE complex and was demonstrated to be independent of nucleotide binding and hydrolysis (Table 1). Modeling of LolCDE based on crystal structures of the MacB ABC transporter and LolC periplasmic domain predicts the likely structural context of the LolA–LolC interaction and implicates mechanotransmission in lipoprotein extraction and delivery to LolA (Fig. 7). The location of mutations that rescue LolCDE from the chemical inhibitors further suggest such compounds work by interfering directly with mechanotransmission, effectively uncoupling cytoplasmic ATP hydrolysis from periplasmic conformational changes necessary to drive lipoprotein transfer. The combined data give essential mechanistic insights into the progression of lipoproteins from the inner membrane to the periplasmic LolA chaperone during lipoprotein trafficking.
The key features of LolC that underpin binding of LolA are the Hook and Pad. Disruption of either causes substantial reduction in the affinity of LolA for LolC and complex formation is abrogated entirely if the Hook is deleted or if R163 of the Pad is replaced with alanine. These experiments demonstrate that the binding interface of LolC is bipartite and that neither Hook nor Pad alone is sufficient to mediate interaction with LolA. Comparison of the structure of the LolA–LolC complex with that of LolA in isolation reveals significant conformational changes that suggest it may represent a “receptive state” for lipoprotein binding. Several studies implicate the mouth of the LolA barrel as a putative site for lipoyl group interaction (25, 27, 46, 47), meaning that both the Hook and lipoprotein may be in competition for the same binding site. If so, it is plausible that lipoprotein binding to LolA may directly cause release from LolC by displacement of the Hook.
The work presented here establishes the interaction of LolA with LolC as independent of ATP binding and hydrolysis by the LolCDE complex. A key question for LolCDE, therefore, is what the role of energy input is, in vivo. Given that nucleotide cycling is not required for LolA binding, the most likely role for ATP binding and hydrolysis is in driving lipoprotein extraction from the inner membrane. Efforts to determine the role of ATP binding and hydrolysis in the release of lipoproteins from LolCDE have been made previously (48, 49), but molecular details of this process remain obscure. One possibility is that ATP-powered extraction of lipoproteins from the inner membrane by the LolCDE complex uses a mechanotransmission mechanism as described for MacB (33). ATP-bound and nucleotide-free states of MacB have been structurally characterized, revealing long-range conformational changes and extensive periplasmic motions driven by ATP binding and hydrolysis. Similar motions in the LolCDE complex may provide the mechanical force needed to “pull” the lipoprotein from the inner membrane.
Our structural model suggests that LolA is bound as much as 60 Å from the inner-membrane surface. Previous work has shown that LolE is the site of lipoprotein binding (29), but fine details of where the interface is located are yet to be determined. Mechanotransmission-driven parting of the periplasmic domains in LolCDE might expose an intermediate lipoprotein binding site between LolC and LolE periplasmic domains that would provide a “stop-off point” between the membrane and chaperone. Additional experiments will be required to test these hypotheses further.
In summary, we have determined the crystal structure of LolA in complex with the periplasmic domain of LolC and probed the physical basis of the interaction using complementary techniques. We find that complex formation between LolA and LolC is independent of the LolCDE ATP binding and hydrolysis cycle and propose a mechanism where recruitment of LolA to the LolC Hook facilitates presentation to newly extracted lipoproteins, possibly pulled from the membrane in an ATP-dependent manner by a mechanotransmission mechanism resembling that of the MacB ABC transporter.
