Significance
Estrogen receptor-positive (ER+) breast cancer is treated with endocrine therapies, although therapeutic resistance almost invariably develops in advanced disease. Using genome-wide CRISPR screens, we identified genes whose loss confers endocrine resistance, as well as synthetic lethal vulnerabilities to overcome such resistance. These findings reveal an estrogen-induced negative feedback loop that constrains the growth of ER+ tumors, thereby limiting the efficacy of therapies that inhibit ER, and suggest a previously unappreciated therapeutic route to overcoming endocrine resistance.
Keywords: breast cancer, endocrine resistance, estrogen receptor, CRISPR screen, CSK
Abstract
Endocrine therapy resistance invariably develops in advanced estrogen receptor-positive (ER+) breast cancer, but the underlying mechanisms are largely unknown. We have identified C-terminal SRC kinase (CSK) as a critical node in a previously unappreciated negative feedback loop that limits the efficacy of current ER-targeted therapies. Estrogen directly drives CSK expression in ER+ breast cancer. At low CSK levels, as is the case in patients with ER+ breast cancer resistant to endocrine therapy and with the poorest outcomes, the p21 protein-activated kinase 2 (PAK2) becomes activated and drives estrogen-independent growth. PAK2 overexpression is also associated with endocrine therapy resistance and worse clinical outcome, and the combination of a PAK2 inhibitor with an ER antagonist synergistically suppressed breast tumor growth. Clinical approaches to endocrine therapy-resistant breast cancer must overcome the loss of this estrogen-induced negative feedback loop that normally constrains the growth of ER+ tumors.
Oncogenic activation of the estrogen receptor (ER) signaling pathway occurs in over 70% of breast cancers (1). This forms the basis of endocrine therapy that employs antiestrogens and aromatase inhibitors for both breast cancer prevention and treatment (2). However, most patients with advanced disease eventually develop resistance to these endocrine therapies, and even for patients treated in the adjuvant setting, there is a consistent risk of relapse that persists indefinitely (3). Notably, approximate 50% of patients with locally advanced or metastatic ER+ breast cancer do not respond to first-line endocrine treatment because of primary, de novo resistance (4). In addition, most patients who initially respond to the therapy eventually develop secondary acquired resistance (5). Despite significant research efforts and discoveries made in recent years, the precise reasons for endocrine therapy failure in patients with ER+ breast cancer remain largely unknown. Published studies have implicated increased expression of ER; mutations in the ESR1 gene; and activated growth factor receptor signaling, especially of the EGFR/HER2 pathway, as important mechanisms of de novo or acquired resistance (6–8). This has led to various targeted combination strategies aiming to combat endocrine resistance (3). For example, recent clinical trials have shown that cotargeting the ER pathway with either the PI3K/Akt/mammalian target of rapamycin pathway or with the CDK4/6 inhibitor significantly improved the progression-free survival of patients with ER+ breast cancer compared with endocrine therapy alone (9–11).
Although clinical trials of combining other treatments with endocrine therapy are ongoing (12), the current choice of combination has been limited to existing therapies in breast cancer. Furthermore, how the downstream oncogenic pathways of ER and growth factors are activated during endocrine therapy and whether there are other intrinsic mechanisms limiting the efficacy of current therapies remain to be elucidated. Therefore, systematic studies to uncover synthetic lethal vulnerabilities and synergistic targets with endocrine therapy are crucial for identifying novel and optimal combination therapies. In addition, biomarkers predictive of clinical response and molecular mechanisms underlying resistance to endocrine therapies are critical for developing better therapeutic strategies and improving patient outcome.
The recent introduction of the clustered, regularly interspaced, short palindromic repeat (CRISPR)-associated nuclease Cas9 system has facilitated pooled loss-of-function genetic screens (13–16). Seminal studies demonstrated the power of CRISPR screens in identifying key genes or biomarkers that mediate drug and toxin resistance (14, 17), immune response (18–21), and tumor metastasis (22). This approach has also shown great promise in identifying synthetic lethal interactions and revealing novel combination therapies for precision cancer medicine (23, 24).
In this study, we conducted genome-wide CRISPR knockout screens on two ER+ breast cancer cell lines to identify key mediators of endocrine resistance. Based on the top tumor suppressor gene identified in the primary screen whose loss induced endocrine therapy resistance, we performed genome-wide synthetic lethal CRISPR screens to discover novel targets for treating endocrine resistance. The gene signature revealed by the CRISPR screen hits is associated with the response to endocrine therapy in numerous clinical studies. Finally, we validated combining endocrine therapy with inhibiting a novel target identified in the synthetic lethal screen to overcome endocrine resistance in cell line and patient-derived tumor xenograft models. Our approach of utilizing a second round of CRISPR screens to identify novel therapeutic targets provides an efficient alternative to conducting primary CRISPR screens on large cell line panels to infer synthetic lethal interactions.
Results
Genome-Wide CRISPR Screens Identified ER+ Breast Cancer-Specific Essential Genes.
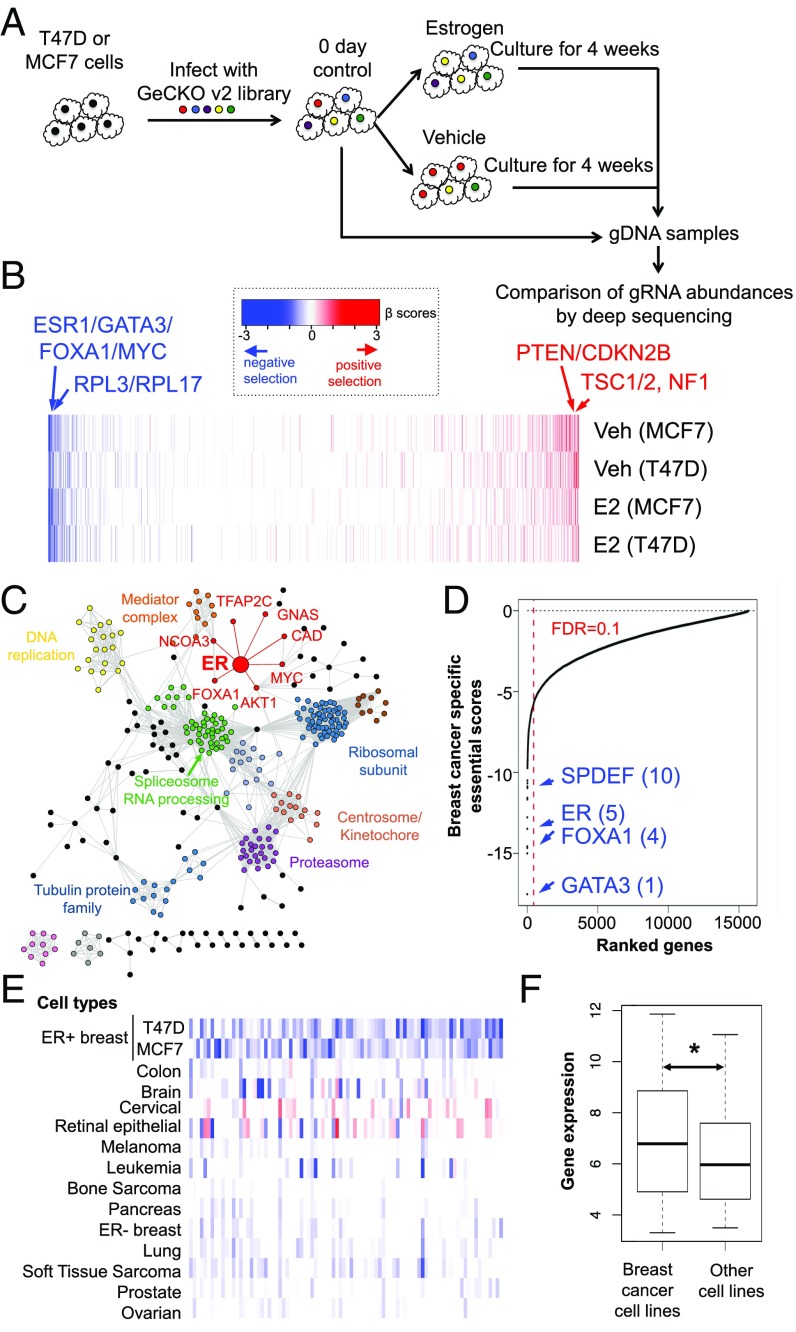
To systematically investigate genes whose loss affects cell viability or potentiates the estrogen-independent growth of ER+ breast cancer cells, we performed genome-wide CRISPR/Cas9 knockout screens in the ER+ breast cancer cell lines MCF7 and T47D using the Genome-Scale CRISPR Knock-Out (GeCKO) v2 library (25). After infection with the lentiviral guide RNA (gRNA) library and selection by puromycin, the cells were cultured in hormone-depleted medium and treated with either estrogen (17β estradiol or E2) or vehicle (Veh) control over 4 wk (Fig. 1A). The sequences encoding the gRNA were PCR-amplified from the transduced cells at day 0 and after 4 wk of culture and quantified by high-throughput sequencing (SI Appendix, Fig. S1 A and B). We then identified negatively and positively selected genes by calculating the gene essentiality score using MAGeCK-VISPR, a statistical algorithm we previously developed for CRISPR screen analyses (26, 27). The MAGeCK-VISPR algorithm compares the gRNA abundance of all of the gRNAs targeting a gene across different conditions and assigns each gene a log fold-change “β-score” of essentiality in each condition compared with the controls. A positive (or negative) β-score indicates the corresponding gene is under positive (or negative) selection in the CRISPR screen (Dataset S1). Overall, most genes are consistently positively or negatively selected across conditions and cell lines (Fig. 1B). Known driver genes for ER+ breast cancers (28, 29), such as ER (ESR1) itself, GATA3, FOXA1, and MYC, are strongly negatively selected (Fig. 1 B and C), while tumor suppressors, such as NF1, TSC1, TSC2, and PTEN, are positively selected (Fig. 1B). The essential genes are enriched in many fundamental biological processes, such as gene expression, RNA processing, and translation (Fig. 1C and SI Appendix, Fig. S1C). In addition, ER and its signaling network genes (MYC, FOXA1, CCND1, PHB2, NCOA3, and AKT1) remain essential in the Veh condition (SI Appendix, Fig. S1D), and the top essential genes significantly overlap between the E2 and Veh conditions (Fisher’s exact test: P < 2.2e-16; SI Appendix, Fig. S1E and Dataset S2). These data indicate that the ER pathway continues to play an essential role even in the absence of estrogen.
