Significance
Phosphorylation of tyrosine residues in the cytoplasmic tail of the epidermal growth factor receptor (EGFR) by its kinase domain propagates a rich variety of information downstream of growth factor binding. The amino acid sequences surrounding each phosphorylation site encode the extent of phosphorylation as well as the extent of binding by multiple effector proteins. By profiling the kinase activity of EGFR alongside the binding specificities of an SH2 domain and a PTB domain for thousands of defined phosphorylation site sequences, we discovered that the sequences surrounding the phosphorylation sites in EGFR are not optimal and that discrimination against phosphorylation by cytoplasmic tyrosine kinases such as c-Src and c-Abl is likely to have shaped the evolution of these sequences.
Keywords: EGFR, SH2, specificity, deep mutational scanning, signaling
Abstract
Upon activation, the epidermal growth factor receptor (EGFR) phosphorylates tyrosine residues in its cytoplasmic tail, which triggers the binding of Src homology 2 (SH2) and phosphotyrosine-binding (PTB) domains and initiates downstream signaling. The sequences flanking the tyrosine residues (referred to as “phosphosites”) must be compatible with phosphorylation by the EGFR kinase domain and the recruitment of adapter proteins, while minimizing phosphorylation that would reduce the fidelity of signal transmission. To understand how phosphosite sequences encode these functions within a small set of residues, we carried out high-throughput mutational analysis of three phosphosite sequences in the EGFR tail. We used bacterial surface display of peptides coupled with deep sequencing to monitor phosphorylation efficiency and the binding of the SH2 and PTB domains of the adapter proteins Grb2 and Shc1, respectively. We found that the sequences of phosphosites in the EGFR tail are restricted to a subset of the range of sequences that can be phosphorylated efficiently by EGFR. Although efficient phosphorylation by EGFR can occur with either acidic or large hydrophobic residues at the −1 position with respect to the tyrosine, hydrophobic residues are generally excluded from this position in tail sequences. The mutational data suggest that this restriction results in weaker binding to adapter proteins but also disfavors phosphorylation by the cytoplasmic tyrosine kinases c-Src and c-Abl. Our results show how EGFR-family phosphosites achieve a trade-off between minimizing off-pathway phosphorylation and maintaining the ability to recruit the diverse complement of effectors required for downstream pathway activation.
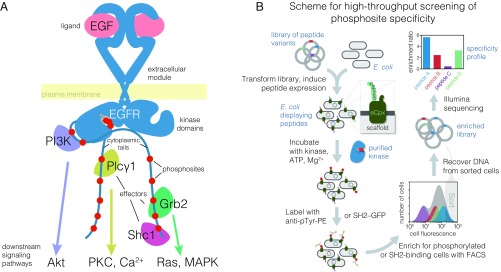
The epidermal growth factor receptor (EGFR) is a tyrosine kinase that couples extracellular ligand binding to the activation of intracellular signaling pathways (1–3). Activation of human EGFR (also called “ErbB1” or “human EGF receptor 1,” Her1), results from ligand-induced homodimerization or heterodimerization with one of three family members, Her2/ErbB2/neu, Her3/ErbB3, or Her4/ErbB4 (4). Allosteric activation of one kinase domain within this dimer by the other kinase domain results in autophosphorylation of multiple tyrosines within the C-terminal tails of both monomers (5). The resulting phosphotyrosine and flanking residues (phosphosites) can then serve as binding sites for intracellular proteins containing Src homology 2 (SH2) or phosphotyrosine-binding (PTB) domains (6, 7). With their enzymatic or scaffolding activities recruited to the plasma membrane, these effector proteins propagate signals inside the cell (Fig. 1A) (8).
Fig. 1.
Overview of EGFR signal transduction at the membrane and a bacterial surface display scheme to analyze the specificity of tyrosine kinases and phosphotyrosine-binding proteins. (A) Illustration of membrane-proximal EGFR signaling components. Autophosphorylation of the tyrosine phosphosites in the C-terminal cytoplasmic tail (red circles) by the activated kinase domain produces binding sites for many downstream effectors, a subset of which are depicted. These effectors go on to activate second-messenger pathways, also depicted. Grb2, growth factor receptor-bound protein 2; MAPK, mitogen-activated protein kinases; PI3K, phosphoinositide 3-kinase regulatory subunit; PKC, protein kinase C; Plcγ1, phospholipase C-gamma-1; Shc1, SH2 domain-containing-transforming protein C1. (B) Workflow for determining phosphosite specificity profiles of tyrosine kinases and phosphotyrosine-binding proteins by bacterial surface display coupled with FACS and deep sequencing. Phosphotyrosine on the surface of the cells is detected either by immunostaining with an anti-phosphotyrosine antibody or, for binding profiles, with either a tandem SH2 or PTB construct fused to GFP. The frequency of each peptide-coding sequence in the highly phosphorylated population, or enrichment, and thus the relative efficiency of phosphorylation or binding for each peptide, is determined by counting the number of sequencing reads for each peptide in the sorted and unsorted populations.
Tyrosine kinases recognize their substrates through the formation of a short, antiparallel, β-stranded interaction between the substrate peptide and the activation loop of the kinase domain (9). The kinase domain provides limited opportunity for stereospecific engagement of substrate side-chains. This fact contributes to the impression that tyrosine kinases are “sloppy enzymes” (10, 11) and is consistent with the failure to develop high-affinity substrate-mimicking inhibitors. The catalytic domains of tyrosine kinases do have intrinsic preferences for some substrate sequences over others, with specificity being determined by the pattern of amino acid residues directly adjacent to the tyrosine (12, 13).
For EGFR-family members, the high local concentration of tail phosphosites with respect to the kinase domains may allow these sites to be phosphorylated without much regard for sequence. This made us wonder about the extent to which each phosphosite in the cytoplasmic tails of EGFR-family members is optimized in its sequence for the recruitment of specific adaptor proteins versus phosphorylation efficiency by the EGFR kinase domain. The subset of tail tyrosines that are phosphorylated during EGFR signaling is thought to define the subset of downstream pathways that are activated, although the extent to which the intrinsic specificity of the EGFR kinase domain allows discrimination between different phosphosites in the tail has not been mapped (14, 15). The properties that determine the efficiency of binding of SH2 or PTB domains to EGFR-family phosphosites are also not completely understood, because several SH2 or PTB domains can bind to a particular phosphosite, and each SH2 and PTB domain can use multiple different phosphosites (16–20).
Another layer of complexity in EGFR signaling is the potential for cross-talk with cytoplasmic tyrosine kinases, particularly the ubiquitously expressed kinases c-Src and c-Abl (21, 22). Direct phosphorylation of EGFR by c-Src might allow transactivation of EGFR in the absence of growth factors (23–25). Recently, c-Src has been shown to supply a priming phosphorylation that improves EGFR catalytic efficiency for a phosphosite with two neighboring tyrosine residues in the adapter protein Shc1 (26). Despite the evident importance of c-Src in EGFR signaling, very few phosphosites in EGFR-family members have been shown to be direct substrates of c-Src (27–29). In particular, the inhibition of Src-family kinases has little effect on the phosphorylation of EGFR tail phosphosites (30). We therefore wondered whether there are mechanisms that insulate phosphosites in the EGFR-family receptors from phosphorylation by c-Src and other cytoplasmic tyrosine kinases.
In this work, we address several questions about the phosphosites in the cytoplasmic tails of EGFR-family receptors. What is the intrinsic specificity of the EGFR kinase domain with respect to all potential substrates? How does the intrinsic specificity of EGFR map onto tail phosphosites, and how is this specificity differentiated from that of c-Src and c-Abl? How has the specificity of SH2 and PTB domains impinged on the evolution of the sequences of EGFR-family tail phosphosites? To answer these questions, we assayed the activity of EGFR against thousands of peptides with defined sequences representing tyrosine phosphorylation sites in the human proteome, using a high-throughput method based on bacterial surface display of peptides and deep sequencing (Fig. 1B). This method was used recently to determine the mechanistic basis for the orthogonal specificity of two kinases, Lck and ZAP-70, in the T cell receptor pathway (31) and to map the specificity of kinases in the Src family (32). To delineate the specificities of SH2 and PTB domains, we augmented the bacterial surface display system to measure protein binding instead of phosphorylation efficiency.
