Abstract
Myotonic dystrophy type 1 (DM1) is a multi-systemic disease resulting in severe muscle weakening and wasting. DM1 is caused by expansion of CTG repeats in the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene. We have developed an inducible, skeletal muscle-specific mouse model of DM1 (CUG960) that expresses 960 CUG repeat-expressing animals (CUG960) in the context of human DMPK exons 11–15. CUG960 RNA-expressing mice induced at postnatal day 1, as well as adult-onset animals, show clear, measurable muscle wasting accompanied by severe histological defects including central myonuclei, reduced fiber cross-sectional area, increased percentage of oxidative myofibers, the presence of nuclear RNA foci that colocalize with Mbnl1 protein, and increased Celf1 protein in severely affected muscles. Importantly, muscle loss, histological abnormalities and RNA foci are reversible, demonstrating recovery upon removal of toxic RNA. RNA-seq and protein array analysis indicate that the balance between anabolic and catabolic pathways that normally regulate muscle mass may be disrupted by deregulation of platelet derived growth factor receptor β signaling and the PI3K/AKT pathways, along with prolonged activation of AMP-activated protein kinase α signaling. Similar changes were detected in DM1 skeletal muscle compared with unaffected controls. The mouse model presented in this paper shows progressive skeletal muscle wasting and has been used to identify potential molecular mechanisms underlying skeletal muscle loss. The reversibility of the phenotype establishes a baseline response for testing therapeutic approaches.
Introduction
Myotonic dystrophy type 1 (DM1) is the most common adult-onset muscular dystrophy that affects multiple organ systems including skeletal and cardiac muscle, neurological, endocrine, gastrointestinal and reproductive functions (1). Despite defects in skeletal muscle, such as myotonia and general muscle weakening, being among the first clinical DM1 presentations (2) and skeletal muscle wasting accounting for 60% of the mortality associated with DM1 (1), the mechanisms that directly underlie skeletal muscle wasting in DM1 remain largely unknown.
DM1 is caused by expansion of CTG tandem repeats in the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene to 50–3000 repeats compared with 5–37 repeats in the general population (3). CUG repeat expansion-containing RNA (CUGexp RNA) transcripts from the expanded allele are retained in the nucleus in discrete foci resulting in sequestration and functional depletion of the Muscleblind-like (MBNL) family of RNA-binding proteins (4–7). CUGexp RNA also induces up-regulation of CUGBP, Elav-like family member 1 (CELF1) protein (8). Both the loss of MBNL and increased CELF1 protein function lead to aberrant mRNA splicing associated with DM1 features (9,10). Sequestration of MBNL proteins has also been demonstrated to alter mRNA localization and transport, mRNA stability, microRNA biogenesis and polyadenylation (11–15) while CELF1 has additional roles in mRNA stability and translation regulation (16).
Many studies have focused on the specific roles played by either MBNL or CELF1 proteins in DM1 pathology (17–21). Skeletal muscle wasting has been demonstrated in mice with combined Mbnl1 and Mbnl2 knockout (17) and in Celf1 over-expressing mice (21). Additionally, CUGexp-expressing mouse models have been developed to study mechanisms of DM1 (22–27). Studies using these mice have shown a variable range of DM1-like phenotypes associated with repeat expression with mice expressing >900 CUG repeats, such as the EpA960 model (24) and the DMSXL model (25,27), showing more severe muscle wasting phenotypes than CUGexp RNA models with shorter expansions or RNA containing five CUGs [HSALR, DM300 and (CTG)5 mouse models] (22,23,26). The variable range of DM1 phenotypes in these models suggests that DM1 pathogenesis may involve multiple pathways (28,29). Since the DMSXL model is not reversible and expression of CUGexp RNA in the EpA960 model has extinguished, this study set out to generate a mouse model with reversible expression of long CUGexp RNA and significant muscle wasting to study the mechanisms of muscle wasting.
We developed a tetracycline-inducible skeletal muscle-specific model of DM1 that expresses RNA containing 960 interrupted repeats in the context of human DMPK exons 11–15. Transgenic mice expressing the CUG960 RNA showed significantly reduced muscle weight, histological myopathy consistent with DM1 and an increase of oxidative muscle fibers. Additionally, the CUG960-expressing mice exhibited nuclear RNA foci with colocalized Mbnl1 protein; however, splicing defects that are typically associated with DM1 were mild, suggesting the possibility that abnormal splicing is not solely responsible for the wasting phenotype observed in these mice. Multiple effects of the CUG960 RNA were reversible. Protein array analysis showed that animals exhibiting severe muscle loss had significantly increased levels of activated AMP-activated protein kinase α (AMPKα) and reduction of the phosphorylated form of the platelet derived growth factor receptor β (PDGFRβ) receptor tyrosine kinase involved in cell survival, with little to no change in these protein levels in animals with only moderate muscle loss. DM1 skeletal muscle showed similar changes to protein expression. These results suggest that pathways activated in response to nutrient or oxidative stress are deregulated, resulting in disruption of the balance between anabolic and catabolic pathways normally responsible for maintaining muscle mass.
Results
An inducible DM1 mouse model shows significant skeletal muscle wasting
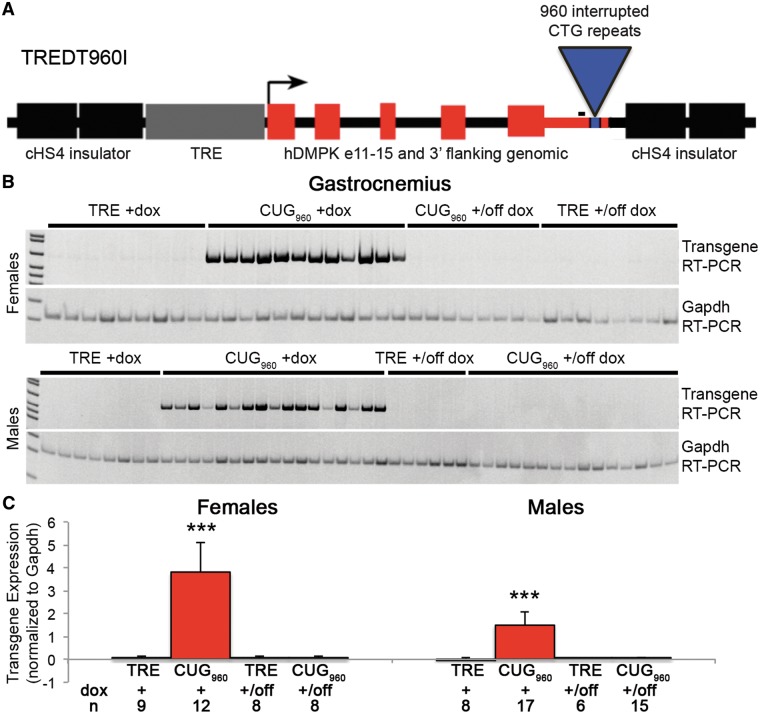
To assess the mechanisms of skeletal muscle wasting in DM1, we developed a tetracycline-inducible transgene, TREDT960I, containing 960 interrupted CTG repeats in the context of human DMPK exons 11–15 (Fig. 1A). Southern blot analysis confirmed that the integrated transgene contained 960 repeats and the repeats remained stable over multiple generations (data not shown). Bi-transgenic animals, designated CUG960, that are homozygous for the TREDT960I transgene (TRE) and hemizygous for a muscle-specific reverse transactivator (MDAFrtTA) (30) were fed doxycycline (dox)-containing chow to induce expression of the CUG960 RNA. Titration of dox indicated that expression of CUG960 RNA and the associated phenotypes increased with increasing dox dose, reaching a plateau between 1 and 2 g dox/kg chow (data not shown). Additionally, initial experiments showed that skeletal muscle abnormalities are observed when animals are fed dox-containing chow beginning in utero, at postnatal day 1 (PN1), and as adults, with little difference between phenotypes of mice started in utero or at PN1 (data not shown). Mice used in this study were fed mouse chow containing 2 g dox/kg chow for 10 weeks, beginning at PN1, to induce transgene expression. CUG960 mRNA levels in skeletal muscle from bi-transgenic animals were assessed by RT-PCR (CUG960 +dox; Fig. 1B) and quantified relative to Gapdh mRNA (Fig. 1C, Supplementary Material, Fig. S1D). Relative expression was compared with littermate control animals (TRE +dox) that were homozygous for TREDT960I and lack the reverse transactivator. No CUG960 RNA was detected in TRE +dox controls, as expected, while all CUG960 +dox animals expressed readily detectible CUG960 RNA (Fig. 1B). There was no difference in relative expression levels of CUG960 RNA between female and male CUG960 +dox animals when assayed simultaneously (Supplementary Material, Fig. S1E). Additionally, we observed comparable expression levels in gastrocnemius muscle induced for 6 and 12 weeks with 2 g/kg dox, beginning at PN1 (Supplementary Material, Fig. S1A and B) with some reduction in CUG960 RNA expression in quadriceps between 6 and 12 weeks (Supplementary Material, Fig. S1C). Transgene expression was not detected in mice that were switched to regular chow for 8 weeks following 10 weeks of dox chow starting at PN1, demonstrating loss of CUG960 RNA upon removal of dox (CUG960 +/off dox compared with TRE +/off dox control, Fig. 1B and C). Similarly, bi-transgenic animals not given dox chow did not express detectable levels of CTG960 indicating that ‘leaky’ expression is not detectable (CTG960 –dox, Supplementary Material, Fig. S1A-C). Finally, we found that CUG960 RNA expression was comparable with RNA expression from the previously published EpA960 model developed in our laboratory (24) using tissue samples obtained prior to extinguished expression of the EpA960 transgene (Supplementary Material, Fig. S1F).
Figure 1.
CUG960 transgene expression. (A) The TREDT960I transgene expresses RNA containing 960 interrupted CUG repeats in the context of human DMPK exons 11–15 in response to induction of a tet-responsive promoter. (B) RT-PCR of transgene expression of RNA from female (top) and male (bottom) gastrocnemius muscle shows CUG960 RNA levels in CUG960 bi-transgenic mice fed dox chow for 10 weeks (CUG960+dox) mice. CUG960 RNA was not detected in TRE+dox controls or in CUG960 or TRE mice fed dox-containing chow for 10 weeks starting at PN1 then fed chow without dox for 8 weeks (+/off dox). RT-PCR of Gapdh was used as a loading control. The amplified region of the TREDT960I mRNA is located 5′ of CUG repeats (black bar in A). (C) Quantification of transgene expression was normalized to Gapdh levels and expression of CUG960+dox mice was compared with TRE+dox and CUG960 and TRE +/off dox mice. ***P<0.001.