Fig. 1.
CRISPR functional screens on two breast cancer cell lines, T47D and MCF7. (A) Outline of the genome-wide CRISPR screen strategy. (B) Positively selected (red) and negatively selected (blue) genes in T47D and MCF7 cells under E2 (10 nM) and Veh (EtOH) treatments in hormone-depleted medium plus 10% dextran-charcoal–treated FBS. The positive (or negative) β values (calculated from the MAGeCK algorithm) indicate positive (or negative) selection of a gene, respectively. (C) Network view of top 1,000 negatively selected genes in T47D and MCF7. Here, nodes represent genes and an edge connecting two genes if both are in the same pathway. ER and its associated genes are highlighted in red. The pathway information is extracted from the GeneMANIA database (69). Six hundred seventy-one unconnected genes are not shown. (D) Scores of breast cancer-specific essential genes. The names and ranks of some known breast cancer-specific genes are marked. The score is based on the screening data of T47D and MCF7 (under E2 treatment), as well as other cell lines collected from public CRISPR screens. (E) β-Scores of breast cancer-specific essential genes in multiple cancer cell types. (F) Expression of breast cancer-specific essential genes is significantly higher in breast cancer cell lines than in other cell lines (Wilcoxon rank-sum test: *P < 0.05).
We next sought to identify genes with a stronger negative selection in ER+ breast cancer cells compared with other cancer cells, as they may be the therapeutic targets for breast cancer. We collected public genome-wide CRISPR screen data from 43 cell lines representing 13 different cell types (14, 16, 30, 31) and compared them with the screening results on T47D and MCF7 (under E2 treatment). We derived a score to quantify whether a gene is ER+ breast cancer-specific essential by comparing this gene with other genes within the same cell line and the same gene in other cancer cells (details are provided in Materials and Methods). This approach identified ∼176 genes with specific essentiality in ER+ breast cancers [false discovery rate (FDR) ≤ 0.1; Fig. 1 D and E and Dataset S3]. Overall, the ER+ breast cancer-specific essential genes tend to have higher expression in T47D and MCF7 cells compared with the other cell lines (Fig. 1F), are amplified or up-regulated in samples from patients with breast cancer (SI Appendix, Fig. S1F), and are enriched in breast cancer-related pathways (Dataset S4). Many of these genes have physical or genetic interactions with ER (SI Appendix, Fig. S1G), confirming the central role of ER in ER+ breast cancer cells. Interestingly, seven of the top 20 specific essential genes are transcription factors (GATA3, TFAP2C, TRPS1, FOXA1, ESR1, SPDEF, and GRHL2). Among these, FOXA1, GATA3, SPDEF, and TFAP2C are known to interact with ER and exert critical functions in breast cancer (32–35). TRPS1 and GRHL2 are also associated with breast cancer progression and have been implicated as oncogenes in ER+ breast cancers (36–38) (SI Appendix, Fig. S1H). The identification of ER and the known components of the ER signaling pathway, as well as other previously identified breast cancer oncogenes, validated the robust nature of the screen.
Loss of C-Terminal SRC Kinase Mediates Estrogen-Independent Growth of ER+ Breast Cancer Cells.
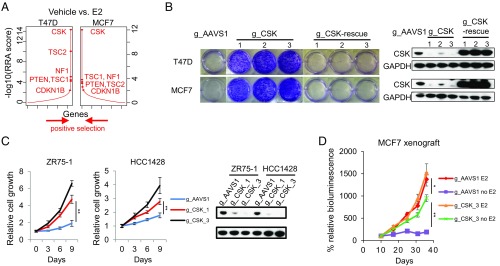
We next searched for key genes that drive estrogen-independent growth by finding genes with stronger positive selection in the Veh condition compared with the E2 condition (Fig. 2A). The hit list (Dataset S5) includes several known tumor suppressor genes, including NF2, TSC2, LATS2, and PTEN, as well as NF1, whose silencing has been previously reported to cause tamoxifen resistance (39). The strongest differential hit in both T47D and MCF7 cells is C-terminal SRC kinase (CSK) (Fig. 2A and SI Appendix, Fig. S2A). CSK has been previously implicated to be involved in fulvestrant-induced cell death in MCF7 cells, yet through a mechanism independent of SRC family kinases (SFKs) (40). All six CSK-targeting gRNAs in the GeCKO2 library are dramatically enriched in both MCF7 and T47D cells in the Veh versus E2 condition (SI Appendix, Fig. S2B). Interestingly, CSK was not found to be positively selected in recent genome-wide shRNA screens of ER+ breast cancer cells cultured in full medium (41) (SI Appendix, Fig. S2C). Given that CSK was the top positively selected gene in both MCF7 and T47D cell lines in estrogen-depleted medium and its known role in inhibiting the function of SFKs (42), we focused on CSK for further analysis.
Fig. 2.
CSK mediates hormone-independent breast cancer cell growth. (A) Top positively selected genes in both T47D and MCF7 cells cultured in hormone-depleted medium plus 10% dextran-charcoal–treated FBS (DCC-FBS) treated with Veh (EtOH) conditions, compared with E2 (10 nM) conditions. The RRA scores obtained by comparing Veh versus E2 conditions from MAGeCK are shown. (B, Left) Effects of cell growth by knocking out CSK in T47D and MCF7 cells using three different gRNAs against CSK and one AAVS1_gRNA as a control. The CSK function was rescued by the expression of three gRNA-resistant CSK cDNAs in these CSK-null cells. Cell growth was measured by crystal violet staining assays, and the cells were cultured in hormone-depleted medium plus 10% DCC-FBS. (B, Right) Immunoblot analysis for indicated proteins of control (gAAVS1), CSK-null, and rescued CSK-null cells. GAPDH was used as a loading control. (C, Left and Center) Estrogen-independent growth of ZR75-1 and HCC1428 cells harboring two different gRNAs against CSK and one gAAVS1 as a control. All of the cells were cultured in hormone-depleted medium plus 10% DCC-FBS. The data are represented as mean ± SD (n = 3; two-tailed Student’s t test: *P < 0.05, **P < 0.005). (C, Right) Immunoblot analysis for indicated proteins of control (gAAVS1) and CSK-null cells. GAPDH was used as a loading control. (D) Estrogen-independent growth of MCF7 xenografts. MCF7 cells harboring either gAAVS1 or gCSK were injected into the ovariectomized nude mice in the presence of estrogen. Mice were assigned randomly (day 7), in groups of eight, to continued estrogen supplementation (E2, 0.1 mg⋅kg−1⋅wk−1) or estrogen withdrawal (−E2). Luminescence values were plotted as an average of percentage of the first measurement (% relative bioluminescence) for each mouse in each respective group. The measurements were performed in intact male mice at days 10, 17, 25, 31, and 37 after tumor cell injection (two-tailed Student’s t test: *P < 0.05, **P < 0.005).