Deep mutational scanning of EGFR phosphosites with selection based on either phosphorylation or adapter protein binding revealed specificity determinants that differ from optimal motifs observed in previous studies (12, 26, 33). We find that phosphosites in the tails of EGFR-family members avoid some sequence features that are consistent with efficient phosphorylation by EGFR and efficient binding of the Shc1 PTB domain and the Grb2 SH2 domain. The sequence features that are avoided in the tails would, if present, promote phosphorylation by c-Src. Thus, our studies of specificity in the context of natural phosphosite sequences have uncovered evidence of evolutionary trade-offs between on-pathway phosphorylation and binding and the suppression of potentially interfering reactivity in EGFR signaling.
Results and Discussion
Specificity Profile of the EGFR Kinase Domain Derived from a Library of Tyrosine Phosphorylation Sites Found in the Human Proteome.
Previous studies of tyrosine kinase specificity have been based primarily on degenerate peptide libraries in which a tyrosine residue is flanked by random amino acid residues except at one defined position (34). This approach assesses the sufficiency of particular types of residues at specific sites to confer phosphorylation by the kinase of interest. To gain an understanding of EGFR kinase specificity in the context of defined sequences, we developed a high-throughput assay based on bacterial surface display, FACS (35, 36), and deep sequencing to screen for efficiently phosphorylated tyrosine phosphosites (Fig. 1B) (31). This multiplexed bacterial surface display kinase assay has been used recently to characterize tyrosine kinase specificity in the T cell receptor-signaling pathway (31) and to analyze tyrosine kinase specificity on substrates spanning the human proteome (32).
We used this method to screen the EGFR kinase against a library of 15-residue tyrosine-containing peptides referred to as the “Human-pTyr library.” This library corresponds to a diverse set of ∼2,600 tyrosine-containing sequences from the human proteome that have been reported as tyrosine kinase substrates in the PhosphoSitePlus (37) or UniProt (38) databases (Methods and ref. 32). Briefly, Escherichia coli cells displaying individual peptides from the Human-pTyr library on their surfaces were subjected to phosphorylation by the purified EGFR kinase. The cells were then labeled with an anti-phosphotyrosine antibody, and the highly phosphorylated population was enriched by FACS. The abundances of each peptide-coding DNA sequence in the sorted and unsorted samples were inferred by their read frequencies in high-throughput sequencing. The ratio of sorted over input read frequency gives an enrichment score for each peptide. This score correlates well with in vitro measurement of the specific activity of the kinase at low peptide concentrations relative to expected KM values across a wide dynamic range (SI Appendix, Fig. S1A), indicating that it is a good measure of catalytic efficiency. The library contains ∼700 sequences with more than one tyrosine residue in a 15-residue stretch, and these were analyzed separately.
A key step in our analysis of EGFR specificity was the use of a soluble, dimeric form of the EGFR intracellular module. The isolated EGFR kinase domain has been shown to have ∼15-fold higher specific activity when forced to dimerize on lipid vesicles compared with the activity of the monomeric kinase domain (5, 39, 40). We designed constructs that consist of the EGFR intracellular module fused C-terminally to the proteins FKBP and FRB, which, when mixed together along with rapamycin, exhibit an ∼30-fold higher specific activity than either protein alone (SI Appendix, Fig. S1B). This soluble construct also includes part of the juxtamembrane region, the kinase domain, and the full-length cytoplasmic tail (see SI Appendix for details). For this synthetically dimerized construct, the values of kcat/KM against the peptides we tested ranged from ∼30–400⋅min−1⋅mM−1, as estimated from steady-state reactions at low peptide concentration (SI Appendix, Fig. S1A). This is comparable to or slightly higher than that reported for detergent-solubilized full-length EGFR bound to EGF (41–43). As discussed below, this construct also has specific activity against preferred substrates comparable to that of the kinase domain of c-Src against its preferred substrates.
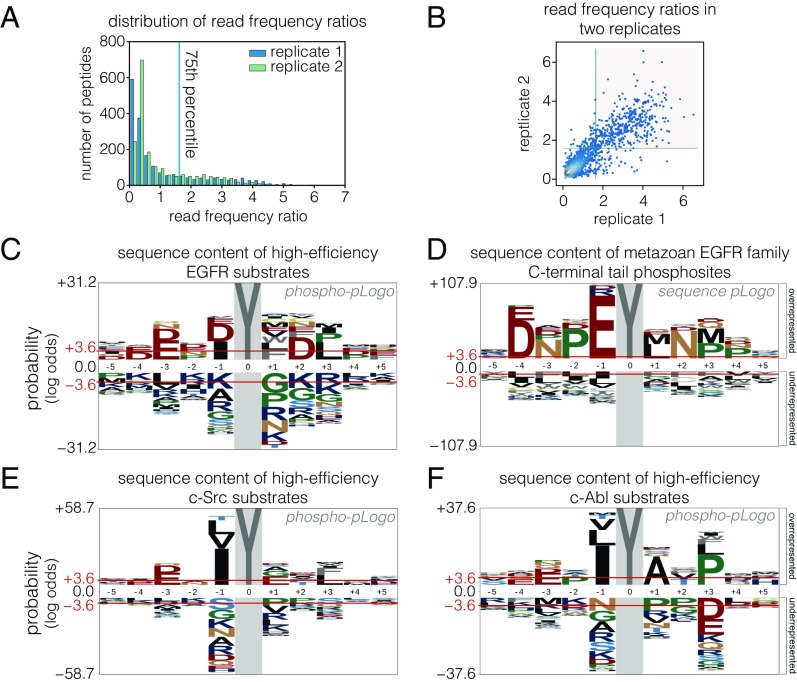
The distribution of phosphorylation enrichment values for single-tyrosine peptides in the Human-pTyr library screened against human EGFR kinase is centered close to zero (Fig. 2A). The distribution has a long tail toward higher values, suggesting that EGFR phosphorylates most sites poorly and phosphorylates a relatively narrow subset efficiently. To determine the sequence features that underlie efficient phosphorylation by EGFR, we analyzed the positional enrichment of amino acid residues in peptides in the top quartile of enrichment ratios in two replicates (Fig. 2B). The data are displayed using a probability sequence logo diagram (pLogo), which compares the frequency of an amino acid residue at each position in the set of efficiently phosphorylated sequences with the frequency of that residue in the same position with respect to tyrosine residues in a reference database of tyrosine-containing sequences (44). For clarity, we refer to pLogos generated from sequences that are filtered by experimental data on phosphorylation efficiency as “phosphorylation-probability Logos” (phospho-pLogos) and those based on a collection of sequences derived from bioinformatics analysis as “sequence-probability logos” (sequence-pLogos).
Fig. 2.
Comparison of intrinsic EGFR and c-Src substrate specificity with EGFR-family phosphosite sequences. (A) Histogram of peptide read frequency ratios from EGFR phosphorylation of a library of human phosphosites obtained by bacterial surface display and deep sequencing. The distribution of ratios of read frequencies for input and sorted samples are plotted from two replicate experiments. (B) Read-frequency ratios for two replicate Human-pTyr library phosphorylation experiments plotted against each other. Peptides with ratios above the 75th percentile in both replicates (gray box) were counted as highly phosphorylated in C. (C) Phospho-pLogo of highly phosphorylated peptides for EGFR in the bacterial surface display experiment. The height of each letter corresponds to the negative log-odds ratio of binomial probabilities of finding a given amino acid residue at a particular sequence position at higher versus lower frequencies than the expected positional frequency for all peptides in the library. Higher values indicate an enrichment of a residue versus the background distribution. Red lines indicate the log-odds ratio values for a significance level of 0.05, as defined in ref. 44. (D) Sequence-pLogo of EGFR-family C-terminal tail tyrosines. Sequence segments surrounding tyrosine were extracted from the regions C-terminal to the kinase domain for metazoan EGFR-family protein sequences. The positional amino acid frequency in these segments was compared with the frequency in metazoan intracellular and transmembrane proteins and was plotted as a pLogo. (E and F) Phospho-pLogo of highly phosphorylated sequences from c-Src phosphorylation (E) and c-Abl phosphorylation (F) of the Human-pTyr library (raw data are from ref. 32). Sequences above the 75th percentile in three replicates are included in the highly phosphorylated set.
