Significance
In addition to functioning as a delayed-rectifier K+ channel, Kv2.1 interacts with the cortical endoplasmic reticulum (ER) in hippocampal neurons to form somatic ER/plasma membrane (ER/PM) junctions. Neuronal activity and insult induce Kv2.1 release from the cortical ER and subsequent ER withdrawal from the PM. Neuronal ER/PM contacts represent >10% of the cell surface and play roles in membrane trafficking, the regulation of burst firing, Ca2+ homeostasis, and control of PM lipid. We report here that Kv2 channel–VAMP-associated protein (VAP) interaction tethers the cortical ER to the PM via a noncanonical FFAT motif contained within the channel C terminus. Since VAPs have a wide-ranging interactome, Kv2-induced ER remodeling and VAP concentration at ER/PM contacts likely play a central role in neuronal physiology.
Keywords: Kv2.1, Kv2.2, subsurface cisternae, VAP, ER/PM junctions
Abstract
Kv2.1 exhibits two distinct forms of localization patterns on the neuronal plasma membrane: One population is freely diffusive and regulates electrical activity via voltage-dependent K+ conductance while a second one localizes to micrometer-sized clusters that contain densely packed, but nonconducting, channels. We have previously established that these clusters represent endoplasmic reticulum/plasma membrane (ER/PM) junctions that function as membrane trafficking hubs and that Kv2.1 plays a structural role in forming these membrane contact sites in both primary neuronal cultures and transfected HEK cells. Clustering and the formation of ER/PM contacts are regulated by phosphorylation within the channel C terminus, offering cells fast, dynamic control over the physical relationship between the cortical ER and PM. The present study addresses the mechanisms by which Kv2.1 and the related Kv2.2 channel interact with the ER membrane. Using proximity-based biotinylation techniques in transfected HEK cells we identified ER VAMP-associated proteins (VAPs) as potential Kv2.1 interactors. Confirmation that Kv2.1 and -2.2 bind VAPA and VAPB employed colocalization/redistribution, siRNA knockdown, and Förster resonance energy transfer (FRET)-based assays. CD4 chimeras containing sequence from the Kv2.1 C terminus were used to identify a noncanonical VAP-binding motif. VAPs were first identified as proteins required for neurotransmitter release in Aplysia and are now known to be abundant scaffolding proteins involved in membrane contact site formation throughout the ER. The VAP interactome includes AKAPs, kinases, membrane trafficking machinery, and proteins regulating nonvesicular lipid transport from the ER to the PM. Therefore, the Kv2-induced VAP concentration at ER/PM contact sites is predicted to have wide-ranging effects on neuronal cell biology.
Kv2.1 and Kv2.2 are abundant voltage-gated K+ channels in the mammalian brain. Kv2.1 is the predominant channel in the hippocampus while both channels are differentially expressed in the cortex (1). Both channels localize to micrometer-sized clusters on the neuronal surface of the soma, proximal dendrites, and axon initial segment (AIS) in vivo and in vitro (2). Clustered Kv2.1 channels disperse in response to ischemic or hypoxic conditions, neuronal activity, and glutamate-induced excitotoxity via calcineurin-dependent dephosphorylation of the channel C terminus (3, 4). While Kv2.1 clustering was first proposed to regulate channel voltage dependence (5), several studies indicate little connection between channel clustering and regulation of conductance (6–8). In fact, our evidence suggests that the freely diffusive channel population provides the voltage-dependent K+ conductance that regulates neuronal electrical activity while clustered channels are nonconducting and have other functions. We previously reported that the clusters represent trafficking hubs where membrane protein insertion and retrieval at the cell surface are localized (9). These findings agree with results from Lotan and coworkers (10) that indicate one nonconducting function of Kv2.1 is to enhance dense core vesicle release from neuroendocrine cells. Recent studies also indicate Kv2.1 clusters regulate insulin exocytosis from pancreatic beta cells (11, 12). Taken together these studies strongly suggest that Kv2.1 clustering plays a structural role related to the cell biology of the neuronal surface. Indeed, we recently determined that the clustered localization pattern is due to Kv2.1 interacting with the cortical endoplasmic reticulum (ER) and inducing stable ER/plasma membrane (ER/PM) contact sites (13). In rat hippocampal neurons this cortical ER remodeling is regulated by activity, for glutamate treatment induces Kv2.1 declustering that is shortly followed by cortical ER retraction from the cell surface (13). While ER/PM contacts are best understood for their role in store-operated calcium entry and nonvesicular lipid transfer from the ER to the cell surface (14), additional research indicates these microdomains regulate neuronal burst firing (15) and plasma membrane PIP2 levels (16). In addition, a recent study from Hess and coworkers (17) reveals that neuronal ER/PM contact sites represent ∼12% of the somatic surface in vivo. Given the abundance and functional significance of neuronal ER/PM contacts, and the likelihood that processes within these domains are influenced by the Kv2.1–ER interaction, it is paramount to understand the mechanisms underlying the activity-dependent interaction between Kv2 channels and the cortical ER.
Our present work demonstrates that Kv2 channels interact with VAMP-associated proteins (VAPs) embedded in the ER membrane. VAPs were first discovered in Aplysia where they are required for fast neurotransmitter release (18). VAPs are now known to be ubiquitous ER scaffolding proteins with a large and growing list of interactors, including AKAPs, protein kinases, Rabs, and lipid transfer proteins (19, 20). Interestingly, single amino acid substitutions in VAP-B cause late-onset spinal muscular atrophy and amyotrophic lateral sclerosis (ALS)-type 8 (21, 22), which is intriguing given that Kv2.1 clustering over the cortical ER also exists in alpha motor neurons (23). Both the clustering of the Kv2 channels and induction of ER/PM junctions occur via a noncanonical VAP-binding motif contained within the Kv2 channel C terminus. This binding motif contains phosphorylation sites that are known to regulate Kv2 clustering and cortical ER remodeling. The balance of phosphorylation/dephosphorylation at these sites likely governs affinity for VAPs, thus explaining the phosphorylation dependence of the Kv2–ER interaction. Since Kv2 channels concentrate VAPs at the ER/PM contact site, the Kv2–VAP interaction summarized in the present work is likely to have a major influence on neuronal physiology.
Results
Identification of VAPs as the Putative Kv2 ER-Binding Partner.
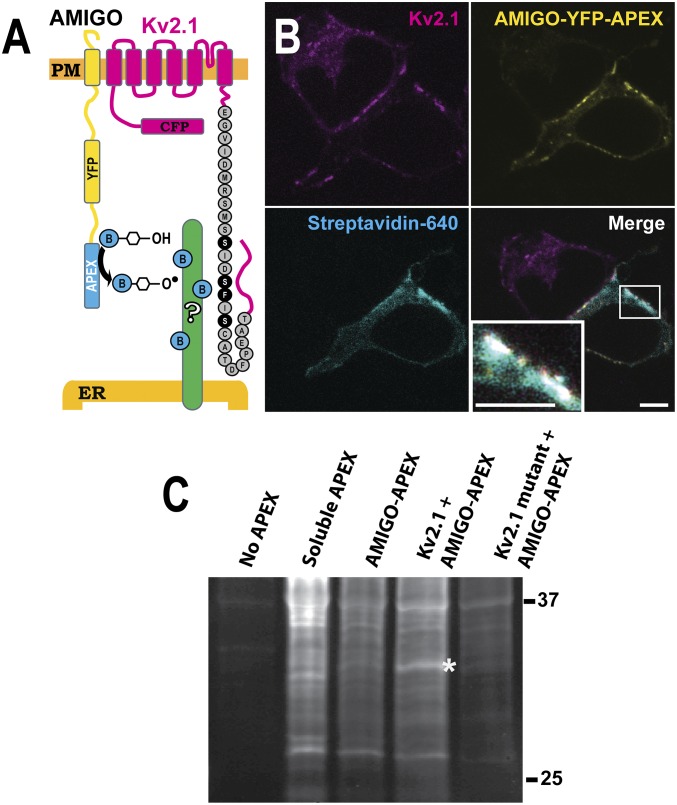
Previous studies employing antibody-based affinity purification of Kv2.1 from either transfected HEK cells or rat brain isolated the channel protein free of any abundant interacting proteins (24). This result is not unexpected since a majority of Kv2.1 is insoluble in the nonionic detergents used for affinity purification (24–26) and macromolecular complexes containing Kv2.1 are likely to reside in this detergent-insoluble fraction. Indeed, when imaging GFP-Kv2.1 clusters in transfected HEK cells during the application of 1% TX-100 at 37 °C, conditions which solubilize putative lipid raft structures (27), the clusters remained intact as the rest of the membrane was solubilized. Since attempts to biochemically purify these detergent-insoluble microdomains using a variety of fractionation procedures were unsuccessful, we used APEX proximity-biotinylation techniques (28) in transfected HEK cells to identify putative Kv2.1-interacting ER-resident proteins responsible for the formation of ER/PM junctions. APEX, in the presence of biotin-phenols and hydrogen peroxide, generates freely diffusing, but short-lived, biotin radicals that nondiscriminately biotinylate nearby proteins. To maximize the biotinylation of neighboring proteins, as opposed to Kv2.1 itself, APEX was appended to the cytosolic end of the Kv2.1 beta subunit AMIGO as diagrammed in Fig. 1A. The C-terminal Kv2.1 sequence known to be involved in the clustering and ER-binding phenotype is indicated with previously described point mutations that abolish soma clustering and ER interaction highlighted in black (29). AMIGO, a single transmembrane cell adhesion molecule, associates with the clustered Kv2.1 channels when coexpressed (30, 31). Importantly, in our HEK cells AMIGO on the PM does not cluster or associate with the cortical ER when expressed alone. As shown in Fig. 1B, coexpression of CFP-Kv2.1 and AMIGO-YFP-APEX induced protein biotinylation in the vicinity of Kv2.1 clusters as indicated by the binding of CF640R-conjugated streptavidin. The streptavidin binding was most prominent at Kv2.1 cell surface clusters, indicating localized biotinylation (Fig. 1B, Lower Right Inset). Next, a Western blot analysis was performed to characterize the biotinylated proteins. Nontransfected cells and cells expressing soluble APEX were examined in addition to cells where AMIGO-YFP-APEX was expressed alone, with Kv2.1, or with the nonclustering Kv2.1(S586A) point mutant (13). Western blots were probed with fluorophore-conjugated streptavidin to detect the biotinylated proteins. As indicated by the asterisk in Fig. 1C, a 33-kDa protein was detected in cells cotransfected with AMIGO and Kv2.1 but not in cells transfected with either AMIGO alone or AMIGO plus the Kv2.1(S586A) point mutant that does not interact with the ER. VAPs are abundant ER proteins of 33 kDa that function in membrane contact site formation between the ER and a variety of organelles (20). Thus, VAPs became an obvious candidate.