We first sought to validate that CSK knockout confers E2-independent growth of ER+ breast cancer cells. We introduced three different gRNAs targeting CSK (one from the GeCKO2 library and two newly designed) and a control gRNA targeting the AAVS1 safe-harbor locus into T47D and MCF7. All three CSK-targeting gRNAs depleted CSK protein and markedly stimulated cell growth in stripped medium without E2 (Fig. 2B), but they had only marginal growth effects in full medium or in stripped medium plus E2 (SI Appendix, Fig. S2D), indicating CSK loss is a specific driver for E2-independent growth of these cells. The same phenotype was also observed in two additional ER+ breast cancer cell lines, ZR75-1 and HCC1428 (Fig. 2C). Extending our cell culture findings in vivo, we also confirmed that depletion of CSK resulted in E2-independent growth of MCF7 xenografts in ovariectomized mice (Fig. 2D). In contrast to the AAVS1-targeted control MCF7 xenografts that showed almost complete dependence on E2 for growth, the CSK-depleted MCF7 xenografts grew robustly in the absence of E2 and were only modestly stimulated by the addition of E2. Moreover, deletion of CSK induced a striking sickle-like morphology in both T47D and MCF7 cells and increased cell migration in a transwell assay (SI Appendix, Fig. S2 E and F), suggesting a more motile phenotype (43). Finally, the estrogen-independent growth and morphological changes in these CSK-null cells could be fully reversed by the overexpression of a human CSK cDNA containing protospacer adjacent motif sequence mutations in each of the targets of the three CSK gRNAs to escape CRISPR/Cas9 cutting (Fig. 2B and SI Appendix, Fig. S2E).
CSK Is Directly Regulated by ER.
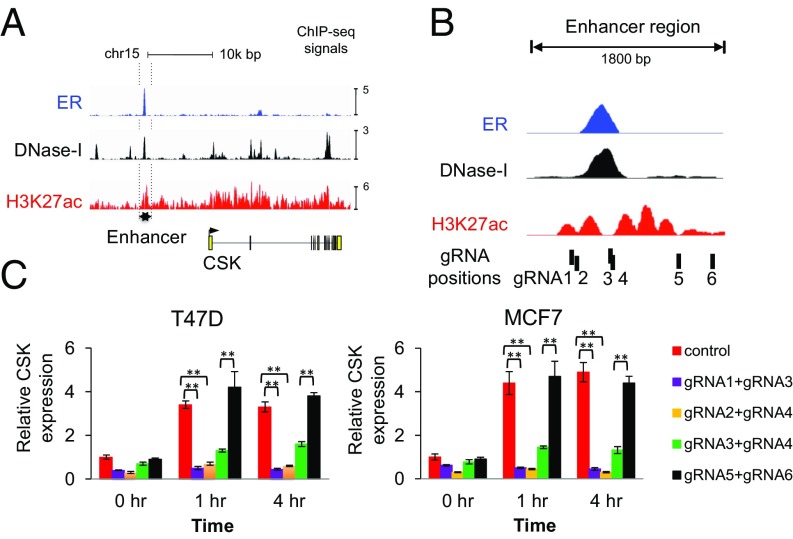
As CSK was differentially selected between the Veh and E2 conditions, we next asked whether estrogen and ER regulated CSK expression. The expression of CSK is up-regulated in a majority of breast cancer cell lines or mouse models treated with E2 in the Nuclear Receptor Signaling Atlas database (44) (SI Appendix, Fig. S3A). Examination of ER and H3K27ac ChIP-sequencing (ChIP-seq) and DNase-sequencing data revealed a putative ER-bound enhancer ∼10 kb upstream of the CSK transcription start site (Fig. 3A). This region contains an ER-binding site as well as an ER-binding motif, and it is largely marked by H3K27ac only in ER+ breast cancer cell lines, suggesting that it is an ER-specific enhancer (SI Appendix, Fig. S3B). In 86% (19 of 22) of patients with ER+ breast cancer, ER was bound to the enhancer of CSK, while in two patients with ER− breast cancer, no ER binding was detected (45) (SI Appendix, Fig. S3C), an indication that the ER-CSK regulation may be widespread in ER+ breast tumors. To test whether ER activates CSK through this putative enhancer, we introduced three pairs of gRNAs together with Cas9 to fully or partially delete the putative enhancer and another pair targeting a region away from the enhancer as a control (Fig. 3B). In the absence of Cas9/gRNA transfection, CSK expression is strongly up-regulated upon E2 treatment. This activation was abrogated when the enhancer was disrupted, while deletion of a nearby region did not affect CSK expression (Fig. 3C and SI Appendix, Fig. S3D). Moreover, the deleted enhancer, but not the nearby region, confers hormone-independent growth of the cells, indicating that this enhancer is required both for the ER regulation of CSK and for the suppression of estrogen-stimulated growth (SI Appendix, Fig. S3E).
Fig. 3.
ER binds to CSK enhancer and regulates CSK expression. (A) ER ChIP-seq, as well as DNA hypersensitivity (DNase-I) and H3K27ac signals on the proximal enhancer region of CSK. (B) Zoomed-in view of the enhancer regions upstream of CSK. The positions of six gRNAs targeting this putative enhancer are also shown. (C) Knockout of the ER-binding site and its flanking region by four pairs of gRNAs against these regions. After hormone depletion for 2 d, the cells were treated with E2 (40 nM) for the indicated times and total RNA was subjected to real-time RT-PCR analysis to measure the mRNA level. The relative gene expression was measured by qRT-PCR after normalizing to the amount of GAPDH signal (mean ± SD for n = 3; two-tailed Student’s t test: *P < 0.05, **P < 0.005).
Changes in Growth Factor Signaling upon CSK Loss Mediate Endocrine Resistance in ER+ Breast Cancer Cells.