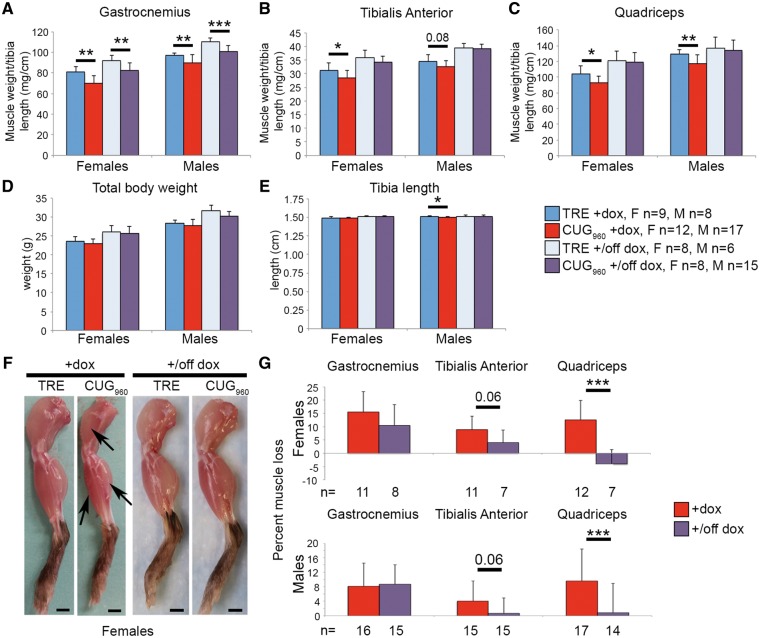
Following 10 weeks of CUG960 RNA expression, significant skeletal muscle wasting is observed in gastrocnemius, quadriceps and tibialis anterior (TA) muscles from CUG960 +dox mice compared with TRE +dox controls (Fig. 2A–C, arrows in F). Quantification of skeletal muscle wasting was based on muscle weight, normalized to tibia length. Tibia length is unaffected in female CUG960 +dox mice but is slightly shorter in male CUG960 +dox mice compared with TRE +dox littermate controls (Fig. 2E). Differences in total body weight of females and males that do or do not express CUG960 RNA are apparent at 10 weeks of CUG960 RNA expression (Fig. 2D) but did not reach significance, even in the event of prolonged CUG960 RNA expression up to 20 weeks (Supplementary Material, Fig. S2F). Evidence of skeletal muscle loss in female CUG960 +dox mice was present as early as 6 weeks of expression compared with non-induced control mice (Supplementary Material, Fig. S2A). By 12 weeks of transgene expression, significant muscle loss was observed in female CUG960 +dox mice compared with TRE +dox and CTG960 –dox control mice (Supplementary Material, Fig. S2A and C). Significant reduction in muscle weight was not observed in flexor digitorum brevis or triceps muscles at 6 or 12 weeks (Supplementary Material, Fig. S2D and E).
Figure 2.
Significant muscle wasting is observed in CUG960 +dox mice. (A–C) Gastrocnemius and quadriceps weight were significantly reduced in CUG960+dox females and males following 10 weeks of CUG960 RNA expression. TA weight was also significantly reduced in female CUG960+dox mice. (D) Total body weight is unaffected by CUG960 RNA expression. (E) Tibia length is unaffected in female CUG960+dox mice and is mildly, yet significantly, reduced in male CUG960+dox mice compared with controls. (F) Muscle loss in CUG960 +dox mice (arrows), compared with TRE+dox is fully rescued in quadriceps muscle and slightly improved in TA muscle (quantified in G) when CUG960 transgene expression is turned off (CUG960+/off dox), but is not improved in gastrocnemius muscle. *P<0.05, **P<0.01, ***P<0.001.
Since CUG960 RNA expression was induced at PN1, it is possible that muscle loss is due to a defect in postnatal development. To determine whether muscle wasting is also observed in animals induced to express CUG960 RNA as adults, we induced transgene expression in mice beginning at 6 weeks of age using 2 g/kg dox chow and performed analysis 18 weeks later. We observed significant skeletal muscle loss in gastrocnemius and TA muscles in both female and male CUG960 +dox mice compared with TRE +dox controls (Supplementary Material, Fig. S3A and B) but no significant wasting in quadriceps (Supplementary Material, Fig. S3C). The percent muscle loss for gastrocnemius and TA muscles were comparable between adult- and PN1-induced CUG960 +dox mice (data not shown), relative to TRE +dox controls, indicating that muscle loss is due to expression of CUG960 RNA rather than effects of dox and is not secondary to disrupted postnatal development.
To determine whether muscle loss in CUG960 +dox mice can be reversed, we first induced transgene expression for 10 weeks, beginning at PN1, then switched mice to standard chow without dox for an additional 8 weeks to turn off transgene expression (CUG960 +/off dox). Littermate TRE mice were also fed dox-containing chow for 10 weeks followed by standard chow for 8 weeks (TRE +/off dox). As noted above, CUG960 +/off dox animals did not express CUG960 RNA (Fig. 1A and B). Turning off repeat expression of CUG960 RNA resulted in full rescue of muscle loss in quadriceps and a strong trend toward rescue in TA in both females and males (Fig. 2B, C, F and quantified in G), however, rescue of muscle weight relative to tibia length was not observed in gastrocnemius in CUG960 +/off dox mice (Fig. 2A and G), possibly because a longer recovery period is required since the percentage of muscle loss is greater than in quadriceps and TA.
CUG960 +dox mice exhibit severe muscle myopathy and RNA foci
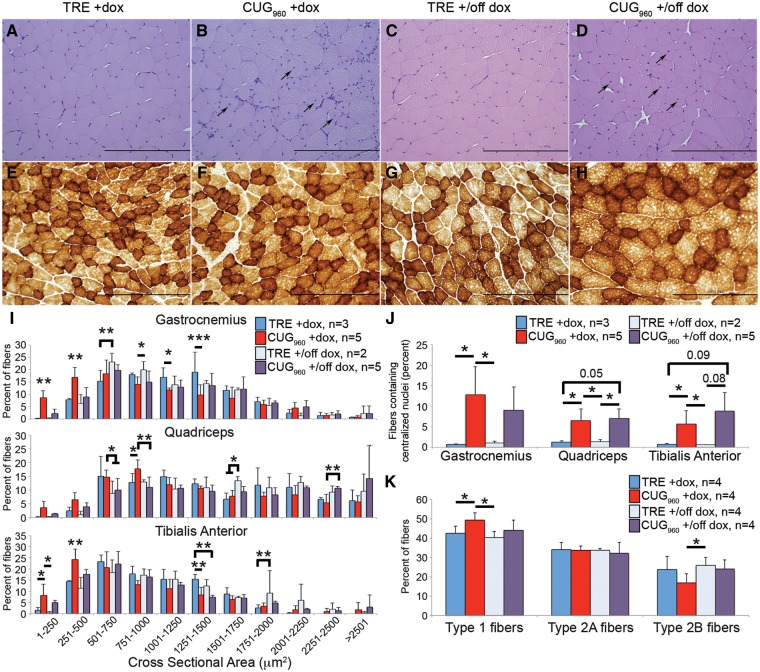
To evaluate myopathy resulting from transgene expression in skeletal muscle as well as reversibility of myopathy, we examined histological sections of CUG960 and TRE +dox and CUG960 and TRE +/off animals. Cross-sections of female gastrocnemius (Fig. 3A–D), quadriceps (not shown) and TA (not shown) muscles were assessed using hematoxylin and eosin staining. Compared with TRE +dox controls (Fig. 3A), CUG960 +dox mice (Fig. 3B) exhibited severe muscle myopathy, with no increase in immune or inflammatory markers (data not shown). Muscle myopathy in CUG960 +dox mice included increased variation in fiber size (gastrocnemius and TA only, Supplementary Material, Table S1), reduced cross-sectional area (CSA) in all three muscle groups (Fig. 3I, Supplementary Material, Table S1) and significantly increased percent of fibers containing centralized nuclei (Fig. 3B and quantified in J). Increased centralized nuclei is a histological feature of regeneration, however, regeneration is not observed in DM1 (31) and therefore appears to result from a different mechanism. Similar to DM1, regeneration is not observed in muscles of CUG960 +dox mice.
Figure 3.
Repeat expressing mice exhibit severe histopathology. (A, B) Severe histopathology, characterized by increased percent of fibers with centralized nuclei [arrows in (B), quantified in (J)] and fiber size variability, was observed in gastrocnemius muscle of female CUG960+dox mice fed dox chow 10 weeks, beginning at PN1, compared with TRE +dox muscles. (C, D). Muscle histology improved when CUG960 transgene was turned off (+/off dox), resembling controls, however, nuclei remain centralized [arrows in (D), quantified in (J)]. Fiber CSA (250 fibers per sample, I) was significantly reduced in all muscle groups for female CUG960+dox animals compared with TRE +dox controls and improved in CUG960 +/off dox muscles. (E–H) Cytochrome oxidase staining for fiber type in female gastrocnemius muscle indicated subtle changes Type 1 oxidative fibers (dark brown) and Type 2B glycolytic fibers (light brown) in CUG960+dox mice compared with controls. (K) Quantification of fiber type percentage indicates increased percentage of oxidative fibers and decreased percentage of glycolytic fibers in CUG960+dox mice compared with controls. Fiber type percentage more closely resembles age-matched controls in CUG960+/off dox muscles. Bar, 200 μm. *P<0.05, **P<0.01, ***P<0.001.
To test whether the change in muscle CSA is due to altered protein synthesis in CUG960 +dox mice, we used a modified SUnSET assay (32,33). Following 2 weeks of CUG960 RNA induction with dox, beginning at 6 weeks of age, mice received an intraperitoneal injection of puromycin 1 h prior to tissue collection. No change in puromycin levels, normalized to total protein levels, was detected in gastrocnemius or quadriceps muscles of CUG960 +dox mice. Taken together, these data suggest that the expression of repeat RNA results in myopathy without affecting total skeletal muscle protein synthesis (Supplementary Material, Fig. S4).
Female CUG960 +dox mice induced as adults show less severe muscle myopathy than do CUG960 +dox animals induced postnatally (Supplementary Material, Fig. S3D-G). In adult-onset CUG960 RNA-expressing mice that did not show muscle loss, histology is similar to TRE +dox controls (Supplementary Material, Fig. S3D and E), while CUG960 +dox females with significant muscle loss exhibited myopathy, including increased fibers with central nuclei (Supplementary Material, Fig. S3F and quantified in G). In CUG960 +/off dox mice (Fig. 3D), muscle histology more closely resembles TRE +/off dox controls (Fig. 3C) than CUG960 +dox histology with regard to CSA (Fig. 3I) and fiber size variation showing a level of phenotype rescue (gastrocnemius and quadriceps, Supplementary Material, Table S1). However, myonuclei remain centralized 8 weeks after animals were switched to regular chow.
