Significance
The φ29 DNA packaging motor can be thought of as a five-cylinder engine, where each cylinder (subunit) catalyzes ATP hydrolysis to power DNA translocation. The motor operates in two phases: first, the subunits load fuel (ATP) sequentially one at a time; second, all subunits rapidly hydrolyze ATP in sequential fashion. How the subunits coordinate their activities remained unknown. Here, we show that the subunits use regulatory features observed in signaling pathways, a very different biological context, to coordinate their activities. Moreover, one of the subunits is stimulated by a contact with the DNA, transforming it into a “timer” element that paces the activity of the remaining cylinders. Similar mechanisms are likely present in other molecular machines.
Keywords: ring ATPases, molecular switch-like regulation, single molecule DNA packaging, ring-shaped molecular motors, intersubunit enzymatic regulation
Abstract
Subunits in multimeric ring-shaped motors must coordinate their activities to ensure correct and efficient performance of their mechanical tasks. Here, we study WT and arginine finger mutants of the pentameric bacteriophage φ29 DNA packaging motor. Our results reveal the molecular interactions necessary for the coordination of ADP–ATP exchange and ATP hydrolysis of the motor’s biphasic mechanochemical cycle. We show that two distinct regulatory mechanisms determine this coordination. In the first mechanism, the DNA up-regulates a single subunit’s catalytic activity, transforming it into a global regulator that initiates the nucleotide exchange phase and the hydrolysis phase. In the second, an arginine finger in each subunit promotes ADP–ATP exchange and ATP hydrolysis of its neighbor. Accordingly, we suggest that the subunits perform the roles described for GDP exchange factors and GTPase-activating proteins observed in small GTPases. We propose that these mechanisms are fundamental to intersubunit coordination and are likely present in other ring ATPases.
Many ATPases self-organize into ring-shaped structures and coordinate their enzymatic activity to efficiently perform biological functions, such as protein degradation, ATP synthesis, DNA strand separation, and chromosomal segregation (1, 2). Although such enzymatic coordination is necessary for proper function of the multimeric complex (3), how it emerges from stochastic processes and local molecular interactions is not well-understood.
The bacteriophage φ29 DNA packaging motor illustrates the highly coordinated enzymatic activity that such macromolecular complexes can attain (4). This motor operates using a two-phase cycle: an ADP-releasing/ATP-binding dwell phase, during which the DNA remains stationary, followed by an ATP-hydrolyzing and translocating phase, or burst phase, during which 10 bp of DNA is rapidly internalized in the capsid. In this paper, we use optical tweezers (Fig. 1), targeted mutagenesis of the conserved arginine finger R146 (5), and cryo-EM reconstruction to elucidate how the coordinated enzymatic activity displayed by the φ29 DNA packaging motor arises from stochastic processes and local interactions.
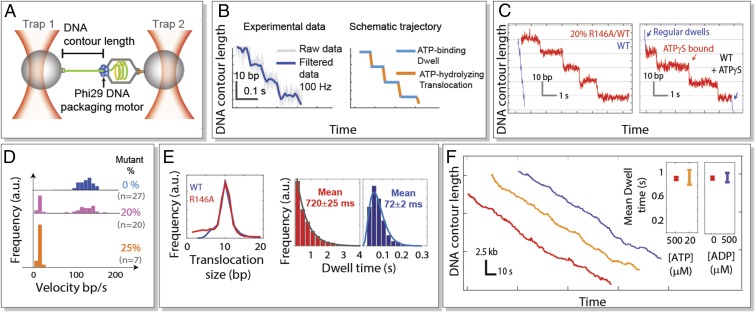
Fig. 1.
