Abstract
OEP7, a 6.7-kDa outer envelope protein of spinach chloroplasts inserts into the outer envelope of the organelle independent of a classical cleavable targeting signal. The insertion of OEP7 was studied to describe the determinants for association with, integration into, and orientation of the protein in the outer envelope of chloroplasts. The insertion of OEP7 into the membrane is independent of outer membrane channel proteins and can be reconstituted with the use of protein-free liposomes. In situ, the binding of OEP7 to the membrane surface is not driven by electrostatic interaction because reduction of phosphatidylglycerol or phosphatidylinositol did not reduce the association with the liposomes. The positively charged amino acids flanking the transmembrane domain at the C terminus are essential to retain the native Nin-Cout orientation during insertion into chloroplasts. OEP7 inserts with reversed orientation into liposomes containing the average lipid composition of the outer envelopes. The native like Nin-Cout orientation is achieved by reduction of the phoshpatidylglycerol concentration mimicking the composition of the outer leaflet of the outer envelope of chloroplasts. We conclude that the unique lipid composition of the outer leaflet due to lipid asymmetry of the outer envelope is essential for the correct topology of OEP7.
INTRODUCTION
Many chloroplast proteins are encoded by the nuclear genome and have to be imported into the organelle. The best-studied translocation pathway is initiated by cytosolic chaperones that transfer the preproteins to a membrane-located complex comprised of translocon at the outer envelope of chloroplast (Toc) proteins. The preprotein will then be transferred to the subunits of the inner envelope import machinery (Tic complex), released into the stroma, and distributed to the subcompartments of the organelle (Keegstra and Cline, 1999; Schleiff and Soll, 2000). However, most proteins of the chloroplast outer envelope were found to insert independently of this classical import pathway in vitro (reviewed by Soll and Tien, 1998). Three proteins studied in some detail are the 14-kDa OEP14 (Li et al., 1991; Tu and Li, 2000), the import receptor Toc34 (Kessler et al., 1994; Seedorf et al., 1995), and a prominent outer envelope protein, the 6.7-kDa OEP7 from spinach (Salomon et al., 1990; Kolke et al., 1998).
OEP14 contains a single transmembrane (TM) domain and has a Nin-Cout orientation. The insertion is independent of ATP and thermolysin-sensitive factors (Li and Chen, 1996). Furthermore, OEP14 specifically inserts into the chloroplast outer envelope but not into microsomal (Li and Chen, 1996) or mitochondrial membranes (Li et al., 1991). The insertion of a heterologously expressed His tag containing protein was found to be N-ethylmaleimide sensitive and saturable but not dependent on cytosolic factors (Tu and Li, 2000). However, nothing is known about the determinants of the topology. Toc34 contains a single C-terminal transmembrane domain with a Cin-Nout orientation (Seedorf et al., 1995). Insertion of Toc34 was found to be stimulated by ATP (Seedorf et al., 1995; Li and Chen, 1997; Tsai et al., 1999) and GTP (Chen and Schnell, 1997; Tsai et al., 1999). The two positive charges flanking the transmembrane domain at the cytosolic site influence the orientation of this protein (May and Soll, 1998). The influence of outer envelope proteins on the insertion or assembly process remains to be further investigated, because protease treatment reduced but did not abolish Toc34 integration (Seedorf et al., 1995; Chen and Schnell, 1997; Tsai et al., 1999). OEP7 also has a single transmembrane domain but with an Nin-Cout orientation flanked by two equally sized soluble domains (Salomon et al., 1990; Waegemann et al., 1992). The insertion of OEP7 is dependent on temperature, but independent of light, ATP, a membrane potential, or thermolysin-sensitive components of the outer envelope (Salomon et al., 1990). Due to its simple structure OEP7 might serve as a model to study the mechanism of protein insertion into the chloroplast outer envelope as well as the determinants that govern OEP7 topology.
The outer envelope of chloroplast exhibits several unique and important features. It contains a lower concentration of phosphatidylcholine (PC) and a higher concentration of phosphatidylglycerol (PG) in the inner than in the outer leaflet of the outer envelope membrane (Dorne et al., 1985). It is also the only membrane facing the cytosol, which contains the nonbilayer lipid monogalactosyldiacylglyceride (MGDG) (Bruce, 1998). Furthermore, MGDG is the only nonbilayer lipid present in the envelopes. MGDG plays an important role during association of the transit sequence of preferredoxin and pre small subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase (SSU) with lipid surfaces (van 't Hof et al., 1991, 1993; Chupin et al., 1994; Pilon et al., 1995). Phosphatidylethanolamine (PE), another nonbilayer lipid, assists protein folding of membrane proteins (Bogdanov and Dowhan, 1998; Bogdanov et al., 1999) and is required for efficient protein transport across the plasma membrane of Escherichia coli (Rietveld et al., 1995). PE was shown to mediate the interaction of the catalytic domain of the leader peptidase with the membrane of E. coli (van Klompenburg et al., 1998). A second class of lipids found to be important for association and insertion of proteins into bilayers is charged (anionic) lipids such as PG and phosphatidylinositol (PI) (van't Hof et al., 1991, 1993). Proteins such as the GTPase FtsY associate with membranes in an anionic lipid stimulated manner (de Leeuw and Luirink, 1997; de Leeuw et al., 2000). In addition, anionic lipids are thought to mediate insertion of peptides with an overall hydrophobicity not sufficient to mediate spontaneous insertion into neutral membranes (Liu and Deber, 1997).
Here we show that the association of OEP7 with the membrane is initiated by the hydrophobicity of the transmembrane region. OEP7 binds to and inserts into the membrane independent of other envelope proteins. The positively charged amino acids of the C terminus flanking the transmembrane domain are the only determinants of the topology within OEP7. However, the topology of OEP7 was inverted when liposomes with an average lipid composition of the outer envelope were used. After reducing the content of charged lipids the same orientation as in situ was observed. We conclude that the asymmetric distribution of PG between both leaflets of the outer envelope is a major determinant for the topology of OEP7.
MATERIALS AND METHODS
OEP7 Mutations
The cDNA coding for OEP7 was cloned into pBluescript (Salomon et al., 1990). OEP7-Δ4 was created by digestion with HincII and HindIII removing the last 21 base pairs of the cDNA followed by insertion of a DNA fragment (CTG AGG ACG TAA) coding for a leucine, arginine, and threonine. cDNA constructs coding for OEP7-Δ12 or OEP7 with single amino acid exchanges were obtained by recombinant polymerase chain reaction. In OEP7-Δ12 the last 12 amino acids are deleted. In OEP7-LM1 two point mutations were introduced; codon GAG (base pairs 34–36) was changed to CAG resulting in a Glu-to-Gln mutation at amino acid 12 (Table 1) and codon TCC (base pairs 40–42) was changed to AAA, resulting in a Gly-to-Lys mutation at amino acid 14. For OEP7-LM2, OEP7 was modified with the use of two primers to introduce a point mutation at base pairs 130–132 (CGA to GAA), resulting in an Arg-to-Glu mutation at amino acid 44. In OEP7-LM3, all point mutations as described for OEP7-LM1 and OEP7-LM2 were combined. Polymerase chain reaction products were cloned into pBluescript (OEP7-Δ12) or pET21b (OEP7 containing point mutations) and mutations were confirmed by sequencing.
Table 1.
Amino acids sequence and charge distribution of OEP7wt and mutants
| OEP7 | mEsvakpattkEgsakqaaivvgvlalgwfaiqvafiplfnkvrgggsDkkDDDvnaftpDt |
| − + +− + TM ++ −++−−− − | |
| OEP7-Δ4 | mEsvakpattkEgsakqaaivvgvlalgwfaiqvafiplfnkvrgggsDkkDDDvlrt |
| − + +− + TM ++ −++−−− + | |
| OEP7-Δ12 | mEsvakpattkEgsakqaaivvgvlalgwfaiqvafiplfnkvrgggsDk |
| − + +− + TM ++ −+ | |
| OEP7-LM1 | mEsvakpattkqgkakqaaivvgvlalgwfaiqvafiplfnkvrgggsDkkDDDvnaftpDt |
| − + + ++ TM ++ −++−−− − | |
| OEP7-LM2 | mEsvakpattkEgsakqaaivvgvlalgwfaiqvafiplfnkvEgggsDkkDDDvnaftpDt |
| − + +− + TM +− −++−−− − | |
| OEP7-LM3 | mEsvakpattkqgkakqaaivvgvlalgwfaiqvafiplfnkvEgggsDkkDDDvnaftpDt |
| − + + ++ TM +− −++−−− − |
| N-terminus ↓ | 4aa ↓ ← | 4aa → ↓ | C-terminus ↓ | ||
|---|---|---|---|---|---|
| OEP7 | +1 | +1 | charge of the flanking regions | +2 | −1 |
| OEP7-Δ4 | +1 | +1 | +2 | +1 | |
| OEP7-Δ12 | +1 | +1 | +2 | +2 | |
| OEP7-LM1 | +3 | +2 | +2 | −1 | |
| OEP7-LM2 | +1 | +1 | ±0 | −3 | |
| OEP7-LM3 | +3 | +2 | ±0 | −3 |
Capital letter, acidic amino acid; italic letter, basic amino acid; underlined letter, predicted TM domain; bold letter (OEP7), charged amino acid; bold letter (OEP7 mutant), point mutation; aa, amino acid.