Fig. 1.
Use of proximity biotinylation to identify potential Kv2.1 interactors. (A) Diagram of the approach. APEX was attached to the C-terminal end of the Kv2.1 beta subunit, AMIGO. (B) Localization of APEX-mediated biotinylation. HEK cells were transfected with Kv2.1 and AMIGO-YFP-APEX, treated with biotin and H2O2, and then fixed using formaldehyde and labeled with CF640R-conjugated streptavidin to visualize biotinylated proteins as imaged with a laser scanning confocal microscope. Biotinylation localized at ER/PM junctions occurs only in cells expressing AMIGO-YFP-APEX and Kv2.1 (see Lower Right Inset for an enhanced view of biotinylation at a Kv2.1 cluster). (Scale bar: 5 μm.) (C) Parallel samples were collected and subjected to Western blot analysis without affinity purification, using streptavidin–horse radish peroxidase to visualize all biotinylated proteins. A 33-kDa band is present when AMIGO-YFP-APEX is coexpressed with the WT Kv2.1 channel (asterisk). This band is absent from the indicated control lanes, i.e., without any APEX transfection, with soluble APEX expression, and when AMIGO-YFP-APEX is expressed alone or expressed with a mutant Kv2.1 channel which is unable to form ER/PM junctions.
VAPs Specifically Redistribute to Kv2-Induced ER/PM Junctions.
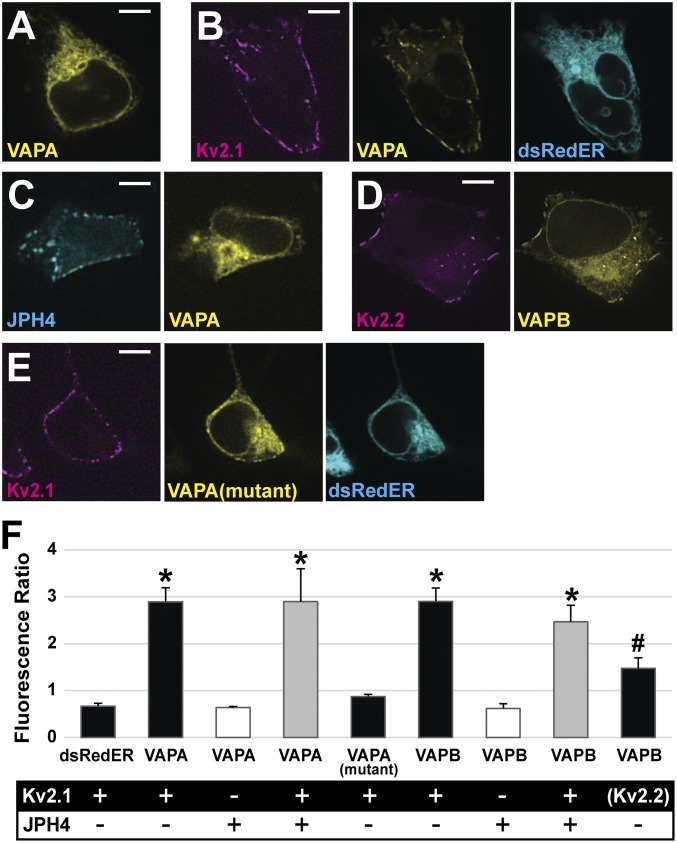
While our previous work (13) demonstrated Kv2.1 induction of ER/PM junctions, no direct binding partner was known at that time. Our first approach to determining whether VAPs interact with Kv2.1 was to perform colocalization experiments. VAPA-GFP or VAPB-GFP was expressed either alone or with a Kv2.1-loopBAD construct that allowed for CF640-streptavidin labeling of only surface channels. Since all cells express endogenous VAPs, the exogenous GFP-tagged VAPs act as a marker for endogenous proteins when expressed at low levels (20). As illustrated in Fig. 2A, when expressed alone, VAPA-GFP displayed a uniform distribution throughout the ER as expected. Fig. 2B shows that in the presence of Kv2.1 the VAPA-GFP localization was dramatically altered, with an obvious redistribution in favor of the Kv2.1-induced ER/PM junctions. We next used junctophilin 4 (JPH4) to induce ER/PM junctions independent of Kv2.1 to rule out the possibility that VAP favors all ER/PM contacts, regardless of molecular composition. JPH4 is an ER membrane protein which induces membrane contacts by binding PM lipid (32). When coexpressed with mCherry-JPH4, the VAPA-GFP remained evenly dispersed throughout the ER, showing no concentration at the JPH4-induced ER/PM contacts (Fig. 2C). Thus, the presence of ER/PM junctions per se had no effect on VAPA-GFP distribution. We also coexpressed GFP-Kv2.2 with VAPB-Ruby2 since Kv2.2 also clusters over the ER (1, 30). Fig. 2D shows that Kv2.2 also concentrates VAPs at its induced ER/PM contact sites. When Kv2.1 and Kv2.2 were coexpressed with VAPB, all three proteins colocalized within the same ER/PM contact sites as illustrated in SI Appendix, Fig. S1A. To determine whether the VAP redistribution to Kv2.1-induced ER/PM contacts is dependent on FFAT motif binding by VAP we coexpressed the K87D/M89D VAPA mutant (20), which is unable to bind FFAT motifs, with Kv2.1 as shown in Fig. 2E. This VAP mutant showed less redistribution to Kv2.1-induced ER/PM junctions but was still slightly enriched at the Kv2.1 clusters. Taken together, these data suggest that VAP concentration at ER/PM junctions is dependent on the presence of Kv2 channels and that this localization is largely dependent on a functional FFAT-binding motif within the VAP protein.
Fig. 2.
Kv2 channels impact localization of VAPA and VAPB in HEK cells. (A) VAPA-GFP expressed alone displays uniform localization across the ER. (B) VAPA-GFP expressed with Kv2.1-loopBAD redistributes to Kv2.1-induced ER/PM junctions. (C) VAPA expressed with the ER/PM junction forming protein, JPH4, does not redistribute to junctions. (D) Kv2.2 coexpressed with VAPB-GFP redistributes this VAP to the induced ER/PM junctions. (E) VAPA(K87D/M89D) has a reduced ability to redistribute to Kv2.1-induced ER/PM junctions. (Scale bars: 5 µm.) (F) Bar graph summarizing VAP redistribution by calculating the ratio of fluorescence at ER/PM junctions to that at ER deeper within the cell when junctions are formed using Kv2 and/or JPH4 as indicated. Only Kv2.1 and Kv2.2 increase the ratio of VAP fluorescence. For analysis, a log transformation was used to satisfy the homogeneity of variance condition and a one-way ANOVA was performed, F(8, 36) = 25.699, P = 1.106 × 10−12 with post hoc pairwise Tukey’s tests. *P < 0.0001, significant difference relative to the dsRedER control,. #P < 0.01 significance. Error bars represent SEM. Twenty-five ROIs from five cells were examined in each case.
The effect of Kv2.1 expression on VAP localization is summarized in Fig. 2F. Here we calculated the ratio of VAP-GFP fluorescence at the PM to the cytoplasmic ER signal located farther within the cell (fluorescence ratio) to quantify the degree to which VAPs were concentrating to ER/PM junctions formed by the various proteins. dsRedER, a soluble ER marker, had a ratio of 0.67, indicating a lower ER intensity at Kv2.1 clusters on the PM compared with signal in the ER deeper within the cell. This ratio was not significantly different from the ratios of VAPA and VAPB when coexpressed with JPH4 (ratio of 0.64, P = 1, for VAPA and ratio of 0.62, P = 0.9996, for VAPB). In contrast to what was observed with JPH4-induced junctions, the VAPs were significantly more concentrated at Kv2.1 ER/PM junctions (ratio of 2.9, P ≤ 0.0001, for VAPA and ratio of 2.9, P ≤ 0.0001, for VAPB). Note that JPH4 and Kv2.1 form ER/PM junctions that are similar in appearance (Fig. 2C) and, in fact, Kv2.1 and JPH4 colocalize within the same junctions when expressed together as illustrated in SI Appendix, Fig. S1B. The presence of Kv2.1 alongside JPH4 at ER/PM contact sites results in significant VAPA redistribution not seen when JPH4 is forming junctions alone (ratio of 2.90 vs. 0.67, P = ≤0.0001, compared with JPH4 alone). Note that while the ratio, 0.87, for VAPA(K87D/M89D) was not statistically significant compared with the dsRedER control, there was a trend toward increased concentration, with this redistribution being visibly noticeable in some cells. These data support the idea that FFAT motif binding is critical for VAP redistribution to Kv2.1-induced ER/PM junctions; however, a secondary mechanism by which VAPs concentrate to Kv2.1-containing ER/PM junctions that is independent of FFAT motif binding may exist.
Förster Resonance Energy Transfer Analysis Supports a Direct Kv2.1–VAP Interaction at Kv2.1-Induced ER/PM Junctions.
