Significance
Schwann cells (SCs), the ensheathing glia of the peripheral nervous system, promote nerve repair/regeneration. Defects in these SC injury responses potentially contribute to the pathogenesis of diabetic neuropathy, the most prevalent form of peripheral neuropathy. Here, we show that O-GlcNAcylation, a posttranslational modification controlled by the metabolic state of the cell, influences SC injury responses and nerve repair. The transcription factor JUN, an essential regulator of the SC injury program, is O-GlcNAcylated at multiple sites, and loss of this modification leads to increased activity and ineffective repair SC function. These results demonstrate that O-GlcNAcylation regulates SC injury responses via modulation of JUN activity and broaden our understanding of how changes in metabolism, such as occur in diabetes, affect nerve repair.
Keywords: OGT, Schwann cells, O-GlcNAcylation, JUN, nerve injury
Abstract
Schwann cells (SCs), the glia of the peripheral nervous system, play an essential role in nerve regeneration. Upon nerve injury, SCs are reprogrammed into unique “repair SCs,” and these cells remove degenerating axons/myelin debris, promote axonal regrowth, and ultimately remyelinate regenerating axons. The AP-1 transcription factor JUN is promptly induced in SCs upon nerve injury and potently mediates this injury-induced SC plasticity; however, the regulation of these JUN-dependent SC injury responses is unclear. Previously, we produced mice with a SC-specific deletion of O-GlcNAc transferase (OGT). This enzyme catalyzes O-GlcNAcylation, a posttranslational modification that is influenced by the cellular metabolic state. Mice lacking OGT in SCs develop a progressive demyelinating peripheral neuropathy. Here, we investigated the nerve repair process in OGT-SCKO mutant mice and found that the remyelination of regenerating axons is severely impaired. Gene expression profiling of OGT-SCKO SCs revealed that the JUN-dependent SC injury program was elevated in the absence of injury and failed to shut down at the appropriate time after injury. This aberrant JUN activity results in abnormalities in repair SC function and redifferentiation and prevents the timely remyelination. This aberrant nerve injury response is normalized in OGT-SCKO mice with reduced Jun gene dosage in SCs. Mechanistically, OGT O-GlcNAcylates JUN at multiple sites, which then leads to an attenuation of AP-1 transcriptional activity. Together, these results highlight the metabolic oversight of the nerve injury response via the regulation of JUN activity by O-GlcNAcylation, a pathway that could be important in the neuropathy associated with diabetes and aging.
Nerve regeneration is a metabolically demanding task and is particularly vulnerable to metabolic disturbances associated with diabetes and aging (1, 2). Schwann cells (SCs), the ensheathing/myelinating glia of the peripheral nervous system, play a key role in nerve repair (3). Upon nerve damage, SCs promptly dedifferentiate into “repair” SCs that remove degenerating axons and myelin debris as well as provide support for axonal regrowth. Subsequently, functional recovery of the injured nerves is achieved only after the repair SCs redifferentiate and ensheathe/myelinate the regenerating axons. JUN, the core component of the AP-1 transcription factor complex, is considered the master regulator of the SC injury response and plays a key role in the generation of the repair SC. Loss of Jun in SCs delays myelin clearance and subsequent nerve repair (4), whereas enforced expression of Jun in SCs inhibits myelination (5, 6). However, it is unclear how the repair function of JUN is regulated in SCs.
O-GlcNAcylation is an O-linked N-acetyl-glucosamine (O-GlcNAc) posttranslational modification (PTM) of nuclear and cytoplasmic proteins (7). This unique PTM is catalyzed by O-GlcNAc transferase (OGT) whose activity is regulated by cellular metabolic/nutrient status (i.e., intracellular glucose concentration). Conversely, O-GlcNAcase (OGA) reverses this process and removes the O-GlcNAc moiety from modified proteins. A key function of O-GlcNAcylation includes epigenetic cellular reprogramming that occurs by modifying transcription factors, histones, and DNA methylation enzymes (8–11). Abnormal O-GlcNAcylation is associated with impaired glucose metabolism, as occurs in diabetes and Alzheimer’s disease (12). In addition, recent studies highlighted important roles of OGT in stem cell renewal, reprogramming, and pluripotency (10, 11).
We previously showed that mice with SC-specific deletion of OGT (OGT-SCKO mice) developed a progressive demyelinating peripheral neuropathy, indicating that SC O-GlcNAcylation is required for myelin maintenance (13). In this study, we subjected the OGT-SCKO mice to sciatic nerve crush injury to investigate a potential role of SC OGT and O-GlcNAcylation in nerve repair processes. We found that SC loss of OGT severely impairs remyelination of regenerating axons but does not prevent the generation of the repair SC or axonal regrowth. Further, we show that OGT modifies JUN, a component of the AP-1 transcription complex, and that this O-GlcNAcylation suppresses JUN-dependent injury gene expression in SCs. In the absence of OGT, JUN is not O-GlcNAcylated, and this results in a prolonged aberrant activation of JUN that prevents repair SCs from redifferentiating into myelinating SCs. The increased constitutive JUN activity severely impedes remyelination of regenerating axons. In accord, the heterozygous deletion of Jun to reduce JUN levels in SCs dramatically restored postinjury remyelination in OGT-SCKO mice. These mechanistic and in vivo observations highlight OGT and O-GlcNAcylation as important regulators of SC nerve injury responses.