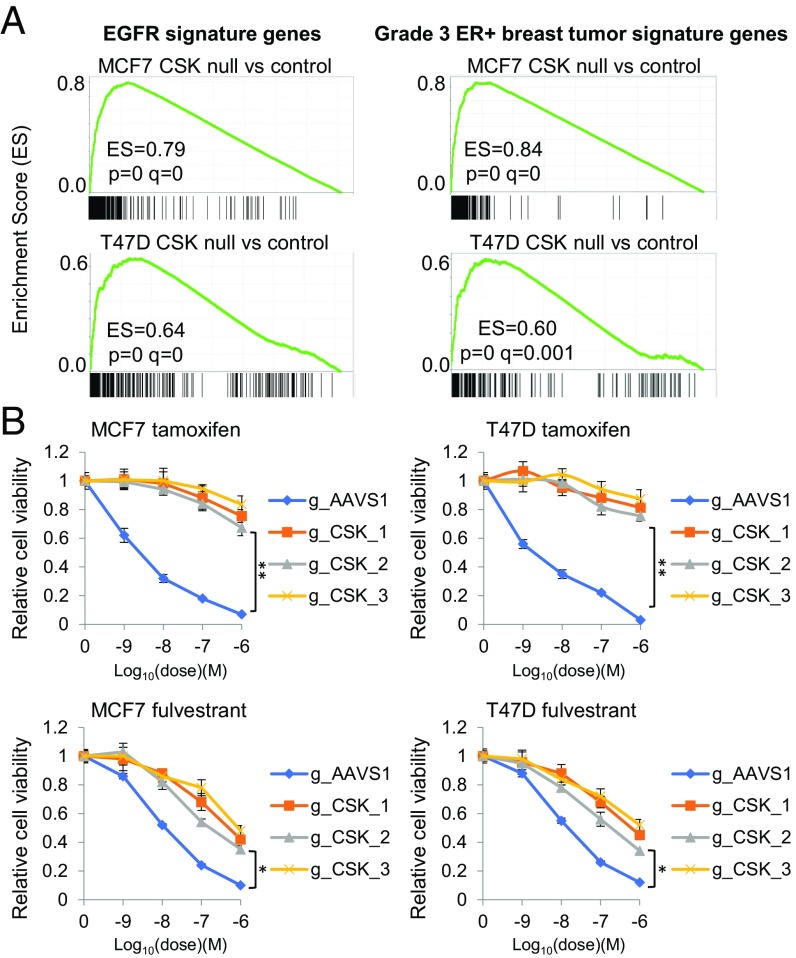
To understand how CSK loss leads to estrogen-independent growth of ER+ breast cancer cells, we performed RNA-seq analysis to identify differentially expressed genes and pathways upon CSK loss in T47D and MCF7 cells. Loss of CSK led to global changes in gene expression (SI Appendix, Fig. S4A and Dataset S6), and we defined a signature of 292 CSK-suppressed genes consistently up-regulated upon CSK knockout in both MCF7 and T47D (log2 fold change > 1 and adjusted P < 0.01; SI Appendix, Fig. S4C and Dataset S7). The expression of EGFR, whose overexpression can elicit tamoxifen resistance (1), was significantly increased after CSK loss (SI Appendix, Fig. S4A). In addition, EGFR signature genes, genes associated with high-grade ER+ breast tumors as well as other oncogenic pathways, such as metastasis, cell cycle, and epithelial-mesenchymal transition, were significantly up-regulated (Fig. 4A and SI Appendix, Fig. S4B and Dataset S7). These results suggest that CSK deletion or expression suppression activates growth factor signaling pathways, which could drive the E2-independent growth of ER+ breast cancer cells and elevate breast tumor grade. Partial or complete ablation of ER significantly decreased cell proliferation in both MCF7 and T47D CSK-null cells (SI Appendix, Fig. S4D), suggesting that ER continues to play an important role in the absence of CSK. Given the essential role of ER in CSK-null cells, we tested whether the growth of CSK-null T47D or MCF7 cells could be inhibited by tamoxifen or fulvestrant. Notably, the CSK-null T47D or MCF7 cells were completely resistant to the ER antagonist tamoxifen but retained partial sensitivity to the ER degrader fulvestrant (Fig. 4B). We observed that loss of CSK only partially inhibits the ability of fulvestrant to degrade ER (SI Appendix, Fig. S4E), consistent with a previous study showing activated SFKs blocking fulvestrant-induced ER down-regulation (46). Additionally, we have previously demonstrated that while tamoxifen is unable to prevent growth factor-stimulated ER signaling, fulvestrant is able to more fully inhibit ER action (47). These results demonstrate that ER remains essential for estrogen-independent growth induced by CSK loss.
Fig. 4.
Growth factor and ER signaling changes induced by CSK loss. (A) Gene set enrichment analysis for EGFR signature genes and genes associated with higher grade (grade 3) ER+ breast tumors. The corresponding gene set IDs in the Molecular Signatures Database (70) are “KOBAYASHI_EGFR_SIGNALING_24HR_DN” and “SOTIRIOU_BREAST_CANCER_GRADE_1_VS_3_UP,” respectively. Genes in the figures (black bars) are ranked based on their differential expression between CSK-null and control cells. (B) Effects of CSK knockout on sensitivity to two ER antagonists, tamoxifen and fulvestrant, in T47D and MCF7 cells. The relative cell viability of control (AAVS1) and CSK-null cells after treatment with indicated compound concentrations for 5 d is shown (mean ± SD for n = 3; two-tailed Student’s t test: *P < 0.05, **P < 0.005). All of the cells were cultured in full medium (DMEM or RPMI 1640) with 10% FBS.
Genome-Wide CRISPR Screen for Genes Synthetically Lethal with CSK Loss.
To identify the key genes that drive hormone-independent growth upon CSK loss, we performed a second round of genome-wide CRISPR screening in the CSK-null T47D cells and control cells cultured in hormone-depleted medium treated with E2 or Veh (SI Appendix, Fig. S5 A–C and Dataset S8). Using the same approach to compare public screening datasets of non-breast cancer cell lines, we identified 685 specific essential genes in CSK-null T47D cells under Veh treatment with statistical significance (FDR ≤ 0.05; Dataset S9). These genes include genes in the HER2 (ERBB2), PI3K-AKT (PIK3R1/2, AKT1), and MAPK signaling pathways [MAPK8, p21 protein-activated kinase 2 (PAK2)] that are known to be activated in endocrine-resistant breast tumors (1) (Fig. 5A). Of note, ER remains essential in the absence of CSK, albeit to a lesser extent compared with the control cells (β = −0.43 and −0.28, ranking = 23 and 629 in the control and CSK-null cells, respectively). The essentiality of ER and genes in the HER2/EGFR signaling pathway in the CSK-null cells is further supported by the up-regulated ER and EGFR signature genes in the CSK-null cells (Fig. 4A and SI Appendix, Fig. S4A). These results suggest that loss of CSK increases cross-talk between ER and growth factor signaling pathways.
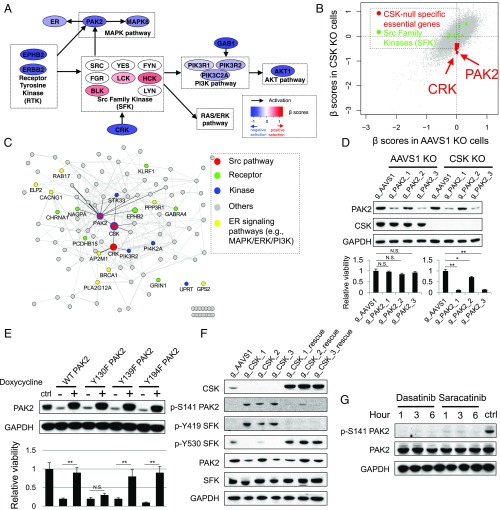
Fig. 5.
PAK2 is synthetic lethal to CSK loss. (A) Essentialities of genes in the SFKs and associated pathways, measured by β-scores from CRISPR screens, in CSK-null cells. Genes are colored based on their β-scores. (B) β-Scores in CSK-null cells versus CSK WT cells. Two Src pathway genes (PAK2 and CRK) and SFKs are marked. (C) Interaction network of genes that are differentially required upon CSK loss. Edges connecting genes indicate possible gene interactions from public datasets. (D) Validation of a synthetic lethal interaction between PAK2 and CSK in T47D cells. Relative viabilities of T47D CSK-null (gCSK3) and control (gAAVS1) cells harboring three gRNAs against PAK2 and one gAAVS1 as a control are shown (mean ± SD for n = 3; two-tailed Student’s t test: *P < 0.05, **P < 0.005). N.S., not significant. The control cells were cultured in hormone-depleted medium [10% dextran-charcoal–treated FBS (DCC-FBS)] plus E2 (10 nM), and the CSK-null cells were cultured in hormone-depleted medium (10% DCC-FBS) plus Veh. Immunoblot analysis indicated the protein level of PAK2 and CSK in the control (AAVS1) and CSK-null T47D cells. GAPDH was used as a loading control. (E) Rescue assay for PAK2 function. Relative viabilities of T47D WT (ctrl) cells, as well as CSK and PAK2 double-KO cells with inducibly rescued WT PAK2 and PAK2 mutants (Y130F, Y139F, and Y194F) are shown [“+” and “−” indicate plus/minus doxycycline (1 μg/mL) treatments respectively]. The T47D CSK-null cells were first transduced with PAK2-encoding lentiviruses to allow inducible overexpression of WT PAK2 and the PAK2 mutants. The endogenous PAK2 was then deleted using the specific gRNA (gPAK2_3) without affecting the exogenous PAK2 (mean ± SD for n = 3; **P < 0.005). All of the cells were cultured in hormone-depleted medium plus 10% DCC-FBS, and GAPDH was used as a loading control. (F) Immunoblot analysis indicated the protein level of phosphorylated PAK2 and SFK as well as the total protein level of CSK, SFK, and PAK2 in the control (gAAVS1), CSK-null, and rescued CSK-null T47D cells. (G) Expression of PAK2 and PAK2S141 upon treatment with two SFK inhibitors, dasatinib and saracatinib, in the CSK-null cells for 1, 3, and 6 h. ctrl, CSK-null cells with Veh treatment for 6 h. All of the cells were cultured in hormone-depleted medium plus 10% DCC-FBS, and GAPDH was used as a loading control.