Cytochrome oxidase staining of gastrocnemius cross-sections (Fig. 3E–H) showed subtle but significant differences in the percentages of oxidative and glycolytic fibers in female CUG960 +dox mice compared with TRE +dox, CUG960 +/off dox and TRE +/off dox muscles. Similar results were obtained using succinate dehydrogenase staining (not shown). Quantification of fiber type percentages based on these staining results (Fig. 3K) revealed significantly increased percentage of Type 1 oxidative fibers and decreased Type 2B glycolytic fibers in CUG960 +dox gastrocnemius muscles, compared with TRE +dox and TRE +/off dox controls. Additionally, the significant difference in Type 1 fibers between CUG960 +dox and TRE +dox was lost between CUG960 +/off dox mice and TRE +/off dox mice, suggesting at least a partial rescue of fiber type differences. Taken together, histological analyses showed significant, but reversible myopathy present in CUG960 +dox mice.
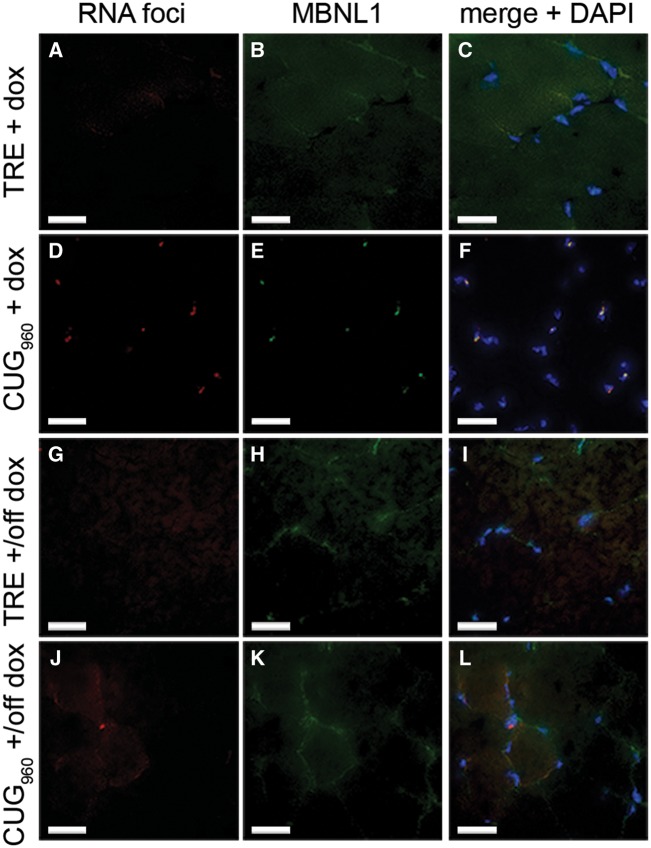
Nuclear RNA foci that colocalize with the MBNL protein family is a key pathogenic feature of DM1 (4,6). To determine whether CUG960 RNA-expressing mice accumulate nuclear foci that colocalize with Mbnl protein, we performed fluorescent in situ hybridization (FISH) using a Tye-563-labeled antisense linked nucleic acid probe (CAG)5, followed by immunofluorescent (IF) staining for Mbnl1. We demonstrate the presence of multiple discrete RNA foci and colocalized Mbnl1 in nuclei of CUG960 +dox gastrocnemius muscles following 10 weeks of induction starting at PN1 (Fig. 4D–F) and no foci present in TRE +dox control nuclei (Fig. 4A–C). When CUG960 mice were switched to regular chow for 8 weeks (+/off dox), nuclear foci and Mbnl1 colocalization (Fig. 4J–L) were not detected, as in littermate TRE +/off dox controls (Fig. 4G–I). The finding showing that foci and colocalized Mbnl1 disperse when transgene expression is turned off indicates that phenotypes and downstream effects can be reversed in the CUG960 +dox model.
Figure 4.
CUG960 +dox muscle contains nuclear RNA foci that colocalize with Mbnl1 protein. (A–F) FISH of CUG repeat RNA, coupled with IF of Mbnl1 protein showed that CUG960 +dox (D–F) gastrocnemius muscle contains nuclear RNA foci that colocalize with Mbnl1 and were absent in TRE +dox (A–C) controls. (G–L) Neither RNA foci nor colocalized Mbnl1 proteins were detected in TRE or CUG960 +/off dox muscles (G–L). Bar, 25 μm.
CUG960 RNA expression has a broad but mild effect on splicing
Thus far, we have presented a mouse model expressing RNA containing 960 CUG repeats that exhibits significant skeletal muscle wasting, myopathy, and the presence of RNA foci that colocalize with Mbnl1 protein, consistent with phenotypes associated with DM1 skeletal muscle pathology. To determine the most significant transcriptome changes contributing to progressive muscle wasting in these mice, we performed RNA-seq analysis on either gastrocnemius or quadriceps muscles from two CUG960 +dox mice, two CTG960 –dox control mice, and one TRE +dox control mouse after 6 and 12 weeks of induction, beginning at PN1 (Supplementary Material, Table S2). Poly-A-selected cDNA libraries were used for 100 bp paired-end reads, 97–147 million reads per sample, with >80% of reads mapping to the mouse genome (Supplementary Material, Table S2). The high quality sequencing data obtained provided us with sufficient depth and quality to assess changes to both alternative splicing (Fig. 5 and Supplementary Material, Fig. S5) and gene expression (Supplementary Material, Fig. S6) that respond to CUG960 RNA expression and correlate with skeletal muscle wasting.
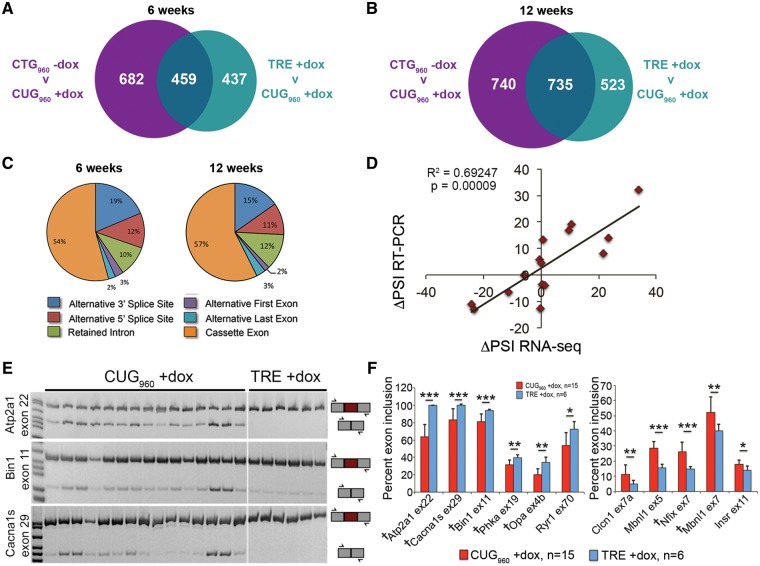
Figure 5.
CUG960 expression broadly but mildly affects splicing of genes involved in cytoskeletal dynamics and GTPase and PI3K activity. (A, B) CUG960 RNA affects a broad number of alternative splicing events at 6 and 12 weeks of induction, compared with bi-transgenic mice not fed dox (CTG960 –dox, purple) and TRE +dox mice (teal). Only splicing events altered by CUG960 RNA relative to both controls (overlapping regions) were used for additional analysis. (C) Greater than 50% of all alternative splicing changes at 6 and 12 weeks affect cassette exons. Validation of RNA-seq results by RT-PCR showed a significant correlation (R2=0.69247, P=0.00009, (D) in the change of percent spliced inclusion (ΔPSI) between TRE +dox and CUG960 +dox mice fed dox chow for 10 weeks compared with ΔPSI from RNA-seq in mice fed dox chow for 12 weeks. (E) Representative RT-PCRs for female CUG960 +dox and TRE +dox mice showing alternative splicing changes in Atp2a1 exon 22, Bin1 exon 11, and Cacna1s exon 29. (F) Quantification of splicing in female CUG960 +dox mice showed significantly reduced inclusion of six exons and increased inclusion of five exons affected in DM1 skeletal muscle. Seven of 12 events correlated with skeletal muscle weakness in DM1 (daggers in F) were significantly altered in CUG960 +dox mice. *P<0.05, **P<0.01, ***P<0.001.
RNA-seq data were evaluated using the mixture of isoforms (MISO) (34) and SpliceTrap (35) algorithms. Events with changes in percent spliced inclusion (ΔPSI) (36) values ≥10% (Bayes factor >5) were considered significantly altered. Events that were altered by the presence of dox alone were filtered out of the dataset. At 6 weeks, 682 splicing events were significantly altered with 740 events at 12 weeks in CUG960 +dox mice compared with CTG960 –dox controls and 437 events at 6 weeks and 523 events at 12 weeks were significantly altered in CUG960 +dox mice compared with TRE +dox controls (Fig. 5A and B). We looked further into only those events that were significantly altered in CUG960 +dox muscles when compared with both CTG960 –dox and TRE +dox controls, as these events were likely to be more significant to muscle wasting. At 6 weeks, 459 events and at 12 weeks, 735 events were significantly changed in CUG960 +dox mice relative to both controls (overlapping regions in Fig. 5A and B). We chose to focus our analysis on cassette exons in protein coding regions. Of the splicing events with ΔPSI ≥ 10% in CUG960 +dox muscles relative to both controls, 54–57% of the altered events reflect changes to cassette exons (Fig. 5C). PSI values from the combined MISO and SpliceTrap datasets showed that splicing of cassette exons was reproducible among biological replicates (Supplementary Material, Fig. S5A and B). Gene Ontology and functional enrichment analyses of alternatively spliced genes in CUG960 +dox mice, using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (37,38), showed significant enrichment of genes involved in cytoskeleton dynamics, GTPase and PI3K activity, and vesicular trafficking at both 6 and 12 weeks of induction. Calcium handling genes were also enriched following 12 weeks of induction (Supplementary Material, Fig. S5E and F).