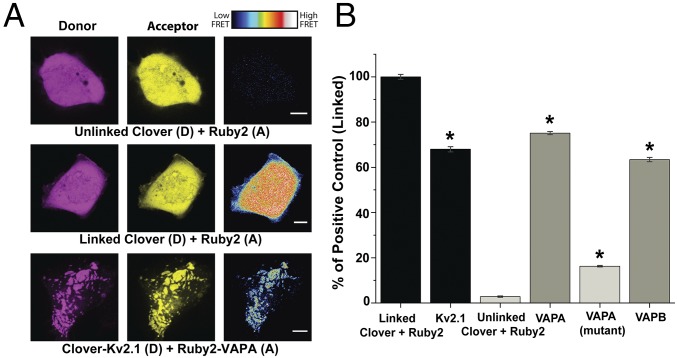
The data presented thus far support a relationship between Kv2 channels and VAPs but do not demonstrate direct binding between the two proteins. Therefore, we used Förster resonance energy transfer (FRET), to determine whether these two proteins are likely in direct contact. We attached the FRET acceptor (mRuby2) and donor (Clover) to the VAP cytoplasmic domains and the N termini of Kv2.1, respectively. A Clover–mRuby2 linked tandem construct was used as a positive control while coexpression of soluble unlinked mRuby2 and Clover served as a negative control. The FRET signals obtained from these two controls, and the FRET observed between Kv2.1 and VAPA, are shown in Fig. 3A, Right. The FRET signals observed in all experiments are summarized in Fig. 3B. We observed significant FRET efficiency between Kv2.1 and both VAPA and VAPB (75% of linked control and 63% of linked control, P ≤ 0.000001 and P ≤ 0.000001 compared with unlinked control, respectively), indicative of protein–protein interaction. By contrast, unlinked Clover and mRuby2 displayed FRET efficiency values that were only 3% of the linked control. An additional positive control examined the FRET efficiency existing between Kv2.1 subunits within a heteromeric channel, i.e., mRuby2- and Clover-Kv2.1 subunits (68% of linked control). The decreased FRET between Kv2.1 subunits, relative to the linked Clover-mRuby2 positive control, is likely due to the random assembly of the channel tetramer. Interestingly, a second negative control, the VAPA(K87D/M89D) mutant, which is incapable of binding FFAT motifs (33), displayed a diminished, but still significant (16% of linked control, P ≤ 0.000001 compared with unlinked control) FRET efficiency. This signal could be due to oligomerization with endogenous VAPs via the transmembrane domain as has been previously described (34). In essence, the VAPA(K87D/M89D) mutant which is incapable of binding Kv2.1 is oligomerizing with endogenous VAPs that are bound to the channel. Such a mechanism would allow for the accumulation of VAPs with available FFAT motif-binding domains within the Kv2.1-induced ER/PM contacts.
Fig. 3.
FRET between Kv2.1 and both VAPs in transfected HEK cells. (A) Representative images of donor, acceptor, and FRET efficiency between the indicated constructs. FRET efficiency magnitude is illustrated by the representative heat maps. (Scale bars: 5 μm.) (B) Quantified FRET efficiency. Here the FRET signals were standardized to that obtained with the linked Clover-Ruby2 positive control. Positive controls are indicated by the black bars, negative controls are in light gray, and the Kv2.1/VAP interactions are in darker gray. A one-way ANOVA was performed, F(5, 481) = 195.7, P = 1.81 × 10−133 with post hoc Tukey’s tests to examine significance. *P < 0.000001, significant difference relative to the unlinked negative control. Error bars represent SEM. n = 109 linked, 104 VAPA, 76 VAPA (mutant), 48 VAPB, 75 Kv2.1, and 58 unlinked cells. Each cell had 15 ROIs examined.
Knockdown of VAP Protein Impacts the Clustering Behavior of Kv2.1.
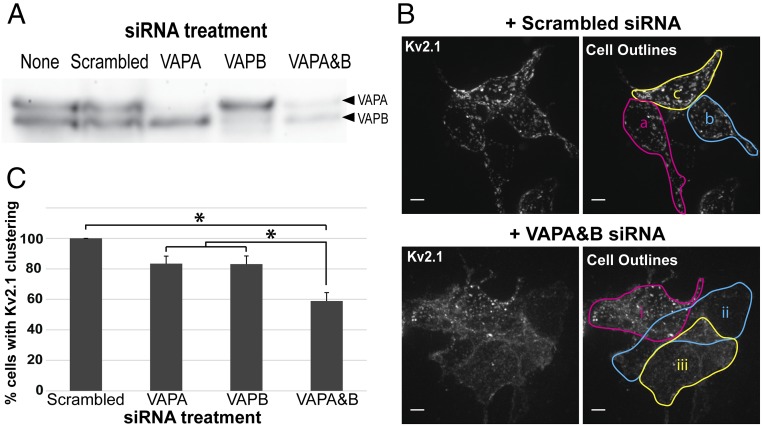
To confirm that VAPs are directly involved in Kv2.1 clustering over the ER we used a siRNA approach to reduce endogenous VAP expression in HEK cells. As illustrated in Fig. 4A, while the scrambled siRNA control had no effect on either VAPA or VAPB expression, combining both siRNAs greatly reduced both VAPA and VAPB. We next examined the effect of VAP knockdown on Kv2.1 clustering as illustrated in Fig. 4B and summarized in Fig. 4C. As shown in Fig. 4B, knocking down both VAPA and VAPB visually decreased the extent of GFP-Kv2.1-loopBAD clustering as imaged with CF640-SA binding to the biotinylated channels on the cell surface. Since Kv2.1 cluster size and intensity are dependent on the Kv2.1 expression levels, we quantitated the effect of VAP knockdown by simply comparing the percentage of cells with clusters observed under the different siRNA treatments. For a cell to be classified as Kv2.1 cluster-free it had to have the homogeneous surface distribution illustrated by the two cells (ii and iii) in Fig. 4B, Lower Right. Cell i was classified as possessing clustered Kv2.1. Using this quantitation approach the effects of reducing VAPA or VAPB levels are summarized in Fig. 4C (83.4% and 83.1%, NS, compared with the scrambled siRNA control). Combining both siRNAs had the greatest effect on Kv2.1 clustering percentage, reducing clustering from 100% to 58.9% (P < 0.00001) compared with the scrambled siRNA control. In summary, the results presented in Fig. 4 indicate both VAP isoforms are involved in Kv2.1 clustering, i.e., binding to the cortical ER, in HEK cells. While not apparent in Fig. 4, VAP siRNA, relative to the scrambled siRNA control, decreased the total surface Kv2.1 to 41% of control and decreased the intensity of Kv2.1 clusters to 40%. GFP intensity was reduced to 55% of control. Thus, VAP knockdown suppressed Kv2.1 surface levels to a somewhat greater extent than the overall Kv2.1 expression. Whether VAP levels are specifically linked to Kv2.1 biosynthesis, trafficking, or stability remains an open question.
Fig. 4.
Effect of siRNA-mediated knockdown of VAPA and VAPB on Kv2.1 clustering. (A) Western blot demonstrating efficacy of VAPA and VAPB siRNA. Protein blot was probed with anti-VAPB antibody which cross-reacts with VAPA. (B) Representative image of GFP-Kv2.1-loopBAD clustering in the presence of scrambled or VAPA and VAPB siRNA taken via spinning-disk microscopy. z-stack maximum intensity projections are shown. In Upper Right image all three cells (a–c) were scored as having clustered Kv2.1. In the Lower Right image only cell i was scored as having clusters. Note that this image is presented simply to illustrate how clustering was defined as opposed to being quantitative with respect to the effect of the siRNA treatment. (Scale bars: 5 µm.) (C) Quantification of the percentage of cells displaying Kv2.1 clustering after various siRNA treatments. Eighty-six cells receiving the scrambled siRNA, 90 cells receiving VAPA siRNA, 61 cells receiving VAPB siRNA, and 144 cells receiving both VAPA and VAPB siRNA were examined within 26, 27, 21, and 41 images, respectively. Error bars represent SEM. For analysis, a one-way ANOVA was performed, F(3, 111) = 13.61, P = 1.27 × 10−7, with post hoc Tukey’s tests. *P < 0.01 significance.
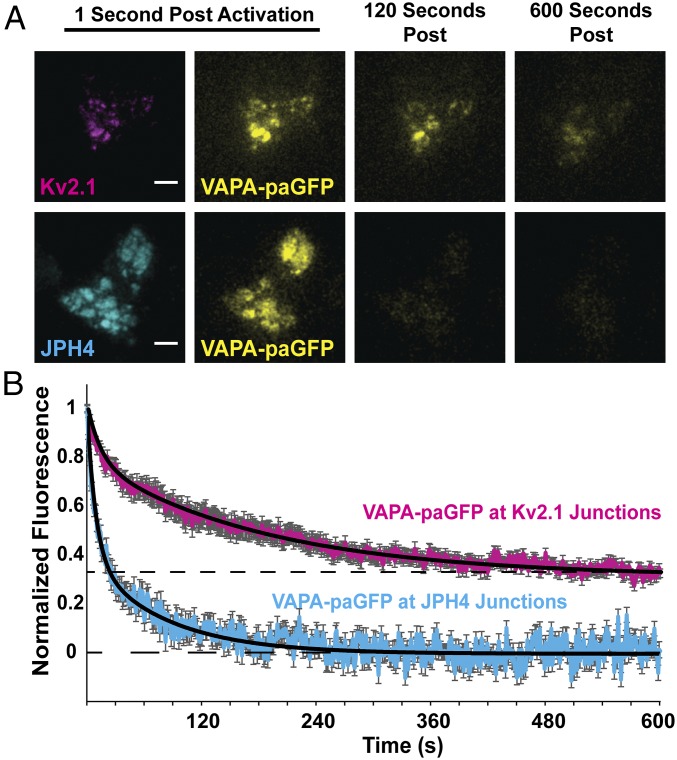
VAP Resident Time at Kv2.1-Induced ER/PM Junctions Is Long Lived.