Results
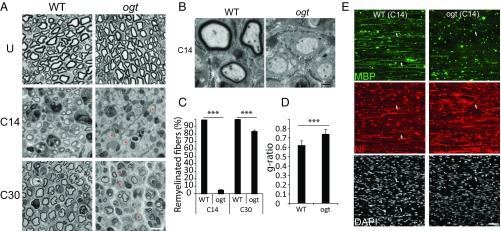
OGT Deficiency in SCs Causes Defective Nerve Remyelination.
OGT catalyzes protein O-GlcNAcylation and, thereby, couples cellular functions to metabolic activity via the hexosamine biosynthetic pathway (HBP) (7). We previously showed that SC-specific Ogt knockout mice (P0-Cre, OgtloxP), called OGT-SCKO mice, develop age-related tomaculous demyelinating neuropathy due to defective myelin maintenance. Nevertheless, OGT is dispensable for SC maturation and myelination during development. Myelination proceeded properly without any significant delay, and myelinated fibers are grossly normal at 6 wk of age in OGT-SCKO nerves (Fig. 1A) (13). We took advantage of these normally developing peripheral nerves in OGT-SCKO mice to investigate a potential role for OGT in regulating the SC injury response. Sciatic nerves in 6-wk-old OGT-SCKO mice were crushed at the sciatic notch with forceps and harvested at various times after injury. By light microscopic examination of semithin sections of injured nerves, we found that newly regenerating axons in wild-type mice were mostly remyelinated at 14 d after injury (C14). In OGT-SCKO nerves, less than 5% of fibers were myelinated (Fig. 1 A and B), indicating that the loss of OGT in SCs impairs postinjury remyelination. This remyelination defect in OGT-SCKO nerves persisted for weeks, with ∼20% of regenerating nerve fibers in mutant animals remaining unmyelinated up to 30 d after injury (Fig. 1 A and C). Furthermore, quantification of myelin thickness using g-ratio analysis indicated that myelinated axons in OGT-SCKO nerves at 30 d after injury (C30) were significantly hypomyelinated compared with those in control nerves (g ratio: 0.62 ± 0.05 in control vs. 0.74 ± 0.05 in OGT-SCKO, P < 0.001) (Fig. 1 A and D). Taken together, these morphometric analyses indicate that loss of OGT in SCs severely hampers peripheral nerve repair processes.
Fig. 1.
Schwann cell O-GlcNAcylation is required for peripheral nerve remyelination after injury. (A) Semithin sections of sciatic nerves: uninjured (U), 14 d (C14), or 30 d (C30) after nerve crush injury. Note unmyelinated/thinly myelinated fibers (asterisks) in OGT-SCKO nerves (ogt) at C14 and at C30. (B) Electron micrographs of injured sciatic nerves at 14 d after injury (C14). (C) Quantification of remyelination (% myelinated axons) in A. ***P < 0.001. n = 3 mice per genotype each injury time point. (D) Analysis of g ratios of remyelinated fibers at C30 in A. ***P < 0.001. n = 3 mice per genotype each injury time point. (E) Immunostaining of injured sciatic nerves at C14 using antibodies to detect myelin (MBP), axons (NF), and nuclei (DAPI). Arrows, myelinated axons. n = 3 mice per genotype. (Scale bars: A, 10 µm; B, 1 µm; E, 20 µm.)
To investigate ultrastructural aspects of the defective remyelination observed in OGT-SCKO mice, we performed electron microscopy on the distal segments of injured nerves. Electron micrographs of C14 nerves showed well myelinated regenerating axons in control nerves, whereas similar or larger size (>1 µm in diameter) axons in OGT-SCKO nerves remained unmyelinated despite their association with SCs (Fig. 1B). Nevertheless, this electron microscopic examination of injured nerves did not reveal any structural abnormality in the regenerating axons or their associated SCs in the OGT-SCKO nerves.
To further evaluate postinjury remyelination, we performed an immunohistochemical analysis on the injured sciatic nerves. At 14 d after injury, we observed numerous myelin basic protein (MBP) labeled axons in wild-type nerves, but these were rarely observed in OGT-SCKO nerves, highlighting the severe defect in postinjury remyelination caused by OGT deficiency (Fig. 1E). The staining of regenerating axons with neurofilament (NF) and the number of DAPI-labeled SC nuclei were comparable between wild-type and OGT-SCKO nerves, suggesting the remyelination defect is not secondary to defects in axon regeneration or postinjury SC proliferation. Overall, these results demonstrate that the loss of OGT in SCs interferes with peripheral nerve repair by inhibiting the remyelination of regenerating fibers.
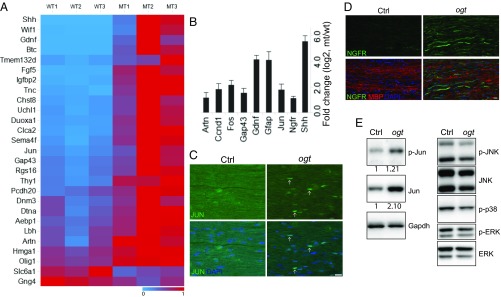
JUN Activity Is Increased in OGT-SCKO Nerves.
