Abstract
Aim:
To investigate the relationship between GRHL3 methylation and the etiology of neural tube defects (NTDs).
Materials & methods:
Analyze data from a genome-wide DNA methylation array. Targeted DNA methylation analysis was performed for 46 cases and 23 controls. At last, grhl3 overexpression and gene depletion experiments were conducted in zebrafish.
Results:
Five hypomethylated CpGs were discovered in the methylation arrays performed on NTD cases. In a validation study, 15 hypomethylated CpGs were found and the overall methylation levels decreased in brain/spinal cord tissue from NTD cases. The knockdown and overexpression of grhl3 in zebrafish damaged embryonic convergent extension processes.
Conclusion:
Hypomethylation of GRHL3 in central nervous tissue is associated with NTDs, further supporting the importance of GRHL3 and methylation in proper neural tube closure.
Keywords: : GRHL3 gene, methylation, neural tube defects, PCP pathway
Neural tube defects (NTDs) are a group of severe malformations that arise following closure failure of the neural tube during early embryonic development, and affect more than 300,000 newborns every year globally [1]. Genetic factors are believed to play key roles in the occurrence of NTDs [2]. More than 300 genes have been identified as NTD-related mutations in mouse models [2,3]. While modest positive associations between gene variants such as the 677C >T of MTHFR gene and an increased risk for NTDs in humans, there are few other confirmed gene variation associations in humans [4]. It is highly likely that there may exist other underlying mechanisms, including epigenetic modifications, that are involved in the etiology of NTDs.
DNA methylation is an epigenetic mechanism that has been shown to play an important role in the normal closure of the neural tube. Folate is a critical component in the one-carbon metabolic pathway by providing methyl groups for a variety of biochemical reactions, including methylation of DNA [5]. Several studies have shown that supplementation with folic acid may prevent NTDs by stimulating DNA methylation [6,7]. Additional investigations have demonstrated that the MTHFR 677C >T polymorphism could influence global DNA methylation status through an interaction with folic acid [8,9]. Recently, our group performed a genome-wide DNA methylation array and found that hypermethylation of the CTNNA1 and MYH2 genes in the tight junction pathway may be associated with an increased risk of NTDs [10]. The NTD-related aberrant methylation of other genes has also been reported, including imprinted genes [11,12], transposon genes [13], DNA repair genes [14], folate receptor genes [15] and HOX genes [16].
The planar cell polarity (PCP) signaling pathway plays a key role in the closure of neural tube, and variants of PCP pathway genes have previously been reported to cause NTDs [17,18]. Epigenetic modification of these genes may also play a role in the formation of NTDs by perturbing their expression. One such study reported that aberration of DNA methylation, rather than mutations of FZD3 gene (one of PCP core genes), increased the risk of spinal bifida [19]. Given the important and well-established roles of PCP pathway genes in the proper closure of neural tube, it is possible that aberrant methylation of other PCP genes may also be involved and worthy of further investigation. GRHL3 gene is an important PCP pathway gene that is involved in establishing dorsal/ventral patterning and subsequent organ development in early stages of mouse embryogenesis [20]. Inactiavation of this gene was produced NTDs in the Grhl3 knockout mice [21] and in the curly tail (Ct) mouse model [22]. Recently, Castro et al. reported that the conditional Grhl3 knockout in the neural plate and node-streak border has minimal effect on closure, suggesting that abnormal functioning of the surface ectoderm is primarily responsible for early failure of spinal neurulation in Grhl3 null embryos [23]. Lemay et al. described eight deleterious variants in GRHL3 in human spina bifida cases [24], demonstrating the importance of this gene in the development of human NTDs. Therefore, an investigation into potential aberrant methylation of the in GRHL3 gene in NTD cases may enhance our understanding of the underlying mechanisms leading to the development of NTDs.
We hypothesized that aberrant DNA methylation of GRHL3 gene is related to the formation of NTDs. To test this hypothesis, we initially examined the methylation changes in the GRHL3 gene in the array data of ten NTD cases and eight controls. We subsequently performed a locus-specific methylation validation study in a larger sample using the MassARRAY platform. At last, we conducted overexpression and morpholino (MO) experiments using a zebrafish model to study the function of ghrl3 gene in the development of the neural tube.
Materials & methods
Participants & samples
The subjects of this study were recruited from five counties in Shanxi Province, China (Pingding, Zezhou, Xiyang, Shouyang and Taigu) from 2011 to 2014, a province long known to have a high prevalence of NTDs [25]. Cases were electively terminated following prenatal diagnosis of an NTD; subjects with defects in other system were excluded. Controls were terminated fetuses with no gross congenital malformations. For anencephalic cases, the residual brain tissues were harvested, while for spina bifida cases, tissues were collected from the anatomical lesion site of the spinal cord. A normal brain or spinal cord tissue sample from nonmalformed control fetuses were also collected at pregnancy termination by experienced pathologists. All samples were stored at -80°C until utilized for various analyses. This study was approved by the institutional review board of Peking University and all of the participating mothers provided written informed consent.
