Significance
The motile flagellum/cilium is found across eukaryotic life, and it performs critical functions in many organisms including humans. A fundamental requirement for a motile flagellum is that it must undergo the appropriate waveform for its specific function. Much is known about the generation of asymmetry in flagellum movement; however, it is unknown how a motile flagellum specifies where waves should start and whether waves should go from base to tip, or from tip to base. We show here in two flagellum model organisms (the human parasites Trypanosoma brucei and Leishmania mexicana) that differences in the outer dynein arms between the distal and proximal regions of the flagellum determine wave propagation direction and are generated and maintained by the flagellum growth machinery.
Keywords: flagellum, motility, outer dynein arm, intraflagellar transport, trypanosomatid
Abstract
The 9 + 2 axoneme structure of the motile flagellum/cilium is an iconic, apparently symmetrical cellular structure. Recently, asymmetries along the length of motile flagella have been identified in a number of organisms, typically in the inner and outer dynein arms. Flagellum-beat waveforms are adapted for different functions. They may start either near the flagellar tip or near its base and may be symmetrical or asymmetrical. We hypothesized that proximal/distal asymmetry in the molecular composition of the axoneme may control the site of waveform initiation and the direction of waveform propagation. The unicellular eukaryotic pathogens Trypanosoma brucei and Leishmania mexicana often switch between tip-to-base and base-to-tip waveforms, making them ideal for analysis of this phenomenon. We show here that the proximal and distal portions of the flagellum contain distinct outer dynein arm docking-complex heterodimers. This proximal/distal asymmetry is produced and maintained through growth by a concentration gradient of the proximal docking complex, generated by intraflagellar transport. Furthermore, this asymmetry is involved in regulating whether a tip-to-base or base-to-tip beat occurs, which is linked to a calcium-dependent switch. Our data show that the mechanism for generating proximal/distal flagellar asymmetry can control waveform initiation and propagation direction.
Eukaryotic flagella/cilia are highly conserved cellular structures and play key roles as both sensory and motile organelles. Motile flagella/cilia undergo different waveforms necessary for their function: a symmetrical sinusoid or helical flagellar-type beat (e.g., human sperm), or an asymmetrical wafting ciliary-type beat (e.g., ciliated epithelia, Chlamydomonas reinhardtii). Many organisms can switch between asymmetric and symmetric waveforms (1–6), and this switch is typically mediated by calcium (4, 6–14). In addition to the waveform shape, the direction of waveform propagation can vary, and these two phenomena are distinct. C. reinhardtii and animal sperm flagella generally undergo base-to-tip waveforms, while other flagella (including Leishmania and Trypanosoma) normally undergo tip-to-base waveforms (15–17). Many flagella, including Leishmania and the sperm of some animal species, can switch the direction of waveform propagation to change the direction of swimming (16, 18–22). Like changes in waveform symmetry, the switching of waveform direction is calcium mediated (18–21, 23, 24). Regardless of whether it can switch between waveforms, every flagellum/cilium must somehow specify where a waveform should start and thus the direction in which it propagates.
The mechanism defining the point at which waveforms are initiated has been generally overlooked, possibly because the most popular model of flagellar waveform propagation, the geometric clutch model, suggests that flagellar beating can start spontaneously (25). Moreover, experimental evidence shows that the beat can initiate not only at the flagellar tip but also in the midflagellum in trypanosomatids (16). However, since the initiation point defines the direction of waveform propagation (proximal initiation for base-to-tip propagation and distal initiation for tip-to-base propagation), determining the mechanisms by which the position and direction of flagellar beat are initiated is critical to our understanding of waveform generation and switching.
Motor proteins in the inner and outer dynein arm complexes (IDAs and ODAs, respectively) are key to the generation and control of flagellar movement. The canonical view is that the IDAs generate and regulate the beat, while ODAs provide the force to generate the final waveform (26). This view is predominantly derived from genetic evidence in C. reinhardtii: Loss of IDAs (of which there are several classes) tends to alter waveform shape (27), while loss of ODAs reduces flagellar beat frequency with a small effect on waveform shape (27, 28). ODA defects are one of the main causes of primary ciliary dyskinesia in humans (29), a recessive genetic disorder characterized by chronic pulmonary disease, randomization of the left/right body axis, and infertility. In Trypanosoma brucei the loss of ODAs eliminates the tip-to-base flagellar beat, preventing forward motion of the parasite (15, 30). C. reinhardtii mutants lacking ODAs move more slowly and cannot swim backward in response to stimulation with light (27, 28, 31).
In recent years studies have revealed that components of the IDAs and ODAs in several organisms are asymmetrically arranged along the length of the flagellum. IDA asymmetries have been identified in C. reinhardtii (32, 33), and ODA asymmetries occur in several model organisms. In humans, this asymmetry differs among cell types. In ciliated epithelia, the outer arm dynein (OAD) DNAH5 localizes to the whole axoneme, while DNAH9 and DNAH11 localize only to the distal axoneme (34–36); in contrast, in sperm DNAH5 localizes to the proximal flagellar axoneme, and DNAH9 localizes to the whole axoneme (34). C. reinhardtii has one microtubule doublet with particularly strong proximal/distal asymmetry (32, 37), and this asymmetry appears to be at least partially due to the proximal-only localization of an ODA-associated complex including ODA5 and ODA10 (38, 39). The function of the proximal/distal asymmetry of ODAs has not previously been analyzed in any detail; however, it appears important: In humans disruption of the asymmetric DNAH proteins is associated with defects in ciliary motility and primary ciliary dyskinesia (34–36). In C. reinhardtii, mutations in the proximal proteins ODA5 and ODA10 are associated with defects in swimming, but this phenotype is complicated by the additional roles of these proteins in ODA assembly (38, 39). How these IDA and ODA asymmetries are generated is also largely unknown.
We hypothesized that proximal/distal molecular asymmetries in the flagellum control where the flagellum waveform starts and thus control the direction of beat propagation and contribute to the control of beat type. Since asymmetry has been observed in the IDAs of C. reinhardtii and in the ODAs in at least two model organisms, we examined the flagella of T. brucei and Leishmania mexicana for similar asymmetries. These organisms are well-characterized models for flagellar motility and are capable of switching between tip-to-base and base-to-tip waveforms. There is some evidence of asymmetry between the proximal and distal regions of the flagellum in T. brucei (40), although the functional relevance of this is not yet clear. We show that in both organisms the proximal and distal regions of the flagellum contain distinct ODA-docking complexes (DCs), with an inherent asymmetry achieved early and maintained throughout flagellum growth. We demonstrate that this asymmetry is produced and maintained by an intraflagellar transport (IFT)-dependent concentration gradient of DC proteins by retrograde transport of proximal DCs. Finally, we show that ODA proximal/distal asymmetry is involved in regulating whether a tip-to-base or base-to-tip beat occurs, likely via a calcium-dependent switch.
Results
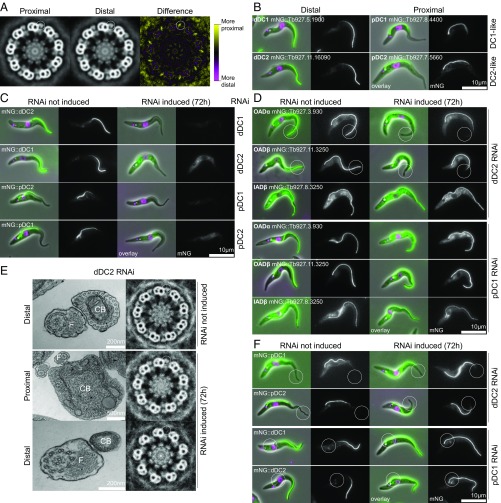
By thin-section transmission electron microscopy (TEM) we observed that the proximal and distal regions of the T. brucei axoneme are not identical. The T. brucei flagellum is laterally attached to the cell for most of its length; therefore, most transverse cross-sections through the flagellum have an attached cross-section through the cell body. This architecture allows unambiguous identification of flagellar axoneme cross-sections as proximal or distal based on the size and ultrastructure of the neighboring cell body. Averaged electron density analysis of distal axoneme cross-sections showed a subtle difference in electron density in the outer dynein arm region between the proximal and distal regions of the axoneme (Fig. 1A). No other differences were detectable by this unbiased analysis; however, this does not exclude smaller proximal/distal differences in other structures.
Fig. 1.