We next sought to identify genes that are specifically essential in the CSK-null cells under the Veh condition, as these would be the key genes that are suppressed by CSK and confer endocrine resistance in the cells upon CSK loss. Applying the same MAGeCK-VISPR method to compare screening results between the control and CSK-null T47D cells, we discovered over 60 genes that are not required in control cells under the E2 condition but are specifically required in the CSK-null cells under the Veh condition (Dataset S10). Several top hits, such as EPHB2, proto-oncogene c-crk (CRK), PAK2, and PIK3R2, are in SFK pathways (Fig. 5A). However, none of the nine SFK members could be identified as an essential gene in the CSK-null cells (Fig. 5B), indicating that paralogs of the SFKs may provide functional redundancy (30).
Two particularly interesting genes, PAK2 and CRK, are directly associated with SFK signaling pathways (Fig. 5C). PAK2 is a serine/threonine kinase whose activity can be stimulated by the small GTPases CDC42 and RAC1 (48) and regulated by the SFKs (49, 50). CRK is a member of an adapter protein family that binds to several tyrosine-phosphorylated proteins and is involved in activating SFKs (51). We focused on PAK2 among the top synthetic lethal candidates of CSK as it is highly expressed in ER+ breast cancer cells and it is a potential therapeutic target with existing small-molecule inhibitors. To confirm the specific requirement of PAK2 in the CSK-null cells, we knocked out PAK2 in the CSK-null cells and control cells using three different gRNAs targeting PAK2 (Fig. 5D). Consistent with our screen data, PAK2 (but not PAK1) is essential only in the CSK-null cells cultured under the Veh condition (SI Appendix, Fig. S5D), and the degree of PAK2 essentiality is correlated with the knockout efficiency (Fig. 5D). This synthetic lethal interaction between PAK2 and CSK is also observed in three additional ER+ breast cancer cells (MCF7, ZR75-1, and HCC1428; SI Appendix, Fig. S5E), supporting its general importance in ER+ breast tumors. In addition, the cell growth inhibition in the CSK-null cells upon PAK2 loss could be rescued by the doxycycline-inducible overexpression of a gPAK2/Cas9-resistant WT PAK2 cDNA (Fig. 5E), confirming the essential role of PAK2 in hormone-independent cells induced by CSK loss.
To further understand how CSK loss leads to PAK2 activation, we investigated the phosphorylation patterns of PAK2 and SFKs. The autophosphorylation site (Serine141) of PAK2, an important marker of PAK2 activation (52), could be distinctly detected in the CSK-null cells but not in the control or CSK-rescued cells (Fig. 5F). More importantly, this differential phosphorylation pattern of PAK2 is correlated with the differential phosphorylation pattern of the SFKs (Fig. 5F), suggesting that PAK2 and SFKs could be simultaneously activated upon CSK loss. To understand whether the activation of PAK2 is SFK-dependent, we treated the CSK-null cells with two SFK inhibitors, dasatinib and saracatinib. The phosphorylation of PAK2 S141 was abrogated upon the inhibitor treatment (Fig. 5G), suggesting that SFKs are involved in PAK2 activation by tyrosine phosphorylation. To uncover the specific tyrosine on PAK2 that is important for PAK2 function, we generated three Y-to-F mutations (Y130F, Y139F, and Y194F) in PAK2 that have been previously implicated in PAK2 function (49). As PAK2 is essential in the CSK-null cells, we first transduced the CSK null cells with PAK2-encoding lentiviruses to allow inducible overexpression of WT PAK2 and the PAK2 mutants. The endogenous PAK2 was then deleted using a specific gRNA, and the cell viability was assayed in the presence or absence of the inducible PAK2 alleles. While the Y139F and Y194F mutants could rescue PAK2 function to similar levels as WT PAK2, the Y130F mutant failed to rescue PAK2 function (Fig. 5E), indicating the critical role of Y130 in SFK-mediated phosphorylation and activation of PAK2.
An Estrogen-Regulated Feedback Loop Limits the Effectiveness of Current Endocrine Therapy.
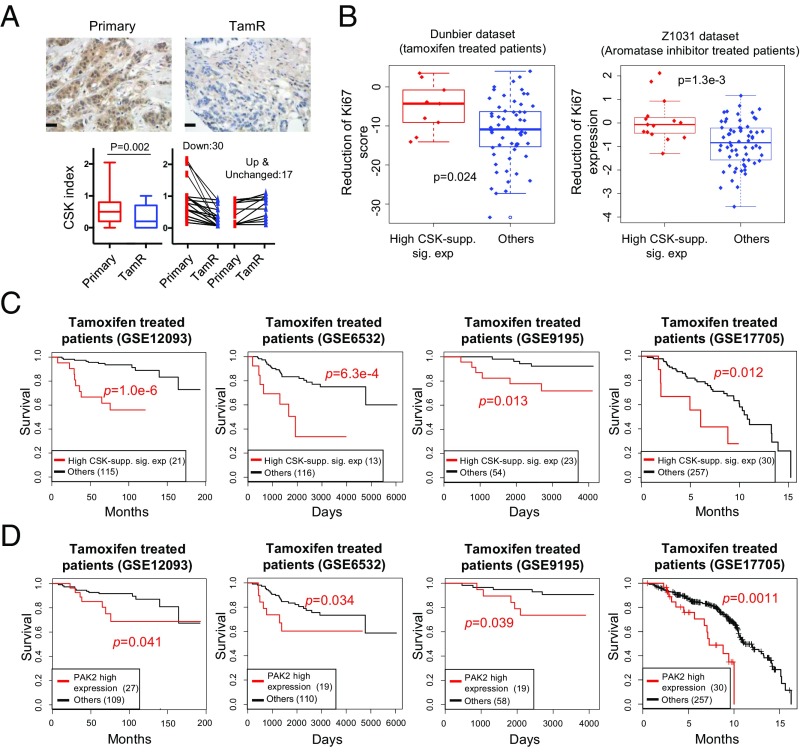
As CSK expression in ER+ breast cancer cells is induced by estrogen, we next asked whether endocrine therapy inhibited CSK expression in these cells. Treatment of MCF7 cells with tamoxifen or fulvestrant for 2, 4, and 6 d gradually decreased CSK protein expression and induced activating phosphorylation of SFKs and PAK2 (SI Appendix, Fig. S6A). This supports the existence of a negative feedback loop, where endocrine therapies that inhibit ER activity repress CSK expression and activate SFKs and PAK2. In addition, CSK expression is decreased in other models of endocrine resistance, including long-term estradiol-deprived (LTED) and tamoxifen- or fulvestrant-resistant cell lines (SI Appendix, Fig. S6B). To investigate the clinical relevance of CSK inhibition as a mechanism of endocrine therapy resistance, we obtained 47 matched pairs of primary and tamoxifen-resistant tumor samples and found CSK to be down-regulated in 64% (30 of 47) of the tamoxifen-resistant tumors by immunohistochemistry (Fig. 6A). These data suggest that down-regulation of CSK may be a frequent mechanism of acquired endocrine resistance.
Fig. 6.
Clinical relevance of an ER-regulated feedback loop. (A) Illustrated images of CSK staining (immunohistochemistry) in matched primary and tamoxifen-resistant (TamR) ER+ breast tumors (Scale bar: 100 μm.). Quantification distribution of CSK staining in 47 matched pairs is shown, including 30 (or 12) that have decreased (or increased) CSK expression at the TamR stage, respectively. The P value is calculated using a two-tailed paired Student’s t test. (B) CSK gene signatures predict patient response to endocrine treatments in two endocrine treatment clinical trials that have matched expression measurements before and after treatment (53, 54). Patients in the “High CSK-supp. sig. exp” group (or patients with high expression of CSK-suppressed signature genes as a surrogate of CSK loss after treatment) have less reduction of Ki67 gene expression, an indication of less efficacy in endocrine treatment. The P value is calculated using the Wilcoxon rank-sum test. More details are provided in Materials and Methods. (C) Higher expression of CSK-suppressed signature genes (High CSK-supp. sig. exp) indicates worse clinical outcome in four expression datasets of patients with breast cancer who were treated with tamoxifen (55–59). Expression data are extracted and processed from the National Center for Biotechnology Information Gene Expression Omnibus under the accession number shown in each figure, respectively. (D) PAK2 overexpression corresponds to worse clinical outcome in a cohort of patients with breast cancer who were treated with tamoxifen.