Tyrosine kinases have a general preference for peptides with negatively charged residues located before the tyrosine (45). Consistent with this, acidic residues are enriched and basic residues are depleted in the phospho-pLogo diagram corresponding to the set of peptides that are phosphorylated efficiently by EGFR (Fig. 2C). The phosphorylation motif determined for EGFR in earlier work using oriented peptide libraries (12), with acidic residues before the tyrosine and a hydrophobic residue in the position immediately after the tyrosine (the +1 position), is also apparent. In our data, peptides containing a −1 acidic/+1 hydrophobic motif are significantly more likely to be in the highly phosphorylated set (P < 10−16, Fisher’s exact test), suggesting that this feature is predictive of efficient phosphorylation by EGFR. Unexpectedly, peptides with a −1 isoleucine or +3 leucine residue are also significantly enriched among sites that are phosphorylated efficiently by EGFR (P < 10−14, Fisher’s exact test). Sequences with a leucine at the +3 position would be compatible with the binding of several SH2 domains, such as those of c-Src and phosphatidylinositol-3′-kinase (46), suggesting convergence in specificity between EGFR phosphorylation and SH2 domain binding in this instance.
When peptides with more than one tyrosine are included in the analysis, tyrosine is enriched significantly at multiple positions in the resulting phospho-pLogo (SI Appendix, Fig. S2). This might reflect, in part, the recently described preference of EGFR for a Tyr–phospho-Tyr motif (26); we do not see an equivalent enrichment of tyrosine residues in the phospho-pLogo diagrams for other tyrosine kinases tested using the Human-pTyr library and bacterial surface display (32). The analysis in this paper is restricted to the set of sequences that contain only one tyrosine residue at the central position. This includes many phosphosites in which a secondary tyrosine is mutated to alanine in the library.
Phosphosites in the Cytoplasmic Tails of EGFR-Family Members Represent a Subset of the Sequence Patterns That Are Phosphorylated Efficiently by EGFR.
We next compared the sequence patterns of the highly phosphorylated peptides in the screen of human phosphosites with the sequence patterns of phosphosites in members of the EGFR family. We analyzed the positional amino acid enrichment in the 10 residues flanking each tyrosine in the tails of a diverse collection of 87 EGFR-family members from across metazoan evolution. This is displayed as a sequence-pLogo diagram in Fig. 2D. The sequence-pLogo diagram for tail phosphosites is different from the phospho-pLogo for efficiently phosphorylated EGFR substrates. Note, however, that the conserved features of the tail sequences comprise a subset of the sequence features that define efficient phosphorylation by the EGFR kinase domain. The major difference between the two logos arises from the enrichment of a central EYL motif in the phosphosites of EGFR-family tails. This EYL motif is compatible with the phospho-pLogo for EGFR (Fig. 2C) as well as with the optimal EEEEYFLVE motif reported for EGFR based on in vitro phosphorylation of degenerate peptide libraries (12, 47). The convergence of EGFR-family phosphosites on the EYL motif implies that efficient phosphorylation by EGFR is an important evolutionary pressure shaping these sequences.
Peptides with an isoleucine residue rather than a glutamate at the −1 position are also phosphorylated efficiently by EGFR, but a hydrophobic residue is very rarely found at the −1 position in the tails of EGFR-family members (compare Fig. 2 C and D). The preferred motifs for phosphorylation by cytoplasmic tyrosine kinases often consist of hydrophobic residues at the −1 and +3 positions (12, 13, 47). The phospho-pLogo diagrams for c-Src and c-Abl phosphorylation of the Human-pTyr library confirm the importance of a large hydrophobic residue at the −1 position for efficient phosphorylation (Fig. 2 E and F) (32). c-Src and c-Abl also disfavor large hydrophobic residues at the +1 position (Fig. 2 E and F), which are overrepresented in the sequence-pLogo for EGFR-family tail phosphosites. These observations suggest that the EYL motif in EGFR-family tails may have arisen as an evolutionary adaptation to minimize phosphorylation by cytoplasmic tyrosine kinases such as c-Src and c-Abl.
Other differences between the phospho-pLogo for efficient EGFR phosphorylation and the sequence-pLogo for EGFR-family tail phosphosites arise from the clear imprint of the binding motifs for SH2 and PTB domains in the tail phosphosites. For example, the binding motifs for the SH2 domains of Grb2 (pYxN, where x is any residue) and phosphatidylinositol-3′-kinases (pYxxΦ, where Φ is any hydrophobic residue) (48, 49), as well as for the PTB domains of Shc and Dok proteins (NPxpY) (7, 50), are highly enriched in the sequence-pLogo (Fig. 2D). Because the sequence-pLogo is derived from EGFR-family sequences spread across the metazoan lineage, this result points to a conservation in the core specificity determinants of SH2 and PTB domains across evolution.
Saturation Mutagenesis of EGFR Tail Phosphosites.
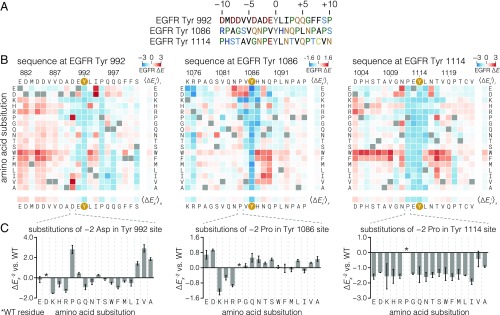
We performed mutational screens to assess the contribution to phosphorylation efficiency of each position in three phosphosites in the EGFR tail (Tyr-992, Tyr-1086, and Tyr-1114) (Fig. 3A). Of these, the sequence flanking Tyr-992 is most similar to the phospho-pLogo derived from the Human-pTyr library (Fig. 2C) and previously published motifs, with an EYL motif and mostly acidic residues upstream of the tyrosine (12, 26). The other two sites, spanning Tyr-1086 and Tyr-1114, both contain an NPxY motif, the signature of PTB-domain binding, as well as a +2 asparagine, which is the main determinant for Grb2 SH2 binding. These two sites differ in their −1 and +1 residues, however. The Tyr-1114 phosphosite contains the consensus EYL motif, while Tyr-1086 diverges from this consensus, with valine and histidine at the −1 and +1 positions, respectively.
Fig. 3.
Effect of single amino acid substitutions on the phosphorylation of three EGFR phosphosite peptides by EGFR. (A) Sequences of three human EGFR C-terminal tail phosphosites. (B) Heat maps showing the effect of all single amino acid substitutions (except tyrosine and cysteine) on the phosphorylation level of three EGFR phosphosite peptides relative to the wild-type peptide upon phosphorylation by EGFR, measured by bacterial surface display and deep sequencing. Squares for each substitution x at each wild-type position i are colored as log-twofold enrichment relative to wild type (∆Exi), calculated from read frequency ratios of sorted and input samples. Wild-type residue squares (∆Ewti = 0 by definition) are indicated by gray squares. The ∆E scales for each peptide, displayed in the top right corner of each heat map, are not directly comparable because different optimized cell-sorting parameters were used for each peptide. Red and blue colors indicate variants that were phosphorylated more or less, respectively, than the wild-type sequence. Row and column mean ∆E values are displayed separately. Data are the variantwise mean of at least two replicates. (C) Enrichment values for the −2 column (∆Ex−2) for each peptide. Error bars indicate the SEM.
We used the surface display/deep sequencing assay to measure the effect of every amino acid substitution along 21-residue stretches spanning these three EGFR phosphosites. The results are presented in Fig. 3B as heat maps of enrichment values relative to the wild-type peptide for each position in the wild-type sequence (rows) and each substitution to one of 17 other amino acids (excluding tyrosine and cysteine; see Methods) (columns). Mean values across columns and rows are shown as separate bars to the right of and below the main heat maps. The column mean denotes the average effect of perturbing a specific position, whereas the row mean indicates the impact of introducing a specific residue type into the peptide. As expected, mutating the central tyrosine to other residues produces low enrichment relative to the wild-type sequence. Expression levels for each mutant in the Tyr-1114 matrix, measured by cell sorting and deep sequencing, varied marginally compared with the differences in enrichment scores attributed to phosphorylation (SI Appendix, Fig. S3).