Analysis of differential gene expression was conducted similarly to alternative splicing analysis using Cufflinks/Cuffdiff (39,40) and Ht-Seq/EdgeR (41,42) pipelines. Gene expression data were highly reproducible among biological replicates (Supplementary Material, Fig. S6A and B). Overall, a limited set of 67 genes at 6 weeks and 163 genes at 12 weeks were significantly differentially expressed (>1.5-fold, P < 0.05) in CUG960 +dox mice relative to both control groups (overlapping regions in Supplementary Material, Fig. S6C and D). After filtering out low abundance genes (RPKM <0.5), 55 and 112 genes (6 and 12 weeks, respectively) remained. Analysis showed that all genes with differential gene expression at 6 weeks were up-regulated, while 107 of 112 differentially expressed genes were up-regulated at 12 weeks (Supplementary Material, Fig. S6E). Genes at both 6 and 12 weeks were enriched for functions in cytoskeleton dynamics associated with actin binding, cell cycle control and cell death. By 12 weeks, enrichment is also observed for genes involved in calcium handling, GTPase activity and vesicular transport (Supplementary MaterialFig. S6F and G). Furthermore, there was little to no overlap in genes with changes to alternative splicing and differential gene expression, suggesting that although the most enriched categories of genes from both the alternative splicing and gene expression analyses overlap, the individual genes altered are independent of each other (Supplementary Material, Fig. S5C and D).
Alternative splicing events from RNA-seq data were validated by RT-PCR comparing the ΔPSI (36) between CUG960 +dox and TRE +dox obtained by RNA-seq at 12 weeks with the ΔPSI from RT-PCR at 10 weeks on dox chow (Fig. 5D–F). By RT-PCR, we tested 16 events known to be alternatively spliced in DM1 skeletal muscle, with emphasis on 12 events previously reported to correlate with muscle weakness (43). To validate both strong and weak splicing changes, 7 of the 16 tested events had a ΔPSI > 10%, while nine had ΔPSI < 10% in the RNA-seq dataset. The results showed a strong correlation between RNA-seq and RT-PCR results (R2=0.69247, P = 0.00009, Fig. 5D).
Of the 16 known DM1 alternatively spliced exons tested by RT-PCR in CUG960 +dox and TRE +dox females, 6 events showed mild, yet significant, reduction in exon inclusion (Fig. 5E and F), while another five events showed significantly increased exon inclusion (Fig. 5F). Seven of the 12 splicing events tested that correlate with muscle weakness in individuals affected by DM1 (43) were affected in female CUG960 +dox mice (daggers in Fig. 5F). Splicing changes were milder in male CUG960 +dox animals than females (not shown), consistent with male CUG960 +dox mice exhibiting more mild muscle loss and histopathology. Taken together, these results suggest that only mild splicing defects exist in CUG960 +dox mice.
CUG960 RNA induces deregulation of signaling pathways involved in cell survival during energetic stress
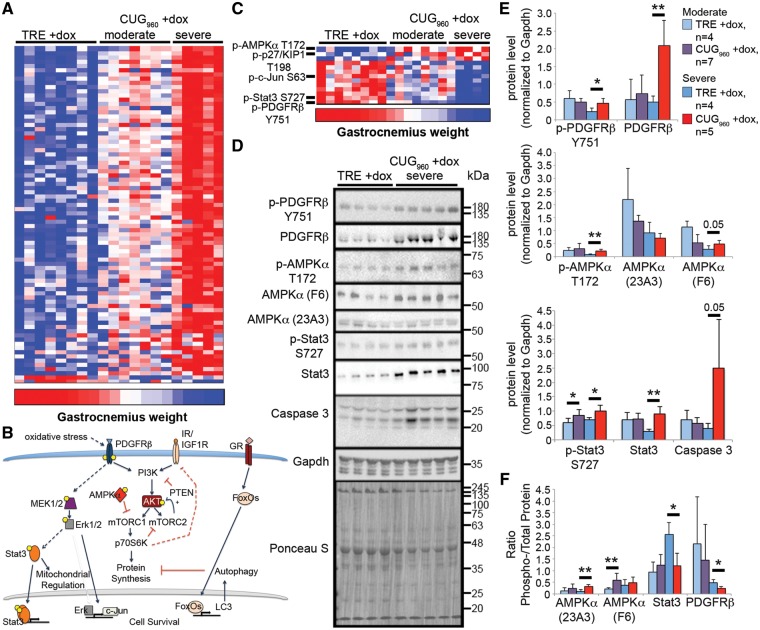
The relatively mild splicing defects in CUG960 +dox mice suggest that spliceopathy might not be the only contributing factor to skeletal muscle wasting in mice expressing CUG960 RNA. To identify altered signaling pathways that significantly correlate with skeletal muscle wasting in CUG960 +dox mice, we performed reverse phase protein array (RPPA) analysis (44) on gastrocnemius muscle of 12 CUG960 +dox mice and eight TRE +dox age-matched mice fed 2 g/kg dox chow for 10 weeks, beginning at PN1. The array measured changes to total protein abundance and post-translational modifications using 216 validated antibodies. RPPA analysis on total protein lysates from gastrocnemius muscle revealed significant up-regulation of 77 proteins and down-regulation of 2 proteins (maximum signal intensity ≥200, fold-change >1.25, P < 0.05, Fig. 6A). Furthermore, the level of protein expression changes tightly segregated with moderate or severe wasting, based on gastrocnemius weight (severe weight <56 mg, moderate weight 60–76 mg, control weight >79 mg, P ≤ 0.001), indicating that the degree of signaling pathway deregulation correlates with the severity of muscle wasting. Ingenuity Pathway Analysis indicated that the top canonical pathways altered in response to CUG960 RNA expression involve PTEN, IL-8, PI3K/AKT and glucocorticoid receptor signaling pathways (Fig. 6B, Supplementary Material, Fig. S7A). Deregulated PTEN/PI3K/AKT signaling is consistent with gene enrichment analysis obtained by RNA-seq and presents an interesting pathway on which to focus future analyses. Up-regulated expression of the glucocorticoid effector protein FoxO1, as well as proteins involved in the autophagy pathway regulated by FoxO1, was observed in CUG960 mice, consistent with previous reports in DM1 models (45,46).
Figure 6.
Signaling pathway analysis reveals deregulation of PTEN/PI3K/AKT pathway. (A) RPPA analysis was performed using total protein lysates from female gastrocnemius muscle with 216 validated antibodies. Normalized fluorescence intensity, grouped according to muscle wasting severity (severe gastrocnemius weight <56 mg, moderate gastrocnemius weight 60–76 mg, control gastrocnemius weight >79 mg, P≤0.001), showed predominantly increased protein expression in CUG960 +dox mice with severe muscle wasting. (B) The PTEN/PI3K, IL-8, and glucocorticoid signaling pathways were the most significantly altered pathways in CUG960 mice. (C) Analysis of the ratio of phosphorylated to total protein level suggests that in CUG960 +dox mice with severe muscle wasting, the PTEN/PI3K pathway antagonist AMPKα is significantly increased while activated PDGFRβ receptor, known to activate PI3K, and activated Stat3, downstream of PDGFRβ, were reduced. (D) Validation by western blot analysis in female gastrocnemius muscle extracts of severely affected animals. (E) Quantification of western blots shown in (D). (F) Quantification of phospho- to total protein ratios for western blots in (D). *P<0.05, **P<0.01.
In addition to overall protein expression changes, we assessed changes in the ratio of phosphorylated to total protein levels in the RPPA dataset. From this analysis, we found that 3 and 10 proteins show significantly up-regulated and down-regulated phospho-to-total protein ratios, respectively (Fig. 6C). The most significantly increased phospho- to total protein ratio is Thr172-phosphorylated AMPKα, which antagonizes PI3K/AKT signaling (47,48), while the protein with the most significantly decreased phospho- to total protein ratio was Tyr751-phosphorylated PDGFRβ (Fig. 6C–F).
Validation of the RPPA results was performed by western blot of gastrocnemius muscle extracts from CUG960 +dox mice exhibiting severe (Fig. 6C) and moderate (Supplementary Material, Fig. S7B) muscle wasting. Total AMPKα abundance was unaffected in both groups of CUG960 +dox mice while Thr172-phosphorylated AMPKα was significantly up-regulated in CUG960 +dox mice with severe muscle loss (Fig. 6D–F). Consistent with activation of AMPKα, RPPA data also showed increased activation of p27/Kip1 at Thr198, which was previously reported to be a phosphorylation target of AMPKα (49). Levels of both phosphorylated and total PDGFRβ were increased (Fig. 6D and E) and the ratio of phospho- to total PDGFRβ was decreased in CUG960 +dox with severe muscle loss (Fig. 6F). PDGFRβ phosphorylation at Tyr751 is known to activate PI3K/AKT, as well as MAPK signaling (50,51). Although MAPK signaling was not enriched in pathway analysis (Supplementary Material, Fig. S7A), several MAPK signaling pathway components were up-regulated in the CUG960 +dox mice (Fig. 6A) and phospho- to total protein ratios of the MAPK downstream targets, Ser63 p-c-Jun (Fig. 6C) and Ser727 p-Stat3 (Fig. 6C–F) involved in regulating cell cycle and survival, were reduced. Additional validation of RPPA by western blot (Fig. 6D and E) showed increased levels of the cell death indicator, caspase 3. Using the same extracts as in Figure 6D we found that Celf1 protein levels were increased in muscle from CUG960 +dox mice exhibiting severe but not moderate muscle wasting (Supplementary Material, Fig. S9).
Similar results were obtained in DM1 muscle, relative to unaffected control muscle by western blot (Supplementary Material, Fig. S8A). Quantification of the phospho- to total protein ratios for PDGFRβ, AMPKα and Stat3 showed similar trends as the ratios of these protein modifications in CUG960 +dox mice; however, owing to high variability between individuals, the phospho- to total protein ratios did not reach a level of statistical significance (Supplementary Material, Fig. S8B). Total levels of the proteins assessed by western blot showed similarly significant up-regulation of Tyr751-phosphorylated PDGFRβ, total PDGFRβ and caspase 3 proteins, while levels of Thr172 p-AMPKα, Ser727 p-Stat3 and total Stat3 showed similar, though not significant, up-regulation in DM1 skeletal muscle relative to unaffected controls (Supplementary Material, Fig. S8C). Similar to what we observed in the mice, the robustness of the signaling changes are likely to correlate with disease severity in affected DM1 skeletal muscle. Taken together, results from the RNA-seq and RPPA analysis suggest that deregulation of PI3K pathway may be involved in inhibition of cell survival and promoting cell death of muscle cells during progressive skeletal muscle wasting in CUG960 +dox mice and in individuals affected by DM1.