Packaging behavior of R146A/WT hybrid motors. (A) Single-molecule DNA packaging experiments. A viral packaging particle is held between two optical traps. (B, Left) Experimental DNA packaging trajectory acquired at 2,500 kHz (gray) and down-sampled to 100 Hz (blue). (B, Right) Schematic packaging biphasic cycle. ATP-binding “dwell” phase: DNA remains stationary (light blue). ATP-hydrolyzing “burst” phase: DNA is translocated into the protein capsid (orange). (C, Left) High-resolution packaging trajectories of WT (blue) and slow R146A/WT hybrid motors from a mixture containing 20% mutant subunits (red). (C, Right) Sample packaging trajectory of WT motors in the presence of the nonhydrolyzable nucleotide ATPγS in a 1:500 ([ATP]:[ATPγS]) ratio. ATPγS-induced pauses are highlighted in red; regular packaging behavior is in blue. (D) Velocity distribution of R146A/WT hybrid motors for different mixing ratios. (E) Analysis of the translocation size (Left) and dwell times (Right) of slow R146A/WT hybrid rings (red) fitted to a single-exponential distribution (gray) vs. WT motors (blue) fitted to a gamma distribution. (F) Sample packaging trajectories of R146A/WT hybrid motors and mean dwell times under different packaging conditions: saturating [ATP] (red; [ATP] = 500 μM), low [ATP] (yellow; [ATP] = 20 μM), and in competition with ADP (purple; [ATP] = [ADP] = 500 μM).
We show that, during the nucleotide exchange phase, the subunits display poor, thermally activated ATPase activities, which are then drastically elevated during the DNA translocation phase. Throughout this process, each subunit uses its arginine finger to promote nucleotide exchange (during the dwell) and to activate ATP hydrolysis (during the burst) in the adjacent ATPase. These features recall the operation of small GTPases, which are regulated by nucleotide exchange factors and activating proteins and behave as molecular switches (6, 7). Therefore, the φ29 ring ATPase can be thought of as five molecular switches arranged in a closed configuration. Moreover, the enzymatic activity of one of the subunits is up-regulated compared with the other four, likely due to a DNA contact, which transforms it into a more efficient ATPase, becoming a global regulator that also uses its arginine finger to, in turn, initiate the nucleotide exchange and ATP hydrolysis phases. We propose that similar mechanisms of intersubunit regulation operate in other ring ATPases.
Results
To disrupt intersubunit coordination, we used mutants of the arginine R146, an essential residue for ATPase activity recently identified as the arginine finger (5, 8). Two different substitutions—R146A and R146K—were screened in a single-molecule optical tweezers assay (Fig. 1 A and B). No tethers were observed with homomeric mutant rings, indicating that such rings either do not package DNA or do not assemble into functional motors. To obtain active mutant motors, we mixed WT subunits with mutants of either type in various ratios. Using this approach, we were able to obtain packaging trajectories of the resulting hybrid ring motors.
The R146A Substitution Is Tolerated in only One Copy.
Mixtures of R146A and WT subunits in a 1:4 ratio (20% mutant) yielded two clearly distinguishable phenotypes: WT-like velocity (>100 bp/s) and very slow motors (∼10 bp/s) (Fig. 1 C and D). Hybrid motors from a 1:3 mixing ratio (25% mutant) displayed only the slow packaging behavior (Fig. 1D). No tethers formed with higher mutant ratios. All packaging trajectories displayed the biphasic dwell–burst cycle described in WT motors (9) (Fig. 1C). However, the slow packaging traces displayed dwells that are 10 times longer (∼720 ms) than those in the WT (∼72 ms) (Fig. 1E), whereas the size and duration of the 10-bp translocation phase remained unaffected (Fig. 1E). It was previously shown that each translocating subunit packages 2.5 bp of DNA per hydrolyzed ATP (9) and that R146A homomeric rings possess no ATPase activity in bulk (5). Thus, the 10-bp translocation size of R146A/WT hybrid rings (Fig. 1E) implies that four subunits must be competent for DNA translocation and ATP hydrolysis. This observation—together with the bimodal velocity distribution (a detailed analysis is in SI Appendix) observed with this mutant (Fig. 1D)—indicates that the slow phenotype corresponds to rings with one R146A and four WT translocating subunits, in agreement with previous results showing that, under normal operation, one of the motor subunits does not translocate DNA (4, 9).
R146A Substitution Does Not Support ATP Hydrolysis.