A readily apparent feature of all three substitution matrices is the preponderance of positive enrichment values at many different positions. This indicates that the wild-type residues at these positions are not optimal for EGFR phosphorylation. For instance, almost any substitution of an acidic residue 5–10 positions before Tyr-992 increases phosphorylation relative to the wild-type sequence. For the Tyr-992 peptide, these acidic residues are part of an “electrostatic hook” element that is implicated in the suppression of kinase activity in the full-length receptor (39, 51, 52). This suggests that the regulatory function of these residues provides an evolutionary constraint on the sequence at this phosphosite, at the expense of phosphorylation efficiency.
The three mutational datasets indicate that the EGFR kinase domain does not have sharply defined specificity. In only a few cases are the majority of substitutions at any one position detrimental, notably at the −2, −1, and +1 positions of the Tyr-1114 phosphosite. Substitutions of the −1 or +1 residues away from glutamate and leucine, respectively, in both the Tyr-992 and Tyr-1114 peptides negatively affect phosphorylation at these sites, in agreement with the emergence of this motif in the Human-pTyr library screen (Fig. 2C). In further agreement with the Human-pTyr screen, substitutions of the −1 residue by isoleucine either increase phosphorylation (Tyr-992 and Tyr-1086) or reduce it only slightly (Tyr-1114).
Phosphorylation by EGFR of Peptides Containing Either Acidic or Large Hydrophobic Residues at the −1 Position May Reflect Alternate Conformations of the Bound Substrate.
The EGFR kinase domain clearly possesses a degree of selectivity at the −1 position of the substrate, as is evident in all three mutational matrices. It is unexpected, however, that both negatively charged residues (aspartic acid, glutamic acid) and hydrophobic residues (isoleucine, methionine, valine) are permitted at the −1 position in EGFR substrates. A clue to the origin of the dual specificity of EGFR for residues at the −1 position in substrates comes from noting that the effect of replacing the residue at the −2 position depends on whether the residue at the −1 position is acidic or hydrophobic (Fig. 3C). When the residue at the −1 position is acidic, as in the Tyr-992 and Tyr-1114 phosphosites, then either proline or a residue with a β-branched side-chain, such as isoleucine or valine, is strongly preferred at the −2 position. For example, the wild-type Tyr-992 phosphosite has an aspartate at the −2 position, and substitution of this residue by valine or proline leads to increased phosphorylation in the mutational screen. Purified peptides corresponding to the Tyr-992 site with substitutions to proline or valine at the –2 position are also phosphorylated more efficiently than the wild-type peptide in an in vitro kinase assay, confirming that the results from the surface display screening assay are not due to differences in the display levels of the mutant peptides (SI Appendix, Fig. S4). If, however, the residue at the −1 position is hydrophobic, as in the Tyr-1086 phosphosite, there is hardly any constraint on the nature of the residue at the −2 position (Fig. 3C, Center).
We used all-atom molecular dynamics simulations to analyze the conformational space sampled by a substrate peptide. We generated a 200-ns trajectory for an isolated peptide in water, with the sequence of the Tyr-1114 phosphosite without the kinase domain. In this simulation, the residues at the +1 to +4 positions with respect to the tyrosine were constrained to be in the β-conformation, corresponding to how these residues interact with the activation loops of tyrosine kinases (9, 53). We docked instantaneous structures sampled from the simulations onto the EGFR kinase domain [Protein Data Bank (PDB) ID code 2GS6] using the residues at the +1 to +4 positions as in ref. 5. We then examined the conformations of the peptide that showed potential interactions with the surface of the kinase domain but did not collide with it.
When we analyzed the backbone torsion angles of the –1 and –2 residues of the modeled peptide over the course of the simulation, we observed that each residue readily adopts both α- and β-conformations (SI Appendix, Fig. S5 A and B). Clustering the conformations based on these torsion angles revealed that the –1 and –2 residues most often share either the α or β region of the Ramachandran diagram (SI Appendix, Fig. S5C). Examining the positioning of the –1 and –2 residue side-chains in each of these top two clusters, we found that they correspond to two principal modes in which the residue at the −1 position can be recognized by the kinase domain (SI Appendix, Fig. S6). When both the –1 and –2 residues of the peptide adopt the β-conformation, the tyrosine side-chain and the side-chains at the −1 and −2 positions point in alternating directions with respect to the direction of the peptide backbone (SI Appendix, Fig. S6A). An example of this binding mode is provided by the crystal structure of a substrate complex of the c-Kit kinase domain (PDB ID code 1PKG) (54). In this conformation, the side-chain of the residue at the −1 position points away from the main tyrosine residue and is located within a surface pocket that contains the positively charged side-chains of Lys-855 and Lys-889. In substrates that are primed by phosphorylation at the +1 position, this pocket recognizes the +1 phosphorylated tyrosine residue (26). The side-chain of the −2 residue points toward a hydrophobic pocket near the active site consisting of Trp-856 and the aliphatic portion of the side-chain of Lys-855. Thus, the β-conformation supports binding of substrates with an acidic residue at the −1 position and a hydrophobic residue at the −2 position.
Alternatively, when the −1 and −2 residues adopt an α-conformation, the side-chain of the residue at the −1 position is pointed in roughly the same direction as the tyrosine residue, toward the same hydrophobic pocket occupied by the −2 residue side-chain in the β-conformation. Examples of this binding mode are provided by structures of substrate complexes of EGFR (PDB ID code 5CZH) (26) and insulin receptor (PDB ID codes 1IR3 and 1GAG) (53 and 55). The side-chain of the residue at the −2 position is exposed toward solvent (SI Appendix, Fig. S6B). Thus, the α-conformation supports recognition by EGFR of peptides with a hydrophobic residue at the −1 position and does not support sharp discrimination at the −2 position.
The importance of proline at the –2 position in EGFR phosphosites appears to be a consequence of having an acidic rather than a hydrophobic residue at the –1 position. We assume that this favors binding of the peptide in a β-conformation, which places the residue at the −2 position in the hydrophobic binding site. For the Tyr-1086 phosphosite, which has a hydrophobic residue at the −1 position, the mutational matrix for phosphorylation by c-Src (Fig. 4D) shows that phosphorylation efficiency is decreased by mutations to either the proline at the −2 position or the valine at the −1 position, suggesting that this peptide is also recognized by c-Src in the β-conformation. In contrast to EGFR, c-Src does not tolerate acidic residues at the −1 position. A plausible explanation for this effect is the replacement of the positively charged Lys-899 in the F–G loop of the EGFR kinase domain by hydrophobic residues in c-Src and other Src-family kinases. Earlier studies have pointed out the importance of the corresponding residue in cytoplasmic tyrosine kinases for differential recognition of the substrate –1 position (31, 32).
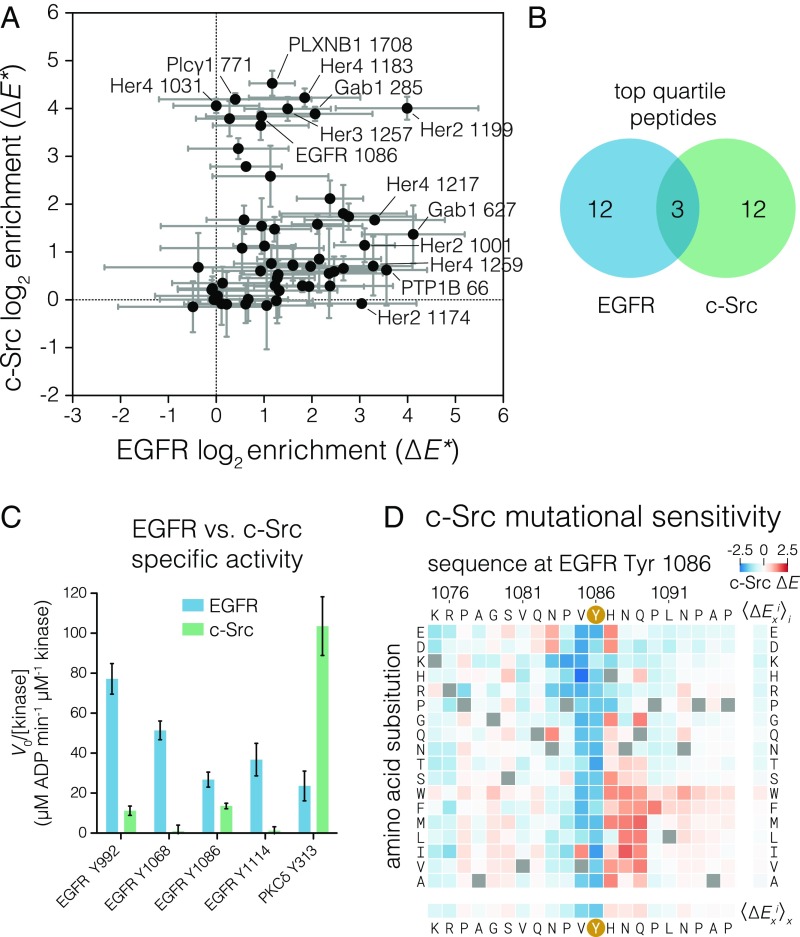
Fig. 4.
