Significance
BRD4, a major BET family protein, regulates gene transcription through coordinated binding of its characteristic tandem bromodomains (BrDs) to lysine-acetylated histones and transcription factors. Studies show that BRD4 function in transcriptional regulation is likely context- and cell-type-dependent, consistent with the observation that pan-BET BrD inhibitors such as JQ1 are much less effective in solid tumors than in hematopoietic cancers. Here, we show that spatially constrained bivalent inhibition of BRD4 BrDs with our BET inhibitor MS645 results in a sustained repression of BRD4 transcriptional activity in solid-tumor cells including triple-negative breast cancer (TNBC) cells. Our study offers a therapeutic strategy to maximally control BRD4 activity required for rapid cell proliferation of the devastating TNBC that lacks targeted therapy.
Keywords: BRD4, bivalent BET inhibitors, gene transcription, TNBC, drug discovery
Abstract
The importance of BET protein BRD4 in gene transcription is well recognized through the study of chemical modulation of its characteristic tandem bromodomain (BrD) binding to lysine-acetylated histones and transcription factors. However, while monovalent inhibition of BRD4 by BET BrD inhibitors such as JQ1 blocks growth of hematopoietic cancers, it is much less effective generally in solid tumors. Here, we report a thienodiazepine-based bivalent BrD inhibitor, MS645, that affords spatially constrained tandem BrD inhibition and consequently sustained repression of BRD4 transcriptional activity in blocking proliferation of solid-tumor cells including a panel of triple-negative breast cancer (TNBC) cells. MS645 blocks BRD4 binding to transcription enhancer/mediator proteins MED1 and YY1 with potency superior to monovalent BET inhibitors, resulting in down-regulation of proinflammatory cytokines and genes for cell-cycle control and DNA damage repair that are largely unaffected by monovalent BrD inhibition. Our study suggests a therapeutic strategy to maximally control BRD4 activity for rapid growth of solid-tumor TNBC cells.
Bromodomains (BrDs) in transcription proteins bind acetyl-lysine in histones and transcription factors to direct gene transcription in biology and disease conditions (1–3). As arguably the best-known BrD and extraterminal (BET) family protein, BRD4 contains two characteristic tandem BrDs and is recognized as a major drug target owing to its implicated functions in oncogenesis and inflammation (4, 5). Specifically, it has been shown that a BRD4-NUT fusion protein is responsible for aggressive carcinoma (6), and BRD4 is identified by RNAi screening as one of the most important oncogenes in acute myeloid leukemia (7). BRD4 relies on BrD/acetyl-lysine binding to recruit transcription factors and mediators to form cis-regulatory element enhancers for spatial and temporal control of gene transcription (8, 9) and the p-TEFb (positive transcription elongation factor) complex to phosphorylate RNA polymerase II and activate transcriptional elongation (10, 11). Thus, pharmacological inhibition of BRD4 BrDs is considered a new therapeutic approach to treat cancer and inflammatory disorders.
Potent pan-BET BrD inhibitors have been developed (12), including JQ1 (13), MS417 (14), and I-BET762 (15), that share the core diazepine scaffold, initially disclosed in a patent by Mitsubishi Tanabe Pharma (16). Selective BET inhibitors were also reported such as diazobenzene MS436 with 10-fold selectivity for the first BrD (BD1) of BRD4 over the second BrD (BD2) (17) and a quinazolone RVX208 with 20-fold selectivity for BD2 over BD1 (18). The notion of simultaneous chemical inhibition of tandem BrDs of BET proteins using bivalent inhibitors, suggested by Arnold et al. in a patent filing (19), was demonstrated with two recently reported bivalent BET BrD inhibitors, triazolopyridazine-based AZD5153 (20–22) and diazepine-based MT1 (23) that exhibit much higher efficacy than monovalent BrD inhibitors in growth inhibition of hematopoietic cancer cells. Notably, while these BET inhibitors validate the therapeutic potential of BRD4 in cancer models, they are mostly effective in hematopoietic cancers, but much less so for solid tumors such as malignant breast and cervix cancers (7, 13, 24–29). In this study, we report a class of bivalent BET BrD inhibitors designed to inhibit the tandem BD1/BD2 of BRD4 in a spatially constrained manner. We show through structural and biophysical analysis that chemical composition and rigidity of a linker of bivalent BET inhibitors play an important role in determining their cellular efficacy in inhibiting BRD4 activity in gene transcription in chromatin, as illustrated in triple-negative breast cancer (TNBC) cells.