To further support the clinical importance of CSK inhibition for endocrine resistance in ER+ breast cancers, we collected public datasets of patients with breast cancer treated with endocrine therapy and linked the gene expression data with clinical outcome (Fig. 6 C and D). As CSK is very highly expressed in lymphoid tissues (SI Appendix, Fig. S6C), its expression in tumors may be biased by the presence of tumor infiltrating lymphocytes. We thus used the increased expression of the 292 CSK-suppressed signature genes as a proxy for CSK inhibition (SI Appendix, Fig. S4C). The expressions of the signature genes are negatively correlated with CSK expression across different tissue types (SI Appendix, Fig. S6 D and E), providing a reliable proxy for estimation of CSK activity in tumor samples. In two clinical trials that include breast tumor expression measurements before/after endocrine therapy (53, 54), tumors with the CSK inhibition signature show significantly less Ki67 reduction upon treatment, indicating that endocrine therapy is less effective in these patients (Fig. 6B). In four additional studies (55–59) of patients treated with tamoxifen, the CSK inhibition signature is significantly correlated with high-grade ER+ tumors and worse survival (Fig. 6C and SI Appendix, Fig. S6F). Moreover, higher expression of PAK2 in these cohorts is also significantly associated with worse clinical outcome after tamoxifen treatment (Fig. 6D). These data support the clinical importance of the negative feedback loop, whereby endocrine therapies inhibit expression of CSK, leading to activation of PAK2, ultimately limiting the effectiveness of the therapy.
Potential Therapeutic Strategies to Overcome Endocrine Resistance.
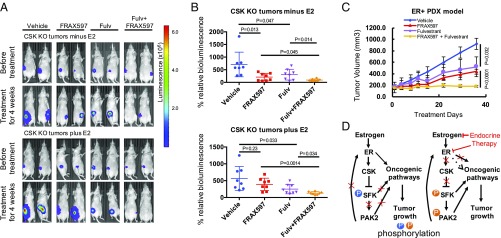
Given the existence of the ER-regulated negative feedback loop involving CSK inhibition and PAK2 activation that limits the effectiveness of endocrine therapy and contributes to endocrine resistance of ER+ breast cancer cells, we tested the effect of simultaneously inhibiting ER and PAK2. Combining the PAK2 inhibitor FRAX597 with fulvestrant or tamoxifen showed strong synergy in both MCF7 CSK-null cells and WT MCF7 cells (SI Appendix, Fig. S7A). To test PAK2 as a potential therapeutic target in endocrine-resistant breast cancer, we treated the endocrine-resistant, CSK-null xenograft tumors in ovariectomized mice with FRAX597. Tumors were more sensitive to PAK2 inhibition in the absence of E2 (Fig. 7 A and B), suggesting the CSK–PAK2 synthetic lethal interaction was more pronounced in hormone-deprived conditions. While fulvestrant alone inhibits the growth of CSK-null tumors to some extent, the combination of fulvestrant with FRAX597 is substantially more effective in blocking the growth of CSK-null tumors with or without E2 (Fig. 7B). We further tested the efficacy of combining FRAX597 with fulvestrant in an ER+ patient-derived xenograft (PDX) model of breast cancer (TM00386; The Jackson Laboratory). While treatment with FRAX597 or fulvestrant alone reduces tumor growth, the combination was synergistic and completely inhibited tumor growth (Fig. 7C and SI Appendix, Fig. S7 B and C). The individual and combined treatments were very well tolerated, as we observed consistent weight gain among the mice on the various treatment arms of the study (SI Appendix, Fig. S7D). These data suggest dual PAK2 and ER inhibition as an effective combination therapy for treating ER+ breast cancer.
Fig. 7.
Potential therapeutic strategies to overcome endocrine resistance. (A) Representative images showing BILs in female, athymic, ovariectomized nude mice plus/minus estrogen (0.1 mg⋅kg−1⋅wk−1) bearing MCF7 CSK-null tumors, which were treated with Veh, FRAX597(60 mg⋅kg−1⋅d−1), fulvestrant (5 mg⋅wk−1), or combinations for 4 wk. (B) Effects of treatments on the CSK-null xenografts. Mice with CSK-null tumors were treated with single or combination treatment with Veh, FRAX597, and fulvestant for 4 wk. Luminescence values were plotted as an average of percentage of the first measurement (% relative BIL) for each mouse in each respective group (n = 8). P values were calculated by the Student’s t test. (C) Single or combination treatments with Veh, FRAX597, and fulvestant in TM00386 PDX (The Jackson Laboratory) tumors for 35 d in each respective group (n = 8). P values are indicated from a two-tailed unpaired t test. Data are represented as mean ± SD. (D) Proposed mechanism of endocrine resistance driven by CSK loss and synthetic lethal vulnerabilities with SFK and PAK2 genes for ER+ breast cancer.
Discussion
Using genome-wide CRISPR/Cas9 screening, we identified essential genes driving the growth of ER+ breast cancer cells, including many transcription factors that are key components of the estrogen signaling pathway. More significantly, these screens revealed a previously unappreciated negative feedback loop that limits the efficacy of ER-targeted therapies and, when disrupted, leads to endocrine resistance (Fig. 7D). We found CSK to be a key component of this feedback loop and an estrogen-stimulated tumor suppressor whose loss drives hormone-independent cell growth. From a secondary genome-wide CRISPR/Cas9 screen, we uncovered a synthetic lethal interaction between CSK and PAK2 in endocrine-resistant breast cancer cells. In the presence of estrogen, ER activates CSK, whose expression represses SFK and PAK2 activity. Inhibition of ER activity by endocrine therapy represses CSK, allowing the activation of SFK and PAK2 independent of ER regulation. Activation of SFK and PAK2, in turn, stimulates oncogenic signaling pathways and promotes estrogen-independent growth. While the continued requirement for ER genomic action upon CSK depletion is supported by the finding of the up-regulation of direct ER target genes in the CSK-null cells versus the control cells in estrogen-depleted medium, we cannot rule out a contribution of nongenomic ER signaling as well.
Our preclinical results both in vitro and in vivo, combined with clinical observations of the impact of CSK loss and PAK2 overexpression on patient survival, strongly support the clinical importance of this negative feedback loop. Our data also suggest PAK2 as a therapeutic target in patients with ER+ breast cancer, both after the development of endocrine resistance and potentially in hormone-sensitive patients, to increase the efficacy of ER-targeted therapy.
Materials and Methods
Breast Cancer Cell Culture.
The MCF-7, T47D, ZR75-1, and HCC1428 human cell lines were grown as described previously (60). Tam-R and Flu-R cells were derived by long-term exposure to tamoxifen and fulvestrant grown under the same conditions as WT MCF-7 and T47D cells (61). T47D/LTED and MCF-7/LTED cells were generated through culture in phenol red-free RPMI 1640 and DMEM supplemented with 10% dextran-charcoal–treated FBS (HyClone) (62).
Computational Analysis of the Screens.
The CRISPR/Cas9 screening data are processed and analyzed using the MAGeCK and MAGeCK-VISPR algorithms we previously developed (26, 27). We use the MAGeCK-VISPR algorithm (27) to compare the gene selections across different conditions and different studies (Fig. 1E), as well as the specific essential genes in CSK-null cells (Dataset S9). The MAGeCK-VISPR algorithm uses a metric β-score to measure gene selections. The definition of the β-score is similar to the term “log fold change” in differential expression analysis, and β > 0 (or β < 0) means the corresponding gene is positively (or negatively) selected, respectively. MAGeCK-VISPR models the gRNA read counts as a negative binomial (NB) variable, whose mean value is determined by the sequencing depth of the sample, the efficiency of the gRNA, and a linear combination of β-scores of the genes. MAGeCK-VISPR then builds a maximum likelihood (MLE) model to model all gRNA read counts of all samples and iteratively estimate the gRNA efficiency and gene β-scores using the expectation-maximization algorithm. A detailed description of the MAGeCK-MLE algorithm can be found in the original study (27).
To identify breast cancer-specific essential genes (Fig. 1E), we make use of public genome-wide CRISPR screening datasets recently published (14, 16, 30, 31). The first dataset (16) includes screens of cells from colorectal carcinoma (DLD1 and HCT116), patient-derived glioblastoma (GBM), cervical carcinoma (HELA), and retinal epithelium (RPE1). The second dataset performs screens on leukemia cell lines (KBM7, K562, JIYOYE, and RAJI) (30), the third dataset is based on one melanoma cell line (A375), and the last dataset includes CRISPR screening data of 33 cell lines from various cancer types (31). For each dataset, we use the MAGeCK-VISPR algorithm to calculate the β-scores of all genes. Breast cancer-specific essential genes are those that (i) are negatively selected in breast cancer cell lines and (ii) have stronger negative selection values in breast cancer cell lines compared with non-breast cancer cell lines. Therefore, for each gene, we define its breast cancer-specific essential score as
where ts is the t-statistics tested on the β-scores of two groups [breast cancer (BC) cells and non-BC cells] and is the rank function (converted to uniform distributed values between [0,1]). A lower score indicates this gene is an essential gene in breast cancer cells [smaller ] and is more essential in breast cancer cell lines compared with non-breast cancer cell lines (smaller ). The P values are calculated from the null distribution of rank product statistics as described before (63, 64). Multiple comparison correction of the P values is performed using the Benjamini–Hochberg method (65).