Interestingly, unlike WT motors—with dwell times that are controlled by four ADP release events and are best described by a gamma distribution (4)—dwell times of R146A/WT hybrid motors are exponentially distributed, indicating that a single step has become rate limiting during this phase (Fig. 1E). To determine whether ADP release or ATP binding by the inactive subunit has become the rate-limiting step, we collected packaging trajectories of R146A/WT hybrid motors at low [ATP] (20 μM) alone or at high [ATP] (500 μM) mixed with ADP (500 μM) (Fig. 1F). Neither of these conditions showed a change in mean dwell time (Fig. 1F). Therefore, we conclude that the dwells are lengthened, because the R146A subunit cannot hydrolyze ATP. Our conclusion agrees with previous results showing that R146A homomeric ring motors do not display ATPase activity in bulk measurements (5) and that monomeric WT subunits do not exhibit ATPase activity (10), indicating that the presence of a transacting residue from the neighboring subunit is necessary for such activity.
R146A Substitution Uncovers the Basal ATPase Activity of the First Translocating Subunit.
The interpretation that R146A substitution abolishes the subunit’s ATPase activity is supported by the strikingly similar behavior observed when a single subunit is temporarily inactivated by the binding of a nonhydrolyzable ATP analog in WT motors (Fig. 1C), a behavior that was reported first in a previous study (4). In that same study, it was shown that, if the ATPγS had bound to any of the four translocating subunits, the motor paused, displaying incomplete bursts (7.5, 5, and 2.5 bp), and remained in the pause until the nucleotide was released or the inactivated subunit adopted the regulatory role (4). The phenotypes of R146A/WT hybrid motors imply that, in the absence of ATP hydrolysis by the nontranslocating subunit, the remaining subunits are able to spontaneously initiate DNA translocation after a long-enough dwell—720 ms on average. We propose that the cycle resumes with the thermally activated hydrolysis by one of the translocating subunits. Furthermore, because R146A/WT motors display consistently 10-bp bursts (Fig. 1E), such subunit must be the first immediately after the regulatory one and not the second, third, or fourth, as those cases would result in 7.5-, 5.0-, and 2.5-bp bursts, respectively. This observation also implies that the rate of spontaneous hydrolysis of the remaining translocating subunits is slower than ∼1.4 s−1. It is not clear at this point what the physical or structural bases are for the faster spontaneous hydrolysis rate of the first translocating subunit. As shown below, however, an asymmetric cryo-EM reconstruction (Fig. 2D) of packaging motors suggests that this difference can be originated by more extensive contacts established between the nontranslocating and the first translocating subunits than between any other pair (grey arrow). Accordingly, the mean dwell time of R146A/WT hybrid rings yields the spontaneous ATP hydrolysis rate of the first translocating subunit (∼1.4 s−1).
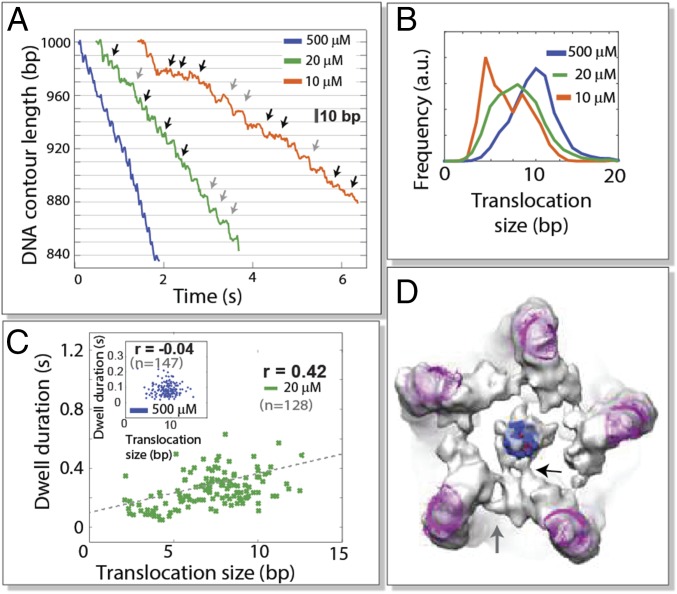
Fig. 2.