Results and Discussion
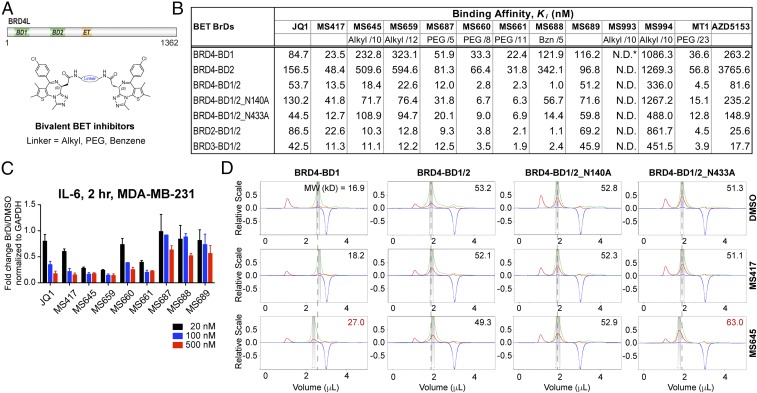
The tandem BrDs in BRD4 interact with multiple lysine-acetylated proteins in a coordinated fashion at different steps during gene transcription in chromatin (5). This dynamic functionality is likely offered by a flexible BD1/BD2 linker sequence of 140–190 residues with high-degree variations among the BET proteins (Fig. 1A). To understand the role of conformational dynamics of this linker sequence in BET protein functions in gene transcription, we designed and synthesized a series of bivalent BrD inhibitors by linking two thienodiazepine-based BET inhibitor MS417 molecules at the amide moiety with an alkyl, PEG, or aromatic benzene linker of varied length and rigidity (Fig. 1B and SI Appendix, Table S1; see the synthesis in SI Appendix).
Fig. 1.
Biophysical characterization of bivalent BrD inhibition of the BET proteins. (A) Domain organization of BRD4 and depiction of bivalent BET inhibitors. (B) Binding affinity of BrD inhibitors to BET BrD proteins as determined by an FP assay. The BET BrD proteins used are BRD4-BD1 (amino acids 44–168), BRD4-BD2 (amino acids 347–460), BRD4-BD1/2 (amino acids 44–477), BRD4-BD1/2_N140A (amino acids 44–477), BRD4-BD1/2_N433A (amino acids 44–477), BRD2-BD1/2 (amino acids 73–473), and BRD3-BD1/BD2 (amino acids 24–434). The chemical composition and length (depicted by number of atoms) for the linker are indicated for bivalent BET inhibitors. (C) Dose-dependent effects of BET inhibitors on transcriptional expression of IL-6 in MDA-MB-231 cells treated with the BrD inhibitors as indicated for 2 h. Results represent at least three independent experiments and error bars denote SEM. (D) Effects of BrD inhibitor binding on protein conformation of BRD4 BD1 alone or tandem BD1/BD2, as assessed by aMW of the protein determined by DLS. Measurements of UV, scattering intensity, and refractive index change are color-coded in green, red, and blue, respectively.
We evaluated binding of our bivalent inhibitors to the individual BD1 and BD2 and the tandem BD1–BD2 of BRD4 using a fluorescence anisotropy-based competition binding assay (14) (Fig. 1B and SI Appendix, Fig. S1A). PEG-linker attachment alone as in MS689 has minimal impact on MS417 binding to the BD1 or BD2 (Fig. 1B). PEG-linker bivalent inhibitors MS660 and MS661 have affinities comparable to that of MS417 for binding to the BD1 or BD2 (Ki of 33.3 nM and 22.4 nM vs. 23.5 nM for BD1), but exhibit an over 10-fold increase in affinity when tested for binding to the tandem BD1–BD2 (Ki of 2.8 nM and 2.3 nM), which are five- to sixfold more potent than MS417 (13.5 nM) (Fig. 1B). The linker length is important, as the shorter linker inhibitor MS687 (with one PEG unit) shows much smaller gain in affinity for bivalent binding. A rigid benzene-linker inhibitor, MS688, shows a striking 120-fold gain in affinity for bivalent binding. Similarly, alkyl-linker inhibitors MS645 and MS659 show a 12- to 28-fold gain in affinity for binding to the tandem BD1–BD2 over the single BD1 or BD2. This synergistic BrD binding of bivalent MS645 and MS659 makes their affinities comparable to that of MS417 to the tandem BD1–BD2, even though their affinities to the single BD1 and BD2 are much weaker than MS417 (Ki of 232.8 and 323.1 nM vs. 23.5 nM for BD1). Notably, the coordinated binding of these bivalent inhibitors to the tandem BD1–BD2 is markedly compromised upon Ala mutation of Asn140 or Asn433, a key conserved residue in the acetyl-lysine binding pocket that forms a hydrogen bond to the triazole moiety of the thienodiazepine scaffold in MS417 (Fig. 1B) (14). As expected, two bivalent control compounds of MS645(S/S), which are composed of one S and one R enantiomer (MS994), or two inactive R enantiomers (MS993), display a drastic reduction or complete loss of binding to the BD1 and BD2 of BRD4, respectively. Finally, these bivalent inhibitors show comparable potency to the tandem BD1–BD2 of BRD2 and BRD3 as to that of BRD4.