Transcription and Translation of OEP7 wt and Variants
In vitro transcription of linearized plasmids encoding for OEP7 and was mutants performed with the use of T7 polymerase (Salomon et al., 1990). Proteins were synthesized in a system containing reticulocyte lysate (Amersham Pharmacia Biotech, Freiburg, Germany) in the presence of either [35S]methionine (1175 Ci/mmol) or [3H]leucine (148 Ci/mmol) for 1.5 h at 30°C. The translation mixture was centrifuged for 1 h at 250,000 × g at 4°C and the postribosomal supernatant was used for import. The in vitro transcription and translation of preSSU was described in (Waegemann and Soll, 1995)
Protein Import into Spinach Chloroplasts
Spinach chloroplasts were isolated by standard procedures and further purified on a Percoll gradient. Chlorophyll concentration was determined (Arnon, 1949; Mourioux and Douce, 1981; Schindler et al., 1987). Standard import into chloroplasts equivalent to 40 μg of chlorophyll was performed in 100 μl of import buffer (10 mM methionine (or leucine), 20 mM potassium gluconate, 10 mM NaHCO3, 3 mM MgSO4, 330 mM sorbitol, 50 mM HEPES/KOH, pH 7.6) containing 1–10% of in vitro-translated 35S- or 3H-labeled proteins. Insertion assays were carried out in the dark. Import was initiated by addition of organelles to import mixture and stopped after the times indicated. Intact chloroplasts were reisolated through a Percoll cushion (40% Percoll in 330 mM sorbitol, 50 mM HEPES/KOH, pH 7.6) washed once in 330 mM sorbitol, 50 mM HEPES/KOH, pH 7.6, 3 mM MgCl2, and used for further treatments.
Chloroplasts were treated with thermolysin (40 μg/20 μg of chlorophyll) for 30 min on ice in 330 mM sorbitol, 50 mM HEPES-KOH, pH 7.6, 3 mM MgSO4, 0.5 mM CaCl2. The reaction was stopped with 10 mM EDTA and chloroplasts were recovered by centrifugation (Joyard et al., 1983). Alkali extraction was performed as described (Salomon et al., 1990). Import products were analyzed by Tricine SDS-PAGE (Schägger and von Jagow, 1987) followed by fluorography (Bonner and Laskey, 1974). Alternatively, emulsifier scintillator 299TM-cocktail (Packard, Groningen, The Netherlands) was added and radioactivity was quantified with the use of a PW 4700 liquid scintillation counter (Philips, Eindhoven, The Netherlands).
Synthesis of Liposomes for Insertion of OEP7wt and Mutants
Purified plant lipids were provided by Nutfield Nurseries (Surrey, United Kingdom). Liposomes with various lipid content (Table 2) were prepared as follows. The lipids were mixed in a glass tube to yield a final concentration of 5 μmol of total lipid content and dried under N2 flow. Lipids were dissolved in 1 ml of trichlormethane followed by N2 drying and complete removal of the organic solvent under vacuum for at least 3 h. The created lipid film was either stored at −80°C under argon or directly dissolved in buffer S (50 mM HEPES-KOH, pH 7.6, 0.2 M sucrose, degassed with the use of N2) for synthesis of S-liposomes or in buffer N (50 mM HEPES-KOH, 125 mM NaCl, degassed) for the synthesis of N-liposomes. The solution was vortexed and freeze-thawed five times. The multilamellar vesicles were extruded 21 times through a 100-nm pore polycarbonate filter mounted in the mini-extruder (Liposofast; Avestin, Ottawa, ONT) to give unilamellar liposomes (MacDonald et al., 1991).
Table 2.
Lipid content (mol%) of different liposomes used in the experiments
| Lipid | Standard | Comp 1 | Comp 2 | Comp 3 | Comp 4 | Comp-PG | Comp-SL | Comp-PI |
|---|---|---|---|---|---|---|---|---|
| MGDG | 17 | 21 | 15 | 13 | 14 | 19 | 18 | 18 |
| DGDG | 29 | 37 | 27 | 22 | 25 | 32 | 31 | 31 |
| SL | 6 | 7 | 5 | 4 | 3 | 6.5 | — | 6 |
| PC | 32 | 16 | 39 | 50 | 50 | 36 | 34 | 34 |
| PG | 10 | 12 | 9 | 7 | 5 | — | 11 | 11 |
| PI | 6 | 7 | 5 | 4 | 3 | 6.5 | 6 | — |
Standard composition according to Bruce, 1998.
Purification of Outer Envelope Lipids
Outer envelopes of spinach chloroplasts were purified as described (Joyard et al., 1983). Lipids were extracted from 1 ml of outer envelope membranes (1 mg of protein/ml) by addition of 2.5 ml of trichlormethane:methanol (2:1, vol/vol). Then, 20 ml of trichlormethane:water (1:1) was added, the mixture vortexed and centrifuged at 3000 × g for 15 min at 4°C. The trichlormethan layer was transferred to a new tube and further centrifuged for 1 min at 3000 × g. Again only the trichlormethane fraction was removed to a new tube and dried under a stream of N2. The lipid film obtained was dissolved in 5 ml of trichlormethane and finally dried under N2 gas. Phospholipid concentration was determined in a Lowry-Tinslay assay (Lowry and Tinsley, 1974) and total concentration calculated according to the mol% of phospholipids present in the outer envelope. No protein contamination was observed as controlled by SDS-PAGE. Liposomes were prepared as described with the use of 5 μmol of outer envelope lipids.
Insertion Assay of OEP7wt and Mutants into Liposomes
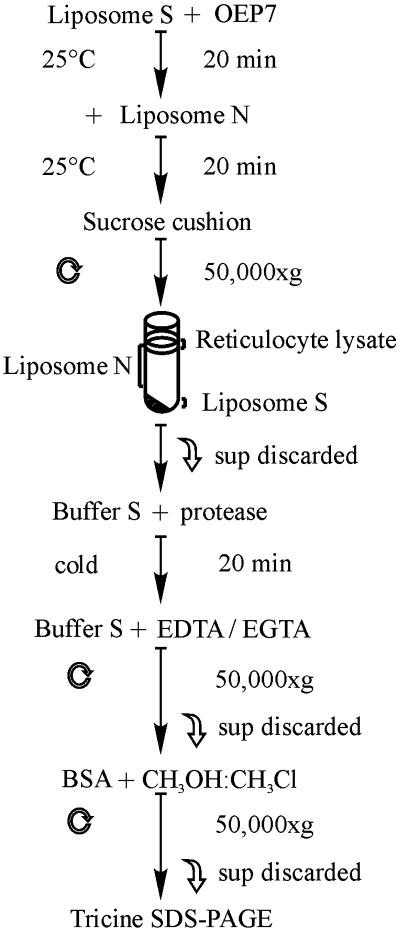
Radioactive labeled translation product of OEP7 and variants were incubated with 1 mM (final lipid concentration) S-liposomes in 100 μl of buffer S (Figure 1) at 25°C and indicated times. After insertion, surface bound OEP7 was removed from S-liposomes by competition with 10 mM (final lipid concentration) N-liposomes in 100 μl of buffer N/100 μl of buffer S for 20 min (Schleiff, et. al., 1999). Both liposomes species were separated by centrifugation through a sucrose cushion (buffer S) for 30 min at 50,000 × g. Separation was tested by addition of 0.05 mol% fluorescent labeled 1,2-dioleyl-sn-glycero-3-phosphoethanolamine-N (18:1) during preparation of N-liposomes or S-liposomes. Fluorescence spectroscopy revealed that maximal 2% N-liposomes were pelleted but at least 82% S-liposomes were recovered from the cushion. The pellet was resuspended in 100 μl of buffer S or for further treatment with thermolysin in 100 μl of 50 mM HEPES-KOH, pH 7.6, 0.2 M sucrose, 0.5 mM MgCl2, 0.1 mM CaCl2. Thermolysin was added to a final concentration of 50 μg and proteolysis was stopped after 30 min with 20 mM each of EDTA and EGTA. Proteoliposomes were recovered by centrifugation for 30 min at 50,000 × g. The pellet was resuspended 100 μl of buffer S containing 30 μg of fatty acid-free bovine serum albumin and adjusted to 1500 μl with methanol:trichlormethane (2:1). Proteins were recovered by centrifugation for 30 min at 50,000 × g and separated by 14% Tricine-SDS-PAGE. Radioactivity was visualized by fluorography or quantified by scintillation counting.
Figure 1.
Outline of the procedure for OEP7 insertion into liposomes. Details are described in MATERIALS AND METHODS.