The trypanosome flagellum has proximal/distal asymmetry arising from proximal- and distal-specific OAD DCs. (A) Ninefold rotational averages of TEM of transverse sections through the T. brucei axoneme. Averages were generated from either the proximal (n = 23) or distal (n = 24) region. A difference map shows differences only in the ODAs. (B) Widefield epifluorescence micrographs of T. brucei cells expressing DC proteins tagged with mNG at the N terminus. Overlays of phase-contrast (gray), DNA (Hoechst 33342, magenta), and mNG (green) images and mNG fluorescence alone are shown. (C) Micrographs of T. brucei RNAi cells targeting dDC1, dDC2, pDC1, or pDC2 ORFs and respectively expressing mNG-tagged dDC2, dDC1, pDC2, or pDC1. Induction of RNAi for 72 h caused loss of fluorescence signal in each case (summarized in Table 1). (D) Micrographs of T. brucei RNAi cell lines targeting dDC2 or pDC1 expressing mNG-tagged OADα, OADβ, or IADβ. Induction of dDC2 RNAi for 72 h caused loss of distal OADα and OADβ (circled), while pDC1 RNAi had no effect on OADα and OADβ fluorescence. (E) TEMs (Left) and ninefold rotational averages (Right) of transverse sections through the axoneme of the T. brucei dDC2 RNAi cell line. ODAs are present in uninduced samples (representative of n = 25) and in the proximal axoneme 72 h after induction of RNAi (representative of n = 11) but are absent in the distal axoneme after 72-h induction of RNAi (representative of n = 7). CB, cell body; F, flagellum. (F) Micrographs of T. brucei RNAi cell lines targeting dDC2 or pDC1 and respectively expressing mNG-tagged pDC1 or dDC2. Seventy-two hour induction of dDC2 RNAi caused distal extension of the pDC1 and pDC2 fluorescent signal (circled), and 72-h induction of pDC1 RNAi caused proximal extension of the dDC1 and dDC2 fluorescent signal (circled) (summarized in Table 1).
We identified candidate proteins that may be responsible for this asymmetry using TrypTag, a project localizing every protein encoded in the T. brucei genome (41). TrypTag identified several proteins with proximal- or distal-only axoneme localizations, including homologs of the ODA DC proteins DC1/ODA3 and DC2/ODA1 in C. reinhardtii, corresponding to CCDC151 and CCDC114, respectively, in humans. One DC1 and one DC2 homolog (Tb927.5.1900 and Tb927.11.16090, respectively) localized to the distal part (approximately half) of the axoneme, and a second DC1 and second DC2 homolog (Tb927.8.4400 and Tb927.7.5660, respectively) localized to the proximal part (Fig. 1B), as determined by N-terminal tagging. We named these proteins “proximal DC1” (pDC1), “proximal DC2” (pDC2), “distal DC1” (dDC1), and “distal DC2” (dDC2), forming the proximal (pDC) and distal (dDC) DC pairs, respectively. Asymmetry is unlikely to be due to aberrant targeting, since both C-terminal and N-terminal tagging gave the same results (SI Appendix, Fig. S1A).
In C. reinhardtii, DC1 and DC2 are predicted to form a coiled-coil heterodimer and are mutually dependent for flagellar localization and function (42, 43). T. brucei DC proteins are rich in predicted coiled coils (SI Appendix, Fig. S1B); therefore, to test if each of the four DC proteins (pDC1, pDC2, dDC1, and dDC2) is mutually dependent on its putative partner for correct localization, we generated inducible RNAi cell lines targeting the ORF of each DC gene. RNAi target sequences were selected ensuring they are not present elsewhere in the genome (44), and spurious knockdown of an incorrect DC protein is unlikely because protein sequence identity is very low (<20%) and there is no identical DNA sequence longer than 11 nt between any pair of DC proteins. For each RNAi cell line, we fluorescently tagged either the same gene or the expected heterodimer partner at the endogenous locus. In cell lines where the same gene was both tagged and targeted for RNAi, the fluorescent signal of the tagged protein was undetectable after 72 h of RNAi induction, confirming effective RNAi knockdown at the protein level (Table 1 and SI Appendix, Fig. S1C). When a given DC gene was targeted for RNAi knockdown in a cell line expressing a tagged copy of its putative partner, RNAi induction led to the loss of the fluorescence signal for the expected partner protein: Tagged dDC2 was undetectable following dDC1 RNAi (and vice versa), and tagged pDC2 was undetectable following pDC1 RNAi (and vice versa) (Fig. 1C and Table 1). Off-target effects of RNAi seem unlikely, as they would not be expected to generate these clear reciprocal phenotypes. Hence, the four T. brucei DC proteins likely make distinct distal (dDC1+dDC2) and proximal (pDC1+pDC2) heterodimers. Therefore, we focused further on one pDC protein and one dDC protein, pDC1 and dDC2, respectively.
Table 1.
Summary of DC protein localization changes upon DC RNAi knockdowns in T. brucei
| Protein tagged | No RNAi, % | dDC1, % | dDC2, % | pDC1,% | pDC2, % |
| dDC1 | Distal 50 | 0 (no signal) | 0 (no signal) | 100 (whole flagellum) | 100 (whole flagellum) |
| dDC2 | Distal 50 | 0 (no signal) | 0 (no signal) | 100 (whole flagellum) | 100 (whole flagellum) |
| pDC1 | Proximal 50 | Proximal 75 | Proximal 75 | 0 (no signal) | 0 (no signal) |
| pDC2 | Proximal 50 | Proximal 75 | Proximal 75 | 0 (no signal) | 0 (no signal) |
To examine whether DC proteins performed their expected function of docking the OADs to the axoneme, we tagged each of the OAD heavy chains (OADα and OADβ), and one inner arm dynein (IAD) heavy chain (IADβ) as a negative control, on the background of RNAi targeting either dDC2 or pDC1. Before RNAi induction, both OAD and IAD fluorescence signals extended along the entire flagellum. Induction of pDC1 RNAi for 72 h had no detectable effect on OAD or IAD fluorescent signal (Fig. 1D); however, induction of dDC2 RNAi for 72 h resulted in a decrease of the OAD (but not IAD) fluorescence signal in the distal ∼25% of the flagellum (Fig. 1D). The loss of ODAs from the distal axoneme only following dDC2 RNAi was confirmed using electron microscopy (Fig. 1E).
To determine how ODAs remain attached to the proximal axoneme in the absence of pDC1, we tested whether the loss of the pDC alters the localization of the dDC and vice versa. We therefore tagged pDC1 and pDC2 on the background of dDC2 RNAi and tagged dDC1 and dDC2 on the background of pDC1 RNAi. Following 72-h induction of dDC2 RNAi, the pDC1 and pDC2 signals remained proximal but extended along the flagellum to ∼75% of the flagellum length (Fig. 1F, Table 1, and SI Appendix, Fig. S1D), matching the length of the OAD signal upon dDC2 RNAi (Fig. 1D and SI Appendix, Fig. S1D). Following 72-h induction of pDC1 RNAi, the dDC1 and dDC2 fluorescence signals extended to cover the entire flagellum (Fig. 1F and SI Appendix, Fig. S1D), indicating that the dDC can dock ODAs to the entire axoneme in the absence of the pDC. RNAi knockdown of dDC1 with tagged pDC1 or pDC2 and RNAi knockdown of pDC2 with tagged pDC1 or pDC2 confirmed this result (Table 1 and SI Appendix, Fig. S1F). Importantly, axoneme asymmetry changed upon RNAi knockdown of components of the pDCs or dDCs, indicating that both of these DCs must be present to generate asymmetry.
While lengthwise asymmetries along the flagellar axoneme have been observed in other organisms, the mechanism that generates this asymmetry is unknown. To address this issue, we considered critically a number of possible models: DCs may attach to an underlying asymmetry, such as another protein or tubulin modification (model 1). This is unlikely, as knockdown of DC proteins altered the asymmetry (Fig. 1 D and F), and thus they cannot purely be clients to an existing asymmetry. Asymmetry may be derived from flagellum growth, with the pDC assembled early and the dDC assembled later (model 2). This is not correct, as proximal/distal asymmetry was achieved early and maintained throughout flagellar growth (see below). We considered the possibility that asymmetry comes from information passed through the lateral attachment of the flagellum to the T. brucei cell body (model 3). This is also unlikely, as knockdown of DC proteins caused asymmetry changes without affecting flagellum/cell body attachment (Fig. 1 D and F). Finally, asymmetry may be generated intrinsically in the flagellum by tip structures or by IFT (model 4). As DC asymmetry extends over the whole flagellum, and the switching point occurs at the midpoint, asymmetry is unlikely to be due to direct interaction with the basal body or flagellum tip structures. However, the IFT system that assembles the axoneme could generate asymmetry by creating a proximal/distal concentration gradient of DCs.