We next assessed cellular activity of these bivalent inhibitors on transcription of BRD4 target gene IL-6. MDA-MB-231 cells were treated with individual BET inhibitors of 20, 100, and 500 nM for 2 h and IL-6 mRNA level was measured by qPCR compared with DMSO control. Notably, the alkyl-linker inhibitors MS645 and MS659 exerted over 70% inhibition of IL-6 expression at 20 nM, much higher than 20–30% inhibition by JQ1 and MS417 (Fig. 1C). In contrast, the PEG-linker inhibitors MS660 and MS661 surprisingly showed only effects comparable to JQ1 and MS417, and the shorter PEG-linker inhibitor MS687 and the rigid benzene-linker inhibitor MS688 have little inhibition on IL-6 expression even at 100–500 nM. These data strongly suggest that interdomain conformational dynamics of the BD1–BD2 of BRD4 in cells plays an important role in BRD4 functions in gene transcription in chromatin, likely through influencing BRD4 interactions with effector proteins. Accordingly, we extended further characterization of bivalent BET inhibitor MS645.
We characterized MS645 bivalent binding effects on protein conformation of the tandem BD1–BD2 module using dynamic light scattering (DLS) technology. In contrast to the monovalent inhibitor MS417 that has little effect on apparent molecular weight (aMW) of the single BD1 of BRD4 compared with DMSO, the bivalent inhibitor MS645 binding resulted in a major increase of aMW (27.0 kDa vs. 16.9 kDa), indicating that it likely binds to two BD1 molecules (Fig. 1D). Strikingly, with BD1–BD2, MS645 binding appears to tighten the tandem module as reflected by a reduction in aMW (49.3 kDa vs. 53.2 kDa) (Fig. 1D). When Asn140 in BD1 is mutated to Ala, aMW is about the same as when treated with DMSO or MS417, whereas mutation of Asn433 to Ala in BD2 results in a noticeable increase in aMW (i.e., 63.0 kDa with MS645 vs. 51.3 kDa and 51.1 kDa with DMSO and MS417, respectively) (Fig. 1D). Similar effects were observed for BD1 and BD1–BD2 binding with JQ1 and the PEG-linker bivalent inhibitor MS660 (SI Appendix, Fig. S1B). Collectively, these results indicate that MS645 likely forms a 1:1 stoichiometric complex with the tandem BD1–BD2, and MS645 binding to the BD1 tends to affect domain–domain orientation of the tandem module. Our NMR spectral analysis further supported the notion that MS645 binds to the BD1 or the tandem BD1–BD2 in a stoichiometric manner and revealed that in comparison with MS417, MS645 binding to BRD4 BD1–BD2 is potent and highly dynamic, as illustrated by severe ligand-induced line broadening of NMR resonances of indole NH of Trp81 and Trp374 that are located in the inhibitor binding site in BD1 and BD2, respectively (SI Appendix, Fig. S1C). Further, when the key Asn140 and Asn433 residues were mutated to Ala, the tandem BD1–BD2 of BRD4 binding to MS645 was significantly reduced.