Since OGT and O-GlcNAcylation play essential roles in transcriptional regulation of many cellular pathways, we hypothesized that the loss of OGT causes abnormalities in the expression of SC genes required for proper nerve remyelination. To uncover abnormally regulated genes in these mutant SCs, we performed gene expression profiling experiments using sciatic nerves from 1-mo-old OGT-SCKO and wild-type mice. We found 1,099 differentially expressed genes (DEGs, ± twofold change, P < 0.05), with 212 down-regulated genes and 887 up-regulated genes in mutant vs. wild-type nerves (Datasets S1 and S2). KEGG pathway analysis of these DEGs indicated that loss of OGT caused decreased expression of genes in metabolic pathways (SI Appendix, Table S1), consistent with the metabolic sensor role of OGT. The up-regulated DEGs were enriched for genes involved in MAPK signaling and inflammatory/immune pathways. Among the most highly up-regulated genes in OGT-SCKO nerves were repair SC markers including Shh, Gdnf, and Gfap. The up-regulation of JUN, a key transcriptional regulator of the repair SC, prompted us to compare the OGT-SCKO DEG dataset with a set of previously identified JUN-dependent SC injury genes (4). We found that many genes whose expression is positively regulated by JUN in SCs were also up-regulated in OGT-SCKO nerves; for example, Artn, Ccnd1, Gdnf, Gfap, Jun, and Shh (Fig. 2A). Conversely, we found that genes normally repressed by JUN, such as Sox13, Apba2, Nes, Slc6a1, and Gng4, were down-regulated in OGT-SCKO nerves, suggesting OGT deficiency in SCs leads to a hyperactive JUN environment. To confirm these gene expression profiling results, we also performed quantitative RT-PCR (qRT-PCR) on a set of JUN target genes, Artn, Ccnd1, Fos, Gdnf, Gfap, and Shh. Consistent with the microarray results, RT-PCR also showed a significant up-regulation of these JUN target genes in OGT-SCKO nerves (Fig. 2B). It is particularly notable that genes such as Gdnf, Gfap, and Shh that are up-regulated in SCs after nerve injury in a JUN-dependent manner are also highly expressed (more than 10-fold increase) in mutant nerves, suggesting a pseudoinjury state in “uninjured” OGT-SCKO nerves. These gene expression analyses provide strong evidence that OGT is essential for proper regulation of JUN activity and hence JUN target gene expression in SCs.
Fig. 2.
Abnormal induction of JUN-regulated “injury response” genes in uninjured OGT-SCKO nerves. (A) Heatmap representation of gene expression analysis of JUN regulated injury response genes as defined by Arthur-Farraj et al. (4). OGT-SCKO nerves (MT1, MT2, MT3) vs. control nerves (WT1, WT2, WT3). Heatmap scale: 1 (high, red), 0 (low, blue). (B) qRT-PCR analysis of JUN-target genes in uninjured control vs. OGT-SCKO nerves from 1-mo-old mice. Data are represented as fold change ± SEM [log2, OGT-SCKO (mt)/control (wt)]; n = 3. (C and D) Immunostaining of uninjured sciatic nerves from 1-mo-old control (Ctrl) and OGT-SCKO (ogt) mice. Increased expression of JUN (arrows) (C) and NGFR (D) in OGT-SCKO (ogt) nerves vs. control (Ctrl) nerves. (Scale bars: 20 µm.) (E) Western blot analyses of uninjured sciatic nerves from 1-mo-old control (Ctrl) and OGT-SCKO mice (ogt). Note JUN expression was increased ∼twofold (normalized by Gapdh) in OGT-SCKO compared with control nerves.
The abnormal transcription of JUN target genes in OGT-SCKO nerves further prompted us to examine the levels of a subset of these gene products. Using immunohistochemical analysis, we were able to detect JUN expression in 1-mo-old uninjured sciatic nerves in OGT-SCKO SCs; however, JUN expression was never observed in SCs of wild-type nerves (Fig. 2C). We also examined NGFR (p75) expression as it is induced after nerve injury in repair SCs in a JUN-dependent manner. We found that NGFR expression was dramatically increased in OGT-SCKO nerves in the absence of injury, suggesting a pseudoinjury state in uninjured OGT-SCKO nerves (Fig. 2D). Western blot analysis showed that JUN levels are increased in OGT-SCKO nerves, consistent with the autoregulation of Jun expression (Fig. 2E). It is notable that increases in expression of JUN and JUN-regulated genes in OGT-SCKO nerves were not accompanied by increased activity (i.e., phosphorylation) of JUN MAPKs (JNK, p38, ERK) (Fig. 2E), suggesting an alternative mechanism for up-regulation of JUN activity in OGT-deficient cells. In summary, these results demonstrate that OGT regulates JUN activity as evidenced by changes in JUN target gene expression in SCs lacking this enzyme, which appears to result in a pseudoinjury state in OGT-KO SCs.