The human study involved two phases. At the discovery phase, pathological brain/spinal cord samples from ten NTD cases (six anencephaly, four spinal bifida) and brain/spinal cord tissue from eight nonmalformed controls recruited between 2011 and 2012 were used in a genome-wide methylation analysis [10]. At the validation phase, 46 NTD cases (20 anencephaly, 26 spinal bifida) and 23 controls which were 2:1 pair matched by gender and gestational weeks and recruited during 2013–2014 were included. Pathological or normal brain/spinal cord tissue samples were used to examine locus-specific methylation differences within GRHL3 gene.
DNA methylation analysis using the Illumina 450 K BeadChip array
Fetal tissue DNA was extracted by using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). The concentration of DNA was measured by using a NanoDrop2000 Ultramicro spectrophotometer (Thermo Fisher Scientific, MA, USA). All of the DNA samples were stored at -80°C until used for specific assays.
Infinium Human Methylation 450 (HM450K) Bead Chip assay (Illumina, CA, USA) was performed to analyze genome-wide DNA methylation level in neural tissues from both cases and controls, as previously described in more detail elsewhere [10]. Briefly, bisulfite conversion of the DNA (500 ng) was performed using the EZ DNA methylation kit (Zymo Research, CA, USA). The bisulfite conversion reaction was performed in duplicate for each sample to minimize potential bias caused by variable conversion efficiency, and pooled bisulfite treated DNA was used for subsequent array analysis. Arrays were processed according to the manufacturer's protocol.
Cluster analysis and component analysis with the methylation array data showed that the NTD case group and the nonmalformed group could be distinguished from each other [10]. The data for the GRHL3 gene was extracted from the genome-wide array.
Methylation of CpGs using the Sequenom EpiTYPER
Target verification of differentially methylated CpG islands in human gDNA was performed using the Sequenom EpiTYPER (Sequenom, CA, USA). Bisulfite conversion of 1 mg DNA was conducted using an EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's instructions. In this step, the unmethylated cytosine is converted into uracil while the methylated cytosine remains unchanged.
The bisulfite treated DNA was amplified by PCR experiment. The PCR primers of GRHL3 gene were designed by the online tool Epidesigner (www.epidesigner.com). Five CpG sites whose methylation intensity was significantly different between NTD cases and controls, were located far away from each other and one amplicon could not cover all five sites. As such, we designed two amplicons to target the two aberrant CpG sites of which differences between cases and controls were higher than 0.2 in HM450K array. Additionally, considering the important role of the CpG island, which is considered to be a region with a high frequency of CpG sites, we designed another two amplicons to target the CpG island of GRHL3 gene in the validation study. The primer sequence was presented in Supplementary Table 1. The reaction volume of PCR was 5 μl. The PCR reaction program was as follows: 94°C for 4 min; 45 cycles of (95°C for 20 s; 56°C for 30 s; 72°C for 60 s); 72°C for 3 min. The PCR product was incubated using shrimp alkaline phosphatase for 20 min at 37°C and then purified at a temperature of 85°C for 5 min. Enzyme digestion was performed using T cleavage enzyme (T Cleavage Mix) at 37°C for 3 h and the product was desalinized using 384 dimple plates. Subsequently, the methylation of the DNA was analyzed by the MassARRAY system. The results were analyzed using EpiTYPER software. To control for the quality of the methylation assay, we used both positive controls (completely methylation) and negative controls (nonmethylation) in our analyses.
grhl3 overexpression & depletion in zebrafish
Tübingen (Tu) strain zebrafish were raised according to standard protocols. The GRHL3 gene is highly conservative between humans and zebrafish. In this study, we used human mRNA to conduct overexpression experiments. To produce GRHL3 mRNA, the ORF cDNA of GRHL3 was cloned into the pCMV6-Entry vector (Origene Technologies, Beijing, China). The cDNA was transfected into Escherichia coli and the plasmid containing the GRHL3 cDNA was extracted and enzyme digested. At last, the capped and polyadenylated GRHL3 mRNA was produced by in vitro transcription using mMESSAGE mMACHINE™ T7 Transcription Kit (Ambion, TX, USA). The synthesized mRNA was diluted in phenol red to different concentrations for microinjection. The MO reagent with an ATG-blocking (initiation codon) grhl3 sequence (5′-TGAGAGCCTCAATCTCCTTGGTCAT-3′) was obtained from GeneTools LLC, Co. (OR, USA) [26].
Different doses of MO reagent (6–14 ng) and mRNA (100–300 pg) were microinjected into each zebrafish embryo at its one-cell stage. Noninjected embryos were used as controls. All embryos were observed using fluorescent stereo microscopy at 24 and 48 h postfertilization (hpf). Images of each phenotype were captured by using the charge coupled device (CCD) image system under bright field. All the embryos were divided into four categories according to the severity of the malformations. All involved embryos were independently observed and counted by two blinded researchers to reduce subjective error.
Statistical analysis
In the discovery stage, potential sources of technical bias were excluded prior to further analysis. Probes were excluded from further analysis if >95% of the samples had a detection value >0.01 [16]. Illumina Genome Studio software (Illumina, CA, USA) was used to extract signal intensities for each probe, perform the initial control quality checks and estimate β-scores and detection p-values. β-score was defined as the proportion of total signal from the methylation-specific probe or color channel. Detection p-value was defined as the 1-p-value computed from the background model characterizing the chance that the target sequence signal was distinguishable from the negative controls. Independent t-tests were used to identify differentially methylated CpG sites. The p-value was adjusted for multiple testing using the Benjamini–Hochberg false discovery rate (FDR) methods to control for the false discovery rate. Differentially methylated CpG sites were identified by the following criteria: the false discovery rate p < 0.05; the absolute β-value difference >0.05 [16]. Some more detailed data analysis was shown in the Supplementary Methods.