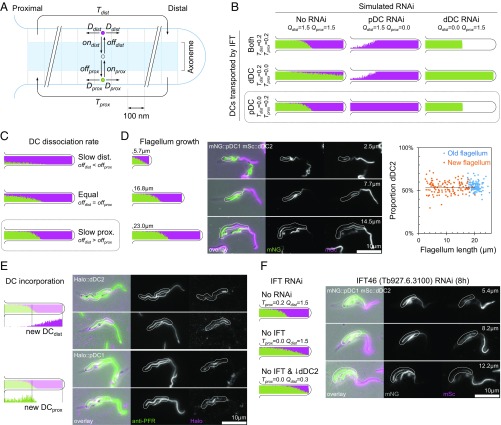
To test and make predictions about the IFT model of asymmetry generation (model 4), we built a quantitative agent-based model of DC binding, diffusion, and IFT transport. The axoneme was simulated in 100-nm sections, and the proximal and distal heterodimers were simulated as single particles which diffuse along the flagellum, attach to and detach from the axoneme, and may be transported by IFT. The key parameters which define the behavior of the model are the probability of DC attachment (on), detachment (off), diffusion to a neighboring section (D), rate of transport by IFT (T), and quantity of DCs (Q) (Fig. 2A). Conceptually, IFT transport of unbound DCs (retrograde transport for the pDCs, anterograde transport for the dDCs, or transport of both) could generate a concentration gradient which drives proximal/distal asymmetry. This assumes that unbound DCs diffuse freely along the axoneme when not transported by IFT and then bind to the next available site. Simulating IFT transport of pDCs, dDCs, or both DCs all generated proximal/distal asymmetry (Fig. 2B).
Fig. 2.
Retrograde movement of pDCs by IFT is responsible for proximal/distal axoneme asymmetry. (A) Outline of the agent-based model of DC distribution by IFT and diffusion, showing key parameters: Dprox and Ddist, the probability of diffusion of pDCs or dDCs to the neighboring axoneme segment; onprox and ondist, the probability of DC binding; offprox and offdist, the probability of DC unbinding; Tprox and Tdist, the capacity for anterograde and retrograde IFT transport; and Qprox and Qdist (not shown in diagram), the quantity of DCs. (B) Simulated distribution of proximal (green) and distal (magenta) DCs in full-length flagella with different rates of IFT (Tprox and Tdist) and quantities of proximal and distal DCs (Qprox and Qdist). The conditions Tprox = 0.2 and Tdist = 0.0 (retrograde transport of proximal DCs) most closely matched the experimental data (Fig. 1F). (C) Simulated distribution of pDCs and dDCs in full-length flagella with different rates of dissociation of DCs (offprox and offdist). The condition offprox < offdist most closely matched the experimental data (Fig. 1B). (D) Comparison of simulated distribution of pDCs and dDCs in growing flagella (Left) with micrographs of dividing cells with a new growing flagellum (Center). In the micrographs the new flagellum (white outlines) is indicated in a T. brucei cell line expressing mNG-tagged pDC1 and mSc-tagged dDC2. The measured proportion of flagellum with a dDC2 signal was always ∼50%, independent of flagellum length (Right). (E) Comparison of simulation of new protein incorporation in a new growing flagellum (Left) with micrographs of T. brucei cells in an equivalent pulse-chase experiment (Right). In the micrographs the new growing flagellum is indicated (white outlines) in a T. brucei cell line expressing HaloTag-tagged dDC2 or pDC1 labeled with a 45-min pulse of tetramethylrhodamine HaloTag ligand. The flagellum was labeled with an anti-PFR antibody. (F) Comparison of the simulated distribution of pDCs and dDCs in growing flagella with or without IFT transport of the dDC and with no IFT transport and reduced quantity of dDC (Left) in comparison with micrographs of dividing cells in a T. brucei cell line expressing N terminally mNG-tagged pDC1 and N terminally mSc-tagged dDC2 8 h after induction of IFT46 RNAi knockdown (Right). In the simulation and the new flagellum of dividing cells reduced IFT caused a shorter region of dDC/dDC2 signal.
Our experiments showed that the effects pDC knockout on dDCs and the effects of dDC knockout on pDCs were different: on pDC knockdown, the dDCs extended along the entire flagellum, while pDCs were still excluded from part of the distal flagellum on dDC knockdown (Fig. 1F). This suggested that only one of the pDCs or dDCs is transported. We used this to constrain our model by specifying whether one or both DCs were transported by IFT and then simulating the resulting flagellum asymmetry in the presence of both DCs, only pDCs, or only dDCs, with the latter two mirroring the RNAi experiments (Fig. 2B). Simulation of retrograde transport of the pDC alone matched the changes in DC localization on RNAi knockdown. Any anterograde transport of dDCs prevented the dDC from extending along the proximal flagellum in the absence of the pDC (compare Fig. 1F and Fig. 2B). Finally, given the retrograde transport of the pDC, a higher binding affinity of pDC was necessary for the simulation to match the observed DC localization (Fig. 1B and Fig. 2C). The model gave qualitatively similar results even with large changes to the estimated parameters (SI Appendix, Fig. S3A). Therefore, our data are consistent with an asymmetry-generation model in which the higher-affinity pDC is restricted to the proximal axoneme by retrograde IFT transport, while the lower-affinity dDC fills the remaining axoneme binding sites.
The IFT-mediated asymmetry model predicts that growing flagella will maintain their proximal distal asymmetry independent of flagellum length (Fig. 2D). T. brucei grows a new flagellum each cell cycle, with each daughter cell inheriting one full-length flagellum. The new growing flagellum is always positioned anterior of the old flagellum (45, 46). Using a cell line in which dDC2 and pDC1 were tagged with different fluorescent proteins, we saw that the new growing flagellum contained proportions of pDC1 and dDC2 similar to those found in the old flagellum (Fig. 2D). Measurements of the dDC2 signal length confirmed that there was no correlation between flagellum length and the proportion of the flagellum with dDC2 signal (Spearman’s rank correlation −0.013, n = 123) (Fig. 2D). Therefore, the DC asymmetry is established early during flagellum growth, eliminating flagellum growth per se as a mechanism of asymmetry generation.
The IFT model also predicts that new dDC molecules would be incorporated at the distal end of growing flagella. In contrast, the pDC would be incorporated more slowly and diffusely, weakly focused at the distal end of the pDC region, which corresponds to the middle of the flagellum (Fig. 2E). We tested this prediction using pulse labeling in cell lines expressing dDC2 or pDC1 tagged with HaloTag (SI Appendix, Fig. S2B). Incubation with a nonfluorescent ligand followed by a pulse with fluorescent ligand allowed us to observe the incorporation of new material into the flagellum; as predicted by the model, new dDC2 was incorporated at the distal end of new growing flagella, and the new pDC1 signal was weaker, focused toward the middle of the growing flagella (Fig. 2E).
Finally, the IFT model predicts that disruption of IFT should alter the proximal/distal asymmetry of the flagellum. The precise effect is hard to predict: IFT disruption also reduces flagellum growth, and IFT-mediated entry of DCs into the flagellum may be affected. The model suggests that the pDC is unlikely to require IFT for entry into the flagellum, as it is actively removed by retrograde transport, while the dDC may require IFT for entry into the flagellum and so may be depleted on IFT disruption. Simulation predicts that reduced IFT will allow the pDC region to extend distally, outcompeting the binding of dDC, and this effect could be exacerbated by a reduction in the quantity of dDC proteins. We tested this prediction using RNAi knockdown of IFT46 (Tb927.6.3100), which is required for anterograde IFT, thus disrupting both anterograde and retrograde IFT. Induction of IFT46 RNAi for 24 h caused the cellular phenotypes of IFT knockdown (47): cells with shorter flagella, cytokinesis defects, and reduced population growth (SI Appendix, Fig. S2C). We tagged dDC2 and pDC1 with different fluorescent proteins in this RNAi cell line and then looked at cells 8 h and 16 h after RNAi induction to examine the earliest effects of IFT46 RNAi. In a minority of dividing cells at 8 h and in a majority at 16 h the new flagellum had the predicted changes to axoneme asymmetry. The region occupied by pDC1 was greatly expanded, with a corresponding reduction in dDC2 signal (Fig. 2F and Table 1). This reduced proportion of dDC2 signal was inherited by one daughter cell, and after 24-h induction cells with a single flagellum and a similar proximal/distal defect were common in the population (SI Appendix, Fig. S2D). This confirms that retrograde transport of pDCs generates a concentration gradient which, combined with different dissociation rates of pDC and dDC, generates the observed asymmetry in DCs.