Behavior of WT motors at very low [ATP]. (A) Sample packaging trajectories of WT motors at different [ATP]. Gray arrows indicate 10-bp translocation events. Black arrows indicate translocation events smaller than 10 bp in size. (B) Translocation size distribution at saturating and sub-Km ATP conditions. (C) Correlation between dwell duration and the following translocation size at low [ATP] (green dots) and at saturating [ATP] (Inset; blue dots); r is the Pearson correlation coefficient. (D) Asymmetric cryo-EM reconstruction. Electron density of viral particles stalled during DNA packaging with ATPγS. ATPase subunits and DNA viewed in a bottom view (from the outside) of the bacteriophage. Atomic-resolution structures of packaging RNA (magenta) and DNA (dark blue) were fitted to the electron density. The black arrow indicates more extensive contacts between one of the subunits and the DNA. The gray arrow indicates the more extensive contacts established by two adjacent subunits.
The Behavior of the WT at Low [ATP] Uncovers the Spontaneous ATPase Activity of the Nontranslocating Subunit.
Next, we use WT motors to determine if the nontranslocating subunit can also hydrolyze ATP spontaneously. Remarkably, we found that the size of the translocation burst becomes smaller as ATP becomes more limited—below the motor’s Km (35 μM) (Fig. 2 A and B). We note that, even for the most limiting [ATP] tested (10 μM), the bursts are generally no smaller than 5 bp (Fig. 2B). This observation suggests that, at low [ATP], the subunits retain a degree of coordination in nucleotide binding and ATP hydrolysis. Indeed, competition experiments with nucleotide analogs confirmed that the subunits continue exchanging nucleotides and hydrolyzing ATP sequentially at low [ATP] (SI Appendix, Fig. S1). Moreover, the dwell duration correlates positively with the size of their subsequent translocation (Fig. 2C), whereas virtually no correlation was observed at saturating [ATP]. These results imply that, at high [ATP], DNA translocation is promptly activated on ring saturation with ATP (after ∼72 ms), whereas at very low [ATP], an alternative stochastic mechanism must initiate DNA translocation (after ∼250 ms) with or without ring saturation. The most parsimonious explanation is that thermal energy spontaneously activates hydrolysis by an ATP-bound subunit. However, the rate of spontaneous hydrolysis of a translocating subunit (∼1.4 s−1 or less) is too slow to account for the average dwell times observed at very low [ATP]. Therefore, we propose that the event that triggers DNA translocation under these conditions is the spontaneous hydrolysis by the nontranslocating subunit, which occurs at a rate of ∼4 s−1.
Ring Asymmetry Is Introduced by DNA Interaction.
Such faster spontaneous ATP hydrolysis rate (4 s−1 compared with 1.4 s−1 in translocating subunits) in an otherwise identical subunit could result from an event that breaks the ring’s structural symmetry. We sought to confirm this inference by imaging packaging WT motor complexes, stalled with ATPγS, using cryo-EM. No fivefold symmetry constraints were imposed in the reconstruction. Significantly, in the resulting ∼12-Å electron density map (Fig. 2D), one of five subunits is seen to interact more extensively with the DNA than the rest (Fig. 2D, black arrow). In addition, the data suggest more extensive contacts between the regulatory and the first translocating subunits than between any other pair.
Multiple Copies of R146K Subunits Are Tolerated in Active Ring Motors.
Next, we proceeded to determine whether the charge-conservative substitution R146K is better tolerated by the translocating subunits than R146A. We mixed R146K and WT subunits in various ratios; all mixtures displayed a broad distribution of packaging velocities (Fig. 3 A and B). Packaging velocities of R146K/WT motors can be clustered in three main groups: fast (WT like; ∼125 bp/s), intermediate (∼85 bp/s), and slow (∼45 bp/s) (Fig. 3B). Increasing the ratio of R146K subunits in the mixture shifted the velocity distribution monotonically toward lower values (Fig. 3B). This behavior is in stark contrast with the results obtained with R146A/WT motors and suggests that motors can tolerate two or more R146K subunits (SI Appendix, Table S1 has a detailed discussion).