Comparison of c-Src and EGFR specificity with respect to EGFR substrates. (A) Relative phosphorylation of EGFR-family phosphosites and reported cytoplasmic EGFR substrates by c-Src versus EGFR. Log-twofold enrichment values relative to a non–tyrosine-containing control peptide were calculated from peptide read frequencies in sorted and input samples. These enrichment values (denoted ∆E*) were corrected by the relative expression level measured for each peptide by cell sorting and deep sequencing. ∆E* values are not comparable on the same scale between kinases. The mean of three replicates with 95% CIs for each kinase is plotted. (B) Venn diagram showing membership of peptides in the top quartile of ∆E* values for each kinase. (C) Specific activities measured for c-Src and EGFR by an NADH-coupled assay against selected peptides at 0.5 mM peptide. Three EGFR C-terminal tail phosphosites and one c-Src substrate, noted below each set of bars, were measured. Error bars indicate the 95% CI of the mean. (D) Heat map showing the effect of single amino acid substitutions on the phosphorylation level of the EGFR Tyr-1086 phosphosite relative to wild type upon phosphorylation by c-Src, as measured by bacterial surface display and deep sequencing. ∆Exi is displayed as a heat map, as described in Fig. 2.
Tyrosine Residues in the Tails of EGFR-Family Members Are Optimized for Selective Phosphorylation by EGFR Rather than by c-Src.
The fact that EGFR-family phosphosites are highly enriched in sequences that conform to the −1 acidic/+1 hydrophobic rule (Fig. 2D) suggests that phosphorylation efficiency is indeed an important constraint on the sequences of these sites. If the efficiency of phosphorylation by EGFR is important, why do the great majority of these sites exclude a hydrophobic residue at the −1 position, since that would also be consistent with efficient phosphorylation? Src-family kinases and c-Abl efficiently phosphorylate sites with hydrophobic residues at the −1 position (Fig. 2 E and F) (12, 13, 32). Given that c-Src has been shown to participate in EGFR signaling and therefore is likely to have access to many of the same phosphosites as EGFR-family kinases, we wondered whether minimization of phosphorylation by Src-family kinases, and potentially other cytoplasmic tyrosine kinases, is a reason for the exclusion of hydrophobic residues at the −1 position in EGFR-family tail phosphosites.
We compared the phosphorylation efficiency of EGFR and c-Src using a focused library of phosphosites in EGFR-family C-terminal tails and reported EGFR substrates (37) using the high-throughput bacterial surface display assay (Fig. 4A). For this experiment, we took the additional step of normalizing the enrichment values based on the expression level of each peptide (Methods) to compare the two kinases quantitatively. In this assay, the sites phosphorylated efficiently by EGFR are phosphorylated poorly by c-Src, and vice versa. Except for the Her2 Tyr-1199 peptide, the highly phosphorylated substrates of EGFR do not include the highly phosphorylated substrates of c-Src (Fig. 4B).
To confirm that EGFR phosphosites are poor substrates for Src-family kinases, we compared the catalytic efficiencies of the dimerized EGFR intracellular module and the c-Src kinase domain for selected EGFR phosphosite peptides using an in vitro kinase assay with purified peptides. We compared the specific activity of these kinases at a fixed peptide concentration that is well below the expected KM values for peptide substrates (32, 42, 56). Under such conditions the catalytic rate is proportional to the catalytic efficiency (kcat/KM). For comparison, we also included a peptide from protein kinase Cδ, spanning Tyr-313, which is a good substrate for c-Src (32). Tail phosphosites were phosphorylated by EGFR with efficiency similar to the phosphorylation of a preferred substrate by c-Src, while c-Src phosphorylated the tail sites poorly (Fig. 4C). This confirms that the c-Src kinase domain is inherently poor at phosphorylating the EGFR tail phosphosites rather than simply being a less active enzyme than the dimerized EGFR kinase.
The Tyr-1086 phosphosite has valine at the −1 position, which is not optimal for EGFR according to the mutational screen (Fig. 3B). On the other hand, previous studies using position-specific oriented peptide library screens (13) and bacterial surface display (32) indicate that c-Src efficiently phosphorylates peptides with a −1 valine. Consistent with this, our results show that Tyr-1086 is among the most efficiently phosphorylated substrates for c-Src in the EGFR tail but is a relatively poor substrate for EGFR. This observation is also consistent with previous studies reporting that Tyr-1086 is directly phosphorylated by c-Src (27). Intriguingly, mutation of Tyr-1086 in EGFR attenuates the phosphorylation of Tyr-845 in the activation loop of EGFR in a cellular assay, although evidence that this effect is connected to phosphorylation of the EGFR activation loop by c-Src is lacking (52).
We determined the site-specific amino acid substitution sensitivity of the Tyr-1086 phosphosite in EGFR to phosphorylation by c-Src (Fig. 4D) and compared it with the sensitivity of this phosphosite to phosphorylation by EGFR (Fig. 3B and SI Appendix, Fig. S7). Although the Tyr-1086 phosphosite sequence is permissive for phosphorylation by c-Src relative to other EGFR-family phosphosites, this phosphosite is not optimal. Substitution of the histidine residue at the +1 position to most other residues increases phosphorylation by c-Src. A histidine residue at the +1 position of the Tyr-1086 site is a conserved feature of mammalian EGFR sequences (SI Appendix, Fig. S8) despite its having no obvious benefit for EGFR phosphorylation (Figs. 2C and 3A) or effector protein binding (see below). This suggests that, although Tyr-1086 provides a potential channel for phosphorylation of EGFR by c-Src, the sequence of this phosphosite limits the efficiency of such phosphorylation.
The Binding of Shc1 and Grb2 to EGFR Phosphosites Can Be Enchanced by Sequence Changes That Would also Increase Phosphorylation by c-Src.
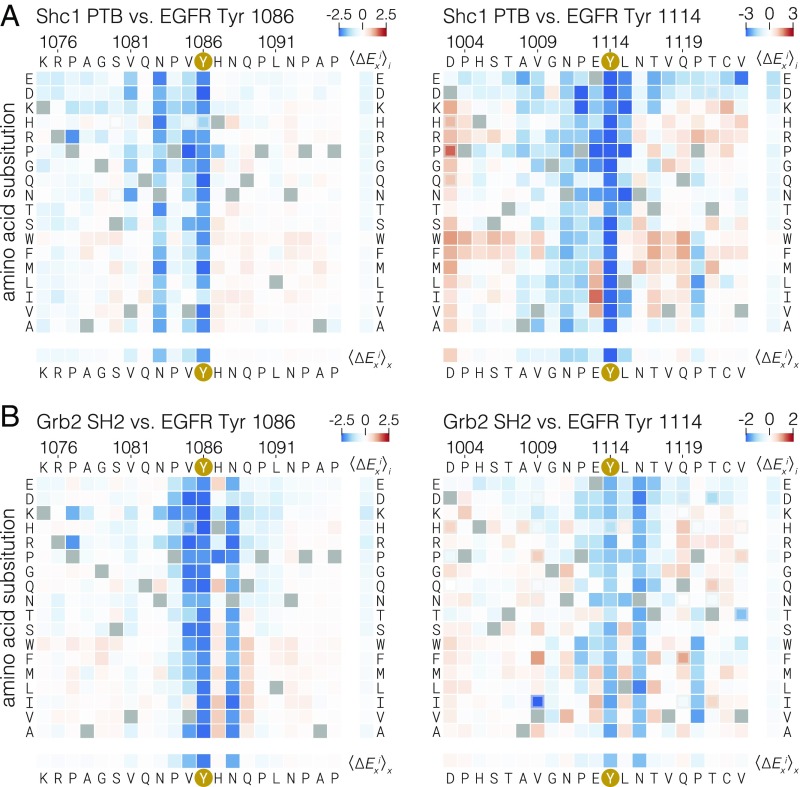
We expanded the bacterial surface display method to test the positional amino acid preferences of the SH2 domain of Grb2 and the PTB domain of Shc1 at two EGFR phosphosites, Tyr-1086 and Tyr-1114. We surmised, based on the presence of known binding motifs as well as binding data from previous studies (19, 49, 57), that the SH2 domain of Grb2 and the PTB domain of Shc1 are prominent adapter proteins that bind to these sites in cells.
