Significance
Proteolysis targeting chimera (PROTAC)-based protein degradation is an emerging field that holds significant promise for targeting the “undruggable” proteome: the vast majority of the proteins that do not exhibit enzymatic activity and are thereby not amenable to classical inhibition. Despite significant progress, a thorough mechanistic characterization of biochemical determinants that underpin efficient PROTAC activity is lacking. Here we address one such question: Is positive cooperativity necessary for potent protein degradation? Through a collection of independent techniques, we show that within a Bruton’s tyrosine kinase/cereblon PROTAC system, potent knockdown correlates with alleviation of steric clashes in the absence of thermodynamic cooperativity. This result broadens the scope of PROTAC applications and affects fundamental design criteria across the field.
Keywords: targeted protein degradation, PROTAC, BTK, catalytic, proteasome
Abstract
Proteolysis targeting chimeras (PROTACs) are heterobifunctional small molecules that simultaneously bind to a target protein and an E3 ligase, thereby leading to ubiquitination and subsequent degradation of the target. They present an exciting opportunity to modulate proteins in a manner independent of enzymatic or signaling activity. As such, they have recently emerged as an attractive mechanism to explore previously “undruggable” targets. Despite this interest, fundamental questions remain regarding the parameters most critical for achieving potency and selectivity. Here we employ a series of biochemical and cellular techniques to investigate requirements for efficient knockdown of Bruton’s tyrosine kinase (BTK), a nonreceptor tyrosine kinase essential for B cell maturation. Members of an 11-compound PROTAC library were investigated for their ability to form binary and ternary complexes with BTK and cereblon (CRBN, an E3 ligase component). Results were extended to measure effects on BTK–CRBN cooperative interactions as well as in vitro and in vivo BTK degradation. Our data show that alleviation of steric clashes between BTK and CRBN by modulating PROTAC linker length within this chemical series allows potent BTK degradation in the absence of thermodynamic cooperativity.
Proteolysis targeting chimeras (PROTACs) (1) are bivalent ligands that recruit an E3 ligase to a target protein thus facilitating polyubiquitination and subsequent target degradation (2–4). A powerful technology, first introduced in 2001 (1), PROTACs remained largely dormant for close to a decade due to physicochemical property limitations of chemical matter and poor cellular penetration. Notable breakthroughs, particularly the departure from poorly cell-penetrant peptides to small-molecule–based ligands, have led to enhanced drug uptake and efficacy both in cultured cells and, most recently, in vivo (5, 6). As such, numerous proteins ranging from gene regulatory proteins [AR (3, 7), ERα (7, 8), ERRα (6), and BRD2/3/4 (5, 9–11)], retinoic acid-binding proteins [CRABPI/II (12, 13) and RAR (7)], to enzymes [prolyl isomerase FKBP12 (5), kinases BCR-ABL/c-ABL (14), and RIPK2 (6)] have successfully been degraded by small-molecule PROTACs. This breadth has led to tremendous excitement in the drug development field, particularly with regard to targeting the “undruggable” proteome (15). There are thousands of predicted protein–protein interactions (PPIs) (16) where long, shallow interfaces are, in theory, sufficient to engage a moderate-affinity PROTAC and potentially mediate the target protein’s degradation. The task of choosing the optimal target–ligase pair that would lead to efficacious target degradation seems daunting given the sheer number of possible targets and ligases (600+) (17) available. Selection could be funneled for target–ligase pairs that colocalize in similar cellular compartments and for targets that have Lys residues accessible for ubiquitination. A second selection criterion en route to efficacious and selective target degradation could be de novo rational design of PROTACs. In a recent report, Gadd et al. (18) describe an elegant model of PROTAC-facilitated target–ligase interactions as the foundation for target selectivity of BRD4 over closely related family members BRD2 and BRD3, suggesting that stabilizing target–ligase PPIs may be a powerful strategy when designing efficient PROTACs.
Here we assess the requirement of cooperativity in PROTAC-mediated degradation of Bruton’s tyrosine kinase (BTK). BTK is a nonreceptor tyrosine kinase that belongs to the tyrosine kinase expressed in hepatocellular carcinoma (Tec) family of kinases and is crucial in normal B cell development (19). Mutations in Btk cause X-linked agammaglobulinemia (20), whereas aberrant regulation of BTK signaling has been linked to numerous B cell malignancies including chronic lymphocytic leukemia (CLL), mantle cell lymphoma, and diffuse large B cell lymphoma (20–22). Ibrutinib is a selective and potent irreversible (covalent) inhibitor of BTK that has affected the treatment landscape for B cell malignancies, particularly CLL (21, 23). What confers ibrutinib its selectivity is the scarcity in other kinases of a Cys residue homologous to C481 in BTK, which forms the covalent adduct (24, 25). However, recent reports on patient relapses as a result of acquired resistance mutations in BTK (C481S) following Ibrutinib treatment have raised the need for new avenues for therapeutic intervention (26).
Targeted degradation of BTK is explored as a modality and serves as a model system to further understand the biochemical underpinnings of efficient, targeted protein degradation. A library of 11 PROTACs of varying linker lengths that engage BTK on one end and cereblon [CRBN, a substrate-binding member of the Cul4 E3 ligase family (27, 28)] on the other are initially used to assess BTK degradation efficacy. Through in vitro, cellular, and in vivo experiments, rapid, efficient, prolonged, and selective BTK degradation is demonstrated. Degradation of BTK is dependent on ternary complex formation but does not rely on energetically cooperative BTK–CRBN interactions. On the contrary, negative BTK–CRBN cooperativity is observed with PROTACs of shorter linkers, and relief of steric clashes is determined to describe the transition between ineffective and potent BTK degradation within this system. We thus describe a case where robust thermodynamic cooperativity appears unnecessary for efficient degradation.
Results
PROTACs Facilitate BTK Degradation in Cultured Cells.