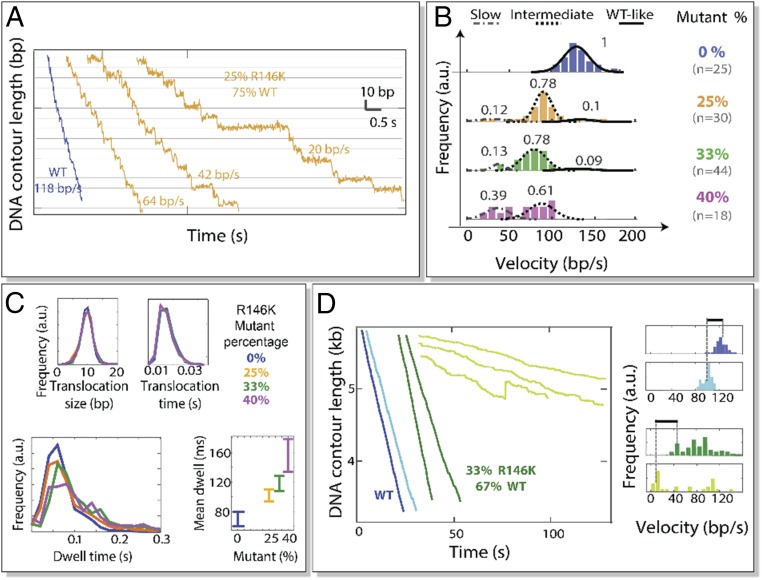
Fig. 3.
Packaging behavior of R146K/WT hybrid motors. (A) High-resolution packaging trajectories with WT (blue) and R146K/WT hybrid rings (gold). (B) Velocity distribution of R146K/WT mixtures for different mixing ratios. Each velocity group was fitted to a Gaussian distribution. The Gaussians were normalized within each mixing condition to represent the fraction of motors belonging to each group (SI Appendix has a detailed analysis). (C) Analysis of the dwell and translocation phases of R146K/WT hybrid rings (25, 33, and 40% mutant) and the WT (0% mutant). (D) ADP inhibition experiments. (Left) Sample packaging trajectories for WT (blue) and R146K/WT hybrid motors (green) and in the presence of ADP (light blue and light green, respectively; [ATP] = [ADP] = 500μM). (Right) Velocity distribution of WT and R146K/WT hybrid rings in the presence and absence of ADP (same color code as in Left). Dotted lines correspond to the mean packaging velocity. Black lines on top indicate the extent of inhibition by ADP in either scenario.
The Arginine Finger Mediates Nucleotide Exchange.
A detailed analysis of the resulting phenotypes shows that the translocation phase—during which four of the ATP hydrolyses events occur (4)—is not affected by the R146K mutation (Fig. 3C). This result may be rationalized by the observation that other ring ATPases utilize a transacting lysine to catalyze ATP hydrolysis (11). We find that, as we increase R146K/WT mixing ratios, the mean dwell time increases relative to the WT (Fig. 3C). Since nucleotide exchange occurs during the dwell phase, we sought to determine which kinetic step is affected by the R146K substitution. To this end, we first considered the following kinetic scheme (Scheme 1) for nucleotide exchange.
Scheme 1.
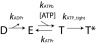
Kinetic scheme.
In Scheme 1, D, E, T, and T* indicate ADP-bound, empty, ATP-bound, and ATP-tight bound states, respectively; and kADPr, kATPb, kATPr, and kATP_tight are the kinetic rates for ADP release, ATP binding, ATP release, and ATP tight binding per subunit, respectively. Given previous results showing that the Hill coefficient of the ring motor at saturating [ATP] equals 1 (nH = 1) (9), this kinetic scheme does not take into consideration subunit cooperativity. Instead, the value of nH = 1 led to a model, in which ATP binding to consecutive subunits is separated by an irreversible transition and the binding of ATP to an empty site induces the release of an ADP bound to the next adjacent subunit (9). As a result, only one subunit is available for binding ATP at any given time, which takes place in an interlaced fashion with the release of ADP. Therefore, sequential nucleotide exchange around the ring can be described by the kinetic scheme successively in each subunit. Next, we derived an analytical expression for the packaging velocity of the motor in terms of its Michaelis–Menten parameters, Km and Vmax:
| [1] |
where
| [2] |
and
| [3] |
Eq. 2 indicates that the reduced Vmax observed with mutant ring motors must be due to a slower rate of ADP release, a slower rate of ATP tight binding, or both. To determine Km and Vmax in a homogeneous population of mutant ring motors, we designed a substitution of the residue adjacent to the arginine finger, F145I, to moderately perturb R146’s local environment. Homomeric rings of F145I mutants displayed reduced ATPase and packaging activities in bulk assays and a similar phenotype (longer dwell times and no effect in the translocation events) as R146K/WT hybrid motors (SI Appendix, Fig. S2).
