Significance
The dissemination of antibiotic resistance in pathogenic bacteria is remarkable, and antivirulence agents with novel mechanisms of action could lead to innovative therapeutic concepts. The simultaneous suppression of multiple virulence factors or pathways using small-molecule compounds is an attractive approach to reducing the virulence of Staphylococcus aureus and may offer promising therapeutic potential in combating multidrug-resistant bacteria such as methicillin-resistant S. aureus. Here we describe a conceptual breakthrough in applying chemical genetics to probe for small-molecule compounds that will suppress the expression of multiple virulence factors in S. aureus simultaneously. Compound M21 has the ability to reverse virulent S. aureus to a nonvirulent state, as demonstrated in a mouse model of infection.
Keywords: MRSA, antivirulence, virulence factors, ClpP, bacterial infection
Abstract
Emerging antibiotic resistance among bacterial pathogens has necessitated the development of alternative approaches to combat drug-resistance-associated infection. The abolition of Staphylococcus aureus virulence by targeting multiple-virulence gene products represents a promising strategy for exploration. A multiplex promoter reporter platform using gfp-luxABCDE dual-reporter plasmids with selected promoters from S. aureus-virulence-associated genes was used to identify compounds that modulate the expression of virulence factors. One small-molecule compound, M21, was identified from a chemical library to reverse virulent S. aureus into its nonvirulent state. M21 is a noncompetitive inhibitor of ClpP and alters α-toxin expression in a ClpP-dependent manner. A mouse model of infection indicated that M21 could attenuate S. aureus virulence. This nonantibiotic compound has been shown to suppress the expression of multiple unrelated virulence factors in S. aureus, suggesting that targeting a master regulator of virulence is an effective way to control virulence. Our results illustrate the power of chemical genetics in the modulation of virulence gene expression in pathogenic bacteria.
As a major human pathogen in both community and hospital settings, Staphylococcus aureus causes a variety of infections, ranging from topical infections to life-threatening conditions (1). Studies have shown that S. aureus has acquired resistance to virtually all antibiotics (2). In addition, multidrug-resistant strains of methicillin-resistant Staphylococcus aureus (MRSA) that combine resistance to methicillin and several other antibiotics are relatively frequent in the hospital setting, and treatment of such infections has become increasingly difficult (3). An array of virulence factors, including hemolysins, leukocidins, immune-modulatory factors, and exoenzymes, work in concert and contribute to the virulent properties of S. aureus (4). As there are no vaccines available on the market yet and attempts to develop such interventions are not yet successful (5), virulence suppression presents an alternative approach for combating S. aureus infection (6, 7).
In contrast to conventional antibiotic strategies, inhibition of the action or production of virulence factors would prevent infection via nonbactericidal pathways. As few virulence factors are essential for the survival of bacteria, in principle, the inhibition of virulence would presumably exert less selective pressure for acquiring resistance to corresponding antibiotics (8).
Several antivirulence attempts have focused on accessory gene regulator (agr) (9), the sortase enzyme system (10), the carotenoid biosynthetic pathway (CrtM and CrtN) (11, 12), and other regulatory pathways (MgrA and SarA) (13) or have directly targeted toxin proteins (14) and the toxins’ receptors (15). As virulence expression is a highly regulated and concerted process modulated by various regulators that are known or yet to be discovered, knocking down one virulence-related pathway may have consequences that affect the expression of other virulence factors. For example, a mutation in agr eliminates α-toxin expression but causes a burst in the expression of protein A (16); as a result, it is difficult to abolish S. aureus virulence by repressing the expression of any single virulence gene or regulator (17). The simultaneous suppression of multiple virulence factors thus represents a promising strategy to control or limit infections caused by S. aureus. As a matter of fact, it has been shown that the combined use of antibodies against four important surface-associated factors in S. aureus leads to a substantial improvement in preventing infections in vaccinated animals (18). Suppressing the expression of multiple virulence factors with small-molecule compounds will further enhance our arsenal for combating bacterial infections.
Virulence factors of microbes are involved in different stages of host–microbe interactions (19–21). The onset and establishment of infections can be seen as a subversion of the host immunity by the invading pathogens. On the basis of the concept of chemical genetics, we have used small-molecule compounds to perturb the complex network of bacterial virulence regulation and have identified several compounds that modulate the expression of various virulence-associated genes. One of our bioactive compounds that exerts suppressive activities in more than 14 virulence factors was further characterized, and its target was mapped to ClpP, a bacterial protease and a major player in virulence regulation (22, 23). We demonstrate here that the small-molecule compound M21, identified via high-throughput screening (HTS), attenuates the virulence of S. aureus by inhibiting ClpP, and that the administration of this compound blocks the establishment of infection in animal models.
Results
Screening for Virulence Repressors.
A platform with a total of 20 gfp-luxABCDE dual-reporter plasmids with selected promoters from S. aureus virulence-associated genes was constructed (24). The luminescence signal, driven by an hla promoter from USA300-pGLhla, was monitored with a multimode plate reader. Only compounds that resulted in significantly low luminescence readings were selected for secondary screening. Overall, 50,240 compounds were tested in triplicate in primary screening, using luminescence from S. aureus USA300-pGLhla as the readout. A total of 670 compounds that repressed 65% of hla promoter activity were selected for secondary screening.