Discussion
We demonstrate that CUG960 RNA expression in CUG960 +dox mice results in significant muscle loss in gastrocnemius, quadriceps, and TA muscles (Fig. 2). Induced expression of CUG960 RNA was started at PN1, however, it is unlikely that the reduced muscle weight is due to delayed development, since CUG960 RNA expression initiated at PN1 did not result in reduced overall growth and induction of CUG960 +dox expression in adults also resulted in skeletal muscle wasting. When CUG960 RNA expression is turned off, muscle loss in the quadriceps and TA muscles was reversed. CUG960 +dox mice exhibited histological features consistent with DM1 histology (Fig. 3). CUG960 +dox mice showed changes to the proportion of fiber types within the muscle, including increased Type 1 oxidative fibers and decreased Type 2B glycolytic fibers. In contrast, Type 1 fiber atrophy is predominant in DM1 (1). It is possible that these effects are partially due to expression of CUG960 RNA limited to Type 2 myofibers since the fast myosin light chain promoter drives rtTA expression in MDAFrtTA mice. However, both the HSALR and DMSXL models also showed increased oxidative and decreased glycolytic fiber percentages (22,25), suggesting the difference in the direction of change in fiber type proportions may be due to differences between mouse and human biology when CUGexp RNA is expressed. Histopathology, reduced CSA, and increased oxidative and decreased glycolytic fiber percentages were reversed in CUG960 +/off dox mice in gastrocnemius, quadriceps, and TA. Overall, induction of muscle loss and histopathology and its rescue indicates that this model can be used to test the effectiveness of therapeutics aimed at reducing muscle loss.
A key feature of DM1 pathology is retention of expanded CUG-containing RNA into nuclear foci (6,7). Here, we show the presence of nuclear foci in gastrocnemius of CUG960 +dox mice (Fig. 4) that are not present in control animals. The number and size of the foci vary between nuclei, consistent with previous reports (52) and are absent in CUG960 +/off dox mice after CUG960 expression has ceased. Additionally, we observed colocalization of Mbnl1 protein in the nuclear foci, consistent with DM1 and other mouse models expressing CUG repeat expansions (4,5,22,23,25,53).
MBNL sequestration is an early molecular event in DM1 that contributes to most of the alternative splicing defects (43). Combined knockout of both Mbnl1 and Mbnl2 led to severe muscle pathology (17). Given these observations, one would expect to see strong splicing defects and reduced physiological function in CUG960 +dox mice that exhibit significant muscle wasting. Myotonia, a clinical hallmark of DM1 pathology, has been directly linked to the functional loss of MBNL proteins (54). However, we detected weak to no myotonia by electromyography (data not shown) and only mildly increased inclusion of Chloride channel 1 (Clcn1) exon 7a (Fig. 5F) in CUG960 +dox mice, which results in myotonia in DM1. The DMSXL mouse model also showed mild splicing abnormalities, including for Clcn1 exon 7a, and mild myotonia (25). In contrast, DM1 mouse models that exhibit strong changes in alternative splicing of Clcn1 exon 7a show strong, sustained myotonia (26,24,22). Mis-splicing known to be regulated by MBNL proteins, such as those in Fig. 6, are correlated with skeletal muscle weakness in individuals affected by DM1 (43). We observed skeletal muscle weakness in one group of CUG960 +dox mice by grip strength (data not shown) but were unable to reproduce the weakness phenotype in a separate independent experiment owing to individual variability. The HSALR and DMSXL models also failed to show muscle weakness by grip assessment following normalization for body mass; however, by in situ isometric contraction and in vivo dynamometric assays, the DMSXL mice were shown to have significant skeletal muscle weakness (22,25).
Significant muscle loss is consistently observed in CUG960 +dox mice despite very mild Mbnl-associated phenotypes suggesting the possibility that factors in addition to Mbnl contribute to the wasting phenotype observed in CUG960 +dox mice. One proposed mechanism for DM1 pathogenesis is up-regulation of CELF1 protein (8,21,55). CUG960 +dox mice showed significant up-regulation of Celf1 in mice with severe muscle wasting (Supplementary Material, Fig. S9). Significant up-regulation of Celf1 was also demonstrated in EpA960 and (CTG)5 mice but HSALR mice do not show increased Celf1 levels (9) and Celf1 was up-regulated in only one of four DMSXL mice (25). Another study showed increased Celf1, active GSK3β and reduced cyclin D3 levels in skeletal muscles from the HSALR DM1 mouse model prior to the onset of skeletal muscle weakness; GSK3β inhibition corrected levels of cyclin D3 and alleviated muscle weakness and myotonia (56). In CUG960 +dox mice, no significant changes to GSK3β or cyclin D3 levels were observed by western blot or RPPA (data not shown).
Ingenuity Pathway Analysis of RPPA results showed the top pathways affected by CUG960 RNA expression in CUG960 +dox mice were the PTEN, IL-8, PI3K/AKT, and glucocorticoid signaling pathways (Fig. 6B; Supplementary Material, Fig. S7A). Phosphatase and tensin homologue (PTEN) is a negative regulator of PI3K signaling (57) and IL-8 has been shown to be activated by the PI3K and MAPK pathways (58–60), both of which were affected by expression of CUG960 RNA. The PI3K/AKT pathway is the primary anabolic pathway that results in muscle hypertrophy (61,62). The glucocorticoid pathway indirectly activates the autophagic/lysosomal pathway, regulating muscle atrophy and is characterized by loss of Type 2 glycolytic fibers, as observed in CUG960 +dox mice (Fig. 3K), consistent with Type 2 fiber loss in HSALR and DMSXL mice (22,25). Furthermore, antagonistic crosstalk between the PI3K and glucocorticoid pathways has been reported (63,64), suggesting that the balance between anabolic and catabolic pathways, which is critical to coordinately regulate skeletal muscle mass, is deregulated in CUG960 +dox mice.
Further analysis of the RPPA data revealed that ratios of phospho- to total protein levels of Thr172 p-AMPKα and Thr198 p-p27/KIP1 were significantly up-regulated in CUG960 +dox mice (Fig. 6C). AMPKα serves as an energy switch in the cell that is activated under conditions of nutrient and energy deprivation. It was demonstrated that glucose uptake is reduced in DM1 (31,65), suggesting a state of chronic metabolic stress owing to reduced energy availability during the progression of the disease.
When activated, AMPKα antagonizes anabolic pathways, such as the PI3K/AKT/mTOR signaling pathway, while activating catabolic pathways (66–68). Following glucose starvation, activated AMPKα, through phosphorylation and stabilization of p27/KIP1, has been reported to induce autophagy. Furthermore, Thr198 p-p27/KIP1 was sufficient to promote autophagy in the absence of nutrient deprivation (49). Increased levels of the autophagy markers Atg7, Atg8a and Atg12 and increased apoptosis by TUNEL assay and increased caspase 3 activity were shown in a fruit fly CUGexp model for DM1 that exhibited skeletal muscle wasting, suggesting that both apoptosis and autophagy catabolic pathways may be involved in DM1 muscle wasting (45). We observed up-regulation in autophagy markers, including Atg7, Beclin-1, LC3A and Atg12, in addition to increased caspase 3 levels (Fig. 6).
It was recently demonstrated that activation of AMPKα and PI3K/AKT signaling in HSALR mice were deregulated in response to starvation, although no changes to AMPKα or PI3K activation were reported between fed control and HSALR mice (46). In contrast, we observed significantly increased AMPKα activation in CUG960 +dox mice in the absence of nutrient deprivation (Fig. 6) and slightly, though not significantly, elevated activation of AMPKα in DM1 skeletal muscle (Supplementary Material, Fig. S7C and D). While metabolic stress has not been directly tested in CUG960 +dox mice, it is possible that the level of metabolic stress required for increased activation of AMPKα in the absence of starvation contributes to the severity of the muscle wasting phenotype in the CUG960 +dox mice, compared with HSALR mice.
PDGFRβ levels are also increased by glucose deprivation (69) and were significantly up-regulated in CUG960 +dox mice and in DM1 skeletal muscle. The ratio of Tyr751 phospho- to total PDGFRβ, which is critical for binding PI3K to the PDGFRβ receptor to mediate signaling (51), was significantly decreased in CUG960 +dox mice and reduced in DM1 skeletal muscle, suggesting reduced PDGFRβ signaling may be involved in PI3K deregulation. PDGFRβ also signals through pathways other than PI3K such as the Ras/MEK/ERK pathway (50). Erk1/2 has been shown to phosphorylate Stat3 on Ser727 in cancer cells in response to Ras signaling, enhancing both its canonical role in transcriptional activity and its non-canonical role in mitochondrial function, including limiting reactive oxygen species (ROS) production in response to oxidative stress and regulation of mitochondrial membrane potential and energy supply (70,71). Furthermore, Ser727 p-Stat3 was shown to protect against ROS accumulation in both a heart damage model and a cancer model (71). We show reduced Ser727 p-Stat3 relative to the total Stat3 pool in CUG960 +dox mice, and to a lesser extent in DM1 skeletal muscle, suggesting its role in mitochondrial regulation may be disrupted in response to CUG960 RNA expression, resulting in energetic stress in the skeletal muscle.
In conclusion, we developed a CUG960 repeat expressing mouse model of DM1 that exhibits significant skeletal muscle wasting and myopathy and provide insight into molecular mechanisms directly underlying progressive skeletal muscle wasting. Molecular analysis points toward pathways activated in response to nutrient deprivation and oxidative stress. Our data implicating the activation of autophagic degradation pathways is consistent with previously published reports in DM1, as well as DM1 mouse and fly models (45,46,72–74). Further mechanistic studies are required to assess the roles that deregulated PI3K and AMPK pathways and PDGFRβ play in progressive skeletal muscle wasting in CUG960 RNA expressing mice.