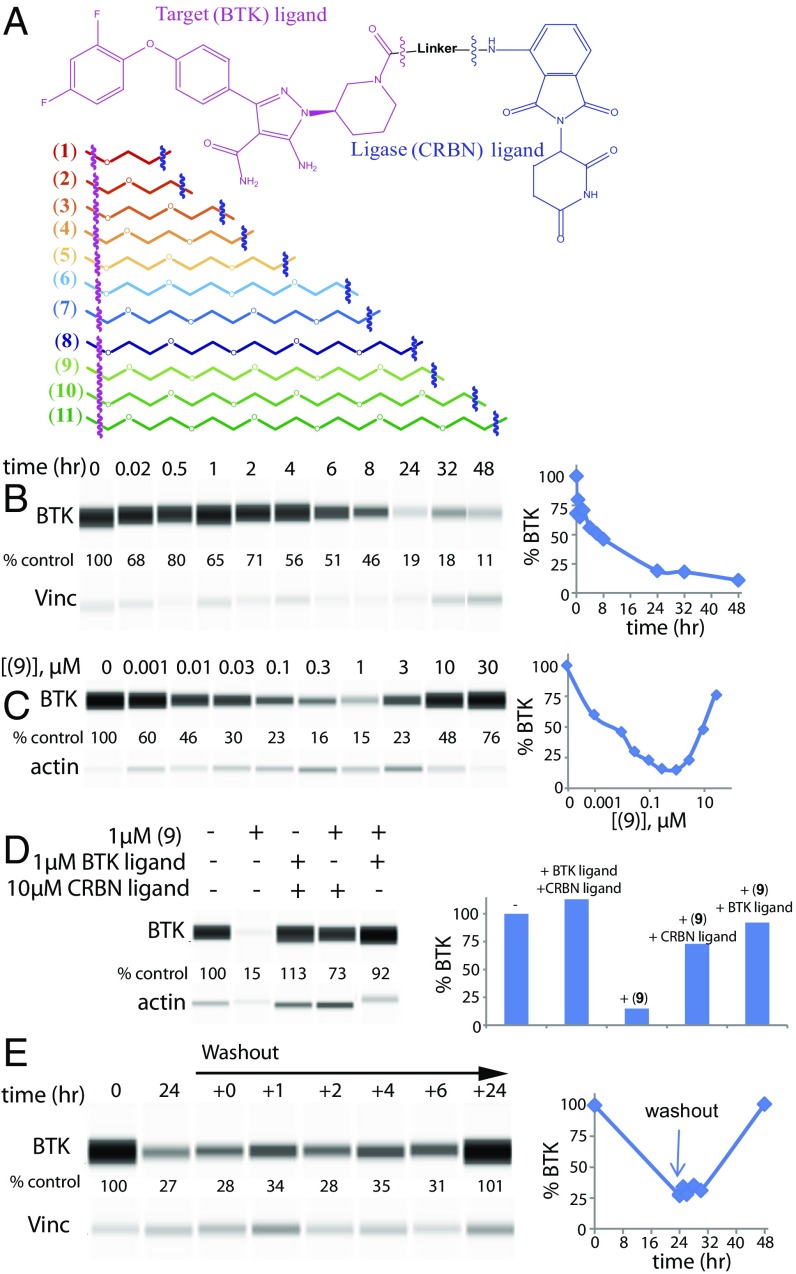
To achieve targeted protein degradation of BTK, an 11-compound library of heterobifunctional ligands was prepared in which a BTK binding scaffold was joined to a CRBN ligand by PEG-linkers of various lengths (Fig. 1A). The BTK binding core is a noncovalent analog derived from a previously disclosed covalent phenyl-pyrazole series (29) (Fig. 1A and SI Appendix, Fig. S1), and we use pomalidomide as the CRBN ligand (30). An initial study of PROTAC (9) (the primary analog of this set) in cultured Ramos cells shows time-dependent depletion of BTK levels as assessed by Western blot using Wes assay plates (Fig. 1B and SI Appendix). The exponential decrease in BTK over time begins as early as 1 h and plateaus after 24 h, at which point BTK levels are reduced to 16 ± 4% of the initial value. Furthermore, knockdown after 24 h is dependent on compound concentration (Fig. 1C), with BTK levels showing a U-shaped dose dependence, with maximal knockdown achieved at 1 µM concentration (DC50 = 5.9 ± 0.5 nM at 24 h; Fig. 1C and SI Appendix, Figs. S2A and S3). This U-shaped dose–response has been previously observed for PROTACs (6, 9, 11), and the reduced potency at higher drug concentrations is a consequence of competitive formation of binary complexes {BTK–PROTAC} and {PROTAC–CRBN} vs. the required ternary complex {BTK–PROTAC–CRBN}. In further support of this mechanism of action, efficacy is markedly reduced by competition with a monofunctional BTK binder, a monofunctional CRBN binder (pomalidomide), or both, suggesting that simultaneous engagement of both BTK and CRBN by the PROTAC is required for BTK degradation (Fig. 1D). It is also confirmed that the monofunctional BTK ligand alone does not significantly affect levels of BTK (SI Appendix, Fig. S3E). Sustained knockdown is also dependent on continued compound exposure because washout leads to recovery of BTK levels within 24 h (19, 31) (Fig. 1E).
Fig. 1.
PROTACs facilitate BTK degradation in cultured cells. (A) Schematic of BTK and CRBN parent molecules (Top) from which 11 PROTACs of varying linker lengths were generated (Bottom). (B) Western blot analysis of time-dependent degradation of BTK in the presence of 0.1 µM PROTAC (9) in cultured Ramos cells. Degradation is observed in as little as 1 h with steady-state BTK degradation reached at 24 h. (C) BTK degradation is responsive to PROTAC (9) concentrations. U-shaped dose–response curve is indicative of the involvement of a ternary complex formation in BTK degradation. Ramos cells were incubated with (9) for 24 h at the indicated concentrations. (D) PROTAC (9) requires simultaneous engagement of BTK and CRBN to effectively degrade BTK. Out-competition of (9) with parent molecules or lack of the linker between them does not affect BTK levels. Ramos cells were incubated with compound for 24 h at the indicated concentrations. (E) BTK degradation is dependent on (9) as compound washout restores BTK levels. After incubation of Ramos cells with 0.1 µM PROTAC (9), cells were washed 3× with PBS and resuspended in fresh media. BTK levels were monitored at the indicated time points. All lysates were analyzed by Western blotting using the ProteinSimple Wes system.