In this assay, a mixture of c-Src, c-Abl, and EGFR kinases was used to phosphorylate E. coli cells expressing surface displayed peptide libraries. The phosphorylation was allowed to proceed to completion, as monitored by flow cytometry with an anti-phosphotyrosine antibody (SI Appendix, Fig. S9). To assay binding, tandem copies of phosphotyrosine-binding protein domains fused to GFP were used in place of the anti-phosphotyrosine antibody in the FACS selection. Tandem versions of these binding domains were required to maintain a stable signal for the duration of the cell-sorting protocol.
The mutational matrices for the binding of the Shc1 PTB domain (Fig. 5A) and the Grb2 SH2 domain (Fig. 5B) have features consistent with previously determined binding motifs for these domains (49). The main determinant of Grb2 SH2 binding is the presence of an asparagine at the +2 position (48, 58). In the mutational matrices, substitution of this residue in both the Tyr-1086 and Tyr-1114 phosphosites is, on average, just as detrimental as substitution of the central tyrosine residue. Similarly, substitution of the asparagine at the −3 position has a uniformly strong negative effect on the binding of the Shc1 PTB domain at these sites. This corresponds to the known specificity of the Shc1 PTB domain (50, 59). For the Tyr-1086 phosphosite, the −2 proline suggested to be important for PTB binding in earlier studies appears to be dispensable for Shc1 binding, while the −2 proline appears to be more important at the Tyr-1114 phosphosite.
Fig. 5.
Effect of single amino acid substitutions on the binding of the Grb2 SH2 domain and Shc1 PTB domain to two EGFR phosphosites. Log-twofold changes in read frequency ratios relative to wild-type (∆Exi) were determined by cell sorting after labeling phosphorylated bacteria displaying peptides with tandem copies of the Shc1 PTB domain (A) or the Grb2 SH2 domain (B) fused to GFP. ∆Exi values for single amino acid substitutions are displayed as heat maps, as described in Fig. 2.
Surprisingly, despite completely different modes for binding to peptides, the Grb2 SH2 and Shc1 PTB domains are both sensitive to substitutions of the −1 valine in the Tyr-1086 phosphosite. Isoleucine, another hydrophobic, β-branched residue, is the only substitution that is tolerated by these domains at this position. Introduction of hydrophobic residues, particularly isoleucine or valine, at the −1 position of the Tyr-1114 phosphosite also improves binding, suggesting that a preference for such residues is a shared feature of these domains in multiple sequence contexts. This specificity determinant has not been described previously, and it is not easily explained by available structural models. It is, however, consistent with an alanine-scanning mutagenesis study of phosphosites in the insulin receptor (50).
The preference exhibited by the Grb2 SH2 domain and the Shc1 PTB domain for a hydrophobic residue at the −1 position is at odds with the conserved sequence features found in EGFR-family tail phosphosites, which are not enriched in valine or isoleucine at the −1 position (Fig. 2D). This preference is, however, aligned with the specificity determinants of Src-family kinases and c-Abl (Fig. 2 E and F) (13, 32). The observation that most EGFR-family tail phosphosites avoid hydrophobic residues at the −1 position, despite such residues being preferred by two of the principal effector proteins that bind to these sites, lends further support to the idea that these sites have evolved to discriminate against phosphorylation by c-Src and, potentially, other cytoplasmic tyrosine kinases.
Conclusions
The ancestral metazoan organism appears to have had a nearly full complement of cytoplasmic tyrosine kinases with recognizable counterparts in modern animals, but orthologs of modern receptor tyrosine kinases were lacking (60, 61). For example, the choanoflagellates, which are among the closest living relatives to true metazoans, have clearly identifiable orthologs of Src- and Abl-family kinases. Choanoflagellates also contain numerous receptor tyrosine kinases, but the extracellular domains of these receptors have no counterparts in modern metazoans. Thus, EGFR-family members emerged as important signaling molecules in the background of preexisting signaling networks that included cytoplasmic tyrosine kinases, such as the Src-family kinases.
This raises the question of whether the tails of the EGFR-family members, in addition to being evolutionarily adapted for autophosphorylation and effector binding, are also adapted to encode minimized phosphorylation by cytoplasmic tyrosine kinases. Our work indicates that the EGFR tail is insulated from efficient phosphorylation by kinases that favor hydrophobic residues at the −1 position, which includes Src-family kinases and c-Abl. While substrates with a hydrophobic residue at −1 can be phosphorylated by EGFR, c-Src, and c-Abl, phosphosites in tails of EGFR-family members generally have an acidic residue at this position. This permits efficient phosphorylation by EGFR but not by c-Src and is likely to be an evolutionary adaptation that preserves the integrity of the growth factor-dependent signals emanating from EGFR-family members.
The phosphosites in the tails of EGFR-family members are presented in cis for phosphorylation by the receptor, potentially allowing these receptors to rely on proximity rather than on sequence specificity for efficient phosphorylation of tail phosphosites. Also, the sequences of EGFR-family tail phosphosites do not conform to the optimal motifs identified for efficient EGFR phosphorylation beyond the −1 and +1 residues (12, 26), making it unclear whether phosphorylation by EGFR is an important parameter constraining the evolution of these sequences. By assaying kinase and binding specificity with respect to discrete, determined sequences, we have discovered that the tail phosphosites do conform to the rules governing efficient phosphorylation by EGFR but that the sequence motifs found in the tails exclude phosphorylation by Src-family kinases and c-Abl while maintaining binding of SH2 and PTB domains. Our findings underscore the importance of amino acid sequence context in determining which phosphosites are efficiently recognized by a particular kinase or adapter protein. The context-dependent recognition of proline at the −2 position found in many EGFR-family phosphosites simultaneously allows efficient phosphorylation by EGFR and binding by the Shc1 PTB domain. The relative lack of importance of the +2 position in determining EGFR efficiency allows specification of Grb2 binding independent of EGFR phosphorylation.
EGFR autophosphorylation takes place in the context of a full-length transmembrane receptor, where distal portions of the tail, as well as other parts of the kinase domain, might affect the docking of short peptide motifs at the active site and thereby the rates and levels of phosphorylation in cells. The tertiary structural interactions that might underlie such a mechanism have been difficult to observe in EGFR-family members (62). The synthetically dimerized EGFR construct developed for this study lends itself well to measuring autophosphorylation rates of individual sites, although this is left to future investigation. The role of substrate docking might also be played by an EGFR-binding partner, as seen in the cyclin-dependent kinase system, where the cyclin bound to the kinase domain directs RxL motif-containing substrates to the active site (63). In addition to the intrinsic specificity of the kinase domain, it will be important to investigate the role of other features of EGFR, as well its membrane environment, in the overall phosphorylation efficiency of each tail site.