Materials and Methods
Transgenic mice
TREDT960I transgenic mice were generated by standard techniques in an FVB background. The TREDT960I transgene contains a human genomic segment containing exons 11–15 of DMPK with 960 interrupted CTG repeats in the natural site of the repeats. The interrupted repeats were generated as described (55) and contain 20 CTGs separated by 5 nucleotide spacer formed by ligation of SalI and XhoI restriction sites. The repeats are highly stable in bacterial plasmids and transgenic mice. The construct includes 307 bp of the DMPK 3’ flanking region containing the natural polyadenylation signals. The transgene contains two copies of the cHS4 insulator flanking the 5’ and 3’ ends of the expression construct to prevent effects of the chromosomal insertion site. The TREDT960I mice are available at The Jackson Laboratory as Stock No. 032050. MDAFrtTA transgenic mice (30) were maintained on a mixed C57BL/6×DBA background. Mice that were homozygous for the TREDT960I transgene and hemizygous for MDAFrtTA were mated and progeny used for this study were either bi-transgenic TREDT960I/MDAFrtTA (CUG960) or homozygous TREDT960I (TRE). Whenever possible, bi-transgenic and single transgenic mice used for the experiments were littermates. Genotype was confirmed by extracting genomic DNA from mouse tails using DirectPCR lysis reagent (Viagen Biotech) and analyzed by multiplex PCR using a β-actin internal control. Primer sequences can be found in Supplementary Material, Table S3. Doxycycline containing chow (2 g doxycycline/kg chow, Bio-Serv) was provided to the mice beginning at PN1 initially through the nursing dam or at 6 weeks of age to induce expression of the CUG960 RNA. All experiments involving mice were conducted in accordance with the NIH Guide for the Use and Care of Laboratory Animals and approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
RNA isolation and RT-PCR
Total RNA was isolated from individual skeletal muscles using TRIzol reagent (Invitrogen) with a Bullet Blender (Next Advance). RNeasy fibrous tissue mini-kit (Qiagen) was used for RNA extraction of skeletal muscle tissues used in RNA-seq experiments according to the manufacturer’s protocol. RNA was resuspended in RNase-free water (Ambion) and treated with DNaseI (Ambion). RT-PCR was performed using PCR Super Master Mix (Bimake). Primers for transgene expression were designed to amplify the region of DMPK exon 15 upstream of the repeat expansion. Gapdh was used as an internal control for the normalization of transgene expression. Primers for analysis of alternatively spliced exons were designed to anneal to flanking constitutive exons. Primer sequences can be found in Supplementary Material, Table S3. PCR products were separated on a 5% polyacrylamide gel and PSI (36) was calculated after ethidium bromide staining using a Kodak E1 Logic 2200 imaging system.
Muscle weight
Individual skeletal muscles were isolated, weighed and muscle weight was normalized to tibia length to account for differences in animal size. Direct comparisons of normalized muscle weight were only made between age matched treatment groups. For comparison of CUG960 mice induced for 10 weeks (+dox) with CUG960 mice induced 10 weeks followed by 8 weeks off dox (+/off dox), statistically determined outliers for each treatment group were removed, followed by normalization of muscle weight/tibia length to the average muscle weight/tibia length of control animals for each treatment (+dox or +/off dox). From these values, percent muscle loss of control-normalized CUG960 +dox or CUG960 +/off dox mice was calculated from average control muscle weight.
Histology
Skeletal muscles were isolated and fixed overnight in 10% formalin, paraffin-embedded, and cut in 10 μm cross-sections or flash frozen in liquid nitrogen-cooled isopentane as previously described (75) and cut in 7 μm cross-sections. Hematoxylin and eosin, cytochrome oxidase and succinate dehydrogenase staining were performed using standard procedures. Images were acquired using an Olympus BX41 microscope with Olympus DP70 camera and analyzed using ImageJ software (National Institutes of Health). Five field-of-view images of each muscle were used for quantification of CSA (50 fibers per field-of-view were measured), percent of fibers containing centralized nuclei (all whole fibers in field-of-view were counted) and fiber typing (all whole fibers in field-of-view were counted).
Detection of puromycin-labeled proteins
Six-week-old mice were fed 2 g/kg dox chow for 2 weeks. Puromycin (0.04 μmol/g body weight, Enzo Life Sciences) was injected intraperitoneally 1 h prior to tissue collection. Muscles were isolated, homogenized in lysis buffer [10 mM HEPES pH 7.5, 0.32 M sucrose, 5 mM EDTA, protease inhibitor cocktail tablet (Roche) and 1% SDS]. BCA protein assay kit (Thermo Scientific) was used to determine the protein concentration. Western blot analysis using rabbit anti-puromycin antibody (1:1000, Kerafast) was performed using 30 μg protein lysate. Puromycin-labeled protein level for each sample was normalized to total protein levels using the same western blots, stained with Ponceau S (Sigma).
Combined FISH/IF
Combined FISH/IF staining was carried out using a modified protocol previously described (4). Gastrocnemius sections were dried 30 min then fixed 30 min in 3% PFA/1× PBS at room temperature, washed five times in 1× PBS for 2 min each, followed by permeabilization in pre-chilled 2% acetone/1× PBS for 10 min. Sections were placed in 30% formamide/2× SSC for 30 min, hybridized with Tye-563-labeled (CAG)5 LNA probe (0.5 ng/μl, Exiquon) for 5 h at 42°C in buffer [30% formamide, 2× SSC, 0.02% BSA, 66 μg/ml yeast tRNA, 2 mM Vanadyl ribonucleoside complex], and washed 30 min in 30% formamide/2× SSC at 42°C. Sections were then washed in 1× SSC for 30 min at room temperature, followed by incubation in monoclonal mouse anti-MBNL1 clone 3A4 antibody (1:25, Santa Cruz Biotechnology) overnight at 4°C, washed five times in 1× PBS for 2 min, incubated in Alexa 488-labeled goat anti-mouse polyclonal antibody (1:500, Invitrogen) and DAPI (0.5 μg/ml) for 2 h, washed five times in 1× PBS for 2 min, and mounted in SlowFade Gold Antifade mountant (Life Technologies). Images were acquired using a DeltaVision Elite (GE Healthcare) and processed using SoftWoRx software (GE Healthcare).
RNA-seq
RNA was isolated from gastrocnemius or quadriceps muscles of two CUG960 +dox mice and one TRE +dox mice after 6 and 12 weeks of induction, beginning at PN1, as well as two CTG960 –dox mice at 6 and 12 weeks of age. It is generally recommended that at least six biological replicates are used for each condition in RNA-seq analysis (76); however, we have consistently found that the data obtained using ≥100 million paired end 100 bp reads is highly reproducible between the biological replicates (Supplement Material, Fig. S5A and B and Fig. S6A and B) (77–80). The Genomic and RNA Profiling Core (GARP) at Baylor College of Medicine (BCM) performed HiSeq library preparation and Illumina sequencing of 100-bp paired-end reads on a HiSeq 2500 sequencer. RNA was poly-A selected, barcoded and run in eight lanes. Low-quality nucleotides were trimmed from both ends of the sequence reads and reads were mapped to the mouse genome (UCSC mm10) using Tophat (81). Alternative splicing events, including skipped exons, alternative 5’/3’ splice sites, alternative first or last exons, mutually exclusive exons and retained introns, were analyzed using MISO and SpliceTrap algorithms (34,35). Alternative splicing was quantified by PSI value, indicating the fraction of transcripts that include a given exon. Events with ΔPSI >10%, when comparing the treatment group with the controls with a Bayes factor >5, were considered significant. Cufflinks/Cuffdiff (39,40) and Ht-Seq/EdgeR (41,42) pipelines were used to count the number of reads mapping to known genes and to detect differentially expressed genes between the treatment group and control groups using a false discovery rate cutoff of 0.05. For both alternative splicing data and gene expression data, events that changed when comparing CTG960 –dox with TRE +dox controls were filtered out of the analysis to rule out any events that were altered owing to the presence of doxycycline when repeats were not expressed. To determine the most significant transcriptome changes in response to expression of the repeats, only events that changed in the experimental treatment group relative to both control groups were considered for analysis using DAVID (37,38). Genes with relatively weak as well as relatively strong splicing changes were selected for validation of RNA-seq results by RT-PCR.
Reverse phase protein array
The Antibody-based Proteomics Core at BCM performed protein isolation and RPPA analysis as previously described with minor modifications (82–85). Protein lysates were isolated from gastrocnemius muscles of 12 CUG960 and eight TRE mice fed 2 g/kg dox chow for 2 weeks beginning at PN1 using modified Tissue Protein Extraction Reagent (TPER) (Pierce) with a cocktail of protease and phosphatase inhibitors (Roche Life Sciences). Protein lysates were diluted into 0.5 mg/ml of total protein in SDS sample buffer, denatured and printed on nitrocellulose-coated slides (Grace Bio-labs) using an Aushon 2470 Arrayer (Aushon BioSystems). A 40 pin (185 μm) configuration was used to spot samples and control lysates using an array format of 960 lysates/slide (2880 spots/slide). Slides were immunolabeled with 216 antibodies against total and phosphorylated proteins using an automated slide stainer Autolink 48 (Dako) as previously described. Each slide was incubated with a specific primary antibody and antibody diluent, rather than primary antibody, was used as a negative control. A biotinylated secondary antibody followed by streptavidin-conjugated IRDye680 fluorophore (LI-COR Biosciences) was used for detection of primary antibody binding. Fluorescent staining with Sypro Ruby-Blot Stain (Molecular Probes) was used to assess total protein for each spotted lysate.
Fluorescence-labeled slides and accompanying negative control slides were scanned on a GenePix 4400 AL scanner at an appropriate PMT to obtain optimal signal for the specific set of samples. Images were analyzed by GenePix Pro 7.0 (Molecular Devices). Local slide background signal was subtracted from the total fluorescence intensity to determine the fluorescence signal of each spot. Fluorescent signal for each spot per sample were normalized for variation in total protein level, background and non-specific labeling using group-based normalization method previously described (82). Each image and its associated normalized data were inspected manually and through control samples to evaluate quality of the data. Significantly changed proteins were assessed using Student’s t-test of the median intensity of triplicate experimental values; the criteria for significance were: maximum signal intensity greater than or equal to 200 across all samples, fold-change exceeding 1.25× in either direction between the comparison groups, and P < 0.05. Phospho- to total protein ratios were calculated by dividing the phosphoprotein expression by its total protein and the significant ratios were identified by t-test (fold-change >1.25 or <1/1.25, P < 0.05). Data are presented as a heat map of median normalized fluorescent signal intensity, using the Python language scientific library script. Muscle wasting in CUG960 +dox samples was classified as moderate or severe based on gastrocnemius weight. Ingenuity pathway analysis was performed for proteins that were significantly changed in CUG960 +dox mice. Protein lysates for CUG960 and TRE +dox mice prepared by the RPPA core were used to validate the RPPA data by western blot.
Western blot
Mouse protein lysates were prepared as detailed above for RPPA analysis. Human skeletal muscle was obtained from the National Disease Research Interchange tissue bank, the University of Miami tissue bank, and Dr Charles Thornton. Total protein lysates from DM1 and unaffected control skeletal muscle were prepared in RIPA buffer and protein concentration was quantified using a BCA protein assay kit (ThermoScientific).