Two observations were made from F145I ring motors. First, we found that the statistical parameter Nmin—defined as <τ>2/(<τ2> − <τ>2), where τ is the dwell time, which represents a lower bound to the number of rate-limiting events underlying the dwell time distribution (12)—is the same in F145I and in WT motors (Nmin equals ∼3.4) (SI Appendix, Fig. S2). ADP release by the four translocating subunits is known to be the slowest event during the dwell phase in WT motors (4). Therefore, the invariance of Nmin suggests that kADPr remains rate limiting in F145I but reduced relative to the WT to account for lengthening of the dwell in these mutants. The alternative scenario, in which kATP_tight alone is reduced, was tested in a Monte Carlo simulation. We discarded such a scenario, as it would lead to an increase in Nmin, which we do not observe experimentally (SI Appendix, Table S2). Second, ATP titration showed that, although Vmax is reduced in F145I, Km is invariant (SI Appendix, Fig. S3). According to Eq. 3, to retain Km invariance, the reduced ADP release rate in F145I must be accompanied by a similar reduction in ATP loose-binding (or docking) rate, kATPb (assuming that the rates for ADP release and ATP release remain comparable, kADPr ∼ kATPr, a reasonable assumption given that both nucleotides are likely to undo similar contacts during release). Indeed, a Monte Carlo simulation confirms that equally reduced kADPr and kATPb decrease Vmax without affecting Km and Nmin (SI Appendix). This simulation also predicts that the mutant rings would be inhibited more in the presence of ADP than WT rings (SI Appendix, Table S2). We tested this prediction by measuring the activities of F145I homomeric rings and R146K/WT hybrid rings (33% mutant) in the presence of equal amounts (500 μM) of ADP and ATP (SI Appendix, Fig. S4 and Fig. 3D, respectively). Indeed, ADP inhibited F145I motors more than WT rings (∼50 vs. ∼16% reduction in mean velocity, respectively) (SI Appendix, Fig. S4), and the slowest population of the R146K/WT hybrid rings (those containing more R146K mutant subunits) also had a greater velocity reduction in the presence of ADP than WT motors (∼66 vs. 16% reduction in mean velocity, respectively) (Fig. 3D). These observations indicate that, during the dwell, the transacting arginine finger plays a direct role in the release of ADP in the adjacent subunit (SI Appendix has a detailed analysis).
Discussion
Our results indicate that, during nucleotide exchange, ATP binding to one subunit induces the release of ADP by the adjacent subunit, a process mediated by the transacting arginine finger. Throughout this process, the subunits’ spontaneous low hydrolysis rates determined here ensure ring saturation with ATP without hydrolysis. However, this ATPase activity is insufficient to account for the duration of the ATP-hydrolyzing/translocating phase (∼0.01 s), which requires catalytic rates >250 s−1 (assuming four subunits hydrolyzing ATP sequentially). Therefore, on ring saturation with ATP, the subunits must switch to a stimulated ATPase activity to support the observed rapid DNA translocation. The crystal structure of the ATPase subunit suggests the physical basis for sequential ATPase stimulation (5): ATP hydrolysis in one subunit causes a power stroke by its DNA translocating loop, which repositions its arginine finger to stimulate ATP hydrolysis in the following catalytic site and so on.