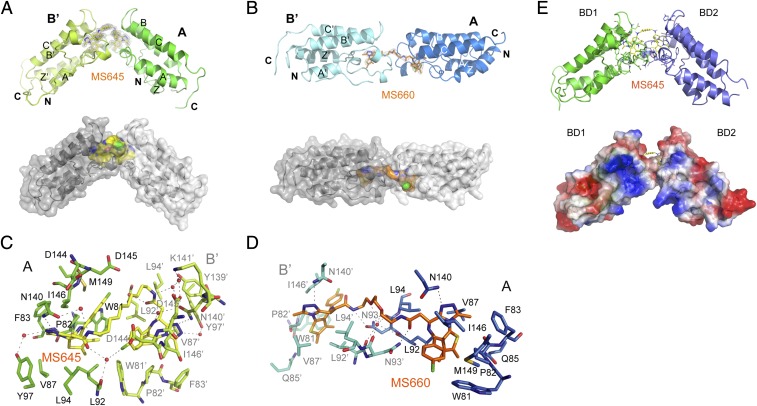
We attempted to cocrystalize bivalent inhibitor in complex with BRD4 BD1–BD2 to understand the structural basis of bivalent BET inhibition. Despite much effort, we succeeded in obtaining only cocrystals of bivalent inhibitors MS645 or MS660 bound to the BD1 resulting from the BD1–BD2 protein in crystallization. The crystals diffracted to 1.46 Å and 2.96 Å, respectively, and structures were solved by molecular replacement using coordinates of the I-BET151/BRD4–BD1 complex (3ZYU) (26) as the search model (SI Appendix, Tables S2 and S3). The crystal structure of the MS645/BD1 complex reveals that each unit cell contains two BD1 molecules and MS645 is bound to two symmetry-related BD1 protomers from two unit cells, and the four-helical bundles of two BD1 protomers arranged in a nearly 100°orientation (Fig. 2A and SI Appendix, Fig. S2A). In contrast, the MS660/BD1 structure shows that MS660 bridges two symmetry-related BD1 molecules, also from two unit cells, in a head-to-head and 180° orientation (Fig. 2B and SI Appendix, Fig. S2A). Despite the difference in protein domain orientation, the core thienodiazepine of the two bivalent inhibitors is bound nearly identically in each BD1 (SI Appendix, Fig. S2B), and similarly to monovalent inhibitor MS417 bound in the BD1. As revealed in the 1.46-Å resolution structure of the MS645/BD1 complex, the residues in the BD1 in the acetyl-lysine binding pocket are projected in the same poses to interact with the bound ligand such as Asn140 that forms a 3.1-Å hydrogen bond between its amide nitrogen and the triazole moiety and two water-mediated hydrogen bonds from its amide oxygen to the inhibitor (Fig. 2C and SI Appendix, Fig. S2C).
Fig. 2.
Structural insights into bivalent BET inhibitor binding to BRD4. (A) Crystal structure of MS645 (yellow)/BRD4 BD1 dimer complex, shown in ribbon and space-filled surface depictions. MS645 is color-coded by atom type and its electron density with Sigma-A-weighted 2mFo-DFc map. (B) Crystal structure of MS660 (orange)/BRD4 BD1 dimer complex, shown in ribbon and space-filled surface depictions. MS660 is color-coded by atom type and its electron density with Sigma-A-weighted 2mFo-DFc map. (C and D) Structural insights of MS645 or MS660 linker recognition at the domain–domain interface in the BRD4 BD1 dimer. Note that two symmetry-related protomers in the BD1 dimer are denoted as A and B′ as two molecules in two different crystallographic unit cells. (E) Structural model of MS645 bound to the BD1–BD2 of BRD4, built based on the crystal structure of the MS645/BRD4 BD1 dimer.
The MS645/BD1 dimer structure reveals a unique water-mediated hydrogen-bonding cluster, formed in a triangular fashion by backbone oxygen of Leu92 of BD1 protomer A, oxygen of the amide moiety projected from the core thienodiazepine bound in BD1 protomer A, and chlorine of the chlorophenyl ring of the thienodiazepine scaffold at the other end of MS645 bound in BD1 protomer B′ (Fig. 2C). Given that the alkyl linker sets the distance between the amide and chlorophenyl groups from two ends of MS645, the structure argues that a 10-carbon linker as in MS645 favors optimal bivalent inhibition. MS660 binding by BD1 also has a water-mediated hydrogen-bond network arranged in a tetrahedral geometry between a PEG oxygen atom in the linker and backbone oxygens of Leu92 and Asn93 in the ZA loop (Fig. 2D and SI Appendix, Fig. S2D). However, unlike MS645, the flexible PEG linker in MS660 does not offer rigidity to constrain the bivalent inhibitor in a defined configuration that would in turn spatially define domain–domain orientation of the BD1 dimer.