Total protein lysates (20 μg) were separated on 10% Tris–glycine SDS-PAGE gels and transferred to Immobilon-P PVDF membranes (EMD Millipore) for western blot analysis. Membranes were stained with Ponceau S to visualize total protein, incubated with primary antibody overnight at 4°C. Membranes were washed three times in PBST (0.1% Tween-20, Sigma) and incubated 2 h in HRP-conjugated goat anti-rabbit (1:5000, Invitrogen) or goat anti-mouse (1:10 000, Cell Signaling Technology) antibody. After washing three times in PBST (0.1% Tween-20) immunoreactivity was detected using West Pico HRP-chemiluminescence system (ThermoScientific). Western blot membranes were imaged on a ChemiDoc XRS+ Imaging system (BioRad) and quantification of protein levels following normalization were calculated using Carestream Molecular Imaging software (Carestream Health). The following primary antibodies were used for western blot analysis: rabbit anti-PDGFRβ clone 28E1 (1:1000, Cell Signaling Technology), rabbit anti-PDGFRβ (Tyr751) (1:1000, Cell Signaling Technology), rabbit anti-AMPKα (Thr172) clone 40H9 (1:1000, Cell Signaling Technology), mouse anti-AMPKα clone F6 (1:1000, Cell Signaling Technology), rabbit anti-AMPKα clone 23A3 (1:1000, Cell Signaling Technology), rabbit anti-Stat3 (Ser727) (1:1000, Cell Signaling Technology), rabbit anti-Stat3 clone D3Z2G (1:1000, Cell Signaling Technology), rabbit anti-Caspase 3 (1:1000, Cell Signaling Technology), rabbit anti-Gapdh (1:7000, Cell Signaling Technology). Protein levels in mice were normalized to Gapdh protein levels for quantification. Human protein levels were normalized to total protein levels from the same blots, stained with Ponceau S (Sigma).
Statistical analysis
Results were presented as mean ± standard deviation. P-Values were calculated using two-tailed Student’s t-test when comparing only two treatment groups. For comparisons of three or more groups, P-values were determined by one-way ANOVA with Bonferroni multiple comparisons. Outliers for assessing percent muscle loss were determined using extreme studentized deviate tests. For analysis of CSA, two-way ANOVA with Tukey’s multiple comparisons was used for distribution and Kruskal–Wallis analysis with Dunn’s multiple comparisons was used to assess significance in median CSA. *P < 0.05, **P < 0.01, ***P < 0.001.
Note added in Proof
Initial studies found that Celf1 levels were unchanged in mice expressing CUG960 RNA. However, upon stratifying the samples based on severity of muscle loss and using the same samples as in Figure 6, we found that Celf1 protein levels are significantly increased only in mice that exhibit severe muscle wasting (Supplementary Material, Fig. S9).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
This project was supported by the Genomic and RNA Profiling Core at Baylor College of Medicine (BCM) and the expert assistance of core director, Dr Lisa D. White, Ph.D. RPPA analysis was performed by the Antibody-Based Proteomics Core at BCM, under the direction of core director Dr Shixia Huang, Ph.D. and research staff Fuli Jia and Danli Wu. Dr. Qianxing Mo performed RPPA data normalization. FISH/IF images were acquired in the Integrated Microscopy Core at BCM under the direction of academic director Dr Michael A. Mancini, Ph.D. and core director Dr Fabio Stossi, Ph.D., with assistance from Hannah Johnson. AccuraScience performed initial bioinformatics analyses for RNA-seq. The authors would like to thank Dr. Corey Reynolds, Ph.D., of the Mouse Metabolic and Phenotyping Core at BCM for assistance with grip strength tests. We also thank Dr. Mendell Rimer for the MDAF-rtTA mice. Special thanks to Donnie Bundman, Arseniy Kolonin, and Roberta Palau for technical assistance.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health [R01AR045653 (to T.A.C.), R01AR060733 (to T.A.C.), and K12GM084897 (to G.R.M.)]; the Muscular Dystrophy Association (T.A.C); and the Myotonic Dystrophy Foundation (G.R.M). Support for the Antibody-Based Proteomics Core at BCM was provided by CPRIT Core Facility Award [RP170005 (to Dean P. Edwards and S.H.)] and P30 Cancer Center Support Grant [NCI-CA125123 (to Dean P. Edwards and S.H.)]. The Mouse Metabolic and Phenotyping Core is supported by the National Institutes of Health [UM1HG006348 and R01DK114356]. The Integrated Microscopy Core at BCM is supported by P30 Cancer Center Support Grant [NCI-CA125123], P30 Digestive Disease Center [NIDDK-56338–13/15], CPRIT [RP170005], and John S. Dunn Gulf Coast Consortium for Chemical Genomics.
References
- 1. Harper P. (2001) Myotonic Dystrophy. W.B. Saunders Company, London. [Google Scholar]
- 2. Thornton C.A. (2014) Myotonic dystrophy. Neurol. Clin., 32, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.-P., Hudson T.. et al. (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell, 68, 799–808. [DOI] [PubMed] [Google Scholar]
- 4. Mankodi A., Urbinati C.R., Yuan Q.P., Moxley R.T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M.S., Thornton C.A. (2001) Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet., 10, 2165–2170. [DOI] [PubMed] [Google Scholar]
- 5. Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. (2004) Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet., 13, 3079–3088. [DOI] [PubMed] [Google Scholar]
- 6. Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. (1995) Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell. Biol., 128, 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. (1997) Expansion of a CUG trinucleotide repeat in the 3' untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. U.S.A., 94, 7388–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuyumcu-Martinez N.M., Wang G.-S., Cooper T.A. (2007) Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell, 28, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A. (2006) Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet., 15, 2087–2097. [DOI] [PubMed] [Google Scholar]
- 10. Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. (2008) A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A., 105, 20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang E.T., Cody N.A.L., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S.. et al. (2012) Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell, 150, 710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masuda A., Andersen H.S., Doktor T.K., Okamoto T., Ito M., Andresen B.S., Ohno K. (2012) CUGBP1 and MBNL1 preferentially bind to 3' UTRs and facilitate mRNA decay. Sci. Rep., 2, 209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rau F., Freyermuth F., Fugier C., Villemin J.-P., Fischer M.-C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D.. et al. (2011) Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol., 18, 840–845. [DOI] [PubMed] [Google Scholar]
- 14. Kalsotra A., Singh R.K., Gurha P., Ward A.J., Creighton C.J., Cooper T.A. (2014) The Mef2 transcription network is disrupted in myotonic dystrophy heart tissue, dramatically altering miRNA and mRNA expression. Cell. Rep., 6, 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A.. et al. (2014) Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell, 56, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vlasova I.A., Tahoe N.M., Fan D., Larsson O., Rattenbacher B., Sternjohn J.R., Vasdewani J., Karypis G., Reilly C.S., Bitterman P.B.. et al. (2008) Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell, 29, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee K.-Y., Li M., Manchanda M., Batra R., Charizanis K., Mohan A., Warren S.A., Chamberlain C.M., Finn D., Hong H.. et al. (2013) Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med., 5, 1887–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanadia R.N., Shin J., Yuan Y., Beattie S.G., Wheeler T.M., Thornton C.A., Swanson M.S. (2006) Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A., 103, 11748–11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chamberlain C.M., Ranum L.P.W. (2012) Mouse model of muscleblind-like 1 overexpression: skeletal muscle effects and therapeutic promise. Hum. Mol. Genet., 21, 4645–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Timchenko N.A., Patel R., Iakova P., Cai Z.-J., Quan L., Timchenko L.T. (2004) Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem., 279, 13129–13139. [DOI] [PubMed] [Google Scholar]
- 21. Ward A.J., Rimer M., Killian J.M., Dowling J.J., Cooper T.A. (2010) CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum. Mol. Genet., 19, 3614–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. (2000) Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science, 289, 1769–1773. [DOI] [PubMed] [Google Scholar]
- 23. Seznec H., Agbulut O., Sergeant N., Savouret C., Ghestem A., Tabti N., Willer J.C., Ourth L., Duros C., Brisson E.. et al. (2001) Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum. Mol. Genet., 10, 2717–2726. [DOI] [PubMed] [Google Scholar]
- 24. Orengo J.P., Chambon P., Metzger D., Mosier D.R., Snipes G.J., Cooper T.A. (2008) Expanded CTG repeats within the DMPK 3' UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A., 105, 2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huguet A., Medja F., Nicole A., Vignaud A., Guiraud-Dogan C., Ferry A., Decostre V., Hogrel J.-Y., Metzger F., Hoeflich A.. et al. (2012) Molecular, physiological, and motor performance defects in DMSXL mice carrying >1, 000 CTG repeats from the human DM1 locus. PLoS Genet., 8, e1003043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahadevan M.S., Yadava R.S., Yu Q., Balijepalli S., Frenzel-McCardell C.D., Bourne T.D., Phillips L.H. (2006) Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet., 38, 1066–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomes-Pereira M., Foiry L., Nicole A., Huguet A., Junien C., Munnich A., Gourdon G. (2007) CTG trinucleotide repeat ‘big jumps’: large expansions, small mice. PLoS Genet., 3, e52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gomes-Pereira M., Cooper T.A., Gourdon G. (2011) Myotonic dystrophy mouse models: towards rational therapy development. Trends. Mol. Med., 17, 506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sicot G., Gourdon G., Gomes-Pereira M. (2011) Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: new findings and future challenges. Hum. Mol. Genet., 20, R116–R123. [DOI] [PubMed] [Google Scholar]
- 30. Ponomareva O.N., Ma H., Vock V.M., Ellerton E.L., Moody S.E., Dakour R., Chodosh L.A., Rimer M. (2006) Defective neuromuscular synaptogenesis in mice expressing constitutively active ErbB2 in skeletal muscle fibers. Mol. Cell. Neurosci., 31, 334–345. [DOI] [PubMed] [Google Scholar]
- 31. Savkur R.S., Philips A.V., Cooper T.A. (2001) Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy JA. Nat. Genet., 29, 40–47. [DOI] [PubMed] [Google Scholar]
- 32. Lee C.S., Georgiou D.K., Dagnino-Acosta A., Xu J., Ismailov I.I., Knoblauch M., Monroe T.O., Ji R., Hanna A.D., Joshi A.D.. et al. (2014) Ligands for FKBP12 increase Ca2+ influx and protein synthesis to improve skeletal muscle function. J. Biol. Chem., 289, 25556–25570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt E.K., Clavarino G., Ceppi M., Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods, 6, 275–277. [DOI] [PubMed] [Google Scholar]