We use MAGeCK (26) to identify genes whose knockout leads to stronger positive selection in Veh conditions compared with E2 conditions in T47D and MCF7 cells (Fig. 2A). The MAGeCK algorithm works as follows. It first collects read counts of all gRNAs in all conditions from fastq files, and then normalizes the read counts of control and treatment conditions using median normalization. After that, MAGeCK builds a linear model to estimate the variance of gRNA read counts, evaluate the gRNA abundance changes between control and treatment conditions, and assign a P value using the NB model. Finally, the selection of genes is evaluated from the rankings of gRNAs (by their P values) using the α-robust rank aggregation (α-RRA) algorithm. For each gene, α-RRA evaluates the rankings of all its gRNAs and assigns a lower score (RRA score) if the distribution is more skewed compared with a uniform distribution. The statistical significance of the RRA score is evaluated by permutation, and the Benjamini–Hochberg method is used for multiple comparison adjustments. To increase the statistical power, genes that have fewer than four gRNAs or genes that have fewer than two significant gRNAs are excluded from the comparison. A detailed description of the MAGeCK algorithm can be found in the original study (26).
Cell Proliferation Assays.
The breast cancer cells were plated in 24-well plates (4–5 × 104 cells per well) and kept under indicated conditions. The cells were trypsinized and collected. The number of viable cells was determined by Trypan blue exclusion and directly counted using a hemocytometer. Data represent mean ± SD from three independent replicates. P values were calculated using an unpaired Student’s t test. ATP-based measurements of cellular proliferation were performed by the CellTiter 96 Non-Radioactive Cell Proliferation Assay (Promega). The cells were plated in 96-well plates (4,000 cells per well) and biologically replicated three times.
Cell Viability Assays.
The activity levels of single agents and combinations were determined by the CellTiter 96 Non-Radioactive Cell Proliferation Assay. The cells were seeded in 96-well plates (4,000 cells per well), cultured 18–24 h before compound addition, and biologically replicated three times. Upon addition of drug, cells were incubated for 5 d.
Cell Migration.
The migratory ability of MCF-7 cells was assessed using a transwell chamber (6.5-mm insert diameter, 8-μm pore size) (catalog no. 353097; Corning) placed in 24-well culture plates. Differently treated MCF-7 cells (5 × 104 per well) were suspended in 10% FBS DMEM and seeded to the upper chambers of the transwell plates, and the lower chambers were filled with 1 mL of DMEM containing 20% FBS. Following 36 h of incubation at 37 °C, cells located on the upper membranes were removed with Q-tips, and the cells that migrated through membranes were fixed with 95% methanol and stained with 0.1% crystal violet solution (HARLECO, catalog no. 65092A95; EMD Millipore) for 15 min. Images of the stained cells that migrated to the lower sides of the filter were captured with an inverted microscope.
ChIP-Seq.
ChIP experiments for H3K27ac in T47D cells were performed as previously described (66), and the antibody for H3K27ac was ab4729 (Abcam). Library construction was performed using the ChIP-seq DNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions, followed by high-throughput sequencing with Illumina Hi-Seq.
RNA-Seq.
Total RNAs were isolated by TRIzol (Invitrogen), followed by library construction using the TruSeq RNA Library Prep Kit (Illumina) for Illumina Hi-Seq.
Immunohistochemistry.
A total of 47 matched pairs of ER+ breast cancer tumors came from patients treated with tamoxifen, including treatment-naive primary tumors and local recurrent tumors that developed after tamoxifen treatment. Details of these tissue samples were previously published (67). The tissue samples were evaluated on tissue microarrays (TMAs) obtained from consenting patients at the Royal Marsden Hospital in London. Immunohistochemical staining of CSK was performed on 4-μm sections of TMAs using the Bond Refine Detection System on a Leica Bond Rx automated immunostainer. The sections were automatically deparaffinized, and antigen retrieval was performed with EDTA buffer (pH 9.0) and processed for 20 min. The slides were incubated with the antibody against CSK (mouse monoclonal, ab54684; Abcam) at a dilution of 1:50 for 2 h at room temperature. WT MCF7 cell pellets were used as positive controls, and CSK knockdown MCF7 cell pellets were used as weak controls. Omission of the primary antibody was used as a blank control. Immunohistochemistry for CSK showed cytoplasmic staining in breast cancer cells. To evaluate immunohistochemical expression of CSK on the TMAs, we utilized a semiquantitative scoring system, whereby stain intensity and percentage of tumor cell positivity were reviewed and scored in a blinded manner by a pathologist. For each core, we assigned an intensity score of 0 for negative staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining, accompanied by the percentage positivity. CSK indices were calculated by multiplying quantity (0–100%) by intensity (0–3).
Tumor Xenograft Study.
The MCF7 cells harboring either gAAVS1 or gCSK infected with lentiviruses encoding a firefly luciferase (68) were injected s.c. into the left and right posterior flanks of 5-wk-old, ovariectomized, nude female mice (Charles River Laboratories) in the presence of estrogen. Mice were assigned randomly (day 7) to continued estrogen supplementation [E2, 0.1 mg/kg of 17 β-estradiol valerate (Sigma) once a week] or estrogen withdrawal (−E2). Bioluminescence (BIL) signal was measured once a week, and tumor volumes were calculated by volume = width2 × length/2. When the tumor reached ∼150–200 mm3, the mice were randomized for drug treatments [Veh (10% PEG400/Tween-80/PVP-K30 at a ratio of 90:5:5, 15% d-α-tocopheryl polyethylene glycol succinate (vitamin E-TPGS), and 75% hydroxypropylcellulose [0.5%] in 50 mM citrate buffer [pH 3.0]), fulvestrant (5 mg⋅wk−1, s.c. [formulated in arachis oil]), and FRAX597 (60 mg/kg, p.o.)]. FRAX597 was formulated in 10% PEG400/Tween-80/PVP-K30 at a ratio of 90:5:5, 15% vitamin E-TPGS, and 75% hydroxypropylcellulose (0.5%) in 50 mM citrate buffer (pH 3.0), and administered by oral gavage. The fold change in BLI signal was normalized to the first measurement (percentage relative BIL) when treatments were initiated for each tumor. The duration of treatment was 28 d for all of the tumor xenografts. The efficacy of FRAX597 and fulvestrant was also assessed in a PDX model of breast cancer that may better represent the response in patients. Nonobese diabetic/SCID gamma mice bearing tumor fragments of the TM00386 PDX breast cancer tumor were sourced from The Jackson Laboratory. The TM00386 PDX model was derived from an ER+/PR+/HER2− invasive ductal carcinoma. When the tumor reached ∼150–200 mm3 (80 d postimplantation), mice were randomized into treatment groups (n = 8 mice per group) and administered Veh [10% PEG400/Tween-80/PVP-K30 at a ratio of 90:5:5, 15% vitamin E-TPGS, and 75% hydroxypropylcellulose (0.5%) in 50 mM citrate buffer (pH 3.0)], fulvestrant (5 mg⋅wk−1, s.c.), and/or FRAX597 (60 mg/kg, p.o.). FRAX597 was formulated in 10% PEG400/Tween-80/PVP-K30 at a ratio of 90:5:5, 15% vitamin E-TPGS, and 75% of hydroxypropylcellulose (0.5%) in 50 mM citrate buffer (pH 3.0). Tumor size and body weight were measured twice per week throughout the duration of the study (35 d). All mouse experiments were carried out at the Dana–Farber Cancer Institute Lurie Family Imaging Center. The animal experiments were carried out under the Lurie Center Institutional Animal Care and Use Committee (IACUC) protocol and were in accordance with the IACUC standards for the welfare of animals.