We next performed mutagenesis of residues at the inhibitor binding site including Trp81, Leu92, and Leu94 in the BD1 and corresponding Trp374, Leu385, and Leu387 in the BD2. As shown by our fluorescence polarization (FP) assay, single mutation of L92 and W81 to Ala, but not L94, in the BD1 of the tandem BD1–BD2 of BRD4 resulted in a marked reduction of protein binding to MS645 (i.e., Ki of 13 nM vs. 404 nM, 808 nM, and 26 nM for WT vs. L92A, W81A, and L94A of BRD4 BD1/2, respectively) (SI Appendix, Fig. S2E). Notably, Ala mutation of L92 and W81 in both BD1 and BD2 of the tandem BD1–BD2 showed even more profound loss in binding affinity to MS645 than the mutations in the BD1 alone [i.e., Ki of 267 nM (L92A/L94A) vs. >100,000 nM (L92A/L94A/L385A/L387A), and 808 nM (W81A) vs. >800,000 nM (W81A/W387A)]. Further, while important, both L92 and W81 appear to be much less critical to MS660 binding than to MS645. Finally, mutation effects for MS688, a rigid benzene-linker bivalent inhibitor, are more similar to MS645 than MS660. Collectively, these mutagenesis results support our structural insights of two distinct modes of BRD4 BD1/2 recognition by MS645 vs. MS660.
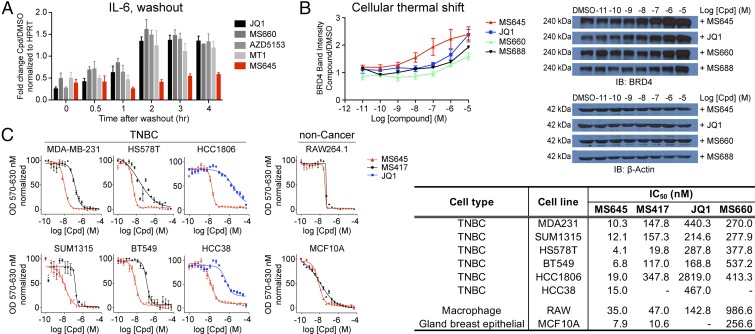
This spatially constrained bivalent BRD4 BrD inhibition by MS645 is attributed to a much slower release of IL-6 transcription inhibition in a washout experiment than that by the PEG-linker inhibitor MS660, monovalent inhibitors JQ1 and MS417, or the reported bivalent inhibitors MT1 and AZD5153 (Fig. 3A). This result is supported by the observation that MS645/BRD4 binding results in a much higher enhancement in cellular protein stability of BRD4 than that by MS660, JQ1, MS417, MT1, or MS688, without affecting stability of nonrelated protein (β-actin) as shown by a cellular thermal shift study (Fig. 3B and SI Appendix, Fig. S3A). This mechanism of action likely explains MS645’s superior potency over JQ1, MS417, and MS660 in growth inhibition of a panel of TNBC cell lines (Fig. 3C) and other cancer cells of ductal breast, prostate, and bladder cancers (SI Appendix, Fig. S3B). MS645 is also consistently more potent than MT1 and AZD5153 in a number of TNBC cells including MDA-MB-231 cells (SI Appendix, Fig. S3C). Further, we found that MS645 has cell growth inhibitory effects similar to MS417 or JQ1 on noncancer cell lines of mouse macrophage RAW cells and nontumorigenic breast epithelial MCF10A (Fig. 3C). These results indicate that MS645’s profound inhibitory activity against fast proliferation of TNBC cell lines is likely not due to nonspecific cell toxicity.
Fig. 3.
Superior cellular efficacy of MS645 in cancer-cell growth inhibition. (A) Persistent transcriptional repression of IL-6 by MS645 over other BET inhibitors in a washout study of MDA-MB-231 cells. The cells were treated with a BET inhibitor (1 μM) or DMSO for 2 h then washed with fresh medium twice and cultured for time periods as indicated. The mRNA level of IL-6 was measured after compound-imposed transcriptional inhibition. The data are plotted from one representative experiment and error bars represent SD of technical repeats. (B) Effects of the BET inhibitors of MS645, MS660, MS688, and JQ1 on protein stability assessed in a cellular thermal shift assay and shown by a representative set of Western blot analyses of BRD4. (C) Effects of MS645, MS660, JQ1, and MS417 on cell growth inhibition of cancer and noncancer cell lines, as assessed in an MTT assay. IC50 values are listed in a table. Results presented in B and C were all from at least three independent experiments and error bars designate SEM.
We further compared MS645 to drugs approved by the Food and Drug Administration or being evaluated in clinical trials to treat breast cancer that include ENMD-2076 (kinase inhibitor) (30), panobinostat (pan-HDAC inhibitor by Novartis) (31), methotrexate (folate metabolism for purine synthesis) (32), and erlotinib (EGFR kinase inhibitor) (33) in cell growth inhibition of MDA-MB-231 and macrophage RAW 264.1 cells. MS645 is much more potent than ENMD-2076 and erlotinib on MDA-MB-231 cell growth inhibition and comparable to methotrexate and panobinostat (SI Appendix, Fig. S3D). These results highlight that MS645 appears to have a clear advantage over panobinostat, as the latter is rather toxic as shown with macrophage RAW cells.