In the replication stage, the differences in proportions of population characteristics between groups were examined with Pearson's χ2 test. The distribution of methylation values of cases and controls were tested by Shapiro–Wilk test. Paired t-test was used to identify CpGs that were differentially methylated between the NTD cases and controls. Binary logistic regression was performed to investigate the relationship between methylation levels of GRHL3 with the risk for NTDs. In order to make it better to explain the regression coefficient, we take the negative value of the methylation data into the model, and whether or not taking folic acid was further adjusted in the regression model. Maternal folate supplementation (yes vs no) was considered a potential confounding factor because folate is a one-carbon unit donor in the methylation pathway. In the zebrafish study, Pearson's χ2 test and the Fisher test were used to analyze the distribution of malformed zebrafish. A two-tailed p-value of <0.05 was considered statistically significant. SPSS 20.0 software (IBM Co., NY, USA) was used for analyzing the result in this study.
Results
DNA methylation analysis of GRHL3 gene detected by microarray: discovery
Methylation data for CpGs in GRHL3 gene in the discovery stage were extracted from the dataset obtained from HM450K array-based genome-wide methylation analysis by using DNA isolated from central nervous tissue from ten NTD cases and eight unrelated nonmalformed controls. No significant differences were found in gestational age and gender distribution between cases and controls [10].
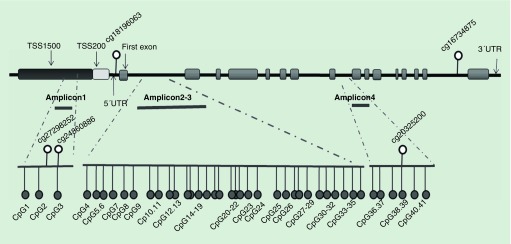
As shown in Table 1, a total of 28 CpGs on the GRHL3 gene were detected by the HM450K array. Of these, 17 CpGs were found to be hypomethylated in NTD fetuses, and five of these 17 CpGs (cg24860886, cg27298252, cg18196063, cg20325200 and cg16734875) were significantly hypomethylated in NTD cases (β-difference of -0.341, -0.054, -0.080, -0.222 and -0.060, respectively). The location of the five significantly aberrant CpGs is shown in Figure 1. Of these five CpGs, two CpG probes (cg24860886 and cg27298252) were located in the TSS1500 region; one CpG probe (cg18196063) was located at 5′UTR; the remaining two CpG probes (cg20325200 and cg16734875) were located within the body of the GRHL3 gene.
Table 1. . Methylation of GRHL3 gene using the Human Methylation 450 Bead Chip assay.
| Target ID | Chr | Mapinfo | Case | Control | Diff | ad-p | Region |
|---|---|---|---|---|---|---|---|
| cg24860886 | 1 | 24645397 | 0.205 | 0.546 | -0.341 | 0.005† | TSS1500 |
| cg27298252 | 1 | 24645380 | 0.876 | 0.930 | -0.054 | 0.048† | TSS1500 |
| cg06376426 | 1 | 24648203 | 0.675 | 0.720 | -0.045 | 0.285 | TSS1500 |
| cg21273275 | 1 | 24648328 | 0.367 | 0.283 | 0.084 | 0.310 | TSS1500 |
| cg09322349 | 1 | 24645184 | 0.532 | 0.601 | -0.069 | 0.438 | TSS1500 |
| cg22176324 | 1 | 24648851 | 0.072 | 0.069 | 0.003 | 0.722 | TSS1500 |
| cg05036846 | 1 | 24648984 | 0.073 | 0.072 | 0.001 | 0.911 | TSS1500 |
| cg14616251 | 1 | 24648696 | 0.076 | 0.076 | 0.000 | 0.995 | TSS1500 |
| cg13987674 | 1 | 24649494 | 0.854 | 0.913 | -0.059 | 0.106 | TSS200 |
| cg09662754 | 1 | 24645805 | 0.081 | 0.099 | -0.018 | 0.188 | TSS200 |
| cg12757705 | 1 | 24645802 | 0.051 | 0.057 | -0.006 | 0.464 | TSS200 |
| cg11709405 | 1 | 24649398 | 0.394 | 0.448 | -0.055 | 0.489 | TSS200 |
| cg15050111 | 1 | 24645850 | 0.075 | 0.073 | 0.002 | 0.817 | TSS200 |
| cg20242797 | 1 | 24645853 | 0.114 | 0.118 | -0.005 | 0.852 | TSS200 |
| cg18196063 | 1 | 24652248 | 0.811 | 0.891 | -0.080 | 0.012† | 5′UTR |
| cg26365938 | 1 | 24645953 | 0.077 | 0.089 | -0.012 | 0.074 | 5′UTR |
| cg21289280 | 1 | 24645885 | 0.049 | 0.058 | -0.010 | 0.166 | 5′UTR |
| cg12405265 | 1 | 24655961 | 0.843 | 0.833 | 0.010 | 0.678 | 5′UTR |
| cg04552500 | 1 | 24645917 | 0.099 | 0.095 | 0.004 | 0.711 | 5′UTR |
| cg23820945 | 1 | 24646022 | 0.145 | 0.153 | -0.008 | 0.906 | 5′UTR |