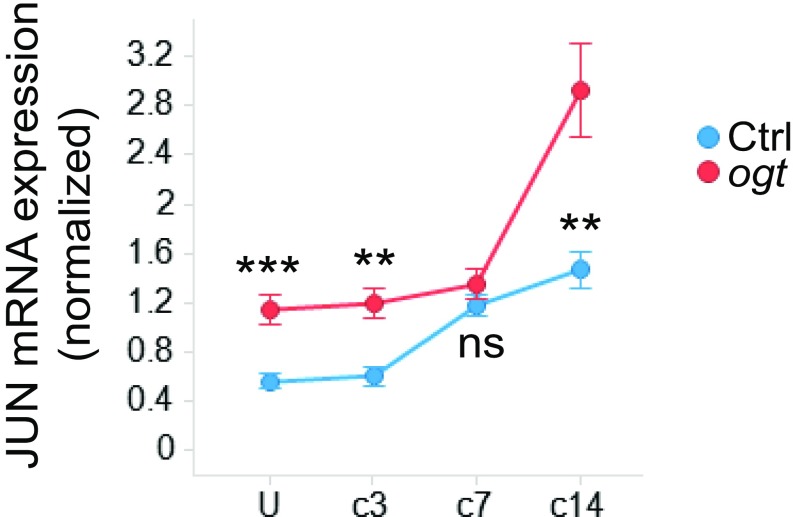
OGT Deficiency Leads to Persistently High Jun Expression After Nerve Injury.
JUN is a crucial regulator of SC gene expression after nerve injury. To investigate whether JUN expression is affected by the loss of OGT during nerve repair, we examined Jun transcripts by qRT-PCR in sciatic nerves after injury (uninjured, and 3, 7, and 14 d after injury). For comparison, we also examined other transcription factors important for SC differentiation and dedifferentiation (e.g., Egr1, Egr2, Sox2, Sox10). We found that, in control nerves, Jun transcription was gradually induced after nerve injury and reached maximal levels 7 d after injury. In contrast, Jun mRNA levels in OGT-SCKO nerves were consitutively elevated before injury and during the entire nerve repair process. Indeed, Jun levels continued to increase 7–14 d after injury in the mutant animals (Fig. 3 and SI Appendix, Fig. S1), indicating that loss of OGT caused an abnormally persistent induction of Jun in SCs. Interestingly, other SC transcription factors examined in this analysis did not show significant changes in expression between control and OGT-SCKO mice during nerve repair, highlighting the impact of O-GlcNAcylation on Jun expression.
Fig. 3.
Persistent induction of JUN in OGT-SCKO nerves after injury. qRT-PCR analysis of control (Ctrl) vs. OGT-SCKO (ogt) sciatic nerves at indicated times after nerve injury. U (uninjured), C3 (3 d after injury), C7 (7 d after injury), C14 (14 d after injury). mRNA expression was normalized to Gapdh. **P < 0.01; ***P < 0.001; n.s., not significant. n = 3∼5 nerves per genotype per injury time point.
JUN Is O-GlcNAcylated at Multiple Sites.
The increased expression of JUN/AP-1 targets in OGT-SCKO mice suggested that JUN itself may be regulated by O-GlcNAcylation in SCs. To test whether JUN is O-GlcNAcylated in vivo, we enriched O-GlcNAcylated proteins from rat sciatic nerves using immunoprecipitation with O-GlcNAc antibody (RL2). Western blot analysis showed JUN is present among the O-GlcNAcylated proteins in distal nerve segments at 3 and 7 d after injury (SI Appendix, Fig. S2). In addition, there is a modest increase both in total O-GlcNAcylation and JUN O-GlcNAcylation at 7 vs. 3 d after injury.
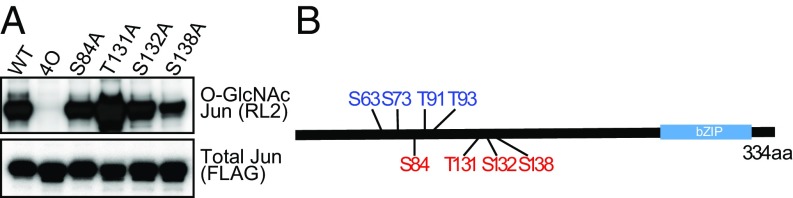
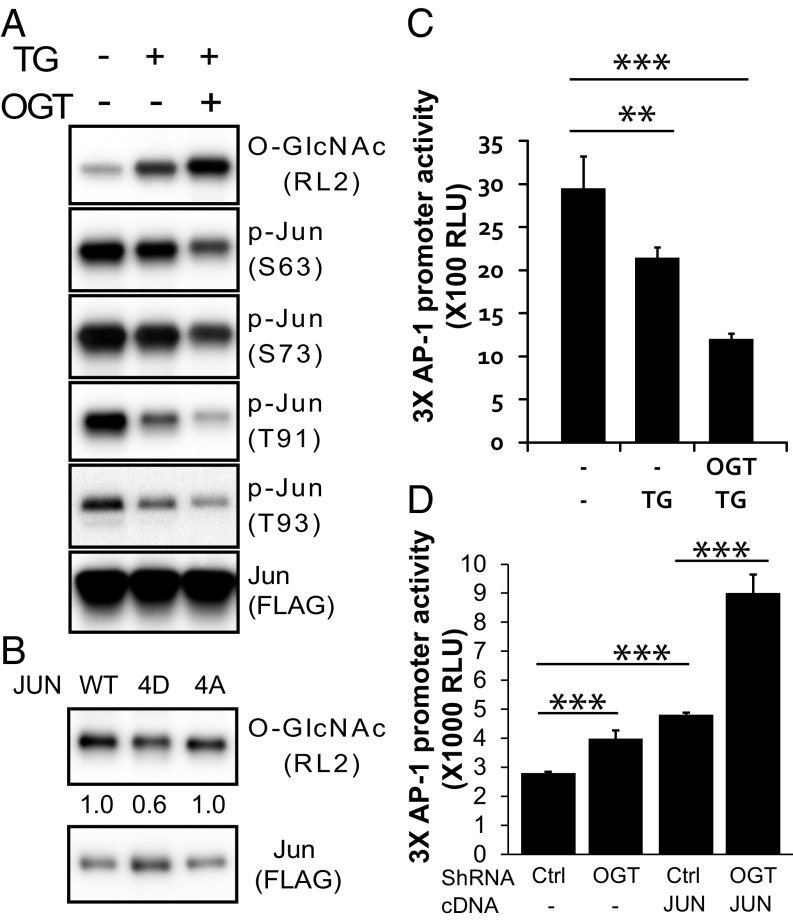
To further characterize JUN O-GlcNAcylation, we turned to a cell culture system where this process could be more easily manipulated. First, we showed that O-GlcNAcylation of ectopically expressed FLAG-tagged JUN in HEK293T cells was readily detectable by Western blot using anti-O-GlcNAc antibody (RL2), and dynamically increased by treatment with Thiamet-G (a potent OGA inhibitor) and OGT overexpression (Figs. 4A and 5A). To determine the sites of O-GlcNAc modification, we performed affinity purified mass spectrometry on JUN ectopically expressed in HEK293T cells that overexpress OGT and were treated with Thiamet-G. The FLAG-tagged JUN protein was enriched by immunoprecipitation, and in-gel proteins were subjected to mass spectrometry/electron transfer dissociation analysis. In the initial analysis we found that trypsin digestion did not provide sufficient peptide coverage to monitor a large portion of JUN. To provide increased peptide coverage of JUN, we introduced an Arg116Lys (R116K) mutation to provide an additional site of protease cleavage. O-GlcNAcylation of this mutant was comparable to that of wild-type JUN (SI Appendix, Fig. S3). FLAG-JUN (R116K) peptides from in-gel digestion using cyanogen bromide (CNBr) and Lys-N protease were analyzed. This led to identification of four O-GlcNAc modified sites: S83/S84 (ambiguous assignment), T131, S132, and S138 (SI Appendix, Fig. S4).