Calculation of Free Energy of Membrane Association and Membrane Insertion
The calculation of the free energy of association and insertion is based on the findings of several studies (Engelman et al., 1986; Kim et al., 1991; van der Goot et al., 1991; Peitzsch et al., 1995; Ben-Shaul et al., 1996; Ben-Tal et al., 1996a,b; Murray et al., 1998; White and Wimley, 1998; Wieprecht et al., 1999; Kessel et al., 2000) and a detailed discussion is given under “Supplementary Material.”
RESULTS
Negatively Charged Region at C Terminus Is not Essential for Topology of OEP7
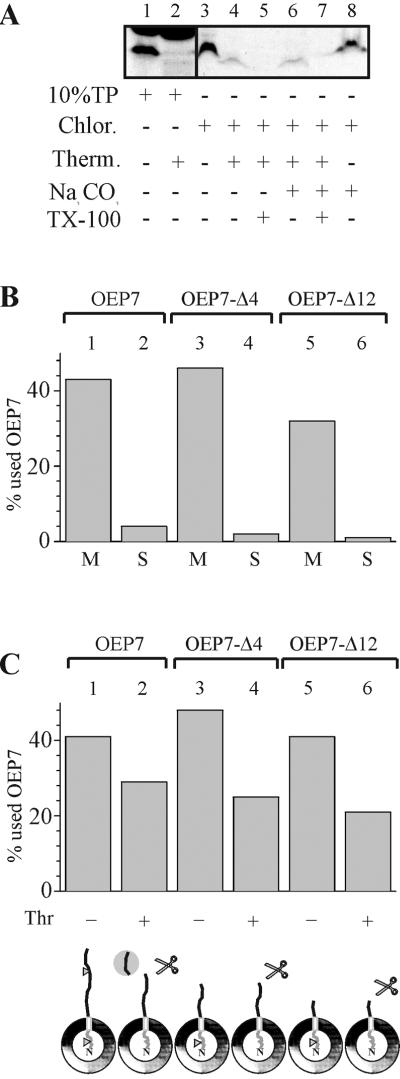
OEP7 contains a centrally located transmembrane domain and inserts into the outer envelope of chloroplasts (Salomon et al., 1990; Figure 2A, lane 3) in a carbonate-resistant manner (Figure 2A, lane 8). The N terminus of OEP7 contains the only methionine of the protein. Therefore, proteolysis can be used as a tool to identify the localization of the N-terminal hydrophilic domain. After protease treatment of inserted OEP7 a smaller labeled proteolytic fragment is detectable (Figure 2A, lane 4). This fragment was not detected when translation product was treated with protease (Figure 2A, lane 2). This suggests that the N terminus of OEP7 has translocated over the outer envelope membrane, whereas the C-terminal region is exposed at the cytosolic side and therefore remains protease sensitive. The 35S-labeled fragment was resistant to extraction at pH11, indicating that it behaved like an integral membrane protein (Figure 2A, lane 6). To further confirm this interpretation, chloroplasts were solubilized with detergent before protease treatment (Figure 2A, lanes 5 and 7), resulting in the loss of the labeled fragment. After establishing conditions for OEP7 insertion into chloroplasts we wanted to investigate the determinants for the topology of OEP7.
Figure 2.
In vitro import of OEP7 and C-terminal deletion mutants into spinach chloroplasts. (A) In vitro synthesized [35S]methionine-labeled OEP 7 was imported into chloroplasts for 30 s (lanes 3–8). Chloroplasts were then treated with thermolysin (lanes 4–7) without (lanes 4 and 6) or with (lanes 5 and 7) lysis of the chloroplasts. After completed treatment chloroplasts were extracted by addition of carbonate (lanes 6–8). Translation product (10%) before (lane 1) and after (lane 2) treatment is shown. (B and C) In vitro-synthesized [35S]methionine-labeled OEP 7 and variants (Table 1) are imported into intact spinach chloroplasts for 60 s (lanes 1–6) followed by proteolysis with the use of thermolysin (B, lanes 2, 4, and 6) or alkali extraction (C). In C, lanes 1, 3, and 5 represent the insoluble membrane fraction (M) and lanes 2, 4, and 6 the supernatant (S) after extraction at pH 11. Quantifications are given in percentage of added OEP7. The entire sample was loaded and not corrected for loss during thermolysin treatment. The cartoon indicates the orientation and possible cleavage sides are indicated by triangle.
OEP7 contains several charged amino acids within both soluble domains flanking the TM region (Table 1). The Nin-Cout topology of OEP7 (Salomon et al., 1990) might be regulated by the charge distribution of the soluble regions. To test whether the negative net charge of the C-terminal region is required to achieve the topology, 35S-methionine–labeled OEP7 and two mutants, which contained deletions of the C-terminal region, i.e., OEP7Δ4 and OEP7Δ12 (Table 1), were incubated with isolated spinach chloroplasts. Insertion was controlled by alkali extraction (Figure 2B). OEP7 is inserted in an Nin-Cout orientation, which results in a smaller radioactive labeled fragment after thermolysin incubation because the [35S]methionine-labeled N terminus is protected against proteolytic cleavage (Figures 2C, lanes 1 and 2; and3A, lanes 2 and 3, [35S]). In OEP7-Δ4 the last seven amino acids are removed and three amino acids where added, including a positively charged arginine to increase the net charge to +1 as in the N-terminal domain (Table 1). In OEP7-Δ12 the last 12 amino acids are removed, resulting in an increase of the C-terminal net charge to +3 (Table 1). The topology was not altered by the deletions because the N-terminal domain remained protease insensitive (Figure 2B, lanes 4 and 6). The protease cleavage site seems to be located within the last 7–10 amino acids of the C-terminal soluble domain because no smaller proteolytic fragment was observed after OEP7-Δ4 insertion.
Charge of Flanking Regions of Transmembrane Domain Is a Determinant for Topology of OEP7 in Chloroplast Outer Envelope
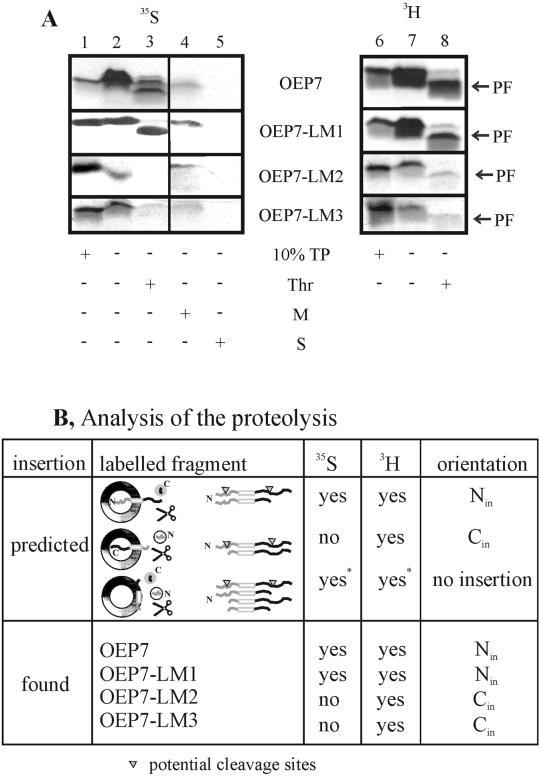
As demonstrated in Figure 2, neither the net charge of the C-terminal region nor the negatively charged cluster at the extreme C terminus is the determinant of the topology of OEP7. Interestingly, the flanking regions of the TM domain also contain a charge divergence, which is opposite compared with the net charge divergence (+1 at the lumenal side and +2 at the cytosolic side; Table 1). Therefore, we created mutants of OEP7 to study the influence of the charges flanking the TM domain. In the mutant OEP7-LM1 a positively charged amino acid was introduced N proximal of the membrane anchor region, whereas a negatively charged amino acid was removed (Table 1). This increase of the N-terminal net charge did not result in a loss of insertion as shown by alkali resistance (Figure 3A, lane 4, [35S]) or change of orientation as shown by protease treatment (Figure 3A, lane 3, [35S]).
Figure 3.
In vitro import of OEP7 and variants into spinach chloroplasts. In vitro synthesized [35S]methionine- (35S) or [3H]leucine (3H)-labeled OEP7 and variants (Table 1) are imported into intact spinach chloroplasts for 30 s (A, lanes 2–5, 7, and 8). The orientation of the proteins was probed by proteolysis with the use of thermolysin (A, lanes 3 and 8 [Thr]). After alkali extraction, soluble (A, lane 5 [S]) and insoluble fractions (A, lane 4 [M]) were separated. Lanes 1 and 6 represent 10% of translation product (TP) used per import experiment. PF indicates the proteolytic fragment. In B, the possible insertion modes as well as the resulting labeled fragments are indicated by yes or no. For association only, partial digest would reveal two 35S-labeled and three 3H-labeled fragments (two of similar size), whereas complete digest would result in no 35S-labeled and one short 3H-labeled fragment. The observed results are given for OEP7, OEP7-LM1, OEP7-LM2, and OEP7-LM3.