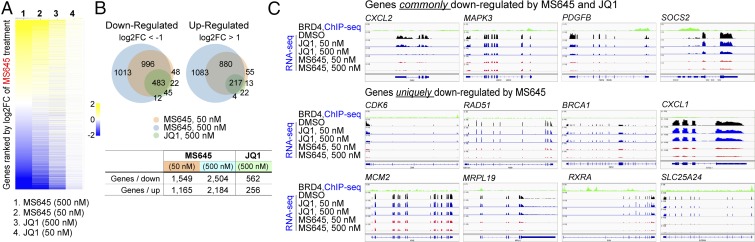
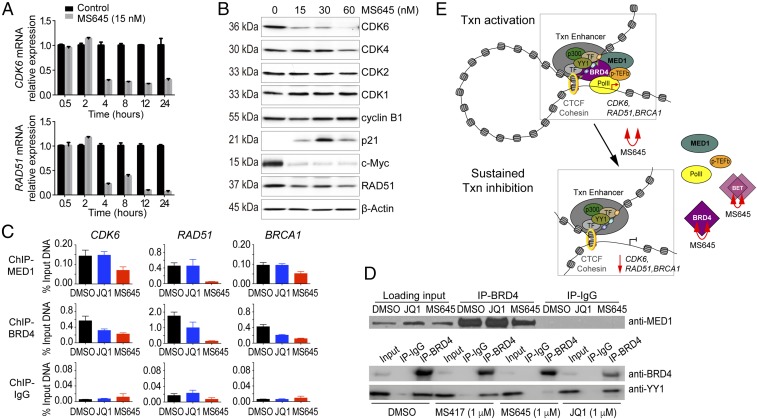
We next performed RNA sequencing (RNA-seq) of MDA-MB-231 cells that were treated with MS645 or JQ1 at 50 nM and 500 nM in an effort to understand how MS645 exerts such a profound cell growth inhibition on cancer cells. While MS645 or JQ1 treatment appear to have a similar trend in altering gene transcription, the former affects many more genes both in categories of down-regulated and up-regulated genes (Fig. 4A, SI Appendix, Fig. S4 A and B, and Dataset S1). Specifically, Venn diagram analyses show that treatment of MS645 at 500 nM or 50 nM resulted in 2,504 and 1,549 genes down-regulated and 2,184 and 1,165 genes up-regulated at least by twofold, respectively, whereas JQ1 at 50 nM shows limited effects, and at 500 nM causes only 562 and 256 genes down- or up-regulated, respectively (Fig. 4B, SI Appendix, Fig. S4 C and D, and Dataset S2). Gene Ontology analyses further revealed that genes important in cytokine signaling, IFN signaling, and the immune system (such as CXCL2, MAPK3, PDGFB, and SOCS2) are among the top enriched gene groups commonly down-regulated by MS645 and JQ1 (SI Appendix, Fig. S4E). Notably, genes that function in cell-cycle control and DNA damage repair as well as mitochondrial translation (such as CDK6, RAD51, and MRPL19) represent the top enriched groups uniquely down-regulated by MS645 (Dataset S3). Examples of RNA-seq tracks showing transcriptional down-regulation of representative genes in the top enriched groups by MS645 vs. JQ1 treatment illustrate the profound cellular activity of this bivalent BET inhibitor over monovalent JQ1 in controlling BRD4 function in gene transcriptional activation (Fig. 4C). We confirmed by qPCR that mRNA transcript levels of CDK6 and RAD51 exhibit a rapid reduction in HCC1806 TNBC cells, as early as 4 h after MS645 treatment (Fig. 5A). This is correlated with an MS645-induced decrease of protein expression levels of CDK6 and RAD51, and to a lesser extent CDK4, but not CDK2, CDC2/CDK1, or cyclin B1 in a dose-dependent manner (Fig. 5B). MS645 treatment also resulted in a dramatic reduction of c-Myc expression and an increase of p21, a tumor suppressor and cell-cycle inhibitor (Fig. 5B).
Fig. 4.