Supplementary Material
Acknowledgments
We thank Peng Zhang and Yiwen Chen for helpful discussion. We thank Kristen Jones and Quang-Dé Nguyen for animal studies. This study was funded by NIH Grants U01 CA180980 (to X.S.L.), P01 CA080111 (to M.B.), and R01 HG008728 (to M.B. and X.S.L.); by Department of Defense Synergistic Idea Development Award PC140817 (to M.B. and X.S.L.); by a Komen SAB Grant (to M.B.); by the Breast Cancer Research Foundation (M.B. and X.S.L.); by the NIHR BRC at the Royal Marsden (M.D. and S.D.); and by the Claudia Adams Barr Award in Innovative Basic Cancer Research from the Dana–Farber Cancer Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-sequencing (RNA-seq) data are deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) for T47D cells [accession nos. SRR3278937 (AAVS1 knockout); SRR3278984 (C-terminal SRC kinase [CSK] knockout using g_CSK1, vehicle treatment [vehicle]); SRR3278985 (CSK knockout using g_CSK2, vehicle); SRR3278992 (CSK knockout using g_CSK3, vehicle)] and MCF7 cells [SRR4293006 (CSK knockout using g_CSK1, vehicle); SRR4293010 (CSK knockout using g_CSK2, vehicle); SRR4293016 (CSK knockout using g_CSK3, vehicle); SRR4293022 (AAVS1 knockout)]. H3K27ac ChIP-sequencing (ChIP-seq) data are deposited in the NCBI SRA for T47D cells [SRR3278995 (replicate 1); SRR3278996 (replicate 2)]. CRISPR screening data are deposited in the NCBI SRA for T47D cells [SRR3278998 (day 0); SRR3278999 (vehicle, week 4); SRR3279002 (estradiol treatment [E2], week 4)] and MCF7 cells [SRR3279335 (day 0); SRR3279336 (vehicle, week 4); SRR3279337 (E2, week 4)]. Secondary CRISPR screening data are deposited in the NCBI SRA for T47D cells with AAVS1 knockout [SRR3279406 (day 0); SRR3279409 (E2); SRR3279410 (E2); SRR3284761 (vehicle); SRR3284767 (vehicle)] or CSK knockout [SRR3279408 (day 0); SRR3279413 (vehicle); SRR3279414 (vehicle); SRR3284781 (E2); SRR3284806 (E2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722617115/-/DCSupplemental.
References
- 1.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 2.Howell A. The endocrine prevention of breast cancer. Best Pract Res Clin Endocrinol Metab. 2008;22:615–623. doi: 10.1016/j.beem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Johnston SRD. Enhancing endocrine therapy for hormone receptor-positive advanced breast cancer: Cotargeting signaling pathways. J Natl Cancer Inst. 2015;107:djv212. doi: 10.1093/jnci/djv212. [DOI] [PubMed] [Google Scholar]
- 4.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer–An overview and update. Mol Cell Endocrinol. 2015;418:220–234. doi: 10.1016/j.mce.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston SRD. Targeted combinations for hormone receptor-positive advanced breast cancer: Who benefits? J Clin Oncol. 2016;34:393–395. doi: 10.1200/JCO.2015.64.0771. [DOI] [PubMed] [Google Scholar]
- 6.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan P, Maximov PY, Curpan RF, Abderrahman B, Jordan VC. The molecular, cellular and clinical consequences of targeting the estrogen receptor following estrogen deprivation therapy. Mol Cell Endocrinol. 2015;418:245–263. doi: 10.1016/j.mce.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn RS, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 11.Turner NC, et al. PALOMA3 Study Group Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 12.Rugo HS, Rumble RB, Burstein HJ. Endocrine therapy for hormone receptor positive metastatic breast cancer: American Society of Clinical Oncology guideline summary. J Oncol Pract. 2016;12:583–587. doi: 10.1200/JCO.2016.67.1487. [DOI] [PubMed] [Google Scholar]
- 13.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 16.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 18.Parnas O, et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel SJ, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burr ML, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017;168:890–903.e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 25.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol. 2015;16:281. doi: 10.1186/s13059-015-0843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehra R, et al. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005;65:11259–11264. doi: 10.1158/0008-5472.CAN-05-2495. [DOI] [PubMed] [Google Scholar]
- 29.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aguirre AJ, et al. Genomic copy number dictates a gene-independent cell response to CRISPR-Cas9 targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Eeckhoute J, et al. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- 34.Buchwalter G, et al. PDEF promotes luminal differentiation and acts as a survival factor for ER-positive breast cancer cells. Cancer Cell. 2013;23:753–767. doi: 10.1016/j.ccr.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang H-J, et al. Differential regulation of estrogen receptor α expression in breast cancer cells by metastasis-associated protein 1. Cancer Res. 2014;74:1484–1494. doi: 10.1158/0008-5472.CAN-13-2020. [DOI] [PubMed] [Google Scholar]
- 36.Chen JQ, et al. Expression and relevance of TRPS-1: A new GATA transcription factor in breast cancer. Horm Cancer. 2011;2:132–143. doi: 10.1007/s12672-011-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner S, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288:22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang X, et al. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 2012;7:e50781. doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendes-Pereira AM, et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci USA. 2012;109:2730–2735. doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh W-L, et al. Fulvestrant-induced cell death and proteasomal degradation of estrogen receptor α protein in MCF-7 cells require the CSK c-Src tyrosine kinase. PLoS One. 2013;8:e60889. doi: 10.1371/journal.pone.0060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcotte R, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada M. Regulation of the SRC family kinases by Csk. Int J Biol Sci. 2012;8:1385–1397. doi: 10.7150/ijbs.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Z, et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat Cell Biol. 2013;15:860–871. doi: 10.1038/ncb2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanz RB, et al. 2006 Nuclear receptor signaling atlas ( www.nursa.org): Hyperlinking the nuclear receptor signaling community. Nucleic Acids Res 34:D221–D226.
- 45.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz LJ, et al. LYN-activating mutations mediate antiestrogen resistance in estrogen receptor-positive breast cancer. J Clin Invest. 2014;124:5490–5502. doi: 10.1172/JCI72573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lupien M, et al. Growth factor stimulation induces a distinct ER(alpha) cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24:2219–2227. doi: 10.1101/gad.1944810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knaus UG, Morris S, Dong HJ, Chernoff J, Bokoch GM. Regulation of human leukocyte p21-activated kinases through G protein–coupled receptors. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- 49.Renkema GH, Pulkkinen K, Saksela K. Cdc42/Rac1-mediated activation primes PAK2 for superactivation by tyrosine phosphorylation. Mol Cell Biol. 2002;22:6719–6725. doi: 10.1128/MCB.22.19.6719-6725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koh W, et al. Formation of endothelial lumens requires a coordinated PKCepsilon-, Src-, Pak- and Raf-kinase-dependent signaling cascade downstream of Cdc42 activation. J Cell Sci. 2009;122:1812–1822. doi: 10.1242/jcs.045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabe H, Okada M, Nakagawa H, Hanafusa H. Activation of c-Src in cells bearing v-Crk and its suppression by Csk. Mol Cell Biol. 1992;12:4706–4713. doi: 10.1128/mcb.12.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung J-H, Traugh JA. Regulation of the interaction of Pak2 with Cdc42 via autophosphorylation of serine 141. J Biol Chem. 2005;280:40025–40031. doi: 10.1074/jbc.M509075200. [DOI] [PubMed] [Google Scholar]
- 53.Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: Clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype–ACOSOG Z1031. J Clin Oncol. 2011;29:2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunbier AK, et al. Molecular profiling of aromatase inhibitor-treated postmenopausal breast tumors identifies immune-related correlates of resistance. Clin Cancer Res. 2013;19:2775–2786. doi: 10.1158/1078-0432.CCR-12-1000. [DOI] [PubMed] [Google Scholar]
- 55.Loi S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci USA. 2010;107:10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loi S, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2009;116:303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 58.Symmans WF, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma X-J, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knowlden JM, et al. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–1044. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 62.Miller TW, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010;120:2406–2413. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 64.Eisinga R, Breitling R, Heskes T. The exact probability distribution of the rank product statistics for replicated experiments. FEBS Lett. 2013;587:677–682. doi: 10.1016/j.febslet.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 65.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 66.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drury SC, et al. Changes in breast cancer biomarkers in the IGF1R/PI3K pathway in recurrent breast cancer after tamoxifen treatment. Endocr Relat Cancer. 2011;18:565–577. doi: 10.1530/ERC-10-0046. [DOI] [PubMed] [Google Scholar]
- 68.Kimbrel EA, Davis TN, Bradner JE, Kung AL. In vivo pharmacodynamic imaging of proteasome inhibition. Mol Imaging. 2009;8:140–147. [PubMed] [Google Scholar]
- 69.Warde-Farley D, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.