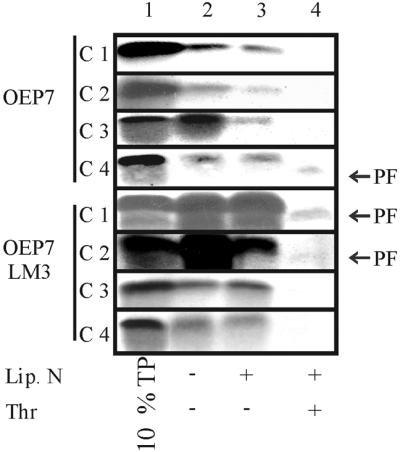
Subsequently, two other mutants were constructed. In OEP7-LM2 a positively charged amino acid C proximal of the TM region was exchanged for a negative one. OEP7-LM3 contained the same exchange as OEP7-LM2 and in addition the mutations introduced into OEP7-LM1 (Table 1). Both, OEP7-LM2 and OEP7-LM3 still inserted into the organellar membrane as shown by alkali resistance (Figure 3A, lane 4, [35S]). On protease treatment no radioactively labeled fragment was detectable when 35S-labeled protein was used (Figure 3A, lane 3, [35S]). This could indicate that insertion occurred with an inverted orientation leading to a nonlabeled and therefore nondetectable fragment upon protease treatment. An increase of the insertion time for OEP-LM2 and OEP-LM3 to 5 min revealed a small amount of 35S-labeled protease protected fragment, indicating that a minor fraction inserts with Nin-Cout orientation (our unpublished data).
To confirm that OEP7-LM2 and OEP7-LM3 had indeed inserted with reversed orientation, all polypeptides were synthesized in the presence of [3H]leucine. For proteins with Nin orientation we would expect an equally sized labeled fragment after protease treatment compared with the 35S-labeled protein (Figure 3B). For proteins with Cin orientation we should now detect a stable fragment of a similar size as seen for the proteins with the Nin orientation. If the proteins only associate with the surface they should be extractable at pH 11. OEP7 as well as the mutants inserted into the outer envelope as deduced from the appearance of a proteolytic fragment (Figure 3A, lane 8, [3H]). Furthermore, all fragments had a similar size and were resistant to extraction at pH 11 (our unpublished data), indicating that all proteins were inserted. Comparison of the yield of the observed proteolytic fragments with the use of either [35S]methionine- or [3H]leucine-labeled proteins was used to determine the orientation. OEP7 was inserted exclusively in Nin-Cout orientation as concluded from the similar yield of [35S]methionine- or [3H]leucine-labeled proteolytic fragment. However, all three mutants, OEP7-LM1, OEP7-LM2, and OEP7-LM3, inserted in both orientations but at very different ratios. OEP7-LM1 was mainly incorporated in Nin-Cout orientation (>70%), but OEP7-LM2 and OEP7-LM3 inserted mainly (>90%) in Nout-Cin orientation. The analysis is sumarized in Figure 3B. The lower binding and/or insertion efficiency of OEP7-LM2 and OEP7-LM3 might be explained by enhanced aggregation behavior of the polypeptides after introduction of the mutations or by a lower ability to associate with the chloroplast membrane. Our results indicate that the orientation is mediated by the amino acids flanking the TM region. Because the translocation of OEP7 was not found to be dependent on protease-sensitive factors of the outer envelope (Salomon et al., 1990), the orientation of OEP7 should be the same when inserted into lipid membranes not containing any proteins.
OEP7 Inserts into Liposomes with Average Lipid Composition of Outer Envelope in Reversed Orientation
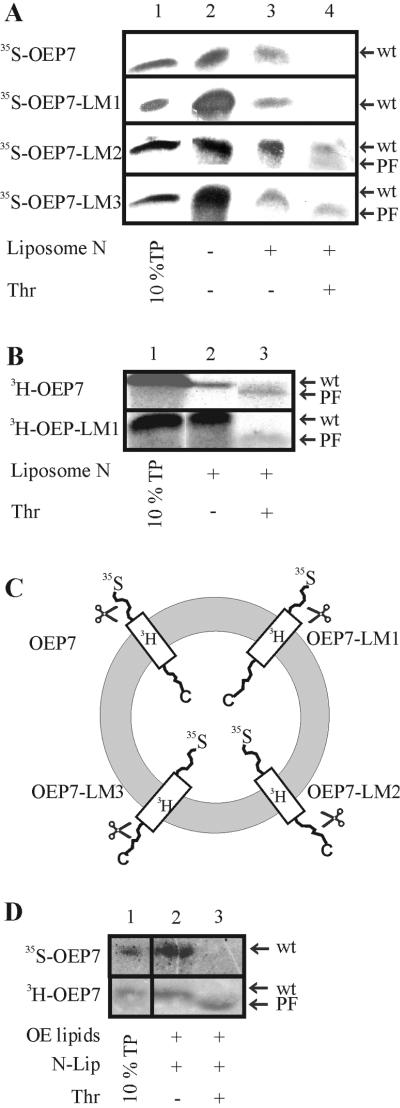
To test whether OEP7 can insert into protein-free membranes we prepared liposomes as described in MATERIALS AND METHODS. We used purified lipids in a ratio corresponding to the lipid content of the chloroplast outer envelope (Table 2) to mimic the properties of this membrane. Wild-type OEP7 and the different mutant proteins inserted into liposomes clearly demonstrating that OEP7 can integrate into a lipid membrane in the absence of other proteinaceous components (Figure 4A, lane 3). On thermolysin treatment no protease-resistant fragment was detected from 35S-labeled OEP7 and OEP7-LM1 (Figure 4A, lane 4), but from 3H-labeled protein (Figure 4B, lane 3), indicating an Nout-Cin orientation. OEP7-LM2 and OEP7-LM3, however, showed an Nin-Cout orientation as indicated by the appearance of a lower molecular weight fragment after proteolysis of the 35S-labeled proteins (Figure 4A, lanes 3 and 4). We conclude that OEP7 and variants insert into liposomes in a reversed orientation compared with the topology in the outer envelope of chloroplasts (compare Figures 3 and 4C). Interestingly, the association with the lipid surface after competition with N-liposomes (Figure 1) is highest for OEP7-LM1 and OEP7-LM2 (Figure 4A, lane 3, and B, lane 2), whereas the amount of protease protected protein is highest for OEP7 (Figure 4B, lane 3). This will be discussed below.
Figure 4.
Insertion of OEP7 and variants into liposomes. [35S]Methionine-labeled OEP7, OEP7-LM1, OEP7-LM2, and OEP7-LM3 (A and D) as well as [3H]leucine-labeled OEP7 and OEP7-LM1 (B and D) were incubated with liposomes composed from purified single lipids (A and B) or lipids purified from spinach outer envelope (D). In A, lane 1 represents 10% translation product (TP), and lane 2 binding and insertion into liposomes before and lane 3 after competition with N-liposomes. Lane 4 represents liposomes after thermolysin (Thr) treatment. In B and D, lane 1 represents 10% TP, and lane 2 binding and insertion into liposomes after competition with N-liposomes. Lane 3 represents liposomes after Thr treatment. In C, the topology of OEP7 and mutants within the liposome bilayer is shown. The position of the radioactive label is indicated. PF, proteolytic fragment; wt, full-length protein).
The reversal of OEP7 insertion into liposomes compared with the insertion into chloroplast outer envelope might have different reasons. For instance, the lipid composition of the outer envelope membrane might differ somewhat from the determined composition (Mazliak, 1977; Bruce, 1998). To test this possibility, lipids from outer envelopes of spinach chloroplast were purified and used to prepare liposomes. When [35S]methionine-labeled OEP7 was inserted no protease-protected fragment could be detected after thermolysin treatment (Figure 4D, lanes 2 and 3, top). However, the protein was inserted into the membrane as confirmed with the use of 3H-labeled OEP7 (Figure 4D, lanes 2 and 3, bottom). We conclude that other factors are involved in controlling the topology of OEP7.
Insertion of OEP7 into Chloroplasts Is not Dependent on Known Channel Activity
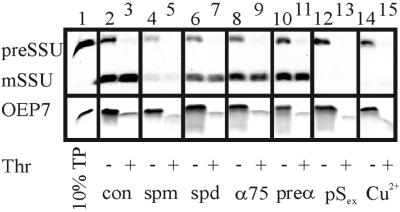
The reversed orientation of OEP7 might be explained by an active or passive transport through a pore present in the outer envelope. After reaching the intermembrane space it might then self-insert from the inside into the outer membrane. Furthermore, a protease insensitive protein on the outer membrane of chloroplast could also mediate the import. Therefore the insertion of OEP7 and the import of the preSSU were tested with the use of intact spinach chloroplasts (Figure 5) under various conditions. Incubation with heterologously expressed and purified preSSU does result in an inhibition of import of 35S-labeled preSSU but not of 35S-labeled OEP7 insertion into the outer envelope (Figure 5, lanes 12 and 13 versus 2 and 3). We further tested the possibility of OEP7 being translocated by Toc75, the postulated preprotein translocation pore in the outer envelope (Tranel et al., 1995; Hinnah et al., 1997). Toc75 was blocked by antibodies, which resulted in a 50% reduction of preSSU import (Figure 5, lane 9 versus 11 and 3), whereas OEP7 insertion remained unaltered (Figure 5, lanes 8–11). Furthermore, the import of preSSU decreased at least 10-fold by addition of spermine (lanes 4 and 5) and twofold by addition of spermidine (lanes 6 and 7) (Hinnah et al., 1997), whereas the insertion of OEP7 was only slightly reduced in the presence of spermine and not affected by the presence of spermidine (Figure 5, lanes 4–7). CuCl2 is able to abolish import most likely by inducing disulfide bridge formation within the Toc complex (Seedorf et al., 1995). Again, preSSU import was completely blocked by CuCl2 but no significant decrease of insertion of OEP7 could be observed (Figure 5, lanes 14 and 15). From these experiments and the observation that OEP7 insertion is not dependent on protease-sensitive proteins of the outer envelope (Salomon et al., 1990) we conclude that OEP7 does not use a channel or helper protein to insert into the outer envelope.