Methods
Complete methods are available in SI Appendix, Supplemental Methods. In brief, structures of LolA bound to the LolC periplasmic domain, the LolA F47E variant, and the LolC ∆Hook periplasmic domain were each determined by X-ray crystallography. Proteins were expressed in E. coli, purified using Ni-IMAC and crystallized using a sitting-drop vapor-diffusion set-up. Crystals of the LolA–LolC complex were obtained in 100 mM Hepes pH 6.5 and 45% (wt/vol) poly(acrylic acid) 2100. LolA F47E was crystallized in 13% (wt/vol) PEG 8000, 20% (vol/vol) glycerol. The periplasmic domain of LolC ΔHook was crystallized in 30% (wt/vol) PEG 2000 MME, 200 mM ammonium sulfate, 150 mM sodium acetate pH 4.6, assisted by seeds from crystals of the wild-type LolC periplasmic domain obtained previously (33). Crystals were cryoprotected before flash-freezing in liquid nitrogen using the reservoir solution supplemented with either 20% ethylene glycol (LolA–LolC) or 25% (vol/vol) glycerol (LolC ΔHook and LolA F47E). X-ray diffraction data were collected remotely at the European Synchrotron Radiation Facility (France) and Diamond (United Kingdom) synchrotrons. Structure determinations used the CCP4 suite (50). Diffraction data were indexed and reduced with iMOSFLM (51), scaled with Aimless (52), and phased by molecular replacement using Phaser (53). Probes for molecular replacement were derived from PDB ID codes 5NAA (33) and 1IWL (25). Model building and refinement used Coot (54) and Refmac (55). Structure validation was assisted by RAMPAGE (56) and Procheck (57). SEC was performed using an Äkta FPLC equipped with a Superdex 75, 10/300 GL column. Typically, 100 μL of protein at 200 μM was analyzed. ITC experiments were performed using a Microcal VP-ITC instrument. A typical titration used LolA in the cell (25 μM) and LolC variant in the syringe (300 or 450 μM) with 30 × 10-μL injections (reference power 25, 300 rpm stirring, 25 °C). LolA binding to His-tagged LolC periplasmic domain or LolCDE immobilized on IMAC resin was performed using microbatch spin columns. Immobilized proteins were incubated with tag-free LolA for ∼5 min, washed three times, then eluted and visualized by SDS/PAGE. ATPase activity of purified LolCDE variants was assessed using the EnzChek phosphate assay kit (Thermofisher) at 1-μM concentration. Purified LolCDE variants used dodecyl maltopyranoside as a stabilizing detergent. The growth-inhibitory effect of extracytoplasmic targeting of the LolC periplasmic domain was assessed by monitoring OD600 of E. coli C43 (DE3) cultures (58) expressing the wild-type or variant domain fused behind an N-terminal Sec secretion signal.
Supplementary Material
Acknowledgments
We thank Entasis Therapeutics for the gift of LolCDE inhibitor, compound 2; and the staff at the European Synchrotron Radiation Facility (France) and Diamond (United Kingdom) synchrotrons for beamline facilities. This work was supported by grants from the UK Medical Research Council (MR/N000994/1) and the Wellcome Trust (101828/Z/13/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 6F3Z (LolA–LolC complex), 6F49 (LolC ΔHook), and 6FHM (LolA F47E variant)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806822115/-/DCSupplemental.
References
- 1.Konovalova A, Kahne DE, Silhavy TJ. Outer membrane biogenesis. Annu Rev Microbiol. 2017;71:539–556. doi: 10.1146/annurev-micro-090816-093754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson JC, et al. The power of asymmetry: Architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol. 2016;70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 3.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 4.Cascales E, Bernadac A, Gavioli M, Lazzaroni J-C, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol. 2002;184:754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu Y, et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 6.Noinaj N, Gumbart JC, Buchanan SK. The β-barrel assembly machinery in motion. Nat Rev Microbiol. 2017;15:197–204. doi: 10.1038/nrmicro.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 8.Dong H, et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature. 2014;511:52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 9.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abellón-Ruiz J, et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol. 2017;2:1616–1623. doi: 10.1038/s41564-017-0046-x. [DOI] [PubMed] [Google Scholar]
- 11.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabowicz M, Silhavy TJ. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci USA. 2017;114:4769–4774. doi: 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakushi T, Tajima T, Matsuyama S, Tokuda H. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol. 1997;179:2857–2862. doi: 10.1128/jb.179.9.2857-2862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuyama Si, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tajima T, Yokota N, Matsuyama S, Tokuda H. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett. 1998;439:51–54. doi: 10.1016/s0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka K, Matsuyama SI, Tokuda H. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J Bacteriol. 2001;183:6538–6542. doi: 10.1128/JB.183.22.6538-6542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narita S, Tanaka K, Matsuyama S, Tokuda H. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J Bacteriol. 2002;184:1417–1422. doi: 10.1128/JB.184.5.1417-1422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buddelmeijer N. The molecular mechanism of bacterial lipoprotein modification—How, when and why? FEMS Microbiol Rev. 2015;39:246–261. doi: 10.1093/femsre/fuu006. [DOI] [PubMed] [Google Scholar]