The regulatory features described here recall those observed in small GTPases, which require exchange factors to release GDP and activating proteins to efficiently hydrolyze GTP (13). In the φ29 DNA packaging motor, each subunit uses its arginine finger to function as an exchange factor—during the dwell phase—and as an activating protein—during the ATP-hydrolyzing phase—for the neighboring ATPase (Fig. 4). The cryo-EM reconstruction presented here (Fig. 2D) shows that one subunit establishes a distinct DNA contact. We propose that the DNA-contacting subunit operates differently from the rest and is possibly up-regulated by such contact. This interpretation is consistent with previous bulk experiments showing that the subunits’ ATPase activities increase in the presence of DNA (10). In this picture, the DNA-contacting subunit releases ADP first—hence initiating the nucleotide exchange phase—and hydrolyzes ATP first—hence initiating the DNA translocation phase (Fig. 4). Altogether, these mechanisms illustrate how global coordination and the quasideterministic behavior of molecular machines can arise from local stochastic chemical interactions.
Fig. 4.
The ring ATPase is a set of five molecular switches arranged in a closed configuration. (1) The DNA-bound subunit releases ADP on interacting with the DNA (red loop in the subunit; red). (2) The subunits display poor ATPase activity during the ATP-binding phase (light blue). Each subunit uses its arginine finger to facilitate ADP release in its neighbor. All of the subunits bind ATP in a sequential manner. Hydrolysis can only take place slowly at a basal spontaneous rate. (3) The subunit contacting the DNA hydrolyzes ATP first (navy). (4) The subunits display efficient ATPase activities during the translocating phase (orange). Each subunit uses its arginine finger to activate ATP hydrolysis in its neighbor, resulting in a rapid hydrolysis cascade. (Center) Schematic representation of each regulatory mechanism in the context of a molecular packaging trajectory.
Small GTPase-like regulation has been proposed to underlie the operation of other ring ATPases and molecular machines (14–16). For instance, in F1-ATPase, the β subunit contains the catalytic pocket, whereas the α subunit is known to not only stimulate the ATPase activity (17) but also, prevent the ADP-inhibited state by facilitating ADP release from the β subunit using its arginine finger (18). Moreover, small GTPases and the subunits of ring ATPases are thought to have evolved from a common ancestor (19). It is tempting to speculate that the mechanisms of global enzymatic regulation described here are conserved in other ring ATPases as well as in other biological contexts.
Materials and Methods
Sample Preparation.
WT and mutant ATPase gp16, genomic DNA, and fiberless proheads were isolated as described previously (5). To avoid high filling conditions, the 19.3-kb genomic DNA was digested with BstEII (New England Biolabs) to produce a 12.5-kb piece of DNA. The 12.5-kb piece of DNA was biotinylated using Klenow exo- (New England Biolabs) to fill in the overhang with biotinylated nucleotides (Invitrogen). Proheads were partially packaged (30–60 s) with biotinylated 12.5-kb genomic DNA and stalled with ATPγS (Roche). The packaging buffer contained 25 mM Tris⋅HCl, pH 7.8, 50 mM NaCl, and 5 mM MgCl2 with various concentrations of nucleotides and nucleotide analogs as specified in the text.
Optical Trapping.
High-resolution packaging measurements were conducted on a dual-trap instrument using a solid-state 1,064-nm laser as described previously (20). Traps were calibrated as previously described (4, 9). Tethers were formed between a 0.90-μm-diameter streptavidin-coated bead and a 0.88-μm-diameter anticapsid antibody-coated bead (Spherotech) held in separate optical traps. Packaging was restarted in an ATP-containing buffer, and DNA translocation by individual motor complexes was determined from the decrease in the bead-to-bead distance. An oxygen scavenging system [100 μg mL−1 glucose oxidase, 5 mg mL−1 dextrose (Sigma-Aldrich), 20 μg mL−1 catalase (Calbiochem)] was included in the buffer to prevent the formation of reactive singlet oxygen. All packaging experiments were conducted as described before in a semipassive mode, such that tension applied to the motor was kept between 7 and 12 pN.
Data Analysis.
Raw 2.5-kHz data were collected and filtered to 100–250 Hz for further analysis. A modified Schwarz Information Criterion algorithm was used to find steps in the packaging traces as described previously (20) in a custom-written Matlab script. Mean values of the dwell duration distributions were computed by boot strapping to normalize the distribution and compute confidence intervals (displayed as errors in the figures). Velocity distributions were built as histograms of the motors’ velocities. Then, the velocity histograms were normalized to compare across different sets of data. The velocity of each motor was determined by fitting a straight line to its packaging trajectory.