Figure 5.
Import of preSSU and OEP7 into intact spinach chloroplast under various conditions. [35S]Methionine-labeled preSSU and OEP7 were imported into spinach chloroplasts (lanes 2–15) under various conditions. Lane 1 shows 10% translation product (TP). In the same experiments chloroplasts were preincubated for 10 min with 10 mM spermine (spm, lanes 4 and 5) or spermidine (spd, lanes 6 and 7), with antibodies against Toc75 (α75, lanes 8 and 9), preimmune serum (preα, lanes 10 and 11), or with 10 μg of purified preSSU (pSex, lanes 12 and 13). Pore activity was also blocked by 1 mM CuCl2+ (Cu2+, lanes 14 and 15). After import, chloroplasts were treated with thermolysin (Thr, lanes 3, 5, 7, 9, 11, 13, and 15). A fluorogram is shown.
Correct Orientation of OEP7 in Liposomes Is Inhibited by a High Content of Phosphatidylglycerol or Phosphatidylinositol
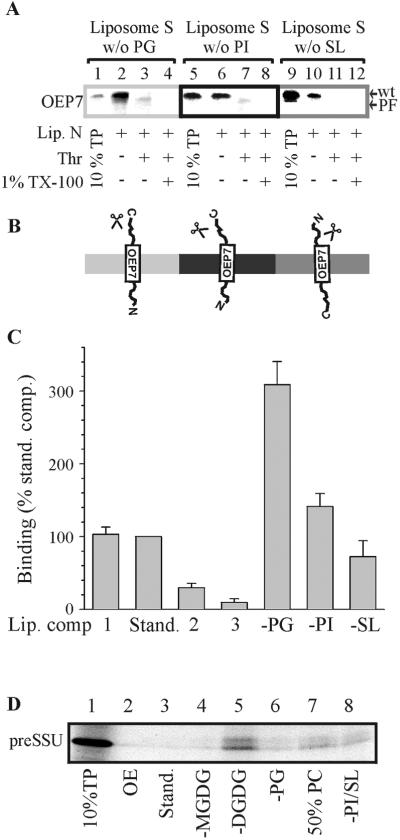
Dorne et al. (1985) have postulated that the content of PC in the outer leaflet of the outer envelope is 50% compared with 6% in the inner leaflet; in contrast, PG seemed to be exclusively present in the inner leaflet. Therefore, we asked whether this asymmetric distribution of the two types of lipids might have an influence on the insertion and orientation of the OEP7 protein. Liposomes were created with various concentrations of PC, whereas the ratio of the other lipids to each other was kept similar in composition 1–3. In composition 1 the PC content was adjusted lower than average (16 mol% compared with 32 mol% of total lipid), in composition 2 slightly higher (39 mol%), in composition 3 drastically higher (50 mol%), and in composition 4 the PC concentration was kept at 50%, whereas the concentration of the charged lipids was reduced by half (Table 2 and Figure 6). Neither of the liposomes with different PC content resulted in the correct orientation of wtOEP7 except when the charged lipid content was reduced (Figure 6, C4, lane 4). Insertion of OEP7-LM3 protein with Nin-Cout topology could be observed with the use of liposomes of composition 1 and 2 but not of composition 3 (Figure 6, lane 4). Previous reports had already shown that the association of the transit peptide of preferredoxin with a membrane surface was drastically reduced when only dioleoyl-PC was used (Pilon et al., 1995). To demonstrate the specificity of OEP7 association with the different liposomes we tested whether the precursor of the soluble preSSU also interacted with the lipid surface (Figure 7D). preSSU did not associate with liposomes of standard composition or made from purified lipids of the outer envelope from spinach chloroplasts (Figure 7D, lanes 2 and 3). Only in the absence of digalactosyldiacylglyceride (DGDG) did we detect a significant interaction of preSSU with liposomes (Figure 7D, lane 5). The association of OEP7 and OEP7-LM3 with the lipid surface containing higher concentration of PC was decreased (compare Figure 6, lanes 1 and 3, and Figure 7C), which is again in contrast to preSSU (Figure 7D). However, reducing the content of charged lipids partly restored the association of OEP7 with the surface and insertion could be demonstrated by the presence of the proteolytic fragment (Figure 6, C4, lane 4).
Figure 6.
Insertion of OEP7 and OEP7-LM3 into liposomes with different lipid compositions. Insertion of [35S]methionine-labeled OEP7 and OEP7-LM3 into liposomes was performed as described in Figure 1. The lipid composition of the liposomes is given in Table 2. Lane 1 shows 10% translation product (TP) used for insertion assay. Lane 2 represents binding to liposomes before and lane 3 after N-liposome competition. Lane 4 represents the determination of OEP7 orientation by thermolysin (Thr) treatment.
Figure 7.
Insertion of OEP7 into liposomes lacking PG, PI, or SL. Insertion of [35S]methionine-labeled OEP7 into liposomes (A) was performed as described with the use of liposomes of standard lipid composition lacking PG (lanes 2–4), PI (lanes 6–8), and SL (lanes 10–12). After competition with N-liposomes (lanes 2, 6, and 10) S-liposomes where treated with thermolysin in the absence (lanes 3, 7, and 11; Thr) or presence of 1% (vol/vol) Triton X-100 (lanes 4, 8, and 12; 1% TX-100). Lanes 1, 5, and 9 show 10% translation product (TP) used for insertion. A model of the orientation of OEP7 within the different bilayers is shown in B. In C, the amount of [35S]methionine-labeled OEP7 bound to liposomes of various lipid compositions was quantified and the results of three independent experiments were averaged. The total amount of bound protein to liposomes of standard lipid composition was set to 100% for comparison. In D, preSSU (10% shown in lane 1) association with liposomes composed of outer envelope lipids (lane 2), an isolated lipid mixture with standard composition (lane 3), without MGDG (lane 4), without DGDG (lane 5), without PG (lane 6), with increased PC content (lane 7), and without PI/SL (lane 8) is shown.
Guided by the idea from Dorne et al. (1985) we investigated whether charged lipids might be the determinant for the orientation of OEP7 during insertion into the lipid bilayer and furthermore, which lipid might stimulate the association with the membrane surface. Therefore, we composed liposomes lacking PG, PI, or sulfoquinovosyl-diacylgycerol (SL) (Table 2). The association of OEP7 with the liposomes containing a reduced amount of charged lipids was increased compared with the liposomes with standard lipid composition (compare Figure 4A, lane 3; Figure 6, C4, lane 3; and Figure 7A, lanes 2 and 6; Figure 7B). Furthermore, insertion of OEP7 in Nin-Cout orientation can be demonstrated by the appearance of the proteolytic fragment after thermolysin treatment when liposomes not containing PG or PI were used. Although insertion was more efficient into liposomes lacking PG (Figure 7C). Insertion of OEP7 into liposomes not containing SL still occurred in a reversed orientation (Figure 7A, compare lanes 3, 7, and 11, orientation indicated in Figure 7B). We conclude that the orientation of OEP7 in liposomes is determined by the content of charged lipids, suggesting that the topology of OEP7 in vivo is sensitive to the lipid asymmetry of PG between the two leaflets of the outer envelope resulting in a low content of charged lipids on the surface of chloroplasts.
DISCUSSION
Association of OEP7 Is Driven by Hydrophobicity of Transmembrane Domain
We could demonstrate that proteins in the outer envelope do not initiate the association of OEP7, because channel proteins (Figure 5) and protease-sensitive factors (Salomon et al., 1990) are not involved in binding of OEP7. We show that OEP7 associates with protein-free liposomes (Figure 4) and the interaction with the lipid surface was decreased when the content of PC was increased (Figure 6 and 7C). Our results indicate that this effect is not due to the reduction of charged lipids, because the association of OEP7 was increased when liposomes were deficient of PI or PG (Figure 7, A and C). Therefore, we conclude that the interaction is driven by the hydrophobicity of the TM domain and possibly galactosyldiacylglycerids such MGDG or/and SL because OEP7 association with liposomes lacking SL was slightly decreased (Figure 7C). This is consistant with the result that the outer envelope protein OEP14 does not insert into mitochondria or microsomes in vitro (Li et al., 1991; Li and Chen, 1996). Both organelles do not contain SL (Douce and Joyard, 1990) and only microsomes might contain a low amount of MGDG (Douce, 1974; Mackender and Leech, 1974).