Our previous analysis of Kv2.1 behavior at the single-molecule level revealed that individual Kv2.1 channels can reside within a cluster for >25 min (35), suggesting a very stable interaction between the Kv2.1 C terminus and its ER-binding partner. To examine the stability of Kv2.1 VAP binding we measured the dissociation kinetics of photoactivatable GFP (paGFP)-tagged VAPA at Kv2.1 clusters following photoactivation. We selectively activated and quantitated VAP-paGFP fluorescence solely within the TIRF field to avoid activating fluorescence removed from the PM as demonstrated in Fig. 5A. Activating the paGFP and then measuring its loss due to diffusion into the cytoplasmic ER allowed us to measure the relative stability of the VAP–Kv2.1 interaction (Fig. 5B). We found that VAPs were demonstrably more stable at Kv2.1 junctions than at JPH4-induced junctions, as expected if Kv2.1 channels and VAPs are interacting to form this junction. As indicated in Fig. 2, although VAPs are not concentrated to these areas, VAPs are present at the JPH4 membrane junctions solely because they exist throughout the ER. The paGFP fluorescence decayed to baseline with the JPH4-induced junctions while in the presence of Kv2.1 approximately one-third of the original VAPA-paGFP was stably retained, suggesting a stable Kv2.1–VAP interaction lasting longer than 10 min. If VAPs do indeed function as scaffolding proteins at ER/PM contacts, this stability allows for the existence of long-lived complexes. A two-exponential decay was required to fit the data, indicating time constants of 9.1 s and 80.5 s for the JPH4 membrane junctions compared with 13.9 s and 186.3 s for the Kv2.1-induced ER/PM contacts. These time constants are likely the result of VAP diffusion into the deeper ER that is removed from the PM. The time constant increases observed in the presence of Kv2.1 could be due to increased molecular crowding in the presence of Kv2.1 compared with JPH4.
Fig. 5.
Photoactivatable-GFP (paGFP)-based analysis of VAPA stability at Kv2.1-induced ER/PM junctions. (A) Representative TIRF microscopy images of VAPA-paGFP at either Kv2.1- or JPH4-induced ER/PM junctions 1 s, 120 s, and 600 s after 405-nm induced photoactivation in TIRF. (Scale bars: 5 µm.) (B) Time course of paGFP-fluorescence loss. Normalized fluorescence measurements were fitted with a two-exponential decay, black lines (). VAPA was significantly less stable at JPH4-induced ER/PM junctions (τ1 = 9.1, τ2 = 80.5) than at Kv2.1 junctions (τ1 = 13.9, τ2 = 186.3), P = 0.0395 at 600 s. Error bars represent SEM. Twenty-five ROIs from 5 Kv2.1-expressing cells and 20 ROIs from 20 JPH4-expressing cells were used.
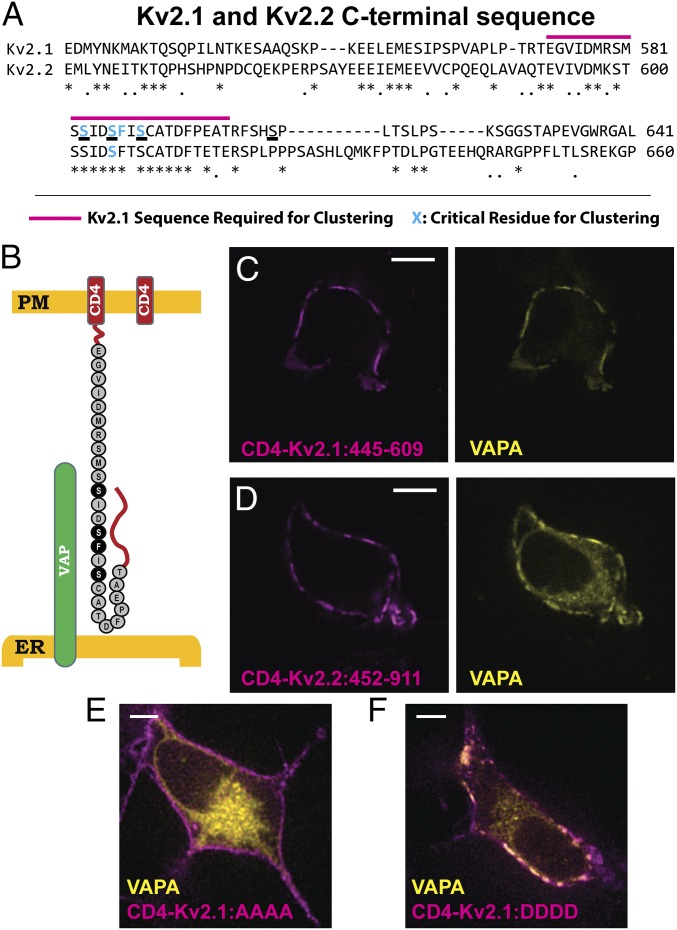
Binding to VAPs Is Mediated by the Kv2.1 and Kv2.2 C Termini.
VAPs bind a loosely defined FFAT motif (two phenylalanines within an acidic tract) contained within their binding partners. Kv2.1 clustering requires a previously defined motif within the channel C terminus as highlighted in Fig. 6A (29), with the underlined serines likely representing phosphorylated amino acids involved in Kv2.1 clustering over the ER (13, 29). However, obvious VAP-binding FFAT motifs, which have a consensus sequence of EFFDAxE (20), are lacking within both the Kv2.1 and -2.2 C termini (compared in Fig. 6A). As a first step toward identifying the sequences within Kv2 channels that are involved in VAP binding we appended amino acids 445–609 of Kv2.1 and amino acids 552–911 of Kv2.2 to the single-pass transmembrane protein CD4 as diagrammed in Fig. 6B. These amino acids were selected to keep the total amino acid length from the PM to the regions known to be required for clustering as close to WT Kv2.1 as possible, as there is reason to believe that ER–PM membrane distance can have a profound effect on protein localization to ER/PM junctions (36). CD4 was chosen because WT CD4 shows a homogenous cell surface distribution in both transfected rat hippocampal neurons and HEK cells. Thus, any clustering or concentration over the cortical ER is readily detected. Since structural studies indicate CD4 forms a dimer (37), we assume each of these chimeric constructs contains two Kv C-terminal domains. The CD4/Kv2 chimeras were then coexpressed with VAPA-GFP in HEK cells and the CD4 localized using CF640-conjugated anti-CD4 monoclonal antibody directed against an extracellular epitope. As shown in Fig. 6C the CD4/Kv2.1 C-terminus chimera clustered on the cell surface and concentrated VAPA similar to the full-length Kv2.1 in Fig. 2B. The same clustering and VAPA redistribution were observed with the CD4/Kv2.2 C-terminal chimera (Fig. 6D). Thus, a noncanonical VAP-binding motif is likely present within these C-terminal sequences.
Fig. 6.
Kv2 channel C terminus is sufficient to form ER/PM junctions through VAP interaction. (A) Sequence comparison of Kv2.1 and Kv2.2 C termini. Note the high degree of conservation in the area required for clustering, but lack thereof elsewhere. Known sequence required for clustering in Kv2.1 is indicated by the magenta line. Known amino acids required for clustering are highlighted in cyan. (B) Schematic of WT CD4, on right, and the CD4-Kv2.1:445–609 construct, on left. Critical amino acids for Kv2.1 clustering are included in black. (C) Appending amino acids 445–609 of Kv2.1 onto the CD4 C terminus results in ER/PM junctions that concentrated VAPA at the PM. (D) The C terminus of Kv2.2 attached to CD4 also results in a construct that clusters and interacts with VAPs. (E) Expression of VAPA-GFP with the CD4-Kv2.1:445–609 construct in which the serines underlined in A were mutated to alanines. (F) Expression of VAPA-GFP with the CD4-Kv2.1:445–609 construct in which the serines underlined in A were mutated to aspartic acids. (Scale bars: 5 µm.)
Since phosphorylation is believed to regulate the Kv2.1–ER interaction and the resulting channel clustering phenotype, we next mutated the underlined serines highlighted in Fig. 6A to either alanines incapable of phosphorylation (CD4-Kv2.1:AAAA) or aspartic acids to create phosphomimetics (CD4-Kv2.1:DDDD). CD4-Kv2.1 C-terminus chimeras containing these substitutions within the appended 445–609 aa of Kv2.1 were then coexpressed with VAPA-GFP and VAPA redistribution was assessed. As illustrated in Fig. 6E alanine substitution prevented both the chimera clustering and the VAPA redistribution while Fig. 6F shows robust clustering and VAPA redistribution with the phosphomimetic substitutions. These data further support the idea that Kv2.1–VAP interaction is regulated by phosphorylation within a small section of the channel C terminus.
Kv2.1 Forms ER/PM Junctions Through Interaction with VAPs via a Noncanonical FFAT Motif.