The architecture of the EGFR-signaling pathway is organized around a central repertoire of enzymes and adapter proteins that are capable of producing a large variety of cellular outcomes depending on the cellular context (1, 14). The timing and extent of phosphorylation and effector recruitment varies depending on which ligands and heterodimerization partners are present (64–67). One explanation for the ability of different EGFR-family ligands to produce alternative phosphorylation and effector recruitment patterns is the intrinsic sequence specificity of EGFR-family kinases, which can determine which sites become phosphorylated and able to support effector binding at different thresholds of kinase activity. It will be interesting to see what role intrinsic kinase specificity plays in determining phosphorylation levels in cells, where substrates are often presented to kinases within the locally dense environment of signaling clusters.
The intrinsic specificity of EGFR and other kinases might be important in the context of activating mutations in the EGFR kinase domain in human cancers (68). In certain cases, such as the L834R substitution and exon 19 deletions in EGFR, the molecular basis of increased activity has been interpreted as a general increase in the maximal substrate turnover (kcat) across the spectrum of substrates (69) due to changes in dimerization propensity (70). Nevertheless, given the multiple alternative specificity-conferring mechanisms that we and others have observed for EGFR substrates, it is reasonable to suspect that changes in both substrate engagement and maximal turnover account for the effects of activating mutations, thus opening the possibility that these mutations also alter the specificity landscape of EGFR. Compellingly, the L834R substitution alters the order of EGFR autophosphorylation in the context of a kinase domain–tail construct (71). By changing specificity along with activity, cancer mutations might affect not only overall phosphorylation levels but also the relative engagement of different downstream signaling pathways and therefore the cellular outcome of EGFR signaling. As more nuances of EGFR-family signaling are discovered, it will be important to consider how the intrinsic specificity of kinases and their binding partners is projected onto the cellular systems they constitute.
Methods
Detailed methods can be found in the SI Appendix. A brief summary is provided here.
Recombinant Proteins.
The sequences of proteins and peptides used in this study are presented in SI Appendix, Tables S1 and S2, respectively. The c-Src kinase domain, tandem Grb2 SH2– and Shc1 PTB–GFP fusions, and peptides were expressed in E. coli. The FKBP- and FRB-EGFR intracellular module proteins were expressed in insect cells. These proteins and peptides were purified by standard liquid chromatography methods, as detailed in SI Appendix.
Bacterial Surface Display and Deep Sequencing.
Quantification of the phosphorylation of tyrosine-containing peptides displayed on bacteria and the construction of site-saturating mutagenesis libraries were performed largely as described previously (31). Details for the construction of the Human-pTyr library can be found in ref. 32 (see SI Appendix for more details). After induction of peptide expression on the N terminus of the engineered surface display scaffold eCpx (35), E. coli cells were subjected to phosphorylation by a purified kinase under conditions determined empirically to give ∼30% maximal phosphorylation, as judged by flow cytometry with anti-phosphotyrosine 4G10 staining. Cells were sorted by FACS based on anti-phosphotyrosine staining into a single bin corresponding to the brightest 25% of events. DNA from the sorted and input cells was amplified and analyzed by Illumina paired-end sequencing. The read frequencies for each peptide-coding DNA sequence from the input and sorted samples from each phosphorylation reaction were compared to give an enrichment ratio for each peptide. For site-saturating mutagenesis libraries, the read frequency ratios of mutant peptides were normalized to that of the wild-type peptide to give a log-relative enrichment value (∆E). For the EGFR substrate library in Fig. 4A, ∆E was calculated relative to a non–tyrosine-containing control peptide. The data in Fig. 4A were additionally corrected for expression level on the basis of anti–Strep-tag immunostaining, using the tag encoded at the C terminus of the surface display scaffold.
Analysis of SH2- and PTB-domain binding to the saturation mutagenesis libraries was performed similarly to the analysis of phosphorylation, but tandem versions of each binding domain fused to GFP were used in place of the anti-phosphotyrosine antibody. The cells used for this analysis were phosphorylated by a mixture of EGFR, c-Src, and c-Abl kinases and were monitored for uniform phosphorylation by anti-phosphotyrosine flow cytometry (SI Appendix, Fig. S9).
Kinase Peptide Activity Assays.
The specific activity of purified kinases against purified peptides was monitored with an enzyme-coupled assay based on NADH absorbance (72). Steady-state reaction rates for kinases at 0.5 µM, peptides at 0.5 mM, and ATP at 0.5 mM at room temperature were converted to specific activity on the basis of the change in NADH absorbance over time.
Generation of Sequence-pLogo of EGFR-Family Phosphosites.
A collection of bona-fide metazoan EGFR-family kinase sequences was extracted from the EggNOG database (orthology group ENOG410XNSR) (73), aligned, and filtered by sequence identity of the kinase domain to avoid oversampling taxa that have relatively high numbers of species in the sequence database. C-terminal tyrosine sites were analyzed for positional amino acid enrichment with the pLogo tool (44), with tyrosines from metazoan transmembrane and intracellular proteins serving as the background distribution. The height of each letter in a pLogo corresponds to the log-odds ratio of the binomial probability of observing an amino acid at least as many times in the foreground set as in the background set, divided by the probability of observing that amino acid that many times or fewer in the background set.
Molecular Dynamics Simulations.
A peptide encompassing the Tyr-1114 phosphosite corresponding to residues 1110–1118 of human EGFR was simulated with NAMD (74) using the CHARMM36 force field (75). The peptide was modeled based on the peptide substrate in the crystal structure of the phosphorylated insulin receptor kinase (PDB ID code 1IR3) (53) and was constrained to have residues 1114–1118 adopt β-conformation backbone dihedral angles throughout the simulation. Trajectories were analyzed by clustering frames on the basis of the backbone dihedral angles of residues 1112 and 1113. Representative frames from each cluster were selected visually.
Supplementary Material
Acknowledgments
We thank Hector Nolla and Alma Valeros of the University of California, Berkeley (UC Berkeley) Cancer Research Laboratory flow cytometry facility for assistance with cell sorting; Bill Russ and Rama Ranganathan at the University of Texas Southwestern Medical Center for assistance with Illumina sequencing and helpful discussions; and Xiaoxian Cao of the J.K. laboratory at UC Berkeley for assistance with protein expression. This work was supported in part by NIH Grants 5R01CA096504-12 and P01AI091580 (to J.K.). N.H.S. was supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.D.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803598115/-/DCSupplemental.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem. 2015;84:739–764. doi: 10.1146/annurev-biochem-060614-034402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzahar E, et al. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Moran MF, et al. Src homology region 2 domains direct protein-protein interactions in signal transduction. Proc Natl Acad Sci USA. 1990;87:8622–8626. doi: 10.1073/pnas.87.21.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kavanaugh WM, Turck CW, Williams LT. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- 8.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 9.Bose R, Holbert MA, Pickin KA, Cole PA. Protein tyrosine kinase-substrate interactions. Curr Opin Struct Biol. 2006;16:668–675. doi: 10.1016/j.sbi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Hunter T. Tyrosine phosphorylation: Thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer BJ. Perspective: Dynamics of receptor tyrosine kinase signaling complexes. FEBS Lett. 2012;586:2575–2579. doi: 10.1016/j.febslet.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Songyang Z, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 13.Deng Y, et al. Global analysis of human nonreceptor tyrosine kinase specificity using high-density peptide microarrays. J Proteome Res. 2014;13:4339–4346. doi: 10.1021/pr500503q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citri A, Yarden Y. EGF-ERBB signalling: Towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 15.Knudsen SLJ, Mac ASW, Henriksen L, van Deurs B, Grøvdal LM. EGFR signaling patterns are regulated by its different ligands. Growth Factors. 2014;32:155–163. doi: 10.3109/08977194.2014.952410. [DOI] [PubMed] [Google Scholar]