Longer Linkers Promote Ternary Complex Formation and Predict PROTAC Efficacy.
Previous work on the mechanism of PROTAC-mediated degradation suggests that PROTACs facilitate formation of a {target–PROTAC–ligase} ternary complex, enabling subsequent target polyubiquitination and proteasomal degradation (Fig. 2A). This requisite ternary complex formation of PROTACs is governed by a well-described equilibrium (32) that predicts a bell-shaped dependence of ternary complex concentration {BTK–PROTAC–CRBN} on the concentration of PROTAC (Fig. 2B). Therefore, it is hypothesized that maximizing PROTAC efficacy is associated with maximizing ternary complex formation.
Fig. 2.
Longer linkers are predictive of in-cell PROTAC efficacy and {BTK–PROTAC–CRBN} ternary complex formation. (A) Proposed model of simultaneous engagement of BTK and CRBN by PROTACs, which leads to BTK ubiquitination and eventual degradation. (B) Schematic of {BTK(target = A)–PROTAC(B)–CRBN(E3 ligase = C)} ternary complex equilibria with predominant species and the respective governing equilibria (KAB and KBC) highlighted. (C) Effect of linker length on efficacious BTK degradation. Ramos cells were incubated with compound at designated concentrations for 24 h, and lysates were analyzed by Western blot. (D) TR-FRET–based assay to evaluate effect of linker length on {BTK–PROTAC–CRBN} ternary complex formation. Two hundred nM biotinylated BTK and 500 nM biotinylated CRBN were incubated with varying PROTAC concentrations for 30 min before endpoint data collection at 620 nm (donor) and 665 nm (acceptor). Curves are shown relative to PROTAC (9), whose maximum is normalized to 1. (E) Three-body modeling of {BTK–PROTAC–CRBN} uses a steric score to identify PROTACs that form lower-energy ternary complex solutions thereby delineating inefficacious (1–4) and the most efficacious PROTACs (7–10) in line with TR-FRET data.
As a first step, we sought to understand the determinants of PROTAC efficacy in cultured cells by using the 11-compound PROTAC library of varied linker lengths (Fig. 2C and SI Appendix, Fig. S3). Whereas PROTACs of longer linker lengths (6–11) potently degrade BTK (DCmax = 1 µM and DC50 = 1–40 nM at 24 h), shorter PROTACs (1–4) are largely ineffective (Fig. 2C). Between these extremes lies an intermediate compound, PROTAC (5), which demonstrates modest knockdown. BTK knockdown appears to depend on choice of E3 because PROTACs of similar length are less effective when engaging the ligases von Hippel–Lindau protein [VHL, which serves as a substrate recognition domain in Cul2-based E3 ligases (33–35)] or inhibitor of apoptosis [IAP, which serves as an E3 ubiquitin ligase (36)], despite all species appearing to colocalize in shared cellular compartments (SI Appendix, Fig. S2 B–D)
To explore whether PROTAC linker length affected the {target–PROTAC–ligase} ternary complex formation and cellular efficacy, an in vitro time-resolved fluorescence resonance energy transfer (TR-FRET) assay was developed. Lumi4-Tb–labeling of BTK and XLA665-labeling of CRBN provide an appropriately matched pair of fluorophores to allow a FRET signal to be detected when BTK and CRBN are brought into proximity. Although unable to provide absolute quantitation, comparison of relative heights and inflection points suggests two distinct classes of PROTACs. These classes agree with results from cellular knockdown in the sense that longer PROTACs (6–11) that yield readily detectable ternary complex by FRET also demonstrate potent cellular knockdown, whereas shorter PROTACs (1–4) that yield only weak/no FRET signal appear ineffective in cells. PROTAC (5) again shows an intermediate behavior (Fig. 2D). Parallel efforts using a computational modeling approach to evaluate linear linkers using a steric scoring scheme also suggest that longer linkers formed more numbers of lower-energy ternary complexes (Fig. 2E and SI Appendix, Fig. S4 A–E). This method was also extended to another ligase, VHL: BTK and VHL were also predicted to engage in a sterically favorable ternary complex mediated by PROTACs (12, 13) (SI Appendix, Fig. S4F).
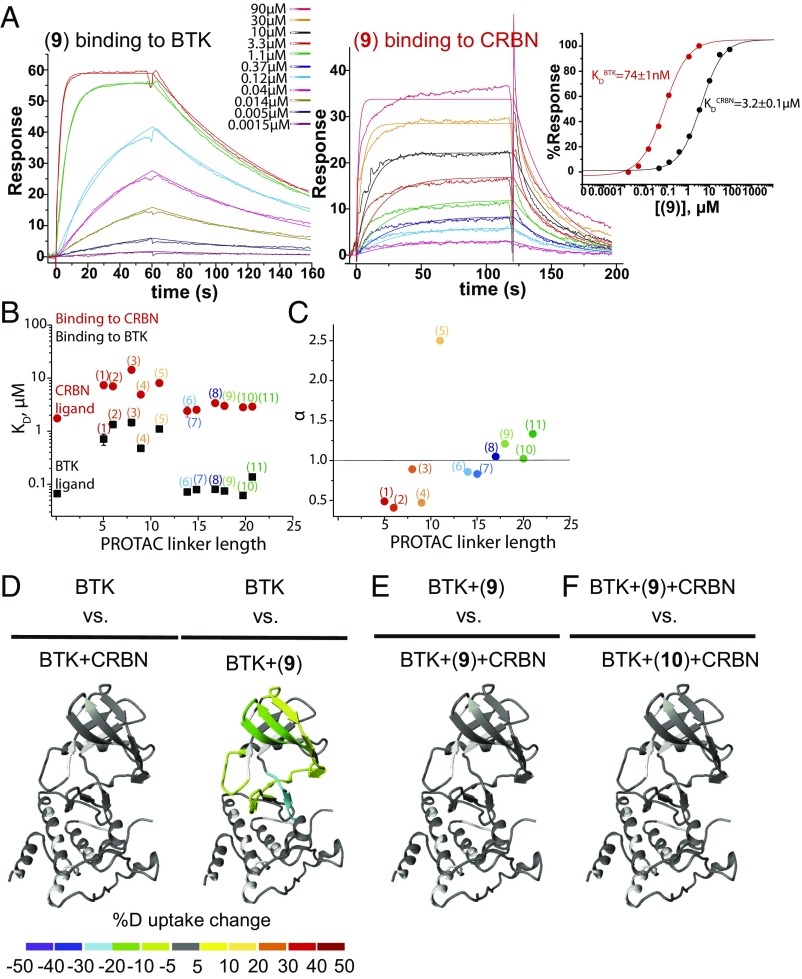
Ternary Complex Formation Lacks Positive Cooperativity.