In secondary screening, the repressive effects of 670 compounds on hla promoter activity were first verified by the disk diffusion method (24). Of the 670 hit compounds, 400 showed reproducible repressive effects on hla promoter activity. Selected compounds were subsequently tested against 14 promoters: agr, cap5, cap8, clfA, fnbA, fnbB, hla, psm, pvl, RNAIII, sae P1, sae P3, spa, and srtA. Finally, 54 compounds were selected for further study.
Our previous chemical genetics studies of viral pathogens suggested that the hit compounds generated by HTS of a diverse chemical library targeting infectivity/virulence should yield specific inhibitors of the crucial components or pathways involved in viral propagation (25). As ClpP has been shown to be a major player in regulating S. aureus virulence, the third round of screening was thus aimed at finding inhibitors of ClpP. ClpP has been shown to degrade protein substrates such as SspB (26) and the fluorescent substrate Suc-LY-AMC, which is a fluorescence-quenched probe with the AMC fluorophores quenched by Suc (27). The successful cleavage of the peptide bond between leucine and tyrosine by ClpP results in increased fluorescence intensity. As the Km value of Suc-LY-AMC was determined to be 133 μM, we chose 100-μM substrate concentration for subsequent screening against 670 selected hla repressors. M21 was the only compound that had both ClpP inhibition effect and targeting multiple virulence factors at the same time. M21 was then used as a candidate compound for evaluation of its capability to attenuate S. aureus virulence via ClpP inhibition.
M21 Represses the Expression of Virulence Factors.
The minimum inhibitory concentration of M21 for S. aureus USA300 was higher than 500 μM (Fig. 1A), and the median toxic concentrations acquired from cytotoxic assay using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay were 133 μM for Vero cells and 277 μM for Hep2 cells.
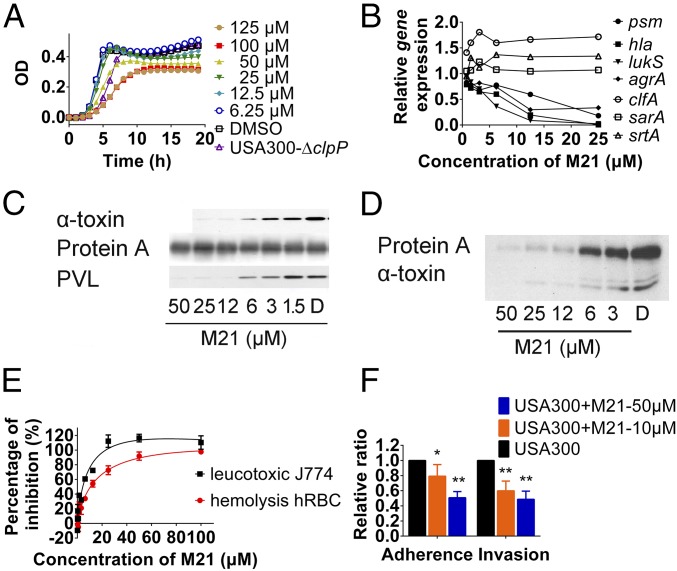
Fig. 1.
M21 affecting the expression and production of selected virulence factors. (A) USA300 growth monitored by OD600 in the presence of different concentrations of M21. (B) Expressions of psm, hla, agrA, lukS, clfA, and sarA in the presence of M21 at different concentrations as monitored by qPCR. (C) Production of protein A, α-toxin, and PVL in the presence of M21 assayed by Western blot in USA300. (D) Production of protein A and α-toxin in the presence of M21 assayed by Western blot in Mu3. (E) M21 inhibition on hemolysis of human erythrocyte caused by S. aureus and on leukotoxic effect of S. aureus against mouse macrophage cell line (J774.1). (F) M21 inhibition on the adherence and invasion effect of S. aureus against A549. Compound pretreated bacteria were used in the adherence assay and invasion assay. All the experiments were repeated at least three times. Data were analyzed by Student t test. All data represent mean values ±SD (*P < 0.05; **P < 0.01; ***P < 0.001). P values were determined using GraphPad Prism.
As shown in Fig. 1B, the expression levels of major virulence factor genes hla and psm were repressed more than 10-fold, whereas pvl was reduced more than 200-fold. For agrB and spa, the decrease was only around twofold, but srtA was nearly unaffected by 20-μM compound M21. Meanwhile, M21 showed a dose-dependent effect on repressing the production of α-toxin in both community-acquired MRSA (CA-MRSA, USA300) and hospital-acquired MRSA (HA-MRSA, mu3) and representing Panton–Valentine leukocidin in USA300. The production of protein A was slightly affected by the addition of M21 in USA300 and was obviously reduced in mu3 (Fig. 1 C and D). This observation may be because USA300 is a high virulent CA-MRSA with protein A hyperexpressed. The protein A in USA300 may be at its maximal expression, and thus becomes less responsive to M21 inhibition.
As the reduced expression of α-toxin and other leukotoxins should reduce hemolysis and the leukotoxic effect of S. aureus, the M21-treated bacterial culture supernatant was applied to test the hemolysis and leukotoxic effects. A dose-dependent inhibition effect of M21 on hemolysis against human erythrocyte and a leukotoxic effect against J774.1 mouse macrophage cell was observed (Fig. 1E). In addition, as the reduced expression of cell-surface-associated virulence factors could affect the adherence and invasion of S. aureus to epithelial cells, M21-treated S. aureus showed a significantly reduced ratio of adherence and internalization in A549 cells (Fig. 1F).