- 16.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005.0008. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 18.Kaushansky A, et al. System-wide investigation of ErbB4 reveals 19 sites of Tyr phosphorylation that are unusually selective in their recruitment properties. Chem Biol. 2008;15:808–817. doi: 10.1016/j.chembiol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hause RJ, Jr, et al. Comprehensive binary interaction mapping of SH2 domains via fluorescence polarization reveals novel functional diversification of ErbB receptors. PLoS One. 2012;7:e44471. doi: 10.1371/journal.pone.0044471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill K, Macdonald-Obermann JL, Pike LJ. Epidermal growth factor receptors containing a single tyrosine in their C-terminal tail bind different effector molecules and are signaling-competent. J Biol Chem. 2017;292:20744–20755. doi: 10.1074/jbc.M117.802553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 22.Tanos B, Pendergast AM. Abl tyrosine kinase regulates endocytosis of the epidermal growth factor receptor. J Biol Chem. 2006;281:32714–32723. doi: 10.1074/jbc.M603126200. [DOI] [PubMed] [Google Scholar]
- 23.Luttrell LM, Della Rocca GJ, van Biesen T, Luttrell DK, Lefkowitz RJ. Gbetagamma subunits mediate Src-dependent phosphorylation of the epidermal growth factor receptor. A scaffold for G protein-coupled receptor-mediated Ras activation. J Biol Chem. 1997;272:4637–4644. doi: 10.1074/jbc.272.7.4637. [DOI] [PubMed] [Google Scholar]
- 24.Moro L, et al. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J Biol Chem. 2002;277:9405–9414. doi: 10.1074/jbc.M109101200. [DOI] [PubMed] [Google Scholar]
- 25.Joo CK, et al. Ligand release-independent transactivation of epidermal growth factor receptor by transforming growth factor-beta involves multiple signaling pathways. Oncogene. 2008;27:614–628. doi: 10.1038/sj.onc.1210649. [DOI] [PubMed] [Google Scholar]
- 26.Begley MJ, et al. EGF-receptor specificity for phosphotyrosine-primed substrates provides signal integration with Src. Nat Struct Mol Biol. 2015;22:983–990. doi: 10.1038/nsmb.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stover DR, Becker M, Liebetanz J, Lydon NB. Src phosphorylation of the epidermal growth factor receptor at novel sites mediates receptor interaction with Src and P85 alpha. J Biol Chem. 1995;270:15591–15597. doi: 10.1074/jbc.270.26.15591. [DOI] [PubMed] [Google Scholar]
- 28.Lombardo CR, Consler TG, Kassel DB. In vitro phosphorylation of the epidermal growth factor receptor autophosphorylation domain by c-src: Identification of phosphorylation sites and c-src SH2 domain binding sites. Biochemistry. 1995;34:16456–16466. doi: 10.1021/bi00050a029. [DOI] [PubMed] [Google Scholar]
- 29.Biscardi JS, et al. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 30.Reddy RJ, et al. Early signaling dynamics of the epidermal growth factor receptor. Proc Natl Acad Sci USA. 2016;113:3114–3119. doi: 10.1073/pnas.1521288113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah NH, et al. An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. eLife. 2016;5:e20105. doi: 10.7554/eLife.20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah NH, Löbel M, Weiss A, Kuriyan J. Fine-tuning of substrate preferences of the Src-family kinase Lck revealed through a high-throughput specificity screen. eLife. 2018;7:e35190. doi: 10.7554/eLife.35190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan PM, Nestler HP, Miller WT. Investigating the substrate specificity of the HER2/Neu tyrosine kinase using peptide libraries. Cancer Lett. 2000;160:159–169. doi: 10.1016/s0304-3835(00)00581-4. [DOI] [PubMed] [Google Scholar]
- 34.Songyang Z. Analysis of protein kinase specificity by peptide libraries and prediction of in vivo substrates. Methods Enzymol. 2001;332:171–183. doi: 10.1016/s0076-6879(01)32200-0. [DOI] [PubMed] [Google Scholar]
- 35.Rice JJ, Daugherty PS. Directed evolution of a biterminal bacterial display scaffold enhances the display of diverse peptides. Protein Eng Des Sel. 2008;21:435–442. doi: 10.1093/protein/gzn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henriques ST, et al. A novel quantitative kinase assay using bacterial surface display and flow cytometry. PLoS One. 2013;8:e80474. doi: 10.1371/journal.pone.0080474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jura N, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Red Brewer M, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y-X, Wong L, Deb TB, Johnson GR. Ligand regulates epidermal growth factor receptor kinase specificity: Activation increases preference for GAB1 and SHC versus autophosphorylation sites. J Biol Chem. 2004;279:38143–38150. doi: 10.1074/jbc.M405760200. [DOI] [PubMed] [Google Scholar]
- 42.Fan Y-X, Wong L, Johnson GR. EGFR kinase possesses a broad specificity for ErbB phosphorylation sites, and ligand increases catalytic-centre activity without affecting substrate binding affinity. Biochem J. 2005;392:417–423. doi: 10.1042/BJ20051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu C, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Shea JP, et al. pLogo: A probabilistic approach to visualizing sequence motifs. Nat Methods. 2013;10:1211–1212. doi: 10.1038/nmeth.2646. [DOI] [PubMed] [Google Scholar]
- 45.Patschinsky T, Hunter T, Esch FS, Cooper JA, Sefton BM. Analysis of the sequence of amino acids surrounding sites of tyrosine phosphorylation. Proc Natl Acad Sci USA. 1982;79:973–977. doi: 10.1073/pnas.79.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Songyang Z, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 47.Miller ML, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Songyang Z, et al. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, et al. Defining the specificity space of the human SRC homology 2 domain. Mol Cell Proteomics. 2008;7:768–784. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Wolf G, et al. PTB domains of IRS-1 and Shc have distinct but overlapping binding specificities. J Biol Chem. 1995;270:27407–27410. doi: 10.1074/jbc.270.46.27407. [DOI] [PubMed] [Google Scholar]
- 51.Pines G, Huang PH, Zwang Y, White FM, Yarden Y. EGFRvIV: A previously uncharacterized oncogenic mutant reveals a kinase autoinhibitory mechanism. Oncogene. 2010;29:5850–5860. doi: 10.1038/onc.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovacs E, et al. Analysis of the role of the C-terminal tail in the regulation of the epidermal growth factor receptor. Mol Cell Biol. 2015;35:3083–3102. doi: 10.1128/MCB.00248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mol CD, et al. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- 55.Parang K, et al. Mechanism-based design of a protein kinase inhibitor. Nat Struct Biol. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- 56.Engel K, Sasaki T, Wang Q, Kuriyan J. A highly efficient peptide substrate for EGFR activates the kinase by inducing aggregation. Biochem J. 2013;453:337–344. doi: 10.1042/BJ20130537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laminet AA, Apell G, Conroy L, Kavanaugh WM. Affinity, specificity, and kinetics of the interaction of the SHC phosphotyrosine binding domain with asparagine-X-X-phosphotyrosine motifs of growth factor receptors. J Biol Chem. 1996;271:264–269. doi: 10.1074/jbc.271.1.264. [DOI] [PubMed] [Google Scholar]
- 58.Rahuel J, et al. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat Struct Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 59.Zhou MM, et al. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]
- 60.Suga H, et al. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci Signal. 2012;5:ra35. doi: 10.1126/scisignal.2002733. [DOI] [PubMed] [Google Scholar]
- 61.Richter DJ, King N. The genomic and cellular foundations of animal origins. Annu Rev Genet. 2013;47:509–537. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 62.Keppel TR, et al. Biophysical evidence for intrinsic disorder in the C-terminal tails of the epidermal growth factor receptor (EGFR) and HER3 receptor tyrosine kinases. J Biol Chem. 2017;292:597–610. doi: 10.1074/jbc.M116.747485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams PD, et al. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krall JA, Beyer EM, MacBeath G. High- and low-affinity epidermal growth factor receptor-ligand interactions activate distinct signaling pathways. PLoS One. 2011;6:e15945. doi: 10.1371/journal.pone.0015945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ronan T, et al. Different epidermal growth factor receptor (EGFR) agonists produce unique signatures for the recruitment of downstream signaling proteins. J Biol Chem. 2016;291:5528–5540. doi: 10.1074/jbc.M115.710087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc Natl Acad Sci USA. 2017;114:E2836–E2845. doi: 10.1073/pnas.1617994114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Freed DM, et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 2017;171:683–695.e18. doi: 10.1016/j.cell.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 71.Kim Y, et al. Temporal resolution of autophosphorylation for normal and oncogenic forms of EGFR and differential effects of gefitinib. Biochemistry. 2012;51:5212–5222. doi: 10.1021/bi300476v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barker SC, et al. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: Autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 73.Huerta-Cepas J, et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang J, et al. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.