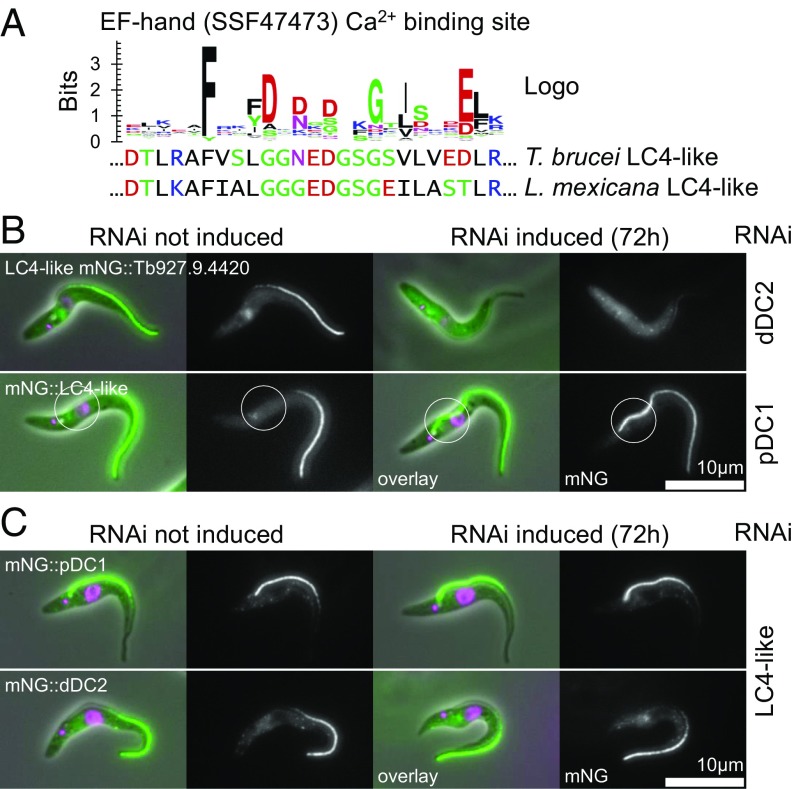
As ODAs are required for flagellar beating, we reasoned there may be regulatory proteins for modulating the site of waveform initiation (and therefore waveform propagation direction) which bind only to the proximal or distal ODAs. Using TrypTag (41), we identified a candidate beat-regulation protein (Tb927.9.4420) based on its localization and predicted domains. It is the only EF-hand/calmodulin domain-containing protein (Fig. 3A) localized specifically to the distal axoneme (Fig. 3B). This protein could plausibly interact with Ca2+, a known beat regulator. The most similar C. reinhardtii protein is LC4 (but LC4 is not a reciprocal best BLAST result), an ODA-binding protein implicated in flagellar beat control (48, 49). We named this protein “LC4-like.” Fluorescently tagged LC4-like localized to the distal axoneme, similar to dDC1/dDC2, and the fluorescence signal was undetectable following induction of dDC2 RNAi knockdown for 72 h, suggesting that LC4-like relies on the dDC for localization (Fig. 3B and Table 2). These results provide further support for the biological plausibility of LC4-like being a distal ODA regulator. LC4-like is not an obligate part of the DC, as 72-h induction of LC4-like RNAi knockdown did not affect the localization of dDC2 (Fig. 3B and Table 2). This is similar to C. reinhardtii, where the DC heterodimer is associated with a calcium-binding protein, DC3 (ODA14) (50); however, we did not find a clear homolog of DC3 by reciprocal best BLAST in T. brucei. RNAi knockdown of pDC1 caused the LC4-like signal to extend to the proximal end of the flagellum (Fig. 3C and Table 2), as does the dDC. In C. reinhardtii LC4 binds the ODAs (49), and therefore LC4-like may bind only to ODAs attached by the dDC, although it may bind directly to the dDCs.
Fig. 3.
A calcium-binding LC4-like protein is a candidate regulator of the flagellar beat. (A) Sequence logo of the Ca2+ binding site of 100 reference EF-hand domains and the aligned T. brucei and L. mexicana LC4-like sequences. (B) Micrographs of a T. brucei cell line with integrated RNAi constructs targeting dDC2 or pDC1 and expressing LC4-like tagged with mNG at the N terminus. Seventy-two hour induction of dDC2 RNAi causes the loss of flagellar LC4-like signal, and 72-h induction of pDC1 RNAi causes proximal extension of the LC4-like signal (circled) (summarized in Table 2). (C) Micrographs of a T. brucei cell line with an integrated RNAi construct targeting LC4-like and expressing dDC2 or pDC1 tagged with mNG at the N terminus. Seventy-two hour induction of LC4-like RNAi causes no change in pDC1 or dDC2 signal (summarized in Table 2).
Table 2.
Summary of evidence for LC4-like localization dependent on distal DCs in T. brucei
| Protein tagged | No RNAi, % | dDC2, % | pDC1, % | LC4-like, % |
| dDC2 | Distal 50 | 0 (no signal) | 100 (whole flagellum) | Distal 50 |
| pDC1 | Proximal 50 | Proximal 75 | 0 (no signal) | Proximal 50 |
| LC4-like | Distal 50 | 0 (no signal) | 100 (whole flagellum) | 0 (no signal) |
We predicted that disruption of proximal/distal asymmetry or LC4-like would alter the direction of flagellum waveform propagation. However, full analysis of flagellar waveforms is complicated in T. brucei due to the lateral attachment of the flagellum to the cell body. To better analyze changes in flagellum movement, we used the related parasite L. mexicana, which does not have a laterally attached flagellum, greatly simplifying the beat waveform analysis. We identified L. mexicana homologs of dDC1, dDC2, pDC1, and pDC2 (LmxM.15.0540, LmxM.31.2900, LmxM.10.0960, and LmxM.06.1040, respectively). Each has localizations comparable to those in T. brucei, except that the pDC occupies ∼20% of the proximal axoneme rather than 50% (SI Appendix, Fig. S3A). Deletion of both alleles of dDC2 on the background of fluorescently tagged pDC1 caused distal extension of the pDC1 signal and loss of ODAs from the distal axoneme, similar to dDC2 RNAi knockdown in T. brucei (compare Fig. 1 E and F and SI Appendix, Fig. S3 B and C). The localization of LC4-like (LmxM.01.0620) in L. mexicana also reflected that of T. brucei, and deletion of both dDC2 alleles caused loss of LC4-like from the axoneme, similar to dDC2 RNAi knockdown in T. brucei (compare Fig. 3B and SI Appendix, Fig. S3D).
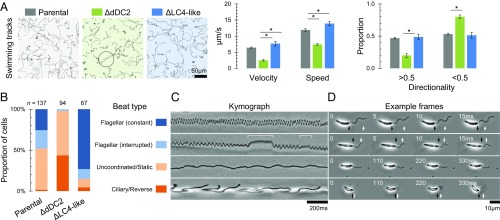
Deletion of both alleles of L. mexicana dDC2 decreased the speed, velocity, and directionality of cell swimming (Fig. 4A), although flagella still moved. We analyzed flagellum movement with 200-Hz high-frame-rate video microscopy. L. mexicana can undergo both a tip-to-base sinusoidal flagellar beat and a base-to-tip asymmetric ciliary type flagellar beat (16). Normal flagellum movement in the parental cell line was mostly a flagellar beat, with occasional pauses or ciliary beats, with a large minority of cells undergoing low-frequency or uncoordinated movement (Fig. 4B and Movie S1). Flagellum movement after dDC2 deletion entirely lacked flagellar beats. Approximately half of the flagella were uncoordinated, and half of the cells underwent an asymmetric base-to-tip reverse beat, far more than in the parental cell line (Fig. 4 C and D and Movie S1). The shape of base-to-tip waveforms was unaffected, having the same asymmetric shape as in the parental cell line. The distal ODAs are therefore required for the tip-to-base flagellar beat to occur, most likely initiating the flagellar beat waveform at the distal end.
Fig. 4.
Proximal/distal flagellum asymmetry contributes to the control of flagellum beat type. (A) Swimming paths, swimming speed, and directionality of L. mexicana cell lines with deletion of both alleles of dDC2 or LC4-like in comparison with the parental cell line. Swimming tracks show slower, less directional, swimming following dDC2 deletion, which occasionally corresponds to tight helical paths (circled). dDC2 deletion caused a significant decrease in speed, velocity, and proportion of highly directional (>0.5) cells, while LC4-like deletion caused a significant increase in speed and velocity (Student’s t test, P < 0.05). (B) The proportion of cells undergoing different types of flagellar movement in L. mexicana cell lines with deletion of both alleles of dDC2 or LC4-like in comparison with the parental cell line. Behaviors were assessed from a 4-s 200-Hz video micrograph. dDC2 deletion caused significantly more ciliary-type and uncoordinated movement, and LC4-like deletion caused significantly more uninterrupted flagellar-type movement (χ2 test, P < 10−9). (C) Examples of kymographs of flagellar movement illustrating the types of flagellar movement used as classes in B. Animated examples are shown in Movie S1. (D) Frames from high-speed videos corresponding to the kymographs in C. Propagation of a flagellum wave over the frames is indicated (white arrows) relative to the cell posterior (black arrows).