Surface plasmon resonance (SPR) is used to generate binary binding affinities of PROTACs to BTK and CRBN (Fig. 3 A and B and SI Appendix, Fig. S5). Based on previous reports, affinities of pomalidomide/lenalidomide for CRBN span a range of <1 to >10 μM and highlight that care must be taken when comparing values measured under different conditions (37, 38). To minimize variation, we performed independent matched measurements for all compounds in the library. Using the monofunctional BTK and CRBN binders as benchmarks for unhindered BTK (KD = 70 nM) or CRBN (KD = 1.8 µM) binding, it is observed that similar binding affinities are maintained for the longest PROTACs (6–11) bound to BTK (KD = 70 nM–140 nM) and CRBN (KD = 2.5 µM–3.6 µM), whereas binding is impaired for the shortest PROTACs by up to ∼20× (Fig. 3B and SI Appendix, Fig. S5). This decrease in potency may be due to binary steric clashes between the shortest PROTAC linkers and BTK/CRBN. To further explore this hypothesis, an adaptation of a recently described three-body mathematical model is used to compare experimental with simulated ternary complex formation data for each PROTAC (32) (SI Appendix). For the shortest PROTACs (1–4), simulations based on binary KD values are unable to reproduce the relative height and shape of experimental curves. However, model fit is improved when solving for cooperativity factor α (Fig. 3C and SI Appendix, Fig. S6A). PROTACs (1–4) (α < 1) and PROTACs (6–11) (α ∼ 1) show negative and noncooperativity within a ternary complex. PROTAC (5) yields modest apparent positive cooperativity, but remains inferior to compounds with longer linkers.
Fig. 3.
Efficacious ternary complex formation is noncooperative. (A) Representative raw SPR sensorgrams of PROTAC (9) binding to either BTK (Left) or CRBN (Middle) with corresponding fits used to generate PROTAC (9) KD values (Right). (B) KD values of all PROTACs and parent molecules binding to either BTK or CRBN derived from equilibrium analysis of sensorgrams. For more details, please refer to SI Appendix, Fig. S5. (C) Negative cooperativity (α < 1) is needed to emulate TR-FRET curves based on SPR PROTACs (1–4) KD values. Intermediate PROTAC (5) shows apparent positive cooperativity, whereas efficacious PROTACs (6–11) do not show appreciable cooperativity. (D) HDX MS perturbation data mapped onto the 3D structure of BTK kinase domain. A color gradient is used to represent the average deuterium uptake differences across all six time points between any two different states of BTK. White indicates regions not covered by the core peptide set. Compared with BTK apo form, PROTAC (9) protects BTK from deuterium exchange in a region of the protein that maps to the compound binding site. Conversely, CRBN does not cause perturbation in BTK whether in the absence (D) or presence of efficacious PROTACs (E) (9) or (F) (10).
It is important to note that because FRET signal is dependent on both distance and conformation, potential differences in ternary complex ensembles or structure could affect the ability to compare signal across this library. In an effort to independently test cooperativity, we selected our most potent cellular PROTAC, compound (10), and performed SPR in which we measured affinity for CRBN either alone (1.3 ± 0.7 μM) or while in complex with BTK (2.2 ± 1.4 μM) (SI Appendix, Fig. S6B). Binding affinity is not enhanced in the presence of BTK, consistent with a low-/no-cooperativity system.
Solution-phase hydrogen/deuterium exchange coupled with mass spectrometry (HDX–MS) analysis is used as an independent assessment of possible long-lived or stable protein–protein interactions within the {BTK–PROTAC–CRBN} ternary complex. HDX–MS uses time-dependent, differential deuterium uptake of a protein in the absence or presence of binding partners, to report on protein–ligand or protein–protein interactions. HDX–MS is thus ideally suited to report on de novo (whether direct or allosteric) contacts, should potent and specific interactions occur. In an effort to ensure a significant population of the ternary complex, concentrations of BTK–PROTAC–CRBN of 10 µM–10 µM–15 µM are used (based on FRET results above, as well as the need for robust signal detection by HDX). The presence of ternary complex was independently confirmed under related concentrations using native state mass spectrometry. This experiment has the added benefit of permitting direct quantitation of ternary complex formed, yielding values consistent with a noncooperative system (SI Appendix, Fig. S6C).
HDX–MS efforts are focused on BTK as our reporter—with a core set of 21 BTK peptides selected from a robust 90% BTK sequence coverage dataset (SI Appendix, Fig. S7A)—to report on differential deuterium uptake of BTK when engaged in various interactions. Although CRBN is present in this experiment and yields a high degree of sequence coverage by mass spectrometry (94%; SI Appendix, Fig. S7B), we did not observe statistically significant protection by any ligands, likely owing to the reduced potency of the CRBN–ligand interaction as well as the small pomalidomide-binding pocket which provides only a single backbone H-bond donor. In the case of BTK, PROTAC (9) leads to as much as 23% protection of deuterium exchange in reporter peptides compared with the apo state of the protein (Fig. 3D). This change maps to the ATP site of BTK where the parent scaffold is known to bind (SI Appendix, Fig. S7C). Monitoring the protection pattern on BTK demonstrates that the presence of CRBN, both alone and in the presence of PROTACs, has no significant effect on the BTK deuterium uptake profile suggesting that there is no single highly populated interface between BTK and CRBN alone or within a PROTAC ternary complex (Fig. 3 D and E). Similar behavior is observed for both PROTAC (9) and (10) with no significant difference in protection when the two profiles are compared (Fig. 3F).