M21 Is an Inhibitor of ClpP.
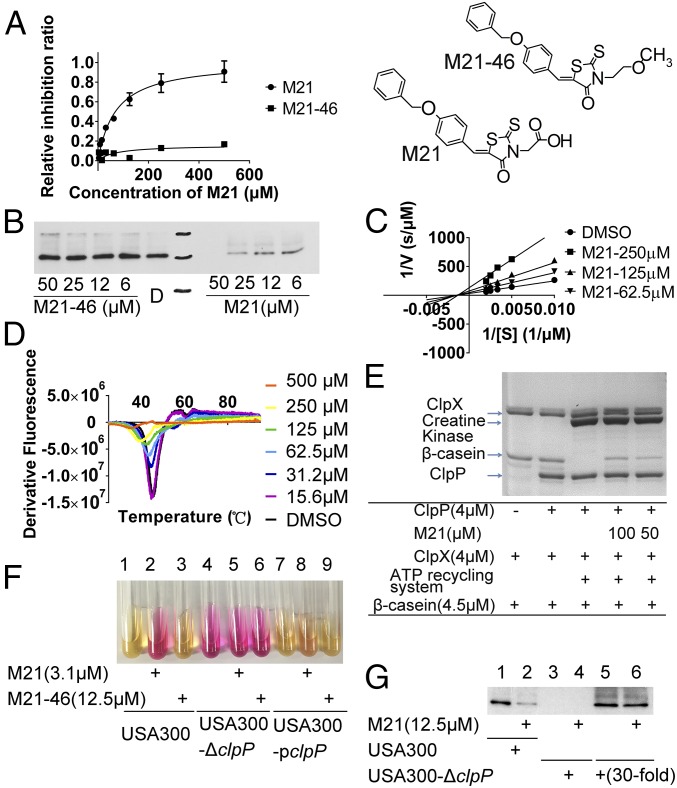
Using the fluorescent probe–based peptidase assay, the IC50 value of M21 on ClpP was determined to be 41.42 μM (Fig. 2A). A small modification of the acetic acid group of M21 to methoxyethyl of M21-46 led to little or no inhibition effect on the peptidase activity of ClpP (Fig. 2A), suggesting that the inhibitory activity of M21 to ClpP is specific and sensitive to slight structural deviations. Western blot analysis of the production of α-toxin indicated that M21 had a robust repression effect on α-toxin production, whereas its structural analog M21-46 had no effect on α-toxin production (Fig. 2B). Further kinetic studies showed that M21 increased the maximum velocity (Vmax), but did not affect the Michaelis constant (Km) of ClpP enzyme reactions (Fig. 2C), indicating that M21 is a noncompetitive inhibitor of ClpP. M21 is a noncompetitive inhibitor of ClpP discovered.
Fig. 2.
M21 Binds to and Inhibits ClpP Activity. (A) M21 inhibits ClpP cleavage of SUc-LY-AMC substrate in a peptidase assay. Concentrations of ClpP and Suc-LY-AMC used were 1 μM and 100 μM, respectively. (B) Western blotting analysis of a-toxin production altered by M21 and its analog. (C) Noncompetitive inhibition of M21 on ClpP cleavage of Suc-LY-AMC. M21 increased the maximum velocity (Vmax), but did not affect the Michaelis constant (Km) of ClpP enzyme reactions. (D) Differential scanning fluorimetry analysis of the binding between different concentrations of M21 and ClpP. The ClpP became instable to heat with the presence of M21. (E) Proteolytic assay confirming M21 protect β-casein from degradation by ClpP/ClpX. (F) Urease production in USA300 was induced by M21, but not M21-46. Overexpression of ClpP lead to the resistance of USA300 to M21-induced urease production. (G) Western blot analysis of the production of α-toxin in wild-type strain and clpP deleted strain. M21 showed no repression effect on the production of α-toxin when clpP was deleted.
To confirm the direct interaction between compound M21 and ClpP, the effect of M21 on the thermal stability of ClpP was analyzed with differential scanning fluorimetry (Fig. 2D). In the presence of increasing amounts of M21, ClpP became thermally unstable, indicating that M21 does interact physically with ClpP.
ATPase-dependent protein degradation by ClpP protein was also investigated using the well-studied substrate β-casein in the presence of ClpX, creatine kinase, and ATP. After a 1-h incubation period, β-casein was completely degraded by ClpP (4 μM), whereas treatment with compound M21 inhibited such proteolytic activity (Fig. 2E), further demonstrating the specific inhibitory effects of M21 on ClpP.
M21 Represses Expression of Virulence Factors via Inhibition of ClpP.
ClpP plays a central role in virulence regulation. It indirectly controls the production of α-toxin and protein A and a number of other virulence factors under the control of agr (28). In agreement with a previous study (23), our experimental results demonstrate that the deletion of ClpP in S. aureus led to strong induction of urease production (Fig. 2F, lanes 1 and 4). Similarly, the inhibition of ClpP by M21 displayed the phenotype of up-regulated urease production (Fig. 2F, lanes 1–3). When ClpP was overexpressed (Fig. 2F, lane 8), the urease induction effect of M21 was diminished. Our results show that M21 induces the production of urease through inhibiting ClpP activity.