Deletion of both LC4-like alleles caused a significant increase in swimming speed and velocity (Fig. 4A). High-frame-rate video of the LC4-like deletion showed that flagellum movement was almost entirely a flagellar beat, with fewer pauses in the beat, less uncoordinated movement, and far fewer ciliary beats than in the parental cell line (Fig. 4B and Movie S1). The frequency of the flagellar beat was also significantly higher in the LC4-like deletion (44.2 ± 8.9 Hz, n = 33) than in the parental cell line (25.4 ± 9.0 Hz, n = 27) (Mann–Whitney u test, P < 10−10), although the waveform showed the same sinusoidal shape. LC4-like therefore appears to be a regulatory protein that inhibits the initiation of tip-to-base flagellar beat waveforms, and the absence of LC4-like has the opposite effect to that of missing distal ODAs.
Discussion
It is becoming increasingly apparent that in many organisms there are molecular asymmetries in the IDAs and ODAs of the axoneme both between the outer microtubule doublets (37, 51) and along the length of the axoneme (32–34, 36). In C. reinhardtii, proximal/distal and doublet–doublet asymmetries in the IDAs have been implicated in controlling whether a flagellum waveform is asymmetric or symmetrical (27). We show here that proximal/distal axoneme asymmetry is also important in controlling the site of flagellum waveform initiation and therefore the direction of waveform propagation. The control of the site of waveform initiation and the control of waveform asymmetry are significantly different phenomena; however, as both involve proximal/distal asymmetries in dynein arms, there may be similarities in the underlying mechanisms.
We have concentrated on the lengthwise axonemal asymmetry and demonstrate that both T. brucei and L. mexicana use distinct pDCs and dDCs to confer proximal/distal asymmetry of the molecular composition of ODAs in the axoneme. The resulting axoneme structure is similar to previous reports of asymmetry of ODAs in human cilia and sperm flagella (34–36), and to proximal/distal asymmetry of one axoneme microtubule doublet in C. reinhardtii (32, 37), which is linked to DC-related accessory proteins (38, 39). It is perhaps surprising that we identified the ODAs, rather than IDAs, as important for specifying the site of waveform initiation in T. brucei and L. mexicana. The canonical view (based primarily on C. reinhardtii) is that ODAs provide the force which drives the beat, while the IDAs initiate and regulate the waveform shape (26). However, we saw no clear proximal/distal asymmetry of IDAs by TEM in T. brucei, while the loss of distal ODA DCs or the ODA-associated LC4-like was sufficient to change the site of waveform initiation. The change in the site of waveform initiation occurred with the normal waveform for tip-to-base (symmetric) and base-to-tip (asymmetric) waveforms. This is consistent with IDAs regulating the shape of the waveform (as in C. reinhardtii) but ODAs regulating the site of wave initiation. While we cannot formally exclude the possibility that IDAs have a parallel or underlying role in controlling whether waveform initiation is distal or proximal, we saw no change in the localization of IDA components during DC knockdown. However, as C. reinhardtii flagella always undergo base-to-tip waveforms, it is also possible C. reinhardtii has lost some proximal/distal ODA asymmetries. Given that this asymmetry occurs in diverse organisms, proximal/distal asymmetry in ODAs may represent a general mechanism for defining proximal and distal regions of the flagellum, although the specific proteins involved may differ through evolution. Guided by this, we identified a similar, previously unrecognized, phenomenon by analyzing previously published data from the unrelated unicellular parasite Giardia lamblia (52). We noticed that this species also has proximal and distal DC1-like proteins (GL50803_13288 and GL50803_16998) but a single DC2 protein (GL50803_114462).
Proximal/distal axoneme asymmetry had been described, but the mechanism of asymmetry generation was unknown, although competition for axoneme binding between distal and proximal components has been suggested for C. reinhardtii and T. brucei (33, 40). We show that asymmetry of the DCs is generated at the very earliest stages of flagellar growth and is maintained as the flagellum elongates. Therefore, the mechanism by which asymmetry is generated is intrinsic to the flagellar growth machinery. By modeling various permutations of the IFT transport of each DC and comparing these with the experimental data, we show that two factors are sufficient to generate the asymmetries we observed: lower-affinity binding of the dDC and retrograde transport of the pDC by IFT. Retrograde transport of the pDC toward the base of the flagellum generates and maintains a concentration gradient of the pDC. The higher affinity of the pDC out-competes binding of the dDC, generating the axoneme asymmetry, and unbound dDC is free to diffuse throughout the flagellum and fills in the remaining spaces. This model is simple, and, in reality, there are likely to be additional complexities, but it precisely matches our experimental observations: (i) maintenance of asymmetry throughout flagellar growth, (ii) the locations at which newly synthesized pDCs and dDCs are incorporated into the growing flagellum, and (iii) the effects on DC distribution when IFT is disrupted.
Diffusion of DCs along the axoneme, filling available binding sites, has previously been observed in C. reinhardtii flagella lacking ODAs; the DC enters the base of the flagellum, binds first to the proximal axoneme, and proceeds distally, filling each unoccupied site (53). Some evidence for diffusion of the ODA protein DNAI1 has also been observed in T. brucei (54). Diffusion alone could initially generate a proximal/distal axoneme asymmetry; for example, diffusion of a limited quantity of high-affinity pDC into the proximal flagellum out-competes a low-affinity dDC. However, our simulation indicated retrograde transport of the pDC by IFT was required to maintain this asymmetry over an extended period. Together, our data indicate that a concentration gradient generated by directional transport of a protein complex can generate and maintain asymmetry in an organelle and may be a fundamental mechanism through which an organelle can generate internal asymmetry/structure, analogous to the concentration gradients driving polarity in cell and tissue development.
We show that proximal/distal asymmetry of ODAs in T. brucei and L. mexicana is involved in the control of the site of initiation of flagellum beat waveforms. The loss of the dDC resulted in the loss of ODAs from only the distal portion of the flagellum, and the subsequent loss of tip-to-base waveforms demonstrates that ODAs in this region are required for initiation and/or propagation of tip-to-base waveforms. This is consistent with previous studies showing that beat initiation occurs in the most distal 2 µm of the flagellum (16). Proteins involved in regulating waveform shape are often associated with the ODAs, with LC1 and LC4 representing well-characterized examples in C. reinhardtii (48, 49, 55), although IDA components are also important. In T. brucei, knockdown of the ODA component LC1 resulted in the loss of tip-to-base waveform generation (15, 29), but this was complicated by the complete loss of ODAs in these mutants. Our data showed that a distal-only LC4-like protein with a Ca2+ binding site is a repressor of distal initiation of tip-to-base waveforms. This reveals that the distal ODAs are an important site for the regulation of initiation of flagellar waveforms, thus controlling whether the waveform travels from tip to base or from base to tip.
Flagellum movement arises from dynein-driven sliding of neighboring axoneme microtubule doublets (56), and there are three different models to explain waveform propagation (56). The arguably leading model is the geometric clutch hypothesis, which states that mechanical distortion of the axoneme as it bends regulates force generation by regulating dynein arm engagement (25, 57, 58). The simplest interpretation of our results in the context of this hypothesis is that the dDC positions the distal ODAs so they are more likely than the proximal ODAs to spontaneously engage and start a flagellar waveform (although more complex interpretations, such as the involvement of the IDAs, are also possible). Loss of distal ODAs prevents distal waveform initiation, allowing spontaneous proximal initiation. Calcium binding to LC4-like could then modulate the likelihood of engagement of the distal ODAs, regulating the site of waveform initiation and therefore the direction of beat propagation. This role for LC4-like is consistent with previous data in C. reinhardtii, in which the switch from flagellar to ciliary beating upon photostimulation is calcium mediated (7, 59).
Given that mutations in both DCs (60) and proteins with an asymmetric localization (36) lead to primary ciliary dyskinesia in humans, a better understanding of the mechanisms by which asymmetry occurs and how it contributes to flagellar motility is essential. Here, we demonstrate that molecular asymmetries within the axoneme can be generated by an IFT-dependent concentration gradient of proteins within the flagellum and that this asymmetry is linked to the control of waveform initiation, defining whether a tip-to-base or base-to-tip beat occurs. This control is mediated by a potentially calcium-responsive protein, which relies on the dDCs for proper localization, enabling T. brucei and L. mexicana parasites to switch the direction of flagellum waveform propagation and control their swimming. It seems likely that proximal/distal asymmetry is a common feature of cilia and flagella and that the true extent and function of this important phenomenon are only beginning to become clear.