PROTACs Degrade BTK with High Specificity both in Cultured Cells and in Vivo.
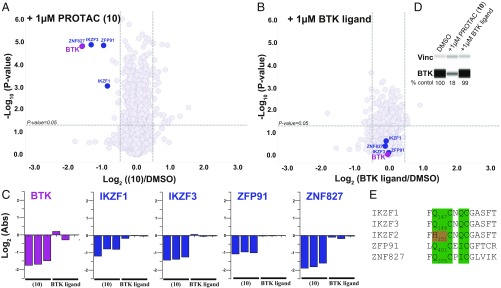
We sought to address the extent of PROTAC specificity by monitoring whole cellular proteome by mass spectrometric (MS) analysis after 24 h treatment of Ramos cells with either DMSO, 1 µM PROTAC (10), or 1 µM BTK parent molecule. Experiments were run in triplicate, compared with DMSO controls (Fig. 4 A–C), and blotted for BTK to confirm knockdown (Fig. 4D). Quantitative proteomics employing tandem mass tags (TMT) chemical labeling coupled with nano-liquid chromatography (LC)/MS/MS enabled the detection of >80,000 unique peptides corresponding to ∼8,000 unique proteins.
Fig. 4.
PROTACs selectively degrade BTK. (A) PROTAC (10) specifically degrades BTK and IMiD-sensitive proteins thought to respond to pomalidomide piece of (10), whereas (B) BTK ligand alone does not lead to any significant protein degradation. Volcano plot shows protein abundance (log2) as a function of significance level (log10). Nonaxial vertical lines denote a fold change of ±1.4, whereas nonaxial horizontal line marks P = 0.05 significance threshold. Down-regulated proteins of significance are found in the upper left quadrant of the plots. Ramos cells were treated for 24 h with either compound [1 µM PROTAC (10) or 1 µM BTK ligand] or DMSO. Lysates were TMT-treated and subjected to LC/MS/MS. Datasets represent an average of n = 3 replicates. A total of 7,877 proteins were identified, and only the ones with at least one uniquely identified peptide are displayed. (C) Individual MS replica showing quantification (log2) of selected proteins from A and B relative to DMSO controls. (D) Half of the MS lysates used in these experiments were saved for Western blots. Representative of one of three MS replicates showing degradation of BTK by PROTAC (10) but not BTK ligand. (E) Multiple sequence alignment of the putative IMiD-binding motif (39, 52), highlighting conserved residues in green and residues shown to reduce binding to IMiDs in orange.
PROTAC (10) (Fig. 4 A and C) and also PROTAC (9) (SI Appendix, Fig. S8 A and C) specifically degrade BTK, whereas the BTK ligand alone (Fig. 4 B and C) or Ibrutinib (SI Appendix, Fig. S8 B and C) do not. TEC, a closely related protein to BTK that potently binds BTK PROTACs (SI Appendix, Fig. S8F), is degraded by PROTAC (9) as well (SI Appendix, Fig. S8 A and C), despite having >100× lower expression in Ramos cells relative to BTK (SI Appendix, Fig. S8E) [TEC is not detected in proteomic study with PROTAC (10) possibly due to its low cellular expression] (Fig. 4 A and C). Lymphoid transcription factors ZFP91, IKZF1, and IKZF3—but not the closely related IKZF2—are specifically degraded in samples treated with PROTAC (9) or (10) (Fig. 4 A and C and SI Appendix, Fig. S8 A and C). However, no degradation is observed when cells were treated with BTK ligand or Ibrutinib (Fig. 4 B and C and SI Appendix, Fig. S8 B and C), suggesting that immunomodulatory imide drugs (IMiD)-based CRBN reprogramming (39–41) is preserved within BTK PROTACs. In addition, a target is identified, ZNF827, that exhibits essential hallmarks of an IMiD substrate (Fig. 4E) and is specifically degraded by pomalidomide containing PROTACs (9) and (10) but not by either BTK ligand alone or Ibrutinib (Fig. 4 A–C and SI Appendix, Fig. S8 A–C).
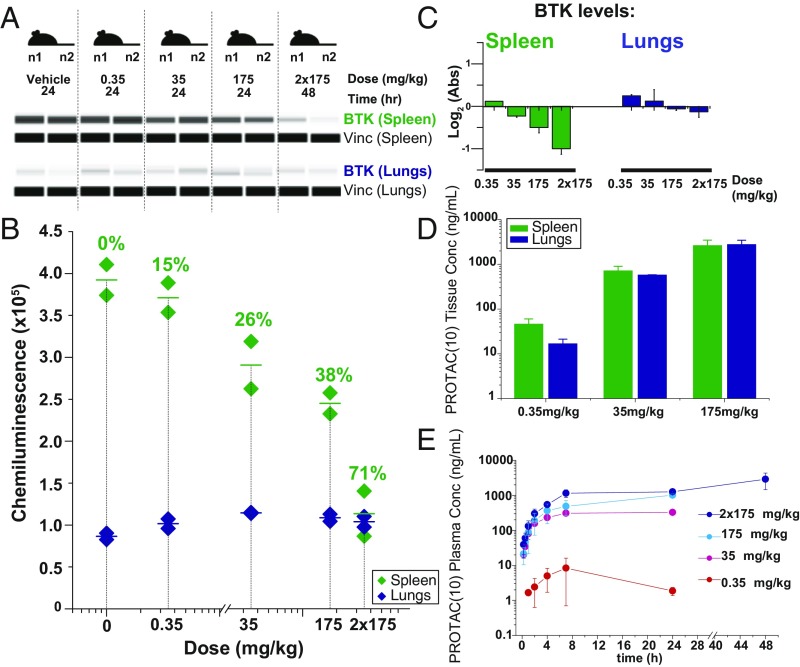
BTK levels were also monitored in spleen and lungs of rats s.c. dosed with PROTAC (10) (Fig. 5 A–C). Increasing concentrations of PROTAC (10) track with efficient BTK knockdown in spleen but not in lungs (Fig. 5 A–C and SI Appendix, Fig. S9B) despite similar tissue uptake and plasma availability of compound across different concentrations (Fig. 5 D and E). A second band, confirmed to be BTK by MS, also tracks with degradation of BTK of the right molecular weight (SI Appendix, Figs. S9A and S10). Altogether, these data show that PROTAC (10) is successfully delivered to tissues and can specifically degrade BTK in vivo, in a dose-dependent and tissue-biased manner.