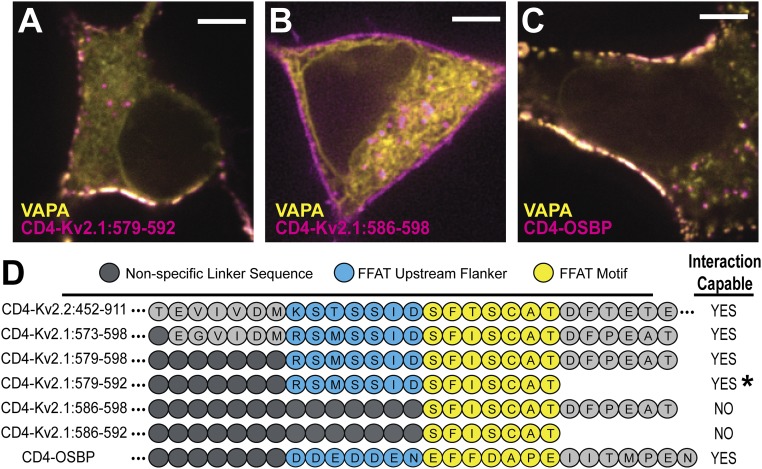
To identify the exact Kv2.1 C-terminal sequence involved in VAP binding we generated a series of CD4 chimeras containing varying amounts of Kv2.1 C-terminal sequence (summarized in Fig. 7D). To provide enough cytoplasmic depth to contact the cortical ER we added β2-microglobulin between the CD4 and channel sequence to create a rigid linker. Variable-length flexible linkers were also inserted between the microglobulin and lengths of short Kv2.1 sequence in an effort to maintain constant spacing between the ER and PM. As a positive control, we also appended the classic FFAT sequence of oxysterol-binding protein (OSBP) with both upstream and downstream flankers (see SI Appendix, Supplementary Materials and Methods for additional details). OSBP is a lipid transfer protein that is a known VAP interactor and has a well-characterized FFAT motif (14, 20, 38). We began by confirming that the amino acid sequence already known to be responsible for the clustering behavior of Kv2.1 channels [amino acids 573–598 (29)] was also responsible for VAP binding. As shown in Fig. 7A a CD4-Kv2.1 C-terminus chimera containing only amino acids 579–592 of Kv2.1 both clustered and concentrated VAPA at the ER/PM junction. In contrast, the chimera with amino acids 586–598 failed to both cluster and redistribute the VAP as illustrated in Fig. 7B. Fig. 7C shows the VAP binding observed with the CD4-OSBP FFAT motif positive control. The sequence and behavior of these and other constructs are summarized in Fig. 7D. For comparison the Kv2.2 C-terminal sequence used in Fig. 6D is included. The comparisons shown in Fig. 7D suggest that there is a noncanonical FFAT motif present in both the Kv2.1 and -2.2 C termini. The 7 aa in yellow represent the core motif where the second expected phenylalanine is absent. The upstream amino acids in blue likely substitute for the upstream acidic tract obvious in the OSBP sequence. In Kv2.1 serine phosphorylation likely provides the negative charge required to guide the Kv2–VAP interaction, similar to other studied FFAT-containing proteins which are known to interact with the VAPs (20). This required phosphorylation explains the calcineurin-induced declustering of Kv2.1 that occurs in response to excitotoxicity or neuronal insult (3, 39, 40). Note that the 579–592 construct clusters only when VAPs are cotransfected, suggesting that the lack of downstream sequence reduces VAP affinity. We cannot say whether these specific amino acids are increasing affinity of binding or whether any random sequence after the FFAT motif would serve this purpose; to our knowledge no FFAT motif has ever been described that is localized at the very end of any protein as is the case with this CD4 chimera.
Fig. 7.
Defining the minimum domain necessary for Kv2.1–VAP interaction. (A) CD4-Kv2.1:579–592 forming ER/PM junctions in HEK cells via VAP interaction. This construct represented the minimal sequence of amino acids we observed capable of this behavior (however, see asterisk in D). (B) CD4-Kv2.1:586–598 was not capable of forming ER/PM junctions through VAP interaction. (C) CD4-OSBP construct demonstrating that ER/PM formation and VAP concentration occur in the presence of a classic FFAT motif containing protein on the PM. (Scale bars: 5 µm.) (D) Schematic displaying amino acid identity and relative position of Kv2.1 sequence fragments appended to the CD4 backbone. Right-hand side indicates which chimeras formed clusters on the membrane with concomitant VAP relocalization to these sites. *, CD4-Kv2.1:579–592 was capable of forming ER/PM junctions if VAP was coexpressed but did not form these microdomains with only the endogenous VAP levels.
Interestingly, 3 aa of the 4 aa already found to be critical for the clustering of Kv2.1 (S586, F587, S589) reside directly within the FFAT motif (13, 29). A fourth critical residue (S583) is located just 3 aa upstream and within the FFAT motif flanker region known to be important for VAP binding (20). The serine at 586 is a known phosphorylation site (41).
Kv2.1 and Kv2.2 Interact with and Regulate the Localization of VAPA and VAPB in Rat Hippocampal Neurons.
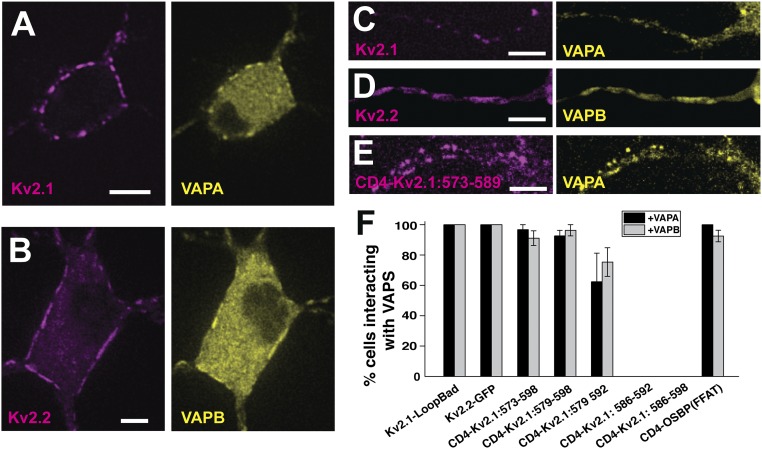
To confirm that the Kv2 VAP interactions also occur in hippocampal neurons Kv2.1-loopBAD was coexpressed with VAPA-GFP in DIV 7 rat hippocampal neurons as illustrated in Fig. 8A. Again, VAPA concentrated at the Kv2.1-induced ER/PM junctions. Fig. 8B shows a similar result when GFP-Kv2.2 and VAPB-mRuby2 were coexpressed. Kv2 channels also form ER/PM junctions within the AIS (42, 43) and Fig. 8 C and D illustrates VAP concentration here in the presence of Kv2.1 and Kv2.2, respectively. Fig. 8E illustrates the interaction of the CD4-Kv2.1: 573–589 chimera with VAPA within the AIS. Recent work indicates that Kv2.1 likely contains two independent AIS localization signals within the C terminus; one is contained within the sequence shown in Fig. 6A and the other one is located distally at amino acids 720–745 (44). The CD4 chimera lacks this secondary AIS localization signal, thus demonstrating that VAP binding alone can localize Kv2.1 to the AIS. Quantitation of Kv2-VAP interaction is summarized in Fig. 8F. Here the percentage of transfected neurons concentrating either VAPA or VAPB to the Kv2-induced ER/PM junctions is indicated. The CD4 chimeras that failed to interact with VAPs in HEK cells (Fig. 7) also failed to bind VAPs in neurons. These data indicate that Kv2–VAP interaction is similar between HEK cells and rat hippocampal neurons, which is not surprising given that HEK cells are of neuronal origin (45) and the calcineurin-dependent regulation of Kv2.1 clustering is conserved between these two cell types (46).
Fig. 8.
Kv2 interaction with VAPs in rat hippocampal neurons. (A) Coexpression of Kv2.1-loopBAD and VAPA-GFP. Surface Kv2.1-loopBAD was visualized with CF640-conjugated streptavidin. (B) Coexpression of GFP-Kv2.2 and VAPB-mRuby2. (C–E) Colocalization of Kv2.1-loopBAD, GFP-Kv2.2, and the CD4-Kv2.1:573–589 chimera with VAPA-GFP, VAPB-mRuby2, and VAPA-GFP, respectively, within the AIS. The AIS was confirmed with anti-neurofascin antibody staining as illustrated in SI Appendix, Fig. S2. (Scale bars: 5 μm.) (F) Summary of the percentage of neurons concentrating VAPs at induced ER/PM junctions. Error bars indicate SEM. P values comparing the interaction of the first three CD4-Kv2.1 chimeras with either VAPA or VAPB to WT Kv2.1 were not significant, with Kruskal–Wallis ANOVA values of P = 0.37 and P = 0.1, respectively.
Discussion
Our data demonstrate that a Kv2 channel–VAP interaction links the PM to cortical ER as summarized in Fig. 9. The formation of this membrane contact site gives rise to Kv2 channel clusters on the neuronal surface. VAPA and VAPB are abundantly expressed in hippocampal, cortical, and motor neurons based on both Western blot and immunostaining approaches and these neuronal types display prominent Kv2.1 clusters on the somatic surface (47). However, no concentration of VAPs into plasma membrane-associated clusters has been previously reported, perhaps because the available antibodies target VAP domains associated with FFAT motif binding, thus preventing immune labeling of VAPs within an assembled complex. While we previously proposed that individual Kv2 channels within these microdomains must be corralled behind a cytoskeletal fence due to their high lateral mobility within the PM (35), both the mobility and clustering are now best explained by the binding to freely diffusing VAPs within the ER. The FRET experiments presented in Fig. 3 indicate Kv2.1 and VAPs reside within 1–10 nm of each other (48), suggesting they are in direct contact. The fluorescence loss kinetics after VAP-paGFP photoactivation (Fig. 5) suggest this interaction is relatively long lived (>10 min), which agrees with our previous studies indicating that individual Kv2.1 channels can remain within a cluster for 25 min or more under resting conditions (35). The siRNA VAP knockdown experiments (Fig. 4) support the idea that VAPs are required for the Kv2.1 clustering and indicate that both VAPA and VAPB participate in this process. However, Kv2.1 surface expression was depressed by 60% upon knockdown of both VAPA and VAPB. Whether this result means VAPs are specifically involved in Kv2.1 biosynthesis, trafficking, or stability remains an open question. However, Kv2.1 channels are delivered to the cell surface at Kv2.1-induced ER/PM junctions (9), making it likely that disruption of this membrane contact site will affect Kv2.1 delivery. Note that when Kv2.1 is first synthesized in a transfected HEK cell, it is delivered to the cell surface at the small and dynamic ER/PM contacts that exist in the absence of Kv2.1 (49). As Kv2.1 accumulates on the surface it begins to bind ER VAPs and form the large and stable membrane junctions.
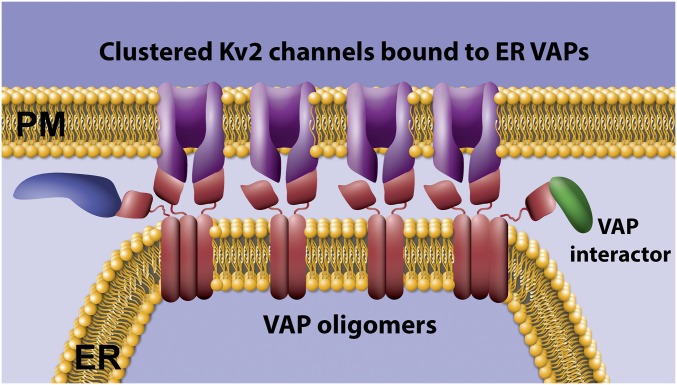
Fig. 9.