Methods
SmOxP9 procyclic T. brucei [derived from TREU 927, expressing T7 RNA polymerase and tetracycline repressor (61)] were grown in SDM79 with 10% FCS (62). Constructs for endogenous mNeonGreen (mNG) or mScarlet (mSc) tagging were generated by PCR and transfected as previously described (63), with the pPOT version 4 series of vectors (specifically pPOT mNG Blast or pPOT mSc Neomycin) used as PCR templates (63). Target sequences were selected and primers were designed using TAGit (www.sdeanresearch.com/cgi-bin/tagitA.cgi) (SI Appendix, Table S1) (63). Constructs for RNAi were generated using the pQuadra system (64). Primers for amplification of the target ORF fragment were designed using RNAit (44). Transfectants were elected with the necessary combination of 5 µg/mL blasticidin S hydrochloride, 5 µg/mL G-418 disulfate, and 10 µg/mL phleomycin (Melford Laboratories) and were cloned by limiting dilution in 96-well plates.
Cas9T7 L. mexicana [derived from WHO strain MNYC/BZ/62/M379, expressing Cas9 and T7 RNA polymerase (65)] were grown in M199 (Life Technologies) supplemented with 2.2 g/L NaHCO3, 0.005% hemin, 40 mM Hepes·HCL (pH 7.4), and 10% FCS. Constructs and single-guide RNA (sgRNA) templates for endogenous mNG-tagging templates were generated by PCR as previously described (65) and were transfected as previously described (63). The pLrPOT series of vectors was used as PCR templates for generating tagging constructs, specifically pLrPOT mNG Blast. These are a variant of pPLOT (63) with T. brucei and Crithidia fasciculata 5′ or 3′ UTRs and intergenic sequences replaced with complete L. mexicana intergenic sequences. The T. brucei actin 5′ UTR was replaced with the L. mexicana actin (LmxM.04.1230) 5′ UTR, the T. brucei aldolase 3′ UTR/C. fasciculata PGKB 5′ UTR fusion was replaced with the L. mexicana histone 2B intergenic sequence (between LmxM.19.0050 and LmxM.19.0030), the C. fasciculata PGKA/B intergenic sequence was replaced with the L. mexicana histone 2A intergenic sequence (between LmxM.08_29.1740 and LmxM.08_29.1730), and the T. brucei aldolase 3′ UTR was replaced with the L. mexicana eukaryotic initiation factor 5 (LmxM.25.0720) 3′ UTR. Constructs and sgRNA templates for ORF deletion were generated by PCR and transfected as previously described, using pT Blast and pT Neo as templates (65). Primers were designed using LeishGEdit (www.leishgedit.net/) (65). Transfectants were selected with the necessary combination of 20 µg/mL puromycin dihydrochloride, 5 µg/mL blasticidin S hydrochloride, 40 µg/mL G-418 disulfate, 50 µg/mL nourseothricin sulfate, and 25 µg/mL phleomycin (Melford Laboratories) and were cloned by limiting dilution in 96-well plates using MM199 as previously described (63).
T. brucei and L. mexicana cultures were grown at 28 °C. Culture density was maintained between 1 × 106 (T. brucei) or 1 × 105 (L. mexicana) and 1 × 107 cells/mL for continued exponential population growth. Culture density was measured using a CASY model TT cell counter (Roche Diagnostics) with a 60-µm capillary.
T. brucei and L. mexicana cell lines expressing fluorescent fusion proteins were imaged live. Cells were washed three times by centrifugation at 800 × g followed by resuspension in PBS supplemented with 10 mM glucose and 46 mM sucrose (vPBS). DNA was stained by including 10 µg/mL Hoechst 33342 in the second washing. Washed cells were settled on glass slides and were observed immediately. To generate cytoskeletons, cells were prepared as for live-cell microscopy, the membrane was solubilized with 0.5% Nonidet P-40 in PEME [100 mM Pipes·NaOH (pH 6.9), 2 mM EGTA, 1 mM MgSO4 and 100 nM EDTA] for 30 s, and then the remaining cytoskeleton was fixed by immersion in −20 °C methanol for 20 min. Cytoskeletons were rehydrated in PBS, mounted in 50 mM phosphate-buffered glycerol (pH 8.0), and imaged. For fluorescent labeling of HaloTag fusion proteins, cells were incubated in culture with fluorophore-conjugated ligands. For labeling of all HaloTag fusion proteins, cells were incubated with tetramethylrhodamine (TMRDirect ligand; Promega) at a 0.1-µM final concentration for 45 min. For pulse labeling of HaloTag fusion protein, cells were incubated with Coumarin ligand (Promega) at a 10-µM final concentration for 45 min, washed three times with medium, then incubated with 0.1 µM TMRDirect ligand for 45 min. We could not detect the expected blue fluorescence of the Coumarin ligand but found it was an effective block, as described previously (66). Widefield epifluorescence and phase-contrast images were captured using a Zeiss Axioimager.Z2 microscope with a 63× numerical aperture (NA) 1.40 oil immersion objective and a Hamamatsu ORCA-Flash4.0 camera. Cell morphology measurements were made in ImageJ (67).
Swimming and flagellar beat behaviors were analyzed for cells in the exponential growth phase in normal culture medium essentially as previously described (17). For cell swimming analysis, a 25.6-s video at five frames/s under darkfield illumination was captured from 5 µL of cell culture in a 250-µm deep chamber using a Zeiss Axioimager.Z2 microscope with a 10× NA 0.3 objective and a Hamamatsu ORCA-Flash4.0 camera. Particle tracks were traced automatically, and mean cell speed, mean cell velocity, and cell directionality (the ratio of velocity to speed) were calculated as previously described (17). For flagellar beat analysis, a 4-s video at 200 frames/s under phase-contrast illumination was captured from a thin film of cell culture between a slide and coverslip using a Zeiss Axiovert.A1 microscope with a 20× NA 0.3 objective and an Andor Neo 5.5 camera. Unlike previously, to reduce cell adhesion to the glass, glass slides and coverslips were blocked with BSA by immersion in 1% BSA for 60 s and then were washed with water and allowed to dry before use. Flagellar beat behaviors for each cell in the 4-s videos were classified manually.
Thin-section TEM samples were prepared as previously described (68, 69). Sections with nominal thicknesses 70 nm were cut, stained with lead citrate, and then observed using an FEI Tecnai 12 transmission electron microscopy with a Gatan UltraScan 1000 CCD camera. Transverse sections through flagella were classified as proximal or distal based on the width of the cell body to which the flagellum was attached: proximal if the cell body was more than ∼500 nm wide and distal if the cell body was less than ∼500 nm wide or if the flagellum was not laterally attached to a cell. Ninefold rotational averages of the axoneme structure (Markham rotations) were generated following perspective correction to ensure a circular axoneme cross-section, as previously described (70–72). Axoneme cross-sections were pooled from negative controls from previous studies and then were stacked and averaged in ImageJ (NIH) (67) to generate average proximal and distal axoneme electron density.
The agent-based simulation of flagellum assembly and pDC/dDC asymmetry was written in JavaScript/Node.js. pDC and dDC complexes were simulated as two particles, and particles could be either attached at a fixed position in the axoneme or detached and free to diffuse. The flagellum was simulated in discrete bins from proximal to distal (segments) and in discrete time steps (intervals). An evaluation interval of 0.1 s and a flagellum segment size of 100 nm were selected for useful granularity of binding/dissociation events and axoneme-binding capacity for DCs. The DC binding capacity of the axoneme was 37 per segment, assuming ninefold axoneme symmetry and a 24-nm repeat of ODAs (73). The probability of detached DC diffusion to an adjacent segment was 0.436 per interval, calculated from the probability of diffusing one segment distance in one interval and assuming a 5-nm DC effective hydrodynamic radius and a flagellar cytoplasm viscosity 670× greater than water [derived from BioNumbers generic ID 108250 (74), the diffusion constant of GFP in water, and the diffusion of rate of GFP in bacterial cytoplasm (75)]. Diffusion occurs equally in both directions; therefore the probabilities of anterograde diffusion and retrograde diffusion (D) were both 0.208 per interval. The probability of DC binding to a free site in the axoneme (on) was set to 1, assuming binding kinetics are fast relative to dissociation and diffusion. The probability of dissociation (off) was initially set to 4 × 10−5, giving a dissociation half-life on the order of 1 h. Flagellum growth rate was constant and set to 10 µm/h, up to a maximum length of 23 µm (76). The quantity of DC protein (Q) is expressed as a factor excess over the number of binding sites in half of the flagellum and was initially set to 2.0. For example Qdist = Qprox = 1 indicates an equal number of DC particles and axoneme-binding sites, half pDC and half dDC. Qdist = Qprox = 2.0 indicates a 100% excess. In every interval in which flagellum growth led to the addition of a new axoneme segment, the necessary number of new detached pDC and dDC particles was added to the base of the flagellum. IFT was simulated by sweeping from proximal to distal (anterograde transport) or distal to proximal (retrograde transport) and moving the first detached dDC particle (for anterograde transport) or pDC particle (for retrograde transport) encountered to the distal or proximal end of the axoneme, respectively. The quantity of IFT (T) was initially set to 0.2 per interval, assuming around five IFT trains/s (77) with one binding site per train. Dissociation probabilities were simulated with the following values: offdist = offprox = 4 × 10−5; offdist = 1 × 10−5 offprox = 1.6 × 10−4; and offdist = 1.6 × 10−4 offprox = 1 × 10−5. The final values were offdist = 1.6 × 10−4 offprox = 1 × 10−5. pDC or dDC knockdowns were simulated with Qprox, Qdist, Tprox, and Tdist as indicated in the text. The final values were Qdist = Qprox = 2.0 and Tprox = 0.2 and Tdist = 0.0.