Cryo-EM Imaging.
The DNA packaging motor was assembled as described in the sample preparation. The reaction was stalled by adding ATPγS after 3 min of initiating packaging with ATP (Roche; ATP concentration is 500 μM). The sample was incubated for 2 min; 1 U of RQ DNase I (Promega) was added and incubated at room temperature for 10 min. The sample was placed on ice until grid preparation for cryo-EM imaging. A prohead:DNA ratio of 2:1 was used to maximize packaging efficiency. Three-minute packaging corresponds to ∼75% head full. Grid preparation and image reconstruction were performed as described previously (5).
Supplementary Material
Acknowledgments
We thank Lisa Alexander, Varsha Desai, Ronen Gabizon, Liang Meng Wee, and Shannon Yan for helpful discussion of the paper. C.B. acknowledges support from the Nanomachines program (KC1203) funded by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy (DOE) contract no. DE-AC02-05CH11231 (development and application of step-finding algorithms). This work is also funded, in part, by National Institute of Health grants R01GM071552 (fluorescent protein FRET characterization) and R01GM032543 (fluorescent protein X-ray characterization), and by the Howard Hughes Medical Institute (fluorescent protein counting). S.T. acknowledges the University of California Institute for Mexico and the United States (UC MEXUS) for a graduate fellowship, S.L. acknowledges support from US NIH Pathway to Independence Award R00-GM107365, and J.P.C. acknowledges the Pew Scholar Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802736115/-/DCSupplemental.
References
- 1.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 2.Abbondanzieri EA, Zhuang X. Molecular biology: Concealed enzyme coordination. Nature. 2009;457:392–393. doi: 10.1038/457392a. [DOI] [PubMed] [Google Scholar]
- 3.Tang WK, Xia D. Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants. J Biol Chem. 2013;288:36624–36635. doi: 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chistol G, et al. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao H, et al. Structural and molecular basis for coordination in a viral DNA packaging motor. Cell Rep. 2016;14:2017–2029. doi: 10.1016/j.celrep.2016.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiner DJ, Lundquist EA. Small GTPases. WormBook. 2016;2016:1–99. doi: 10.1895/wormbook.1.67.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall A, Nobes CD. Rho GTPases: Molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos Trans R Soc Lond B Biol Sci. 2000;355:965–970. doi: 10.1098/rstb.2000.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd J, Thielman B, Wendell D. Detailed kinetic analysis of the φ29 DNA packaging motor providing evidence for coordinated intersubunit ATPase activity of gp16. Virology. 2012;432:370–375. doi: 10.1016/j.virol.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Greenleaf WB, Shen J, Gai D, Chen XS. Systematic study of the functions for the residues around the nucleotide pocket in simian virus 40 AAA+ hexameric helicase. J Virol. 2008;82:6017–6023. doi: 10.1128/JVI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffitt JR, Bustamante C. Extracting signal from noise: Kinetic mechanisms from a Michaelis-Menten-like expression for enzymatic fluctuations. FEBS J. 2014;281:498–517. doi: 10.1111/febs.12545. [DOI] [PubMed] [Google Scholar]
- 13.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 14.Vale RD. Switches, latches, and amplifiers: Common themes of G proteins and molecular motors. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 16.Lebbink JHG, et al. Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS. J Biol Chem. 2010;285:13131–13141. doi: 10.1074/jbc.M109.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komoriya Y, et al. Principal role of the arginine finger in rotary catalysis of F1-ATPase. J Biol Chem. 2012;287:15134–15142. doi: 10.1074/jbc.M111.328153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yukawa A, Iino R, Watanabe R, Hayashi S, Noji H. Key chemical factors of arginine finger catalysis of F1-ATPase clarified by an unnatural amino acid mutation. Biochemistry. 2015;54:472–480. doi: 10.1021/bi501138b. [DOI] [PubMed] [Google Scholar]
- 19.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, et al. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014;157:702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.