| cg14167629 | 1 | 24646046 | 0.117 | 0.105 | 0.011 | 0.659 | First exon |
| cg20325200 | 1 | 24667622 | 0.590 | 0.812 | -0.222 | 0.000† | Body |
| cg16734875 | 1 | 24678010 | 0.883 | 0.943 | -0.060 | 0.011† | Body |
| cg19697558 | 1 | 24646085 | 0.070 | 0.068 | 0.002 | 0.916 | Body |
| cg16155382 | 1 | 24646191 | 0.129 | 0.101 | 0.028 | 0.373 | Body |
| cg01119512 | 1 | 24646205 | 0.230 | 0.201 | 0.029 | 0.408 | Body |
| cg02984188 | 1 | 24646392 | 0.364 | 0.482 | -0.118 | 0.240 | Body |
| cg11155431 | 1 | 24664451 | 0.886 | 0.890 | -0.004 | 0.830 | Body |
Target ID is identified according to HM450k. The nucleotide position is based on NCBI build 37/hg19. Region is defined relative to the nearest open reading frame: within 1500 (TSS1500) or 200 bp (TSS200) of a transcription start site, in the 5′ UTR, the first exon of a transcript (exon) and in the body of gene (body). Differentially methylated CpG sites were identified by two criterions: the false discovery rate <0.05, which was analyzed by independent t-tests with multiple comparison tests; the absolute β-value difference >0.0516.
†The CpG site was aberrant methylated in case group.
ad-p: Adjusted p-value; Chr: Chromosome; diff: Difference; Mapinfo: Nucleotide position; UTR: Untranslated region.
Figure 1. . Locations of the CpG sites detected by HM450k and Sequenom EpiTYPER.
In the validation study with the Sequenom EpiTYPER assay, four DNA amplicons were involved; amplicon 1 covered three CpG sites (CpG1 to CpG3) which were located in the TSS1500 region of the GRHL3 gene. Amplicon 2 and amplicon 3 covered the CpG island region of GRHL3 predicted using UCSC (chr1:24648202–24648985). These two amplicons included 32 analytical CpG sites (CpG4 to CpG35). Amplicon 4 covered six CpG sites (CpG36 to CpG41) within the gene body. Nucleotide positions accord to the NCBI build 37/hg19. The differentially methylated CpG units identified by 450 K Array (cg27298252, cg24860886, cg18196063, cg20325200 and cg16734875) are also indicated.
Targeted DNA methylation analysis of GRHL3 gene in a larger sample: validation
To verify the relationship between NTDs and GRHL3 hypomethylation identified in the discovery phase, 46 NTD cases and 23 gestational age and gender-matched controls were included in the validation study using the Sequenom EpiTYPER technology. The demographic characteristics of the subjects are depicted in Table 2. As shown in Figure 1, we completely amplified the four DNA amplicons. Amplicon 1 covered three CpG sites (CpG1 to CpG3), in which the CpG3 was the same as the cg24860886 CpG site identified to be differentially hypomethylated in the discovery phase. Amplicon 2 and 3 covered the CpG island of GRHL3, including 32 analytical CpG sites (CpG4 to CpG35). Amplicon 4 covered six CpG sites (CpG36 to CpG41) within the gene body, in which the CpG39 was the same as the hypomethylated CpG site of cg20325200 identified in the discovery phase.
Table 2. . Characteristics of subjects included in the analysis of Sequenom EpiTYPER.
| Characteristic | Total | Case (n = 46) | Control (n = 23) | p-value |
|---|---|---|---|---|
| Gestational week | 24.23 ± 5.50 | 24.07 ± 5.38 | 24.65 ± 5.83 | 0.679 |
| Gender: | 1.000 | |||
| – Male | 27 (39.1) | 18 (39.1) | 9 (39.1) | |
| – Female | 42 (60.9) | 28 (60.9) | 14 (60.9) | |
| Occupation: | ||||
| – Farmer | 52 (75.4) | 32 (76.2) | 20 (74.1) | 0.842 |
| – Others | 17 (24.6) | 10 (23.8) | 7 (25.9) | |
| Educational level: | ||||
| – Primary school or lower | 7 (10.1) | 4 (9.5) | 3 (11.1) | 0.810 |
| – Junior school | 44 (63.8) | 28 (66.7) | 16 (59.3) | |
| – High school or higher | 18 (26.1) | 10 (23.8) | 8 (29.6) | |
| Folate supplementation: | ||||
| – No | 37 (53.6) | 23 (54.8) | 14 (51.9) | 0.813 |
| – Yes | 32 (46.4) | 19 (45.2) | 13 (48.1) | |
Pearson's χ2 test for gender, occupation, educational level and folate supplementation. Independent samples test for gestational week.