- 34. Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu J., Akerman M., Sun S., McCombie W.R., Krainer A.R., Zhang M.Q. (2011) SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics, 27, 3010–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature, 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang D.W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 38. Huang D.W., Sherman B.T., Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res., 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol., 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Anders S., Pyl P.T., Huber W. (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson M.D., McCarthy D.J., Smyth G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakamori M., Sobczak K., Puwanant A., Welle S., Eichinger K., Pandya S., Dekdebrun J., Heatwole C.R., McDermott M.P., Chen T.. et al. (2013) Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol., 74, 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spurrier B., Ramalingam S., Nishizuka S. (2008) Reverse-phase protein lysate microarrays for cell signaling analysis. Nat. Protoc., 3, 1796–1808. [DOI] [PubMed] [Google Scholar]
- 45. Bargiela A., Cerro-Herreros E., Fernandez-Costa J.M., Vilchez J.J., Llamusi B., Artero R. (2015) Increased autophagy and apoptosis contribute to muscle atrophy in a myotonic dystrophy type 1 Drosophila model. Dis. Model Mech., 8, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brockhoff M., Rion N., Chojnowska K., Wiktorowicz T., Eickhorst C., Erne B., Frank S., Angelini C., Furling D., Rüegg M.A.. et al. (2017) Targeting deregulated AMPK/mTORC1 pathways improves muscle function in myotonic dystrophy type I. J. Clin. Invest., 127, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M.J., Thompson C.B. (2005) AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell, 18, 283–293. [DOI] [PubMed] [Google Scholar]
- 48. Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K.. et al. (2006) TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell, 126, 955–968. [DOI] [PubMed] [Google Scholar]
- 49. Liang J., Shao S.H., Xu Z.-X., Hennessy B., Ding Z., Larrea M., Kondo S., Dumont D.J., Gutterman J.U., Walker C.L.. et al. (2007) The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol., 9, 218–224. [DOI] [PubMed] [Google Scholar]
- 50. Li Q.-L., Gu F.-M., Wang Z., Jiang J.-H., Yao L.-Q., Tan C.-J., Huang X.-Y., Ke A.-W., Dai Z., Fan J.. et al. (2012) Activation of PI3K/AKT and MAPK pathway through a PDGFRβ-dependent feedback loop is involved in Rapamycin resistance in hepatocellular carcinoma. PLoS One, 7, e33379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kazlauskas A., Cooper J.A. (1990) Phosphorylation of the PDGF receptor b subunit creates a tight binding site for phospatidylinositol 3 kinase. EMBO J., 9, 3279–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mankodi A., Teng-Umnuay P., Krym M., Henderson D., Swanson M., Thornton C.A. (2003) Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann. Neurol., 54, 760–768. [DOI] [PubMed] [Google Scholar]
- 53. Michel L., Huguet-Lachon A., Gourdon G. (2015) Sense and antisense DMPK RNA foci accumulate in DM1 tissues during development. PLoS One, 10, e0137620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mankodi A., Takahashi M.P., Jiang H., Beck C.L., Bowers W.J., Moxley R.T., Cannon S.C., Thornton C.A. (2002) Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell, 10, 35–44. [DOI] [PubMed] [Google Scholar]
- 55. Philips A.V., Timchenko L.T., Cooper T.A. (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- 56. Jones K., Wei C., Iakova P., Bugiardini E., Schneider-Gold C., Meola G., Woodgett J., Killian J., Timchenko N.A., Timchenko L.T. (2012) GSK3beta mediates muscle pathology in myotonic dystrophy. J. Clin. Invest., 122, 4461–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maehama T., Dixon J.E. (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem., 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 58. Kumar A., Knox A.J., Boriek A.M. (2003) CCAAT/enhancer-binding protein and activator protein-1 transcription factors regulate the expression of interleukin-8 through the mitogen-activated protein kinase pathways in response to mechanical stretch of human airway smooth muscle cells. J. Biol. Chem., 278, 18868–18876. [DOI] [PubMed] [Google Scholar]
- 59. Wysoczynski M., Shin D.-M., Kucia M., Ratajczak M.Z. (2010) Selective upregulation of interleukin-8 by human rhabdomyosarcomas in response to hypoxia: therapeutic implications. Int. J. Cancer, 126, 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Amir Levy Y., Ciaraldi T.P., Mudaliar S.R., Phillips S.A., Henry R.R. (2015) Excessive secretion of IL-8 by skeletal muscle in type 2 diabetes impairs tube growth: potential role of PI3K and the Tie2 receptor. Am. J. Physiol. Endocrinol. Metab., 309, E22–E34. [DOI] [PubMed] [Google Scholar]
- 61. Hoffman E.P., Nader G.A. (2004) Balancing muscle hypertrophy and atrophy. Nat. Med., 10, 584–585. [DOI] [PubMed] [Google Scholar]
- 62. Egerman M.A., Glass D.J. (2014. Jan-Feb) Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol., 49, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shimizu N., Yoshikawa N., Ito N., Maruyama T., Suzuki Y., Takeda S.-I., Nakae J., Tagata Y., Nishitani S., Takehana K.. et al. (2011) Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab., 13, 170–182. [DOI] [PubMed] [Google Scholar]
- 64. Stitt T.N., Drujan D., Clarke B.A., Panaro F., Timofeyva Y., Kline W.O., Gonzalez M., Yancopoulos G.D., Glass D.J. (2004) The IGF-1/PI3K/Akt pathway prevents short article expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell, 14, 395–403. [DOI] [PubMed] [Google Scholar]
- 65. Furling D., Marette A., Puymirat J. (1999) Insulin-like growth factor I circumvents defective insulin action in human myotonic dystrophy skeletal muscle cells. Endocrinology, 140, 4244–4250. [DOI] [PubMed] [Google Scholar]
- 66. Woods A., Vertommen D., Neumann D., Türk R., Bayliss J., Schlattner U., Wallimann T., Carling D., Rider M.H. (2003) Identification of phosphorylation sites in AMP-activated protein kinase (AMPK) for upstream AMPK kinases and study of their roles by site-directed mutagenesis. J. Biol. Chem., 278, 28434–28442. [DOI] [PubMed] [Google Scholar]
- 67. Hardie D.G. (2008) AMPK: a key regulator of energy balance in the single cell and the whole organism. Int. J. Obes. (Lond.), 32(Suppl 4), S7–S12. [DOI] [PubMed] [Google Scholar]
- 68. Mihaylova M.M., Shaw R.J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell. Biol., 13, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Graham N.A., Tahmasian M., Kohli B., Komisopoulou E., Zhu M., Vivanco I., Teitell M.A., Wu H., Ribas A., Lo R.S.. et al. (2012) Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Mol. Syst. Biol., 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gough D.J., Koetz L., Levy D.E. (2013) The MEK-ERK Pathway Is Necessary for Serine Phosphorylation of Mitochondrial STAT3 and Ras-Mediated Transformation. PLoS One, 8, e83395.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zouein F.A., Altara R., Chen Q., Lesnefsky E.J., Kurdi M., Booz G.W. (2015) Pivotal importance of STAT3 in protecting the heart from acute and chronic stress: new advancement and unresolved issues. Front. Cardiovasc. Med., 2, e24352–e24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Beffy P., Del Carratore R., Masini M., Furling D., Puymirat J., Masiello P., Simili M. (2010) Altered signal transduction pathways and induction of autophagy in human myotonic dystrophy type 1 myoblasts. Int. J. Biochem. Cell. Biol., 42, 1973–1983. [DOI] [PubMed] [Google Scholar]
- 73. Denis J.A., Gauthier M., Rachdi L., Aubert S., Giraud-Triboult K., Poydenot P., Benchoua A., Champon B., Maury Y., Baldeschi C.. et al. (2013) mTOR-dependent proliferation defect in human ES-derived neural stem cells affected by myotonic dystrophy type 1. J. Cell. Sci., 126, 1763–1772. [DOI] [PubMed] [Google Scholar]
- 74. Loro E., Rinaldi F., Malena A., Masiero E., Novelli G., Angelini C., Romeo V., Sandri M., Botta A., Vergani L. (2010) Normal myogenesis and increased apoptosis in myotonic dystrophy type-1 muscle cells. Cell. Death Differ., 17, 1315–1324. [DOI] [PubMed] [Google Scholar]
- 75. Kumar A., Accorsi A., Rhee Y., Girgenrath M. (2015) Do’s and Don’ts in the preparation of muscle cryosections for histological analysis. J Vis. Exp., 10, e52793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schurch N.J., Schofield P., Gierlinski M., Cole C., Sherstnev A., Singh V., Wrobel N., Gharbi K., Simpson G.G., Owen-Hughes T., Blaxter M., Barton G.J. (2016) How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA, 22, 839-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Giudice J., Xia Z., Wang E.T., Scavuzzo M.A., Ward A.J., Kalsotra A., Wang W., Wehrens X.H., Burge C.B., Li W., Cooper T.A. (2014) Alternative splicing regulates vesicular trafficking genes in cardiomyocytes during postnatal heart development. Nat. Commun., 5, 3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Singh R.K., Xia Z., Bland C.S., Kalsotra A., Scavuzzo M.A., Curk T., Ule J., Li W., Cooper T.A. (2014) Rbfox2-coordinated alternative splicing of Mef2d and Rock2 controls myoblast fusion during myogenesis. Mol. Cell. 55, 592-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Giudice J., Xia Z., Li W., Cooper T.A. (2016) Neonatal cardiac dysfunction and transcriptome changes caused by the absence of Celf1. Sci. Rep., 6, 35550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brinegar A.E., Xia Z., Loehr J.A., Li W., Rodney G.G., Cooper T.A. (2017) Extensive alternative splicing transitions during postnatal skeletal muscle development are required for calcium handling functions. Elife., 6, e27192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chang C.-H., Zhang M., Rajapakshe K., Coarfa C., Edwards D., Huang S., Rosen J.M. (2015) Mammary stem cells and tumor-initiating cells are more resistant to apoptosis and exhibit increased DNA repair activity in response to DNA damage. Stem Cell Rep., 5, 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Creighton C.J., Huang S. (2015) Reverse phase protein arrays in signaling pathways: a data integration perspective. Drug Des. Dev. Ther., 9, 3519–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Holdman X.B., Welte T., Rajapakshe K., Pond A., Coarfa C., Mo Q., Huang S., Hilsenbeck S.G., Edwards D.P., Zhang X.. et al. (2015) Upregulation of EGFR signaling is correlated with tumor stroma remodeling and tumor recurrence in FGFR1-driven breast cancer. Breast Cancer Res., 17, 141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Welte T., Kim I.S., Tian L., Gao X., Wang H., Li J., Holdman X.B., Herschkowitz J.I., Pond A., Xie G.. et al. (2016) Oncogenic mTOR signalling recruits myeloid-derived suppressor cells to promote tumour initiation. Nat. Cell. Biol., 18, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.