Working model where Kv2 channels concentrate ER VAPs by both direct binding via the C-terminal noncanonical FFAT motif and VAP oligomerization via the VAP transmembrane domain.
VAPs bind FFAT motifs via a positively charged surface located in their major sperm protein-like domain (20). The FFAT motif core is a sequence of 7 aa that are extended, typically upstream, by an acidic tract. VAP-FFAT motif binding is initiated through the nonspecific negatively charged amino acids upstream of the core motif, as illustrated in Fig. 7 by the classic FFAT motif of our CD4-OSBP(FFAT) construct. While the originally defined sequence for FFAT domains is EFFDAxE, this motif can tolerate a high degree of variability. For example, the two phenylalanines are not present in all VAP-binding sequences; for instance, protrudin has an FFAT-like motif with a lysine at position 3 (compared with isoleucine in Kv2.1) (20). The Kv2.1 and -2.2 VAP-binding domains identified in our present work, SFISCAT and SFTSCAT, respectively, again demonstrate that the two phenyalanines are not essential for binding and that phosphorylated serines likely can substitute for acidic residues. Importantly, the Kv2 sequences adhere to a minimal requirement for FFAT-like motifs: F/Y at position 2, negative residue at position 4, small residue at position 5, and negative flank. Nevertheless, criteria suggested for finding FFAT motifs would place the Kv2 sequences below the established threshold (20). The feature that made these motifs detectable is the previous precise mapping across Kv2.1’s C terminus (29).
Clustering of Kv2.1, and to a lesser extent Kv2.2, is regulated by phosphorylation (1, 3, 50) and phosphorylation of serine residue 586 at the beginning of the noncanonical FFAT motif, confirmed by mass spectrometry and phospho-specific antibody binding (41, 50), likely generates negative charge necessary to facilitate VAP binding. Phosphorylation of residues surrounding FFAT motifs to facilitate binding is already known to occur in other VAP interactors (20). It is currently unknown whether serine 583 within the upstream linker, required for Kv2.1 clustering (29), or serine 589, a critical FFAT motif residue, is phosphorylated. What is known is that neuronal insults, such as ischemia and neuronal activity, result in calcineurin-dependent dephosphorylation of the channel, dispersal of channel clusters, and retraction of the cortical ER (3, 13, 39, 40). How calcineurin accesses the phosphorylated serines involved in VAP binding is unclear. Perhaps the individual FFAT motif–VAP interactions are dynamic enough to allow calcineurin access over the 2-min period required for significant glutamate-induced declustering and ER retraction (13). It is important to note that Kv2 channels are tetrameric. This current study has not investigated the stoichiometry involved in Kv2–VAP interaction but it is likely that one Kv2 tetramer can bind up to four VAPs. Individual VAPs unbinding and quickly rebinding different α-subunits both within a single channel and across channels could explain transient calcineurin FFAT access and would still be consistent with the general long-term stability of proteins within these domains. Clearly, future research is required to address the exact mechanisms underlying the phosphorylation-dependent VAP interaction.
Data presented in Figs. 2 and 3 suggest the VAPA(K87D/M89D) mutant, which is unable to bind FFAT motifs, still localizes to Kv2 channel-induced ER/PM junctions, although to a reduced extent relative to WT VAPA. Since VAPs can form homomeric and heteromeric oligomers, possibly through a transmembrane GxxxG motif (34), the mutant GFP-tagged VAPA may be assembling into oligomers with endogenous WT VAPs that are bound to Kv2 channels at junctions. Such a mechanism would allow for the localization of VAPs to these microdomains which possess FFAT-binding motifs available to interact with additional partners apart from Kv2 channels (see Fig. 9 for depiction). VAPs have a growing list of interactors, including AKAPs, protein kinases, kinase regulators, transcription factors, Rabs, and lipid transfer proteins (19, 20, 38) and any concentration of these proteins to ER/PM contact sites should be physiologically significant. Given that the Kv2–VAP interaction is likely directly regulated by phosphorylation within, and adjacent to, the Kv2 C-terminal FFAT motif, it is possible that the kinases and phosphatases involved are VAP tethered. However, to the best of our knowledge, known Kv2.1-modifying kinases (CDK5, p38 MAPK, src) (51–53) and phosphatases (calcineurin) (3, 40) have not been confirmed to be part of the VAP interactome (19). However, FFAT motif-containing proteins are involved in the nonvesicular transfer of ceramide, cholesterol, and phosphotidylinositols between the ER and late secretory organelles, including TGN and PM. Kv2 channels may establish an ER/PM junction where the concentrated VAPs function as a scaffolding hub, making these membrane contact sites not only functionally distinct from ER/PM contacts such as those induced by STIM1 or the extended synaptotagmins (14), but also regulated by neuronal activity and sensitive to insult. In addition, it is possible that the converse is true, where the VAP-mediated concentration of Kv2 channels imparts specific functions onto the ER/PM junction due to domains contained within the channel itself. Kv2.1 contains a syntaxin-binding region (10, 54), an ion pore, and a voltage-sensing domain that, even in the nonconducting channel, responds to membrane potential (8). In addition to localized SNARE protein binding or K+ conductance, perhaps Kv2.1 communicates neuronal electrical activity to functions occurring at the ER/PM contacts.
ER/PM contact sites represent ∼12% of the somatic surface in vivo but not all junctions are created equal (17). Junctions can be formed by extended synaptotagmins, STIM proteins, junctophilins, and other proteins (14) in addition to Kv2 channels and an obvious question deals with the dynamic composition of these membrane contacts at any particular time. It is likely that multiple-junction–forming proteins can be present and that these components will differentially recruit specific interactors as proposed here for the Kv2 recruitment of VAPs. Kv2.1 and Kv2.2 are differentially expressed in cell types throughout the brain and exhibit different functional properties, including response to stimuli such as acute hypoxia, with Kv2.2 being less responsive in terms of both declustering and altered voltage dependence (1). It seems probable that these differences in junction location and stability as well as functionality are significant with respect to different cell types. In addition, Kv2.1 exhibits different behavior, depending on the subcellular compartment it is located in. Kv2.1 channels on the AIS are more resistant to glutamate-induced declustering compared with channels on the soma (44, 55). While our data indicate VAP-based tethering occurs in both compartments, there is a domain downstream of the FFAT motif (amino acids 720–745) that is sufficient for AIS-specific localization (44). Perhaps during evolution the addition of a second ER-targeting motif ensured that neuronal activity does not alter Kv2.1–ER contacts within the AIS.
As with many ion channels, mutations in Kv2.1 that alter conductance are linked to human disease, with mutations that alter ion selectivity and voltage sensing being associated with epilepsy and developmental delay (56, 57). However, three recently described Kv2.1 mutations result in premature stop codons that are predicted to not alter Kv2.1 conductance (58). These mutations occur downstream of the conserved channel domains, falling between the last transmembrane domain and the noncanonical FFAT motif identified in the present work. One of these mutations truncates Kv2.1 at the arginine residue immediately upstream from the FFAT motif flanker region (R571). All three mutations result in developmental delay and all three are expected to abolish the ER/PM junctions formed by Kv2.1. Thus, mutations that specifically interfere with Kv2.1–VAP binding are likely to be involved in human disease.
Materials and Methods
DNA Constructs.
Plasmids encoding fluorescent protein- and biotin acceptor domain-tagged Kv2.1 have been described previously (25, 35, 59). Briefly, the fluorescent proteins are attached to the channel N terminus and the biotin acceptor domain (BAD) inserted into the extracellular loop between the first and second transmembrane domains. Cotransfection with the BirA biotin ligase in the pSec vector was performed as previously described to biotinylate a specific lysine within the BAD sequence (35, 60, 61). Kv2.1 contains two methionine residues five residues apart, at the beginning of the coding sequence, each of which has been selected as the starting amino acid in different publications. Thus, there are two separate numbering systems currently in use in the Kv2.1/KCNB1 literature. For the sake of consistency between this paper and previously published work concerning Kv2 sequence critical for clustering (29, 41), we have opted to retain the original amino acid numbering which sets the second methionine as amino acid 1. Synthetic full-length Kv2.2 sequence was obtained from Genewiz and inserted into the peGFP-C1 expression vector (Clontech). AMIGO in pDONR221 was obtained from DNASU Plasmid Repository (plasmid ID HsCD00296150) and from this AMIGO-YFP was created by using ApaI and XhoI cut sites and placing the AMIGO fragment into a peYFP-N1 vector. Additional details regarding plasmid construction are presented in SI Appendix, Supplementary Materials and Methods.
Cell Culture, Transfection, and Labeling of Surface Kv2.1 and CD4 Chimeras.
HEK 293 cells [American Type Culture Collection (ATCC), passage 45–48] were cultured, transfected with the indicated DNA constructs via electroporation, and plated onto Matrigel-coated 35-mm glass bottom coverslip dishes (Matsunami Glass Corporation) as previously described (13). BirA cotransfection was used to induce biotinylation of the BAD containing Kv2.1 constructs, e.g., GFP-Kv2.1-loopBAD. Imaging was performed 24 h after transfection. To label the surface Kv2.1-loopBAD, cells were incubated with a 1:1,000 dilution of CF640-conjugated streptavidin (CF640-SA) (Biotium) for 10 min in HEK physiological imaging saline [146 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 0.6 mM MgSO4, 1.6 mM NaHCO3, 0.15 mM NaH2PO4, 0.1 mM ascorbic acid, 8 mM glucose, and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 7.4]. Unbound CF640-SA was removed with imaging saline washes. CD4 chimeras on the cell surface were specifically detected by incubating the transfected cells with a 1:1,000 dilution of CF640-conjugated anti-CD4 antibody targeting an extracellular epitope for 10 min.