In USA300, the deletion of clpP led to reduced production of α-toxin (Fig. 2G, lanes 1 and 3). The effects of treatment with M21 on α-toxin production by the S. aureus USA300 and USA300-ΔclpP strains were analyzed by Western blotting. The presence of M21 could also alter the production level of α-toxin in USA300 (Fig. 2G, lanes 1 and 2). Furthermore, in the USA300-ΔclpP strain, the production of α-toxin was greatly reduced but not totally abolished, and its presence could be detected after the supernatant was concentrated 30-fold. However, the production of α-toxin was not affected by the presence of M21 (Fig. 2G, lanes 5 and 6) in this ΔclpP variant. When the target was deleted, M21 showed no effect on α-toxin production; this finding supports our notion that M21 represses S. aureus virulence in a ClpP-dependent manner.
Molecular Modeling and Mutagenesis Study.
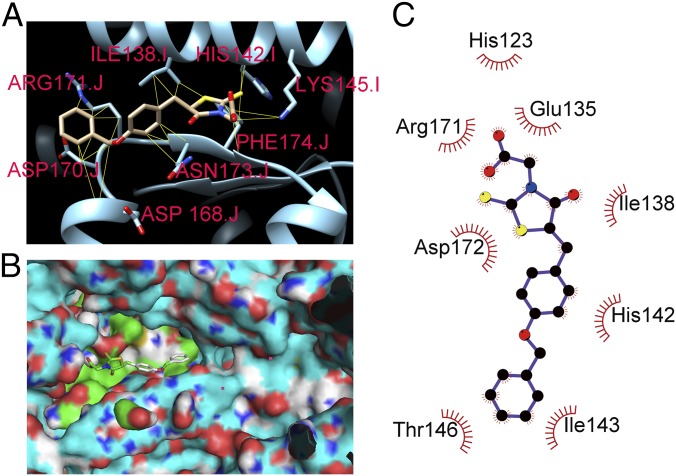
Computer-simulated docking using a grid box surrounding the entire crystal structure of ClpP predicted that M21 could fit into two pockets near H142 or Q35, respectively (SI Appendix, Fig. S1), as both sites are adjacent to the active site, but H142 was found to be more important for ClpP enzyme activity (29). A highest-affinity binding conformation of such docking around H142 is shown in Fig. 3A, with the contacts between M21 and ClpP represented by yellow lines. To further define the possible binding conformations and to mimic the real dynamics of a ligand exiting, we attempted flexible docking by modeling flexible sidechains from amino acids 116 to 122, from 170 to 177 in chain J, and from 135 to 148 in chain I. An alternate binding pocket resulting from our flexible binding model is shown in Fig. 3B. In one conformation (Fig. 3C), direct hydrophobic interactions with residues His123, Glu135, Ile138, His-142, Ile-143, Thr146, Asp172, and Arg171 are indicated.
Fig. 3.
Molecular modeling of ClpP-M21 binding. (A) Predicted highest-affinity binding conformation of M21 to ClpP. The ClpP of S. aureus (PDB entry 3v5e) are labeled in cyan. M21 is predicted to bind in close proximity to the catalytically important amino acid residue H142. The side chains of the predicted interacting amino acid residues that participate in the hydrophobic contacts with M21 are shown. (B) Surface representation of ClpP is labeled in cyan. The green color area is the binding pocket, and the cyan area is the active site. (C) 2D schematic diagram of ClpP-M21 interactions is shown. Hydrophobic contacts are represented by an arc with spokes radiating toward the M21 atoms that they contact. The contacted atoms in M21 are shown with spokes radiating back.
On the basis of the potential binding sites predicted by our docking models, we made site-directed mutations along the predicted binding pocket by replacing the original amino acid residues from sites 116–123, 135–148, and 170–177 with either an alanine or a glycine residue (Table 1).
Table 1.
Mutagenesis analysis of ClpP
| Mutant # | Amino acids | Activity | M21 inhibition |
| 1 | P116A | + | C |
| 2 | N117A | +/− | |
| 3 | A118G | ++ | R |
| 4 | E119A | — | |
| 5 | V120A | ++ | C |
| 6 | M121A | / | |
| 7 | I122A | +/− | |
| 8 | H123A | — | |
| 9 | E135A | — | |
| 10 | I136A | — | |
| 11 | E137A | +++ | C |
| 12 | I138A | +++++ | S |
| 13 | A139G | ++++ | C |
| 14 | A140G | — | |
| 15 | N141A | — | |
| 16 | H142A | — | |
| 17 | I143A | — | |
| 18 | L144A | — | |
| 19 | K145A | +++ | C |
| 20 | T146A | +++ | R |
| 21 | R147A | — | |
| 22 | E148A | ++++++ | C |
| 23 | D170A | — | |
| 24 | R171A | — | |
| 25 | D172A | — | |
| 26 | N173A | +/− | S |
| 27 | F174A | — | |
| 28 | L175A | / | |
| 29 | T176A | +++ | S |
| 30 | A177G | + | C |
| 31 | Wild-type | +++ | C |
C, similar as wild-type ClpP control; S, sensitive than wild-type ClpP control; R, resistance than wild-type ClpP control.
+With enzyme activity. The more pluses, the higher activity.
−No enzyme activity.
/Did not get mutant.