RNA-seq analysis of modulation of gene transcription by MS645 vs. JQ1. (A) Global changes in gene transcription of MDA-MB-231 cells after MS645 or JQ1 treatment at 50 or 500 nM, as shown in log twofold change scale. mRNA was collected for RNA-seq analysis from the MDA-MB-231 cells after treatment with DMSO, MS645, or JQ1 for 18 h. (B) Venn diagram analysis showing genes with over twofold changes after MS645 or JQ1 treatment as indicated. (C) Representative RNA-seq tracks highlighting the profoundly superior activity of MS645 over JQ1 in down-regulation of transcriptional expression of a select number of genes that are important for oncogenesis. The BRD4 ChIP-seq tracks shown as reference for BRD4 target genes were generated with MCF7 breast cancer cells and are available from the Gene Expression Omnibus database (accession no. GSE55921) (35).
Fig. 5.
Control of TNBC cell proliferation by bivalent BET inhibitors. (A) Time-course measurements of mRNA levels of CDK6 and RAD51 in HCC1806 cells after MS645 treatment. (B) Effects of MS645 on expression of proteins important for cell-cycle control in HCC1806 cells, as assessed by Western blot analysis. (C) ChIP analysis of effects of MS645 vs. JQ treatment on BRD4 dissociation from target genes. Results were from at least three independent experiments and error bars designate SEM. (D) Coimmunoprecipitation experiment showing that MS645, but not JQ1 or MS417, dissociates BRD4 from its interaction with MED1 (Upper) or transcription factor YY1 (Lower). (E) Schematic illustration of the mechanism of action of bivalent BET BrD inhibitor MS645 in blocking BRD4 activity in gene transcriptional activation: MS645 binding to the tandem BD1–BD2 of BRD4 exerts spatially constrained inhibition of BRD4, which locks BRD4 in an inactive state and blocks it from binding to effect proteins, leading to a sustained down-regulation of gene transcription. Note that the study cannot exclude the possibility of MS645 inhibition of other BET proteins or engaging in a bimolecular target protein inhibition in cells.
We postulated that spatially constrained BD1/BD2 inhibition by MS645 may allosterically disrupt BRD4 association with transcription factors and enhancer/mediator proteins required for accelerated tumor-cell proliferation (8, 9, 34). It was reported that a significant gain of superenhancers in JQ1-resistant SUM159R TNBC cells resulted from strong association of MED1, a key component in the enhancer/mediator complex, with hyperphosphorylated BRD4 and BrD-independent BRD4 recruitment to chromatin (34). Indeed, we observed by ChIP PCR that MS645 not only dissociates BRD4 but also MED1 from CDK6, RAD51, and BRCA1 gene loci, whereas JQ1 only affects BRD4 but not MED1 (Fig. 5C). Our coimmunoprecipitation study further confirmed that only MS645, but not JQ1 or MS417, inhibits BRD4 association with MED1 or transcription factor YY1 (Fig. 5D). Collectively, these results show that MS645 effectively inhibits TNBC cell growth through its sustained inhibition of BRD4 activity in transcriptional activation of genes of key cellular pathways including cell-cycle control and DNA damage repair required to sustain cancer cell rapid growth.
In this study, we report a class of bivalent BrD inhibitors designed to simultaneously target the tandem BrDs of BRD4. We show that spatially constrained binding of BRD4 BD1–BD2 preferentially by our lead bivalent BET BrD inhibitor MS645 affords a sustained inhibition of BRD4 by blocking BRD4 interactions with transcription enhancer/mediator proteins MED1 and YY1 that are required for accelerated proliferation of solid-tumor TNBC cells (Fig. 5E). We demonstrate that the linker length, composition, and rigidity of a bivalent BET inhibitor are determinant factors in dictating its inhibitory capability toward BRD4 transcriptional activity in the functional context in cells. The recently reported PEG-linker bivalent inhibitor MT1 imitates cellular efficacy of MS645 to some extent, whereas AZD5153 is much less effective, or almost ineffective, against solid-tumor TNBC cells (similar to our rigid, benzene-linker bivalent inhibitor MS688). Importantly, our study highlights the notion that the molecular mechanism of BRD4 function in gene transcription in chromatin is far more complex than the current simplistic view of its BrD binding to acetylated histones and transcription proteins and likely varies in a cell-type and context-dependent manner. The latter is defined and influenced by a distinct set of transcription factors and chromatin regulatory proteins that BRD4 is associated with in gene transcription. It should be noted that our study cannot exclude a possibility of MS645 inhibition of other BET proteins, or to a lesser extent engaging in a bimolecular target protein inhibition in cells. Collectively, our study reported here suggests a therapeutic strategy to maximally control BRD4 transcriptional activity required for rapid solid-tumor cancer-cell proliferation such as the devastating, heterogeneous TNBC that lacks targeted therapy.