Hippocampal neurons were isolated from embryonic (E18) animals deeply anesthetized using isoflurane in accordance with a protocol approved by the Institutional Animal Care and Use Committee of Colorado State University (protocol ID: 15-6130A). Embryos of both sexes were collected and thus the neuronal cultures contain a mixed population of male and female cells. Neurons were dissociated and cultured as previously described (13, 60, 61). On DIV7 rat hippocampal neuron dishes were washed with neuronal imaging saline (NIS) (126 mM NaCl, 4.7 mM KCl, 2.5 CaCl2, 0.6 mM MgSO4, 0.15 mM NaH2PO4, 0.1 mM ascorbic acid, 8 mM glucose, and 20 mM Hepes, pH 7.4), followed by incubation with either CF640-conjugated streptavidin or anti-CD4 antibody as described above. Anti-neurofascin monoclonal antibodies were used to identify the axon initial segment when required. Here anti-NF186 antibody (Neuromab) was used at a 1:1,000 dilution for 10 min, followed by two rinses with NIS and a 10-min incubation with fluorescent (Alexa 594 or 647) goat anti-mouse secondary antibody diluted 1:1,000. Dishes were rinsed three times and imaged immediately.
APEX-Based Proximity Biotinylation.
Promiscuous biotinylation of ER/PM junction components was accomplished by transfecting HEK cells with AMIGO-YFP-APEX, in reality APEX2 (62), with or without other plasmids as indicated. At 24 h posttransfection the cells were incubated with 500 µM biotin phenol in DMEM + 10% FBS for 30 min, treated with 1 mM H2O2 for 1 min, and then fixed in 4% formaldehyde for 15 min before labeling with 2 ng/mL CF640-conjugated streptavidin (CF640-SA) and subsequent imaging. For Western blot analysis unfixed cells were scraped from plates in PBS containing Complete Mini protease inhibitor (Roche) and centrifuged, and the cell pellet was resuspended in SDS gel Laemmli sample buffer (Biorad) containing β-mercaptoethanol. Following sonication and boiling for 10 min the nonpurified samples were fractionated by standard SDS-polyacrylamide gel electrophoresis on a 10% gel. After transfer to nitrocellulose membranes, the proteins were probed with either mouse anti-Kv2.1 (Neuromab) at a 1:1,000 dilution or mouse anti-AMIGO antibody (Neuromab) at 1:1,000 dilution. Antibody binding was detected with LiCor IRDye800CW goat anti-mouse antibody. Biotinylated proteins were detected with LiCor IRDye 680RD streptavidin used at 1:10,000. Imaging was performed using a dual-color Odyssey CLx LiCor system. In this manner any biotinylated ER-resident proteins were visually separated from any potential Kv2.1 or AMIGO protein degradation products.
Microscopy.
Laser scanning and spinning-disk microscopy were employed in addition to TIRF imaging. Unless noted otherwise in the figure legends the spinning-disk system was used. Additional details regarding microscopy are presented in SI Appendix, Supplementary Materials and Methods and our previously published studies (13, 60, 61).
FRET.
Sensitized-emission FRET imaged in living cells employed Clover-Ruby2 pairs analyzed as described by ref. 63. HEK 293 cells were transfected using Lipofectamine 2000 (2 µL; Invitrogen) and 100 µL OptiMEM (Life Technologies) per dish, using the following DNAs: 1 µg Clover-Kv2.1, 1 µg Ruby2-Kv2.1, 200 ng pcDNA3.1-Clover-Ruby2 (tandem), 200 ng mClover-C1, 200 ng mRuby2-C1, 600 ng Ruby2-VAPA, 600 ng Ruby2-VAPAmut, and 600 ng Ruby2-VAPB. FRET images were obtained on the Olympus/Andor spinning-disk confocal microscope. For each cell, four images were collected: (i) excitation with 488 nm paired with a 500/25 bandpass filter (Donor image), (ii) excitation with 488 nm paired with a 600/50 bandpass filter (FRET image), (iii) excitation with 561 nm paired with a 600/50 bandpass filter (Acceptor image), and (iv) a DIC image. Using ImageJ, 15 3-pixel by 3-pixel regions of interest (ROIs) were placed on Kv2.1 clusters (or randomly in the case of tandem and soluble conditions) and the fluorescence intensity of each channel was measured. Cells expressing only the donor (Clover) or acceptor (Ruby2) constructs alone were imaged to calculate bleed-through coefficients for the FRET efficiency calculations. Bleed-through coefficients (BTClover or BTRuby2) were calculated as the average intensity of the FRET channel (IFRET) divided by the average intensity of the donor or acceptor (IClover or IRuby2). For our experimental conditions, BTClover = 11% and BTRuby2 = 4.3%. Although there are many options for the calculation of FRET, we decided to use NFRET, due to its correction for expression levels of donor and acceptor and its utility in the study of intermolecular protein interactions (64). To calculate FRET (NFRET), the following relationship was used as previously described (63):
FRET efficiency images were created using the image calculator in ImageJ and applying mathematical transformations to FRET and donor and acceptor images as described in the equation above. After all transformations were performed, the royal lookup table (LUT) in ImageJ was applied to each image.
siRNA-Based Knockdown of VAPA and VAPB.
HEK 293 cells plated in either 100-mm tissue culture dishes or 35-mm coverslip dishes were transfected with 250 nM siRNA (Dharmacon), using DharmaFECT transfection reagent per the manufacturer’s directions. After 24 h the cells on the 35-mm coverslip dishes were transfected again with the siRNA and with the GFP-Kv2.1-loopBAD and BirA plasmids, 1 µg and 0.5 μg, respectively. The cells on the 100-mm dishes did not receive the Kv2.1-encoding plasmid DNA during the second round of siRNA transfection. After another 24 h the cells on 35-mm coverslip dishes were imaged to assess Kv2.1 clustering while the cells on 100-mm plates were collected as described above for Western blot analysis of VAP expression. Incubation with anti-VAPA and -VAPB mouse antibodies (1:1,000 and 1:2,000 dilutions, MAB5820 and MAB58551, respectively; R&D Systems) followed by HRP-conjugated goat anti-mouse antibody and detection with SuperSignal West Dura (product 34075; Thermo Scientific) were used to assess VAP expression in the presence of various siRNAs.
Image Processing and Analysis.
Image processing was performed with ImageJ. Images were pseudocolored, cropped, and adjusted for contrast and brightness. Image analysis was completed using either ImageJ or Volocity analysis software. Details specific to each experiment are presented in Results or figure legends. Unless noted otherwise, single confocal planes are presented.
Experimental Design and Statistical Analysis.
The majority of our experiments were performed in HEK 293 cells as opposed to cultured hippocampal neurons since these cells are well suited for demonstrating protein–protein interactions that occur only within specific compartments of living cells. In addition, HEK 293 cells lack ion channel subunits that could assemble with the expressed constructs and are less heterogeneous than neuronal cultures. Our primary concern with respect to experimental design focused on overexpression issues, for high-level expression could induce protein interactions that are otherwise nonexistent. The levels of Kv2.1 expressed in both transfected HEK 293 cells and neurons are similar to the level of the endogenous Kv2.1 as previously described (7, 9, 59) where immunostaining of transfected cells was compared with that against the endogenous channel in cultured hippocampal neurons. The CD4-chimera expression levels were similar to those of the endogenous channels. We did not use higher expression levels to avoid potential artifacts and our goal was to express just enough fluorescent protein-tagged VAP to act as a tracer for the endogenous proteins, especially since any VAP redistribution is lost with overexpression. In addition, VAP overexpression alters ER morphology as previously described (33), something we did not observe with our transfected VAP expression levels. Note that high expression levels were not necessary given the sensitivity of our TIRF and spinning-disk microscopes, which are both capable of imaging single molecules.
For statistical analysis, one-way analysis of variance (ANOVA) with ad hoc Tukey’s tests were performed using Origin 2018b software. For the data presented in Fig. 8, Kruskal–Wallis ANOVAs were performed. Number of ROIs and cells examined and P values are indicated in Results.
Supplementary Material
Acknowledgments
We thank Emily Eden (University College London) and Diego Krapf (Colorado State University) for helpful discussions and Ann Hess (Graybill Statistical Laboratory, Colorado State University) for help with statistical analysis. This work was supported by National Institutes of Health Grant R01GM109888 (to M.M.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7849.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805757115/-/DCSupplemental.