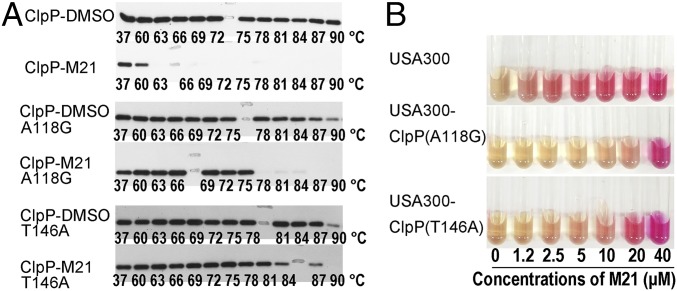
Bacterial lysates of Escherichia coli Rosetta (DE3) expressing wild-type ClpP, our mutant variants and a negative control carrying an empty vector, pET28a, were used to evaluate variation in ClpP activity. Compared with Rosetta (DE3)-pET28aclpP, the negative control showed no activity on the cleavage of Suc-LY-AMC. Mutants with retained activities were further evaluated for potential resistance to M21 inhibition. Two mutants (#3, A118G, and #19, T146A) showed slightly increased resistance to M21 inhibition, whereas M21 at 500 µM completely inhibited wild-type ClpP activity and mutants A118G and T146A retained nearly 20% of their enzymatic activity in the presence of the same amount of M21. Cellular thermal shift assay (30) was used to further examine the binding affinity and interactions of wild-type ClpP and the two mutant variants with M21 (Fig. 4A). The ClpP binding interactions in the bacterial lysate were stable at temperatures reaching 87 °C, but addition of M21 to the lysate resulted in precipitation and unfolding at temperatures as low as 60 °C. The presence of M21 did not affect the stability of the two mutants to the same extent as wild-type ClpP, indicating that the affinity of M21 to A118G and T146A mutants is greatly reduced.
Fig. 4.
Cellular thermal shift assay to verify the interaction of M21 with ClpP. (A) Western blot data from cellular thermal shift assay experiments determining the heat stability of ClpP in bacterial lysate after a 1 h preincubation period in the presence of DMSO. The numbers under each band indicate the temperature in degree Celsius of sample treatment. The shift of denatured temperature of ClpP on treatment with M21 indicated the binding of M21 to the enzyme. The reduced heat sensitivity of mutants of A118G and T146A in the presence of M21 suggested that these amino acid residues should be involved in the binding of M21 on ClpP. (B) M21 showed different effects on urease productions in wild-type and mutants at different concentrations.
To evaluate the phenotypic response of the two mutations to M21 in S. aureus, we replaced wild-type clpP with mutant genes that encoded the A118G or T146A mutations. The A118G and T146A mutation did not influence the effects of ClpP on urease production. However, unlike the wild-type ClpP, the induction effect of M21 on urease production by inhibiting ClpP activity was diminished in the two mutants (Fig. 4B), which suggests that A118 and T146 are crucial amino acid residues involved with M21 binding and further demonstrates that that ClpP is indeed the biological target of M21.
M21 Reduces S. aureus Pathogenicity in Mouse Model of Infection.
By conducting a pharmacokinetic study on M21 in mice, we determined that at a dose of 19.25 mg/kg (0.385 mg per dose), serum concentration of M21 was highest at 175–200 μM 10–30 min postdose. The serum concentration of M21 remained greater than 140 μM for 2 h and decreased to 32 μM after 4 h (SI Appendix, Fig. S2). To confirm the in vivo effect of M21 is a result of antivirulence activities, we used a lower dose of M21 and detected M21-serum concentration at 30 min postdose. When dosed at 3.85 mg/kg (0.077 mg per dose), the serum concentration of M21 was 25.8 μM, and when dosed at 1.925 mg/kg (0.0385 mg per dose), the serum concentration was 8.5 μM (SI Appendix, Table S1). Notably, M21 showed little to no inhibitory effects on S. aureus growth in vitro.
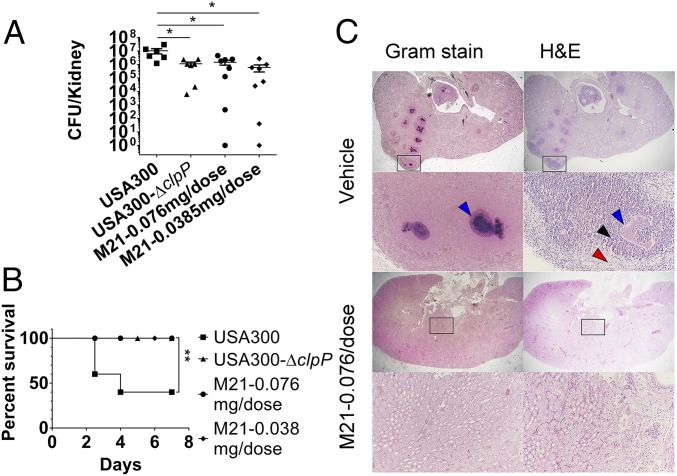
To evaluate the in vivo efficacy of compound M21, we used a mouse bacteremia model to assess S. aureus virulence. For infection of mice with S. aureus USA300 at low inoculum, mice infected with the ClpP deletion mutant and mice treated with two different M21 dosages had the same level of bacteria recovered from the kidneys (Fig. 5A). However, the results between the treatment and control groups were significant (P = 0.0393 for wild-type and clpP deletion mutant, 0.0335 for vehicle and M21 at 0.077 mg per dose, and 0.0208 for vehicle and M21 at 0.0385 mg per dose).
Fig. 5.