The free energy of the association energy between the different domains of OEP7 or its mutants with the membrane surface was calculated. The association of the N and C termini with the lipid surface is energetically unfavorable for all proteins investigated because ΔGass is >0 kcal/mol (Table 3), whereas the energy for the TM domain is ΔGass(TM) = −1.97 kcal/mol. Furthermore, a coiled-helix transition is expected for the 23 amino acids of the TM domain during membrane association, resulting in an additional energy term of −3.22 kcal/mol (Wieprecht et al., 1999). This results in a free binding energy of the TM region of ΔGbin = −5.19 kcal/mol.
Table 3.
Association energy ΔGass between domains of OEP7 and the chloroplast outer envelope and transfer energy ΔGtrans into or across the outer envelope not including the charges
| Protein | ΔGass (kcal/mol)
|
ΔGtrans (kcal/mol)
|
||||
|---|---|---|---|---|---|---|
| N-Term | TM | C-Term | N-Term | TM | C-Term | |
| OEP7 | 10.36 | −1.97 | 11.12 | 17.47 | −11.38 | 20.78 |
| OEP7-Δ4 | 10.36 | −1.97 | 8.98 | 17.47 | −11.38 | 20.88 |
| OEP7-Δ12 | 10.36 | −1.97 | 6.57 | 17.47 | −11.38 | 19.24 |
| OEP7-LM1 | 9.02 | −1.97 | 11.28 | 20.47 | −11.38 | 20.78 |
| OEP7-LM2 | 10.36 | −1.97 | 13.50 | 17.47 | −11.38 | 19.08 |
| OEP7-LM3 | 9.02 | −1.97 | 13.50 | 20.47 | −11.38 | 19.08 |
N-term, N-terminal domain; C-term, C-terminal domain.
The free energy of Pf3 coat protein association with the membrane without consideration of a coiled-helix transition was calculated to be ΔGbin = −1.60 kcal/mol (Kiefer and Kuhn, 1999). The authors suggest a hydrophobic-driven interaction. Therefore, the ΔGass = −1.97 kcal/mol found for OEP7 suggests an even stronger hydrophobic interaction. The experimental and theoretical results indicate that an association of the TM region of OEP7 with the membrane surface is possible without assistance of other proteins.
Lipid Asymmetry Influences OEP7 Topology
The association between OEP7 and the lipid surface is strong enough for a direct insertion of the protein. However, the charges of soluble regions or the charge balance at the flanking regions of the TM anchor might have an influence on the orientation of OEP7.
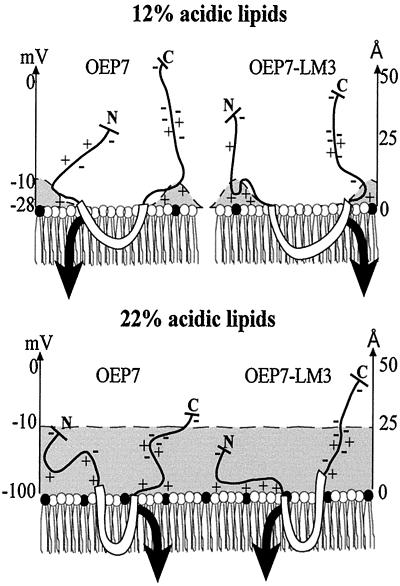
Flanking amino acids were found to influence the orientation of bacterial plasma membrane proteins (Monne et al., 1998). Such influence of the charges would decrease with an increase of the distance from the TM region. As shown in Figure 8, the electrostatic potential considering a concentration of charged lipids of 12 mol% (discussed below) would be sufficient to influence 3–4 amino acids next to the hydrophobic α-helical region. This charge distribution is represented in intact chloroplasts and in liposomes without PG and results in the correct orientation of OEP7. Liposomes made from a standard lipid composition or purified outer envelope membrane lipids contain 22 mol% charged surface lipids. The electrostatic potential of such a membrane creates equipotential surfaces, which is still at −10mV 25Å above the membrane surface (“Supplementary Material”; Figure 8, dashed line). Therefore, not only the charges directly flanking the TM domain will influence the insertion; at least eight amino acids counted from the last amino acid interacting with the membrane surface will be influenced by the potential (Figure 8). This results in a reversed Nout-Cin orientation of OEP7. The lipid asymmetry of the outer envelope of chloroplasts as suggested by Dorne et al. (1985) and as mimicked in our experiments (PG in Figure 7) have ∼12 mol% anionic surface lipids and no equipotential surfaces will be formed (Figure 8, dashed line). Therefore, the insertion data of OEP7 and mutants into chloroplasts can be explained in the following way.
Figure 8.
Model of the association of OEP7 and OEP7-LM3 with the membrane surface. A model for the association of OEP7 (left) and OEP7-LM3 (right) with the membrane surface containing 12% (top) and 22% (bottom) charged lipids is shown. The distance of the electrostatical potential of −10 mV is indicated as dashed line and the affecting electrostatic field by a gray area. The electrostatic potential over the surface is given in millivolts and the distance over the membrane in angstroms. The black arrow indicates direction of insertion.
Within the four flanking amino acids, we altered the charge distribution in OEP7-LM1 from +1/+2 (N-terminal/C-terminal site) to + 2/+2, resulting in an equal distribution of charges at the N- and C-terminal site of the membrane. As expected, OEP7-LM1 inserts in both directions but with preference for Nin-Cout. The preference of OEP7-LM1 to insert in Nin-Cout orientation can be explained by the stronger affinity of arginine for the charged lipid surface than of lysine (“Supplementary Material”). In OEP7-LM2 the distribution was altered from +1/+2 to +1/0 and a reversed orientation was observed after insertion into chloroplast outer envelopes (Figure 3B). OEP7-LM3 with an even more drastic alteration of the charge distribution at the flanking region from +1/+2 to +2/0 results in the same orientation as OEP7-LM2 (Figure 3B). These results confirm that the positive charges at position 2 and at position 4 following the TM domain in OEP7 act as the signal for topology in situ by prohibition of insertion of this C-terminal domain. This is consistent with the observation that the positive charges at the cytosolic site of the TM domain of Toc34 (May and Soll, 1998) are essential for the correct orientation of this protein. Therefore, the membrane contact of the hydrophobic TM region initiates insertion, whereas orientation is driven by the strength of the interaction between the positively charged amino acids flanking the TM domain and the negatively charged head groups of the lipids. This explains why the flanking amino acids of the TM domain have a stronger influence on the orientation of the protein than the overall net charges of the cytosolic and intermembrane space regions.
Further investigations revealed that the content of the negatively charged lipids PG and PI in the liposomes are crucial for the correct orientation of OEP7 (Figure 7). The third charged lipid, SL, does not seem to be important for the orientation but for association of OEP7 (Figure 7). This might be due to the location of the charge more distant from the surface than the charge located in PI and PG. As seen in Table 3, the energy required for translocation of the N- or C-terminal regions not considering electrostatic effects varies only slightly for all mutants. The reduction of the charged lipid content by the amount of PG results in a decrease of the distance of the electrostatic potential influencing the charged amino acids of the protein to 10 Å (∼4 amino acids). This also explains why OEP7, OEP7-Δ4, and OEP7-Δ12 insert with an identical orientation (Figure 2A). For all three proteins the charges C-terminally flanking the transmembrane domain are able to interact with the charged surface. For OEP7, the required energy considering the charges of the four flanking amino acids is 18.87 kcal/mol or 24.28 kcal/mol (N and C terminus, respectively; see “Supplementary Material”), which is consistent with the observed topology. The insertion of a positive charge at the N-terminal side of the TM region within OEP7-LM1 increases the required energy to 23.27 kcal/mol. Therefore, the energy to transfer each of the soluble region is almost even with a preference for the transfer of the N-terminal domain, which is consistent with the observation in Figure 3B. The additional introduction of a negatively charged amino acid in OEP7-LM3 results in an inhibition of the interaction between the lysine 42 with the charged lipid head group possibly by salt bridge formation as discussed above and the required energy to transfer the C terminus is 19.08 kcal/mol. This results in a transfer of the C-terminal domain of OEP7-LM3 where 23.27 kcal/mol is required for the transfer of the N-terminal domain. For OEP7-LM2, the energy to transfer the N-terminal region was calculated to be 18.87 kcal/mol. Therefore, for the transfer of the N- and C-terminal soluble domain of OEP7-LM2 over the membrane a similar energy was calculated. The comparison with the experimental results strengthens our conclusion that the energy to disrupt the interaction of a charged amino acid with a charged lipid head group has to be larger than only the association energy as discussed above.