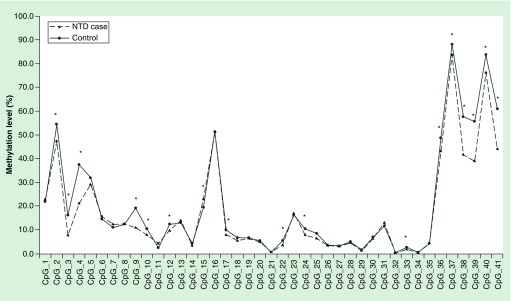
As shown in Figure 2, among the 41 detected CpG sites, 29 CpGs showed hypomethylation in NTD cases, 15 of which were significantly different between the cases and controls. In the validation phase, both the HM450K array-matched CpG sites CpG3 (cg24860886) and CpG39 (cg20325200) showed significant hypomethylation in cases with a β-difference of 7.2 and 16.7%, respectively. CpG15 showed significant hypermethylation in cases with a smaller β-difference of 3.5% (p = 0.027). After the Benjanmini–Hochberg correction, CpG38, CpG39 (cg20325200) and CpG40 still demonstrated significant hypomethylation in the NTD cases. The details are represented in Supplementary Tables 2–4.
Figure 2. . Methylation pattern of GRHL3 gene by using Sequenom EpiTYPER.
Methylation level for each CpG unit between NTD cases and control group within the amplicons were analyzed. Paired t-test was performed.
*The methylation of CpG site had significant difference between case and control group (p < 0.05).
NTD: Neural tube defect.
The average methylation level of GRHL3 gene in the case group was 16.5% (95% CI: 14.0, 19.0%), significantly lower than the control group (20.1%; 95% confidence interval [CI]: 16.9, 23.3%). The logistic regression showed that hypomethylation of GRHL3 gene was a risk factor for NTDs (ad-OR of 1.657; 95% CI: 1.279–2.146), indicating that as the average methylation level is reduced by one unit, the risk of NTDs increases by 65.7%. We further analyzed the association between different subtypes of NTDs, namely spina bifida and anencephaly, with the average methylation level of GRHL3 gene. As shown in Table 3, the average methylation levels of both subgroups were significantly lower than those of the control group and hypomethylation could increase the risk of spina bifida and anencephaly.
Table 3. . Mean methylation level of GRHL3 gene in neural tube defect cases and controls.
| Group | n | Mean methylation (%) | OR† (95% CI) | Adjusted-OR (95% CI) |
|---|---|---|---|---|
| NTDs | 46 | 16.5 (14.0,19.0) | 1.552 (1.232–1.954) | 1.657 (1.279–2.146) |
| Controls | 23 | 20.1 (16.9,23.3) | 1.000 | 1.000 |
| Spinal bifida | 26 | 15.9 (13.4,18.4) | 1.514 (1.121–2.045) | 1.562 (1.140–2.140) |
| Controls_1 | 13 | 19.3 (15.9,22.7) | 1.000 | 1.000 |
| Anencephaly | 20 | 17.4 (15.0,19.8) | 1.800 (1.165–2.780) | 2.114 (1.178–3.792) |
| Controls_2 | 10 | 21.1 (18.5,23.7) | 1.000 | 1.000 |
Using logistic regression to investigate the relationship between methylation of GRHL3 with NTDs and subtypes of NTDs.
†In order to make it better to explain the regression coefficient, we take the negative value of the methylation data into the model.
Adjusted-OR: Adjusted by folic acid supplementation; CI: Confidence interval; control_1: The control matched with spinal bifida case; control_2: The control matched with anencephaly case; NTD: Neural tube defect; OR: Odds ratio.
GRHL3 mRNA overexpression & grhl3 downregulation in zebrafish
Functional studies of the GRHL3 gene (Gene ID: 57822) were performed using the zebrafish model. Grhl3 gene (Gene ID: 794613) is well-conserved between humans and zebrafish, which indicates a high similarity of encoded amino acids between these two species [22]. We performed gene overexpression and gene knockdown experiments by microinjecting synthetic GRHL3 mRNA and a spliced grhl3 MO reagent, respectively.
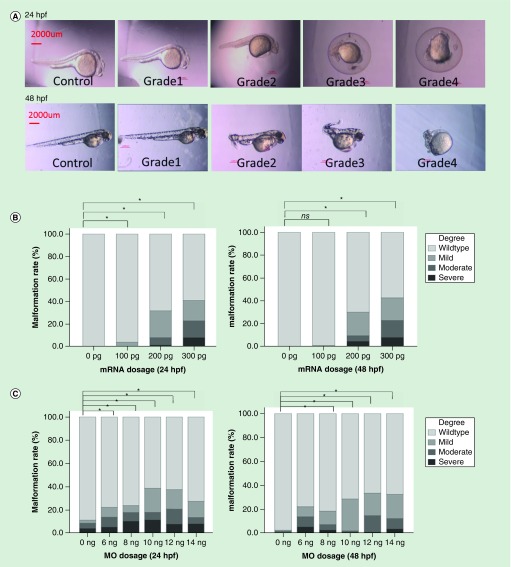
After injection of different concentrations of GRHL3 mRNA into one-cell stage embryos, the convergence extension process of the embryos was damaged at 24 and 48 hpf, including shortening and bending of the body axis. Based on the severity of the observed phenotype, we divided the embryos into four grades (grade 1: wild-type like; grade 2: mild; grade 3: moderate; grade 4: severely abnormal) (Figure 3A). The distributions of these four grades at the different concentration groups at both 24 and 48 hpf were examined. Compared with the control group, the rate of abnormal embryos was significantly higher in the groups receiving 100, 200 and 300 pg mRNA. The malformation rate was increased with increasing concentrations of mRNA at both 24 and 48 hpf (p for trend <0.05) (Figure 3B). In addition, those embryos injected with different dosages ranging from 6 to 14 ng of grhl3-MO showed higher malformation rates than the control group, and the malformed rate increased with the increasing MO dosages at both 24 and 48 hpf (Figure 3C).