Fig. 4.
JUN is O-GlcNAcylated at Ser84, Thr131, Ser132, Ser138. (A) Western blot analysis of JUN O-GlcNAcylation. Flag-tagged versions of JUN wild-type (WT) and Ala substitution mutants (S84A, T131A, S132A, S138A, and 4O; all four residues converted to Ala) were expressed in HEK 293T cells expressing exogenous OGT and treated with Thiamet-G. Immunoblot analysis of immunoprecipitated FLAG-JUN for O-GlcNAcylation (anti-O-GlcNAc RL2) and total JUN (anti-FLAG). (B) Schematic representation of JUN O-GlcNAcylation. Phosphorylation sites (blue), four O-GlcNAcylation sites (red), and basic leucine zipper domain (bZIP) were depicted.
Fig. 5.
O-GlcNAcylation attenuates JUN phosphorylation and JUN/AP-1 transcription. (A and B) An antagonistic relationship in JUN between O-GlcNAcylation and phosphorylation. (A) Western blot analysis of immunoprecipitated FLAG-tagged wild-type JUN in HEK293 cells treated with 1 µM Thiamet-G (TG) and/or overexpressing OGT as indicated. (B) Western blot analysis of immunoprecipitated FLAG-JUN wild-type (WT), dephosphorylation mimetic (4A: S63A, S73A, T91A, T93A), or phosphorylation mimetic (4D: S63D, S73D, T91D, T93D) in HEK293T cells expressing OGT and treated with Thiamet-G. Numeric values denote quantification of O-GlcNAc RL2 normalized by total FLAG-JUN. (C) O-GlcNAcylation negatively affects AP1 transcriptional activity. Luciferase reporter assay using construct containing 3× AP-1 binding sites in HEK 293T cells expressing OGT and/or treated with TG as indicated. RLU, relative luciferase unit. **P < 0.01, triplicate; ***P < 0.001, triplicate. (D) Ogt knockdown enhances AP-1 transcription and JUN transactivation in HEK293T cells. Scrambled shRNA (sh-Ctrl), Ogt shRNA (sh-OGT) with or without JUN overexpression (JUN). ***P < 0.001.
To confirm that these four candidate sites were indeed O-GlcNAc modified, we generated a series of FLAG-JUN variants with alanine substitutions at these sites. We expressed these mutants in OGT-overexpressing HEK293T cells treated with Thiamet-G and examined them for O-GlcNAcylation by Western blotting using anti-O-GlcNAc antibody (RL2). We found that a JUN mutant (JUN 4O) with alanine substitutions at S84, T131, S132, and S138 abolished JUN O-GlcNAcylation (Fig. 4A), indicating that we have identified the predominant O-GlcNAcylated sites. Similar analyses showed that S84, but not S83, was the O-GlcNAcylated site that had been ambiguously assigned to both S83 and S84 using MS analysis (SI Appendix, Fig. S5). However, JUN mutants with individual alanine substitutions at any of these four sites (S84A, T131A, S132A, S138A) were still significantly O-GlcNAcylated, indicating multiple sites of O-GlcNAcylation in JUN (Fig. 4B). Taken together, we conclude that JUN is O-GlcNAcylated at S84, T131, S132, and S138.