In membranes exposing 22 mol% negatively charged lipids, the negatively charged amino acids within at least eight amino acids flanking the transmembrane helix have also to be transferred into the electrostatic potential of the membrane. The charge repulsion results in a decrease of association compared with the association to a membrane containing only 12% charged lipids (Figure 7C). The association nevertheless is initiated by the positively charged amino acids flanking the TM region. However, energy is required for protonation of the negatively charged amino acids because the amino acids are transferred over the membrane in an uncharged form (Kessel et al., 2000). This energy is in the same range as the energy required for disruption of the interaction between positively charged amino acids and negatively charged lipid head groups (see “Supplementary Material”). Therefore, the two positive charges C proximal of the TM domain in OEP7 result in rapid association with the membrane (Figure 8), allowing the start of the insertion. This subsequently results in a transfer of the C-terminal domain of OEP7. The disruption of the association of positively charged amino acids with negatively charged head groups seems to require a higher energy than only the reversal of the association energy. Therefore, the disruption of the association of the four positive charges of the N-terminal region of OEP7-LM1 would require more energy than the transfer of the negative charges of the C-terminal region over the membrane once the negative charges are within the range of the electrostatic potential of the surface. This explains the identical topology of OEP7-LM1 and OEP7. In OEP7-LM2 and OEP7-LM3 the net charge of the C-terminal region is drastically decreased. If we assume a salt bridge between Lys42 and Glu 44, no positive charge is flanking the TM domain and no association of the C-terminal region with the membrane will occur. Furthermore, the charge repulsion will cause an asymmetric insertion of the TM domain (indicated in Figure 8 for OEP7-LM1 by a dented TM domain). The insertion will now be initiated at the N-proximal side of the TM due to the positive charges. The association of the flanking regions is considered to be the essential step for insertion; it is not surprising that OEP7-LM3 inserts with Nin-Cout topology. Therefore, under the artificial conditions of 22% negatively charged lipids, the insertion becomes dependent on the total net charge of the soluble domain as well as on the concentration of the positive charges flanking the TM domain initiating the contact with the membrane.
We conclude that the topology of OEP7 depends on the charge distribution of the flanking region and the low content of charged lipids in the outer leaflet as created by the asymmetry of PG between outer (low concentration) and inner (high concentration) leaflet of the outer envelope (Dorne et al., 1985). But how does OEP7 sense that it approaches the surface of a chloroplast and not another membrane within the plant cell. As discussed above, it is tempting to assume that this selectivity is achieved by the unique presence of galactolipids, i.e., MGDG, DGDG, and SL in the chloroplastic outer envelope.
SUPPLEMENTARY MATERIAL
Calculation of Free Energy of Membrane Association
The free energy for the interaction of a protein with a lipid surface ΔGass can be described by the free energy for the transfer of interacting domain from solution to the lipid interface ΔGwif, a term for the electrostatic attraction between charged amino acids and lipids ΔGelc and a term describing the immobilization energy during membrane association ΔGimm (Ben-Tal et al., 1996a).
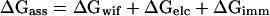 |
1 |
ΔGwif was calculated by summarizing the ΔGwif values for each residue of the corresponding domain. The values for the single residues were derived from a study of the transfer energy of peptides to the palmitoyloleoyl-PC interface (White and Wimley, 1998). The immobilization energy for a polypeptide ΔGimm was estimated to be 3.7 kcal/mol (Ben-Shaul et al., 1996). The free electrostatic energy for the interaction of lysine with a membrane containing 100, 22, or 12% acidic lipids in the presence of 100 mM monovalent salt was found to be −1.4, −0.76, or −0.38 kcal/mol/residue, respectively (Kim et al., 1991; Ben-Tal et al., 1996b). The free electrostatic energy for association of arginine was found to be −2.1 (100% acidic lipids), −1.14 (22%), or −0.57 (12%) kcal/mol/residue (Kim et al., 1991; Ben-Tal et al., 1996b). At equal distribution of lipids in the outer envelope 22% charged lipids is present. This number was used for the calculations of the maximal association energy (Table 3).
Calculation of Free Energy of Membrane Insertion
A protein will directly insert into a lipid bilayer when the energy for the transfer of the TM domain (ΔGtrans − TM) into the lipid bilayer is larger than the required energy to transfer of the intermembrane space domain over the membrane (ΔGtrans − IMSD). Therefore, the total free energy of this process can be calculated by
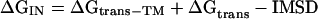 |
2 |
The transfer of the TM domain into the bilayer can be dissected into the interference of the solute with the conformational freedom of the lipid chains ΔGlip calculated to be +2.3 kcal/mol (Ben-Tal et al., 1996a), the conformational change of the polypeptide (ΔGcon), and the transfer of amino acids into a hydrophobic medium (ΔGwif-bulk). This transfer energy for single amino acids was determined from the transfer of peptides into an n-octanol (White and Wimley, 1998). Based on these values the energy ΔGwif-bulk for the TM domain of OEP7 was calculated to be −10.46 kcal/mol. The conformational free energy ΔGcon was determined to be −0.14 kcal/mol/residue (Wieprecht et al., 1999), resulting in a maximal conformational energy for the TM domain of OEP7 of −3.22 kcal/mol. Therefore, the total insertion energy of the TM region is ΔGtrans –−TM = −11.38 kcal/mol.
The free energy for the soluble domains ΔGtrans − IMSD can be divided into the energy of the transfer of amino acids into a hydrophobic medium ΔGwif-bulk, the free energy required for the transfer of a terminus over the membrane ΔGterm, and a retention energy ΔGret resulting from the influence of the electrostatic field of the charged lipids on the charged amino acids.
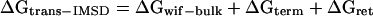 |
3 |
The free energy to translocate the N terminus is 5.0 kcal/mol and to transfer the C terminus 4.3 kcal/mol (Engelman et al., 1986). The energy ΔGwif-bulk was calculated as discussed for the TM domain and the values for ΔGwif-bulk + ΔGterm = ΔGtrans* are given in Table 3.
The retention energy ΔGret can be divided into the energy resulting from the attraction of the positive amino acids to the negatively charged lipid surface ΔGint and the energy resulting from the retention of the negatively charged amino acid to enter the negatively potential of the surface ΔGpKa:
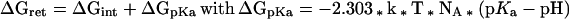 |
4 |
where k is the Boltzman constant, T the temperature, and NA the Avogadro constant (Kessel et al., 2000). The association energy of a single arginine and lysine with a charged head group was described above (−2.1 and −1.4 kcal/mol, respectively) and has to be reversed to transfer the amino acid across the membrane. The influence of the electrostatic potential on the negatively charged amino acids is dependent on the concentration of charged lipids. No constant equipotential surface is formed over a membrane containing 12% charged lipids but over a membrane containing 22% charged lipids (Peitzsch et al., 1995). The electrostatic potential can be calculated based on the Gouy-Chapman Theory:
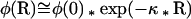 |
5 |
where κ−1 is the Debye length. The Debye length was found to be 10Å in a 100 mM monovalent salt solution (Ben-Tal et al., 1996b). It can be assumed that a potential lower than −10 mV does not result in a considerable electrostatic attraction (Murray et al., 1998)). The potential will drop down to −10 mV after 23Å for 22% acidic lipids and after 10Å for 12% acidic lipids. As seen in Eq. 5 ΔGpKa is dependent on the local pH over the membrane surface, which depends on the potential on the surface (van der Goot et al., 1991):
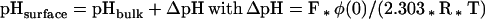 |
6 |
where F is the Faraday constant, R the gas constant, and T the temperature. For a membrane containing 22% acidic lipids an initial potential of −100 mV can be assumed (Peitzsch et al., 1995; for 25% acidic lipids). This results in a ΔpH = −1.7 and subsequently in pHsurface = 5.9. With the use of this values, we calculated for ΔGpKa(E) = 2.0 kcal/mol and for ΔGpKa(D) = 2.4 kcal/mol.
ACKNOWLEDGMENTS
We thank T. Beilharz for carefully reading the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Human Frontier Science Program.