Fig. 5.
PROTACs selectively degrade BTK in rats. (A) Ten rats were randomly distributed into five groups (n = 2 per group) and s.c. dosed with either 0, 0.35, 35, or 175 mg/kg PROTAC (10) for 24 h or 2 × 175 mg/kg for 48 h. Raw Western blot data showing BTK and vinculin loading control levels for spleen or lungs at the indicated experimental conditions were (B) quantified by normalizing to the vinc loading controls with absolute chemiluminescence shown on the y axis and percentage knockdown for spleen BTK shown above data points. (C) MS-based proteomics quantification of BTK abundance in spleen and lungs with respect to vehicle loading controls for the indicated dosage and incubation times. (D) Rat tissue profile for PROTAC (10). Mean (n = 2 animals per dose group) tissue concentration at the 24 h time point following single SC administration of PROTAC (10) in male Wister Han rats at doses 0.35, 35, or 175 mg/kg. (E) Rat pharmacokinetic profile for PROTAC (10). Mean (n = 2 animals per dose group) plasma concentration vs. time profiles of PROTAC (10) in male Wistar Han rats following single or twice a day SC administration of PROTAC (10) at doses 0.35 mg/kg (red), 35 mg/kg (magenta), 175 mg/kg (light blue), or 175 mg/kg twice a day (dark blue).
Discussion
As PROTAC technology has matured, early proof-of-concept experiments have spawned mechanistic-based questions related to PROTAC optimization for therapeutic use. Of critical importance is the need to understand the parameters that govern both potency and selectivity. Within the PROTAC mechanism of action, the design and optimization of the linker is a unique feature that must be explored. We illustrate an example of these parameters with BTK/CRBN as a test case with the observations below.
First, rapid, efficient, potent, and prolonged knockdown of BTK is achieved by PROTACs (Figs. 1 and 2 and SI Appendix, Fig. S3) and follows the hallmarks of described three-body interactions (32). This library of compounds fall into two classes: short PROTACs (1–4) that are largely ineffective in cellular and in vitro assays and longer PROTACs (6–11) that are both effective and potent (Fig. 2C and SI Appendix, Fig. S3). The transition occurs at compound (5), which appears to lie in intermediate space. Surprisingly, these classes of compounds correlate with ternary complex sterics: shorter PROTACs appear unable to support simultaneous binding of BTK and CRBN (Fig. 2 and SI Appendix, Figs. S4 and S6). In agreement with this model is a recent report of a BTK-specific PROTAC based on a shorter linker but connected to a BTK-binding scaffold that is considerably longer and protruding farther out of the ligand-binding pocket than our BTK binder, thereby possibly providing an initial relief of steric hindrance (42). Although the ternary complex assay allows us to properly bin our compounds into active and inactive groups, it is not sufficient for strict rank ordering of potency. Whereas compounds (9) and (11) appear to yield the greatest peak height and most potent complex formation, respectively, it is compound (10) that gives the most potent cellular knockdown of BTK. This may reflect limitations of the FRET assay discussed above or may reflect subtle differences in physical properties or cell permeability, which are implicit factors in the cellular assays.
Proteomic experiments confirm knockdown of BTK and reveal that the related kinase, TEC, is also degraded. Because TEC is a potent off-target of this BTK scaffold (SI Appendix, Fig. S8F), it is likely that TEC degradation is occurring by the PROTAC mechanism as well. Although TEC is detected in only one of two proteomic experiments [PROTAC (9) but not PROTAC (10)], it is likely that TEC is degraded in both instances, but the low abundance of TEC relative to BTK results in more challenging mass spectrometry-based detection. Close inspection reveals a set of nonkinase IMiD targets [IKZF1, IKZF3, and ZFP91 (39–41)] that are also degraded in the proteomic studies, suggesting that the neosubstrate nature of the pomalidomide moiety is not abolished by the presence of linker and BTK scaffold. In addition, ZNF827 is degraded by both PROTAC (9) and (10) (Fig. 4 A and C and SI Appendix, Fig. S8 A and C). ZNF827 is a zinc-finger protein of partially unknown function (43) but appears to retain a consensus motif found in IKZF1, IKZF3, and ZFP91 (Fig. 4E). As such, we propose that ZNF827 likely represents an additional IMiD-catalyzed degradation substrate.
Although PROTACs (6–11) appear potent, we did not detect appreciable positive cooperativity within the ternary complex. This is true from both mathematical modeling (comparing curve shape and inflections of simulation and experiment) and SPR, in which the affinity of CRBN for PROTAC (10) is similar in both the presence and absence of BTK (Figs. 2 and 3 and SI Appendix, Fig. S6). In addition, the steep nature of the cellular hook effect is consistent with a low-cooperativity system because it demonstrates the ability of binary interactions to efficiently compete with ternary interactions (32). The significant improvement in potency for relatively long (6–11) vs. short PROTACs (1–4), appears primarily due to the relief of negative cooperativity that is achieved as linkers are extended. In an extreme test of this hypothesis, a considerably longer PROTAC (16), which contains a 29-atom linker, was shown to still retain potent BTK knockdown (SI Appendix, Fig. S3D).