- 19.Nickerson NN, et al. A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in Gram-negative bacteria. Antimicrob Agents Chemother. 2018;62:e02151-17. doi: 10.1128/AAC.02151-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nayar AS, et al. Novel antibacterial targets and compounds revealed by a high-throughput cell wall reporter assay. J Bacteriol. 2015;197:1726–1734. doi: 10.1128/JB.02552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLeod SM, et al. Small-molecule inhibitors of gram-negative lipoprotein trafficking discovered by phenotypic screening. J Bacteriol. 2015;197:1075–1082. doi: 10.1128/JB.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 23.Masuda K, Matsuyama S, Tokuda H. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc Natl Acad Sci USA. 2002;99:7390–7395. doi: 10.1073/pnas.112085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol. 2000;2:212–218. doi: 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 25.Takeda K, et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 2003;22:3199–3209. doi: 10.1093/emboj/cdg324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukahara J, Mukaiyama K, Okuda S, Narita S, Tokuda H. Dissection of LolB function—Lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 2009;276:4496–4504. doi: 10.1111/j.1742-4658.2009.07156.x. [DOI] [PubMed] [Google Scholar]
- 27.Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA. 2009;106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakada S, et al. Structural investigation of the interaction between LolA and LolB using NMR. J Biol Chem. 2009;284:24634–24643. doi: 10.1074/jbc.M109.001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizutani M, et al. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex. FEBS Lett. 2013;587:23–29. doi: 10.1016/j.febslet.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 30.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS., Jr Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol. 2015;197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- 32.Khwaja M, Ma Q, Saier MH., Jr Topological analysis of integral membrane constituents of prokaryotic ABC efflux systems. Res Microbiol. 2005;156:270–277. doi: 10.1016/j.resmic.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Crow A, Greene NP, Kaplan E, Koronakis V. Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc Natl Acad Sci USA. 2017;114:12572–12577. doi: 10.1073/pnas.1712153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183:5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol. 2008;190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada U, et al. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat Commun. 2017;8:1336. doi: 10.1038/s41467-017-01399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitzpatrick AWP, et al. Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat Microbiol. 2017;2:17070. doi: 10.1038/nmicrobiol.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greene NP, Kaplan E, Crow A, Koronakis V. Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front Microbiol. 2018;9:950. doi: 10.3389/fmicb.2018.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuda M, Iguchi-Yokoyama A, Matsuyama S, Tokuda H, Narita S. Membrane topology and functional importance of the periplasmic region of ABC transporter LolCDE. Biosci Biotechnol Biochem. 2009;73:2310–2316. doi: 10.1271/bbb.90451. [DOI] [PubMed] [Google Scholar]
- 40.Imperi F, Tiburzi F, Visca P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2009;106:20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang DC, et al. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mavrici D, et al. Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc Natl Acad Sci USA. 2014;111:8037–8042. doi: 10.1073/pnas.1321812111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuda S, Watanabe S, Tokuda H. A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins. FEBS Lett. 2008;582:2247–2251. doi: 10.1016/j.febslet.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Miyamoto A, Matsuyama S, Tokuda H. Dominant negative mutant of a lipoprotein-specific molecular chaperone, LolA, tightly associates with LolCDE. FEBS Lett. 2002;528:193–196. doi: 10.1016/s0014-5793(02)03305-7. [DOI] [PubMed] [Google Scholar]
- 45.Ito Y, Matsuzawa H, Matsuyama S, Narita S, Tokuda H. Genetic analysis of the mode of interplay between an ATPase subunit and membrane subunits of the lipoprotein-releasing ATP-binding cassette transporter LolCDE. J Bacteriol. 2006;188:2856–2864. doi: 10.1128/JB.188.8.2856-2864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oguchi Y, et al. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release. J Biol Chem. 2008;283:25414–25420. doi: 10.1074/jbc.M804736200. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe S, Oguchi Y, Takeda K, Miki K, Tokuda H. Introduction of a lethal redox switch that controls the opening and closing of the hydrophobic cavity in LolA. J Biol Chem. 2008;283:25421–25427. doi: 10.1074/jbc.M804737200. [DOI] [PubMed] [Google Scholar]
- 48.Ito Y, Kanamaru K, Taniguchi N, Miyamoto S, Tokuda H. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol Microbiol. 2006;62:1064–1075. doi: 10.1111/j.1365-2958.2006.05378.x. [DOI] [PubMed] [Google Scholar]
- 49.Taniguchi N, Tokuda H. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J Biol Chem. 2008;283:8538–8544. doi: 10.1074/jbc.M800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lovell SC, et al. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 57.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 58.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.