O-GlcNAcylation Attenuates JUN Activities.
Proteins modified by O-GlcNAcylation often show altered patterns of phosphorylation (7). JUN phosphorylation is important for its transcriptional activity and all four JUN O-GlcNAcylation sites are proximal to these critical N-terminal phosphorylation sites (Fig. 4B), so we therefore examined this potential interrelationship in JUN. We found that increased JUN O-GlcNAcylation by OGT overexpression and Thiamet-G treatment was correlated with decreased phosphorylation at the N-terminal phosphorylation sites S63, S73, T91, and T93 (Fig. 5A). This indicates that O-GlcNAcylation exerts a negative effect on JUN N-terminal phosphorylation. We also examined whether JUN O-GlcNAcylation is affected by N-terminal phosphorylation. We found that the O-GlcNAcylation of a JUN phosphomimetic mutant in which all four phosphorylation sites S63, S73, T91, and T93 were changed to Asp (JUN 4D) was lower than that of wild-type JUN (Fig. 5B). Conversely, when these residues are mutated to Ala (JUN 4A), there was no change in O-GlcNAcylation. Taken together, these results indicate an antagonistic interaction between O-GlcNAcylation and phosphorylation in JUN.
JUN is a core component of the AP-1 transcription complex that regulates gene expression in response to extracellular stimuli. To explore whether O-GlcNAcylation affects JUN/AP-1 transcriptional activity, we performed luciferase reporter assays in HEK293T cells using a reporter containing three canonical AP-1 binding sites (TGACTCA). We found that increasing O-GlcNAcylation either by Thiamet-G treatment alone or together with OGT overexpression significantly reduced AP-1 activity by ∼28% or ∼63%, respectively (Fig. 5C). Conversely, decreasing O-GlcNAcylation by shRNA-mediated Ogt knockdown caused a significant increase (∼34%) in AP1 transcriptional activity (Fig. 5D, sh-Ctrl vs. sh-OGT). We also performed experiments using JUN overexpression to boost AP-1 reporter activity (Fig. 5D, sh-Ctrl vs. sh-Ctrl/JUN). In this scenario, decreasing O-GlcNAcylation by Ogt knockdown further enhanced JUN-mediated luciferase reporter activity by an additional 80% (Fig. 5D, sh-Ctrl/JUN vs. sh-OGT/JUN), suggesting a direct effect of O-GlcNAcylation on JUN-dependent regulation of AP-1 transcription (Fig. 5D). Taken together, these results demonstrated that O-GlcNAcylation functions to negatively modulate JUN activity including JUN phosphorylation and JUN/AP-1–mediated transcription.
Decreased Jun Gene Dosage Restores Remyelination of Damaged Axons in OGT-SCKO Nerves.
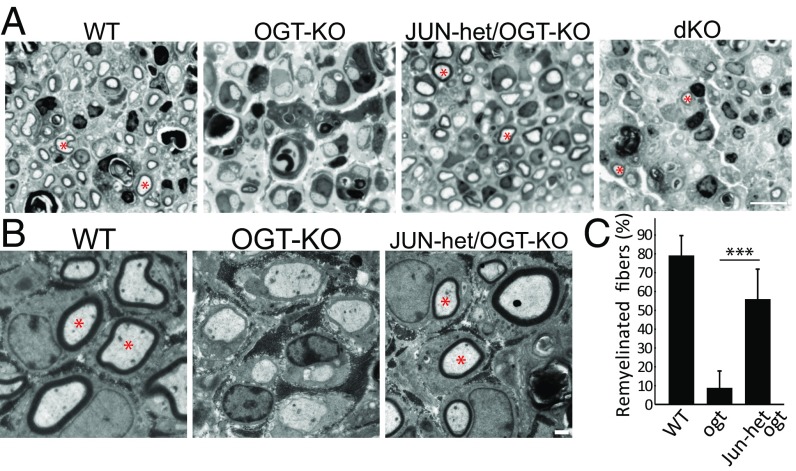
The enforced expression of JUN in SCs in both in vitro myelinating cocultures and in mouse models interferes with myelin development (5, 6). The high Jun expression in OGT-deficient SCs that persists even 14 d after injury (Fig. 3) strongly suggests that abnormally elevated JUN activity is responsible for the defective remyelination observed after nerve injury in OGT-SCKO mice. If this is the case, decreasing Jun expression might be expected to reestablish normal remyelination in these mutant mice. To test this hypothesis, we mated OGT-SCKO mice to mice harboring a floxed Jun allele to produce OGT-SCKO mice lacking one {called JUN-het/OGT-KO (P0-Cre−/+, Ogt loxP/loxP, Jun loxP/+) or both Jun alleles [dKO (P0-Cre−/+, Ogt loxP/loxP, Jun loxP/loxP)]}. We performed nerve crush injury on these mice and examined semithin sections of the sciatic nerves 14 d later to assess nerve remyelination. Notably, we found that nerves from JUN-het/OGT-KO mice showed many remyelinated axons. Remyelinated axons were rarely observed in nerves of OGT-SCKO mice (Fig. 6A). This remarkable recovery of postinjury remyelination in JUN-het/OGT-SCKO mice was confirmed using electron microscopy in which we observed comparable amounts of compact myelin surrounding regenerating axons in JUN-het/OGT-SCKO and control animals, whereas OGT-SCKO nerves showed minimal myelin at this stage of repair (Fig. 6B). Quantification of the myelinated fibers observed in these postinjury sciatic nerve electromicrographs showed that remyelination of JUN-het/OGT-SCKO nerves approached that observed in wild-type nerves and was vastly superior to that of OGT-SCKO mice (Fig. 6C). In keeping with the loss of one JUN allele, qRT-PCR analysis of injured nerve showed a normalization of expression of JUN and a number of JUN target genes in JUN-het/OGT-SCKO nerve compared with OGT-SCKO nerve (SI Appendix, Fig. S6). These results provide strong evidence that the abnormal increase in JUN activity observed in OGT-SCKO mice is a major cause of the aberrant nerve repair in these animals. However, the complete loss of JUN (dKO mice) caused an abnormal repair phenotype with generally atrophic axons and few remyelinated fibers, consistent with the reported roles of SC JUN in axonal regrowth and motorneuron survival (4, 14). In conclusion, these results demonstrate that constitutively high JUN activity in SCs results in abnormal nerve repair and indicates that metabolism (via OGT) plays an important role in regulating JUN activity and its role in promoting nerve injury responses.