In a recent report describing targeted degradation of BRD4 using the ligase VHL, the authors reveal an elegant mechanism by which novel protein–protein interactions between the target and E3 are promoted in the ternary complex (18). These new interactions result in substantial positive cooperativity (α up to 17.6), which leads to a high degree of both potency and selectivity in knockdown among BRD family members (18). Although positive cooperativity has the potential to affect multiple aspects of PROTAC function (including increased activity, broadening the hook effect, and potentially selectivity), the present work on BTK suggests that it is not fundamentally required to achieve potency.
It is important to note that a lack of positive cooperativity does not require or imply that target and E3 ligase do not interact within a ternary complex. Indeed, it is possible that such interactions occur but that the energy gained through protein–protein contacts is offset by the entropic cost of reduced ligand flexibility. Such energetic compensation points to a limitation in what can be inferred from cooperativity measurements in that they may not directly inform on structure. In our case, HDX experiments suggest that our PROTACs are unlikely to lead to a single, stable, rigid ternary conformation, although BTK and CRBN may still interact within the ternary complex, perhaps with a limited ensemble of poses.
Strikingly, potency is intimately related to the identity of the E3 ligase recruited because CRBN PROTACs are remarkably efficient in degrading BTK, whereas VHL PROTACs lead to no degradation, all other variables remaining constant (inhibitor warhead, linker length and composition, and target–ligase cellular colocalization). This is despite the fact that BTK and VHL are expected to form a sterically allowed ternary complex with our tested PROTACs (12, 13) (SI Appendix, Figs. S2B and S4F) and highlights the observation that additional factors are likely involved in appropriate E3–target pairing. Indeed, recent studies using PROTACS derived from promiscuous kinase binders have revealed similar examples of unexpected degradation selectivity and point to additional learnings aside from simple sterics (42, 44).
Consistent with previous reports on BRD4 (5, 9, 10) and Abl/Bcr-Abl (14), BTK now represents a third example of CRBN’s distinctive breadth in targeted degradation as opposed to VHL’s specificity. A possible explanation, first put forth in a recent review by Lai and Crews (45), could be the greater torsional and rotational flexibility of the CRBN-associated E3 ligase scaffold (45–48). This flexibility would not only allow for nestling of diverse substrates within the E3 ligase core but would also provide access to a wider range of surface Lys to E2 active sites that are in large part responsible for substrate degradation (49). Such valuable spatial sampling may be constrained in VHL E3 ligase complexes, which lack the β-propeller domains of CRBN-bound CRL4 E3 ligases (50). XIAP is a noncullin E3 RING ligase (51) that may emulate VHL with regard to limited spatial sampling relative to CRBN.
Although not strictly linked to positive cooperativity, PROTAC potency remains context-dependent as Ramos cells appear more sensitive to our PROTACs than THP-1s (SI Appendix, Fig. S3). Comparison with a primary cell line (PBMC) shows an intermediate sensitivity between Ramos and THP-1 but is much closer to Ramos (SI Appendix, Fig. S2A). Extension to in vivo data reveals a similar puzzle: upon s.c. dosing of PROTAC (10) in rats, dose-dependent knockdown of BTK is observed in spleen but not lung (Fig. 5 A and B). This apparent selectivity occurs despite the fact that compound reaches similar levels of exposure between these two tissues (Fig. 5D). The reasons for these differences are not known but may derive from differences in expression of target or E3, differences in deubiquitinase activity, or a host of as yet unknown determinants. An in-depth study of these determinants will prove critical to better understand and predict differential potency across tissues, which will be important for understanding the efficacy and safety profiles of targeted protein degraders.
Materials and Methods
Expanded methods are available within SI Appendix.
Cell Culture and Immunoblotting.
Ramos and THP-1 cells (ATCC) for dose-dependent studies were cultured in 12-well plates at 37 °C in the presence of compound for 24 h. After lysis, 5 µL of 0.2 µg/µL total protein lysate was loaded onto a 12- to 230-kDa Wes assay plate (ProteinSimple) where 400 nL sample was withdrawn through a capillary, subjected to electrophoretic separation of proteins by size, and followed by HRP-based detection of proteins of interest using an HRP-conjugated secondary antibody.
Ternary Complex Assay.
Biotinylated BTK was labeled with 80 nM Streptavidin-tagged Terbium Cryptate Lumi4-Tb (610SATLB; Cisbio), whereas 2,000 nM (4×) biotinylated CRBN was labeled with 1,000 nM (4×) Streptavidin-tagged XLA665 (610SAXLB; Cisbio). Dose–response curves were obtained by adding increasing concentrations of PROTACs to donor–BTK ([BTK]final = 200 nM, 5 µL per well) and acceptor–CRBN ([CRBN]final = 500 nM, 5 µL per well) conjugates in 20 µL final assay volume in a 384-well low-volume plate.
Biophysical Binding Studies.
On a Biacore SA Chip, BAP–BTK was immobilized on flow cells Fc2 and Fc3 at 4,300 resonance units (RUs) and 4,000 RU, respectively, whereas Fc4 was used as reference. Similarly, in a duplicate set of experiments, BAP–CRBN was immobilized on Fc2 and Fc3 at 4,600 RU and 4,200 RU, respectively. The highest concentration of each compound tested was 90 µM (for CRBN-binding studies) or 10 µM (for BTK-binding studies). Analysis was performed using Scrubber 2.0 (BioLogic Software).
Computational Modeling.
A computational workflow was developed that, given a target, E3 ligase, and PROTAC, will generate an ensemble of multiple possible ternary complexes that is likely to be ubiquitinated. A simple count of the number of possible solutions obtained was then prioritized over other comparable systems that had fewer or no possible solution. Additional modeling details are included in SI Appendix.
Supplementary Material
Acknowledgments
We thank Ravi Kurumbail, Jane Withka, Ann Aulabaugh, Parag Sahasrabudhe, and Mariano Oppikofer for discussions and guidance related to biophysics and structure and Ye Che for discussions and insight on modeling and mechanism. We thank Hua Xu, Theresa Dickinson, and Robin Nelson for initial guidance and support on cellular assays. We acknowledge Joerg Bussenius, Jin Wu, Xiaolin Wu, Limei Zhang, Changwei Zhai, and Zhinguang Chen from WuXi AppTech for additional synthesis support.