M21 reduces S. aureus pathogenicity in mouse model of intravascular infection. (A) Bacterial load of S. aureus USA300 or USA300-∆clpP in the kidney of mice after i.v. infection in the presence or absence of M21 treatment. (B) Bacteremia lethal model on mice with infection of S. aureus USA300 and USA300-∆clpP in the presence or absence of M21 treatment. (C) Histopathology of staphylococcal abscess communities. BALB/c mice were infected with S. aureus Newman via retro-orbital injection. Gram-stained and H&E-stained tissues of infected kidneys on day 10 after infection were analyzed by light microscopy, and images were captured. In the graph, each dot represents a mouse. The group treated with injection buffer is indicated as ‘‘Vehicle.’’ The group treated with compound M21 is indicated as ‘‘M21.’’ The statistical difference was calculated by Student’s two-tailed t test. Survival data were calculated by Log-rank (Mantel-Cox) test. All data represent mean values ±SEM (*P < 0.05; **P < 0.01). P values were determined using GraphPad Prism, using an unpaired parametric t test with Welch’s correction and log-rank (Mantel-Cox) test.
For the lethal infection model, mice infected with USA300 had a 40% survival rate, whereas all mice infected with strains USA300-∆clpP survived. Compared with the vehicle group, M21 was protective against S. aureus for all the mice in the treatment groups (Fig. 5B), with significant differences between the treatment and control groups [P = 0.0057, by log-rank (Mantel-Cox) test]. For the delayed treatment model, treatment started 3 d postinfection, and M21 failed to show an effect on reducing bacterial load in kidneys at both dosages (SI Appendix, Fig. S3).
The mice that survived in the lethal model were visually inspected for abscess formation in the kidneys. Randomly selected kidneys were fixed in formalin, embedded, thin-sectioned, and stained with H&E as well as Gram stain. For each kidney, four sagittal sections at 200-μm intervals were viewed by microscopy (Fig. 5C). On day 10, staphylococcal abscess communities developed as a central nidus (dark blue in gram stain). Staphylococci were enclosed by an amorphous, eosinophilic pseudocapsule (blue arrowhead) and surrounded by a zone of polymorphonuclear leukocytes (black arrowhead), separated through an eosinophilic layer from healthy kidney tissue (red arrowhead). Reduction or complete absence of abscess formation was observed for the M21-treated group.
Discussion
S. aureus has developed resistance mechanisms against nearly all known antibiotics, which necessitates the identification of new therapeutic targets and the development of novel strategies to combat S. aureus-related infections (7, 8, 30). The virulence of S. aureus arises from the combination of surface-associated virulence factors, exotoxins, enterotoxins, and superantigens (1). S. aureus exotoxins, α-toxin, β-toxin, δ-toxin, PVL, and phenol-soluble modulins (PSMs) all contribute to the lysis of leukocytes (31), and α-toxin and PSMs may also contribute to the formation of biofilms (32, 33). The surface-associated virulence factors, protein A, fibronectin-binding protein A/B, and envelope-associated proteins contribute to the adherence and invasion of S. aureus to epithelial cells and to the formation and persistence of abscesses in the host tissues (17). As it is difficult to abolish S. aureus virulence by repressing the expression of any single virulence gene or regulator (17), the simultaneous suppression of multiple virulence factors may offer effective and promising therapeutic potential. Thus, if nonantibiotic compounds can be identified to repress all or most of the important virulence factors in S. aureus, these compounds may actualize a paradigm in the treatment of staphylococcal diseases by abolishing the expression of bacterial virulence.
We used HTS to identify compounds that could repress the luminescence signal of several promoters simultaneously, thus validating the feasibility of suppressing multiple virulence factors, using a small-molecule compound approach. We used a chemical library with 50,240 small-molecule compounds for HTS and selected 670 primary hits for secondary screening. The hit rate during primary screening was ∼1.9%, similar to previous studies from our laboratory (34, 35). For secondary screening, 14 promoters were used to assess the potency and spectrum of the primary hits, and 54 compounds were selected. This group of compounds may help us to develop new therapeutic strategies or discover new pathways by which to regulate virulence. These compounds may function by inhibiting major virulence regulators or multiple targets and may also repress the expression of other unknown or untested virulence factors.
The proteolytic activity of ClpP is critical for the virulence of S. aureus (36). S. aureus mutants clpX and clpP were severely attenuated in a murine abscess model, whereas ClpC showed significant defects in long-term survival and in the intracellular replication of this pathogen (37, 38). The expression of the global regulatory gene agr was decreased in the clpP mutant strain, leading to reduced production of α-toxin and induction of urease activity. The virulence was reduced in the clpP mutant strain, most likely because of the repression of agr-regulated virulence genes (36). Moreover, the global regulator MgrA, which is involved in antibiotic resistance and virulence, was found to be significantly down-regulated by the deletion of clpP. Therefore, ClpP may control the transcription of numerous virulence factors, such as the urease operon and hla, by modulating the MgrA level (13, 28). In addition, there are indications that Rot (repressor of toxin) in complex with RNAIII is a substrate of serine protease sspA under the control of Clp-dependent degradation (39). These reports suggest that Clp chaperones and proteases are central in regulating the virulence of S. aureus.