Fig. 6.
Decreased Jun gene dosage restores remyelination of regenerating axons in OGT-SCKO nerves. (A) Semithin sections of injured sciatic nerves at 14 d after injury. Note that while myelinated regenerating axons (asterisks) are absent from OGT-SCKO, they are present in nerves from JUN-het/OGT-KO, WT, and dKO mice. (B) Electron micrographs of injured sciatic nerves at 14 d after injury. Myelination of regenerated axons is apparent (asterisks) in wild-type and JUN-het/OGT-SCKO, but not in OGT-SCKO nerves. (C) Quantification of remyelinated axons in B. ***P < 0.001, n = 3 mice per genotype. (Scale bars: A, 10 µm; B, 1 µm.)
Discussion
In this study, we demonstrated that OGT promotes peripheral nerve remyelination after nerve injury. We found that loss of OGT causes abnormally persistent activation of JUN/AP-1 transcriptional activity in OGT-SCKO nerves. Importantly, we showed that OGT catalyzes JUN O-GlcNAcylation and that this modification attenuates JUN/AP-1 transcriptional activity. Using mice lacking one Jun allele, we showed that decreased Jun gene dosage dramatically restored postinjury remyelination in OGT-SCKO mice, establishing the causality between abnormal JUN activity and impaired nerve remyelination in OGT-SCKO.
We identified a robust O-GlcNAcylation of JUN at Ser84, Thr131, Ser132, and Ser138. None of the four sites have been reported to be phosphorylated, suggesting that a direct competition between O-GlcNAcylation and phosphorylation is unlikely in this protein. Notably, phosphorylation at Thr91 and Thr93, which are proximal to all four O-GlcNAcylation sites, was more significantly affected than that at Ser63 and Ser-73. These findings suggest that an additive steric hindrance of the bulky O-GlcNAc moiety may underlie the antagonizing effect of O-GlcNAcylation on phosphorylation. Contrary to the C-terminal basic leucine zipper (bZIP) domain for DNA binding and AP-1 dimerization, the JUN N-terminal structure is not well defined and often referred as “disordered” region, a target of multiple posttranslational modifications and mediator of protein–protein interactions (15). Thus, in demonstrating multiple O-GlcNAcylation events in this intrinsically disordered N-terminal interface that influences activity, we propose that OGT is a crucial modulator of JUN function.
Our analyses of the SC injury responses in OGT-SCKO mice designate OGT as a negative regulator of JUN that is critical to the timely progression into the later phases of nerve repair such as remyelination. Importantly, our data demonstrate the causality between persistent elevated JUN activity in SCs lacking OGT and defective remyelination. Collectively, our results are in line with a recent study in which SC-specific overexpression of JUN caused delay of postinjury remyelination in a mouse model (5). Along with this defect in remyelination, they also showed that myelination during development was unaffected, despite a sixfold increase in JUN expression in SCs (5). OGT-SCKO mice show a similar dichotomy, with normal developmental myelination yet defective remyelination after nerve injury (13). These studies add to the evidence that myelination during development and during nerve repair after injury are regulated by fundamentally distinct mechanisms.
A growing number of studies have highlighted the role of the metabolic sensor OGT in coordinating stem cell renewal and reprogramming with cellular metabolic status (10, 11). Our studies of OGT in SCs are consistent with these reports and offer ideas with regard to the metabolic regulation of SC injury responses via epigenetic O-GlcNAc posttranslation modifications. This is particularly significant since aberrant SC injury responses potentially contribute to the pathogenesis of diabetic neuropathy (1), the most prevalent form of peripheral neuropathy. We propose that abnormal O-GlcNAcylation, triggered by the metabolic anomalies associated with diabetes (12, 16), causes impaired SC injury responses that contribute to the axonal damage associated with this disorder.