Figure 3. . Phenotype analysis of GRHL3-overexpression and grhl3-MO in zebrafish embryos.
(A) Phenotype analysis of zebrafish embryos treated with GRHL3 mRNA: based on the phenotype severity, we divided the phenotypes into four grades: grade 1: no difference comparing with wild-type, no shortening of the body axis; grade 2: mild deformity, curling body axis with the length reduced about 1/4 relative to the wild-type; grade 3: moderate deformity, severe curvature and body axis shortened nearly 1/2; grade 4: severe deformity, no body axis. (B) Phenotype analysis of zebrafish embryos after microinjection with GRHL3 mRNA ranging from 100 to 300 pg. The abnormal rates of overexpressed groups were significantly higher than control group. With an increase of the mRNA, the malformation rate showed a rising tendency. (C) Phenotype analysis of MO experiment. Distributions of these four grades of the different dose groups at 24 hpf and 48 hpf were analyzed separately.
*p < 0.05 by χ2 test.
MO: Morpholino.
Discussion
Using a two-phase design and functional studies in zebrafish models, we found that hypomethylation of the GRHL3 gene may be involved in the etiology of some NTDs. In the discovery phase, we found five hypomethylated CpGs of the GRHL3 gene in NTD cases. Subsequently, we detected 15 hypomethylated CpGs in NTD cases in the validation phase with a larger case–control sample. The zebrafish experiment demonstrated that the depletion of functional grhl3 gene and the overexpression of GRHL3 mRNA could severely compromise the convergent extension process, leading to NTD-related phenotypes.
GRHL3 is essential to the formation of dorsal/ventral and subsequent organ development in early embryonic stage [20]. Most animal studies to date have focused on gene mutations including deletion or loss of function in the grhl3 gene. A previously published study has shown that MO-mediated knockdown in zebrafish of the grhl3 led to severe hypoplasia of the lower jaw cartilages [26]. All the Grhl3-/- mutant mice showed spinal bifida and curled tail, and 2% of them had a defect analogous to anencephaly. The Grhl3-/- mice also showed resistance to folate and inositol [21]. Lemay et al. recently reported eight deleterious variants in GRHL3 in human spina bifida cases [24]. In our study, we found that most CpGs of GRHL3 were hypomethylated in the NTD cases and the mean methylation level in NTDs was lower than in nonmalformed controls. Further subgroup analysis suggest that the hypomethylation of GRHL3 gene was present in both spinal bifida and anencephaly subtypes, the two major subtypes of human NTDs. Our study provides novel evidence on the role of epigenetic modifications of the GRHL3 gene in human NTDs.
DNA methylation in mammals is a postreplication modification that is predominantly found in cytosine residues of the dinucleotide sequence CpG. Methylation is now recognized to be a major contributor to the stability of gene expression, and it is essential for vertebrate development [27]. The loss of methylation may lead to apoptosis in embryos [28]. Hypomethylation may upregulate the expression of mRNA of genes which is suggestive for GRHL3 overexpression. Furthermore, in the present study, GRHL3 overexpression experiment was performed in zebrafish to examine possible biological mechanisms of GRHL3 hypomethylation underlying the development of NTDs. The zebrafish experiments showed that the overexpression of GRHL3 mRNA could lead to a disturbance of convergent extension which created different degrees of NTD-related phenotypes, including bending and curling of the body axis. These findings suggest that the hypomethylation of GRHL3 gene may cause abnormal closure of neural tube by secondary to overexpression GRHL3 mRNA in humans.
Transcription start site (TSS) controls the levels of gene expression [29]. Extensive previous work on DNA methylation focused on the CpG island at TSS, and showed that aberrant methylation in TSS plays an important role in regulation of gene expression [30]. Additionally, CpG islands within the gene body, defined as the entire gene from the TSS to the end of the transcript, were also important for the gene's function [31], and several studies reported the abnormal methylated CpG islands within gene body associated with NTDs [16,32]. In the GRHL3 gene, the CpG island is located in the gene body. To investigate the methylation level of GRHL3 gene, we not only detected the CpG sites that showed significant hypomethylation within TSS1500 region and gene body identified by microarray, but also detected the CpG island within GRHL3 gene body. We found that the CpG sites in both TSS region and gene body in GRHL3 gene showed significantly decreased methylation levels in NTD cases, implying that hypomethylation of GRHL3 gene may be a risk factor for NTDs.