Using the concept of chemical genetics (25, 35), compound M21 has been identified as an inhibitor of ClpP, which is supported by our in vitro binding and enzymatic cleavage assays. Different from Beta-lactones (covalent binding inhibitor of ClpP) (40) and AV145 (competitive ClpP inhibitor) (41), M21 affects ClpP activity by noncompetitive inhibition. The IC50 of M21 against ClpP is in the middle micromolar range. A slight modification of M21’s structure has led to analog M21-46, which was found to be ineffective against ClpP; this further suggests that M21 inhibition of ClpP is very specific and that its structure is critical for its inhibitory activity. In a proteolytic assay in which β-casein was used as the substrate, M21 also effectively inhibited the proteolytic activity of ClpP, indicating that M21 is a genuine inhibitor of its target ClpP. Two strategies were applied to further prove the specificity of M21 in inhibiting ClpP: overexpression of the target protein to increase the resistance of S. aureus to the antivirulence effect of M21, and deletion of the target protein to abolish the antivirulence effect of M21. First, the overexpression of ClpP in S. aureus decreased the inhibition effect of M21 against ClpP and the induction effect on urease; second, our newly constructed ClpP-deficient mutant USA300-ΔclpP displayed no response to M21 inhibition of the production of α-toxin and urease in S. aureus. These observations further strengthen our proposal that ClpP is the molecular target of M21.
Virtual docking suggests that the binding pocket of M21 on ClpP is located beside the α-helix (amino acids 135–147) and the β-sheet (amino acids 170–177) of the adjacent monomer. Of particular interest is receptor flexibility, which is essential to capturing the protein conformational changes on ligand binding. The binding model has guided our mutagenesis study to systematically replace each amino acid in the binding pocket of ClpP. Only 14 of 30 mutants retained enzymatic activities, indicating that the amino acids around the binding pocket are important for ClpP enzyme activity and that any alteration in the amino acid residues in this region would have probably inactivated the enzyme. On the basis of enzymatic assay and cellular thermal shift assay, we confirmed that two amino acids are related to the binding of M21 on ClpP. After the introduction of site mutation on clpP in the chromosome, the effects of M21 on ClpP mutant strains validated the interaction between M21 and ClpP in vivo. This again proved that ClpP is the target of M21. The virulence attenuation effect of M21 was further confirmed in animal bacteremia models. In general, forward chemical genetics has been employed to identify compounds that produce the phenotype of interest in an organism without knowing the target first; the target is identified by biophysical, biochemical, or genetics methods. Reverse chemical genetics has been employed to identify compounds that bind to or interact with certain gene products (usually proteins) without knowing the phenotype to be altered; the phenotype induced by the compound will then be observed when the organism is treated with the compound. Both methods have their own advantages and disadvantages. Forward chemical genetics can identify compounds with biological functions in cell- or organism-based assays, but the target identification is the bottleneck. Reverse chemical genetics can readily identify compounds interacting with the gene product, but the biological effects of the selected compound exerted on the cell or the organism may not be apparent, and most potent hit compounds cannot illustrate corresponding biological activities in cell-based or in vivo assays.
In our study, we have combined the use of forward and reverse chemical genetics for the rapid identification of compounds and corresponding molecular targets. We started with forward chemical genetics to acquire a small pool of compounds with defined biological functions (virulence suppression). We then employed reverse chemical genetics using the selected pool of 670 biologically active compounds to identify inhibitors of a known virulence regulator (ClpP). Through this strategy, we have minimized the disadvantages of both methods and shown that M21 specifically targets ClpP to suppress S. aureus virulence.
Experimental Procedures
High-Throughput Screening.
Strain USA300-pGLhla was used in high-throughput screening to identify repressors of hla promoter activity (SI Appendix, Table S2). Briefly, 50 µL brain heart infusion medium was aliquoted into 384-well plates (Thermo Scientific), and 100 nL compound at 10 mg/mL was added to each well by robotic pins to a final concentration of 20 µg/mL. Bacteria were diluted in brain–heart infusion medium to 5E8 CFU/mL, and 100 nL was added to each well to a final concentration of 1E7 CFU/mL. The plates were then placed into a Cytomat incubator (Thermo Scientific) and incubated at 37 °C with CO2 for 14 h. The plates were then transferred to a DTX multimode plate reader (Beckman Coulter) to monitor luminescence, followed by OD620 readings to monitor bacterial growth. Compounds were screened in triplicate, as previously described in our published study (35).
Other Procedures.
Detailed procedures are available in SI Appendix, SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank K. Y. Yuen for his continuous support of the project. We are grateful for the involvement of S. M. Leung, W. W. N. Mak, and C. K. W. Wong in histology studies and R. E. Wan for critical reading of the manuscript. The initial support by Strategic Research Theme fund from HKU URC and partial support from RGC of the Hong Kong SAR Project No. AoE/P-705/16 (to R.Y.T.K.) are acknowledged. The authors also acknowledge the assistance of the University of Hong Kong Li Ka Shing Faculty of Medicine Faculty Core Facility. This work was supported by the Research Fund for the Control of Infectious Diseases Commissioned Study Project Grants HK-09-01-14 and HK-09-01-15 and HMRF Commissioned Study Project Grant HKM-15-M11 (to R.Y.T.K.). We are grateful for Victor Torres and Taeok Bae for their generous gifts of plasmids pOS1hrtAB and pKOR1, respectively. The Providence Foundation is acknowledged for its support of the establishment of LC-MS facilities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720520115/-/DCSupplemental.