Materials and Methods
Animal Studies.
All animal experiments were carried out in compliance with institutional animal protocols (Washington University in St. Louis, no. 20170030). Ogt loxP/loxP (17), Jun loxP/loxP (18), and P0-Cre (19) were crossed to generate the following genotypes of mice: OGT-SCKO (P0-Cre−/+, Ogt loxP/loxP), JUN-het/OGT-KO (P0-Cre−/+, Ogt loxP/loxP, Jun loxP/+), dKO (P0-Cre−/+, Ogt loxP/loxP, Jun loxP/loxP), and Ctrl littermates (P0-Cre−/−, Ogt loxP/loxP). Both female and male mice were used in these experiments. Mating and genotyping were carried out as previously described (13).
Nerve Injury.
The sciatic nerve was crush injured at the sciatic notch.
Nerve Histology, Morphometry, Immunostaining, and Fluorescent Microscopy.
See SI Appendix, Supplemental Materials and Methods for details.
qRT-PCR.
mRNA qRT-PCR was performed using a SYBR green-based detection system on a 7900 HT Sequence Detector instrument (Applied Biosystems) as described previously (20). See details and list of primers in SI Appendix, Supplemental Materials and Methods.
Microarray and Computational Analysis.
See SI Appendix, Supplemental Materials and Methods for details. Briefly, 1,500 ng of each amplified RNA were hybridized onto Agilent Mouse 4 × 44 K mouse V2 Expression Beadchips (Agilent-026655). Differentially expressed genes with at least 2.0-fold differential regulation between OGT-SCKO and Ctrl nerves at a false discovery rate (FDR) of 0.5% were selected for further analysis. Gene enrichment analysis was performed using WebGestalt (21). The microarray data were deposited at NIH/GEO (GSE115333).
Cell Culture, Transfection, and Luciferase Reporter Assay.
Luciferase reporter assays were performed using Dual-Glo Luciferase Assay System (Promega) according to manufacturer’s protocols. See SI Appendix, Supplemental Materials and Methods for a list of plasmids used in the study.
Statistics.
Data are represented as mean ± SEM unless otherwise specified. Statistical tests were performed with Spotfire (TIBCO) and Excel 2010 (Microsoft). Groups of means were compared using one-way ANOVA, and comparisons between two means were performed using Student’s t test. Significance was as *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Supplementary Material
Acknowledgments
We thank S. P. Jones, R. Libby, L. Wrabetz, and A. Messing for mutant mice and Genome Technology Access Center for microarray analysis. Mass spectrometry was provided by Bio-Organic Biomedical Mass Spectrometry Resource at the University of California, San Francisco (A.L.B., Director) supported by Biomedical Technology Resource Center program National Institute of General Medical Sciences 8P41GM103481, Dr. Miriam and Sheldson Adelson Medical Research Foundation, and HHMI. This work was also supported by NIH Grants T32GM108539 (to S.K.), P50 AG05681 (PILOT 34.2, to S.K.), NS087306 (to J.M.), AG13730 (to J.M.), R56 NS099314 (to J.M.), and R01NS105645 (to J.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in NCBI Gene Expression Omnibus (GEO) (accession no. GSE115333).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805538115/-/DCSupplemental.
References
- 1.Kennedy JM, Zochodne DW. Impaired peripheral nerve regeneration in diabetes mellitus. J Peripher Nerv Syst. 2005;10:144–157. doi: 10.1111/j.1085-9489.2005.0010205.x. [DOI] [PubMed] [Google Scholar]
- 2.Painter MW, et al. Diminished Schwann cell repair responses underlie age-associated impaired axonal regeneration. Neuron. 2014;83:331–343. doi: 10.1016/j.neuron.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016;594:3521–3531. doi: 10.1113/JP270874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur-Farraj PJ, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75:633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fazal SV, et al. Graded elevation of c-Jun in Schwann cells in vivo: Gene dosage determines effects on development, re-myelination, tumorigenesis and hypomyelination. J Neurosci. 2017;37:12297–12313. doi: 10.1523/JNEUROSCI.0986-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson DB, et al. c-Jun is a negative regulator of myelination. J Cell Biol. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 10.Jang H, et al. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer’s disease. Mol Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, et al. Schwann cell O-GlcNAc glycosylation is required for myelin maintenance and axon integrity. J Neurosci. 2016;36:9633–9646. doi: 10.1523/JNEUROSCI.1235-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontana X, et al. c-Jun in Schwann cells promotes axonal regeneration and motoneuron survival via paracrine signaling. J Cell Biol. 2012;198:127–141. doi: 10.1083/jcb.201205025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csizmok V, et al. Multivalent interactions with Fbw7 and Pin1 facilitate recognition of c-Jun by the SCFFbw7 ubiquitin ligase. Structure. 2018;26:28–39.e2. doi: 10.1016/j.str.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee PS, Ma J, Hart GW. Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc Natl Acad Sci USA. 2015;112:6050–6055. doi: 10.1073/pnas.1424017112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrens A, et al. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. EMBO J. 2002;21:1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feltri ML, et al. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci. 1999;883:116–123. [PubMed] [Google Scholar]
- 20.Viader A, et al. Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron. 2013;77:886–898. doi: 10.1016/j.neuron.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT anaLysis toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.