Abbreviations used:
- MGDG
monogalactosyldiacylglyceride
- OEP
outer envelope protein
- PC
phosphatidylcholine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- SSU
small subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase
- TM
transmembrane
- Toc/Tic
translocon at the outer/inner envelope of chloroplasts
REFERENCES
- Arnon DJ. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shaul A, Ben-Tal N, Honig B. Statistical thermodynamic analysis of peptide and protein insertion into lipid membranes. Biophys J. 1996;71:130–137. doi: 10.1016/S0006-3495(96)79208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tal N, Ben-Shaul A, Nicholls A, Honig B. Free-energy determinants of α-helix insertion into lipid bilayers. Biophys J. 1996a;70:1803–1812. doi: 10.1016/S0006-3495(96)79744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J. 1996b;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J. 1998;17:52555–55264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bruce BD. The role of lipids in plastid protein transport. Plant Mol Biol. 1998;38:223–246. [PubMed] [Google Scholar]
- Chen D, Schnell DJ. Insertion of the 34-kDa chloroplast protein import component, IAP34, into the chloroplast outer membrane is dependent on its intrinsic GTP-binding capacity. J Biol Chem. 1997;272:6614–6620. doi: 10.1074/jbc.272.10.6614. [DOI] [PubMed] [Google Scholar]
- Chupin V, van't Hof R, de Kruijff B. The transit sequence of a chloroplast precursor protein reorients the lipids in monogalactosyl diglyceride containing bilayers. FEBS Lett. 1994;350:104–108. doi: 10.1016/0014-5793(94)00734-9. [DOI] [PubMed] [Google Scholar]
- de Leeuw E, Luirink J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- de Leeuw E, te Kaat K, Moser C, Menestrina G, Demel R, de Kruijff B, Oudega B, Luirink J, Sinning I. Anionic phospholipids are involved in membrane association of FttsY, and stimulate its GTPase activity. EMBO J. 2000;19:531–541. doi: 10.1093/emboj/19.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorne AJ, Joyard J, Block MA, Douce R. Localization of phosphatidylcholine in outer envelope membrane of spinach chloroplast. J Cell Biol. 1985;100:1690–1697. doi: 10.1083/jcb.100.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R. Site of biosynthesis of galactolipids in spinach chloroplasts. Science. 1974;183:852–853. doi: 10.1126/science.183.4127.852. [DOI] [PubMed] [Google Scholar]
- Douce R, Joyard J. Biochemistry and function of the plastide envelope. Annu Rev Cell Biol. 1990;6:173–216. doi: 10.1146/annurev.cb.06.110190.001133. [DOI] [PubMed] [Google Scholar]
- Engelman DM, Steitz TA, Goldmann A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biphys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Hinnah SC, Hill K, Wagner R, Schlicher T, Soll J. Reconstitution of a chloroplast protein import channel. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Billecocq A, Bartlett SG, Block MA, Chua NH, Douce R. Localization of polypeptides to the cytosolic side of the outer envelope membrane of spinach chloroplasts. J Biol Chem. 1983;258:10000–10006. [PubMed] [Google Scholar]
- Keegstra K, Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel A, Cafiso DS, Ben-Tal N. Continuum solvent model calculations of alamethicin-membrane interactions. Thermodynamic aspects. Biophys J. 2000;78:571–583. doi: 10.1016/S0006-3495(00)76617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Kiefer D, Kuhn A. Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 1999;18:6299–6309. doi: 10.1093/emboj/18.22.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Mosior M, Chung LA, Wu H, McLaughlin S. Binding of peptides with basic residues to membrane containing acidic phospholipids. Biophys J. 1991;60:135–148. doi: 10.1016/S0006-3495(91)82037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolke H, Yoshio M, Kashino Y, Satoh K. Polypeptide composition of envelopes of spinach chloroplasts: two major proteins occupy 90% of outer envelope membranes. Plant Cell Physiol. 1998;39:526–532. doi: 10.1093/oxfordjournals.pcp.a029400. [DOI] [PubMed] [Google Scholar]
- Li HM, Chen LJ. Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell. 1996;8:2117–2126. doi: 10.1105/tpc.8.11.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HM, Chen LJ. A novel chloroplastic outer membrane-targeting signal that functions at both termini of passenger polypeptides. J Biol Chem. 1997;272:10968–10974. [PubMed] [Google Scholar]
- Li HM, Moore T, Keegstra K. Targeting of proteins to the outer envelope membrane uses a different pathway then transport into chloroplasts. Plant Cell. 1991;3:709–717. doi: 10.1105/tpc.3.7.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L-P, Deber CM. Anionic phospholipids modulate peptide insertion into membranes. Biochemistry. 1997;36:5476–5482. doi: 10.1021/bi970030n. [DOI] [PubMed] [Google Scholar]
- Lowry RR, Tinsley IJ. A simple, sensitive method for lipid phosphorus. Lipids. 1974;9:491–492. doi: 10.1007/BF02534277. [DOI] [PubMed] [Google Scholar]
- MacDonald RC, MacDonald RI, Menco BPM, Takeshita K, Subbarao NK, Hu L-R. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim Biophys Acta. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- Mackender LRO, Leech RM. The galactolipid, phospholipid and fatty acid composition of the chloroplast envelope membranes of Vicia faba. Plant Physiol. 1974;53:496–502. doi: 10.1104/pp.53.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J. Positive charges determine the topology and functionality of the transmembrane domain in the chloroplastic outer envelope protein Toc34. J Cell Biol. 1998;141:895–904. doi: 10.1083/jcb.141.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazliak P. Glyco- and Phospholipids of Biomembranes in Higher Plants. Berlin: Springer Verlag; 1977. [Google Scholar]
- Monne M, Nilsson IM, Johansson M, Elmhed N, von Heijne G. Positively and negatively charged residues have different effects on the position in the membrane of a model transmembrane helix. J Mol Biol. 1998;284:1177–1183. doi: 10.1006/jmbi.1998.2218. [DOI] [PubMed] [Google Scholar]
- Mourioux G, Douce R. Slow passive diffusion of orthophosphate between intact isolated chloroplasts and suspending medium. Plant Physiol. 1981;67:470–473. doi: 10.1104/pp.67.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D, Hermida-Matsumoto L, Buser CA, Tsang J, Sigal T, Ben-Tal N, Honig B, Resh MD, McLaughlin S. Electrostatics and membrane association of Src: theorie and experiment. Biochemistry. 1998;37:2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- Peitzsch RM, Eisenberg M, Sharp KA, McLaughlin S. Calculations of the electrostatic potential adjacent to model phospholipid bilayers. Biophys J. 1995;68:729–738. doi: 10.1016/S0006-3495(95)80253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Wienk H, Sips W, de Swaaf M, Talboom I, van't Hof R, de Korte-Kool G, Demel R, Weisbeek P, de Kruijff B. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J Biol Chem. 1995;270:3882–3893. doi: 10.1074/jbc.270.8.3882. [DOI] [PubMed] [Google Scholar]
- Rietveld AG, Koorengevel MC, de Kruijff B. Non-bilayer lipids are required for efficient protein transport across the plasma membrane of Escherichia coli. EMBO J. 1995;14:5506–5513. doi: 10.1002/j.1460-2075.1995.tb00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Fischer K, Flugge UI, Soll J. Sequence analysis and protein import studies of an outer chloroplast envelope polypeptide. Proc Nat Acad Sci USA. 1990;87:5778–5782. doi: 10.1073/pnas.87.15.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schindler C, Hracky R, Soll J. Protein transport in chloroplasts: ATP is prerequisite. Z Naturforsch. 1987;42c:103–108. [Google Scholar]
- Schleiff E, Silvius JR, Shore GC. Direct membrane insertion of voltage-dependent anion-selective channel protein catalyzed by mitochondrial Tom20. J Cell Biol. 1999;145:973–978. doi: 10.1083/jcb.145.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J. Traveling of proteins through membranes. Translocation into chloroplasts. Planta. 2000;211:449–456. doi: 10.1007/s004250000357. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Soll J, Tien R. Protein translocation into and across the chloroplastic envelope membranes. Plant Mol Biol. 1998;38:191–207. [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K. A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 1995;14:2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LY, Tu SL, Li HM. Insertion of atToc34 into chloroplastic outer membrane is assisted by at least two proteinaceous components in the import system. J Biol Chem. 1999;274:18735–18740. doi: 10.1074/jbc.274.26.18735. [DOI] [PubMed] [Google Scholar]
- Tu S-L, Li H-M. Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceaous components of chloroplasts. Plant Cell. 2000;12:1951–1959. doi: 10.1105/tpc.12.10.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot FG, Gonzales-Manas JM, Lakey JH, Pattus F. A 'molten-globule'membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- van Klompenburg W, Paetzel M, de Jong JM, Dalbey RE, Demel RA, von Heijne G, de Kruijff B. Phosphatidylethanolamine mediates insertion of the catalytic domain of leader peptidase in membranes. FEBS Lett. 1998;431:75–79. doi: 10.1016/s0014-5793(98)00733-9. [DOI] [PubMed] [Google Scholar]
- van't Hof R, Demel RA, Keegstra K, de Kruijff B. Lipid-peptide interactions between fragments of the transit peptide of ribulose-1,5-bisphosphate carboxylase/oxygenase and chloroplast membrane lipids. FEBS Lett. 1991;291:350–354. doi: 10.1016/0014-5793(91)81318-3. [DOI] [PubMed] [Google Scholar]
- van't Hof R, van Klompenburg W, Pilon M, Kozubek A, de Korte-Kool G, Demel RA, Weisbeek PJ, de Kruijff B. The transit sequence mediates the specific interaction of the precursor of ferredoxin with chloroplast envelope membrane lipids. J Biol Chem. 1993;268:4037–4042. [PubMed] [Google Scholar]
- Waegemann K, Eichacker S, Soll J. Outer envelope membranes from chloroplasts are isolated as right-side-out vesicles. Planta. 1992;187:89–94. doi: 10.1007/BF00201628. [DOI] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Hydrophobic interactions of peptides with membrane interfaces. Biochim Biophys Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- Wieprecht T, Apostolov O, Beyermann M, Seelig J. Thermodynamics of the α-helix-coil transition of amphipathic peptides in a membrane environment: implications for the peptide membrane binding equilibrium. J Mol Biol. 1999;294:785–794. doi: 10.1006/jmbi.1999.3268. [DOI] [PubMed] [Google Scholar]