References
- 1.Crossley KB. Staphylococci in Human Disease. 2nd Ed. Wiley-Blackwell; Chichester, UK: 2010. p. xii. [Google Scholar]
- 2.Lowy FD. Antimicrobial resistance: The example of Staphylococcus aureus. J Clin Invest. 2003;111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot SI, Vandewoude KH, Hoste EA, Colardyn FA. Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch Intern Med. 2002;162:2229–2235. doi: 10.1001/archinte.162.19.2229. [DOI] [PubMed] [Google Scholar]
- 4.Peacock SJ, et al. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002;70:4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pozzi C, et al. Vaccines for Staphylococcus aureus and target populations. Curr Top Microbiol Immunol. 2017;409:491–528. doi: 10.1007/82_2016_54. [DOI] [PubMed] [Google Scholar]
- 6.Escaich S. Antivirulence as a new antibacterial approach for chemotherapy. Curr Opin Chem Biol. 2008;12:400–408. doi: 10.1016/j.cbpa.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon CP, Williams P, Chan WC. Attenuating Staphylococcus aureus virulence gene regulation: A medicinal chemistry perspective. J Med Chem. 2013;56:1389–1404. doi: 10.1021/jm3014635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens DL, et al. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195:202–211. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 10.Uddin R, Lodhi MU, Ul-Haq Z. Combined pharmacophore and 3D-QSAR study on a series of Staphylococcus aureus Sortase A inhibitors. Chem Biol Drug Des. 2012;80:300–314. doi: 10.1111/j.1747-0285.2012.01403.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu CI, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao P, Davies J, Kao RYT. Dehydrosqualene desaturase as a novel target for anti-virulence therapy against Staphylococcus aureus. MBio. 2017;8:e01224-17. doi: 10.1128/mBio.01224-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun F, et al. Targeting MgrA-mediated virulence regulation in Staphylococcus aureus. Chem Biol. 2011;18:1032–1041. doi: 10.1016/j.chembiol.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick CC, Caballero AR, Balzli CL, Tang A, O’Callaghan RJ. Chemical inhibition of alpha-toxin, a key corneal virulence factor of Staphylococcus aureus. Invest Ophthalmol Vis Sci. 2009;50:2848–2854. doi: 10.1167/iovs.08-3157. [DOI] [PubMed] [Google Scholar]
- 15.Alonzo F, 3rd, et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature. 2013;493:51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Stewart GC. Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J Bacteriol. 2004;186:3738–3748. doi: 10.1128/JB.186.12.3738-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng AG, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stranger-Jones YK, Bae T, Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci USA. 2006;103:16942–16947. doi: 10.1073/pnas.0606863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes-Robles T, et al. Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell Host Microbe. 2013;14:453–459. doi: 10.1016/j.chom.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers ME, Becker RE, Sailer A, Turner JR, Bubeck Wardenburg J. Synergistic action of Staphylococcus aureus α-Toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe. 2015;17:775–787. doi: 10.1016/j.chom.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres VJ, et al. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel A, et al. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol. 2006;188:5783–5796. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frees D, Gerth U, Ingmer H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int J Med Microbiol. 2014;304:142–149. doi: 10.1016/j.ijmm.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Gao P, et al. Construction of a multiplex promoter reporter platform to monitor Staphylococcus aureus virulence gene expression and the identification of usnic acid as a potent suppressor of psm gene expression. Front Microbiol. 2016;7:1344. doi: 10.3389/fmicb.2016.01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao RY, et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J, et al. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res. 2013;12:547–558. doi: 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- 27.Geiger SR, Böttcher T, Sieber SA, Cramer P. A conformational switch underlies ClpP protease function. Angew Chem Int Ed Engl. 2011;50:5749–5752. doi: 10.1002/anie.201100666. [DOI] [PubMed] [Google Scholar]
- 28.Frees D, et al. New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J Proteome Res. 2012;11:95–108. doi: 10.1021/pr200956s. [DOI] [PubMed] [Google Scholar]
- 29.Gersch M, List A, Groll M, Sieber SA. Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein. J Biol Chem. 2012;287:9484–9494. doi: 10.1074/jbc.M111.336222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 31.Löffler B, et al. Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010;6:e1000715. doi: 10.1371/journal.ppat.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson MJ, et al. Alpha-toxin promotes Staphylococcus aureus mucosal biofilm formation. Front Cell Infect Microbiol. 2012;2:64. doi: 10.3389/fcimb.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012;8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao RY, et al. Characterization of SARS-CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence-based assay. FEBS Lett. 2004;576:325–330. doi: 10.1016/j.febslet.2004.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kao RY, et al. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol. 2010;28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol. 2003;48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 37.Frees D, et al. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee I, et al. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J Bacteriol. 2005;187:4488–4496. doi: 10.1128/JB.187.13.4488-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frees D, Sørensen K, Ingmer H. Global virulence regulation in Staphylococcus aureus: Pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect Immun. 2005;73:8100–8108. doi: 10.1128/IAI.73.12.8100-8108.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Böttcher T, Sieber SA. Beta-lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J Am Chem Soc. 2008;130:14400–14401. doi: 10.1021/ja8051365. [DOI] [PubMed] [Google Scholar]
- 41.Pahl A, et al. Reversible inhibitors arrest ClpP in a defined conformational state that can be revoked by ClpX association. Angew Chem Int Ed Engl. 2015;54:15892–15896. doi: 10.1002/anie.201507266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.