CpG methylation is thought to be dynamic and tissue specific [29], so it is critically important to detect the methylation levels of DNA from pathological central nervous tissue, rather than lymphocyte, as is commonly done in the study of human NTDs. Prior to our study, only one genome-wide microarray analysis was performed to detect the aberrant methylation in NTD cases. However, DNA from blood lymphocytes was used for methylation profiling [16,32], which may have different methylation profiles compared with the more biologically relevant central nervous tissue. Our team performed genome-wide methylation microarray analysis using DNA isolated form the cells harvested close to the NTD lesion in affected fetuses, and from brain/spinal cord tissue from nonmalformed controls [10]. In the present study, the methylation experiments were all performed using specific lesion site neural tissue, which provided direct evidence of methylation changes of the target tissue/organ. The limitation of our study was that we did not investigate the expression of GRHL3 gene in NTD cases and controls because the RNA extracted from the neural tissues failed to pass the quality control to enable us to successfully perform RNAseq studies. However, in our study, we used the zebrafish model to perform the functional analyses of the GRHL3 gene, which provided us with unique insight on the association of GRHL3 mRNA expression with the development of NTDs. In addition, the view that the methylation status assayed in this study reflects to the real status when NTD happens is challenging, for methylation is an ever-changing process. Given that the nearly impossible goal of obtaining the neural tissue when neural tube is closing, which happens during 21st to 28th day postconception, the continuum of neural tube development at about 5 months of gestation makes the current study the best approximation possible.
Conclusion & future perspective
In conclusion, we are able for the first time to provide evidence that the GRHL3 gene hypomethylation in central nervous tissue is associated with the failure of neural tube closure. Our findings also provide novel insight into the mechanisms of PCP pathway genes underlying the development of NTDs. Further studies are warranted to examine how abnormal methylation of GRHL3 gene interrupts neural tube development, and investigate whether the aberrant methylation of other PCP pathway genes have causative relationships with the etiology of NTDs.
Summary points.
Although the importance of planar cell polarity (PCP) pathway genes to the occurrence of neural tube defects (NTDs) has been previously reported, very few studies have investigated whether aberrant methylation of PCP pathway genes could lead to NTDs. Our study showed that hypomethylation of GRHL3 gene, which is a PCP pathway gene, was positively associated with an increased risk for NTDs.
Most of the previous research on the association between DNA methylation and NTD risks used samples from blood lymphocytes. Considering that DNA methylation is dynamic and tissue specific, for the present study, the methylation experiments were all performed using specific lesion site neural tissue, which provided direct evidence on methylation changes of the target organ.
We screened five hypomethylated CpGs of GRHL3 gene in NTD cases by using genome-wide DNA methylation analysis, and we detected 15 hypomethylated CpGs in NTD cases in the validation phase by using targeted methylation analysis.
We found that the CpG sites in both transcription start site and gene body in GRHL3 gene showed significantly decreased methylation levels in NTD cases.
The subgroup analysis suggests that the hypomethylation of GRHL3 gene was present in both spina bifida and anencephaly subtypes, the two major subtypes of human NTDs.
We demonstrated that the overexpression of GRHL3 mRNA could lead to disturbance of convergent extension and appeared different degrees of NTD-related phenotypes in zebrafish, including bending and curling of the body axis.
Supplementary Material
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2018-0016
Footnotes
Financial & competing interests disclosure
This work was supported by grants from the National Natural Science Foundation of China (Grants No. 81472987 and 81773441, 81371264); Beijing Natural Science Foundation (Grant No. 7162094); and the National Key Research and Development Program, Ministry of Science and Technology, PR China (Grant No. 2016YFC1000501). Peking University Health Science Center Interdisciplinary Research Seed Fund (No. BMU20140446). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
This study was approved by the institutional review board of Peking University and the mothers provided written informed consent.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Copp AJ, Stanier P, Greene ND. Neural tube defects: recent advances, unsolved questions and controversies. Lancet Neurol. 2013;12(8):799–810. doi: 10.1016/S1474-4422(13)70110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A wide summary of the occurrence and development of neural tube defects (NTDs).
- 2.Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Res. Part A Clin. Mol. Teratol. 2007;79(3):187–210. doi: 10.1002/bdra.20333. [DOI] [PubMed] [Google Scholar]
- 3.Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic and environmental contributions to neural tube closure. Annu. Rev. Genet. 2014;48:583–611. doi: 10.1146/annurev-genet-120213-092208. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A review of the risk factors in the occurrence of NTDs.
- 4.Yan L, Zhao L, Long Y, et al. Association of the maternal MTHFR C677T polymorphism with susceptibility to neural tube defects in offsprings: evidence from 25 case–control studies. PLoS ONE. 2012;7(10):e41689. doi: 10.1371/journal.pone.0041689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam. Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 6.Rochtus A, Jansen K, Van Geet C, Freson K. Nutri-epigenomic studies related to neural tube defects: does folate affect neural tube closure via changes in DNA methylation? Mini Rev. Med. Chem. 2015;15(13):1095–1102. doi: 10.2174/1389557515666150909144828. [DOI] [PubMed] [Google Scholar]
- 7.Blom HJ. Folic acid, methylation and neural tube closure in humans. Birth Defects Res. Part A Clin. Mol. Teratol. 2009;85(4):295–302. doi: 10.1002/bdra.20581. [DOI] [PubMed] [Google Scholar]
- 8.Friso S, Choi SW, Girelli D, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl Acad. Sci. USA. 2002;99(8):5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lalouschek W, Aull S, Serles W, et al. The relation between erythrocyte volume and folate levels is influenced by a common mutation in the methylenetetrahydrofolate reductase (MTHFR) gene (C677T) J. Investig. Med. 2000;48(1):14–20. [PubMed] [Google Scholar]
- 10.Wang L, Lin S, Zhang J, Tian T, Jin L, Ren A. Fetal DNA hypermethylation in tight junction pathway is associated with neural tube defects: a genome-wide DNA methylation analysis. Epigenetics. 2017;12(2):157–165. doi: 10.1080/15592294.2016.1277298. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A study reporting that hypermethylation in tight junction pathway is associated with neural tube defects by using genome-wide methylation array on center neural system tissue.