References
- 1.Bishop HI, et al. Distinct cell- and layer-specific expression patterns and independent regulation of Kv2 channel subtypes in cortical pyramidal neurons. J Neurosci. 2015;35:14922–14942. doi: 10.1523/JNEUROSCI.1897-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarmiere PD, Weigle CM, Tamkun MM. The Kv2.1 K+ channel targets to the axon initial segment of hippocampal and cortical neurons in culture and in situ. BMC Neurosci. 2008;9:112. doi: 10.1186/1471-2202-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misonou H, et al. Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci. 2004;7:711–718. doi: 10.1038/nn1260. [DOI] [PubMed] [Google Scholar]
- 4.Romer SH, Deardorff AS, Fyffe RE. Activity-dependent redistribution of Kv2.1 ion channels on rat spinal motoneurons. Physiol Rep. 2016;4:e13039. doi: 10.14814/phy2.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surmeier DJ, Foehring R. A mechanism for homeostatic plasticity. Nat Neurosci. 2004;7:691–692. doi: 10.1038/nn0704-691. [DOI] [PubMed] [Google Scholar]
- 6.Baver SB, O’Connell KMS. The C-terminus of neuronal Kv2.1 channels is required for channel localization and targeting but not for NMDA-receptor-mediated regulation of channel function. Neuroscience. 2012;217:56–66. doi: 10.1016/j.neuroscience.2012.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox PD, Loftus RJ, Tamkun MM. Regulation of Kv2.1 K(+) conductance by cell surface channel density. J Neurosci. 2013;33:1259–1270. doi: 10.1523/JNEUROSCI.3008-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Connell KMS, Loftus R, Tamkun MM. Localization-dependent activity of the Kv2.1 delayed-rectifier K+ channel. Proc Natl Acad Sci USA. 2010;107:12351–12356. doi: 10.1073/pnas.1003028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutsch E, et al. Kv2.1 cell surface clusters are insertion platforms for ion channel delivery to the plasma membrane. Mol Biol Cell. 2012;23:2917–2929. doi: 10.1091/mbc.E12-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinshreiber L, et al. Non-conducting function of the Kv2.1 channel enables it to recruit vesicles for release in neuroendocrine and nerve cells. J Cell Sci. 2010;123:1940–1947. doi: 10.1242/jcs.063719. [DOI] [PubMed] [Google Scholar]
- 11.Fu J, et al. Kv2.1 clustering contributes to insulin exocytosis and rescues human β-cell dysfunction. Diabetes. 2017;66:1890–1900. doi: 10.2337/db16-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greitzer-Antes D, et al. Kv2.1 clusters on β-cell plasma membrane act as reservoirs that replenish pools of newcomer insulin granule through their interaction with syntaxin-3. J Biol Chem. 2018;293:6893–6904. doi: 10.1074/jbc.RA118.002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox PD, et al. Induction of stable ER-plasma-membrane junctions by Kv2.1 potassium channels. J Cell Sci. 2015;128:2096–2105. doi: 10.1242/jcs.166009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saheki Y, De Camilli P. Endoplasmic reticulum-plasma membrane contact sites. Annu Rev Biochem. 2017;86:659–684. doi: 10.1146/annurev-biochem-061516-044932. [DOI] [PubMed] [Google Scholar]
- 15.Irie T, Trussell LO. Double-nanodomain coupling of calcium channels, ryanodine receptors, and BK channels controls the generation of burst firing. Neuron. 2017;96:856–870.e4. doi: 10.1016/j.neuron.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickson EJ, et al. Dynamic formation of ER-PM junctions presents a lipid phosphatase to regulate phosphoinositides. J Cell Biol. 2016;213:33–48. doi: 10.1083/jcb.201508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, et al. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci USA. 2017;114:E4859–E4867. doi: 10.1073/pnas.1701078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skehel PA, Martin KC, Kandel ER, Bartsch D. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science. 1995;269:1580–1583. doi: 10.1126/science.7667638. [DOI] [PubMed] [Google Scholar]
- 19.Huttlin EL, et al. The BioPlex network: A systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy SE, Levine TP. VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Chattopadhyay D, Sengupta S. First evidence of pathogenicity of V234I mutation of hVAPB found in amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2014;448:108–113. doi: 10.1016/j.bbrc.2014.04.102. [DOI] [PubMed] [Google Scholar]
- 22.Kabashi E, et al. Investigating the contribution of VAPB/ALS8 loss of function in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22:2350–2360. doi: 10.1093/hmg/ddt080. [DOI] [PubMed] [Google Scholar]
- 23.Muennich EA, Fyffe RE. Focal aggregation of voltage-gated, Kv2.1 subunit-containing, potassium channels at synaptic sites in rat spinal motoneurones. J Physiol. 2004;554:673–685. doi: 10.1113/jphysiol.2003.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung JJ, Li M. Biochemical characterization of the native Kv2.1 potassium channel. FEBS J. 2005;272:3743–3755. doi: 10.1111/j.1742-4658.2005.04802.x. [DOI] [PubMed] [Google Scholar]
- 25.O’Connell KM, Tamkun MM. Targeting of voltage-gated potassium channel isoforms to distinct cell surface microdomains. J Cell Sci. 2005;118:2155–2166. doi: 10.1242/jcs.02348. [DOI] [PubMed] [Google Scholar]
- 26.Scannevin RH, Murakoshi H, Rhodes KJ, Trimmer JS. Identification of a cytoplasmic domain important in the polarized expression and clustering of the Kv2.1 K+ channel. J Cell Biol. 1996;135:1619–1632. doi: 10.1083/jcb.135.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 28.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim ST, Antonucci DE, Scannevin RH, Trimmer JS. A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron. 2000;25:385–397. doi: 10.1016/s0896-6273(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 30.Bishop HI, et al. Kv2 ion channels determine the expression and localization of the associated AMIGO-1 cell adhesion molecule in adult brain neurons. Front Mol Neurosci. 2018;11:1. doi: 10.3389/fnmol.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peltola MA, Kuja-Panula J, Lauri SE, Taira T, Rauvala H. AMIGO is an auxiliary subunit of the Kv2.1 potassium channel. EMBO Rep. 2011;12:1293–1299. doi: 10.1038/embor.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garbino A, et al. Molecular evolution of the junctophilin gene family. Physiol Genomics. 2009;37:175–186. doi: 10.1152/physiolgenomics.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser SE, et al. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Leal SS, Ben Halevy D, Gomes CM, Lev S. Structural requirements for VAP-B oligomerization and their implication in amyotrophic lateral sclerosis-associated VAP-B(P56S) neurotoxicity. J Biol Chem. 2010;285:13839–13849. doi: 10.1074/jbc.M109.097345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamkun MM, O’connell KM, Rolig AS. A cytoskeletal-based perimeter fence selectively corrals a sub-population of cell surface Kv2.1 channels. J Cell Sci. 2007;120:2413–2423. doi: 10.1242/jcs.007351. [DOI] [PubMed] [Google Scholar]
- 36.Várnai P, Tóth B, Tóth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, Kwong PD, Hendrickson WA. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 38.Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: From cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Misonou H, et al. Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J Neurosci. 2006;26:13505–13514. doi: 10.1523/JNEUROSCI.3970-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misonou H, Mohapatra DP, Menegola M, Trimmer JS. Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J Neurosci. 2005;25:11184–11193. doi: 10.1523/JNEUROSCI.3370-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park KS, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 2006;313:976–979. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- 42.Johnston J, et al. Initial segment Kv2.2 channels mediate a slow delayed rectifier and maintain high frequency action potential firing in medial nucleus of the trapezoid body neurons. J Physiol. 2008;586:3493–3509. doi: 10.1113/jphysiol.2008.153734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sánchez-Ponce D, DeFelipe J, Garrido JJ, Muñoz A. Developmental expression of Kv potassium channels at the axon initial segment of cultured hippocampal neurons. PLoS One. 2012;7:e48557. doi: 10.1371/journal.pone.0048557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen CS, et al. Trafficking of Kv2.1 channels to the axon initial segment by a novel nonconventional secretory pathway. J Neurosci. 2017;37:11523–11536. doi: 10.1523/JNEUROSCI.3510-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 46.Mohapatra DP, Trimmer JS. The Kv2.1 C terminus can autonomously transfer Kv2.1-like phosphorylation-dependent localization, voltage-dependent gating, and muscarinic modulation to diverse Kv channels. J Neurosci. 2006;26:685–695. doi: 10.1523/JNEUROSCI.4620-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teuling E, et al. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci. 2007;27:9801–9815. doi: 10.1523/JNEUROSCI.2661-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J. A guide to fluorescent protein FRET pairs. Sensors. 2016;16:E1488. doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fox PD, et al. Plasma membrane domains enriched in cortical endoplasmic reticulum function as membrane protein trafficking hubs. Mol Biol Cell. 2013;24:2703–2713. doi: 10.1091/mbc.E12-12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cobb MM, Austin DC, Sack JT, Trimmer JS. Cell cycle-dependent changes in localization and phosphorylation of the plasma membrane Kv2.1 K+ channel impact endoplasmic reticulum membrane contact sites in COS-1 cells. J Biol Chem. 2015;290:29189–29201. doi: 10.1074/jbc.M115.690198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerda O, Trimmer JS. Activity-dependent phosphorylation of neuronal Kv2.1 potassium channels by CDK5. J Biol Chem. 2011;286:28738–28748. doi: 10.1074/jbc.M111.251942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redman PT, Hartnett KA, Aras MA, Levitan ES, Aizenman E. Regulation of apoptotic potassium currents by coordinated zinc-dependent signalling. J Physiol. 2009;587:4393–4404. doi: 10.1113/jphysiol.2009.176321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Redman PT, et al. Apoptotic surge of potassium currents is mediated by p38 phosphorylation of Kv2.1. Proc Natl Acad Sci USA. 2007;104:3568–3573. doi: 10.1073/pnas.0610159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer-Lahat D, et al. K+ channel facilitation of exocytosis by dynamic interaction with syntaxin. J Neurosci. 2007;27:1651–1658. doi: 10.1523/JNEUROSCI.4006-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King AN, Manning CF, Trimmer JS. A unique ion channel clustering domain on the axon initial segment of mammalian neurons. J Comp Neurol. 2014;522:2594–2608. doi: 10.1002/cne.23551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiffault I, et al. A novel epileptic encephalopathy mutation in KCNB1 disrupts Kv2.1 ion selectivity, expression, and localization. J Gen Physiol. 2015;146:399–410. doi: 10.1085/jgp.201511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torkamani A, et al. De novo KCNB1 mutations in epileptic encephalopathy. Ann Neurol. 2014;76:529–540. doi: 10.1002/ana.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Kovel CGF, et al. Neurodevelopmental disorders caused by de novo variants in KCNB1 genotypes and phenotypes. JAMA Neurol. 2017;74:1228–1236. doi: 10.1001/jamaneurol.2017.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connell KM, Rolig AS, Whitesell JD, Tamkun MM. Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J Neurosci. 2006;26:9609–9618. doi: 10.1523/JNEUROSCI.1825-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akin EJ, Solé L, Dib-Hajj SD, Waxman SG, Tamkun MM. Preferential targeting of Nav1.6 voltage-gated Na+ channels to the axon initial segment during development. PLoS One. 2015;10:e0124397. doi: 10.1371/journal.pone.0124397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akin EJ, et al. Single-molecule imaging of Nav1.6 on the surface of hippocampal neurons reveals somatic nanoclusters. Biophys J. 2016;111:1235–1247. doi: 10.1016/j.bpj.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hung V, et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoppe A, Christensen K, Swanson JA. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys J. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.