Footnotes
Conflict of interest statement: All authors are or were employees of Pfizer, Inc.
This article is a PNAS Direct Submission.
Data deposition: Proteomic data have been deposited in the ProteomeXchange via the PRIDE Archive proteomics data repository, https://www.ebi.ac.uk/pride/archive/. Reference to this database is included in the supplementary information for the mass spec proteomics work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803662115/-/DCSupplemental.
References
- 1.Sakamoto KM, et al. Protacs: Chimeric molecules that target proteins to the Skp1-cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh Y, et al. Development of target protein-selective degradation inducer for protein knockdown. Bioorg Med Chem. 2011;19:3229–3241. doi: 10.1016/j.bmc.2011.03.057. [DOI] [PubMed] [Google Scholar]
- 3.Schneekloth AR, Pucheault M, Tae HS, Crews CM. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter GE, et al. Drug development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bondeson DP, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh Y, Kitaguchi R, Ishikawa M, Naito M, Hashimoto Y. Design, synthesis and biological evaluation of nuclear receptor-degradation inducers. Bioorg Med Chem. 2011;19:6768–6778. doi: 10.1016/j.bmc.2011.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Demizu Y, et al. Design and synthesis of estrogen receptor degradation inducer based on a protein knockdown strategy. Bioorg Med Chem Lett. 2012;22:1793–1796. doi: 10.1016/j.bmcl.2011.11.086. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raina K, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zengerle M, Chan KH, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh Y, Ishikawa M, Naito M, Hashimoto Y. Protein knockdown using methyl bestatin-ligand hybrid molecules: Design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J Am Chem Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- 13.Okuhira K, et al. Specific degradation of CRABP-II via cIAP1-mediated ubiquitylation induced by hybrid molecules that crosslink cIAP1 and the target protein. FEBS Lett. 2011;585:1147–1152. doi: 10.1016/j.febslet.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Lai AC, et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: Small molecules, big challenges. Nat Rev Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 16.Zhang QC, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadd MS, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohamed AJ, et al. Bruton’s tyrosine kinase (Btk): Function, regulation, and transformation with special emphasis on the PH domain. Immunol Rev. 2009;228:58–73. doi: 10.1111/j.1600-065X.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 20.Vihinen M, Mattsson PT, Smith CI. BTK, the tyrosine kinase affected in X-linked agammaglobulinemia. Front Biosci. 1997;2:d27–d42. doi: 10.2741/a172. [DOI] [PubMed] [Google Scholar]
- 21.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ML, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369:507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddocks K, Blum KA. Ibrutinib in B-cell Lymphomas. Curr Treat Options Oncol. 2014;15:226–237. doi: 10.1007/s11864-014-0274-8. [DOI] [PubMed] [Google Scholar]
- 24.Burger JA. Bruton’s tyrosine kinase (BTK) inhibitors in clinical trials. Curr Hematol Malig Rep. 2014;9:44–49. doi: 10.1007/s11899-013-0188-8. [DOI] [PubMed] [Google Scholar]
- 25.Lanning BR, et al. A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat Chem Biol. 2014;10:760–767. doi: 10.1038/nchembio.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woyach JA, et al. BTKC481S-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J Clin Oncol. 2017;35:1437–1443. doi: 10.1200/JCO.2016.70.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angers S, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 28.Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP. A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology. 2004;63:1927–1931. doi: 10.1212/01.wnl.0000146196.01316.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rankin AL, et al. Selective inhibition of BTK prevents murine lupus and antibody-mediated glomerulonephritis. J Immunol. 2013;191:4540–4550. doi: 10.4049/jimmunol.1301553. [DOI] [PubMed] [Google Scholar]
- 30.Lentzsch S, et al. S-3-Amino-phthalimido-glutarimide inhibits angiogenesis and growth of B-cell neoplasias in mice. Cancer Res. 2002;62:2300–2305. [PubMed] [Google Scholar]
- 31.Evans EK, et al. Inhibition of Btk with CC-292 provides early pharmacodynamic assessment of activity in mice and humans. J Pharmacol Exp Ther. 2013;346:219–228. doi: 10.1124/jpet.113.203489. [DOI] [PubMed] [Google Scholar]
- 32.Douglass EF, Jr, Miller CJ, Sparer G, Shapiro H, Spiegel DA. A comprehensive mathematical model for three-body binding equilibria. J Am Chem Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai W, Yang H. The structure and regulation of cullin 2 based E3 ubiquitin ligases and their biological functions. Cell Div. 2016;11:7. doi: 10.1186/s13008-016-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwai K, et al. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 36.Silke J, Meier P. Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb Perspect Biol. 2013;5:a008730. doi: 10.1101/cshperspect.a008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Girona A, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akuffo AA, et al. Ligand-mediated protein degradation reveals functional conservation among sequence variants of the CUL4-type E3 ligase substrate receptor cereblon. J Biol Chem. 2018;293:6187–6200. doi: 10.1074/jbc.M117.816868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An J, et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4CRBN ubiquitin ligase. Nat Commun. 2017;8:15398. doi: 10.1038/ncomms15398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krönke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang HT, et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol. 2017;25:88–99.e6. doi: 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conomos D, Reddel RR, Pickett HA. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat Struct Mol Biol. 2014;21:760–770. doi: 10.1038/nsmb.2877. [DOI] [PubMed] [Google Scholar]
- 44.Bondeson DP, et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 2018;25:78–87.e5. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai AC, Crews CM. Induced protein degradation: An emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer ES, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 49.Wu G, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: Destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 50.Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagano T, Hashimoto T, Nakashima A, Kikkawa U, Kamada S. X-linked inhibitor of apoptosis protein mediates neddylation by itself but does not function as a NEDD8-E3 ligase for caspase-7. FEBS Lett. 2012;586:1612–1616. doi: 10.1016/j.febslet.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 52.Petzold G, Fischer ES, Thomä NH. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.