Materials and Methods
Detailed procedures for purification, biochemical and structural analyses of the BET proteins, cell growth and gene transcription assays, genomic sequencing analysis, and chemical synthesis of various BET BrD inhibitors are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Vivian Stojanoff and Silvia Russi and the staff of 14-1 beamline at the Stanford Synchrotron Radiation Light Source for assisting with X-ray data collection and the staff of the Genomics Sequencing Core at the Icahn School of Medicine at Mount Sinai and the Epigenomics Core of Weill Cornell Medical College for generating RNA-seq data. We also thank Drs. Alaa Abdine, Rinku Jain, Zhenzhen Wang, and Iban Ubarretxena for technical help for the DLS experiment, X-ray crystal structure analysis, and mathematical analysis, and Dr. Nadejda Tsankova for providing glioblastoma cell line U87. This work was supported in part by the research fund from the First Hospital of Jilin University, Jilin Province Science and Technology Development Program Grant 20160101076JC (to S.F.), and research grants from the Breast Cancer Alliance, Department of Defense Breast Cancer Research Program Breakthrough Award W81XWH15-1-0043, and the National Institutes of Health (to M.-M.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors for the BRD4-BD1 dimer in complex with MS645 or MS660 have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6DJC and 6DNE, respectively). All RNA-sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE115550).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720000115/-/DCSupplemental.
References
- 1.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 2.Zeng L, Zhou MM. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 3.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: Selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 5.Smith SG, Zhou MM. The bromodomain: A new target in emerging epigenetic medicine. ACS Chem Biol. 2016;11:598–608. doi: 10.1021/acschembio.5b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French CA, et al. BRD4-NUT fusion oncogene: A novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 7.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Smith SG, Zhou MM. Discovery of chemical inhibitors of human bromodomains. Chem Rev. 2015;115:11625–11668. doi: 10.1021/acs.chemrev.5b00205. [DOI] [PubMed] [Google Scholar]
- 13.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, et al. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012;287:28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi S, Ooike S, Iwata K, Hikawa H, Sugaraha K. 2009. Antitumor agent. International Patent PCT/JP2008/073864 (US2010/0286127 A1)
- 17.Zhang G, et al. Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J Med Chem. 2013;56:9251–9264. doi: 10.1021/jm401334s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picaud S, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci USA. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold LD, Foreman KW, Jin M, Wanner J, Werner D. 2016. Bivalent Bromodomain Ligands, and Methods of Using Same, WO Application WO2015081284A1 (June 4, 2016)
- 20.Bradbury RH, et al. Optimization of a series of bivalent triazolopyridazine based bromodomain and extraterminal inhibitors: The discovery of (3R)-4-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-1,3-dimethyl-piperazin-2-one (AZD5153) J Med Chem. 2016;59:7801–7817. doi: 10.1021/acs.jmedchem.6b00070. [DOI] [PubMed] [Google Scholar]
- 21.Waring MJ, et al. Potent and selective bivalent inhibitors of BET bromodomains. Nat Chem Biol. 2016;12:1097–1104. doi: 10.1038/nchembio.2210. [DOI] [PubMed] [Google Scholar]
- 22.Rhyasen GW, et al. AZD5153: A novel bivalent BET bromodomain inhibitor highly active against hematologic malignancies. Mol Cancer Ther. 2016;15:2563–2574. doi: 10.1158/1535-7163.MCT-16-0141. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka M, et al. Design and characterization of bivalent BET inhibitors. Nat Chem Biol. 2016;12:1089–1096. doi: 10.1038/nchembio.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaidos A, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123:697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 28.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J, et al. Disrupting the interaction of BRD4 with diacetylated twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond JR, et al. Predictive biomarkers of sensitivity to the aurora and angiogenic kinase inhibitor ENMD-2076 in preclinical breast cancer models. Clin Cancer Res. 2013;19:291–303. doi: 10.1158/1078-0432.CCR-12-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tate CR, et al. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res. 2012;14:R79. doi: 10.1186/bcr3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allegra CJ, Fine RL, Drake JC, Chabner BA. The effect of methotrexate on intracellular folate pools in human MCF-7 breast cancer cells. Evidence for direct inhibition of purine synthesis. J Biol Chem. 1986;261:6478–6485. [PubMed] [Google Scholar]
- 33.Tan AR, et al. Evaluation of biologic end points and pharmacokinetics in patients with metastatic breast cancer after treatment with erlotinib, an epidermal growth factor receptor tyrosine kinase inhibitor. J Clin Oncol. 2004;22:3080–3090. doi: 10.1200/JCO.2004.08.189. [DOI] [PubMed] [Google Scholar]
- 34.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagarajan S, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.