- 11.Wu L, Wang L, Shangguan S, et al. Altered methylation of IGF2 DMR0 is associated with neural tube defects. Mol. Cell Biochem. 2013;380(1–2):33–42. doi: 10.1007/s11010-013-1655-1. [DOI] [PubMed] [Google Scholar]
- 12.Bai B, Zhang Q, Liu X, et al. Different epigenetic alterations are associated with abnormal IGF2/Igf2 upregulation in neural tube defects. PLoS ONE. 2014;9(11):e113308. doi: 10.1371/journal.pone.0113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Wang F, Guan J, et al. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am. J. Clin. Nutr. 2010;91(5):1359–1367. doi: 10.3945/ajcn.2009.28858. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Wang Z, Li Y, et al. Association of genomic instability, and the methylation status of imprinted genes and mismatch-repair genes, with neural tube defects. Eur. J. Hum. Genet. 2012;20(5):516–520. doi: 10.1038/ejhg.2011.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farkas SA, Bottiger AK, Isaksson HS, Finnell RH, Ren A, Nilsson TK. Epigenetic alterations in folate transport genes in placental tissue from fetuses with neural tube defects and in leukocytes from subjects with hyperhomocysteinemia. Epigenetics. 2013;8(3):303–316. doi: 10.4161/epi.23988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochtus A, Izzi B, Vangeel E, et al. DNA methylation analysis of Homeobox genes implicates HOXB7 hypomethylation as risk factor for neural tube defects. Epigenetics. 2015;10(1):92–101. doi: 10.1080/15592294.2014.998531. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Study of the association between HOXB7 gene hypomethylation and NTDs by measuring the methylation level of blood samples in a case–control study.
- 17.Juriloff DM, Harris MJ. A consideration of the evidence that genetic defects in planar cell polarity contribute to the etiology of human neural tube defects. Birth Defects Res. Part A Clin. Mol. Teratol. 2012;94(10):824–840. doi: 10.1002/bdra.23079. [DOI] [PubMed] [Google Scholar]
- 18.Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res. Part A Clin. Mol. Teratol. 2010;88(8):653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- 19.Shangguan S, Wang L, Chang S, et al. DNA methylation aberrations rather than polymorphisms of FZD3 gene increase the risk of spina bifida in a high-risk region for neural tube defects. Birth Defects Res. Part A Clin. Mol. Teratol. 2015;103(1):37–44. doi: 10.1002/bdra.23285. [DOI] [PubMed] [Google Scholar]; •• The first study of the association between methylation of planar cell polarity pathway gene and NTDs.
- 20.Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human sister-of-mammalian grainyhead (SOM) expands the grainyhead-like family of developmental transcription factors. Biochem. J. 2003;370:953–962. doi: 10.1042/BJ20021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ting SB, Wilanowski T, Auden A, et al. Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nat. Med. 2003;9(12):1513–1519. doi: 10.1038/nm961. [DOI] [PubMed] [Google Scholar]
- 22.Gustavsson P, Copp AJ, Greene ND. Grainyhead genes and mammalian neural tube closure. Birth Defects Res. Part A Clin. Mol. Teratol. 2008;82:728–735. doi: 10.1002/bdra.20494. [DOI] [PubMed] [Google Scholar]
- 23.De Castro SC, Hirst CS, Savery D, et al. Neural tube closure depends on expression of Grainyhead-like 3 in multiple tissues. Dev. Biol. 2018;435(2):130–137. doi: 10.1016/j.ydbio.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemay P, Marco PD, Emond A, et al. Rare deleterious variants in GRHL3 are associated with human spina bifida. Hum. Mutat. 2018;38(6):716–724. doi: 10.1002/humu.23214. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Zhang L, Li Z, et al. Prevalence and trend of neural tube defects in five counties in Shanxi province of Northern China, 2000 to 2014. Birth Defects Res. Part A Clin. Mol. Teratol. 2016;106(4):267–274. doi: 10.1002/bdra.23486. [DOI] [PubMed] [Google Scholar]
- 26.Dworkin S, Simkin J, Darido C, et al. Grainyhead-like 3 regulation of endothelin-1 in the pharyngeal endoderm is critical for growth and development of the craniofacial skeleton. Mech. Dev. 2014;133:77–90. doi: 10.1016/j.mod.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 28.Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10(16):1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 30.Farthing CR, Ficz G, Ng RK, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4(6):e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rochtus A, Winand R, Laenen G, et al. Methylome analysis for spina bifida shows SOX18 hypomethylation as a risk factor with evidence for a complex (epi)genetic interplay to affect neural tube development. Clin. Epigenetics. 2016;108(8):4–12. doi: 10.1186/s13148-016-0272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Study of the association between SOX18 gene hypomethylation and NTDs by measuring the methylation level of blood samples in a case–control study.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.