The localizations of the T. brucei DC proteins were initially identified from TrypTag.org (41). T. brucei and L. mexicana protein and genome sequences were accessed using TriTrypDB.org (78). L. mexicana orthologs of T. brucei proteins were identified using synteny and orthogroup data from TriTrypDB.org (78) and were confirmed manually by reciprocal best BLAST. C. reinhardtii, G. lamblia, and human orthologs were identified using orthogroups as found by OrthoFinder (79) run on 45 diverse eukaryote genomes and then checked manually by BLAST search. We used the same set of 45 ciliated and nonciliated organism genomes as in previous studies concerning evolution of flagellar/ciliary components (80). Coiled coils were predicted using Coils v2.2 with default parameters (81). Key residues in the EF-hand fold [specifically 1a03 A (S100 protein set) for SSF47473] were derived from Superfamily v1.75 (82) and were visualized using WebLogo (83).
Acknowledgments
We thank Heather Jeffrey and Eva Gluenz for generating the L. mexicana LC4-like deletion cell line, Helen Farr and Emily Poon for contributing electron microscopy images, Catherine McEnhill for contributing to flagellum beat data collection, and Samuel Dean and the other coprincipal investigators of TrypTag. This work was supported by Wellcome Trust Grants 103261/Z/13/Z, 108445/Z/15/Z, and 104627/Z/14/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805827115/-/DCSupplemental.
Change History
July 26, 2021: The article text has been updated and the SI file has been replaced; please see accompanying Correction for details.
References
- 1.Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueki N, Matsunaga S, Inouye I, Hallmann A. How 5000 independent rowers coordinate their strokes in order to row into the sunlight: Phototaxis in the multicellular green alga Volvox. BMC Biol. 2010;8:103. doi: 10.1186/1741-7007-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holwill MEJ. The motion of Euglena viridis: The role of flagella. J Exp Biol. 1966;44:579–588. doi: 10.1242/jeb.44.3.579. [DOI] [PubMed] [Google Scholar]
- 4.Diehn B, Fonseca JR, Jahn TL. High speed cinemicrography of the direct photophobic response of Euglena and the mechanism of negative phototaxis. J Protozool. 1975;22:492–494. [Google Scholar]
- 5.Kung C, Chang SY, Satow Y, Houten JV, Hansma H. Genetic dissection of behavior in paramecium. Science. 1975;188:898–904. [PubMed] [Google Scholar]
- 6.Naitoh Y. Ionic control of the reversal response of cilia in Paramecium caudatum. A calcium hypothesis. J Gen Physiol. 1968;51:85–103. doi: 10.1085/jgp.51.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessen M, Fay RB, Witman GB. Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J Cell Biol. 1980;86:446–455. doi: 10.1083/jcb.86.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyams JS, Borisy GG. Flagellar coordination in Chlamydomonas reinhardtii: Isolation and reactivation of the flagellar apparatus. Science. 1975;189:891–893. doi: 10.1126/science.1098148. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt JA, Eckert R. Calcium couples flagellar reversal to photostimulation in Chlamydomonas reinhardtii. Nature. 1976;262:713–715. doi: 10.1038/262713a0. [DOI] [PubMed] [Google Scholar]
- 10.Hyams JS, Borisy GG. Isolated flagellar apparatus of Chlamydomonas: Characterization of forward swimming and alteration of waveform and reversal of motion by calcium ions in vitro. J Cell Sci. 1978;33:235–253. doi: 10.1242/jcs.33.1.235. [DOI] [PubMed] [Google Scholar]
- 11.Doughty MJ, Diehn B. Photosensory transduction in the flagellated alga, Euglena gracilis I. Action of divalent cations, Ca2+ antagonists and Ca2+ ionophore on motility and photobehavior. Biochim Biophys Acta. 1979;588:148–168. doi: 10.1016/0304-4165(79)90380-5. [DOI] [PubMed] [Google Scholar]
- 12.Iwadate Y. Photolysis of caged calcium in cilia induces ciliary reversal in Paramecium caudatum. J Exp Biol. 2003;206:1163–1170. doi: 10.1242/jeb.00219. [DOI] [PubMed] [Google Scholar]
- 13.Iwadate Y, Nakaoka Y. Calcium regulates independently ciliary beat and cell contraction in Paramecium cells. Cell Calcium. 2008;44:169–179. doi: 10.1016/j.ceca.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Shiba K, Inaba K. Inverse relationship of Ca2+-dependent flagellar response between animal sperm and prasinophyte algae. J Plant Res. 2017;130:465–473. doi: 10.1007/s10265-017-0931-7. [DOI] [PubMed] [Google Scholar]
- 15.Baron DM, Kabututu ZP, Hill KL. Stuck in reverse: Loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J Cell Sci. 2007;120:1513–1520. doi: 10.1242/jcs.004846. [DOI] [PubMed] [Google Scholar]
- 16.Gadelha C, Wickstead B, Gull K. Flagellar and ciliary beating in trypanosome motility. Cell Motil Cytoskeleton. 2007;64:629–643. doi: 10.1002/cm.20210. [DOI] [PubMed] [Google Scholar]
- 17.Wheeler RJ. Use of chiral cell shape to ensure highly directional swimming in trypanosomes. PLOS Comput Biol. 2017;13:e1005353. doi: 10.1371/journal.pcbi.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiba K, Shibata D, Inaba K. Autonomous changes in the swimming direction of sperm in the gastropod Strombus luhuanus. J Exp Biol. 2014;217:986–996. doi: 10.1242/jeb.095398. [DOI] [PubMed] [Google Scholar]
- 19.Holwill MEJ, McGregor JL. Control of flagellar wave movement in Crithidia oncopelti. Nature. 1975;255:157–158. doi: 10.1038/255157a0. [DOI] [PubMed] [Google Scholar]
- 20.Johnston DN, Silvester NR, Holwill MEJ. An analysis of the shape and propagation of waves on the flagellum of Crithidia oncopelti. J Exp Biol. 1979;80:299–315. [Google Scholar]
- 21.Sugrue P, Hirons MR, Adam JU, Holwill ME. Flagellar wave reversal in the kinetoplastid flagellate Crithidia oncopelti. Biol Cell. 1988;63:127–131. doi: 10.1016/0248-4900(88)90051-2. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Lu X. Drosophila sperm motility in the reproductive tract. Biol Reprod. 2011;84:1005–1015. doi: 10.1095/biolreprod.110.088773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay AG, Dey CS. Reactivation of flagellar motility in demembranated Leishmania reveals role of cAMP in flagellar wave reversal to ciliary waveform. Sci Rep. 2016;6:37308. doi: 10.1038/srep37308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay AG, Dey CS. Role of calmodulin and calcineurin in regulating flagellar motility and wave polarity in Leishmania. Parasitol Res. 2017;116:3221–3228. doi: 10.1007/s00436-017-5608-6. [DOI] [PubMed] [Google Scholar]
- 25.Lindemann CB. A “geometric clutch” hypothesis to explain oscillations of the axoneme of cilia and flagella. J Theor Biol. 1994;168:175–189. [Google Scholar]
- 26.Brokaw CJ. Control of flagellar bending: A new agenda based on dynein diversity. Cell Motil Cytoskeleton. 1994;28:199–204. doi: 10.1002/cm.970280303. [DOI] [PubMed] [Google Scholar]
- 27.Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell DR, Rosenbaum JL. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papon JF, et al. A 20-year experience of electron microscopy in the diagnosis of primary ciliary dyskinesia. Eur Respir J. 2010;35:1057–1063. doi: 10.1183/09031936.00046209. [DOI] [PubMed] [Google Scholar]
- 30.Branche C, et al. Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci. 2006;119:3443–3455. doi: 10.1242/jcs.03078. [DOI] [PubMed] [Google Scholar]
- 31.Kamiya R, Okamoto M. A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J Cell Sci. 1985;74:181–191. doi: 10.1242/jcs.74.1.181. [DOI] [PubMed] [Google Scholar]
- 32.Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yagi T, Uematsu K, Liu Z, Kamiya R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci. 2009;122:1306–1314. doi: 10.1242/jcs.045096. [DOI] [PubMed] [Google Scholar]
- 34.Fliegauf M, et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panizzi JR, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dougherty GW, et al. DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am J Respir Cell Mol Biol. 2016;55:213–224. doi: 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dean AB, Mitchell DR. Late steps in cytoplasmic maturation of assembly-competent axonemal outer arm dynein in Chlamydomonas require interaction of ODA5 and ODA10 in a complex. Mol Biol Cell. 2015;26:3596–3605. doi: 10.1091/mbc.E15-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirschell M, et al. Oda5p, a novel axonemal protein required for assembly of the outer dynein arm and an associated adenylate kinase. Mol Biol Cell. 2004;15:2729–2741. doi: 10.1091/mbc.E03-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subota I, et al. Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localization and dynamics. Mol Cell Proteomics. 2014;13:1769–1786. doi: 10.1074/mcp.M113.033357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dean S, Sunter JD, Wheeler RJ. TrypTag.org: A trypanosome genome-wide protein localisation resource. Trends Parasitol. 2017;33:80–82. doi: 10.1016/j.pt.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koutoulis A, et al. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takada S, Wilkerson CG, Wakabayashi K, Kamiya R, Witman GB. The outer dynein arm-docking complex: Composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol Biol Cell. 2002;13:1015–1029. doi: 10.1091/mbc.01-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redmond S, Vadivelu J, Field MC. RNAit: An automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol. 2003;128:115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler RJ, Scheumann N, Wickstead B, Gull K, Vaughan S. Cytokinesis in Trypanosoma brucei differs between bloodstream and tsetse trypomastigote forms: Implications for microtubule-based morphogenesis and mutant analysis. Mol Microbiol. 2013;90:1339–1355. doi: 10.1111/mmi.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gull K, et al. The cell cycle and cytoskeletal morphogenesis in Trypanosoma brucei. Biochem Soc Trans. 1990;18:720–722. doi: 10.1042/bst0180720. [DOI] [PubMed] [Google Scholar]
- 47.Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakato M, King SM. Calcium regulates ATP-sensitive microtubule binding by Chlamydomonas outer arm dynein. J Biol Chem. 2003;278:43571–43579. doi: 10.1074/jbc.M305894200. [DOI] [PubMed] [Google Scholar]
- 49.Sakato M, Sakakibara H, King SM. Chlamydomonas outer arm dynein alters conformation in response to Ca2+ Mol Biol Cell. 2007;18:3620–3634. doi: 10.1091/mbc.E06-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casey DM, Yagi T, Kamiya R, Witman GB. DC3, the smallest subunit of the Chlamydomonas flagellar outer dynein arm-docking complex, is a redox-sensitive calcium-binding protein. J Biol Chem. 2003;278:42652–42659. doi: 10.1074/jbc.M303064200. [DOI] [PubMed] [Google Scholar]
- 51.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagen KD, et al. Novel structural components of the ventral disc and lateral crest in Giardia intestinalis. PLoS Negl Trop Dis. 2011;5:e1442. doi: 10.1371/journal.pntd.0001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Owa M, et al. Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. Proc Natl Acad Sci USA. 2014;111:9461–9466. doi: 10.1073/pnas.1403101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincensini L, et al. Flagellar incorporation of proteins follows at least two different routes in trypanosomes. Biol Cell. 2018;110:33–47. doi: 10.1111/boc.201700052. [DOI] [PubMed] [Google Scholar]
- 55.Patel-King RS, King SM. An outer arm dynein light chain acts in a conformational switch for flagellar motility. J Cell Biol. 2009;186:283–295. doi: 10.1083/jcb.200905083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindemann CB, Lesich KA. Flagellar and ciliary beating: The proven and the possible. J Cell Sci. 2010;123:519–528. doi: 10.1242/jcs.051326. [DOI] [PubMed] [Google Scholar]
- 57.Lindemann CB. Testing the geometric clutch hypothesis. Biol Cell. 2004;96:681–690. doi: 10.1016/j.biolcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Lindemann CB, Lesich KA. The geometric clutch at 20: Stripping gears or gaining traction? Reproduction. 2015;150:R45–R53. doi: 10.1530/REP-14-0498. [DOI] [PubMed] [Google Scholar]
- 59.Wakabayashi K, King SM. Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J Cell Biol. 2006;173:743–754. doi: 10.1083/jcb.200603019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hjeij R, et al. UK10K Consortium CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poon SK, Peacock L, Gibson W, Gull K, Kelly S. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2012;2:110037. doi: 10.1098/rsob.110037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brun R, Schönenberger M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 63.Dean S, et al. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biol. 2015;5:140197. doi: 10.1098/rsob.140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inoue M, et al. The 14-3-3 proteins of Trypanosoma brucei function in motility, cytokinesis, and cell cycle. J Biol Chem. 2005;280:14085–14096. doi: 10.1074/jbc.M412336200. [DOI] [PubMed] [Google Scholar]
- 65.Beneke T, et al. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R Soc Open Sci. 2017;4:170095. doi: 10.1098/rsos.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dean S, Moreira-Leite F, Varga V, Gull K. Cilium transition zone proteome reveals compartmentalization and differential dynamics of ciliopathy complexes. Proc Natl Acad Sci USA. 2016;113:E5135–E5143. doi: 10.1073/pnas.1604258113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43(1 Suppl):25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 68.Höög JL, Gluenz E, Vaughan S, Gull K. Ultrastructural investigation methods for Trypanosoma brucei. Methods Cell Biol. 2010;96:175–196. doi: 10.1016/S0091-679X(10)96008-1. [DOI] [PubMed] [Google Scholar]
- 69.Gluenz E, Wheeler RJ, Hughes L, Vaughan S. Scanning and three-dimensional electron microscopy methods for the study of Trypanosoma brucei and Leishmania mexicana flagella. Methods Cell Biol. 2015;127:509–542. doi: 10.1016/bs.mcb.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gadelha C, Wickstead B, McKean PG, Gull K. Basal body and flagellum mutants reveal a rotational constraint of the central pair microtubules in the axonemes of trypanosomes. J Cell Sci. 2006;119:2405–2413. doi: 10.1242/jcs.02969. [DOI] [PubMed] [Google Scholar]
- 71.Wheeler RJ, Gluenz E, Gull K. Basal body multipotency and axonemal remodelling are two pathways to a 9+0 flagellum. Nat Commun. 2015;6:8964. doi: 10.1038/ncomms9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Markham R, Frey S, Hills GJ. Methods for the enhancement of image detail and accentuation of structure in electron microscopy. Virology. 1963;20:88–102. [Google Scholar]
- 73.Hughes LC, Ralston KS, Hill KL, Zhou ZH. Three-dimensional structure of the Trypanosome flagellum suggests that the paraflagellar rod functions as a biomechanical spring. PLoS One. 2012;7:e25700. doi: 10.1371/journal.pone.0025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moran U, Milo R, Jorgensen PC, Weber GM, Springer M. 2009. BioNumbers—The database of key numbers in molecular and cell biology. Available at https://dash.harvard.edu/handle/1/8063390. Accessed October 20, 2017.
- 75.Mullineaux CW, Nenninger A, Ray N, Robinson C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J Bacteriol. 2006;188:3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyler KM, Matthews KR, Gull K. Anisomorphic cell division by African trypanosomes. Protist. 2001;152:367–378. doi: 10.1078/1434-4610-00074. [DOI] [PubMed] [Google Scholar]
- 77.Fort C, Bonnefoy S, Kohl L, Bastin P. Intraflagellar transport is required for the maintenance of the trypanosome flagellum composition but not its length. J Cell Sci. 2016;129:3026–3041. doi: 10.1242/jcs.188227. [DOI] [PubMed] [Google Scholar]
- 78.Aurrecoechea C, et al. EuPathDB: The eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017;45:D581–D591. doi: 10.1093/nar/gkw1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodges ME, Wickstead B, Gull K, Langdale JA. Conservation of ciliary proteins in plants with no cilia. BMC Plant Biol. 2011;11:185. doi: 10.1186/1471-2229-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 82.Wilson D, et al. SUPERFAMILY—Sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]